ABSTRACT
Achromobacter spp. are nonfermenting Gram-negative bacilli mainly studied among cystic fibrosis (CF) patients. The identification of the 19 species within the genus is time-consuming (nrdA-sequencing), thus data concerning the distribution of the species are limited to specific studies. Recently, we built a database using MALDI-TOF mass spectrometry (MS) (Bruker) that allows rapid and accurate species identification and detection of the multiresistant epidemic clones: A. xylosoxidans ST137 spreading among CF patients in various French and Belgium centers, and A. ruhlandii DES in Denmark. Here, we first assessed whether species identification could be achieved with our database solely by analysis of MS spectra without availability of isolates. Then, we conducted a multicentric study describing the distribution of Achromobacter species and of the clone ST137 among French CF centers. We collected and analyzed with our local database the spectra of Achromobacter isolates from 193 patients (528 samples) from 12 centers during 2020. In total, our approach enabled to conclude for 502/528 samples (95.1%), corresponding to 181 patients. Eleven species were detected, only five being involved in chronic colonization, A. xylosoxidans (86.4%), A. insuavis (9.1%), A. mucicolens (2.3%), A. marplatensis (1.1%) and A. genogroup 3 (1.1%). This study confirmed the high prevalence of A. xylosoxidans in chronic colonizations and the circulation of the clone A. xylosoxidans ST137 in France: four patients in two centers. The present study is the first to report the distribution of Achromobacter species from CF patients samples using retrospective MALDI-TOF/MS data. This easy approach could enable future large-scale epidemiological studies.
KEYWORDS: Achromobacter, cystic fibrosis, epidemiology, Maldi-TOF, mass spectrometry, identification, ST137, DES
INTRODUCTION
Achromobacter spp. are Gram–negative nonfermenting bacilli frequently reported in the respiratory samples of cystic fibrosis (CF) patients. To date, 19 officially validated species can be identified on the pubMLST database (https://pubmlst.org/achromobacter/) within the genus (1). The distribution of the species is variable among CF centers and countries (2–14), A. xylosoxidans being the most prevalent one. The pathogenicity of these bacteria remains controversial in CF patients, but chronic colonization has been associated with higher rates of mortality and transplantation among these patients (15). Particular attention should be paid to the A. xylosoxidans ST137 clone detected so far in five French centers and three Belgian centers and the A. ruhlandii Danish epidemic strain (DES) responsible for epidemics in Denmark. These clones are multi-drug resistant and also responsible for chronic colonizations (7, 12, 16).
However, because of time-consuming methods of identification, epidemiological data remain scarce, and more studies are needed to help determine which species or which strains might be of clinical importance. Therefore, it is important to describe the distribution of the species and of these clones among CF patients (2–14, 17, 18). In France, the prevalence of Achromobacter spp. in CF patients raised from 2.7% in 2001 to 6.9% in 2019 (Registre Français de la Mucoviscidose, www.vaincrelamuco.org) and the only study reporting the different species detected was based on isolates collected in 2014 (7).
The current methods of reference for Achromobacter species identification are based on housekeeping gene sequencing and therefore not performed by routine diagnosis laboratories (nrdA-sequencing or multilocus sequence typing [MLST]) (2, 19, 20). Indeed, the databases of commercially available mass spectrometry (MS) systems do not include all the species described to date in the genus and sometimes misidentify species within the genus (21). We recently developed at Dijon center a database for accurate Achromobacter species identification by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF/MS, Bruker Daltonics). This database allows the identification of 19 species and also the detection of the multiresistant epidemic clones A. xylosoxidans ST137 and A. ruhlandii DES (21, 22). The spectra obtained during MS analysis are automatically recorded in the instrument and the files can be exported to other laboratories using the same technology for further analysis.
This prompted us to evaluate the feasibility and reliability of a retrospective spectrum analysis in order to conduct a multicentric epidemiological study describing the distribution of the various species of Achromobacter in sporadic and chronic colonizations and detecting the eventual presence of the A. xylosoxidans ST137 and A. ruhlandii DES clones in various CF centers in France.
MATERIALS AND METHODS
Evaluation of the feasibility and reliability of retrospective spectrum analysis.
For this purpose, we collected the spectra from all isolates (recovered from various anatomic sites, from non-CF patients and sputum from CF patients) (n = 164 isolates from 83 patients) identified in our clinical laboratory (Dijon) as Achromobacter spp. by MALDI-TOF MS (Bruker Daltonics) using Bruker database (MBT-IVD-DB-7712) during 2019. The corresponding strains had all been identified to the species level by a reference method (MLST; n = 36), nrdA gene sequencing (n = 29), or MS using our database from a sheep-blood agar spot (n = 99) (2, 19–21). The spectra were retrospectively analyzed by MALDI-TOF/MS using our database as already described (21). We deliberately chose an operator who was not involved in the construction of the database and did not have access to samples and patients information. During this first step, three situations were observed: i) the spectrum was directly “interpretable” if the first two best matches were obtained with a log score ≥ 2 with the same species, ii) directly “uninterpretable” if the log score was < 2, and iii) interpretation was considered “uncertain” if the first two best matches were obtained with a log score ≥ 2 corresponding to two different species or to A. ruhlandii (because of the risk of misidentification, as already described) (21).
We were able to conclude to a reliable retrospective species identification (i.e., “directly interpretable” spectra) for 92.7% of the isolates, which allowed us to initiate the retrospective study, by collecting spectra from collaborating centers.
Application to the analysis of spectra from 12 CF centers.
For this study, only laboratories using MALDI-TOF/MS (Bruker Daltonics) for routine identification according to the manufacturer’s instructions for in vitro diagnostics were retained: Toulouse, Suresnes-Foch, Giens, Caen, Roscoff, Versailles, La Réunion-Saint-Denis, La Réunion-Saint Pierre, Rennes, Strasbourg, Tours, and Dijon.
Each laboratory provided all the spectra matching with the genus Achromobacter using Bruker database from 1 January 2020 to 31 December 2020 for CF patients sputum isolates. Each center sent the spectra by email to Dijon center with the following informations: CF patient number, sample number, type of colonization (chronicity being defined according to the criteria of Pereira: “when at least three positive cultures in 1 year were obtained, with a minimum 1-month interval between them, for at least 2 years”) (17).
A total of 988 spectra corresponding to 528 samples from 193 patients were included. All the spectra were analyzed in Dijon center by MALDI-TOF/MS using our database for species level identification as already described (21). In case of uninterpretable or uncertain interpretation of the spectra, the sending laboratory was asked for the availability of the isolate. The isolates were subcultured on sheep-blood agar and re-analyzed by MALDI-TOF/MS with our database and, if not conclusive, by nrdA gene sequencing as already described (2). If the isolate was not available, the sample was classified as uninterpretable.
Each spectrum identified as A. xylosoxidans was visually analyzed in flexAnalysis software (Bruker Daltonics) as already described (22) to search for the specific absence of any peaks between m/z 6,640 – 6,700 for A. xylosoxidans ST137.
RESULTS AND DISCUSSION
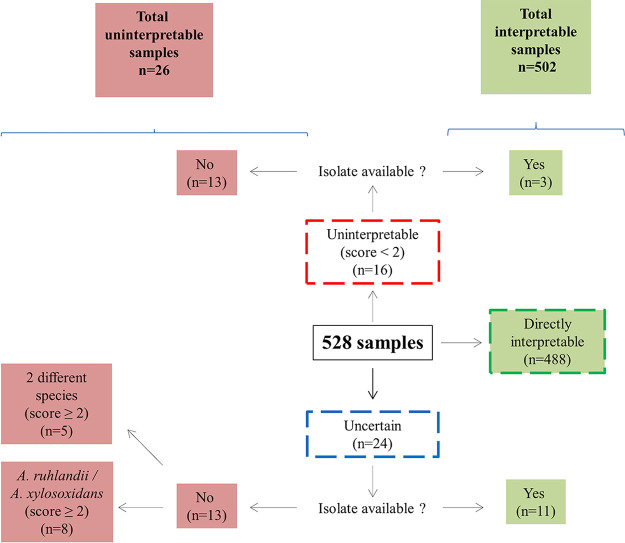
The present study is the first retrospective multicentric study to describe the distribution of Achromobacter species directly from MALDI-TOF/MS data obtained in various laboratories. This approach has the advantage of being easier, faster, and less costly than nrdA-sequencing or MLST currently performed for species identification in the available epidemiological studies. It allowed the quick analysis of 988 spectra corresponding to 528 samples from 193 patients attending 12 CF centers in 2020 (Fig. 1). For 488/528 (92.6%) samples, the spectrum was directly interpretable for species identification. For 16/528 and 24/528 samples, the spectra were respectively uninterpretable or uncertain, probably mostly because of the poor quality of the original spot, and in 8 cases because of the A. ruhlandii/A. xylosoxidans discrimination issue (21). However, we cannot exclude that some isolates belonged to novel species not characterized yet and not included in our database. For example, in one case, the nrdA-analysis of the available isolate enabled us to identify a putative novel species belonging to genogroup 3. In total, our approach enabled to conclude for 502/528 samples (95.1%), corresponding to 181 patients (Fig. 1). The number of patients may have been slightly overestimated due to anonymized data, in the case of patients attending different centers. We always ensured the coherence of the species between the different spectra of the same sample. For example, we were able to detect, for two patients, the presence of two different species on the same sample. Overall, the prevalence of Achromobacter was of 7.9% and varied from 3.1 to 18.8% in the 12 CF centers.
FIG 1.
Analysis process of the isolates.
A total of 11 species were identified, A. xylosoxidans being the most prevalent species in each CF center (74.6% patients), as already reported (2–11, 13, 14, 23), followed by A. insuavis (12.1%), A. mucicolens (3.2%), A. marplatensis (3.2%), A. dolens (2.1%), A. aegrifaciens (1.1%), A. insolitus (1.1%), A. genogroup 20 (1.1%) (identified by nrdA gene sequencing), A. genogroup 3 (0.5%) (identified by nrdA gene sequencing), A. animicus (0.5%), and A. deleyi (0.5%). As already noticed in our former studies in France, we did not detect any isolate belonging to A. ruhlandii in this study although this species is the second most frequent species reported in United States, Brazil, Argentina and Denmark. (2, 9, 11, 14) (Fig. 2A and Table 1).
FIG 2.
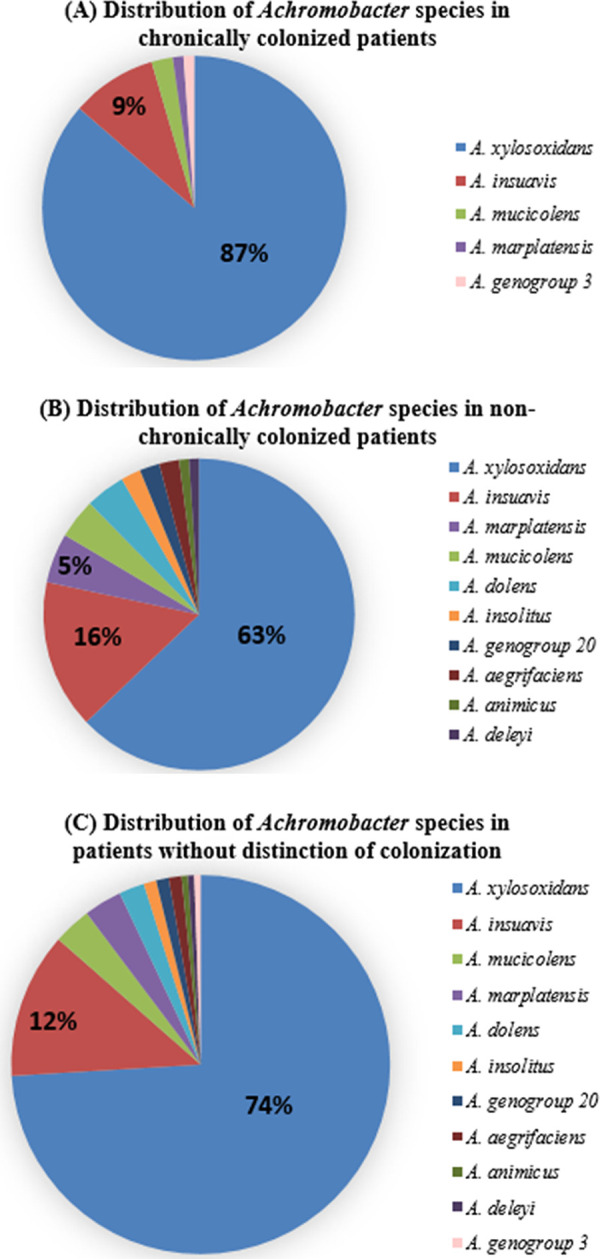
Distribution of Achromobacter species among the 181 CF patients (% of patients): (A) chronic colonized patients, (B) nonchronic colonized patients and (C) without distinction of colonization. Only values > 5% are shown.
TABLE 1.
Distribution of Achromobacter species among the patients of the 12 French centers in 2020a
| Center |
A. xylosoxidans
|
A. insuavis
|
A. mucicolens
|
A. marplatensis
|
A. insolitus
|
A. aegrifaciens
|
A. genogroup 20 |
A. genogroup 3 |
A. animicus
|
A. deleyi
|
A. dolens
|
Total patients |
Total patients in each center | Achromobacter prevalence in each center (%) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | NC | C | NC | C | NC | C | NC | C | NC | C | NC | C | NC | C | NC | C | NC | C | NC | C | NC | C | NC | T | TT | |||
| Roscoff | 8 | 1 | 0 | 1b | 1b | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9b | 5b | 13 | 13 | 185 | 7 |
| Caen | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 4 | 4 | 128 | 3.1 |
| Dijon | 4 | 4 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 5 | 8 | 13 | 13 | 129 | 10.1 |
| Suresnes-Foch | 13 | 11 | 2 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 14 | 30 | 32 | 508 | 6.3 |
| La Réunion St Pierre | 4 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 4 | 12 | 16 | 16 | 85 | 18.8 |
| La Reunion St Denis | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 9 | 9 | 11 | 64 | 17.2 |
| Rennes | 6 | 8 | 1 | 1 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 12 | 19 | 19 | 228 | 8.3 |
| Strasbourg | 6 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 4 | 10 | 14 | 318 | 4.4 |
| Giens | 10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 2 | 13 | 17 | 191 | 8.9 |
| Toulouse | 11c | 9d | 1d | 7 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1c | 0 | 0 | 14c,d | 19c,d | 31 | 31 | 294 | 10.5 |
| Tours | 10 | 3 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 5 | 16 | 16 | 223 | 7.2 |
| Versailles | 1 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1e | 0 | 1e | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 6e | 7 | 7 | 70 | 10 |
| Total | 76 | 61 | 8 | 15 | 2 | 4 | 1 | 5 | 0 | 2 | 0 | 2 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 4 | 88 | 97 | 181 | 193 | 2423 | 7.9 |
C: chronic; NC: nonchronic; T: total patients of the center with Achromobacter identified at the species rank, TT: total patients of the center with Achromobacter (includes patients with isolates unidentified to the species rank).
One patient chronically colonized with A. mucicolens had a positive sample with A. insuavis.
One patient chronically colonized with A. xylosoxidans had a positive sample with A. deleyi.
One patient chronically colonized with A. insuavis had a positive sample with A. xylosoxidans.
One patient had a positive sample with A. insolitus and A. aegrifaciens.
Among the 181 patients, 88 (48.6%) were chronically colonized. This result is in accord with previous studies (Table S1 in the supplemental material) (5, 6, 8, 10, 12). The number of chronic patients might have been underestimated since some consultations did not take place in the year 2020 because of COVID-19. Despite the predominance of A. xylosoxidans, we found a greater diversity of species during nonchronic colonization than in chronic colonization (total of 10 versus 5 species) (Fig. 2B, Table 1, and Table S1 in the supplemental material), as previously reported in France, Canada, and Denmark (5, 12, 23).
Among chronically colonized patients, as expected, A. xylosoxidans was the most predominant species. Noteworthy, 3 species never reported to date in chronic colonization were also detected (Table S1 in the supplemental material) (4–6, 8–10, 12, 14, 23, 24): A. genogroup 3, A. mucicolens, and A. marplatensis.
Among the 181 patients, the epidemic clone A. xylosoxidans ST137 was detected in four patients from two centers: three in Foch CF center and one in Giens CF center. The presence of ST137 had not been documented yet in these patients and no patient carrying the ST137 clone was previously known in Giens center. Each time the strain was responsible for chronic colonization, and multiresistant as in previous descriptions (7, 12, 16). These data show that the clone with epidemic potential continues to spread in new centers. It should be noted that our approach allowed us to detect this clone easily and quickly, and that this method will help in patients monitoring and management of segregation measures.
In conclusion, this study showed that retrospective analyses by our MALDI-TOF/MS database of spectra collected from samples from various centers was possible and led to excellent Achromobacter species identification. It allowed the detection of the multiresistant epidemic clone A. xylosoxidans ST137. Our database is currently only available in our center (Dijon) or by contacting the corresponding author (21). These easy-to-use MALDI-TOF/MS retrospective studies could be used on a large scale for enrichment of epidemiological data concerning the distribution of Achromobacter species and the survey of the circulation of epidemic clones.
Footnotes
Supplemental material is available online only.
Contributor Information
Lucie Amoureux, Email: lucie.amoureux@chu-dijon.fr.
Nathan A. Ledeboer, Medical College of Wisconsin
REFERENCES
- 1.Dumolin C, Peeters C, Ehsani E, Tahon G, De Canck E, Cnockaert M, Boon N, Vandamme P. 2020. Achromobacter veterisilvae sp. nov., from a mixed hydrogen-oxidizing bacteria enrichment reactor for microbial protein production. Int J Syst Evol Microbiol 70:530–536. 10.1099/ijsem.0.003786. [DOI] [PubMed] [Google Scholar]
- 2.Spilker T, Vandamme P, LiPuma JJ. 2013. Identification and distribution of Achromobacter species in cystic fibrosis. J Cyst Fibros 12:298–301. 10.1016/j.jcf.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Coward A, Kenna DTD, Perry C, Martin K, Doumith M, Turton JF. 2016. Use of nrdA gene sequence clustering to estimate the prevalence of different Achromobacter species among cystic fibrosis patients in the UK. J Cyst Fibros 15:479–485. 10.1016/j.jcf.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Coward A, Kenna DTD, Woodford N, Turton JF, Armstrong M, Auckland C, Bowler I, Burns P, Cargill J, Carroll M, Flight W, Graver M, Green H, Horner C, Jones A, Jones AM, Jones G, Mayell S, Orendi J, Perry A, Robb A, Tucker N, Waine D, Winstanley T, Withers N, and members of the UK CF Surveillance Working Group., The UK CF Surveillance Working Group comprised . 2020. Structured surveillance of Achromobacter, Pandoraea and Ralstonia species from patients in England with cystic fibrosis. J Cyst Fibros 19:388–393. 10.1016/j.jcf.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Edwards BD, Greysson-Wong J, Somayaji R, Waddell B, Whelan FJ, Storey DG, Rabin HR, Surette MG, Parkins MD. 2017. Prevalence and outcomes of Achromobacter species infections in adults with cystic fibrosis: a North American cohort study. J Clin Microbiol 55:2074–2085. 10.1128/JCM.02556-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veschetti L, Sandri A, Patuzzo C, Melotti P, Malerba G, Lleo MMY. 2021. Genomic characterization of Achromobacter species isolates from chronic and occasional lung infection in cystic fibrosis patients. Microb Genom 7:000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amoureux L, Sauge J, Sarret B, Lhoumeau M, Bajard A, Tetu J, Bador J, Neuwirth C, Caillon J, Cardot-Martin E, Cattoir V, Doléans-Jordheim A, Ferroni A, Guet-Revillet H, Héry-Arnaud G, Segonds C, Thomas E, Plésiat P, Vu-Thien H, MucoMicrobes group . 2019. Study of 109 Achromobacter spp. isolates from 9 French CF centres reveals the circulation of a multiresistant clone of A. xylosoxidans belonging to ST 137. J Cyst Fibros 18:804–807. 10.1016/j.jcf.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Barrado L, Brañas P, Orellana MÁ, Martínez MT, García G, Otero JR, Chaves F. 2013. Molecular characterization of Achromobacter isolates from cystic fibrosis and non-cystic fibrosis patients in Madrid, Spain. J Clin Microbiol 51:1927–1930. 10.1128/JCM.00494-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereira RHV, Leão RS, Carvalho-Assef AP, Albano RM, Rodrigues ERA, Firmida MC, Folescu TW, Plotkowski MC, Bernardo VG, Marques EA. 2017. Patterns of virulence factor expression and antimicrobial resistance in Achromobacter xylosoxidans and Achromobacter ruhlandii isolates from patients with cystic fibrosis. Epidemiol Infect 145:600–606. 10.1017/S0950268816002624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filipic B, Malesevic M, Vasiljevic Z, Lukic J, Novovic K, Kojic M, Jovcic B. 2017. Uncovering differences in virulence markers associated with Achromobacter species of CF and non-CF origin. Front Cell Infect Microbiol 7:224. 10.3389/fcimb.2017.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papalia M, Steffanowski C, Traglia G, Almuzara M, Martina P, Galanternik L, Vay C, Gutkind G, Ramírez MS, Radice M. 2020. Diversity of Achromobacter species recovered from patients with cystic fibrosis, in Argentina. Rev Argent Microbiol 52:13–18. 10.1016/j.ram.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Gade SS, Nørskov-Lauritsen N, Ridderberg W. 2017. Prevalence and species distribution of Achromobacter sp. cultured from cystic fibrosis patients attending the Aarhus centre in Denmark. J Med Microbiol 66:686–689. 10.1099/jmm.0.000499. [DOI] [PubMed] [Google Scholar]
- 13.Rodrigues ERA, Ferreira AG, Leão RS, Leite CCF, Carvalho-Assef AP, Albano RM, Marques EA. 2015. Characterization of Achromobacter species in cystic fibrosis patients: comparison of blaOXA-114 PCR amplification, multilocus sequence typing, and matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 53:3894–3896. 10.1128/JCM.02197-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabrielaite M, Bartell JA, Nørskov-Lauritsen N, Pressler T, Nielsen FC, Johansen HK, Marvig RL. 2021. Transmission and antibiotic resistance of Achromobacter in cystic fibrosis. J Clin Microbiol 59:e02911-20. 10.1128/JCM.02911-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somayaji R, Stanojevic S, Tullis DE, Stephenson AL, Ratjen F, Waters V. 2017. Clinical outcomes associated with Achromobacter species infection in patients with cystic fibrosis. Ann Am Thorac Soc 14:1412–1418. 10.1513/AnnalsATS.201701-071OC. [DOI] [PubMed] [Google Scholar]
- 16.Cools P, Ho E, Vranckx K, Schelstraete P, Wurth B, Franckx H, Ieven G, Van Simaey L, Van Daele S, Verhulst S, De Baets F, Vaneechoutte M. 2016. Epidemic Achromobacter xylosoxidans strain among Belgian cystic fibrosis patients and review of literature. BMC Microbiol 16:122. 10.1186/s12866-016-0736-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereira RHV, Carvalho-Assef AP, Albano RM, Folescu TW, Jones MCMF, Leão RS, Marques EA. 2011. Achromobacter xylosoxidans: characterization of strains in Brazilian cystic fibrosis patients. J Clin Microbiol 49:3649–3651. 10.1128/JCM.05283-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voronina OL, Kunda MS, Ryzhova NN, Aksenova EI, Sharapova NE, Semenov AN, Amelina EL, Chuchalin AG, Gintsburg AL. 2018. On Burkholderiales order microorganisms and cystic fibrosis in Russia. BMC Genomics 19:74. 10.1186/s12864-018-4472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ridderberg W, Wang M, Nørskov-Lauritsen N. 2012. Multilocus sequence analysis of isolates of Achromobacter from patients with cystic fibrosis reveals infecting species other than Achromobacter xylosoxidans. J Clin Microbiol 50:2688–2694. 10.1128/JCM.00728-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spilker T, Vandamme P, LiPuma JJ. 2012. A multilocus sequence typing scheme implies population structure and reveals several putative novel Achromobacter species. J Clin Microbiol 50:3010–3015. 10.1128/JCM.00814-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrigos T, Neuwirth C, Chapuis A, Bador J, Amoureux L, Andre E, Barbier E, Caillon J, Cardot ME, Cattoir V, Doléans JA, Echahidi F, Ferroni A, Guet RH, Héry AG, Lipuma J, Nørskov LN, Peeters C, Pierard D, Segonds C, Thomas E, Plésiat P, Vandamme P, Verroken A, Vu TH, Collaborators . 2021. Development of a database for the rapid and accurate routine identification of Achromobacter species by matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS). Clin Microbiol Infect 27:126.e1–126.e5. 10.1016/j.cmi.2020.03.031. [DOI] [PubMed] [Google Scholar]
- 22.Garrigos T, Dollat M, Magallon A, Chapuis A, Varin V, Bador J, Makki N, Cremet L, Persyn E, Cardot-Martin E, Echahidi F, Peeters C, Pierard D, Vandamme P, Verroken A, Neuwirth C, Amoureux L. 2021. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid detection of isolates belonging to the epidemic clones Achromobacter xylosoxidans ST137 and Achromobacter ruhlandii DES from cystic fibrosis patients. J Clin Microbiol 59:e0094621. 10.1128/JCM.00946-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amoureux L, Bador J, Bounoua Zouak F, Chapuis A, de Curraize C, Neuwirth C. 2016. Distribution of the species of Achromobacter in a French cystic fibrosis centre and multilocus sequence typing analysis reveal the predominance of A. xylosoxidans and clonal relationships between some clinical and environmental isolates. J Cyst Fibros 15:486–494. 10.1016/j.jcf.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Dupont C, Michon A-L, Jumas-Bilak E, Nørskov-Lauritsen N, Chiron R, Marchandin H. 2015. Intrapatient diversity of Achromobacter spp. involved in chronic colonization of Cystic Fibrosis airways. Infect Genet Evol 32:214–223. 10.1016/j.meegid.2015.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jcm.02422-21-s0001.pdf, PDF file, 0.2 MB (195KB, pdf)