ABSTRACT
Streptococcus pyogenes is a major human pathogen with high genetic diversity, largely created by recombination and horizontal gene transfer, making it difficult to use single nucleotide polymorphism (SNP)-based genome-wide analyses for surveillance. Using a gene-by-gene approach on 208 complete genomes of S. pyogenes, a novel whole-genome multilocus sequence typing (wgMLST) schema was developed, comprising 3,044 target loci. The schema was used for core-genome MLST (cgMLST) analyses of previously published data sets and 265 newly sequenced draft genomes with other molecular and phenotypic typing data. Clustering based on cgMLST data supported the genetic heterogeneity of many emm types and correlated poorly with pulsed-field gel electrophoresis macrorestriction profiling, superantigen gene profiling, and MLST sequence type, highlighting the limitations of older typing methods. While 763 loci were present in all isolates of a data set representative of S. pyogenes genetic diversity, the proposed schema allows scalable cgMLST analysis, which can include more loci for an increased resolution when typing closely related isolates. The cgMLST and PopPUNK clusters were broadly consistent in this diverse population. The cgMLST analyses presented results comparable to those of SNP-based methods in the identification of two recently emerged sublineages of emm1 and emm89 and the clarification of the genetic relatedness among isolates recovered in outbreak contexts. The schema was thoroughly annotated and made publicly available on the chewie-NS online platform (https://chewbbaca.online/species/1/schemas/1), providing a framework for high-resolution typing and analyzing the genetic variability of loci of particular biological interest.
KEYWORDS: outbreak, Streptococcus pyogenes, bioinformatics, genomics, group A Streptococcus, molecular epidemiology, molecular subtyping, population genetics, surveillance studies, typing
INTRODUCTION
Streptococcus pyogenes (Lancefield group A Streptococcus [GAS]) remains a significant cause of global morbidity and ranks among the top 10 infectious causes of death (1). In 2018, the World Health Organization highlighted the importance of developing a GAS vaccine and set out priority activities to reach this goal, including a better characterization of the epidemiology of GAS infections and the identification of appropriate candidate antigens (2).
In recent decades, sequence-based typing of the hypervariable region of the emm gene, encoding the M protein, was the most frequently used method to identify GAS lineages (3). However, complementary methods have long been used, which, together with emm typing, allow finer discrimination of the circulating strains, including serotyping of the major backbone pilus protein (T antigen), pulsed-field gel electrophoresis (PFGE) macrorestriction profiling, multilocus sequence typing (MLST), and profiling of superantigen (SAg)-coding genes (4–6).
Whole-genome sequencing analysis allowed the identification of emerging intra-emm clones with increased fitness or virulence that were otherwise indistinguishable by other typing methods. Such is the case of emm89 clade 3, which emerged during the 2000s and quickly outcompeted other emm89 lineages (7–9). Isolates from this lineage lack the genes encoding hyaluronic acid capsule biosynthesis and carry a high-expression promoter in the operon encoding streptolysin O and NAD-glycohydrolase (Pnga-3) (8, 10). More recently, an emm1 lineage (M1UK), differing from the contemporary globally disseminated emm1 lineage (M1global) by 27 single nucleotide polymorphisms (SNPs), was identified in the United Kingdom (11) and subsequently reported in The Netherlands, the United States, and Canada (12–14).
Whole-genome sequencing has been decisive in clarifying the molecular and evolutionary mechanisms underlying the success of long-term-circulating lineages (15, 16) and has proven useful in the identification of outbreak-related cases (17, 18). Genomic data have the additional potential benefit of providing information on the variability of candidate vaccine antigens and genes involved in antimicrobial resistance (19, 20), further supporting the use of high-throughput sequencing (HTS) in GAS surveillance.
Most genome-wide analyses performed on GAS have been based on the comparison of SNPs between isolates. This usually involves mapping short-read sequence data or aligning de novo-assembled sequences to a selected reference genome (8–11, 15–19). However, the choice of an appropriate reference is challenging when simultaneously comparing diverse lineages (21, 22), such as in population-based studies of S. pyogenes infection isolates. SNP-based phylogenetic analysis also requires the removal of regions of recombination, which are an important source of diversity in GAS (19, 21–23). These limitations can be largely overcome by the use of gene-by-gene approaches like whole-genome MLST (wgMLST) or core-genome MLST (cgMLST) (24), which do not require comparison to a reference genome and which intrinsically dampen the effect of recombination (21, 22, 25). Minimum-spanning-tree (MST)-like downstream analyses further facilitate the use of wg/cgMLST. Additionally, wg/cgMLST schemas can be curated and maintained in centralized databases, providing a standardized nomenclature and ensuring reproducibility and comparison of results across laboratories (21, 24, 26, 27). Indeed, similarly to SNP-based approaches, cgMLST schemas have been successfully used for both outbreak identification and population-based surveillance of multiple pathogens (22, 25–30). However, it is important to remember that wg/cgMLST is not designed to interrogate noncoding regions of the genome and therefore would have been unable to detect the polymorphisms in the nga-ifs-slo promoter present now in the M1global lineage (11).
The aims of this study were to define a publicly available annotated wgMLST schema for S. pyogenes and evaluate its suitability for high-resolution typing and documenting the variability of loci encoding proteins of biological relevance.
MATERIALS AND METHODS
Bacterial strains and data sets.
A collection of 265 nonduplicate GAS strains isolated from pharyngitis, skin and soft tissue infections, and normally sterile sites in Portugal between 2001 and 2009 was selected for HTS and comparison of cgMLST with other typing methods (see supplemental Data Set 1 in reference 31). These isolates were previously characterized regarding emm type, T type, PFGE profile, SAg gene profile, and antimicrobial resistance (5, 32–35) and represent four emm types: emm1, emm3, emm4 (including erythromycin-resistant and -susceptible isolates), and emm89 (including isolates carrying Pnga-1, Pnga-2, and Pnga-3) (7).
In order to evaluate the performance of the proposed wgMLST schema in more diverse collections, outbreak recognition, and the identification of recently emerged intra-emm lineages of interest, publicly available data sets from three previous publications were also included (11, 18, 19). Data Set 2 comprises 2,006 assemblies from a collection of isolates previously selected to represent the genetic, geographic, temporal, and clinical diversity of GAS (19). Data Set 3 consists of 119 isolates associated with 21 outbreaks recorded in England from 2010 to 2015 and 170 contemporaneous sporadic isolates with the same emm types (18). Data Set 4 comprises 135 assemblies from noninvasive emm1 isolates recovered in the United Kingdom from 2009 to 2016 (11) and the MGAS5005 complete genome that was used as a reference. The United Kingdom assemblies include 123 isolates carrying 27 SNPs characteristic of the recently emerged M1UK lineage and 5 intermediate isolates carrying 13 or 23 of those SNPs (11). Data Set 5 includes all the emm89 assemblies included in Data Sets 1 to 4 (n = 194) and the 7 complete genomes of emm89 that were used to create the schema.
The majority of the strains included in Data Sets 2 to 4 (31) were retrieved from collections of publicly available genome assemblies (36, 37). For strains for which it was not possible to retrieve a public genome assembly, the raw sequencing data were downloaded from the European Nucleotide Archive (ENA) and subsequently assembled. All assemblies were filtered according to assembly quality, emm type, and multilocus sequence type (ST) criteria, as detailed below.
High-throughput sequencing.
Genomic DNA was extracted from cultures of GAS grown overnight in Todd-Hewitt broth (Oxoid, Basingstoke, UK) using the PureLink genomic DNA minikit (Invitrogen, Carlsbad, CA, USA). The initial bacterial lysis step was carried out in the presence of 45 U of mutanolysin (Sigma-Aldrich, St. Louis, MO, USA) and 86 μg of hyaluronidase (Sigma-Aldrich, St. Louis, MO, USA). Whole-genome sequencing libraries were generated using the Nextera DNA library preparation kit (Illumina, San Diego, CA, USA). The libraries were sequenced in an Illumina MiSeq or NextSeq instrument.
Sequencing data analysis.
Raw sequence reads were assembled with INNUca v4.2.2 (38), with the following parameters: -s Streptococcus pyogenes, -g 2, –estimatedMinimumCoverage 10, –trueCoverageProceed, and –fastQCproceed. Samples that failed any of the quality control steps related to sequence quality or assembly coverage were excluded from the data sets. Assemblies are available as supplemental material (31).
In silico ST prediction was performed using MLST v2.19.0 (39) with default parameters and the PubMLST database updated on 11 March 2021. Genome assemblies with partial matches to any of the MLST genes or for which it was not possible to identify at least one of the MLST genes were excluded, except for ST293, ST403, ST404, and ST688, which lack the yqiL gene, and ST1087, which lacks the xpt gene. Strains with a predicted ST that was inconsistent with the classification reported in the original study were also excluded.
The emm type was determined using emmTyper v0.2.0 (40) with verbose mode and the CDC M-type-specific sequence databases updated on 11 March 2021. Genome assemblies without an identified emm type, with matches only for alleles flagged in the CDC database as possible emm-like genes, or with a predicted emm type that was inconsistent with the classification reported in the original study were excluded from the data sets. Assemblies classified with multiple emm types were also excluded (multiple subtypes of the same emm type were accepted), except for emm34/emm230 (emm34 corresponds to the enn gene, and the emm type is 230), emm13L/emm13 (these two types correspond to the same sequence), and other cases that were inspected in Geneious v8.1.9 to validate matches to the emm gene.
Variant calling to determine the set of SNPs in each assembled genome from Data Set 4 (31) was performed with Snippy v4.6.0 (41) with default parameters and the complete genome of strain MGAS5005 (RefSeq accession no. GCF_000011765.3) as the reference strain.
The Pnga variant was determined with SeqTyper v2.3 (42) with default parameters. A Fasta file with the sequences for all variants was given as the input to the blast module, followed by variant calling with the assembly module.
Schema creation, annotation, and curation.
The complete genomes available in the NCBI RefSeq database as of 20 July 2020 were downloaded to select a set of 208 genome assemblies (see Table S1 in the supplemental material) (31) for schema creation with chewBBACA v2.7.0 (43). Assemblies with a status of suppressed in the NCBI database were excluded, except for accession no. GCF_001535505.1, GCF_001547815.1, GCF_000013525.1, and GCF_900636425.1, whose status was changed to suppressed after the schema creation and allele-calling processes. Loci originating from these four genomes were inspected to ensure their validity. This initial schema seed, composed of 3,318 distinct loci, was populated through the inclusion of allelic variants from all assemblies included in the data sets and sourced from public databases (36, 37). For schema annotation, the chewBBACA UniprotFinder process and custom scripts (44) were used to create a file with locus coordinates and annotation terms selected from the UniProt database, prioritizing the selection of terms from Swiss-Prot over terms from TrEMBL, and from matches against the translated coding sequences in the GenBank files of the genomes used for schema creation. Some product and gene names were further complemented with relevant literature references. The annotated schema was thoroughly curated to identify and remove spurious loci such as gene fusions, truncated genes, and paralogous loci. These loci were identified based on the retrieved annotations, the inspection of the genomic context, and the list of paralogous loci reported by the chewBBACA AlleleCall process and a custom script evaluating interlocus similarity (44). Due to the minimum sequence length parameter enforced during schema creation, the sagA gene, present in the streptolysin S-encoding operon, was not in the initial schema. Given the importance of this gene for GAS pathogenesis and the potential interest in its variability, a locus was added representing the sagA gene. The full list of changes applied to the schema is available in Table S2 (31).
The schema was uploaded to chewie-NS (45), where a more detailed description of schema creation, annotation, and curation can be found (https://chewbbaca.online/species/1/schemas/1).
cgMLST analysis.
Allelic profiles of the core loci (shared by 100% of the isolates under analysis [cgMLST-100]) were used to create MSTs with the goeBURST algorithm in the desktop or online version of PHYLOViZ (46, 47). Groups of isolates linked by up to n different loci in the MST were determined using the desktop version. The genes present in 95% (cgMLST-95), 99% (cgMLST-99), and 100% (cgMLST-100) of the isolates were identified with the chewBBACA ExtractCgMLST process for Data Set 2. The lists of genes for each gene presence threshold are available as supplemental material (31).
Intracluster and intercluster pairwise distances were determined using custom scripts (44).
PFGE cluster definition.
Previously generated SmaI/Cfr9I macrorestriction PFGE patterns (5, 32, 34, 35) were used to create a UPGMA (unweighted pair group method with arithmetic means) dendrogram with BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). The Dice similarity coefficient was used, with optimization and position tolerance settings of 1.0 and 1.5, respectively. PFGE clusters were defined based on ≥80% relatedness on the dendrogram (4).
Statistical analysis.
The results of cgMLST-100 and other typing methods were compared using Simpson’s index of diversity (SID), the adjusted Wallace (AW) coefficient, and the adjusted Rand (AR) coefficient (4, 48), calculated with an online tool (http://www.comparingpartitions.info/). For comparison with other typing methods, groups were defined by cutting MSTs at a suitable allelic difference to have an SID similar to that of the method to which it was being compared.
Data availability.
The annotated wgMLST schema and a detailed description of its development are publicly available in chewie-NS (45) at https://chewbbaca.online/species/1/schemas/1. The genome assemblies and allele-calling results for each data set, a static version of the wgMLST schema, the list of loci in each subschema, the pairwise distances computed for Data Sets 2 and 3, and Tables S1, S2, S5, and S9 can be found in the supplemental material (31). Raw sequencing data and sample metadata for the 265 isolates included in Data Set 1 have been deposited in the European Nucleotide Archive (ENA) under project accession number PRJEB49967. The custom scripts used for schema annotation, curation, and result analyses are part of the Schema Refinery repository (44).
RESULTS
Development of the wgMLST schema for S. pyogenes.
The final annotated wgMLST schema comprises 3,044 loci with 371,549 alleles. Out of these, 1,096 (36%) loci presented low variability, presenting 1 to 19 DNA alleles (see Fig. S1 in the supplemental material). These correspond essentially to genes that were identified in a minority of assemblies, mostly associated with prophages and other mobile genetic elements. The exception is sagA, encoding the streptolysin S precursor peptide, which presented only 13 alleles despite being ubiquitous among S. pyogenes isolates. The short length of this locus (162 bp) may be partly responsible for the limited number of alleles.
On the opposite extreme, among the 10 most variable loci (>750 alleles) are genes encoding well-known surface-exposed virulence factors but also transcriptional regulators known to play a major role in GAS pathogenesis and virulence, namely, CovS, RopB, and Mga. For these loci, the diversity of DNA alleles also results in a very large number of protein variants (range, 535 to 1,695) (Fig. S2).
Specific analyses can be performed through the identification and creation of subschemas for smaller sets of biologically relevant loci, such as genes encoding virulence factors and transcriptional regulators, for which subschemas are provided as supplemental material (31).
Comparison of cgMLST with other typing methods.
To compare cgMLST analysis with conventional typing methods, a collection of 265 infection isolates with previous information on emm type, ST, T type, PFGE profile, PCR profile of 11 SAg genes, and antimicrobial resistance was used (see Data Set 1 in reference 31). This data set includes isolates of emm types 1, 3, 4, and 89; 15 distinct STs; 6 T types (17 isolates were nontypeable); 19 SAg profiles; and 16 PFGE clusters (Table 1).
TABLE 1.
Simpson’s index of diversity and 95% confidence intervals for the typing methods used to characterize 265 S. pyogenes isolates recovered in Portugal
| Typing method | No. of partitions | SID (CI95%)c |
|---|---|---|
| emm type | 4 | 0.742 (0.727–0.756) |
| ST | 15 | 0.826 (0.800–0.852) |
| T typea | 6 | 0.744 (0.720–0.768) |
| SAg profile | 19 | 0.835 (0.813–0.857) |
| PFGE | 16 | 0.792 (0.766–0.817) |
| MST1000b | 5 | 0.743 (0.728–0.758) |
| MST45b | 15 | 0.807 (0.779–0.835) |
| cgMLST-100 | 245 | 0.999 (0.998–1.000) |
The SID for T type was calculated for the subset of 248 isolates with a defined T type (17 isolates were nontypeable).
Groups of isolates linked by up to n different loci in the MST (MSTn).
SID, Simpson’s index of diversity; CI95%, 95% confidence interval.
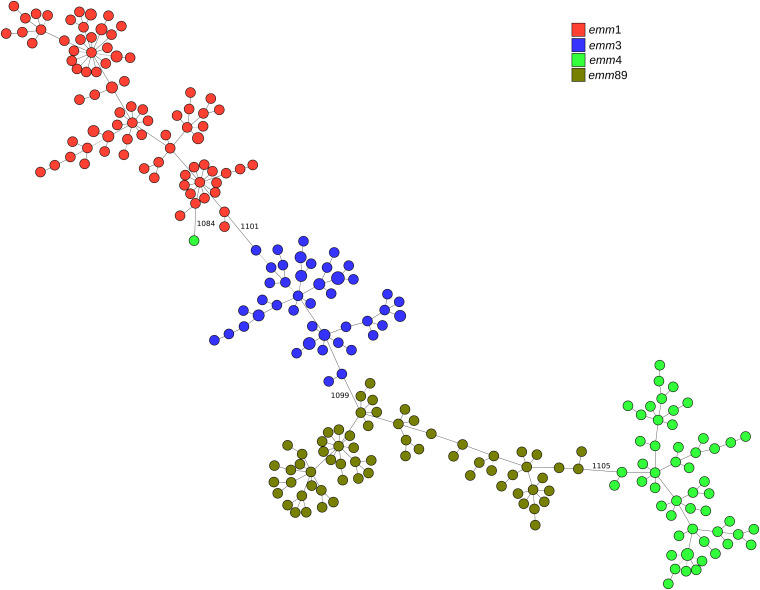
Allele calling using the wgMLST schema followed by cgMLST-100 analysis generated 245 different profiles representing 1,230 loci. The resulting MST separated the isolates according to emm type, except for one emm4 isolate that did not cluster with the others (Fig. 1). The minimum distance between clusters of different emm types varied between 1,084 and 1,105 allelic differences, while those among isolates of the same emm type were ≤28 for emm1, ≤100 for emm3, ≤157 for emm4 (excluding the distantly related isolate), and ≤225 for emm89. Clustering of isolates at a cutoff of 1,000 differences created four groups separating the four emm types and one singleton, corresponding to the emm4 isolate, resulting in high concordance between the MST groups linked by up to 1,000 different loci and emm types (Table 1; see also Tables S3 and S4 in the supplemental material).
FIG 1.
Minimum-spanning tree generated with the goeBURST algorithm for the cgMLST-100 profiles of 265 S. pyogenes isolates recovered in Portugal (see Data Set 1 in reference 31). The size of each node is proportional to the number of isolates with that particular cgMLST-100 profile on a logarithmic scale. Nodes are colored according to emm type. Link distances of ≥1,000 allelic differences are labeled (from a total of 1,230 compared loci).
A lower congruence was obtained between the distributions of isolates in the MST and the remaining typing methods (ST, PFGE, SAg profiling, and T type) (Fig. S3 to S6). The AW and AR values between T types and MST groups linked by up to 1,000 different loci were only slightly lower than those of emm types (Tables S3 and S4), but T type had a lower typeability since 17 isolates were nontypeable. Although MST groups linked by up to 45 different loci resulted in a number of partitions and SIDs comparable to those of ST, SAg profiling, and PFGE (Table 1), the AW coefficient between MST groups linked by up to 45 different loci and these typing methods was lower than that between MST groups linked by up to 1,000 different loci and emm type (Table S4). This means that MST groups linked by up to 45 different loci could not confidently predict the ST, PFGE cluster, or SAg profile, or the converse, which was also reflected in lower AR values (<0.900) (Table S3).
The use of a wgMLST schema instead of a universally defined cgMLST-100 set of loci allows scalable analysis in which higher resolution can be obtained by including larger numbers of common loci when analyzing closely related isolates. As an example, the cgMLST-100 obtained exclusively for the emm4 isolates grouped into the same MST group linked by up to 1,000 different loci (n = 54) comprises 52 profiles of 1,382 cgMLST-100 loci. The emm4 isolates presenting the M phenotype of macrolide resistance (erythromycin resistant and clindamycin susceptible) shared ST39 and an SAg profile with most susceptible isolates (see Data Set 1 in reference 31), rendering these two methods unable to differentiate macrolide-resistant isolates. One PFGE cluster was associated with macrolide resistance (49), although it also included two susceptible isolates. Similarly, one of the MST groups linked by up to 33 different loci comprised exclusively all but two of the macrolide-resistant isolates (Fig. S7). Not surprisingly, the set of 46 loci that were present universally and exclusively in the subset of erythromycin-resistant isolates (list available in the supplemental material in reference 31) represents mostly phage-related genes, including mef(A) and msr(D), the genes most commonly associated with the M phenotype in GAS (50, 51).
Performance of the wgMLST schema on a large and genetically diverse data set.
The genetic structure of the GAS population is known to vary temporally and geographically, with an associated impact on the disease spectrum and incidence (52, 53). To evaluate the performance of the proposed wgMLST schema on the analysis of genetically diverse data sets, we used a large collection of isolates previously selected to represent the genetic, geographic, temporal, and clinical diversity of GAS (19). A total of 2,006 assemblies were included in the data set, comprising 140 emm types and 443 STs and organized into 292 phylogroups defined by PopPUNK (19, 54) (see Data Set 2 in reference 31).
We defined 1,321-locus cgMLST-95, 1,204-locus cgMLST-99, and 763-locus cgMLST-100 schemas (available in the supplemental material in reference 31).
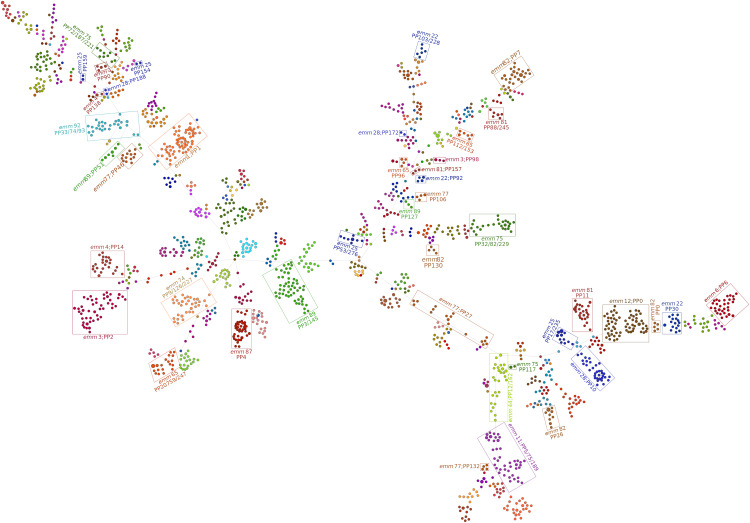
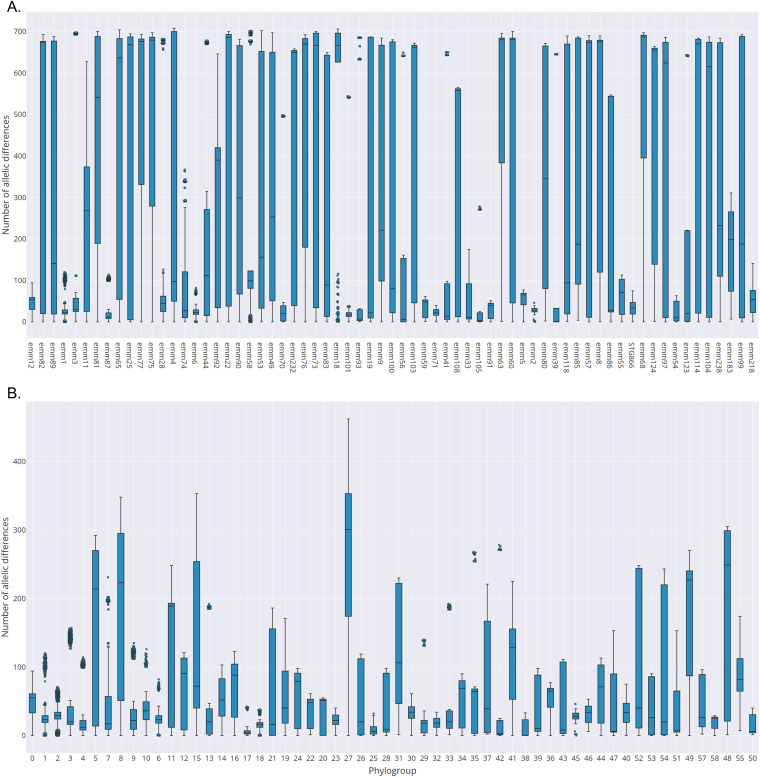
Allele call results identified 1,700 cgMLST-100 profiles. The resulting MST indicates that many emm types include diverse genetic lineages, with 12 of the 19 most prevalent emm types (>30 isolates) comprising isolates distributed in multiple tree regions (Fig. 2). Accordingly, 50 of the 67 emm types comprising ≥10 isolates included assemblies that differed in >50% of the 763 cgMLST-100 loci (up to 708 differences [93%] in emm4) (Fig. 3A). In 31 of these emm types, the mean intra-emm allelic difference was larger than the smaller difference from another emm type (Table S5). This is possibly due to the diversity of geographic and temporal origins of the isolates in this data set and is in line with a previous report of genetic heterogeneity within emm types (19). It is also reflected in a low congruence between emm types and MST groups linked by up to 450 different loci despite similar SID values (Tables S6 to S8).
FIG 2.
Minimum-spanning tree generated with the goeBURST algorithm for the cgMLST profiles of 2,006 genetically diverse S. pyogenes isolates recovered worldwide (19) (see Data Set 2 in reference 31). The size of each node is proportional to the number of isolates with that particular cgMLST profile on a logarithmic scale. Nodes are colored according to emm type. Groups of clustered emm types represented by >30 isolates are highlighted inside rectangles and labeled with the respective emm types and PopPUNK (PP) phylogroup numbers (for simplicity, isolated nodes of emm types 4, 22, 44, 65, 75, 77, 81, and 92 are not highlighted). A total of 763 core loci were compared.
FIG 3.
Box-and-whisker plots for the pairwise distances of the assemblies from Data Set 2 (19, 31) included in each emm type with ≥10 isolates (A) or in each PopPUNK phylogroup with ≥10 isolates (B). The distances were calculated based on the allele call results for the 763 cgMLST-100 loci of the 2,006 assemblies (interactive versions of these plots are available as supplemental material in reference 31).
The overall congruence between STs and MST groups linked by up to 50 different loci was poor although slightly higher than that observed for the less diverse Data Set 1 (AR coefficients of 0.810 and 0.709, respectively) (Table S7). In contrast, there was good congruence, with high AW and AR values, between PopPUNK phylogroups and MST groups linked by up to 200 different loci (Tables S7 and S8). Still, PopPUNK phylogroups can be rather diverse, including multiple STs and isolates differing in up to 61% of the core 763 loci (phylogroup 27) (Fig. 3B; Table S9), highlighting the advantage of using multiple methods for analyzing the evolution of GAS lineages.
Performance of the wgMLST schema in an outbreak context.
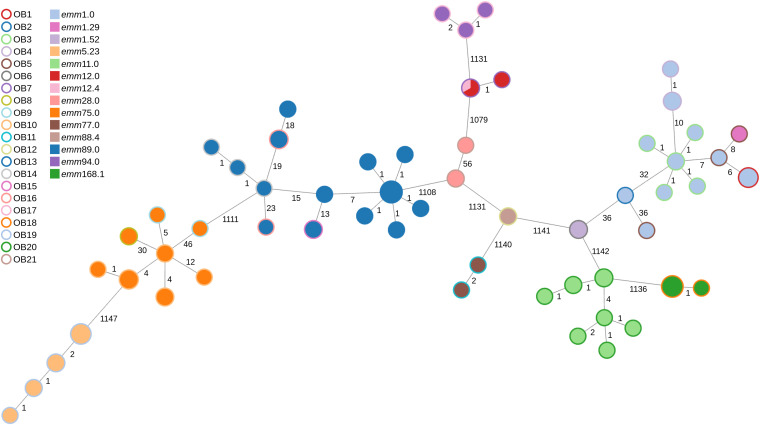
To evaluate the potential contribution of the proposed wgMLST schema for outbreak recognition, we used a previously published data set comprising isolates from 21 outbreaks in England and contemporaneous nonrelated isolates with the same emm types (18). A total of 119 outbreak isolates and 170 sporadic isolates were included (see Data Set 3 in reference 31). Allele calling for the 119 outbreak isolates identified 58 profiles of 1,263 cgMLST-100 loci. In agreement with the SNP-based clustering presented previously (18), the MST clustered the isolates according to emm type, with a minimum distance of 1,079 allelic differences between isolates of different emm types, while isolates of different subtypes or outbreaks of the same emm type were more closely related (Fig. 4).
FIG 4.
Minimum-spanning tree generated with the goeBURST algorithm for the cgMLST-100 profiles of 119 outbreak S. pyogenes isolates recovered in the United Kingdom (18) (see Data Set 3 in reference 31). The size of each node is proportional to the number of isolates with that particular cgMLST profile on a logarithmic scale. The nodes are colored according to the emm type, and the outer ring is colored according to the outbreak number. Link distances are labeled as the number of allelic differences between nodes (from a total of 1,263 compared loci).
Individual MSTs were created for emm types 1, 5, 11, 28, 75, 89, and 94, including outbreak and sporadic isolates (Fig. S8 to S14). Since these MSTs included only isolates sharing the same emm type, they comprised larger sets of cgMLST-100 loci (1,384 to 1,547 loci), potentially allowing higher resolution in the discrimination of outbreak isolates. Ten isolates with epidemiological links could be excluded from the respective outbreaks because they did not cluster with isolates of the same outbreak or differed by too many loci (Table S10 and Fig. S8 and S11 to S13). These isolates also matched the outbreak exclusion criteria based on SNP analysis (18).
Except for these 10 excluded isolates, outbreak isolates linked in the MSTs shared >99.5% of their core genome (maximum link distance of 6 allelic differences), and the mean distance within a given outbreak was much lower than the mean distance among sporadic isolates of the same emm type (Table 2). However, in emm types 1 and 5, there were sporadic isolates with cgMLST profiles very similar to those of outbreak 1 (OB1) and OB19, respectively (0 to 2 allelic differences), indicating that these outbreak strains were also present in the community (Table 2; Fig. S8 and S9).
TABLE 2.
Distances (numbers of allelic differences) among outbreak isolates and between each outbreak and sporadic isolates of the same emm type determined by cgMLST-100 analysis for each emm type, using a collection of isolates from the United Kingdoma
| emm type | No. of loci in cgMLST | Subset (no. of isolates) | Mean distance within subset (range) | Mean distance to sporadic isolates (range) |
|---|---|---|---|---|
| 1 | 1,488 | OB1 (6) | 0.6 (0–1) | 17.9 (2–59) |
| OB3 (6) | 1.7 (1–2) | 18.4 (8–53) | ||
| OB4 (4) | 0.5 (0–1) | 24.8 (15–60) | ||
| OB6 (3) | 0.7 (0–1) | 50.1 (46–55) | ||
| Sporadic (30) | 25.7 (3–63) | NA | ||
| 5 | 1,485 | OB19 (14) | 1.6 (0–4) | 124.1 (0–174) |
| Sporadic (27) | 112.0 (0–175) | NA | ||
| 11 | 1,384 | OB20 (10) | 3.8 (0–8) | 89.8 (35–592) |
| Sporadic (26) | 118.6 (1–597) | NA | ||
| 28 | 1,510 | OB21 (2) | 0 | 44 (12–67) |
| Sporadic (11) | 51.0 (0–74) | NA | ||
| 75 | 1,547 | OB8 (2) | 0 | 20.1 (14–65) |
| OB10 (11) | 5.4 (0–11) | 19.6 (12–70) | ||
| Sporadic (39) | 19.5 (0–76) | NA | ||
| 89 | 1,392 | OB13 (17) | 0.93 (0–2) | 28.4 (11–42) |
| OB14 (3) | 1.3 (1–2) | 29.0 (16–41) | ||
| OB15 (3) | 0 (0–0) | 32.5 (16–47) | ||
| OB16 (4) | 0.5 (0–1) | 33.1 (14–45) | ||
| Sporadic (31) | 31.7 (0–50) | NA | ||
| 94 | 1,506 | OB17 (3) | 2.7 (1–4) | 29.6 (10–48) |
| Sporadic (6) | 31.4 (2–50) | NA | ||
Performance of the wgMLST schema in the identification of recently emerged lineages.
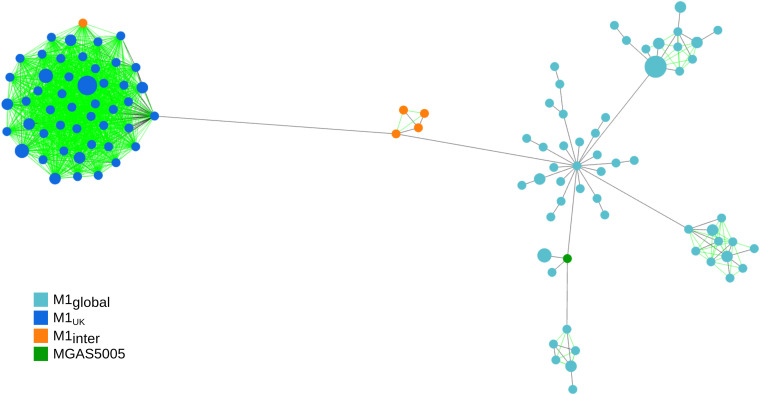
We tested if the proposed wgMLST schema has enough discriminatory power to identify two recently emerged intra-emm lineages that were originally identified by whole-genome SNP analysis, namely, M1UK and emm89 clade 3 (8, 9, 11). Allele calling was performed for the 135 assemblies from noninvasive emm1 isolates (11) together with the complete genome of strain MGAS5005, a reference representative of the M1global lineage (see Data Set 4 in reference 31). The graph in Fig. 5 represents the resulting MST with all links of up to 19 differences depicted. All M1UK isolates were tightly clustered, together with an intermediate isolate (M1inter) carrying 23 of the 27 SNPs characteristic of the M1UK lineage (11). The MST links within this cluster ranged between 0 and 13 differences, while the closest links to the M1inter cluster (13 SNPs) and an M1global isolate were 20 and 31 differences, respectively. M1global isolates presented higher genomic diversity, with MST links of up to 49 differences.
FIG 5.
Graph representation of the relationships between the cgMLST-100 profiles of 135 noninvasive emm1 isolates recovered in the United Kingdom (11) and reference strain MGAS5005 (see Data Set 4 in reference 31), depicting all links with ≤19 allelic differences (from a total of 1,404 compared loci). The size of each node is proportional to the number of isolates with that particular cgMLST-100 profile on a logarithmic scale. Nodes are colored according to the M1 lineage, with MGAS5005 (reference genome for the M1global lineage) in green. Links that would not be present in the standard MST are shown in green. Links shown in black represent the MST links and may represent distances with >19 allelic differences.
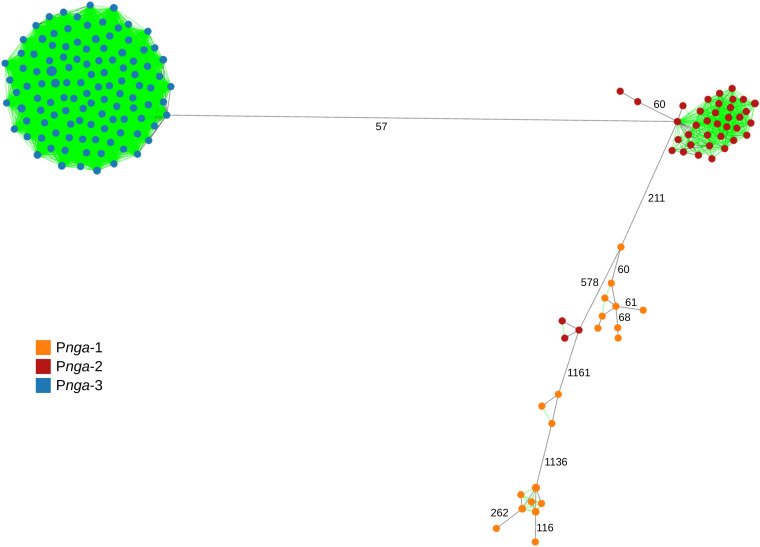
Allele calling was performed for all emm89 assemblies included in the four data sets described above and all the complete emm89 genomes used to create the schema (n = 201) (see Data Set 5 in reference 31). In addition, the Pnga variant was determined for all isolates. The absence of the hasA gene of the capsule locus was confirmed in all Pnga-3 isolates, while all other isolates carried this gene, except for two ST568 isolates that have an internal nonsense codon in hasA. The graph depicting all links of up to 55 differences (Fig. 6) showed limited diversity in the isolates carrying Pnga-3, which clustered closely, with MST links with 0 to 27 differences, while the shortest link to a Pnga-2 isolate was 57 differences. The Pnga-2 and, especially, Pnga-1 isolates were more diverse, presenting fewer links with up to 55 differences and comprising multiple sublineages associated with different STs (Fig. 6; Fig. S15). Both the wider geographic range and collection time span may contribute to this higher diversity. As previously reported (7), MLST was not suitable for discriminating Pnga-3 isolates from those carrying Pnga-2 since ST101 was prevalent among both lineages (Fig. S15). Analysis of the emm89 isolates from Data Set 1 showed that Pnga-3 isolates and most Pnga-2 isolates were also grouped into the same PFGE cluster, and some of them shared the same SAg profile, while the T serotype B3264 was ubiquitous, except for the single Pnga-1 isolate (T11) and one Pnga-3 isolate that was nontypeable (Fig. S16 to S18). PopPUNK clustering also could not discriminate Pnga-3 isolates, which were clustered with isolates carrying Pnga-1 and Pnga-2 in phylogroup 3 (see Data Set 5 in reference 31).
FIG 6.
Graph representation of the relationships between the cgMLST-100 profiles of 201 emm89 isolates (see Data Set 5 in reference 31) depicting all links with ≤55 allelic differences (from a total of 1,279 compared loci). The size of each node is proportional to the number of isolates with that particular cgMLST-100 profile on a logarithmic scale. Nodes are colored according to the variant of the nga promoter (Pnga). Links that would not be present in the standard MST are shown in green. Links shown in black represent the MST links and may represent distances with >55 allelic differences (labeled links).
DISCUSSION
The reduced costs of HTS have facilitated a wider application of whole-genome data to the epidemiological surveillance of multiple pathogens. This leads to a requirement for standardized analysis pipelines producing reproducible and portable results that can be easily compared across laboratories and with those of previously used typing methods (55). Here, we propose a wgMLST schema for S. pyogenes, consisting of 3,044 loci. Hard-defined cgMLST schemas comprising the subsets of loci present in 95% (1,321 loci), 99% (1,204 loci), and 100% (763 loci) of the assemblies of a collection representing the genetic diversity of S. pyogenes (19) are also presented. However, the use of a wgMLST schema from which the cgMLST loci are selected according to the specific data set under analysis has the advantage of allowing the inclusion of larger subsets of loci and, hence, increased resolution when comparing closely related isolates (21). This can be particularly important to track the emergence of intra-emm-type sublineages or identify outbreak-related isolates.
The application of the schema proposed here to previously published data sets and analysis of the resulting MSTs showed a performance comparable to that of SNP-based methods in distinguishing recently emerged intra-emm-type sublineages as well as in identifying clusters of epidemiologically and genetically related isolates associated with local, short-term outbreaks (8, 11, 18). Analyses based on wg/cgMLST build upon the strengths of gene-by-gene approaches, which do not require a reference genome or the removal of regions of recombination (21, 22, 25). This is particularly important when analyzing collections of genetically diverse lineages, such as in long-term surveillance studies, particularly in organisms where mobile genetic elements and recombination play major roles in genomic plasticity and evolution, such as S. pyogenes (19, 23). Moreover, gene-by-gene approaches constitute a framework that has been widely used in surveillance, which can facilitate the transition to wg/cgMLST by reference laboratories involved in surveillance activities.
Comparison of cgMLST-based clustering with other typing methods used for S. pyogenes revealed poor concordance, although in temporally and geographically restricted data sets, the groups defined by emm typing were also supported by cgMLST. By including a much higher number of loci, cgMLST was expected to present a higher discriminatory power than the traditional seven-gene MLST schema and to further discriminate isolates sharing the same ST (22, 27). However, such a simplistic expectation was not universally borne out by the data, which highlights the limitations of the seven-gene MLST schema to correctly identify GAS lineages based on broader genomic information. It is worth noting that from the seven genes included in traditional MLST, two (gtr and yqiL) were excluded from the wgMLST schema because they shared alleles with paralogous genes, and one (xpt) was absent in at least one GAS lineage and therefore was not always included in the cgMLST analysis.
In contrast, a good correlation was found between cgMLST clustering and PopPUNK (19, 54), another whole-genome-based clustering method. However, the flexibility of wg/cgMLST allows increased resolution by lowering the number of allelic differences used to define clusters and a dynamic cgMLST definition, providing further discrimination within PopPUNK clusters.
The proposed wgMLST schema is publicly available on the chewie-NS platform (45), where multiple statistics regarding the whole schema and individual loci can be visualized (https://chewbbaca.online/species/1/schemas/1). The close integration with the chewBBACA suite (43) facilitates its use in surveillance and epidemiological studies and the maintenance of a common nomenclature across different studies. By virtue of the comprehensive annotation, the database can be used to obtain relevant data for basic research, such as the variability of genes of interest (virulence factors, antimicrobial resistance genes, candidate vaccine antigens, and transcriptional regulators, etc.) (19, 20) in addition to its use for typing purposes.
ACKNOWLEDGMENTS
R.M. was supported by the Fundação para a Ciência e Tecnologia (FCT) (grant 2020.08493.BD). Partial support was received from the ONEIDA project (LISBOA-01-0145-FEDER-016417), cofunded by the FEEI (Fundos Europeus Estruturais e de Investimento) from the Programa Operacional Regional de Lisboa, Portugal, 2020 (POR Lisboa 2020), and by national funds from the FCT and the LISBOA-01-0145-FEDER-007391 project, cofunded by FEDER through POR Lisboa 2020 and the Fundação para a Ciência e a Tecnologia.
Footnotes
Supplemental material is available online only.
Contributor Information
M. Ramirez, Email: ramirez@fm.ul.pt.
Daniel J. Diekema, University of Iowa College of Medicine
REFERENCES
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect Dis 5:685–694. 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 2.Vekemans J, Gouvea-Reis F, Kim JH, Excler J-L, Smeesters PR, O’Brien KL, Van Beneden CA, Steer AC, Carapetis JR, Kaslow DC. 2019. The path to group A Streptococcus vaccines: World Health Organization research and development technology roadmap and preferred product characteristics. Clin Infect Dis 69:877–883. 10.1093/cid/ciy1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall B, Facklam R, Thompson T. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol 34:953–958. 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carriço JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, Almeida JS, Ramirez M. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J Clin Microbiol 44:2524–2532. 10.1128/JCM.02536-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friães A, Pinto FR, Silva-Costa C, Ramirez M, Melo-Cristino J. 2013. Superantigen gene complement of Streptococcus pyogenes—relationship with other typing methods and short-term stability. Eur J Clin Microbiol Infect Dis 32:115–125. 10.1007/s10096-012-1726-3. [DOI] [PubMed] [Google Scholar]
- 6.Enright MC, Spratt BG, Kalia A, Cross JH, Bessen DE. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect Immun 69:2416–2427. 10.1128/IAI.69.4.2416-2427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friães A, Machado MP, Pato C, Carriço J, Melo-Cristino J, Ramirez M. 2015. Emergence of the same successful clade among distinct populations of emm89 Streptococcus pyogenes in multiple geographic regions. mBio 6:e01780-15. 10.1128/mBio.01780-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner CE, Abbott J, Lamagni T, Holden MTG, David S, Jones MD, Game L, Efstratiou A, Sriskandan S. 2015. Emergence of a new highly successful acapsular group A Streptococcus clade of genotype emm89 in the United Kingdom. mBio 6:e00622-15. 10.1128/mBio.00622-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu L, Olsen RJ, Nasser W, Beres SB, Vuopio J, Kristinsson KG, Gottfredsson M, Porter AR, DeLeo FR, Musser JM. 2015. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest 125:3545–3559. 10.1172/JCI82478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu L, Olsen RJ, Nasser W, de la Riva Morales I, Musser JM. 2015. Trading capsule for increased cytotoxin production: contribution to virulence of a newly emerged clade of emm89 Streptococcus pyogenes. mBio 6:e01378-15. 10.1128/mBio.01378-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynskey NN, Jauneikaite E, Li HK, Zhi X, Turner CE, Mosavie M, Pearson M, Asai M, Lobkowicz L, Chow JY, Parkhill J, Lamagni T, Chalker VJ, Sriskandan S. 2019. Emergence of dominant toxigenic M1T1 Streptococcus pyogenes clone during increased scarlet fever activity in England: a population-based molecular epidemiological study. Lancet Infect Dis 19:1209–1218. 10.1016/S1473-3099(19)30446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rümke LW, de Gier B, Vestjens SMT, van der Ende A, van Sorge NM, Vlaminckx BJM, Witteveen S, van Santen M, Schouls LM, Kuijper EJ. 2020. Dominance of M1UK clade among Dutch M1 Streptococcus pyogenes. Lancet Infect Dis 20:539–540. 10.1016/S1473-3099(20)30278-4. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Nanduri SA, Van Beneden CA, Beall BW. 2020. M1UK lineage in invasive group A Streptococcus isolates from the USA. Lancet Infect Dis 20:538–539. 10.1016/S1473-3099(20)30279-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demczuk W, Martin I, Domingo FR, MacDonald D, Mulvey MR. 2019. Identification of Streptococcus pyogenes M1UK clone in Canada. Lancet Infect Dis 19:1284–1285. 10.1016/S1473-3099(19)30622-X. [DOI] [PubMed] [Google Scholar]
- 15.Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K, Caugant DA, Steinbakk M, Low DE, McGeer A, Darenberg J, Henriques-Normark B, Van Beneden CA, Hoffmann S, Musser JM. 2014. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci USA 111:E1768–E1776. 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beres SB, Carroll RK, Shea PR, Sitkiewicz I, Martinez-Gutierrez JC, Low DE, McGeer A, Willey BM, Green K, Tyrrell GJ, Goldman TD, Feldgarden M, Birren BW, Fofanov Y, Boos J, Wheaton WD, Honisch C, Musser JM. 2010. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci USA 107:4371–4376. 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner CE, Bedford L, Brown NM, Judge K, Török ME, Parkhill J, Peacock SJ. 2017. Community outbreaks of group A Streptococcus revealed by genome sequencing. Sci Rep 7:8554. 10.1038/s41598-017-08914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coelho JM, Kapatai G, Jironkin A, Al-Shahib A, Daniel R, Dhami C, Laranjeira AM, Chambers T, Phillips S, Tewolde R, Underwood A, Chalker VJ. 2019. Genomic sequence investigation Streptococcus pyogenes clusters in England (2010-2015). Clin Microbiol Infect 25:96–101. 10.1016/j.cmi.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Davies MR, McIntyre L, Mutreja A, Lacey JA, Lees JA, Towers RJ, Duchêne S, Smeesters PR, Frost HR, Price DJ, Holden MTG, David S, Giffard PM, Worthing KA, Seale AC, Berkley JA, Harris SR, Rivera-Hernandez T, Berking O, Cork AJ, Torres RSLA, Lithgow T, Strugnell RA, Bergmann R, Nitsche-Schmitz P, Chhatwal GS, Bentley SD, Fraser JD, Moreland NJ, Carapetis JR, Steer AC, Parkhill J, Saul A, Williamson DA, Currie BJ, Tong SYC, Dougan G, Walker MJ. 2019. Atlas of group A streptococcal vaccine candidates compiled using large-scale comparative genomics. Nat Genet 51:1035–1043. 10.1038/s41588-019-0417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beres SB, Zhu L, Pruitt L, Olsen RJ, Faili A, Kayal S, Musser JM. 2022. Integrative reverse genetic analysis identifies polymorphisms contributing to decreased antimicrobial agent susceptibility in Streptococcus pyogenes. mBio 13:e03618-21. 10.1128/mbio.03618-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maiden MCJ, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann B, Prior K, Bender JK, Harmsen D, Klare I, Fuchs S, Bethe A, Zühlke D, Göhler A, Schwarz S, Schaffer K, Riedel K, Wieler LH, Werner G. 2019. A core genome multilocus sequence typing scheme for Enterococcus faecalis. J Clin Microbiol 57:e01686-18. 10.1128/JCM.01686-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGregor KF, Spratt BG, Kalia A, Bennett A, Bilek N, Beall B, Bessen DE. 2004. Multilocus sequence typing of Streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. J Bacteriol 186:4285–4294. 10.1128/JB.186.13.4285-4294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carriço JA, Rossi M, Moran-Gilad J, Van Domselaar G, Ramirez M. 2018. A primer on microbial bioinformatics for nonbioinformaticians. Clin Microbiol Infect 24:342–349. 10.1016/j.cmi.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgins PG, Prior K, Harmsen D, Seifert H. 2017. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS One 12:e0179228. 10.1371/journal.pone.0179228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bletz S, Janezic S, Harmsen D, Rupnik M, Mellmann A. 2018. Defining and evaluating a core genome multilocus sequence typing scheme for genome-wide typing of Clostridium difficile. J Clin Microbiol 56:e01987-17. 10.1128/JCM.01987-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdel-Glil MY, Chiaverini A, Garofolo G, Fasanella A, Parisi A, Harmsen D, Jolley KA, Elschner MC, Tomaso H, Linde J, Galante D. 2021. A whole-genome-based gene-by-gene typing system for standardized high-resolution strain typing of Bacillus anthracis. J Clin Microbiol 59:e02889-20. 10.1128/JCM.02889-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinto M, González-Díaz A, Machado MP, Duarte S, Vieira L, Carriço JA, Marti S, Bajanca-Lavado MP, Gomes JP. 2019. Insights into the population structure and pan-genome of Haemophilus influenzae. Infect Genet Evol 67:126–135. 10.1016/j.meegid.2018.10.025. [DOI] [PubMed] [Google Scholar]
- 30.Bardenstein S, Gibbs RE, Yagel Y, Motro Y, Moran-Gilad J. 2021. Brucellosis outbreak traced to commercially sold camel milk through whole-genome sequencing, Israel. Emerg Infect Dis 27:1728–1731. 10.3201/eid2706.204902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friães A, Mamede R, Ferreira M, Melo-Cristino J, Ramirez M. 2022. Supplemental material for “An annotated whole-genome multilocus sequence typing schema for scalable high resolution typing of Streptococcus pyogenes”. Zenodo. 10.5281/zenodo.5901775. [DOI] [PMC free article] [PubMed]
- 32.Friães A, Pinto FR, Silva-Costa C, Ramirez M, Melo-Cristino J, Portuguese Group for the Study of Streptococcal Infections. 2012. Group A streptococci clones associated with invasive infections and pharyngitis in Portugal present differences in emm types, superantigen gene content and antimicrobial resistance. BMC Microbiol 12:280. 10.1186/1471-2180-12-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pato C, Melo-Cristino J, Ramirez M, Friães A, Portuguese Group for the Study of Streptococcal Infections. 2018. Streptococcus pyogenes causing skin and soft tissue infections are enriched in the recently emerged emm89 clade 3 and are not associated with abrogation of CovRS. Front Microbiol 9:2372. 10.3389/fmicb.2018.02372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friães A, Lopes JP, Melo-Cristino J, Ramirez M, Portuguese Group for the Study of Streptococcal Infections. 2013. Changes in Streptococcus pyogenes causing invasive disease in Portugal: evidence for superantigen gene loss and acquisition. Int J Med Microbiol 303:505–513. 10.1016/j.ijmm.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Pato CTC. 2011. Streptococcus pyogenes como agente de infecção da pele e tecidos moles. MSc thesis. Faculdade de Ciências, Universidade de Lisboa, Lisbon, Portugal. [Google Scholar]
- 36.Blackwell GA, Hunt M, Malone KM, Lima L, Horesh G, Alako BTF, Thomson NR, Iqbal Z. 2021. Exploring bacterial diversity via a curated and searchable snapshot of archived DNA sequences. PLoS Biol 19:e3001421. 10.1371/journal.pbio.3001421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, et al. 2016. Reference Sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bioinformatics @ Molecular Microbiology and Infection Unit. 2021. INNUca (v4.2.2). GitHub. https://github.com/B-UMMI/INNUca/releases/tag/v4.2.2. Accessed 7 July 2021.
- 39.Seemann T. 2021. mlst (v2.19.0). GitHub. https://github.com/tseemann/mlst/releases/tag/v2.19.0. Accessed 11 March 2021.
- 40.Microbiological Diagnostic Unit Public Health Laboratory. 2021. emmtyper—emm automatic isolate labeller (v0.2.0). GitHub. https://github.com/MDU-PHL/emmtyper/releases/tag/v0.2.0. Accessed 11 March 2021.
- 41.Seemann T. 2021. Snippy (v4.6.0). GitHub. https://github.com/tseemann/snippy/releases/tag/v4.6.0. Accessed 31 July 2021.
- 42.Bioinformatics @ Molecular Microbiology and Infection Unit. 2021. SeqTyper (v2.3). GitHub. https://github.com/B-UMMI/seq_typing/tree/gbs_types. Accessed 29 July 2021.
- 43.Silva M, Machado MP, Silva DN, Rossi M, Moran-Gilad J, Santos S, Ramirez M, Carriço JA. 2018. chewBBACA: a complete suite for gene-by-gene schema creation and strain identification. Microb Genom 4:e000166. 10.1099/mgen.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bioinformatics @ Molecular Microbiology and Infection Unit. 2022. Schema Refinery (v0.1.0). GitHub. https://github.com/B-UMMI/Schema_Refinery/releases/tag/v0.1.0. Accessed 1 February 2022.
- 45.Mamede R, Vila-Cerqueira P, Silva M, Carriço JA, Ramirez M. 2021. Chewie Nomenclature Server (chewie-NS): a deployable nomenclature server for easy sharing of core and whole genome MLST schemas. Nucleic Acids Res 49:D660–D666. 10.1093/nar/gkaa889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nascimento M, Sousa A, Ramirez M, Francisco AP, Carriço JA, Vaz C. 2017. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 33:128–129. 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- 47.Ribeiro-Gonçalves B, Francisco AP, Vaz C, Ramirez M, Carriço JA. 2016. PHYLOViZ Online: Web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res 44:W246–W251. 10.1093/nar/gkw359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Severiano A, Pinto FR, Ramirez M, Carriço JA. 2011. Adjusted Wallace coefficient as a measure of congruence between typing methods. J Clin Microbiol 49:3997–4000. 10.1128/JCM.00624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silva-Costa C, Friães A, Ramirez M, Melo-Cristino J, Portuguese Group for the Study of Streptococcal Infections. 2012. Differences between macrolide-resistant and -susceptible Streptococcus pyogenes: importance of clonal properties in addition to antibiotic consumption. Antimicrob Agents Chemother 56:5661–5666. 10.1128/AAC.01133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva-Costa C, Friães A, Ramirez M, Melo-Cristino J. 2015. Macrolide-resistant Streptococcus pyogenes: prevalence and treatment strategies. Expert Rev Anti Infect Ther 13:615–628. 10.1586/14787210.2015.1023292. [DOI] [PubMed] [Google Scholar]
- 51.Iannelli F, Santoro F, Santagati M, Docquier J-D, Lazzeri E, Pastore G, Cassone M, Oggioni MR, Rossolini GM, Stefani S, Pozzi G. 2018. Type M resistance to macrolides is due to a two-gene efflux transport system of the ATP-binding cassette (ABC) superfamily. Front Microbiol 9:1670. 10.3389/fmicb.2018.01670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. 2009. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis 9:611–616. 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 53.Barnett TC, Bowen AC, Carapetis JR. 2018. The fall and rise of group A Streptococcus diseases. Epidemiol Infect 147:e4. 10.1017/S0950268818002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lees JA, Harris SR, Tonkin-Hill G, Gladstone RA, Lo SW, Weiser JN, Corander J, Bentley SD, Croucher NJ. 2019. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res 29:304–316. 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabat AJ, Budimir A, Nashev D, Sa-Leao R, van Dijl JM, Laurent F, Grundmann H, Friedrich AW, ESCMID Study Group of Epidemiological Markers (ESGEM). 2013. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill 18:20380. 10.2807/ese.18.04.20380-en. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S18 and Tables S1 to S10. Download jcm.00315-22-s0001.pdf, PDF file, 2.2 MB (2.3MB, pdf)
Data Availability Statement
The annotated wgMLST schema and a detailed description of its development are publicly available in chewie-NS (45) at https://chewbbaca.online/species/1/schemas/1. The genome assemblies and allele-calling results for each data set, a static version of the wgMLST schema, the list of loci in each subschema, the pairwise distances computed for Data Sets 2 and 3, and Tables S1, S2, S5, and S9 can be found in the supplemental material (31). Raw sequencing data and sample metadata for the 265 isolates included in Data Set 1 have been deposited in the European Nucleotide Archive (ENA) under project accession number PRJEB49967. The custom scripts used for schema annotation, curation, and result analyses are part of the Schema Refinery repository (44).