Abstract
The emergence of the recent SARS-CoV-2 global health crisis introduced key challenges for epidemiological research and clinical testing. Characterized by a high rate of transmission and low mortality, the COVID-19 pandemic necessitated accurate and efficient diagnostic testing, particularly in closed populations such as residential universities. Initial availability of nucleic acid testing, like nasopharyngeal swabs, was limited due to supply chain pressure which also delayed reporting of test results. Saliva-based reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) testing has shown to be comparable in sensitivity and specificity to other testing methods, and saliva collection is less physically invasive to participants. Consequently, we developed a multiplex RT-qPCR diagnostic assay for population surveillance of Clemson University and the surrounding community. The assay utilized open-source liquid handling robots and thermocyclers instead of complex clinical automation systems to optimize workflow and system flexibility. Automation of saliva-based RT-qPCR enables rapid and accurate detection of a wide range of viral RNA concentrations for both large- and small-scale testing demands. The average turnaround for the automated system was < 9 h for 95% of samples and < 24 h for 99% of samples. The cost for a single test was $2.80 when all reagents were purchased in bulk quantities.
Introduction
Severe acute respiratory syndrome-associated coronavirus-2 (SARS-CoV-2), a novel coronavirus, emerged in late 2019 and rapidly spread throughout the global populations1. The SARS-CoV-2 infection causes coronavirus disease 2019 (COVID-19), a highly contagious disease with potentially severe respiratory and inflammatory symptoms. High transmissibility coupled with low mortality indicated that the virus would spread rapidly through populations and would require increased diagnostic testing2,3. Public health recommendations encouraged wide-scale population screening to isolate cases and subsequently reduce transmission rates4,5,6. Furthermore, models of population surveillance revealed that increasing testing frequency and decreasing reporting time had a greater effect on reducing transmission than increasing test sensitivity7. This is likely because infected individuals could be quarantined earlier, thereby breaking chains of infection.
The original nucleic acid amplification testing (NAAT) standard was nasopharyngeal (NP) swabs processed by RT-qPCR8. However, complications arise with this form of testing for very large populations, such as increased relative cost and exacerbated supply chain pressure9,10. Moreover, both specimen collection and processing of common NAAT methods (including NP swabs, oropharyngeal swabs, mid-turbinate swabs, and nasal swabs) are reliant on specialized equipment, reagents, and medical personnel9,10.
An adequate substitute for NP swab RT-qPCR testing is saliva-based testing, which is an accurate diagnostic tool for SARS-CoV-2 detection11,12,13,14. Directly performing RT-qPCR on saliva samples yields similar sensitivity and specificity as NP swabs15. One major advantage saliva testing has over NP swab testing is that it allows for self-collection of specimens16. This minimizes the need for medical personnel and maximizes ease of sample collection for patients by being less invasive than NP swabs. Additionally, since saliva samples do not require buffers to remove the sample from a swab (as in the case of NP samples), saliva-based tests can utilize heat-based ribonucleic acid (RNA) extraction directly, which decreases testing costs by removing the need for additional buffers, transport media, and/or RNA extraction reagents14,17.
The Clemson University Research and Education in Disease Diagnostics and Intervention (REDDI) Lab was established to address the university’s needs for COVID-19 testing and surveillance. In closed populations, including universities, frequent surveillance testing coupled with social distancing produced the most favorable outcomes in epidemiological models of disease prevalence18. The consolidated CDC 2019-nCOV RT-qPCR19 and SalivaDirect14 protocols were adapted, and automation was utilized in the clinical workflow to decrease cost and improve turnaround time. Previous groups had used open-source liquid handling robots for SARS-CoV-2 RNA extraction steps20,21, but we maximized the use of the robots to prepare test plates and load specimens22. Here, we show that the adapted protocol and the utilization of open-source liquid handling systems (Figure 1) allows for quick and accurate saliva-based RT-qPCR and is an effective strategy for large-scale public health surveillance.
Figure 1: Laboratory workflow utilizing the saliva-based RT-qPCR diagnostic system.
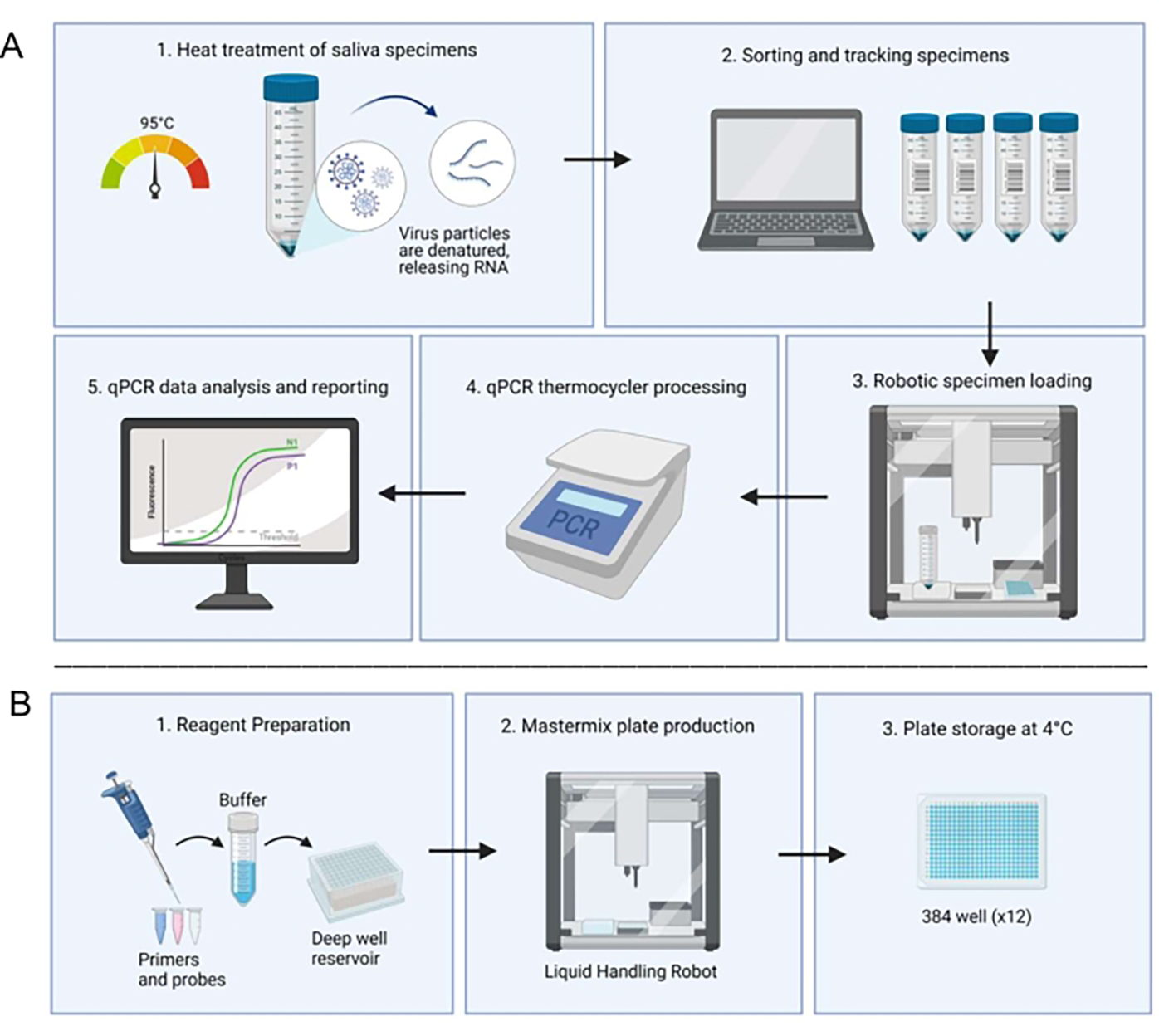
(A) Samples are collected and heat-treated at 95 °C for 30 min. Treated samples are sorted and tracked with patient information through an in-house spreadsheet system. A liquid handling robot loads samples into duplicate wells of prepared master mix plates. A technician manually loads the controls, seals the plate, and places the plate in a thermocycler for processing. Results are analyzed through an automated computer system and verified by a technician. (B) A technician prepares reagents for the master mix which are added to a deep well reservoir in a sterile biosafety cabinet. Filled deep well reservoirs are loaded into a dedicated liquid handling robot. Completed plates are sealed with foil, labeled, and stored at 4 °C.
Protocol
All research was performed in compliance with Clemson University and Prisma Health Institutional Review Boards (Prisma Health IRB # Pro00099491, July 1, 2020).
1. Setup of open-source liquid handling robot
Install high-efficiency particulate air (HEPA) filter modules (see Table of Materials) to the top of each liquid handling robot as per manufacturer’s instructions.
Attach an 8-channel P20 pipette to the left mount of the master mix plate preparation robot(s) as per manufacturer’s instructions.
Attach a P20 pipette to the right mount of the sample loading robot(s) as per manufacturer’s instructions.
Download the custom Python scripts (Supplemental File 1 and Supplemental File 2) on the appropriate computers.
Open TigerSaliva Full 384 Loading.py in the desktop application at the sample loading robot computers. Click Calibrate and set up both the pipette and the program as per software directions.
Open 12 Full Plates.py in the desktop application at the master mix robot computer. Click Calibrate and set up both the pipette and the program as per software directions.
Print custom sample racks with a fused deposition modeling three-dimensional (3D) printer using a computer-aided design (CAD) file (https://www.myminifactory.com/object/3d-print-141363). Print 16 racks per sample loading robot, for two sets of eight racks.
Materials.
| Name | Company | Catalog Number | Comments |
|---|---|---|---|
| 100% EtOH | Fisher scientific | 22-032-601 | |
| 20 uL Filtered Pipette Tips | Opentrons | 20uL tips | |
| 2mL Microcentrifuge Tubes | Fisher Scientific | 14-666-313 | Alternate product may be used |
| Armadillo PCR Plate, 384-well, clear, white wells | Thermo Scientific | AB3384 | Alternate product may be used |
| Celltreat 2mL 96 Deep Well Plates | Fisher Scientific | 50-828-743 | For mastermix preparation |
| Clear PCR Sealing Sheets | Thermo Scientific | AB0558 | Alternate product may be used |
| DPEC Treated Water | Ambion (Thermo Scientific) | AM9916 | |
| Flip Cap 50 mL Conical Tubes | VWR | 75845-210 | For sample collection |
| Foil PCR Sealing Sheets | Thermo Scientific | AB0626 | For storage of mastermix plates, Alternate product may be used |
| HS_RPP30 Synthetic DNA | Integrated DNA Technologies | 299788131 | P1 positive control |
| Luna Buffer Probe One-Step Reaction | New England Biolabs | M3006B | |
| Luna WarmStart RT Enzyme Mix | New England Biolabs | M3002B | |
| nCOV_N1 Forward Primer, 100 nmol | Integrated DNA Technologies | 10006830 | |
| nCOV_N1 Probe Aliquot, 50 nmol | Integrated DNA Technologies | 10006832 | Probe can be synthesized by other vendors with SYBR or FAM fluophores |
| nCOV_N1 Reverse Primer, 100 nmol | Integrated DNA Technologies | 10006831 | |
| Opentron HEPA Filter Module | Opentrons | N/A | Not required, but useful to reduce contamination |
| Opentron Multichannel Attachment, P20 | Opentrons | 999-00005 | For mastermix preparation |
| Opentron OT-2 Liquid Handling Robot | Opentrons | OT-2 | |
| Opentron Pipette Attachment, P20 | Opentrons | 999-0000215 | For sample loading |
| Oven | Memmert | UF450 PLUS 208V-3PH | |
| PCR Tubes (rnase, dnase free) | Fisher Scientific | 14-230-225 | For aliquots of positive and neg controls |
| PolarSafe Aluminum Cooling Block, 15-Well (1.5/2.0 mL Tubes) | VWR | 10808-952 | |
| PolarSaf Aluminum Cooling Block, 24-Well (0.5mL tubes) | VWR | 10808-956 | |
| RNAse P (ATTO 647) Probe, 50 nmol | Integrated DNA Technologies | 10007062 | Probe can be synthesized by other vendors with Cy5 fluorophore |
| RNAse P Forward Primer, 100nmol | Integrated DNA Technologies | 10006836 | |
| RNAse P Reverse Primer, 100 nmol | Integrated DNA Technologies | 10006837 | |
| Sars-CoV-2 Synthetic RNA Control 2 | Twist Biosciences | 102024 / 103907 / 103909 | N1 Positive Control |
| Scanners | Code | CR1500 | Only required when scaling up |
| Small HEPA Filtered Hood | Erlab | Captair Bio 321 | For mastermix preparation |
| Thermocycler CFX384 Touch | Biorad | CFX384 Touch | Alternate models can be used, e.g. CFX384 Opus |
| X-acto Knife Set | Staples | N/A | To cut foil for keeping control wells covered |
2. Preparation of 20× multiplex N1+P1 probe/primer mix
Prepare a batch of 20× multiplex N1+P1 probe/primer mix (total volume of 20 mL, Table 1) in a 50 mL conical tube in a sterile environment away from synthetic SARS-CoV-2 RNA or patient specimens.
Aliquot 1.6 mL of the mix using a serological pipette into sterile 2.0 mL centrifuge tubes and label appropriately.
Store aliquots in a −20 °C freezer until ready to use.
Table 1:
Components of N1+P1 probe/primer mix.
| Component | Sequence (5’→3’) | Stock Concentration | Volume |
|---|---|---|---|
| 2019-nCoV-N1 Probe | /5FAM/ACCCCGCAT/ZEN/TACGTTTGGTGGACC/3IABkFQ | 50 μM | 500 μL |
| 2019-nCoV-N1-For | GACCCCAAAATCAGCGAAAT | 100 μM | 2000 μL |
| 2019-nCoV-N1-Rev | TCTGGTTACTGCCAGTTGAATCTG | 100 μM | 2000 μL |
| Hs RPP30 Cy5 Probe | /5Cy5/TTCTGACCT/ZEN/GAAGGCTCTGCGCG/3IABkFQ | 50 μM | 500 μL |
| Hs-RPP30-For | AGATTTGGACCTGCGAGCG | 100 μM | 2000 μL |
| Hs-RPP30-Rev | GAGCGGCTGTCTCCACAAGT | 100 μM | 2000 μL |
| Water | - | - | 11000 μL |
3. Preparation of positive control mix
NOTE: Positive control mix should not be made in the same sterile environment as probe/primer mix or other master mix components. A separate container of nuclease-free water should be used.
-
Dilute SARS-CoV-2 synthetic RNA (N1) from 1,000,000 gene copies/μL (cpμ) to 10,000 cpμ by adding 10 μL of stock solution to 990 μL of nuclease-free water. Aliquot 25 μL of 10,000 cpμ into sterile 0.2 mL tubes and label appropriately. Store unused aliquots at −80°C.
NOTE: Synthetic RNA must be stored at −80°C to prevent degradation.
Dilute Hs_RPP30 synthetic DNA (P1) from 200,000 cpμ to 10,000 cpμ by adding 50 μL of stock solution to 950 μL nuclease-free water. Aliquot 25 μL of 10,000 cpμ into sterile 0.2 mL tubes and label appropriately. Store unused aliquots at −80°C.
Dilute each component to a final concentration of 200 cpμ by adding 20 μL of both SARS-CoV-2 and Hs_RPP30 10,000 cpμ stocks to 960 μL of nuclease-free water, for a total of 1000 μL. Aliquot 20 μL of mixed 200 cpμ positive control into 0.2 mL tubes and label appropriately. Store aliquots at −80°C until ready for use.
4. Preparation of master mix plates
NOTE: Make master mix in a sterile environment away from synthetic SARS-CoV-2 RNA or patient specimens. All components must be completely thawed before adding to the mixture; without proper thawing, the concentrations may be incorrect. Inadequate thawing is indicated by the presence of ice or uneven color of reagents. Store on a freezer block while preparing the mixture.
Prepare a batch of multiplex master mix (total volume of 48 mL, Table 2) in a 50 mL conical tube.
Homogenize the mixture by turning the tube over 3x. Do not mix by pipetting up and down or vortexing as this will damage the enzyme.
Fill columns 1–4 of a sterile 96-well deep well reservoir by transferring 1.48 mL of master mix into each well. One 50 mL conical fills four columns, enough for 12 plates.
Cover the deep well reservoir with a foil seal and place it in the dedicated master mix liquid handling robot. Place the filled deep well reservoir on deck 10, place six empty 384-well plates on decks 1–6, and the P20 tip box on deck 11. Uncover the deep well plate and tip boxes and close the robot.
Initialize the custom Python operating protocol by clicking Start Run in the robot desktop application.
After 40 min, the run will pause. Cover the filled 384 well plates with foil seals while they remain in the robot and press down with the roller to ensure adherence. Label the edge of each plate with a batch identifier.
Place six new empty 384-well plates on decks 1–6 and resume operating protocol by clicking Resume Run. After the run is complete, cover the final set of 384 well plates with foil seals.
Pipette 2 μL of the remaining master mix into columns 1–3 and 22–24 of an empty 384-well plate for batch quality control. Seal with an optically clear seal and run on the thermocycler (section 10.1–10.2). If any wells have N1 threshold cycle (Ct) values or if more than 10 wells have P1 Ct values, the batch is contaminated and cannot be used.
Store the prepared master mix plates at 4 °C and use them within 7 days of preparation.
Table 2:
Components of multiplex SARS-CoV-2 master mix.
| Component | Stock Concentration | Volume per reaction | Final Concentration | Batch Volume |
|---|---|---|---|---|
| Luna WarmStart RT Enzyme Mix | 20X | 0.5 μL | 1X | 3 mL |
| Luna Buffer Reaction Mix | 2X | 5.0 μL | 1X | 30 mL |
| N1+P1 Primer/Probe Mix | nCoV N1 F: 10 μM | 0.5 μL | 500 nM | 3 mL |
| nCoV N1 R: 10 μM | 500 nM | |||
| Probe nCoV N1: 2.5 μM | 125 nM | |||
| RPP_30 P1 F: 10 μM | 500 nM | |||
| RPP_30 P1 R: 10 μM | 500 nM | |||
| Probe RPP_30 P1: 2.5 μM | 125 nM | |||
| Nuclease Free Water | --- | 2 μL | --- | 12 mL |
| Subtotal | --- | 8 μL | --- | 48 mL |
| Template | 2 μL |
5. Sample collection, intake, and heat treatment
Instruct participants to avoid eating, drinking, smoking, or conducting dental hygiene 30 min prior to the saliva collection. Instruct participants to collect at least 1 mL of saliva that naturally pools in the mouth and deposit it into a sterile 50 mL conical tube without preservatives, then cap the tube (Supplemental File 3).
Decontaminate the outside of saliva collection tubes with 70% ethanol or disinfecting wipes and transfer them to the laboratory for testing.
Record sample arrival by scanning each sample barcode into the daily intake spreadsheet (Supplemental File 4).
Heat-treat the scanned samples for 30 min in a 95 °C oven. Remove samples while wearing heat-resistant gloves.
NOTE: Untreated samples are stable at room temperature (23 °C) for up to 72 h. Once heat-treated, samples must be stored at 4 °C, if not being processed immediately.
6. Sample Assignment
Open the daily sample loading spreadsheets for each sample loading robot (Supplemental File 5) on the computer at the sample assignment station.
Assign 188 samples to each 384-well plate as follows: Samples 1–48 are considered quarter 1, samples 49–96 are considered quarter 2, samples 97–144 are considered quarter 3, and samples 145–188 are considered quarter 4. Sample are analyzed in duplicate.
Label trays with plate name, date, and quarter number. Scan the samples in order into the sample loading spreadsheet.
NOTE: Samples from earlier plates may need to be manually run, as defined in Figure 3. Refer to sections 8.1–8.3 for manual sample assignment and loading instructions.
Figure 3: Sample resulting flowchart.
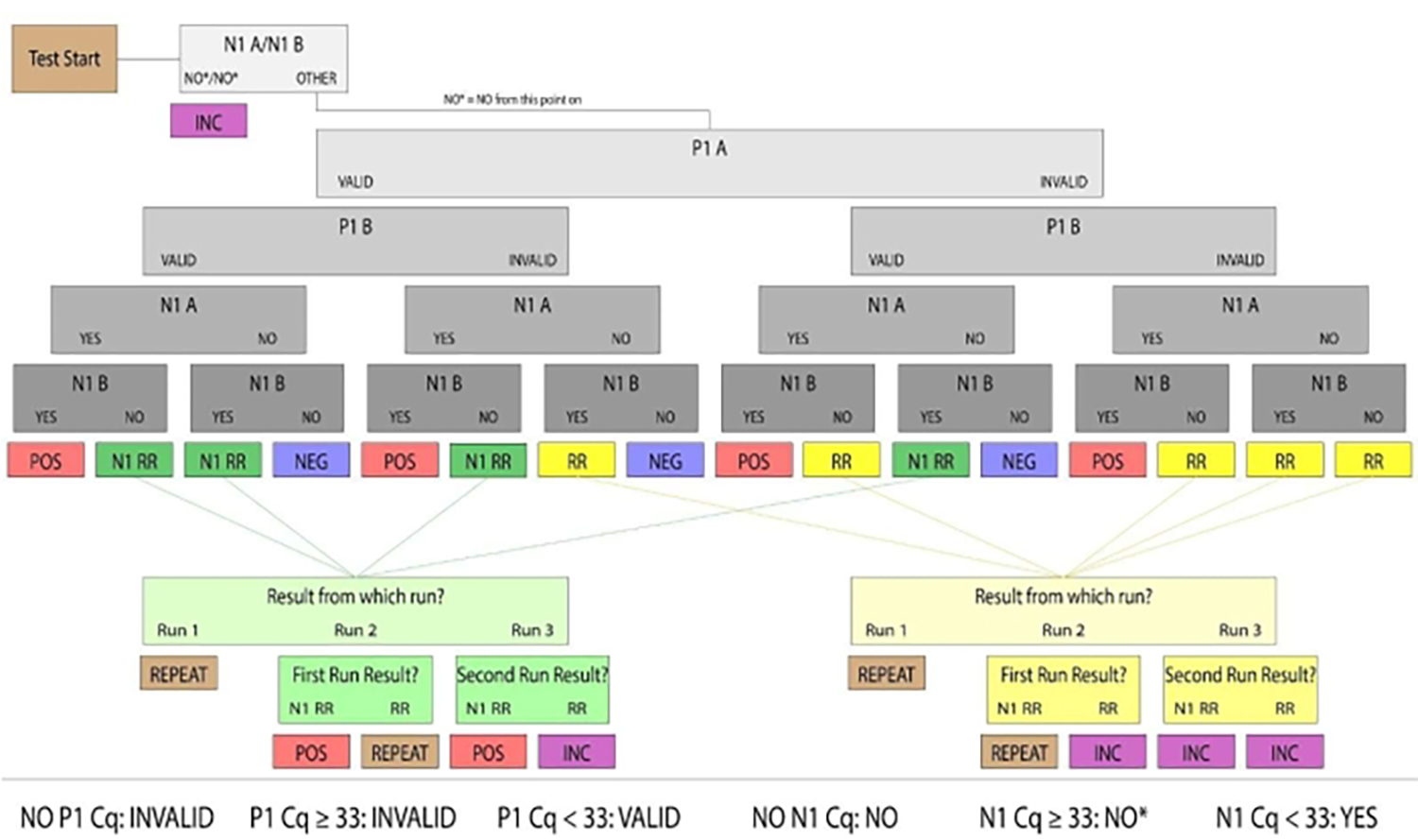
Samples with valid P1 and positive N1 were determined to be human saliva samples positive for SARS-CoV-2. Valid and positive/negative sample results were considered conclusive. Samples that did not produce conclusive results in the first run were categorized as Rerun (denoted RR) or N1 Rerun (denoted N1 RR). Rerun samples had no valid P1 amplification, and N1 Rerun samples had positive N1 amplification in a single replicate. If no valid P1 amplification could be produced by a subsequent manual run, or both replicates had N1 Ct values above the positive threshold (Ct >33), the sample results were considered inconclusive. For clinical purposes, patient samples that did not arrive at the lab, had an insufficient quantity of saliva to pipette or were damaged were considered invalid.
7. Operating sample loading robots
At the sample loading station, line up two full sets of eight 3D printed racks corresponding to the deck placement in the robot.
-
Uncap quarter 1 tubes and place in 3D printed racks, starting with position A1 in rack 1. Fill each rack from left to right and top to bottom. Continue this loading pattern in rack 2, then proceed to racks 4 and 5 (refer to Figure 2C).
NOTE: Racks are not numbered consecutively due to the robot program parameters.
Place loaded quarter 1 sample racks on decks 1, 2, 4, 5, 7, 8, 10, 11. Place P20 tips on decks 3 and 9. To simplify the set-up process, load materials from back to front into the robot.
Take a premade master mix plate from the 4 °C, label it with plate name and use a sharp blade to cut a line in the foil around the control wells (N23/24, O23/24, and P23/24).
Place the master mix plate on deck 6 and peel away the foil cover, leaving behind the small rectangle covering the control wells. Uncover the tip boxes and close the robot.
Initialize the custom Python operating protocol by clicking Start Run through the robot desktop application. Each quarter takes 24.5 min to load onto the plate; set a timer as a reminder.
While the robot is running, uncap and load quarter 2 sample tubes into the second set of 3D printed racks as described in section 7.2.
When the robot pauses, remove quarter 1 racks, and replace them with quarter 2 racks. Click Resume Run in the desktop application.
Recap quarter 1 sample tubes and store them in a 4 °C refrigerator while awaiting results. Repeat this loading process for quarters 3 and 4.
Transfer the loaded plate to a biosafety cabinet. To minimize contamination, keep plate covered during transfer.
Figure 2: Layouts used for the liquid handling robot.
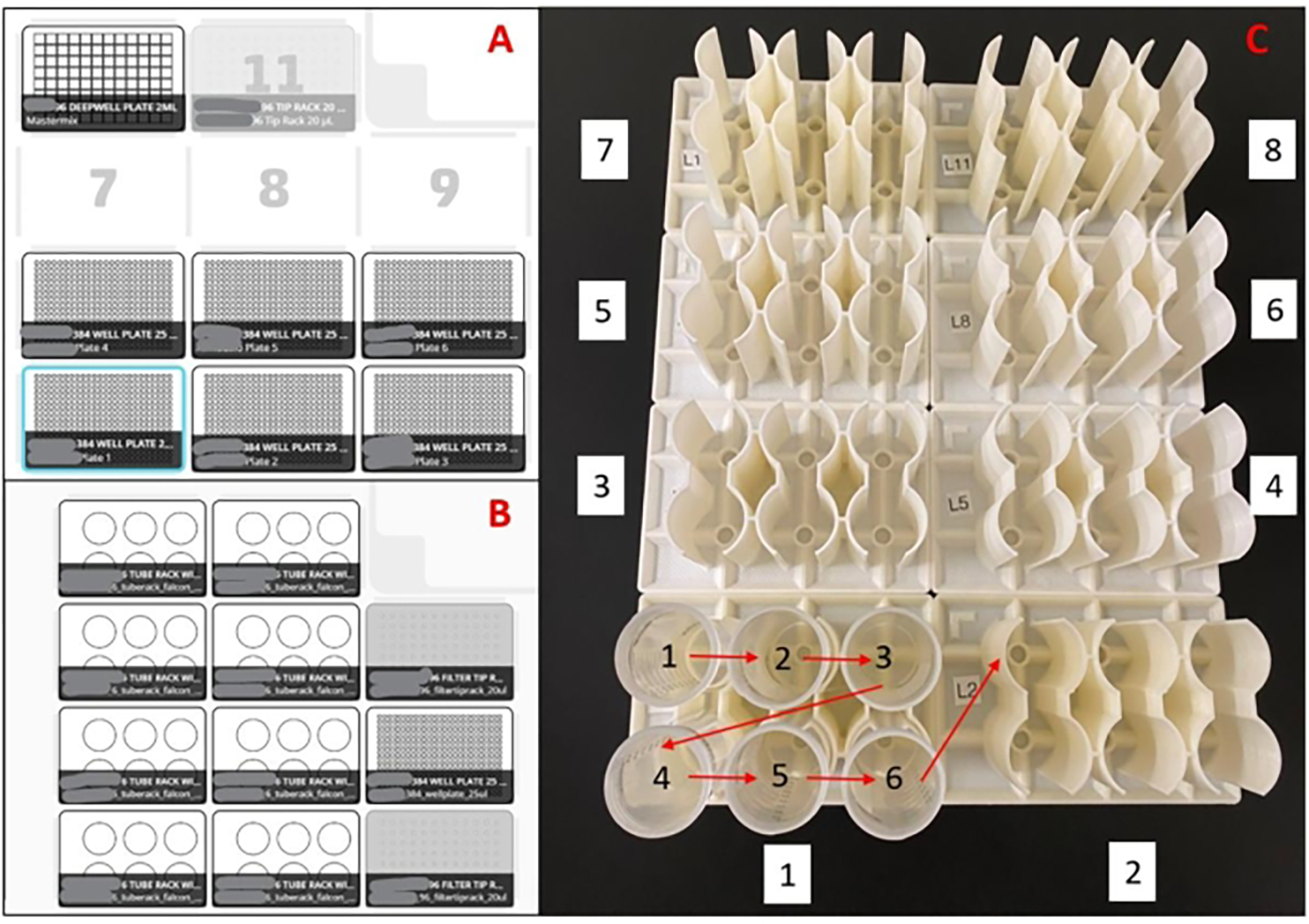
(A) Deck layout for master mix plate preparation robot(s). With an eight-channel pipette, the robot is programmed to pick up pipette tips, aspirate master mix from a 96-well deep well reservoir, dispense master mix into empty 384-well plates, and eject the pipette tips into a waste bin. This is repeated for six plates per run. (B) Deck setup for sample loading robot(s). With a single-channel pipette, the robot is programmed to pick up a pipette tip, aspirate a saliva sample, dispense a saliva sample into duplicate wells of a 384-well master mix plate, and eject the pipette tip into a waste bin. This is repeated for 48 samples per run. (C) Sample tube loading order for 3D printed racks. Red arrows indicate loading order within a rack, and the white boxed numbers indicate the loading order of the entire set of racks. The entire setup will load 188 samples in duplicate into a 384-well plate.
8. Manual sample loading
NOTE: Perform a single manual run on repeat samples (N1 Rerun or Rerun, see Figure 3) in case of inadequate robotic loading.
Gather any repeat samples and assign them as the last samples in quarter 4 (see section 6.2). Number samples, scan the barcodes, and enter the original sample location and result into the sample loading spreadsheet.
Transfer repeat samples to the biosafety cabinet. Do not load repeat sample tubes into the robot loading racks.
Pipette 2 μL of each repeat sample to the correct wells following the plate layout diagram (Supplemental File 6). Use a designated pipette for adding patient samples. Keep control wells covered with foil while adding samples to minimize contamination.
9. Addition of controls to testing plates
Peel away foil cover over control wells using forceps.
Pipette 2 μL of nuclease-free water (no template control) to wells N23-N24 and 2 μL of 200 cpμ mixed positive control (refer to section 3) to wells O23-O24. Pipette 2 μL of a confirmed positive patient sample control into wells M23-M24 as an additional control. Leave wells P23-P24 empty to monitor master mix batch quality.
Cover the plate with an optically clear seal and use the applicator roller to adhere seal to all wells. Vortex the plate at 2500 rpm for 30 sec to mix thoroughly. Centrifuge the plate at 500 × g for 1 min.
10. Performing RT-qPCR
Create a protocol program in the thermocycler software according to the conditions described (Table 3). Save the protocol for future plates. Place the sealed plate in the thermocycler and run the protocol.
-
Export Ct values as a .xslx file and copy the values into the sample loading spreadsheet (Supplemental File 5).
NOTE: These sheets were custom-designed for Ct output files from manufacturer software and may need modification to accept other formats.
Table 3:
Touchdown RT-qPCR protocol.
| Stage | Temperature (°C) | Duration | Number of Cycles |
|---|---|---|---|
| Reverse Transcription | 55 | 10 min | 1 |
| Initial Denaturation | 95 | 1 min | 1 |
| Touchdown | 95 | 10 sec | 3 |
| 72 | 30 sec | ||
| 95 | 10 sec | 3 | |
| 69 | 30 sec | ||
| 95 | 10 sec | 3 | |
| 66 | 30 sec | ||
| Main Amplification | 95 | 10 sec | 40 |
| 65 | 30 sec |
Thermocycling conditions for one-step RT-qPCR SARS-CoV-2 diagnostic assay.
11. Determining plate validity
-
Validate both the positive control and/or known positive samples and the negative control to consider the plate results as valid. Assess the control wells with the following criteria.
For positive control, check if at least one positive control well (O23/O24) produces Ct values of between 22–28 for both P1 and N1 probes. Alternatively, the known positive sample wells (M23/M24) produce P1 and N1 Ct values <33 on the P1 and N1 probes.
For negative control, check that there are no N1 or P1 Ct values in either of the two negative control well (N23/N24). Confirm that Ct values have valid amplification curves before invalidating the plate.
12. Interpreting sample results
-
Determine the patient result following the diagram (Figure 3) and report resolved samples.
-
Assess the P1 result as VALID or INVALID. If P1 produces a result of Ct <33, consider the well VALID and proceed to result from N1. If P1 produces a result of Ct >=33 or no Ct value, consider the well INVALID.
Assess the N1 Result as YES, NO, or NO*. If N1 produces a result of Ct <33, the well is YES. If N1 does not produce a Ct value, the well is NO. If N1 produces a Ct >=33, the well is NO*. Confirm that all N1 Ct values are associated with a real amplification curve. If a Ct value for N1 has no amplification curve, the well is NO.
Identify repeat samples (N1 Rerun or Rerun), label them with an internal sample number and sample type and return them to the loading workflow (section 8.1–8.3).
-
13. Laboratory cleanup
-
Liquid handling robots
Clean all sides with intermediate-level disinfectant. Do not use ethanol as it will degrade the plastic.
Gently wipe pipette tip end and waste bin with an alcohol (70% ethanol or 100% isopropanol) wipe. Wipe down the keyboard and mouse.
-
Biosafety cabinet
Clean all surfaces with intermediate-level disinfectant. Turn on the UV light for 15 min.
Representative Results
We determined the range of detection for RT-qPCR probes and primers for synthetic nucleic acid content for both SARS-CoV-2 (N1) and Hs_RPP30 (P1). A 10-fold serial dilution of known concentrations of combined synthetic SARS-CoV-2 RNA and synthetic Hs_RPP30 DNA in water was done. The following formula was used to convert molecular weight to gene copy number
and RT-qPCR was performed. After carrying out RT-qPCR, linear curves for N1 detection (Figure 4A) and P1 detection (Figure 4B) showed good correlation coefficients across a wide range of gene copy concentrations (R2= 0.9975 and R2= 0.9884, respectively). This result indicates that the combination of primer and probe sets is not inhibitory and can accurately detect SARS-CoV-2 RNA at one gene copy/μL (Cq=33). One gene copy is roughly equivalent to one viral copy; however, we did not determine quantitative viral copy numbers in saliva due to the semi-quantitative nature of RT-qPCR. We attempted to simulate positive saliva samples by spiking synthetic SARS-CoV-2 RNA of known concentrations into virus-free saliva (both heat-treated and non-heat treated) but were unable to produce N1 amplification at low concentrations of RNA (Data not shown). This might be due to RNase degradation or other confounding factors.
Figure 4: RT-qPCR detection of N1 (SARS-CoV-2) synthetic RNA and P1 (Hs_RPP30) synthetic DNA.
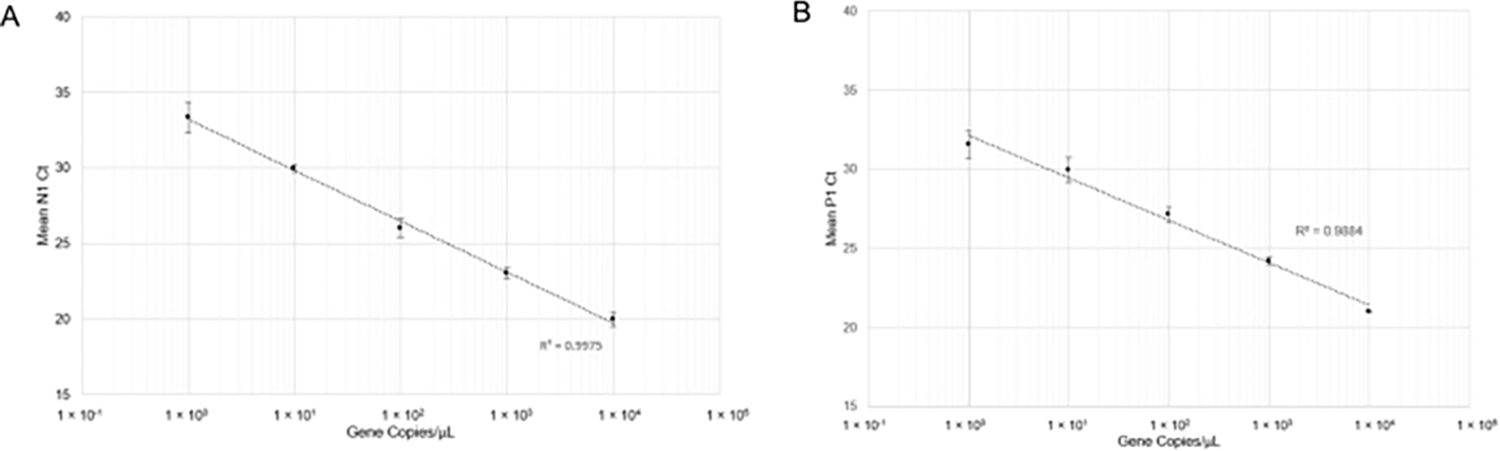
Standard curves were plotted with standard deviations to determine the range of accurate detection using this probe/primer combination. (A) The mean Ct values (n =4) obtained in respective dilutions were plotted against the estimated quantity of synthetic RNA (1×100 to 1×104 RNA copies in 10 μL of RT-qPCR reaction). (B) The mean Ct values (n =3) obtained in respective dilutions were plotted against the estimated quantity of synthetic DNA (1 × 100 to 1 × 104 gene copies in 10 μL of RT-qPCR reaction).
The inter-and intra-assay variability between automated and manual sample loading methods was also assessed. To evaluate inter-assay variability, 20 unique positive samples were loaded using the manual (described in section 8.1–8.3) and automated (described in section 7.1–7.11) methods. N1 Ct values were compared to determine if liquid handling robots and manual sample loading produced equivalent results (Figure 5A). The linear relationship between manual and automated methods produced a high correlation coefficient (R2= 0.9088), indicating that both methods are functionally equivalent. As N1 Ct values increased, variability of Ct values also increased. This trend is likely due to the heterogeneous distribution of viral particles within the saliva, which is more pronounced when fewer particles are present. To evaluate intra-assay variability, a comparison between the N1 Ct values from replicate wells of unique saliva samples using both methods of sample loading was done (Figure 5B). The linear relationship between replicates of automated sample loading (R2= 0.9622) produced a slightly higher correlation coefficient than that of manual loading (R2= 0.9589), indicating high reproducibility of SARS-CoV-2 detection for both loading methods.
Figure 5: Comparison between manual and automated saliva transfer SARS-CoV-2 (N1) Ct values.
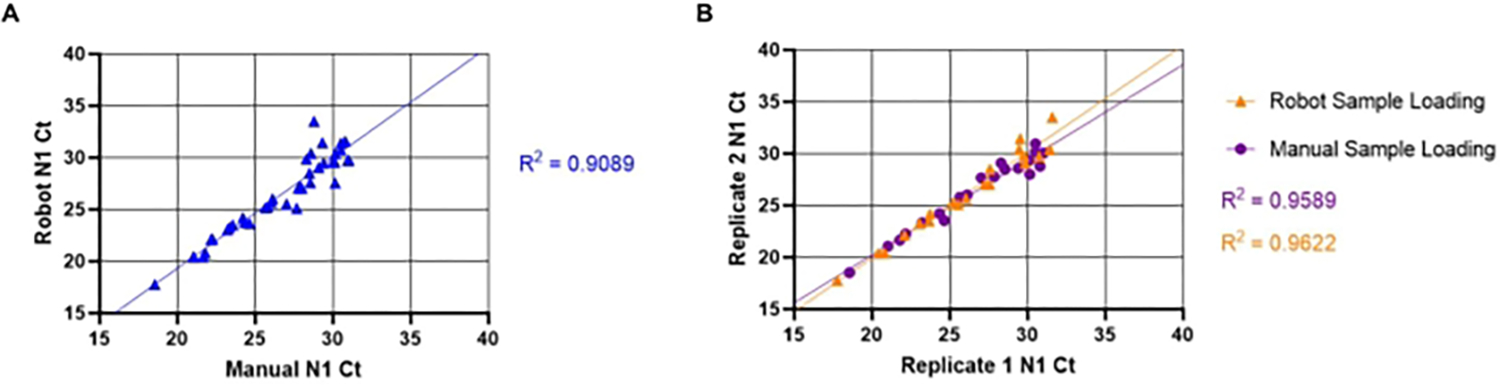
The known SARS-CoV-2 positive saliva samples (n =20) were loaded in duplicate into an RT-qPCR master mix plate by a liquid handling robot. The samples have a Ct value ranging from 18–32 for N1. The same samples were then manually loaded into duplicate wells in a different plate location. (A) N1 Ct values obtained from unique samples using both the robot and manual sample loading were transposed to determine inter-assay variability between manual and robot loading. (B) Intra-assay variability was also determined by using transposed replicate of N1 Ct values obtained from both robot and manual sample loading.
Finally, an evaluation of saliva viscosity reduction with respect to the heat treatment methods was done (Figure 6). Saliva was obtained from a single source to eliminate sample variability. Greater variability in P1 Ct values within one heat treatment method may be indicative of higher sample viscosity as viscous saliva cannot be aspirated and dispensed precisely. Both 30 min and 60 min heat treatment methods produced significantly decreased sample variability when compared to no treatment control (p = 0.0006 and p = 0.0429, respectively). There was no significant difference between 30 min and 60 min treatments (p = 0.2245); therefore, the 30-min heat treatment method was implemented to reduce processing time.
Figure 6: Evaluation of heat treatment methods for viscosity reduction in saliva.
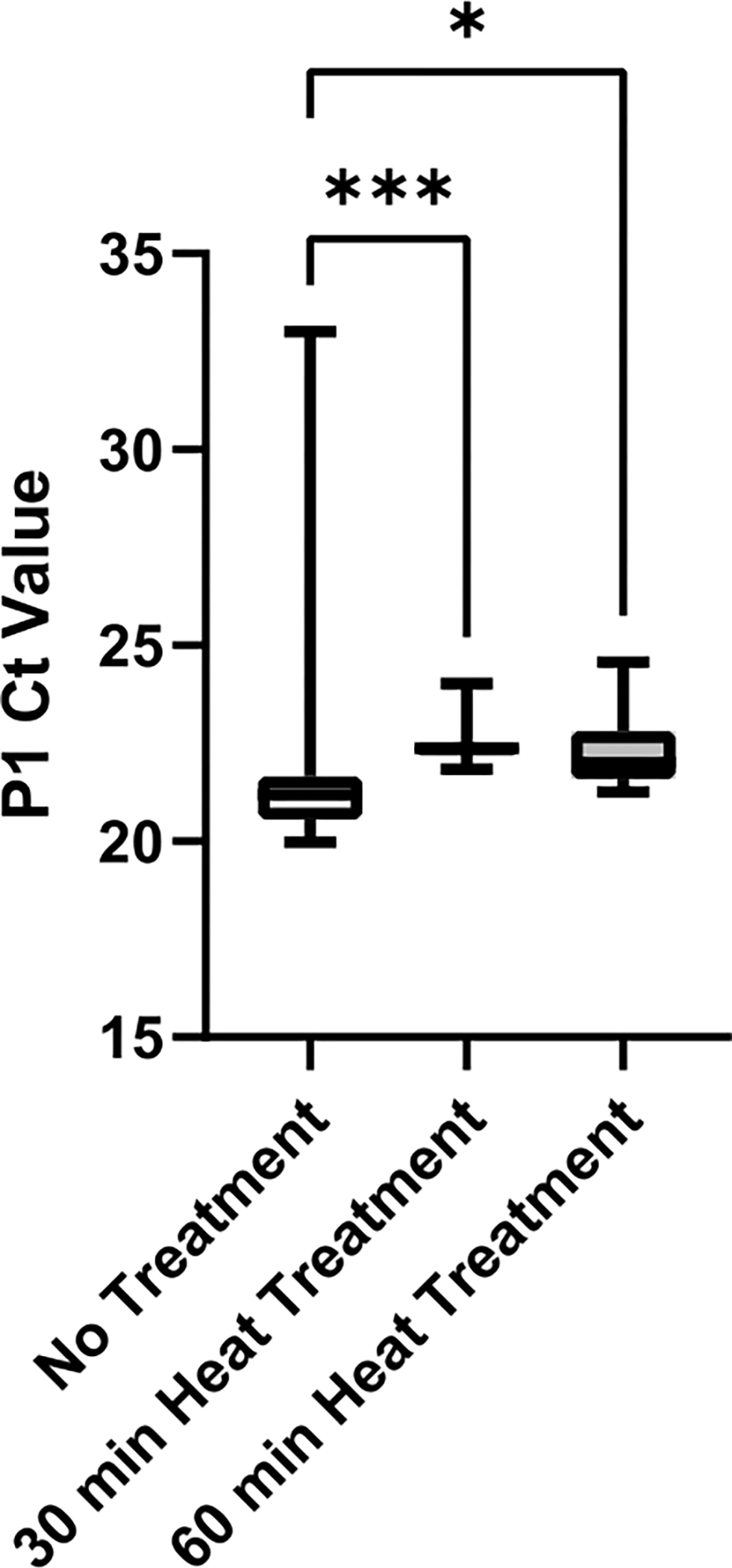
SARS-CoV-2 negative saliva was collected from a single source and aliquots were heat-treated for either 0 min, 30 min, or 60 min at 95 °C. P1 Ct values from technical replicates (n =12) of each condition were plotted to determine variability between treatment methods. Pairwise comparisons between groups were evaluated with an unpaired t-test (*** indicates p <0.001, * indicates p <0.05).
Discussion
The assay described in the protocol was assessed by an independent validation study. It was found that the assay had 98.9% specificity (1.1% false positive) and 90.0% sensitivity (10.0% false negative) when evaluated against paired nasopharyngeal swabs taken at the same time (n =837; 817 negatives, 20 positive). Importantly, three participants who tested positive with TigerSaliva and negative with a nasopharyngeal swab were retested with swabs 48 h later and returned positive results, indicating that TigerSaliva may be able to detect SARS-CoV-2 infections earlier in the course of illness.
We simulated positive saliva samples by spiking virus-free saliva (both heat-treated and non-heat treated) with known concentrations of SARS-CoV-2 synthetic RNA and performed a 10-fold dilution to determine the Ct limit in saliva. The N1 gene was not detectable below 10,000 gene copies (approximately Ct = 28) in simulated positive samples. We suspect this is due to RNase degradation or other confounding factors. However, the interaction of saliva RNases with naked synthetic RNA is likely different from the interaction with viral particles, even after they have been denatured by heat. Positive saliva samples have been identified with Ct >30 and external labs obtained SARS-CoV-2 genetic sequence data from these samples. We speculate that the viral proteins provide protection from RNA degradation in patient saliva samples.
The most critical step in the protocol is the implementation of automation for master mix preparation and saliva sample processing (sections 4 and 7 respectively). This allows for overlapping task processes, which drastically reduces turnaround time. Another critical step is clinical result interpretation (sections 11 and 12). Establishing intermediate result categories (Rerun and N1 Rerun) also minimized the occurrence of inconclusive test results.
We demonstrated that variation between manual and automated saliva sample loading methods is negligible (Figure 5A) and that automation may improve the reproducibility of SARS-CoV-2 detection (Figure 5B). Automation should be favored to facilitate testing when designing and expanding clinical labs25. Laboratory workflow is improved with the implementation of robot-automated tasks26. Open-source capabilities of the liquid handling robots allow for the implementation of custom scripting for protocol design. This makes liquid handling robots an inexpensive and highly modifiable system compared to traditional clinical automation methods. It is also an ideal strategy for executing highly repetitive laboratory tasks. The high level of customizability of the system translates to freedom to alter labware (e.g., collection tubes, pipette tips, or 384-well plates) in case of shortages. Therefore, automation by using liquid handling robots is viable for both large-scale and small-scale surveillance and research.
A major advantage of this testing strategy is a much shorter turnaround time relative to other clinical labs. The utilization of automated liquid handling robots plays a key role in reducing turnaround time, but concurrent use of robots and thermocyclers is also instrumental in maximizing testing efficiency. One robot and thermocycler should be operated as a pair, where both machines are used in tandem for uninterrupted sample loading and sample result analysis. Once a steady flow of assigned samples is established, all machine pairs can be operated simultaneously. Constant concurrent use of robots and thermocyclers drastically increases testing capacity and efficiency, which is crucial to accommodate the high testing volume.
In contrast with other established SARS-CoV-2 RT-qPCR protocols, we included a touchdown step in the thermocycler protocol to improve annealing of the probe and primer sets to the target genes27, reducing the risk of failed amplification. The results demonstrated that the touchdown improved the detection of positive samples without risking the loss of specific primer binding (Table 4). We determined that a wide range of both SARS-CoV-2 RNA copies (Figure 4A) and Hs_RPP30 DNA copies (Figure 4B) can be simultaneously detected by the RT-qPCR assay.
Table 4:
Comparison of touchdown Ct values for five positive samples against no touchdown Ct values.
| Touchdown Step | No Touchdown Step | |||
|---|---|---|---|---|
| Mean N1 Ct | Mean P1 Ct | Mean N1 Ct | Mean P1 Ct | |
| Sample 1 | 19.65 | 22.7 | 27.8 | 28.3 |
| Sample 2 | 22.24 | 24.9 | 28.77 | 30.5 |
| Sample 3 | 18.85 | 19.2 | 24.65 | 25.9 |
| Sample 4 | 25.56 | 22.8 | 31.93 | 29.2 |
| Sample 5 | 22.34 | 24.8 | 38.48 | 40.0 (Failed detection) |
One limitation of liquid handling robots is the possibility of cross-contamination from positive samples during saliva transfer. Saliva is a viscoelastic fluid28 and may string across adjacent wells after being dispensed from the pipette tip. Furthermore, the heterogeneity of saliva29 may cause uneven distribution of viral particles throughout the sample. This increases the possibility of both false positives and negatives, necessitating the designation of N1 Rerun and Rerun samples. However, 14.1% of samples initially designated as N1 Rerun resolved as positive for SARS-CoV-2 and were more than 30 times more likely than Rerun samples to resolve as positive after retesting. Consequently, differentiating Rerun from N1 Rerun (Figure 3) allowed for more accurate separation of potentially positive samples, increasing the sensitivity and specificity of our diagnostic assay. Other resulting parameters for diagnostic saliva testing did not make this distinction12,14,24,30,31.
Saliva specimens can be difficult to pipette due to heterogeneity and viscosity32. Heat treatment adequately denatures proteins in the saliva biomatrix, reducing viscosity and eliminating the need for RNA extraction reagents9, which were scarce during the early stages of the pandemic10. Extended heat treatment also inactivates present viruses33 which allows for laboratory processing at lower biosafety levels. Consequently, a heat-based RNA extraction (described in section 5.4) was implemented to decrease viscosity through protein denaturation (Figure 6). Based on the results, we postulate that heat treatment may also homogenize saliva samples in addition to denaturing the protein biomatrix. Other groups combined heat treatment and proteinase K treatment to increase homogeneity9,14,34. We chose to not implement this step as it may denature virion proteins at a rate that leaves viral RNA exposed to heat degradation35. Furthermore, sample dilution with proteinase K may mask positive samples containing fewer viral particles thus, decreasing sensitivity. In addition, the assay results were compared to a commercially available saliva-based SARS-CoV-2 assay (Logix Smart COVID-19) which uses magnetic bead RNA extraction (Table 5). It was found that the current assay was better suited at detecting weak positive samples as compared to the commercially available assay.
Table 5:
Comparison of TigerSaliva Ct results and commercially available saliva-based SARS-CoV-2 assay results.
| Sample | TigerSaliva | Commercially available saliva-based SARS-CoV-2 assay | ||
|---|---|---|---|---|
| N1 Ct | P1 Ct | Covid-19 Value | RNaseP Value | |
| D11 | 16.4 | 18.1 | 20.86 | 23.4 |
| E11 | 18.9 | 19.1 | 25.6 | 21.2 |
| F11 | 19.5 | 18.4 | 22.8 | 22.2 |
| G11 | 22.2 | 19.1 | 23.7 | 22.9 |
| H11 | 26.4 | 21.3 | 32.2 | 26.7 |
| A12 | 14.8 | 16.5 | 29.15 | 19 |
| B12 | 24 | 19.6 | 31.05 | 21.35 |
| C12 | 14.9 | 17.5 | 20.84 | 18.9 |
Both assays were performed on the same saliva samples (n =8).
It is difficult to quantify virus copy number in saliva using only RT-qPCR, because qPCR is semi-quantitative. There is inherent variation between Ct values that originates from technical limitations. Gene copy number can be determined from Ct values (Figure 4) and is roughly equivalent to viral copy number. One possible solution to determine the viral copy number in saliva samples is ddPCR, which provides hard quantification of gene copies in the reaction. However, we believe that it is adequate to provide qualitative results to clinicians and relative viral content can be compared across samples processed with our methods.
Despite some limitations that arise when using saliva, the SARS-CoV-2 assay by saliva-based RT-qPCR proves to be an effective method for quick and reliable viral RNA detection at any scale of testing. This is especially true when coupled with the utilization of open-source liquid handling systems. This testing approach can be modified to detect other nucleic acid sequences relevant to diagnostics, such as infectious disease agents, disease markers, or other viruses. This makes the assay applicable for both clinical and research diagnostic efforts.
Supplementary Material
Supplemental Figure 1: Comparison of N1 Ct in low P1 Ct saliva samples. The positive samples with low P1 Ct were selected and compared with the N1 Ct (n =106). The N1 Ct values ranged from 14–33, indicating the assay has a dynamic range in saliva samples that is comparable to the standard curve.
Supplemental File 1: Custom script for robot master mix plate creation.
Supplemental File 2: Custom script for saliva processing on sample loading robots.
Supplemental File 3: Instructions for self-collection of high-quality saliva samples from participants. Further details can be found in the short video description of the testing process available at https://www.clemson.edu/centers-institutes/reddilab/index.html.
Supplemental File 4: Sample intake spreadsheet.
Supplemental File 5: Sample loading spreadsheet.
Supplemental File 6: Sample 384-well plate layout diagram.
Acknowledgments
The authors thank Clemson’s administration, medical staff, and clinical lab employees at the REDDI Lab who helped implement and manage SARS-CoV-2 testing. We thank Dr. Phillip Buckhaults and Dr. Carolyn Bannister from the University of South Carolina for initial project consulting and industry contacts for equipment procurement. We thank many students, professors, and staff for their assistance in sample collection. Thank you to Creative Inquiry students for standard curve data collection. Funding for this study was received from the National Institutes of Health grant P20GM121342 (awarded to DD and LGP), Clemson Athletic Department, Clemson University’s Vice President for Research, and the South Carolina Governor & Joint Bond Review Committee.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/63395.
Disclosures
The authors have nothing to disclose. The assay described in the protocol falls under the SalivaDirect EUA filed by Yale School of Public Health.
References
- 1.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. New England Journal Medicine. 382 (8), 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes and Infection. 22 (2), 69–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen E et al. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. The Lancet. Infectious Diseases. 20 (9), e238–244 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honein MA et al. Summary of Guidance for Public Health Strategies to Address High Levels of Community Transmission of SARS-CoV-2 and Related Deaths, December 2020. MMWR. Morbidity and Mortality Weekly Report. 69 (49), 1860–1867 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Surveillance strategies for COVID-19 human infection: interim guidance, 10 May 2020. World Health Organization. https://apps.who.int/iris/handle/10665/332051 (2020). [Google Scholar]
- 6.Centers for Disease Control. Screening testing for early detection of SARS-CoV-2 infection. Division of viral diseases. 2 July 2020. Centers for Disease Control. https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html#PublicHealthSurveillance (2020). [Google Scholar]
- 7.Larremore DB et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Science Advances. 7 (1), eabd5393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sethuraman N, Jeremiah SS, Ryo A Interpreting Diagnostic Tests for SARS-CoV-2. JAMA. 323 (22), 2249–2251 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Chu AW et al. Evaluation of simple nucleic acid extraction methods for the detection of SARS-CoV-2 in nasopharyngeal and saliva specimens during global shortage of extraction kits. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 129, 104519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan D et al. Global supply-chain effects of COVID-19 control measures. Nature Human Behaviour. 4 (6), 577–587 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Bastos ML, Perlman-Arrow S, Menzies D, Campbell JR The Sensitivity and Costs of Testing for SARS-CoV-2 Infection With Saliva Versus Nasopharyngeal Swabs: A Systematic Review and Meta-analysis. Annals of Internal Medicine. 174 (4), 501–510 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasomsub E et al. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clinical Microbiology and Infection: The Official Publication of The European Society of Clinical Microbiology and Infectious Diseases. 27 (2), 285.e1–285.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.To KK et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clinical Infectious Diseases: An Official Publication of The Infectious Diseases Society of America. 71 (15), 841–843 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogels CB et al. SalivaDirect: A simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med (New York, N.Y.). 2 (3), 263–280.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griesemer SB et al. Evaluation of Specimen Types and Saliva Stabilization Solutions for SARS-CoV-2 Testing. Journal of Clinical Microbiology. 59 (5), e1418–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wehrhahn MC et al. Self-collection: An appropriate alternative during the SARS-CoV-2 pandemic. Journal of Clinical Virology: The Official Publication of The Pan American Society for Clinical Virology. 128, 104417. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barza R, Patel P, Sabatini L, Singh K Use of a simplified sample processing step without RNA extraction for direct SARS-CoV-2 RT-PCR detection. Journal of Clinical Virology: The Official Publication of The Pan American Society for Clinical Virology. 132, 104587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paltiel AD, Zheng A, Walensky RP Assessment of SARS-CoV-2 Screening Strategies to Permit the Safe Reopening of College Campuses in the United States. JAMA Network Open. 3 (7), e2016818 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centers for Disease Control. 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Primers and Probes. 2020. May 29. U.S. Department of Health and Human Services. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html. Accessed May 25, 2021 (2020). [Google Scholar]
- 20.Cresswell K, Ramalingam S, Sheikh A Can Robots Improve Testing Capacity for SARS-CoV-2? Journal of Medical Internet Research. 22 (8), e20169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villanueva-Cañas JL et al. Implementation of an open-source robotic platform for SARS-CoV-2 testing by real-time RT-PCR. PLoS One, 16 (7), e0252509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.University of South Carolina College of Pharmacy. Robot Boosts COVID-19 Testing Efficiency. https://sc.edu/study/colleges_schools/pharmacy/about/news/2020/robot-boosts-testing-efforts.php (2020).
- 23.Matic N et al. Practical challenges to the clinical implementation of saliva for SARS-CoV-2 detection. European Journal of Clinical Microbiology & Infectious Diseases: Official Publication of The European Society of Clinical Microbiology. 40 (2), 447–450 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahajpal NS et al. SalivaSTAT: Direct-PCR and Pooling of Saliva Samples Collected in Healthcare and Community Setting for SARS-CoV-2 Mass Surveillance. Diagnostics (Basel, Switzerland). 11 (5), 904 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genzen JR et al. Challenges and Opportunities in Implementing Total Laboratory Automation. Clinical Chemistry. 64 (2), 259–264 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Archetti C, Montanelli A, Finazzi D, Caimi L, Garrafa E Clinical Laboratory Automation: A Case Study. Journal of Public Health Research. 6 (1), 881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hecker KH, Roux KH High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. BioTechniques. 20 (3), 478–485 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Bhat PP et al. Formation of beads-on-a-string structures during break-up of viscoelastic filaments. Nature Physics. 6, 625–631 (2010). [Google Scholar]
- 29.Miller CS et al. Current developments in salivary diagnostics. Biomarkers in Medicine. 4 (1), 171–189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno-Contreras J et al. Saliva Sampling and Its Direct Lysis, an Excellent Option To Increase the Number of SARS-CoV-2 Diagnostic Tests in Settings with Supply Shortages. Journal of Clinical Microbiology. 58 (10), e01659–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 323 (18), 1843–1844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landry ML, Criscuolo J, Peaper DR Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. Journal of Clinical Virology: The Official Publication of The Pan American Society for Clinical Virology. 130, 104567 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lista MJ et al. Resilient SARS-CoV-2 diagnostics workflows including viral heat inactivation. PLoS One. 16 (9), e0256813 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brotons P et al. Validation and implementation of a direct RT-qPCR method for rapid screening of SARS-CoV-2 infection by using non-invasive saliva samples. International Journal of Infectious Diseases: IJID: Official Publication of The International Society for Infectious Diseases. 110, 363–370 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batéjat C, Grassin Q, Manuguerra JC, Leclercr I Heat inactivation of the severe acute respiratory syndrome coronavirus 2. Journal of Biosafety and Biosecurity. 3 (1), 1–3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Comparison of N1 Ct in low P1 Ct saliva samples. The positive samples with low P1 Ct were selected and compared with the N1 Ct (n =106). The N1 Ct values ranged from 14–33, indicating the assay has a dynamic range in saliva samples that is comparable to the standard curve.
Supplemental File 1: Custom script for robot master mix plate creation.
Supplemental File 2: Custom script for saliva processing on sample loading robots.
Supplemental File 3: Instructions for self-collection of high-quality saliva samples from participants. Further details can be found in the short video description of the testing process available at https://www.clemson.edu/centers-institutes/reddilab/index.html.
Supplemental File 4: Sample intake spreadsheet.
Supplemental File 5: Sample loading spreadsheet.
Supplemental File 6: Sample 384-well plate layout diagram.


