SUMMARY
Parasites belonging to the Apicomplexa phylum are among the most successful pathogens known in nature. They can infect a wide range of hosts, often remain undetected by the immune system, and cause acute and chronic illness. In this phylum, we can find parasites of human and veterinary health relevance, such as Toxoplasma, Plasmodium, Cryptosporidium, and Eimeria. There are still many unknowns about the biology of these pathogens due to the ethical and practical issues of performing research in their natural hosts. Animal models are often difficult or nonexistent, and as a result, there are apicomplexan life cycle stages that have not been studied. One recent alternative has been the use of three-dimensional (3D) systems such as organoids, 3D scaffolds with different matrices, microfluidic devices, organs-on-a-chip, and other tissue culture models. These 3D systems have facilitated and expanded the research of apicomplexans, allowing us to explore life stages that were previously out of reach and experimental procedures that were practically impossible to perform in animal models. Human- and animal-derived 3D systems can be obtained from different organs, allowing us to model host-pathogen interactions for diagnostic methods and vaccine development, drug testing, exploratory biology, and other applications. In this review, we summarize the most recent advances in the use of 3D systems applied to apicomplexans. We show the wide array of strategies that have been successfully used so far and apply them to explore other organisms that have been less studied.
KEYWORDS: apicomplexan parasites, host-parasite interaction, organoids, organ-on-a-chip, 2D culture, 3D cultures
INTRODUCTION
The in vitro culture of pathogens has allowed great advances in microbiological research. These two-dimensional (2D) cultures are relatively cheap, easy to maintain, and facilitate the analysis of a target microorganism for a wide array of experimental procedures (Fig. 1). Nevertheless, 2D culture lacks certain compounds, extracellular microenvironments, barriers, signaling molecules, and architectures, which makes the propagation of specific protozoan parasites difficult or different from the natural niche (1). Animal models have been useful to facilitate research addressing cell culture limitations. While animal models can provide us with key information about the pathology of human diseases or the natural host (2, 3), there is pressure to reduce the number of animals used in research. Despite the development of new techniques for animal model replacement, in vivo models are still used for antibody production, diagnosis, drug testing, dissemination experiments, and vaccine challenges.
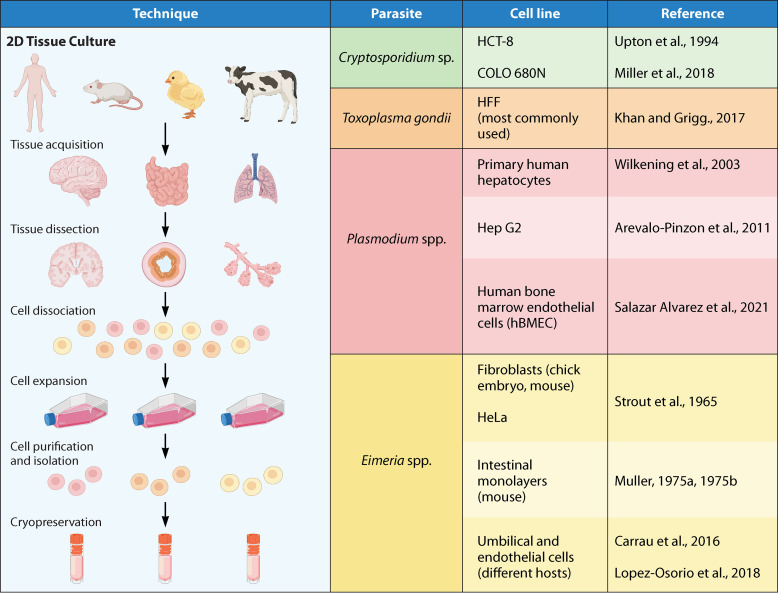
FIG 1.
2D tissue culture systems: list of the most commonly used cell lines for in vitro culture of apicomplexan parasites. The sources shown are references 38 (Upton et al., 1994), 41 (Miller et al., 2018), 62 (Khan and Grigg, 2017), 119 (Arévalo-Pinzón et al., 2011), 121 (Salazar Alvarez et al., 2021), 122 (Wilkening et al., 2003), 158 (Strout et al., 1965), 159 (Müller, 1975a), 160 (Müller, 1975b), 164 (López-Osorio et al., 2018), and 166 (Carrau et al., 2016).
In the last decade, researchers have focused on developing new systems to recapitulate whole body host and pathogen interactions. One widely used system is organoid technology (Fig. 2). Organoids are multicellular in vitro cultures that resemble the whole architecture and most of the function of a specific tissue, such as epithelial polarization, environmental cues, and biomechanical forces (4, 5). These three-dimensional (3D) structures are self-organized and contain organ-specific cells that are grown from primary stem cells (embryonic stem cells or induced pluripotent stem cells) and adult stem cells (6, 7). These structures need to be supplemented with an extracellular matrix and a combination of growth factors to maintain the stem cell niche (8). As a result, organoids are a better representation of an organ created in vitro with the advantage of mimicking most features of the in vivo tissue. The resemblance of the organoids to natural organs was a breakthrough in the microbiology field. Organoids have allowed the study of organisms that were difficult to grow in vitro, like rotaviruses, norovirus, and other pathogens (9–12).
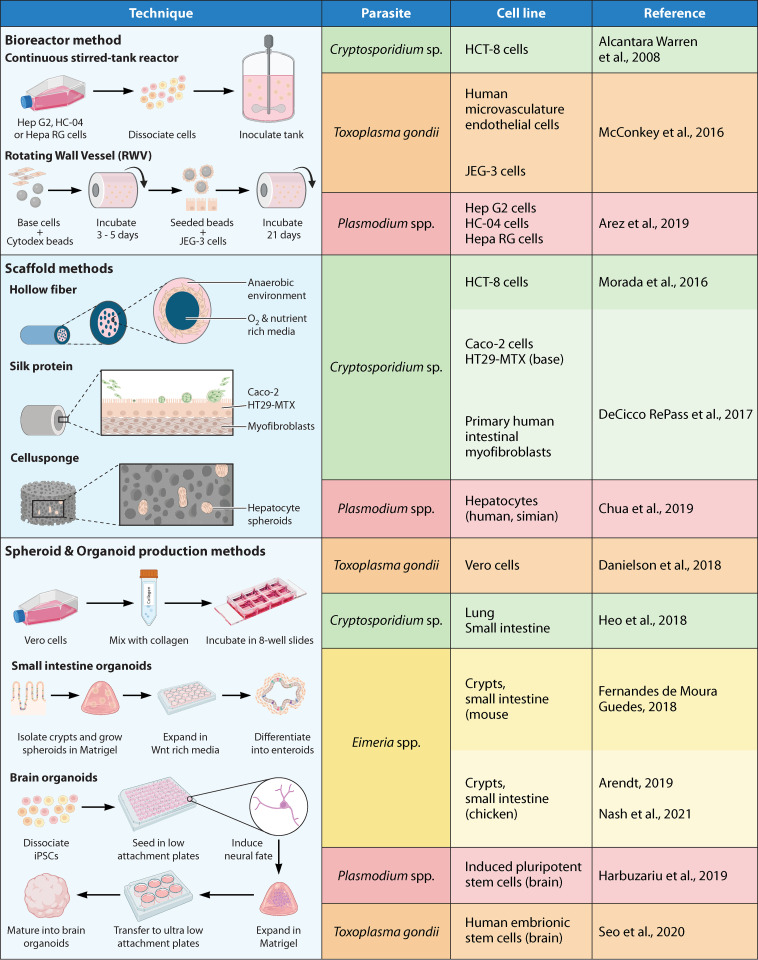
FIG 2.
3D tissue culture systems: list of cell lines and tissues most commonly used for the generation of in vitro 3D structures in apicomplexan research. The sources shown are references 14 (Chua et al., 2019), 32 (DeCicco RePass et al., 2017), 45 (Alcantara Warren et al., 2008), 48 (Morada et al., 2016), 52 (Heo et al., 2018), 70 (McConkey et al., 2016), 71 (Danielson et al., 2018), 84 (Seo et al., 2020), 125 (Arez et al., 2019), 126 (Harbuzariu et al., 2019), 168 (Fernandes de Moura Guedes, 2018), 169 (Arendt, 2019), and 170 (Nash et al., 2021).
There are classifications for 3D cultures, and confusion between organoids and spheroids is quite common. Spheroids are stabilized from immortalized cell lines, primary cells, or fragments of tissue (13–15) and are generated using one cell type or a multicellular mixture of cells (14–16). Organoids originate from adult or embryonic stem cells, physically resemble a portion of the organ, and can maintain the population of stem cells (17, 18). As organoids have the potential to be isolated and cultured from a variety of tissues, a wide range of organoids can be expanded and used to model adult functional tissue, such as stomach, lung, gut, liver, pancreas, and brain (6, 19). This advantage has been used to study host-parasite interactions for many microorganisms, such as bacteria, viruses, and parasites, including pathogens that are difficult to grow using in vitro models (20–23). In this review, we discuss the applicability of organoids for apicomplexan parasite research, with a focus on the study of host-parasite interactions of medical and veterinary relevance. We describe advantages, disadvantages, challenges, and future perspectives for the use of organoid models to study apicomplexan parasites.
CRYPTOSPORIDIUM STUDIES IN 3D SYSTEMS
Cryptosporidium Overview
Cryptosporidium is a genus of intracellular apicomplexan parasites that cause diarrhea and gastroenteritis in many species, including humans. They are the leading cause of child mortality worldwide. The latest report from the Centers for Disease Control and Prevention estimates that 750,000 cases of cryptosporidiosis occur each year in the United States (24). A recent meta-analysis estimates the global prevalence of the parasite at 7.6% (25). Populations at risk of infection include children under the age of 5 (26), immunocompromised individuals (27), and even healthy individuals during outbreaks (28). Clinical manifestations of infection with Cryptosporidium include watery diarrhea, abdominal cramps, nausea, vomiting, and wasting. The main route of transmission is fecal-oral by consuming food or water contaminated with the resistant oocyst form of the parasite. Once consumed, the parasites invade the epithelial cells in the small intestine and undergo their full life cycle within a single host (29). Cryptosporidium has received increased interest in the medical community due to its high resistance to common disinfectants, its infectivity, and the paucity of effective treatments. In contrast to other apicomplexan parasites, like Toxoplasma gondii, long-term passage and genetic manipulation of Cryptosporidium have not been available until recently (30–32). In the following sections, we cover some of the challenges associated with the maintenance of the parasite in vitro, breakthroughs, and perspectives on future exploration.
Challenges and Initial Tissue Culture Systems
While the first description of Cryptosporidium was made more than a century ago (33), studies on the parasite started in the 1980s, when its clinical importance was recognized during the AIDS/HIV epidemic. In the United States, symptoms of cryptosporidiosis were often used to diagnose AIDS even before the real causative agent, HIV, was determined (34). Despite its importance, research in Cryptosporidium has been hindered by obstacles in the methods of culture and expansion, inability to cryopreserve infectious oocysts, and difficulty in genetically modifying the parasite. This contrasts with the progress that has been achieved in other apicomplexan parasites, such as T. gondii (35).
One of the first successful attempts to overcome the challenge of culturing and expanding Cryptosporidium was performed in 1984 by Current and Haynes (36). They observed the complete development of the parasite in cell culture, opening the path for other scientists to study this microorganism. In their research, oocysts isolated from a patient with chronic infection were used to infect human fetal lung cells, chicken kidney cells, and porcine kidney cells. An excystation protocol was used to induce the release of sporozoites to store them in growth medium before infection. The different stages of the life cycle of Cryptosporidium were first observed in human fetal lung cells with the presence of sporulated oocysts 72 h after infection. Their viability was assessed by infecting 5-day-old Swiss-Webster mice, where the full life cycle of Cryptosporidium was recapitulated, confirming the infectivity of the cell-cultured oocysts. In contrast to cell culture, the number of parasites was more abundant in mice, likely due to the initiation of the asexual cycle and proliferation in the ileum of the mice. Infection and proliferation of Cryptosporidium were also achieved in chicken and porcine kidney cells, demonstrating a lack of host specificity in cell culture conditions (36, 37).
The next advance in the study of Cryptosporidium came with the human colonic tumor cell line (HCT-8) (Fig. 1). While several attempts had been performed in different cell lines to expand and maintain the parasite (38, 39), the HCT-8 line proved to have the necessary factors to sustain infection for up to 25 days. This propagation was likely due to the generation of thin-walled oocysts that were able to reinfect the remaining cells in the tissue culture (40), a feature that had not been achieved in other cell lines up to this point. Successful maintenance of the Cryptosporidium life cycle was attained by continuous passage of 5-day-old infected monolayers into fresh HCT-8 cells. A constant pH and monitoring of cell growth (avoiding sloughing, overgrowth, and cell debris) were key to the production of the infective thin-walled oocyst structure. Despite the ability to maintain a continuous passage, completion of the full life cycle was difficult to achieve in this cell line.
The latest development in 2D culture for Cryptosporidium compared human esophageal squamous cell carcinoma (COLO-680N) to other human cancer cell lines, including HCT-8 cells, for their ability to produce infective oocysts in vitro (41). They found that the COLO-680N cell line was able to produce enough infective oocysts to sustain continuous cultivation of the parasites (Fig. 1). This cell line was also able to survive for weeks, requiring only a weekly medium change, and produced 20 times the number of oocysts produced by the established HCT-8 cell line. The cell line also enabled the cryopreservation of the parasite at –80°C. This work addressed important challenges in the field (42, 43) and provided a culture system that was easy to use and sustain, diminishing the dependence on animal models to expand the parasite. This development offered a platform to dig deeper into the parasite’s biology and to screen drugs to treat cryptosporidiosis.
Initial Cryptosporidium Studies in 3D Scaffolds and Organoids
One of the challenges of using tissue culture is the inability to represent the complex 3D structure found in animal models (44). One of the first studies done in 3D used HCT-8 cells grown in a low-gravity environment induced by a rotating wall vessel (RWV) (45) (Fig. 2). Previous studies had shown that cell cultures grown at lower gravity induced the formation of structures found in vivo (46, 47). In HCT-8 cells, intercellular junctions, columns, and villus-like structures were developed with this technique, similar to those observed in normal human samples. After inoculation with the parasite, disruption of the epithelial layer was observed, and the intracellular localization of Cryptosporidium was confirmed by immunolabeling and transmission electron microscopy. This system was able to replicate the morphology and mechanical forces found in the small intestine and made it possible to study the parasite under more natural conditions.
Hollow fiber technology for Cryptosporidium research.
Further advances used a protein scaffold to improve the growth of small intestinal cells, enabling sustained infection of Cryptosporidium for up to 6 months (48). The concept for this approach was borne out of studies performed in Plasmodium (49). However, unlike the environment where Plasmodium resides, the lumen of the small intestine where Cryptosporidium thrives has different oxygen and nutrient concentrations. To address the challenges of this unique environment, Bakshi et al. (49) designed a biphasic system that provided an oxygen-rich environment at the basal side of the cells and a rich nutrient-low oxygen environment at the top layer of the cells to support parasite growth (Fig. 2). This system imitated part of the complex environment found in the small intestine, something that cannot be achieved in a 2D tissue culture system, and provided a platform to test drugs against the parasite. One of the limitations for the widespread use of this technology is the use of specialized equipment that is not readily available to most research laboratories (48).
Silk protein scaffold for Cryptosporidium research.
Another 3D model for Cryptosporidium research is the use of silk protein as a 3D scaffold. Cryptosporidium parasites were able to be cultured for up to 15 days using silk protein as a scaffold to create a 3D tissue model of the small intestine (32). The researchers reconstituted silk cocoons in a solution and poured them into polydimethylsiloxane (PDMS) molds holding a Teflon-coated stainless steel wire to create a hollow cylinder scaffold. The inner part of the cylinder was seeded with the human adenocarcinoma cell lines Caco-2 and HT29-MTX, while the bulk porous part of the cylinder was used to hold primary human intestinal myofibroblasts. This setup allowed the myofibroblasts to support the growth and expansion of the Caco-2 and HT29-MTX cells by secreting growth factors (Fig. 2). This system allowed for the recapitulation of the complete cell cycle of the parasite, which was confirmed by scanning electron microscopy. In contrast to the hollow fiber technology, the silk protein system has a smaller scale, which presents a series of advantages and disadvantages. It provides a lower output of oocysts than the hollow fiber technology but requires less specialized equipment, making it more accessible to other research laboratories. Given its small scale, this technology can be used to study host-pathogen interactions and drug target screening, as well as a platform to select CRISPR-modified parasites (50).
Mouse colon explants for Cryptosporidium carcinogenesis research.
Previous studies with immunodeficient mice (SCID) treated with dexamethasone showed that a low inoculum of 105 oocysts generated lesions similar to those found in adenocarcinoma at 46 days postinfection (51). To study the development of cancer-related lesions caused by Cryptosporidium, a specific set of requirements was needed (31). First, the system required a noncancerous primary cell line, because previous work on Cryptosporidium relied heavily on cancer cell lines such as Caco-2 and HCT-8 to maintain infection. Thus, colon explants of SCID mice were used as the base for this 3D model. To test the viability of the system, colon explants were cultured on membrane inserts for 35 days and compared via histology against fresh colon samples (Fig. 3). Differentiation of epithelial cells and cytotoxicity were also evaluated, proving that the explants were viable for up to 35 days. The ability to sustain Cryptosporidium infection was assessed by infecting the system with 25 or 250 oocysts. Not only was the infection successfully sustained, but at 27 days postinfection, intraepithelial lesions were observed, confirming the results observed in the animal model. This work showed that a primary cell 3D culture can be used and maintained over time to study host-pathogen interactions and the effects of chronic infection in vitro (31).
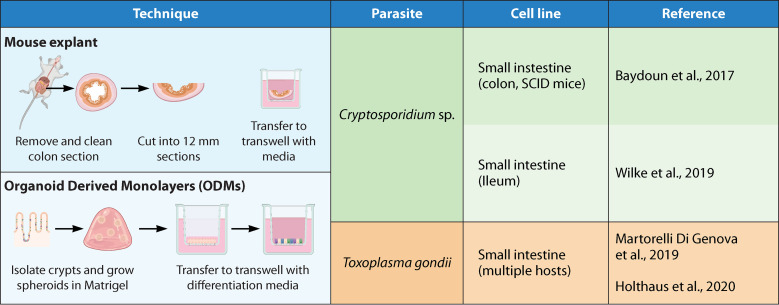
FIG 3.
2D-3D hybrid models: these are techniques that are neither 3D culture nor 2D culture but a combination of the two. Here, we find the use of mouse explants and organ-derived monolayers (ODMs). The sources shown are references 31 (Baydoun et al., 2017), 50 (Wilke et al., 2019), 76 (Martorelli Di Genova et al., 2019), and 89 (Holthaus et al., 2020).
Human small intestinal and lung organoids for Cryptosporidium research.
In 2018, Cryptosporidium was cultured in human intestinal and lung organoids (52). Cryptosporidium parvum oocysts and sporozoites were microinjected in human small intestine organoids in both expansion and differentiation media. Results from this technique showed that replication was 10-fold more efficient in differentiated organoids and that all the stages of its life cycle were recapitulated. This research is the first to demonstrate that Cryptosporidium can infect human bronchial airway-derived organoids (Fig. 2). Gene expression analysis in both intestinal and lung organoids showed the upregulation of the interferon type I response following C. parvum infection, allowing study of the host immune response in an in vitro system. The oocysts generated in this 3D system were infectious to mice. This system was able to support in vitro culture of Cryptosporidium for up to 28 days, but the number of parasites diminished over time and the number of parasites produced was lower than that in the in vivo model.
Air-liquid interface cultivation for Cryptosporidium research.
A system of plating stem cells on Transwells and removing medium from the top chamber to form an air-liquid interface (ALI) was developed (53). Adapting the ALI methodology for Cryptosporidium research allowed for the development of long-term in vitro culture that enables transgenic modifications (50). Mouse intestinal epithelial cells in spheroids were plated on top of Matrigel-coated Transwells covered with a layer of irradiated 3T3-J2 cells. After 7 days in culture, the medium was removed and ALI conditions were established for 3 days, which allowed the intestinal stem cells to differentiate into enterocytes and secretory, goblet, Paneth, and enteroendocrine cells, verified by transcriptional markers (Fig. 3). Cow-derived oocysts were able to replicate continually in this system for more than 20 days, generating all the asexual and sexual stages of the parasite. The Cryptosporidium oocysts produced in ALI conditions were infectious to mice, and in vitro cross-fertilization was also possible. The benefits of ALI conditions in Cryptosporidium research are as follows: (i) microinjection is not needed, (ii) more oocysts are produced than in previous systems, and (iii) ALI conditions eliminate the need for the use of immunocompromised animals for the selection and propagation of the transformed parasites. The advantages of an ALI system in Cryptosporidium in vitro culture rely on the intracellular-extracytoplasmic localization of the parasite for gene expression, metabolism, differentiation, and hypothetically the oxygen level conditions.
Future Perspectives in Cryptosporidium Research
The use of 3D systems in Cryptosporidium research has opened new possibilities to fill the knowledge gaps in its biology. Because the 3D system resembles the natural host microenvironment, it could be used to study the host metabolism changes caused by the parasite. Animal models could be replaced by a 3D system for the production of oocysts and the selection of genetically modified parasites. Human organoids can be used for studies of Cryptosporidium infectivity and anti-Cryptosporidium drug testing (54). Finally, the intestine-on-a-chip system, which is discussed at the end of this review, could be implemented for Cryptosporidium studies controlling factors such as peristalsis, the flow of nutrients, the presence of another microorganism, or microbiota.
T. GONDII STUDIES IN 3D SYSTEMS
T. gondii Overview
T. gondii is an obligate intracellular coccidian parasite that infects warm-blooded vertebrates and is the causative agent of toxoplasmosis. T. gondii prevalence depends on a variety of factors, such as eating habits, the presence of the definitive host (felines), and environmental conditions (55–57). T. gondii is a parasite that establishes a largely silent, lifelong chronic infection in healthy humans. In immunocompromised individuals, toxoplasmic encephalitis occurs due to recrudescence of the chronic infection as cellular immune surveillance is lost (58).
T. gondii is also harmful in pregnant women, as the parasite can cross the placental barrier and infect the fetus, causing retinal inflammation, encephalitis, mental and physical disabilities, and death (59). Examination of T. gondii seroprevalence in females of reproductive age or pregnant women shows great variation depending on the location, from 0.8% in Korea to 77.5% in Brazil (60). T. gondii can invade any nucleated cell of warm-blooded animals, including humans, rodents, birds, and livestock (61), so many cell lines are useful for propagation (Fig. 1). T. gondii in 2D systems offers many advantages, such as low cost, easy manipulation, and quick experimental setup (62). However, it lacks the microbiota, cellular diversity, biological barriers, and immune system present in in vivo models. Mice are often used as the in vivo model, but ethical concerns to reduce the number of vertebrate animals used in research has driven scientists to seek in vitro systems. Additionally, cats are the definitive host of T. gondii and are used to study the sexual stage of the parasite (63), but the costs of maintenance and the public perception of companion animals used in research has made their use extremely limited (64). Therefore, 3D systems offer a solution for modeling T. gondii infection, because it allows research in the human, cat, or other host-derived organoids, simulates the physiological microenvironment, and allows the interaction with immune cells. 3D cell culture systems have been used to study T. gondii motility, migration, mechanisms by which the parasite crosses the placental barrier, replication, egress, and sexual development. Four different 3D systems have been developed In T. gondii research: collagen or Matrigel-based matrices, the RWV bioreactor, organoids from pluripotent stem cells (PSC), and organoid-derived monolayers (ODMs) (Fig. 2 and 3).
T. gondii Motility in 2D versus 3D Environments
T. gondii, like other apicomplexan parasites, uses the cytoskeleton for motility instead of specialized organelles. The motility of T. gondii was quantified in a spheroid system by using a suspension of the medium, Matrigel, and tachyzoites in Pitta chambers (65). Pitta chambers are small platforms designed with glass coverslips holding up to 10 μL of internal volume (Fig. 4). The tachyzoites presented a corkscrew movement in the 3D system, different from the helical and circular gliding motility observed in the 2D systems. The corkscrew movement was also observed in ex vivo experiments of tachyzoites injected in the mouse earflap and imaged with two-photon microscopy. A collagen fibrous matrix was used to analyze the mechanism of tachyzoite gliding motility by biophysical techniques, such as quantitative traction force, reflection interference contrast, and time-lapse video microscopy (66). This matrix allows the analysis of tachyzoite adhesion-deadhesion cycles to the substrate in a realistic fibril environment (66) (Fig. 3). The motility of protozoan parasites is associated with virulence because it is essential to invade and egress from the host cell, cross biological barriers, and spread through tissues (67). 3D systems have demonstrated different kinds of motility of T. gondii tachyzoites, such as counterclockwise and helical and circular gliding, as well as the corkscrew movement. However, these two 3D systems lack cell-cell interactions and do not have diverse phenotypes of host cells as present in vivo models.
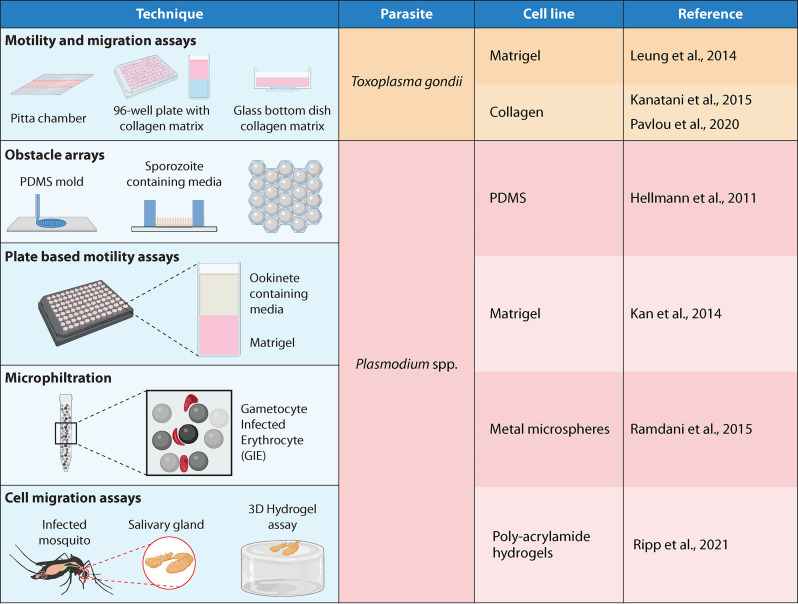
FIG 4.
Other 3D systems: here, we find gel-based and mechanical systems devoid of any cell line, except for the organism of study. These include hydrogels, 3D printed arrays, and filtration systems. The sources shown are references 65 (Leung et al., 2014), 66 (Pavlou et al., 2020), 68 (Kanatani et al., 2015), 114 (Hellmann et al., 2011), 116 (Ripp et al., 2021), 134 (Ramdani et al., 2015), and 137 (Kan et al., 2014).
The Original Trojan Horse: T. gondii-Infected Immune Cell Migration
Three-dimensional systems have been used to analyze the migration of T. gondii-infected dendritic cells (DCs). While DCs are important for the host response against T. gondii, T. gondii-infected DCs also act as a Trojan horse that potentiates parasite dissemination. Hypermigration of T. gondii-infected DCs was seen by depositing DCs on top of the polymerized layer of collagen (68). Parasite-infected DCs had a higher migration rate than uninfected DCs. This system allowed the identification of various migration patterns between different strains of T. gondii. This 3D system allowed the visualization of increased migration distance and dissemination in T. gondii-infected DCs by enabling quantification of individual cells as well as populations of cells (Fig. 4). However, it requires sophisticated imaging systems and analysis software. As Kanatani et al. (68) recognized, the system would underestimate the migration of individual cells. Finally, while the system has high reproducibility between the technical replicates of individual experiments, there is variability added when different human donors are analyzed.
Use of 3D Placental Models for the Study of Congenital Toxoplasmosis
During pregnancy, T. gondii can be transmitted from mother to fetus, causing several complications, such as hydrocephalus, malformations, and miscarriages. How T. gondii crosses the placental barrier is not fully understood. The human placenta has a maternal-fetal interface that is composed primarily of two cell layers: syncytiotrophoblasts and extravillous trophoblasts (69). Syncytiotrophoblasts act as a barrier to infection, but T. gondii can invade the extravillous trophoblasts. To study T. gondii crossing the placental barrier, a 3D placenta was created in a cylindrical bioreactor with a slow-turning lateral RWV (70). Each lateral vessel contains Cytodex beads coated with a coculture of human microvasculature endothelial cells and JEG-3, a trophoblast cell line (Fig. 2). At 48 h postinfection (hpi), the percentage of T. gondii was evaluated by immunofluorescence in JEG-3 cells in 2D monolayers and in 3D relative to syncytiotrophoblasts from a terminal placenta. T. gondii invaded only the JEG-3 2D system. Neither the JEG-3 3D system nor the syncytiotrophoblasts were infected (70). Thus, the JEG-3 3D system was shown to be an excellent model to study resistance of syncytiotrophoblasts to T. gondii infection. It is important to note that monotypic cell cultures of trophoblast cell lines are not compatible with the RWV bioreactor because they show a low rate of fusion and low attachment to the beads. The placenta RWV works well only in coculture with other cells, such as human foreskin fibroblasts (HFFs), human brain microvascular endothelial cells (hBMECs), and RL95-2 cells. It is also unknown if the shear forces induced in the RWV are similar to those in the in vivo placenta, because the precise levels of shear forces have not been measured in vivo yet.
Differences in T. gondii Replication and Egress in 2D versus 3D Systems
The replication and egress of apicomplexan parasites are key mechanisms for the survival and establishment of an infection in the host. These mechanisms were studied using T. gondii infection of Vero cells resuspended in a collagen type I matrix to create spheroids (71). While the rate of replication after 24 hpi was not significantly different between 2D and 3D Vero cells, the structure of the parasitophorous vacuole (PV) was semiflat in 2D but globular in the 3D system. When the path of egress of the parasite from the PV was evaluated, it was found that in the 2D system, the parasite egresses perpendicularly to the bottom of the plate, but in the 3D system, the parasite egresses radially in all directions. This 3D system represented a more physiologically relevant environment for research on the replication and egress of T. gondii without the pressure and tension imposed by the flat-bottom dishes in a 2D system (71).
Bovine, murine, and porcine enteroids have also been established for T. gondii infection. To develop enteroids, crypts were isolated from jejunum sections, cleaned, and resuscitated in 70% Matrigel. The protocols for propagating, cryopreserving, and resuscitating these organoids are well established (72). Pilot infection assays were performed by disrupting the 3D enteroid structure to expose the luminal surface and by incubating the enteroid fragments with tachyzoites from the RH T. gondii strain for 1 h at 37°C (72). Immunofluorescence images showed successful infection at 24 hpi, but it is not clear how long the T. gondii infection was viable in the enteroids and if the replication rate was superior to that of traditional monolayer culture.
In 2019, three different mouse jejunum-derived enteroid models were described for T. gondii infection and replication (73). The first approach used these murine enteroids, which were seeded in Matrigel and then fragmented by pipetting to expose the apical epithelial surface. They were then incubated with T. gondii before plating. After 24 hpi, viable tachyzoites were observed in different kinds of cells within the enteroid structure. The second approach evaluated the microinjection of T. gondii tachyzoites inside the luminal space of the enteroid as described previously for C. parvum (52). However, the rate of infection was low (73). The third approach was enteroid-derived collagen-supported epithelial sheets (Fig. 2). 3D enteroids previously generated were washed from the Matrigel, fragmented, and seeded on collagen, creating monolayers that retain some of the characteristics of the original 3D epithelium. T. gondii was able to proliferate and invade successfully in this new in vitro system as early as 1 hpi. This system was also used to evaluate the host protein response to the parasite by mass spectrometry and the effect of host cholesterol inhibition by atorvastatin (73). In the future, it will be interesting to compare T. gondii invasion of the host cell in 2D versus 3D systems to characterize the targets of invasion and egress. Both potential drugs that block invasion and egress and genetic mutants in T. gondii proteins that play a role in invasion may be more effective at inhibiting invasion in a 3D system.
Use of Cat Intestinal Organoids To Promote T. gondii Sexual Development
Enteroid-derived epithelial sheets and monolayers (73) and cat and mouse intestinal organoids (74, 75) have been applied to study the sexual stages of T. gondii. Martorelli di Genova et al. (76) generated organoid-derived monolayers with intestinal epithelial properties (Fig. 3). When supplemented with linoleic acid, they supported the sexual cycle of T. gondii (76). Cats do not express intestinal delta-6-desaturase, which is the rate-limiting step in the conversion of linoleic acid to arachidonic acid, giving them a high level of linoleic acid in their gut. Inhibition of delta-6-desaturase in mice and supplementation with linoleic acid induced T. gondii to sexually develop in mice (76). In the future, a continuous T. gondii in vitro culture of the sexual cycle could be standardized in mouse intestinal organoids with inhibition of delta-6-desaturase enzyme and supplementation with linoleic acid.
Brain in a Dish: Using Organoids for the Study of Cerebral Toxoplasmosis
Cerebral and retinal organoids have been used to study T. gondii infection of the brain, retina, and spinal cord. Cerebral organoids have been developed using patient-derived human pluripotent stem cells (hPSC) (77, 78). Retinal organoids have been developed using mouse (79, 80) and human embryonic stem cells (81, 82). While cerebral and retinal organoids present the natural cytoarchitecture and 3D microenvironment, they lack some specialized cells, have limited maturity and size, and lack interaction with the immune system. However, significant advances have been achieved with Zika virus using brain organoids (83). Human embryo-derived brain organoids were recently used to model human T. gondii infection (84). These stem cells were used to generate embryoid bodies, induce neuroectoderm growth, and differentiate cerebral organoids. Differentiated organoids were then seeded in a Matrigel matrix with vitamin A and orbital rotation to obtain a brain organoid (Fig. 2). At 4 hpi, different stages and structures of the asexual cycle were observed, including tachyzoites and bradyzoite cysts. Gene expression analysis showed evidence of T. gondii infection in this system, and viability was confirmed by infecting mice, suggesting that a 3D brain system could fill the knowledge gaps in cerebral toxoplasmosis.
3D Systems To Study the Effects of T. gondii on Tight Junctions
Organoid technology is useful to analyze T. gondii dissemination. For T. gondii, the dissemination process includes parasite-host interactions, host immune evasion, and the crossing of host biological barriers. It has been reported that proteases secreted by T. gondii (isolated from mice) influence the dissemination process, affecting the tight junctions on MDCK monolayers (85). A Transwell model to study transmigration in T. gondii tachyzoites was recently employed (86). In this model, MDCK monolayers were seeded on the upper chamber of a Transwell system until they reached confluence and had consistent transepithelial electrical resistance (TEER) values. Monolayers were then challenged with T. gondii excretory/secretory products (ESP; a fraction rich in proteases) in the presence or absence of protease inhibitors to test the capability of ESP to affect the tight junctions. While the treatment of monolayers with ESP decreased the TEER, when a metalloprotease inhibitor was added to ESP, it did not change the TEER. These results suggest that ESP contains a metalloprotease that affects the tight junctions. To test transmigration capability, confluent monolayers in Transwells were pretreated with ESP and then infected with tachyzoites. While tachyzoites were able to transmigrate after 2 h of incubation, parasite transmigration was enhanced by pretreating the monolayer with ESP (86). Although the effect of protease inhibitors was not considered in this assay, the results suggest that ESP promotes parasite migration through host monolayers in the Transwell system.
Two-dimensional ODMs (87, 88) in Transwells were developed from different mammalian species relevant for T. gondii transmission (Fig. 3). Human, mouse, pig, and chicken ODMs were infected with T. gondii alone or in coinfection with Giardia duodenalis, and barrier dysfunction was evaluated (89). The tissue culture-derived RH tachyzoites were able to replicate in the four species of ODMs, but they did not affect tight junction integrity, as evidenced by TEER values (89). In contrast to these culture-derived parasites, when MDCK and ARPE-19 monolayers were infected with T. gondii isolated from mice, decreased TEER values and morphological changes were observed by 4 hpi (85, 86, 90). These differences open the possibility that in vivo infection makes tachyzoites more virulent or allows T. gondii to express molecules that influence tight junctions, but in vitro culture reduces the production of these molecules. In the future, it will be interesting to evaluate the effect of different strains of T. gondii isolated from mice on ODMs and how they affect the integrity of tight junctions.
Shortcomings and Future Perspectives of T. gondii in 3D Systems
Collagen- or Matrigel-based matrices (Fig. 2), RWV bioreactors, organoids from PSC, and ODMs (Fig. 3) have been used to study T. gondii. The pros and cons of the organoid technologies are discussed in other reviews (91–94). We would like to highlight additional disadvantages to consider when using 3D systems to model T. gondii infection. Microinjection of T. gondii into 3D systems can produce low parasite infection rates, likely due to mucus, antimicrobial peptides, or damage to the parasites due to injection pressure (73). When work is being performed with samples from outbred organisms, genetic variability needs to be considered between different host donors (68). Moreover, each 3D system developed for T. gondii has a different host cell stability and longevity. Finally, the lack of standardized 3D system culture conditions between laboratories can affect reproducibility. For future studies, these 3D systems will need to add in microbiota and immune cells to recapitulate a more realistic extracellular environment. At the end of this review, we propose the use of brain, eye, and placental organs-on-a-chip for advances in T. gondii research.
PLASMODIUM STUDIES IN 3D SYSTEMS
Plasmodium Overview
Plasmodium is a parasite belonging to the Apicomplexa phylum and is the causative agent of malaria. In 2018, over 200 million cases of malaria were estimated and around 400,000 deaths were reported worldwide. Five species of Plasmodium infect humans, but infection with Plasmodium falciparum and Plasmodium vivax represent most of the cases and are known for causing multiorgan complications, anemia, cerebral malaria, and death (95–98). Children are the most vulnerable population, comprising 67% of deaths (World Health Organization, 2019). Although P. falciparum is responsible for most deaths, P. vivax remains the most widespread (99).
Challenges in the Eradication of Malaria
Malaria eradication campaigns make use of antimalarial drugs and insecticides (100, 101). As a result, a significant reduction in malaria transmission and morbidity has been observed in the last decade (102–104). Effective vaccines have only recently been developed and deployed. In October 2021, the WHO recommended the use of RTS,S/AS01 antimalaria vaccine for children living in sub-Saharan Africa. Advanced tools are needed to develop additional strategies to further disease eradication. One recent aim has been the study of the Plasmodium liver stage in vitro. The preerythrocytic stage, including sporozoites injected by a mosquito bite, is one of the most critical phases to study due to the low numbers of parasites needed for infection, the time needed to migrate toward hepatocytes, and the surface proteins exposed as vaccine targets (105, 106) The culture of preerythrocytic stages in vitro remains challenging, which is why many questions about this stage remain unanswered. To study the mechanisms of parasite-host interactions in Plasmodium, a system where hepatocytes can be readily infected and analyzed is needed. It is difficult to model Plasmodium spp. due to their species specificity and the multiple host niches during the life cycle of the parasite (107, 108). To study the liver stages, a hepatoma-derived cell line that maintains hepatocyte markers has been used extensively (109, 110).
3D Platforms for Studying Plasmodium Sporozoite Motility and Migration
Different 3D systems have been widely used in the last decade, in addition to organoids or spheroids, to understand the migration capability of Plasmodium sporozoites during infection and to better understand vector-host transmission. These 3D systems resemble a matrix in which the sporozoites can be monitored for mechanisms of locomotion and protein dynamics during migration (Fig. 4).
The sporozoite is a highly motile stage of the parasite. They are injected into the skin of the host by mosquito bite and travel through the blood to reach hepatocytes. There, they differentiate into blood cell-infecting merozoites (111). Previously, the motility and behavior of Plasmodium berghei sporozoites were examined in isolated salivary glands and ducts, suggesting that the parasites can switch between different movement patterns that help to colonize the ducts. The ejection of parasites was also analyzed and quantified during saliva discharge through the proboscis in immobilized live Anopheles mosquitoes (112). These studies suggest that parasite load in the salivary cavity did not correlate with the number of ejected sporozoites, as mosquitoes with high numbers of sporozoites in the salivary cavities did not necessarily discharge sporozoites, even during prolonged salivation. This study also shows that sporozoite motility might play a bigger role in the mosquito stage to spread more efficiently inside the host.
Adhesion dynamics and the locomotion machinery of sporozoites during gliding were also analyzed in cultured hepatocytes on a flexible elastic substrate, as well as in the salivary ducts of mosquitoes and the ear skin of mice (113). Sporozoites seemed to move in a stop-and-go fashion regulated by distinct adhesion sites. Using traction force microscopy on elastic substrates with marker beads, it was shown that the central part of the parasite applies perpendicular and longitudinal forces at the end of the parasite body. In this system, the role of actin-myosin and the transmembrane protein thrombospondin-related anonymous protein (TRAP) was also described. It suggests that both proteins are essential during parasite movement; actin seems to have a role during adhesion and slipping, but only TRAP is needed for slipping and is not essential for parasite adhesion (113).
The migration and movement of P. berghei sporozoites were also studied in vivo in the tail and ear skin of mice after the bite of infected mosquitoes (114). Sporozoites migrating in the dermis in the tail showed more linear trajectories than sporozoites migrating in the ear. In the dermis of the ear, the movement showed random migration patterns and meandering motility. To test whether changes that arise in different tissues are associated with cellular architecture and chemical cues, the researchers analyzed sporozoite movement in microfabricated pillar substrates (obstacle arrays) (Fig. 4) with different architecture. Results revealed that the parasites change their movements depending on the obstacle and that they follow the same pattern of movements (linear or meandering patterns) that were seen in the skin (114). These results suggest that the parasite can adapt to different environments during transmission for an optimal host spread (114).
Most recently, pillar substrates were used to study how sporozoites invade capillaries during the skin stage of the infection cycle. To do this, PDMS pillars of different diameters were generated to imitate the variety of diameters of blood vessels found on the skin. This setup was used to test the hypothesis that the curvature of the sporozoite influenced which blood vessel the parasite would invade (115). Results from this experiment showed that P. berghei parasites tended to adhere to columns between 8 and 12 μm in diameter, which are close to the diameter of blood capillaries found on the skin. Experiments using an array of square and pentagon shapes showed that while the sporozoites attached to the pillars regardless of the shape, some sporozoites changed their shape to adapt to the geometry of the pillar. This result suggests that the parasite can adapt to vessels of different sizes but prefers to attach to surfaces that closely match their natural curvature to be able to infect blood capillaries in the skin (115).
To study sporozoite and ookinete motility, tunable polyacrylamide hydrogels were recently used (116). These systems have an advantage over Matrigel or collagen in that they can be manufactured with specific elasticity ranges and pore sizes to mimic different environments (Fig. 4). Using these hydrogels, it was possible to characterize sporozoite movement in 3D gels when salivary glands were infected and placed between a glass coverslip and hydrogel (116).
Migrating from 2D to 3D Systems To Study the Liver Stage of Plasmodium
Rodents are the natural hosts for several Plasmodium species, making them excellent models for Plasmodium infections. However, human Plasmodium species only infect human hepatocytes, making it difficult to recapitulate these infections in rodent models. To establish models for human Plasmodium species, 2D cultures of primary human hepatocytes, humanized mice, and stem cell-derived progeny have all been used (Fig. 1). Of note, valuable findings were also obtained by infection of HeLa cells, a nonnatural host cell for Plasmodium liver stage (117–120), or human bone marrow endothelial cells (hBMECs) for the study of gametocyte stages (121). Primary hepatocytes and cell lines are unable to recapitulate in vivo conditions due to uncontrolled cell proliferation, shortages of available liver material, abnormal liver-specific functions, and poor infection efficiency (122).
In the last decade, many attempts have been made to obtain the most natural liver environment in vitro. In the case of Plasmodium, liver organoids could be useful to study the life cycle of the parasite and discover new antimalarial drugs. Extensive analysis has demonstrated that liver organoids preserve their genetic integrity for months (123). Furthermore, liver organoids can secrete albumin and bile acid salts, show cytochrome activity, accumulate glycogen, and recapitulate the typical hepatocyte function in vitro in contrast to the commonly used cell line HepG2 (17, 123). Human liver organoids are also positive for molecular markers, such as the hepatocyte nuclear factor 4 (HNF4a), albumin, and the tight junction marker ZO-1 (17). Morphological features of liver hepatocytes, such as prominent nucleoli, decondensed chromatin, many mitochondria, tight junctions, peroxisomes, lysosomes, and multivesicular bodies, have been observed in organoids, which have the advantage of being cultured for several months while maintaining these features (124). Liver organoids are formed with a variety of liver cell types and are more sensitive than conventional in vitro models, making them advantageous to the study of sporozoite biology. The complete development of P. falciparum and P. vivax into schizonts was demonstrated in 2D cultures that maintained functional characteristics of native in situ hepatocytes, such as high albumin expression, biliary metabolites, and active mitochondria, which remained stable for 30 days (8, 17), which suggests that organoids are a promising model for the study of Plasmodium biology (17).
Liver spheroids have enabled the growth of blood-infective merozoites from the liver stage of the rodent-specific parasite P. berghei (125). The spheroids were generated from three hepatic cell lines, HepG2, HC-04, and HepaRG, in a stirred-tank bioreactor system and maintained for up to 30 days (Fig. 2). These structures displayed epithelial polarization, production of albumin, HNF4a, and liver proteins in vitro. This 3D platform was used to evaluate the antiplasmodial drugs M5717 and atovaquone (125), demonstrating the efficacy of the system. However, infection efficiency with P. berghei was lower in the spheroids than in 2D cells. The formation of 3D spheroids was also achieved with simian and human hepatocytes using a Cellusponge platform, a biomaterial designed to recapitulate mechanical and chemical properties that maintain the hepatocyte function (Fig. 2), as well as maturation and differentiation capabilities (14). Spheroids grown using this platform as a scaffold supported the complete live stage of P. cynomolgi and P. vivax from sporozoites to merozoites in vitro (14).
Advances in the Study of Cerebral Malaria using 3D Systems
In recent years, the study of cerebral malaria using organoids has emerged as an innovative model. Brain cortical organoids can be obtained by reprogramming CD34+ human umbilical cord blood cells (126). During human cerebral malaria, elevated heme accompanies inflammation, apoptosis, and damage at the blood-brain barrier (127–130). Due to these observations, the effects of heme in pluripotent cells and 3D cortical organoids were assayed. Heme treatment resulted in apoptosis and structural damage with a disorganization of the cortical organoids (126). These results show that this ex vivo model could play a crucial role in future approaches for cerebral malaria research.
Microdevices for the Study of Plasmodium Gametocytes
During P. falciparum and P. vivax infection, gametocytes can be found in high abundance in the bone marrow, suggesting that this tissue acts as a site for gametocytogenesis (121, 131, 132). To study cytoadherence of gametocytes in bone marrow and the role of endothelial receptors, researchers have used hBMEC and CHO (Chinese hamster ovary) cell lines, respectively. CHO cells do not naturally express endothelial receptors, but they were successfully transfected to express ICAM1, VCAM, or CD36 receptors (121). These results showed that gametocytes can adhere ex vivo to endothelial cells and that expression of ICAM1 enhances adherence to CHO cells, suggesting an important role of ICAM1 in cytoadhesion of gametocytes.
During transmission, gametocyte-infected erythrocytes (GIEs) are sequestered in the bone marrow, where they mature. Once mature, they are released within the blood, where they can be taken up by mosquitoes (133). Morphological changes of GIEs are associated with the migration of the parasite through the spleen. To mimic splenic filtration of GIEs, a microdevice consisting of a matrix composed of metal microspheres was used (134). GIEs were perfused using a pump, and the retention rates were determined. In this system, the authors tested whether cAMP modulated GIE mechanical properties associated with deformability (Fig. 4). Results showed that a decrease of cAMP levels increases the deformability of GIEs and flow through the device (134).
3D Platforms for Studying Plasmodium Ookinete Motility and Migration
The ookinete is a motile stage of the Plasmodium life cycle formed during the mosquito blood meal. It is essential for transmission back to the mosquito vector. Ookinetes reach the midgut of the mosquito, where they transform into sporozoites which then invade the salivary glands. To convert to oocytes, the ookinete must then overcome obstacles such as dense blood boluses, the thickness of the peritrophic matrix, and proteolytic activities of the gut (135). The motility of ookinetes has been studied in the past using Matrigel (136, 137), where it was possible to observe the helical gliding path and speed with the same parameters that were observed within the mosquito blood meal (138). Matrigel was used to check the shape and gliding movements of ookinete mutants. The results of this experiment showed that the balance of the production and degradation of cGMP by different enzymes is necessary to maintain the normal ookinete shape and function (136). The ookinete motion has also been recorded by “infecting” 96-well plates containing Matrigel with green fluorescent protein (GFP)-expressing parasites (Fig. 4). The reconstruction showed left helical motion paths with no apparent bias in directionality, similar to those found in vivo using explants of infected mosquito midgut. The left-handed helical trajectory seems to be the result of the architecture conformation of the microtubular cytoskeleton (137).
Recently, polyacrylamide hydrogels were used for the study of ookinete motility for up to 20 h. The advantage of hydrogels over Matrigel or collagen is the ability to fine-tune the pore size and stiffness of the polymer just by adjusting the concentration of cross-linker. By adjusting the stiffness of the polymer, it was possible to observe the distinct behavior of ookinetes under different conditions. During the assays, ookinetes were able to change their migration behavior from slow or helical movements in softer substrates to faster and straight directional dissemination in stiffer substrates. It was hypothesized that these changes in the migration pattern imitate what happens in vivo when the ookinetes move from the blood bolus to the stiffer midgut epithelium (116).
Shortcomings and Future Perspectives in Plasmodium Research
Organoid research is still developing, and its optimization for the culture of different species and stages of Plasmodium is needed. To date, only Plasmodium berghei, P. cynomolgi, and P. vivax have been studied by using liver organoids and other 3D systems (14, 113, 114, 116, 125). Analysis of the hypnozoite or liver dormant stage of Plasmodium is another understudied area where liver organoids will likely be useful. These systems for the study of Plasmodium still have some limitations, such as poor and inconsistent infectivity rates, the lack of immune cells or microbiota, and parasites becoming trapped in the matrices (139). Optimized organoid systems for Plasmodium research could be helpful to study protein expression of parasite stages, antigenic presentation, host-parasite communication, parasite dynamics, and vaccine development. In the last section of this review, we describe organ-on-a-chip for Plasmodium research as a future perspective.
Mosquito midgut-derived cell lines or organoids to understand the mosquito restricted stages have been considered (140) but not yet developed. The use of mammalian intestinal organoids as insect guts has also been proposed (141). The fruit fly Drosophila melanogaster is the insect model that has been studied the most and is the closest relative of the Anopheles mosquito. Recently, D. melanogaster intestinal stem cells were used as a model for studying epithelial homeostasis and signaling molecules (142). Information from these signaling molecules could help future endeavors to create Anopheles gut and salivary gland organoids that would be used to model the sexual development of Plasmodium. To date, functional salivary gland organoids can be successfully generated from murine embryonic/adult stem cells or adult human stem cells (143–145).
In addition to the 3D platforms already in use, the 3D bioprinting technique will likely become useful for the apicomplexan field. This technique uses functional biomaterials to print biomimetic architecture (146). Printing with stem cells can produce tissue architecture fidelity, mechanical stability, and cytocompatibility, including tissue-like cell migration and proliferation (147). The use of this system has been reported in medicine for personalized drug delivery as well as the production of complex tissues such as corneal and neural tissue for transplants (148, 149). For Plasmodium research, 3D bioprinting could be used to create organs that have been difficult to culture, such as mosquito salivary glands and midgut. The application of 3D bioprinting to Plasmodium research could fill critical knowledge gaps for this parasite.
EIMERIA STUDIES IN 3D SYSTEMS
Eimeria Overview
Eimeria spp. are the causative agents of coccidiosis in livestock. They are intestinal parasites that infect the intestinal epithelium and cause hemorrhagic enteritis. More than 100 species of Eimeria, which infect vertebrates such as horses, cats, rabbits, dogs, cattle, sheep, pigs, turkeys, and chickens, are known. During Eimeria infection, parasites can infect different regions of the intestine at the same time (150). Eimeria infections are especially detrimental to poultry production. Around 30 billion chickens are infected worldwide, costing the poultry industry approximately US$2.4 billion annually (151). Eimeria infection is controlled by good husbandry protocols, anticoccidial drugs, and live vaccines. Avian coccidiosis has been controlled by drugs with three different mechanisms of action: (i) destroying membrane integrity with polyethers such as monensin, maduramicin, salinomycin, narasin, semduramycin, and lasalocid, (ii) inhibiting protein synthesis using drugs like clindamycin, spiramycin, clarithromycin, and paromomycin, and (iii) inhibiting DNA synthesis with sulfonamides, nicarbazin, decoquinate, methylbenzoquate, clopidol, robenidin, ethopabate, pyrimethamine, diaveridine, ormothroprim, epiroprim, and amprolium (152, 153). However, prolonged use of these anticoccidiosis drugs has resulted in drug-resistant strains, increasing the global burden of Eimeria.
In addition to drugs, there are live virulent and attenuated vaccines available against Eimeria. The vaccines Coccivac and Immucox are based on unmodified oocysts of wild-type Eimeria. Because these kinds of nonattenuated vaccines are virulent, they have inherent risks associated with them, and they are not licensed in some countries, especially in Europe. There are also live-attenuated vaccines such as Paracox, Livacox, Hipracox, HuveGuard, and EimeriaVax 4m (154–157). The seven known species of Eimeria have been attenuated by serial passage, which resulted in impaired asexual development (156, 157).
Research on 2D Cultures in Eimeria
Many species of Eimeria have been cultured in different in vitro culture systems showing various degrees of development. Eimeria acervulina has been cultured in vitro in chicken embryo fibroblasts, mouse fibroblasts, and HeLa cells using sporozoites as the inoculum, but only trophozoite stages were observed (158) (Fig. 1). Eimeria contorta has been grown in vitro using mouse intestinal monolayers (159, 160). Eimeria tenella sporozoites have been cultured in vitro in primary chicken kidney cells but with low production of oocysts (161). The bovine strains Eimeria bovis and E. zuernii have been cultured in vitro in bovine fetal gastrointestinal, colonic epithelial, and umbilical vein endothelial cells. E. bovis culture enabled parasites to reach the first generation of merozoites and meront II stages, while E. zuernii reached merozoite stage I (162–164) (Fig. 1). The ovine strains Eimeria ninakohlyakimovae and E. ovinoidalis grew better in caprine umbilical vein endothelial cells than in bovine colonic epithelial and umbilical vein endothelial cells. In caprine cells, parasites reached the macromeront and merozoite I stage but did not complete the cycle (165, 166). Despite these advances, until now, Eimeria in vitro 2D cultures have not reproduced the full parasite life cycle, especially gametocyte and oocyte production. For this reason, the 3D system in vitro culture appears to be a good solution to bridge this knowledge gap in Eimeria research.
Research on Enteroids in Eimeria
The first step toward the use of organoid technology in Eimeria started when intestinal primary cells isolated from rat fetuses were seeded on fibronectin- or gelatin-coated slides and infected with Eimeria nieschulzi (167). The development of macrogamonts and oocysts was seen only with the formation of crypt-like structures. This methodology has been the only one able to produce four rounds of asexual cycles, gametocyte stages, and oocyst in vitro production of E. nieschulzi. Mouse intestinal organoids were infected with Eimeria vermiformis or E. falciformis sporozoites. These mouse intestinal organoids showed self-renewing capabilities, crypt-villus structure, and different kinds of specialized cells and were viable for more than 6 months. For infection, this protocol used 3 to 4 postpassage organoids in which the Matrigel was disintegrated by passing it through a 21-gauge needle. The free organoids were washed and incubated with sporozoites at 37°C for 5 min. After infection, organoids were resuspended in 50% Matrigel. Half of the cells in the organoids were infected in just 4 h (168). The organoids remained infected for at least 7 days to produce a second generation of merozoites for E. vermiformis and possibly a third generation of merozoites for E. falciformis.
Another advance in organoids for Eimeria research was the use of 19-day-old chicken embryos and 3-week-old chick intestine to isolate intestinal crypts (Fig. 2). After washing, the crypt cells were seeded in Matrigel and intestinal organoids were developed (169). The 19-day-old embryos were difficult to use, because a pure epithelial cell culture could not be obtained, and fibroblast contamination was problematic. Due to the inability to avoid fibroblast contamination, intestinal epithelial organoids could not be kept longer than three passages. In contrast, it was easier to obtain pure epithelial cells without fibroblast contamination from 3-week-old chick intestines, but they were more susceptible to bacterial contamination. Contamination was avoided by the mechanical removal of intestinal mucus using a sterile slide and several cold saline washes. To test this system, the Advent coccidiosis vaccine containing E. acervulina, E. maxima, and E. tenella was used as a source of sporulated oocysts. After excystation and incubation in monolayers from chicken intestinal organoids, the parasites were able to replicate. Another research group recently used chicken duodenal crypts from the intestines of a 14-day-old female chicken to establish ODMs (89). This system has not been tested yet for Eimeria infection but has shown promising results in T. gondii and Giardia infection, as previously discussed (Fig. 3).
Additionally, a method to propagate Eimeria has been described using floating avian intestinal organoids derived from embryonic villi or mature chicken crypts (170). This system, which does not require matrices like Matrigel, has the in vivo architecture, cell diversity, polarity, barrier function, and epithelial functionality of the natural chicken intestine. This system has two unique characteristics: an inside-out phenotype, with the apical brush border facing the medium, and the innate presence of leukocytes. The inside-out conformation phenomenon facilitated infection with E. tenella by simply adding the microorganism to the medium. Thus, E. tenella sporozoites were able to infect the epithelium and undergo both schizogony and gametogonia (170).
Future Directions in Eimeria 3D Systems
The use of organoids in Eimeria research is in the early steps of development, but it is a promising technology to advance our knowledge of this parasite. It would be useful to have intestinal and macrophage cocultures to study the immune response against Eimeria. 3D culture could also increase effectiveness and replace the use of live chickens to produce Eimeria oocysts to manufacture vaccines. The benefits of using a 3D cell culture rather than chickens in oocyst production are that there is no bacterial contamination, animal use is reduced, and a consistent product that is not affected by host immune selection could be generated.
LATEST ADVANCES IN 3D CULTURE
Organ-on-a-Chip
While 3D culture systems offer many advantages, they still lack many factors that resemble the natural host model. Some in vivo factors missing from organoids include microvasculature, peristalsis, mechanical forces due to fluid flow, microenvironment, and mechanical forces that occur during infection. Additionally, animal models sometimes produce results that are not translatable to human patients. Organ-on-a-chip (OOAC) technology appears as the next step in tissue culture research to solve these challenges. OOAC is a system of microengineering that mimics the physiological environment of an organ where different factors can be controlled such as gradient concentration, nonstatic conditions, shear force, cell patterning, tissue boundaries, and tissue organ interactions (171, 172). OOAC has been used for personalized medicine and cancer research, generating tumors-on-a-chip from different organs. The use of OOAC in apicomplexan research is just starting, but here we briefly analyze the early steps and possibilities that this new technology presents for studies of Apicomplexa.
Skin-on-a-chip.
Artificial skin systems are useful models for wound healing, dermatological, and infectious disease research. Artificial skin is of particular importance to infectious disease research, because many pathogen vectors feed on host skin and simultaneously inject and transmit microorganisms. To establish a standard testing method, protocols to develop full-thickness skin equivalents (FTSE) have been created. This system begins with the in vitro culture of a mix of human dermal fibroblasts (hDF) with collagen. Then, human epidermal keratinocytes (hEK) are added on top and kept on an air-liquid interface culture (173). These human skin equivalents have already been used to evaluate various infection models such as Candida albicans (174), Staphylococcus aureus (175), and helminths (176).
A human artificial skin model was developed for trypanosome infection (177). A high-density skin equivalent was created with a high concentration of collagen and reduced water content to increase mechanical stability. Normal human dermal fibroblasts (NHDF) were mixed with reconstituted collagen and compressed 7-fold to generate high-density dermal equivalents (hdDEs). A computer-assisted compressor system was used to compress the dermal component. Also, normal human epidermal keratinocytes (NHEK) were added. Tsetse flies, the Trypanosoma vector, were able to penetrate artificial skins, inject the parasite, and infect the mature high-density skin equivalents (177). This model resembles anatomic, cellular, and functional characteristics of the human natural skin. Artificial skin may be useful to study the interactions between ticks, mammals, and Babesia, as well as the mosquito-human-Plasmodium axis. Using this validated skin infection model, it might be possible to study motility, parasite stage differentiation, invasion of host cells, and secretion of compounds in the wound site, among other important processes in apicomplexan microorganisms and their skin stage.
Liver-on-a-chip.
Liver-on-a-chip was initially developed by coculture of primary rat hepatocytes with fibroblasts in a microfluidic system that allows for the production of albumin and urea with a continuous supply of culture medium and oxygen (178). Liver-on-a-chip was later developed in two separate channels with a PDMS membrane, the top channel was seeded with hepatocytes, and the bottom channel was seeded with liver sinusoidal endothelial cells. This liver OOAC was combined with confocal microscopy for the evaluation of cytotoxicity of benzbromarone in hepatocytes (179). A microliver multiwell device of primary human hepatocytes supported by stroma cells (180) was used as a platform for the Plasmodium liver stage by using cryopreserved hepatocytes from donors (181). Cryopreserved Plasmodium parasites were able not only to invade hepatocytes but also to progress through the complete Plasmodium liver stage. This platform was used to evaluate attenuated Plasmodium parasites and anti-Plasmodium drug screening (181). RNA silencing in human microlivers reduced the expression of essential enzymes such as cytochromes CYP3A4 and CYP2D6 (182). Additionally, the CD81 host entry factor was silenced in hepatocytes, which is relevant for Plasmodium because this gene edition reduced the infection with Plasmodium, demonstrating the efficacy of these microdevices in infectious diseases.
Splenon-on-a-chip.
The microfluidic device called splenon-on-a-chip was designed as the minimal functional unit of the red pulp of the spleen that can maintain its filtering functions (183). In a normal spleen, the blood coming from the splenic artery goes to the closed-fast microcirculation to directly reach the splenic venules or the open-slow microcirculation to filtration beds to eliminate unhealthy red blood cells (RBCs) by macrophages. Splenon-on-a-chip was designed with two microfluidic channels that imitate the closed-fast and the open-slow microcirculation under physiological flow rate. Splenon-on-a-chip was used to study the role of the spleen in Plasmodium infections where splenomegaly is a collateral side effect of this infection (183). This device allowed the measurement and live imaging of individual cell deformability in real time of mouse reticulocytes infected with Plasmodium yoelii, demonstrating that infected cells are more deformable than uninfected cells. A limitation of this microdevice is that the slit size of the splenon-on-a-chip is larger than the slit size of the human spleen microvasculature. To better represent the fluctuating flow of the human bloodstream, the author recommended adding a pulsatile pump for future experiments.
Intestine-on-a-chip.
Intestinal or gut OOACs have been designed, and protocols for their fabrication are currently available (184–190). Duodenum OOAC has been developed from human adult-derived intestinal organoids cocultured with microvascular endothelial cells separated by a PDMS membrane (191). This system showed cytoarchitecture, cell-cell interactions, permeability parameters, and gene expression more like the human intestine than organoids. The characteristics obtained by this technology allow for more biologically relevant drug delivery and pharmacokinetic studies. These microfluidic systems can be used and adapted for research in enteric microorganisms, including the enteric apicomplexans covered in this review.
Eye-on-a-chip.
Various retina-on-a-chip (ROC) systems are microfluidic platforms generated from human induced pluripotent stem cells that generate different retinal cell types (192–194). One of these systems has already been tested in Plasmodium research (192). This ROC was designed with vascular perfusion, which allows the in vitro interaction of photoreceptors with retinal pigment epithelium. All of these unique retina-on-a-chip characteristics allowed the evaluation of important biological processes such as calcium flux and phagocytosis. It was also used to study the very well known side effects of the anti-Plasmodium drug chloroquine, showing a reduction in cell viability. ROC will likely be useful in the future to study ocular toxoplasmosis.
Brain-on-a-chip.
Many brain-on-a-chip platforms are being currently developed (195–197). One microfluidic device described consists of two parallel culture chambers separated by three medium channels. The chambers were seeded with brain organoids, the middle medium channel was used as a fluid flow passageway, and the two side channels were filled with cell culture medium (196). To evaluate the effects of nicotinamide on neurogenesis, the system was exposed to nicotinamide continuously and then examined by immunohistochemistry and PCR (196). The results showed that nicotinamide affected neuronal differentiation and brain organization like nicotinamides affect the human fetal brain during gestation. Another kind of brain-on-a-chip uses a multicompartment microdevice of neurospheres. It used human induced pluripotent neural stem cells which were differentiated into four different neuronal phenotypes, one for each compartment of the microdevice. The different neuronal phenotypes were connected to simulate multiple neurotransmission regions (197). These microdevices may enable studies of T. gondii in human brain-derived organs-on-a-chip as well as Plasmodium brain cytoadherence in a continuous flow. This platform could help solve the problem of genetic variation between animal models and human neurological diseases.
In summary, OOAC is a complementary technology to existing 3D systems because OOACs offer the advantage of stem cells plus all the mechanical forces found in real tissues of live organisms. Research in apicomplexan parasites will benefit from these engineered microdevices to develop studies that take advantage of the parameters that can be controlled using organs on chips. These advantages can provide a more accurate and biologically relevant picture of how these apicomplexans affect human health.
ACKNOWLEDGMENTS
We thank Skala lab members at the Morgridge Institute for Research, Bruno Martorelli di Genova, and Maria Arendt for teaching us about the organoid techniques and 3D systems that we use in the lab today. We also thank Nicole M. Davis and Carolina Mendoza Cavazos for their editing and helpful comments. Figures were created using Biorender.com and edited using Adobe Illustrator. We thank Patrick Lane from ScEYEnce Studios for improving our figures.
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Conceptualization, A.M.T.-P., G.M.G.-L., C.J.R.-F., and L.J.K.; funding acquisition, L.J.K.; investigation, G.M.G.-L., C.J.R.-F., and A.M.T.-P.; methodology, G.M.G.-L., A.M.T.-P., and C.J.R.-F.; resources, L.J.K.; supervision, L.J.K.; visualization, A.M.T.-P.; writing, G.M.G.-L., C.J.R.-F., and A.M.T.-P.; writing – review & editing, A.M.T.-P., G.M.G.-L., C.J.R.-F., and L.J.K. All authors read and approved the final manuscript.
This work was supported by the National Institutes of Health (R01AI144016-01 for L.J.K.) and the Morgridge Metabolism Interdisciplinary Fellowship from the Morgridge Institute for Research (G.M.G.-L.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Biographies

Carlos J. Ramírez-Flores graduated with honors as a biologist from Universidad Veracruzana-Xalapa (Veracruz, Mexico). He obtained his M.Sc. and Ph.D. degrees in biochemistry from CINVESTAV-IPN (Mexico City, Mexico). During graduate school, he applied his knowledge to understand the role of proteases released by Toxoplasma gondii modifying tight junctions, focusing on the transmigration process. After graduate school, he joined Laura Knoll’s lab as a postdoctoral fellow at University of Wisconsin–Madison. He is currently studying the role of parasite enzymes and their products associated with host immune evasion and as a signal that triggers the sexual stage of T. gondii. Carlos has already been working for more than 10 years with T. gondii, a model that he would like to continue to explore in his future academic life.

Andrés M. Tibabuzo Perdomo obtained his B.Sc. in Biology from Universidad de los Andes (Bogotá, Colombia), studying the phospholipases present in snake venom. He obtained his Ph.D. in Chemistry from Purdue University (West Lafayette, IN). There, he worked to identify key proteins found in the adhesive produced by marine shellfish by using proteomics. During this period, he also completed an internship at the Vaccine Business Unit at Takeda Pharmaceuticals (Boston, MA), working with the regulatory affairs team. After completing his graduate studies, he joined Laura Knoll’s lab as a postdoc, where he has applied his interdisciplinary knowledge to answer the question “what triggers Toxoplasma gondii to undergo sexual development in cats?” Outside the lab, he applies what he has learned throughout the years to many interests of his, including scientific illustration, programming, molecular gastronomy, bread baking, and entrepreneurship.

Gina M. Gallego-López obtained her biology degree from the Universidad del Tolima (Colombia) and a Ph.D. in veterinary sciences with an emphasis on immunology and infectious diseases from Washington State University. She is currently a postdoctoral fellow at Morgridge Institute for Research and the University of Wisconsin. Gina has been working in apicomplexan parasites for 15 years. She has participated in research projects studying Plasmodium, Babesia, Toxoplasma, and Cryptosporidium. Additionally, Gina has received training in organoid technologies during her postdoctoral studies, using different cancer-, mouse-, and farm animal-derived organoids. Gina studies the molecular and metabolic mechanisms of parasitic infections to develop novel therapeutic strategies.

Laura J. Knoll graduated from St. Olaf College with a B.A. in Chemistry and Biology. Her thesis research was in lipid metabolism with Dr. Jeffrey Gordon at Washington University in St. Louis. Laura did her postdoctoral research with Dr. John Boothroyd at Stanford University, where she used molecular genetic techniques to study the parasite Toxoplasma gondii. In 2001, she moved to the University of Wisconsin–Madison to join the Medical Microbiology and Immunology Department. The Knoll lab focuses largely on T. gondii: understanding how it forms a chronic infection within the host, the intestinal immune responses to oral infection, and the species specificity of sexual development.
REFERENCES
- 1.Visvesvara GS, Garcia LS. 2002. Culture of protozoan parasites. Clin Microbiol Rev 15:327–328. 10.1128/CMR.15.3.327-328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Justice MJ, Dhillon P. 2016. Using the mouse to model human disease: increasing validity and reproducibility. Dis Model Mech 9:101–103. 10.1242/dmm.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murillo-Cuesta S, Artuch R, Asensio F, de la Villa P, Dierssen M, Enríquez JA, Fillat C, Fourcade S, Ibáñez B, Montoliu L, Oliver E, Pujol A, Salido E, Vallejo M, Varela-Nieto I. 2020. The value of mouse models of rare diseases: a Spanish experience. Front Genet 11:583932. 10.3389/fgene.2020.583932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayir E, Sendemir A, Missirlis YF. 2019. Mechanobiology of cells and cell systems, such as organoids. Biophys Rev 11:721–728. 10.1007/s12551-019-00590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Co JY, Margalef-Català M, Li X, Mah AT, Kuo CJ, Monack DM, Amieva MR. 2019. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep 26:2509–2520.e4. 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huch M, Koo B-K. 2015. Modeling mouse and human development using organoid cultures. Development 142:3113–3125. 10.1242/dev.118570. [DOI] [PubMed] [Google Scholar]
- 7.Clevers H. 2016. Modeling development and disease with organoids. Cell 165:1586–1597. 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 8.Roth A, Maher SP, Conway AJ, Ubalee R, Chaumeau V, Andolina C, Kaba SA, Vantaux A, Bakowski MA, Thomson-Luque R, Adapa SR, Singh N, Barnes SJ, Cooper CA, Rouillier M, McNamara CW, Mikolajczak SA, Sather N, Witkowski B, Campo B, Kappe SHI, Lanar DE, Nosten F, Davidson S, Jiang RHY, Kyle DE, Adams JH. 2018. A comprehensive model for assessment of liver stage therapies targeting Plasmodium vivax and Plasmodium falciparum. Nat Commun 9:1837. 10.1038/s41467-018-04221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramírez-Flores CJ, Knoll LJ. 2021. Breakthroughs in microbiology made possible with organoids. PLoS Pathog 17:e1010080. 10.1371/journal.ppat.1010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkbeiner SR, Zeng X-L, Utama B, Atmar RL, Shroyer NF, Estes MK. 2012. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio 3:e00159-12. 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng X-L, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353:1387–1393. 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engevik MA, Yacyshyn MB, Engevik KA, Wang J, Darien B, Hassett DJ, Yacyshyn BR, Worrell RT. 2015. Human Clostridium difficile infection: altered mucus production and composition. Am J Physiol Gastrointest Liver Physiol 308:G510–G524. 10.1152/ajpgi.00091.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gural N, Mancio-Silva L, He J, Bhatia SN. 2018. Engineered livers for infectious diseases. Cell Mol Gastroenterol Hepatol 5:131–144. 10.1016/j.jcmgh.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chua ACY, Ananthanarayanan A, Ong JJY, Wong JY, Yip A, Singh NH, Qu Y, Dembele L, McMillian M, Ubalee R, Davidson S, Tungtaeng A, Imerbsin R, Gupta K, Andolina C, Lee F, Tan KS-W, Nosten F, Russell B, Lange A, Diagana TT, Rénia L, Yeung BKS, Yu H, Bifani P. 2019. Hepatic spheroids used as an in vitro model to study malaria relapse. Biomaterials 216:119221. 10.1016/j.biomaterials.2019.05.032. [DOI] [PubMed] [Google Scholar]
- 15.Schimek K, Frentzel S, Luettich K, Bovard D, Rütschle I, Boden L, Rambo F, Erfurth H, Dehne E, Winter A, Marx U, Hoeng J. 2020. Human multi-organ chip co-culture of bronchial lung culture and liver spheroids for substance exposure studies. Sci Rep 10:7865. 10.1038/s41598-020-64219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell CC, Hendriks DFG, Moro SML, Ellis E, Walsh J, Renblom A, Fredriksson Puigvert L, Dankers ACA, Jacobs F, Snoeys J, Sison-Young RL, Jenkins RE, Nordling Å, Mkrtchian S, Park BK, Kitteringham NR, Goldring CEP, Lauschke VM, Ingelman-Sundberg M. 2016. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci Rep 6:25187. 10.1038/srep25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leboucher G. 1989. Maternal behavior in normal and androgenized female rats: effect of age and experience. Physiol Behav 45:313–319. 10.1016/0031-9384(89)90133-9. [DOI] [PubMed] [Google Scholar]
- 18.Mellin R, Boddey JA. 2020. Organoids for liver stage malaria research. Trends Parasitol 36:158–169. 10.1016/j.pt.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Koo B-K, Knoblich JA. 2020. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol 21:571–584. 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radtke AL, Wilson JW, Sarker S, Nickerson CA. 2010. Analysis of interactions of Salmonella type three secretion mutants with 3-D intestinal epithelial cells. PLoS One 5:e15750. 10.1371/journal.pone.0015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutta D, Clevers H. 2017. Organoid culture systems to study host-pathogen interactions. Curr Opin Immunol 48:15–22. 10.1016/j.coi.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delgado Betancourt E, Hamid B, Fabian BT, Klotz C, Hartmann S, Seeber F. 2019. From entry to early dissemination—Toxoplasma gondii’s initial encounter with its host. Front Cell Infect Microbiol 9:46. 10.3389/fcimb.2019.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klotz C, Aebischer T, Seeber F. 2012. Stem cell-derived cell cultures and organoids for protozoan parasite propagation and studying host-parasite interaction. Int J Med Microbiol 302:203–209. 10.1016/j.ijmm.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. 10.3201/eid1701.p11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong S, Yang Y, Wang Y, Yang D, Yang Y, Shi Y, Li C, Li L, Chen Y, Jiang Q, Zhou Y. 2020. Prevalence of Cryptosporidium infection in the global population: a systematic review and meta-analysis. Acta Parasitol 65:882–889. 10.2478/s11686-020-00230-1. [DOI] [PubMed] [Google Scholar]
- 26.Khalil IA, Troeger C, Rao PC, Blacker BF, Brown A, Brewer TG, Colombara DV, De Hostos EL, Engmann C, Guerrant RL, Haque R, Houpt ER, Kang G, Korpe PS, Kotloff KL, Lima AAM, Petri WA, Platts-Mills JA, Shoultz DA, Forouzanfar MH, Hay SI, Reiner RC, Mokdad AH. 2018. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob Health 6:e758–e768. 10.1016/S2214-109X(18)30283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter PR, Nichols G. 2002. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin Microbiol Rev 15:145–154. 10.1128/CMR.15.1.145-154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mac Kenzie WR, Schell WL, Blair KA, Addiss DG, Peterson DE, Hoxie NJ, Kazmierczak JJ, Davis JP. 1995. Massive outbreak of waterborne cryptosporidium infection in Milwaukee, Wisconsin: recurrence of illness and risk of secondary transmission. Clin Infect Dis 21:57–62. 10.1093/clinids/21.1.57. [DOI] [PubMed] [Google Scholar]
- 29.Laurent F, McCole D, Eckmann L, Kagnoff MF. 1999. Pathogenesis of Cryptosporidium parvum infection. Microbes Infect 1:141–148. 10.1016/S1286-4579(99)80005-7. [DOI] [PubMed] [Google Scholar]
- 30.Vinayak S, Pawlowic MC, Sateriale A, Brooks CF, Studstill CJ, Bar-Peled Y, Cipriano MJ, Striepen B. 2015. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523:477–480. 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baydoun M, Vanneste SB, Creusy C, Guyot K, Gantois N, Chabe M, Delaire B, Mouray A, Baydoun A, Forzy G, Chieux V, Gosset P, Senez V, Viscogliosi E, Follet J, Certad G. 2017. Three-dimensional (3D) culture of adult murine colon as an in vitro model of cryptosporidiosis: proof of concept. Sci Rep 7:17288. 10.1038/s41598-017-17304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeCicco RePass MA, Chen Y, Lin Y, Zhou W, Kaplan DL, Ward HD. 2017. Novel bioengineered three-dimensional human intestinal model for long-term infection of Cryptosporidium parvum. Infect Immun 85:e00731-16. 10.1128/IAI.00731-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyzzer EE. 1907. A sporozoan found in the peptic glands of the common mouse. Exp Biol Med 5:12–13. 10.3181/00379727-5-5. [DOI] [Google Scholar]
- 34.Tzipori S, Widmer G. 2008. A hundred-year retrospective on cryptosporidiosis. Trends Parasitol 24:184–189. 10.1016/j.pt.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dubey JP. 2008. The history of Toxoplasma gondii—the first 100 years. J Eukaryot Microbiol 55:467–475. 10.1111/j.1550-7408.2008.00345.x. [DOI] [PubMed] [Google Scholar]
- 36.Current WL, Haynes TB. 1984. Complete development of Cryptosporidium in cell culture. Science 224:603–605. 10.1126/science.6710159. [DOI] [PubMed] [Google Scholar]
- 37.Widmer G, Köster PC, Carmena D. 2020. Cryptosporidium hominis infections in non-human animal species: revisiting the concept of host specificity. Int J Parasitol 50:253–262. 10.1016/j.ijpara.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Upton SJ, Tilley M, Brillhart DB. 1994. Comparative development of Cryptosporidium parvum (Apicomplexa) in 11 continuous host cell lines. FEMS Microbiol Lett 118:233–236. 10.1111/j.1574-6968.1994.tb06833.x. [DOI] [PubMed] [Google Scholar]
- 39.Arrowood MJ. 2002. In vitro cultivation of Cryptosporidium species. Clin Microbiol Rev 15:390–400. 10.1128/CMR.15.3.390-400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hijjawi N, Meloni B, Morgan U, Thompson RC. 2001. Complete development and long-term maintenance of Cryptosporidium parvum human and cattle genotypes in cell culture. Int J Parasitol 31:1048–1055. 10.1016/s0020-7519(01)00212-0. [DOI] [PubMed] [Google Scholar]
- 41.Miller CN, Jossé L, Brown I, Blakeman B, Povey J, Yiangou L, Price M, Cinatl J, Xue W-F, Michaelis M, Tsaousis AD. 2018. A cell culture platform for Cryptosporidium that enables long-term cultivation and new tools for the systematic investigation of its biology. Int J Parasitol 48:197–201. 10.1016/j.ijpara.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Striepen B. 2013. Parasitic infections: time to tackle cryptosporidiosis. Nature 503:189–191. 10.1038/503189a. [DOI] [PubMed] [Google Scholar]
- 43.Checkley W, White AC, Jaganath D, Arrowood MJ, Chalmers RM, Chen X-M, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA, Priest JW, Roos DS, Striepen B, Thompson RCA, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER. 2015. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis 15:85–94. 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duval K, Grover H, Han LH, Mou Y, Pegoraro AF, Fredberg J, Chen Z. 2017. Modeling physiological events in 2D vs. 3D cell culture. Physiology (Bethesda) 32:266–277. 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 45.Alcantara Warren C, Destura RV, Sevilleja JEAD, Barroso LF, Carvalho H, Barrett LJ, O'Brien AD, Guerrant RL. 2008. Detection of epithelial-cell injury, and quantification of infection, in the HCT-8 organoid model of cryptosporidiosis. J Infect Dis 198:143–149. 10.1086/588819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unsworth BR, Lelkes PI. 1998. Growing tissues in microgravity. Nat Med 4:901–907. 10.1038/nm0898-901. [DOI] [PubMed] [Google Scholar]
- 47.Goodwin TJ, Schroeder WF, Wolf DA, Moyer MP. 1993. Rotating-wall vessel coculture of small intestine as a prelude to tissue modeling: aspects of simulated microgravity. Proc Soc Exp Biol Med 202:181–192. 10.3181/00379727-202-43525. [DOI] [PubMed] [Google Scholar]
- 48.Morada M, Lee S, Gunther-Cummins L, Weiss LM, Widmer G, Tzipori S, Yarlett N. 2016. Continuous culture of Cryptosporidium parvum using hollow fiber technology. Int J Parasitol 46:21–29. 10.1016/j.ijpara.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Bakshi RP, Nenortas E, Tripathi AK, Sullivan DJ, Shapiro TA. 2013. Model system to define pharmacokinetic requirements for antimalarial drug efficacy. Sci Transl Med 5:205ra135. 10.1126/scitranslmed.3006684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilke G, Funkhouser-Jones LJ, Wang Y, Ravindran S, Wang Q, Beatty WL, Baldridge MT, VanDussen KL, Shen B, Kuhlenschmidt MS, Kuhlenschmidt TB, Witola WH, Stappenbeck TS, Sibley LD. 2019. A stem-cell-derived platform enables complete Cryptosporidium development in vitro and genetic tractability. Cell Host Microbe 26:123–134.e8. 10.1016/j.chom.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Certad G, Ngouanesavanh T, Guyot K, Gantois N, Chassat T, Mouray A, Fleurisse L, Pinon A, Cailliez J-C, Dei-Cas E, Creusy C. 2007. Cryptosporidium parvum, a potential cause of colic adenocarcinoma. Infect Agents Cancer 2:22. 10.1186/1750-9378-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heo I, Dutta D, Schaefer DA, Iakobachvili N, Artegiani B, Sachs N, Boonekamp KE, Bowden G, Hendrickx APA, Willems RJL, Peters PJ, Riggs MW, O'Connor R, Clevers H. 2018. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol 3:814–823. 10.1038/s41564-018-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Yamamoto Y, Wilson LH, Zhang T, Howitt BE, Farrow MA, Kern F, Ning G, Hong Y, Khor CC, Chevalier B, Bertrand D, Wu L, Nagarajan N, Sylvester FA, Hyams JS, Devers T, Bronson R, Lacy DB, Ho KY, Crum CP, McKeon F, Xian W. 2015. Cloning and variation of ground state intestinal stem cells. Nature 522:173–178. 10.1038/nature14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tolboom JJ. 1996. The infectivity of Cryptosporidium parvum in healthy volunteers. J Pediatr Gastroenterol Nutr 23:201–202. 10.1097/00005176-199608000-00020. [DOI] [PubMed] [Google Scholar]
- 55.Robert-Gangneux F, Dardé M-L. 2012. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev 25:264–296. 10.1128/CMR.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shapiro K, Bahia-Oliveira L, Dixon B, Dumètre A, de Wit LA, VanWormer E, Villena I. 2019. Environmental transmission of Toxoplasma gondii: oocysts in water, soil and food. Food Waterborne Parasitol 15:e00049. 10.1016/j.fawpar.2019.e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paris L. 2020. Toxoplasmosis, p 803–813. In Eduard R, Solomon T, Endy TE, Hill DR, Aronson NE (ed), Hunter’s tropical medicine and emerging infectious diseases, 10th ed. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 58.Luft BJ, Remington JS. 1992. Toxoplasmic encephalitis in AIDS. Clin Infect Dis 15:211–222. 10.1093/clinids/15.2.211. [DOI] [PubMed] [Google Scholar]
- 59.Bollani L, Strocchio L, Stronati M. 2013. Congenital toxoplasmosis. Early Hum Dev 89:S70–S72. 10.1016/S0378-3782(13)70107-5. [DOI] [Google Scholar]
- 60.Pappas G, Roussos N, Falagas ME. 2009. Toxoplasmosis snapshots: global status of Toxoplasma gondii seroprevalence and implications for pregnancy and congenital toxoplasmosis. Int J Parasitol 39:1385–1394. 10.1016/j.ijpara.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Lindsay DS, Dubey JP. 2014. Toxoplasmosis in wild and domestic animals, p 193–215. In Toxoplasma gondii, 2nd ed. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 62.Khan A, Grigg ME. 2017. Toxoplasma gondii: laboratory maintenance and growth. Curr Protoc Microbiol 44:20C.1.1–20C.1.17. 10.1002/cpmc.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dubey JP, Lindsay DS, Speer CA. 1998. Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin Microbiol Rev 11:267–299. 10.1128/CMR.11.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.National Research Council (US) Committee on Scientific and Humane Issues in the Use of Random Source Dogs and Cats in Research. 2009. Scientific and humane issues in the use of random source dogs and cats in research. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 65.Leung JM, Rould MA, Konradt C, Hunter CA, Ward GE. 2014. Disruption of TgPHIL1 alters specific parameters of Toxoplasma gondii motility measured in a quantitative, three-dimensional live motility assay. PLoS One 9:e85763. 10.1371/journal.pone.0085763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pavlou G, Touquet B, Vigetti L, Renesto P, Bougdour A, Debarre D, Balland M, Tardieux I. 2020. Coupling polar adhesion with traction, spring, and torque forces allows high-speed helical migration of the protozoan parasite Toxoplasma. ACS Nano 14:7121–7139. 10.1021/acsnano.0c01893. [DOI] [PubMed] [Google Scholar]
- 67.Sibley LD. 2004. Intracellular parasite invasion strategies. Science 304:248–253. 10.1126/science.1094717. [DOI] [PubMed] [Google Scholar]
- 68.Kanatani S, Uhlén P, Barragan A. 2015. Infection by Toxoplasma gondii induces amoeboid-like migration of dendritic cells in a three-dimensional collagen matrix. PLoS One 10:e0139104. 10.1371/journal.pone.0139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robbins JR, Zeldovich VB, Poukchanski A, Boothroyd JC, Bakardjiev AI. 2012. Tissue barriers of the human placenta to infection with Toxoplasma gondii. Infect Immun 80:418–428. 10.1128/IAI.05899-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McConkey CA, Delorme-Axford E, Nickerson CA, Kim KS, Sadovsky Y, Boyle JP, Coyne CB. 2016. A three-dimensional culture system recapitulates placental syncytiotrophoblast development and microbial resistance. Sci Adv 2:e1501462. 10.1126/sciadv.1501462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Danielson JJ, Perez N, Romano JD, Coppens I. 2018. Modelling Toxoplasma gondii infection in a 3D cell culture system in vitro: comparison with infection in 2D cell monolayers. PLoS One 13:e0208558. 10.1371/journal.pone.0208558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Derricott H, Luu L, Fong WY, Hartley CS, Johnston LJ, Armstrong SD, Randle N, Duckworth CA, Campbell BJ, Wastling JM, Coombes JL. 2019. Developing a 3D intestinal epithelium model for livestock species. Cell Tissue Res 375:409–424. 10.1007/s00441-018-2924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luu L, Johnston LJ, Derricott H, Armstrong SD, Randle N, Hartley CS, Duckworth CA, Campbell BJ, Wastling JM, Coombes JL. 2019. An open-format enteroid culture system for interrogation of interactions between Toxoplasma gondii and the intestinal epithelium. Front Cell Infect Microbiol 9:300. 10.3389/fcimb.2019.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Múnera JO, Sundaram N, Rankin SA, Hill D, Watson C, Mahe M, Vallance JE, Shroyer NF, Sinagoga KL, Zarzoso-Lacoste A, Hudson JR, Howell JC, Chatuvedi P, Spence JR, Shannon JM, Zorn AM, Helmrath MA, Wells JM. 2017. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell 21:51–64.e6. 10.1016/j.stem.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zou WY, Blutt SE, Crawford SE, Ettayebi K, Zeng X-L, Saxena K, Ramani S, Karandikar UC, Zachos NC, Estes MK. 2019. Human intestinal enteroids: new models to study gastrointestinal virus infections. Methods Mol Biol 1576:229–247. 10.1007/7651_2017_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martorelli Di Genova B, Wilson SK, Dubey JP, Knoll LJ. 2019. Intestinal delta-6-desaturase activity determines host range for Toxoplasma sexual reproduction. PLoS Biol 17:e3000364. 10.1371/journal.pbio.3000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lancaster MA, Renner M, Martin C, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501:373–379. 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Logan S, Arzua T, Yan Y, Jiang C, Liu X, Yu L, Liu Q, Bai X. 2020. Dynamic characterization of structural, molecular, and electrophysiological phenotypes of human-induced pluripotent stem cell-derived cerebral organoids, and comparison with fetal and adult gene profiles. Cells 9:1301. 10.3390/cells9051301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. 2011. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472:51–56. 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 80.Völkner M, Zschätzsch M, Rostovskaya M, Overall RW, Busskamp V, Anastassiadis K, Karl MO. 2016. Retinal organoids from pluripotent stem cells efficiently recapitulate retinogenesis. Stem Cell Rep 6:525–538. 10.1016/j.stemcr.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y. 2012. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10:771–785. 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 82.Capowski EE, Samimi K, Mayerl SJ, Phillips MJ, Pinilla I, Howden SE, Saha J, Jansen AD, Edwards KL, Jager LD, Barlow K, Valiauga R, Erlichman Z, Hagstrom A, Sinha D, Sluch VM, Chamling X, Zack DJ, Skala MC, Gamm DM. 2019. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development 146:dev171686. 10.1242/dev.171686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. 2016. Zika virus impairs growth in human neurospheres and brain organoids. Science 352:816–818. 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 84.Seo H-H, Han H-W, Lee S-E, Hong S-H, Cho S-H, Kim SC, Koo SK, Kim J-H. 2020. Modelling Toxoplasma gondii infection in human cerebral organoids. Emerg Microbes Infect 9:1943–1954. 10.1080/22221751.2020.1812435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramírez-Flores CJ, Cruz-Mirón R, Arroyo R, Mondragón-Castelán ME, Nopal-Guerrero T, González-Pozos S, Ríos-Castro E, Mondragón-Flores R. 2019. Characterization of metalloproteases and serine proteases of Toxoplasma gondii tachyzoites and their effect on epithelial cells. Parasitol Res 118:289–306. 10.1007/s00436-018-6163-5. [DOI] [PubMed] [Google Scholar]
- 86.Ramírez-Flores CJ, Cruz-Mirón R, Lagunas-Cortés N, Mondragón-Castelán M, Mondragon-Gonzalez R, González-Pozos S, Mondragón-Flores R. 2021. Toxoplasma gondii excreted/secreted proteases disrupt intercellular junction proteins in epithelial cell monolayers to facilitate tachyzoites paracellular migration. Cell Microbiol 23:e13283. 10.1111/cmi.13283. [DOI] [PubMed] [Google Scholar]
- 87.VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, Tarr PI, Ciorba MA, Stappenbeck TS. 2015. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64:911–920. 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moon C, VanDussen KL, Miyoshi H, Stappenbeck TS. 2014. Development of a primary mouse intestinal epithelial cell monolayer culture system to evaluate factors that modulate IgA transcytosis. Mucosal Immunol 7:818–828. 10.1038/mi.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Holthaus D, Delgado-Betancourt E, Aebischer T, Seeber F, Klotz C. 2020. Harmonization of protocols for multi-species organoid platforms to study the intestinal biology of Toxoplasma gondii and other protozoan infections. Front Cell Infect Microbiol 10:610368. 10.3389/fcimb.2020.610368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nogueira AR, Leve F, Morgado-Diaz J, Tedesco RC, Pereira MCS. 2016. Effect of Toxoplasma gondii infection on the junctional complex of retinal pigment epithelial cells. Parasitology 143:568–575. 10.1017/S0031182015001973. [DOI] [PubMed] [Google Scholar]
- 91.Kapałczyńska M, Kolenda T, Przybyła W, Zajączkowska M, Teresiak A, Filas V, Ibbs M, Bliźniak R, Łuczewski Ł, Lamperska K. 2018. 2D and 3D cell cultures—a comparison of different types of cancer cell cultures. Arch Med Sci 14:910–919. 10.5114/aoms.2016.63743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Corrò C, Novellasdemunt L, Li VSW. 2020. A brief history of organoids. Am J Physiol Cell Physiol 319:C151–C165. 10.1152/ajpcell.00120.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garreta E, Kamm RD, Chuva de Sousa Lopes SM, Lancaster MA, Weiss R, Trepat X, Hyun I, Montserrat N. 2021. Rethinking organoid technology through bioengineering. Nat Mater 20:145–155. 10.1038/s41563-020-00804-4. [DOI] [PubMed] [Google Scholar]
- 94.Velasco V, Shariati SA, Esfandyarpour R. 2020. Microtechnology-based methods for organoid models. Microsystems Nanoeng 6:76. 10.1038/s41378-020-00185-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Naing C, Whittaker MA, Nyunt Wai V, Mak JW. 2014. Is Plasmodium vivax malaria a severe malaria?: a systematic review and meta-analysis. PLoS Negl Trop Dis 8:e3071. 10.1371/journal.pntd.0003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hirako IC, Ataide MA, Faustino L, Assis PA, Sorensen EW, Ueta H, Araújo NM, Menezes GB, Luster AD, Gazzinelli RT. 2016. Splenic differentiation and emergence of CCR5+ CXCL9+ CXCL10+ monocyte-derived dendritic cells in the brain during cerebral malaria. Nat Commun 7:13277. 10.1038/ncomms13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Howes RE, Battle KE, Mendis KN, Smith DL, Cibulskis RE, Baird JK, Hay SI. 2016. Global epidemiology of Plasmodium vivax. Am J Trop Med Hyg 95:15–34. 10.4269/ajtmh.16-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dayananda KK, Achur RN, Gowda DC. 2018. Epidemiology, drug resistance, and pathophysiology of Plasmodium vivax malaria. J Vector Borne Dis 55:1–8. 10.4103/0972-9062.234620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Battle KE, Karhunen MS, Bhatt S, Gething PW, Howes RE, Golding N, Van Boeckel TP, Messina JP, Shanks GD, Smith DL, Baird JK, Hay SI. 2014. Geographical variation in Plasmodium vivax relapse. Malar J 13:144. 10.1186/1475-2875-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hemingway J, Shretta R, Wells TNC, Bell D, Djimdé AA, Achee N, Qi G. 2016. Tools and strategies for malaria control and elimination: what do we need to achieve a grand convergence in malaria? PLoS Biol 14:e1002380. 10.1371/journal.pbio.1002380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dhiman S. 2019. Correction to: Are malaria elimination efforts on right track? An analysis of gains achieved and challenges ahead. Infect Dis Poverty 8:19. 10.1186/s40249-019-0527-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dabaro D, Birhanu Z, Yewhalaw D. 2020. Analysis of trends of malaria from 2010 to 2017 in Boricha District, Southern Ethiopia. Malar J 19:88. 10.1186/s12936-020-03169-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Manel Yapabandara AMG, do Rosario de Fatima Mota M, Sarmento R, Bosco JD, Wickremasinghe R. 2020. From malaria control to elimination within a decade: lessons learned from Timor Leste, a newly independent country. Malar J 19:104. 10.1186/s12936-020-03162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pava Z, Puspitasari AM, Rumaseb A, Handayuni I, Trianty L, Utami RAS, Tirta YK, Burdam F, Kenangalem E, Wirjanata G, Kho S, Trimarsanto H, Anstey NM, Poespoprodjo JR, Noviyanti R, Price RN, Marfurt J, Auburn S. 2020. Molecular surveillance over 14 years confirms reduction of Plasmodium vivax and falciparum transmission after implementation of artemisinin-based combination therapy in Papua, Indonesia. PLoS Negl Trop Dis 14:e0008295. 10.1371/journal.pntd.0008295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vaughan AM, Aly ASI, Kappe SHI. 2008. Malaria parasite pre-erythrocytic stage infection: gliding and hiding. Cell Host Microbe 4:209–218. 10.1016/j.chom.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Molina-Franky J, Cuy-Chaparro L, Camargo A, Reyes C, Gómez M, Salamanca DR, Patarroyo MA, Patarroyo ME. 2020. Plasmodium falciparum pre-erythrocytic stage vaccine development. Malar J 19:56. 10.1186/s12936-020-3141-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Déchamps S, Maynadier M, Wein S, Gannoun-Zaki L, Maréchal E, Vial HJ. 2010. Rodent and nonrodent malaria parasites differ in their phospholipid metabolic pathways. J Lipid Res 51:81–96. 10.1194/jlr.M900166-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Venugopal K, Hentzschel F, Valkiūnas G, Marti M. 2020. Plasmodium asexual growth and sexual development in the haematopoietic niche of the host. Nat Rev Microbiol 18:177–189. 10.1038/s41579-019-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pewkliang Y, Rungin S, Lerdpanyangam K, Duangmanee A, Kanjanasirirat P, Suthivanich P, Sa-Ngiamsuntorn K, Borwornpinyo S, Sattabongkot J, Patrapuvich R, Hongeng S. 2018. A novel immortalized hepatocyte-like cell line (imHC) supports in vitro liver stage development of the human malarial parasite Plasmodium vivax. Malar J 17:50. 10.1186/s12936-018-2198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tweedell RE, Tao D, Hamerly T, Robinson TM, Larsen S, Grønning AGB, Norris AM, King JG, Law HCH, Baumbach J, Bergmann-Leitner ES, Dinglasan RR. 2019. The selection of a hepatocyte cell line susceptible to Plasmodium falciparum sporozoite invasion that is associated with expression of glypican-3. Front Microbiol 10:127. 10.3389/fmicb.2019.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Frischknecht F, Matuschewski K. 2017. Plasmodium sporozoite biology. Cold Spring Harb Perspect Med 7:a025478. 10.1101/cshperspect.a025478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Frischknecht F, Baldacci P, Martin B, Zimmer C, Thiberge S, Olivo-Marin JC, Shorte SL, Ménard R. 2004. Imaging movement of malaria parasites during transmission by Anopheles mosquitoes. Cell Microbiol 6:687–694. 10.1111/j.1462-5822.2004.00395.x. [DOI] [PubMed] [Google Scholar]
- 113.Münter S, Sabass B, Selhuber-Unkel C, Kudryashev M, Hegge S, Engel U, Spatz JP, Matuschewski K, Schwarz US, Frischknecht F. 2009. Plasmodium sporozoite motility is modulated by the turnover of discrete adhesion sites. Cell Host Microbe 6:551–562. 10.1016/j.chom.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 114.Hellmann JK, Münter S, Kudryashev M, Schulz S, Heiss K, Müller AK, Matuschewski K, Spatz JP, Schwarz US, Frischknecht F. 2011. Environmental constraints guide migration of malaria parasites during transmission. PLoS Pathog 7:e1002080. 10.1371/journal.ppat.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Muthinja MJ, Ripp J, Hellmann JK, Haraszti T, Dahan N, Lemgruber L, Battista A, Schütz L, Fackler OT, Schwarz US, Spatz JP, Frischknecht F. 2017. Microstructured blood vessel surrogates reveal structural tropism of motile malaria parasites. Adv Healthc Mater 6:201601178. 10.1002/adhm.201601178. [DOI] [PubMed] [Google Scholar]
- 116.Ripp J, Kehrer J, Smyrnakou X, Tisch N, Tavares J, Amino R, Ruiz de Almodovar C, Frischknecht F. 2021. Malaria parasites differentially sense environmental elasticity during transmission. EMBO Mol Med 13:e13933. 10.15252/emmm.202113933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carnevale A. 1986. [Contributions of a genetics service to the study of patients in a pediatric hospital]. Gac Med Mex 122:149–156. (In Spanish.) [PubMed] [Google Scholar]
- 118.Arévalo-Pinzón G, Garzón-Ospina D, Pulido FA, Bermúdez M, Forero-Rodríguez J, Rodríguez-Mesa XM, Reyes-Guarín LP, Suárez CF, Patarroyo MA. 2020. Plasmodium vivax cell traversal protein for ookinetes and sporozoites (CelTOS) functionally restricted regions are involved in specific host-pathogen interactions. Front Cell Infect Microbiol 10:119. 10.3389/fcimb.2020.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arévalo-Pinzón G, Curtidor H, Muñoz M, Patarroyo MA, Patarroyo ME. 2011. Synthetic peptides from two Pf sporozoite invasion-associated proteins specifically interact with HeLa and HepG2 cells. Peptides 32:1902–1908. 10.1016/j.peptides.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 120.Niklaus L, Agop-Nersesian C, Schmuckli-Maurer J, Wacker R, Grünig V, Heussler VT. 2019. Deciphering host lysosome-mediated elimination of Plasmodium berghei liver stage parasites. Sci Rep 9:7967. 10.1038/s41598-019-44449-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Salazar Alvarez LC, Vera Lizcano O, da Silva Barros DKA, Baia-da-Silva DC, Monteiro WM, Pimenta PFP, de Lacerda MVG, Costa FTM, Lopes SCP. 2021. Plasmodium vivax gametocytes adherence to bone marrow endothelial cells. Front Cell Infect Microbiol 11:614985. 10.3389/fcimb.2021.614985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wilkening S, Stahl F, Bader A. 2003. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos 31:1035–1042. 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]
- 123.Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MMA, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, Sasaki N, Boers SJ, Kemperman H, de Jonge J, Ijzermans JNM, Nieuwenhuis EES, Hoekstra R, Strom S, Vries RRG, van der Laan LJW, Cuppen E, Clevers H. 2015. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160:299–312. 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hu H, Gehart H, Artegiani B, LÖpez-Iglesias C, Dekkers F, Basak O, van Es J, Chuva de Sousa Lopes SM, Begthel H, Korving J, van den Born M, Zou C, Quirk C, Chiriboga L, Rice CM, Ma S, Rios A, Peters PJ, de Jong YP, Clevers H. 2018. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175:1591–1606.e19. 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 125.Arez F, Rebelo SP, Fontinha D, Simão D, Martins TR, Machado M, Fischli C, Oeuvray C, Badolo L, Carrondo MJT, Rottmann M, Spangenberg T, Brito C, Greco B, Prudêncio M, Alves PM. 2019. Flexible 3D cell-based platforms for the discovery and profiling of novel drugs targeting plasmodium hepatic infection. ACS Infect Dis 5:1831–1842. 10.1021/acsinfecdis.9b00144. [DOI] [PubMed] [Google Scholar]
- 126.Harbuzariu A, Pitts S, Cespedes JC, Harp KO, Nti A, Shaw AP, Liu M, Stiles JK. 2019. Modelling heme-mediated brain injury associated with cerebral malaria in human brain cortical organoids. Sci Rep 9:19162. 10.1038/s41598-019-55631-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tripathi AK, Sullivan DJ, Stins MF. 2007. Plasmodium falciparum-infected erythrocytes decrease the integrity of human blood‐brain barrier endothelial cell monolayers. J Infect Dis 195:942–950. 10.1086/512083. [DOI] [PubMed] [Google Scholar]
- 128.Dalko E, Das B, Herbert F, Fesel C, Pathak S, Tripathy R, Cazenave P-A, Ravindran B, Sharma S, Pied S. 2015. Multifaceted role of heme during severe Plasmodium falciparum infections in India. Infect Immun 83:3793–3799. 10.1128/IAI.00531-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu M, Dickinson-Copeland C, Stiles J, Hassana S. 2016. Plasmodium-infected erythrocytes (pRBC) induce endothelial cell apoptosis via a heme-mediated signaling pathway. Drug Des Devel Ther 10:1009-18. 10.2147/DDDT.S96863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nishanth G, Schlüter D. 2019. Blood-brain barrier in cerebral malaria: pathogenesis and therapeutic intervention. Trends Parasitol 35:516–528. 10.1016/j.pt.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 131.Joice R, Nilsson SK, Montgomery J, Dankwa S, Egan E, Morahan B, Seydel KB, Bertuccini L, Alano P, Williamson KC, Duraisingh MT, Taylor TE, Milner DA, Marti M. 2014. Plasmodium falciparum transmission stages accumulate in the human bone marrow. Sci Transl Med 6:244re5. 10.1126/scitranslmed.3008882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Obaldia N, Meibalan E, Sa JM, Ma S, Clark MA, Mejia P, Moraes Barros RR, Otero W, Ferreira MU, Mitchell JR, Milner DA, Huttenhower C, Wirth DF, Duraisingh MT, Wellems TE, Marti M. 2018. Bone marrow is a major parasite reservoir in Plasmodium vivax infection. mBio 9:e00625-18. 10.1128/mBio.00625-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gardiner DL, Trenholme KR. 2015. Plasmodium falciparum gametocytes: playing hide and seek. Ann Transl Med 3:45. 10.3978/j.issn.2305-5839.2015.01.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ramdani G, Naissant B, Thompson E, Breil F, Lorthiois A, Dupuy F, Cummings R, Duffier Y, Corbett Y, Mercereau-Puijalon O, Vernick K, Taramelli D, Baker DA, Langsley G, Lavazec C. 2015. cAMP-signalling regulates gametocyte-infected erythrocyte deformability required for malaria parasite transmission. PLoS Pathog 11:e1004815. 10.1371/journal.ppat.1004815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shahabuddin M. 1998. Plasmodium ookinete development in the mosquito midgut: a case of reciprocal manipulation. Parasitology 116(Suppl):S83–S93. 10.1017/s0031182000084973. [DOI] [PubMed] [Google Scholar]
- 136.Moon RW, Taylor CJ, Bex C, Schepers R, Goulding D, Janse CJ, Waters AP, Baker DA, Billker O. 2009. A cyclic GMP signalling module that regulates gliding motility in a malaria parasite. PLoS Pathog 5:e1000599. 10.1371/journal.ppat.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kan A, Tan Y-H, Angrisano F, Hanssen E, Rogers KL, Whitehead L, Mollard VP, Cozijnsen A, Delves MJ, Crawford S, Sinden RE, McFadden GI, Leckie C, Bailey J, Baum J. 2014. Quantitative analysis of Plasmodium ookinete motion in three dimensions suggests a critical role for cell shape in the biomechanics of malaria parasite gliding motility. Cell Microbiol 16:734–750. 10.1111/cmi.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vlachou D, Zimmermann T, Cantera R, Janse CJ, Waters AP, Kafatos FC. 2004. Real-time, in vivo analysis of malaria ookinete locomotion and mosquito midgut invasion. Cell Microbiol 6:671–685. 10.1111/j.1462-5822.2004.00394.x. [DOI] [PubMed] [Google Scholar]
- 139.Kulkeaw K. 2021. Next-generation human liver models for antimalarial drug assays. Antibiot (Basel) 10:642. 10.3390/antibiotics10060642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.malERA Refresh Consultative Panel on Basic Science and Enabling Technologies. 2017. malERA: an updated research agenda for basic science and enabling technologies in malaria elimination and eradication. PLoS Med 14:e1002451. 10.1371/journal.pmed.1002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Swevers L, Denecke S, Vogelsang K, Geibel S, Vontas J. 2021. Can the mammalian organoid technology be applied to the insect gut? Pest Manag Sci 77:55–63. 10.1002/ps.6067. [DOI] [PubMed] [Google Scholar]
- 142.Doupé DP, Marshall OJ, Dayton H, Brand AH, Perrimon N. 2018. Drosophila intestinal stem and progenitor cells are major sources and regulators of homeostatic niche signals. Proc Natl Acad Sci USA 115:12218–12223. 10.1073/pnas.1719169115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tanaka J, Ogawa M, Hojo H, Kawashima Y, Mabuchi Y, Hata K, Nakamura S, Yasuhara R, Takamatsu K, Irié T, Fukada T, Sakai T, Inoue T, Nishimura R, Ohara O, Saito I, Ohba S, Tsuji T, Mishima K. 2018. Generation of orthotopically functional salivary gland from embryonic stem cells. Nat Commun 9:4216. 10.1038/s41467-018-06469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Maimets M, Rocchi C, Bron R, Pringle S, Kuipers J, Giepmans BNG, Vries RGJ, Clevers H, de Haan G, van Os R, Coppes RP. 2016. Long-term in vitro expansion of salivary gland stem cells driven by Wnt signals. Stem Cell Rep 6:150–162. 10.1016/j.stemcr.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pringle S, Maimets M, van der Zwaag M, Stokman MA, van Gosliga D, Zwart E, Witjes MJH, de Haan G, van Os R, Coppes RP. 2016. Human salivary gland stem cells functionally restore radiation damaged salivary glands. Stem Cells 34:640–652. 10.1002/stem.2278. [DOI] [PubMed] [Google Scholar]
- 146.Zhu W, Ma X, Gou M, Mei D, Zhang K, Chen S. 2016. 3D printing of functional biomaterials for tissue engineering. Curr Opin Biotechnol 40:103–112. 10.1016/j.copbio.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 147.Jamieson C, Keenan P, Kirkwood D, Oji S, Webster C, Russell KA, Koch TG. 2020. A review of recent advances in 3D bioprinting with an eye on future regenerative therapies in veterinary medicine. Front Vet Sci 7:584193. 10.3389/fvets.2020.584193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liaw C-Y, Guvendiren M. 2017. Current and emerging applications of 3D printing in medicine. Biofabrication 9:024102. 10.1088/1758-5090/aa7279. [DOI] [PubMed] [Google Scholar]
- 149.Afsana Jain V, Haider N, Jain K. 2018. 3D printing in personalized drug delivery. Curr Pharm Des 24:5062–5071. 10.2174/1381612825666190215122208. [DOI] [PubMed] [Google Scholar]
- 150.Quiroz-Castañeda RE, Dantán-González E. 2015. Control of avian coccidiosis: future and present natural alternatives. Biomed Res Int 2015:430610. 10.1155/2015/430610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shirley MW, Smith AL, Tomley FM. 2005. The biology of avian Eimeria with an emphasis on their control by vaccination. Adv Parasitol 60:285–330. 10.1016/S0065-308X(05)60005-X. [DOI] [PubMed] [Google Scholar]
- 152.Greif G, Harder A, Haberkorn A. 2001. Chemotherapeutic approaches to protozoa: Coccidiae—current level of knowledge and outlook. Parasitol Res 87:973–975. 10.1007/s004360100403. [DOI] [PubMed] [Google Scholar]
- 153.Wunderlich F, Al-Quraishy S, Steinbrenner H, Sies H, Dkhil MA. 2014. Towards identifying novel anti-Eimeria agents: trace elements, vitamins, and plant-based natural products. Parasitol Res 113:3547–3556. 10.1007/s00436-014-4101-8. [DOI] [PubMed] [Google Scholar]
- 154.Jeffers TK. 1975. Attenuation of Eimeria tenella through selection for precociousness. J Parasitol 61:1083–1090. 10.2307/3279381. [DOI] [PubMed] [Google Scholar]
- 155.Crouch CF, Andrews SJ, Ward R, Francis MJ. 2003. Protective efficacy of a live attenuated anticoccidial vaccine administered to 1-day-old chickens. Avian Pathol 32:297–304. 10.1080/10307945031000097912. [DOI] [PubMed] [Google Scholar]
- 156.McDonald V, Shirley MW. 2009. Past and future: vaccination against Eimeria. Parasitology 136:1477–1489. 10.1017/S0031182009006349. [DOI] [PubMed] [Google Scholar]
- 157.Rathinam T, Gadde U, Chapman HD. 2016. Attenuation of a drug-sensitive strain of a turkey protozoan parasite Eimeria meleagrimitis by selection for precocious development. Vet Parasitol 216:1–3. 10.1016/j.vetpar.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 158.Strout RG, Solis J, Smith SC, Dunlop WR. 1965. In vitro cultivation of Eimeria acervulina (Coccidia). Exp Parasitol 17:241–246. 10.1016/0014-4894(65)90064-0. [DOI] [PubMed] [Google Scholar]
- 159.Müller BE. 1975. Ultrastructural development of first- to second-generation merozoites in Eimeria contorta Haberkorn, 1971. Z Parasitenkd 47:91–101. 10.1007/BF00382632. [DOI] [PubMed] [Google Scholar]
- 160.Müller BE. 1975. In vitro development from sporozoites to first-generation merozoites in Eimeria contorta Haberkorn, 1971. A fine structural study. Z Parasitenkd 47:23–34. 10.1007/BF00418062. [DOI] [PubMed] [Google Scholar]
- 161.Hofmann J, Raether W. 1990. Improved techniques for the in vitro cultivation of Eimeria tenella in primary chick kidney cells. Parasitol Res 76:479–486. 10.1007/BF00931053. [DOI] [PubMed] [Google Scholar]
- 162.Hermosilla C, Barbisch B, Heise A, Kowalik S, Zahner H. 2002. Development of Eimeria bovis in vitro: suitability of several bovine, human and porcine endothelial cell lines, bovine fetal gastrointestinal, Madin–Darby bovine kidney (MDBK) and African green monkey kidney (VERO) cells. Parasitol Res 88:301–307. 10.1007/s00436-001-0531-1. [DOI] [PubMed] [Google Scholar]
- 163.Hermosilla C, Stamm I, Menge C, Taubert A. 2015. Suitable in vitro culture of Eimeria bovis meront II stages in bovine colonic epithelial cells and parasite-induced upregulation of CXCL10 and GM-CSF gene transcription. Parasitol Res 114:3125–3136. 10.1007/s00436-015-4531-y. [DOI] [PubMed] [Google Scholar]
- 164.López-Osorio S, Silva LMR, Taubert A, Chaparro-Gutiérrez JJ, Hermosilla CR. 2018. Concomitant in vitro development of Eimeria zuernii- and Eimeria bovis-macromeronts in primary host endothelial cells. Parasitol Int 67:742–750. 10.1016/j.parint.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 165.Ruiz A, Behrendt JH, Zahner H, Hermosilla C, Pérez D, Matos L, del Carmen Muñoz M, Molina JM, Taubert A. 2010. Development of Eimeria ninakohlyakimovae in vitro in primary and permanent cell lines. Vet Parasitol 173:2–10. 10.1016/j.vetpar.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 166.Carrau T, Silva LMR, Pérez D, Ruiz de Ybáñez R, Taubert A, Hermosilla C. 2016. First description of an in vitro culture system for Eimeria ovinoidalis macromeront formation in primary host endothelial cells. Parasitol Int 65:516–519. 10.1016/j.parint.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 167.Chen H, Wiedmer S, Hanig S, Entzeroth R, Kurth M. 2013. Development of Eimeria nieschulzi (Coccidia, Apicomplexa) gamonts and oocysts in primary fetal rat cells. J Parasitol Res 2013:591520. 10.1155/2013/591520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Fernandes De Moura Guedes JP. 2018. Host-pathogen interactions at the intestinal epithelial barrier. University of Cambridge, Cambridge, UK. [Google Scholar]
- 169.Arendt MK. 2019. Role of intestinal interleukin-10 in Eimeria infection of chickens. University of Wisconsin, Madison, WI. [Google Scholar]
- 170.Nash TJ, Morris KM, Mabbott NA, Vervelde L. 2021. Inside-out chicken enteroids with leukocyte component as a model to study host-pathogen interactions. Commun Biol 4:377. 10.1038/s42003-021-01901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang Z, He X, Qiao H, Chen P. 2020. Global trends of organoid and organ-on-a-chip in the past decade: a bibliometric and comparative study. Tissue Eng Part A 26:656–671. 10.1089/ten.TEA.2019.0251. [DOI] [PubMed] [Google Scholar]
- 172.Wu Q, Liu J, Wang X, Feng L, Wu J, Zhu X, Wen W, Gong X. 2020. Organ-on-a-chip: recent breakthroughs and future prospects. Biomed Eng Online 19:9. 10.1186/s12938-020-0752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Reuter C, Walles H, Groeber F. 2017. Preparation of a three-dimensional full thickness skin equivalent. Methods Mol Biol 1612:191–198. 10.1007/978-1-4939-7021-6_14. [DOI] [PubMed] [Google Scholar]
- 174.Kühbacher A, Henkel H, Stevens P, Grumaz C, Finkelmeier D, Burger-Kentischer A, Sohn K, Rupp S. 2017. Central role for dermal fibroblasts in skin model protection against Candida albicans. J Infect Dis 215:1742–1752. 10.1093/infdis/jix153. [DOI] [PubMed] [Google Scholar]
- 175.Reddersen K, Wiegand C, Elsner P, Hipler UC. 2019. Three-dimensional human skin model infected with Staphylococcus aureus as a tool for evaluation of bioactivity and biocompatibility of antiseptics. Int J Antimicrob Agents 54:283–291. 10.1016/j.ijantimicag.2019.06.022. [DOI] [PubMed] [Google Scholar]
- 176.Jannasch M, Groeber F, Brattig NW, Unger C, Walles H, Hansmann J. 2015. Development and application of three-dimensional skin equivalents for the investigation of percutaneous worm invasion. Exp Parasitol 150:22–30. 10.1016/j.exppara.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 177.Reuter C, Imdahl F, Hauf L, Vafadarnejad E, Fey P, Finger T, Walles H, Saliba A-E, Groeber-Becker F, Engstler M. 2021. Vector-borne Trypanosoma brucei parasites develop in artificial human skin and persist as skin tissue forms. bioRxiv 10.1101/2021.05.13.443986. [DOI] [PMC free article] [PubMed]
- 178.Kane BJ, Zinner MJ, Yarmush ML, Toner M. 2006. Liver-specific functional studies in a microfluidic array of primary mammalian hepatocytes. Anal Chem 78:4291–4298. 10.1021/ac051856v. [DOI] [PubMed] [Google Scholar]
- 179.Peel S, Corrigan AMM, Ehrhardt B, Jang K-J, Caetano-Pinto P, Boeckeler M, Rubins JEE, Kodella K, Petropolis DBB, Ronxhi J, Kulkarni G, Foster AJJ, Williams D, Hamilton GAA, Ewart L. 2019. Introducing an automated high content confocal imaging approach for organs-on-chips. Lab Chip 19:410–421. 10.1039/C8LC00829A. [DOI] [PubMed] [Google Scholar]
- 180.Khetani SR, Bhatia SN. 2008. Microscale culture of human liver cells for drug development. Nat Biotechnol 26:120–126. 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 181.March S, Ng S, Velmurugan S, Galstian A, Shan J, Logan DJ, Carpenter AE, Thomas D, Sim BKL, Mota MM, Hoffman SL, Bhatia SN. 2013. A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax. Cell Host Microbe 14:104–115. 10.1016/j.chom.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mancio-Silva L, Fleming HE, Miller AB, Milstein S, Liebow A, Haslett P, Sepp-Lorenzino L, Bhatia SN. 2019. Improving drug discovery by nucleic acid delivery in engineered human microlivers. Cell Metab 29:727–735.e3. 10.1016/j.cmet.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rigat-Brugarolas LG, Elizalde-Torrent A, Bernabeu M, De Niz M, Martin-Jaular L, Fernandez-Becerra C, Homs-Corbera A, Samitier J, del Portillo HA. 2014. A functional microengineered model of the human splenon-on-a-chip. Lab Chip 14:1715–1724. 10.1039/c3lc51449h. [DOI] [PubMed] [Google Scholar]
- 184.Huh D, Kim HJ, Fraser JP, Shea DE, Khan M, Bahinski A, Hamilton GA, Ingber DE. 2013. Microfabrication of human organs-on-chips. Nat Protoc 8:2135–2157. 10.1038/nprot.2013.137. [DOI] [PubMed] [Google Scholar]
- 185.Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, Scholl W, Zhang C, Rickner H, Richmond CA, Li H, Breault DT, Ingber DE. 2018. Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci Rep 8:2871. 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shin YC, Shin W, Koh D, Wu A, Ambrosini YM, Min S, Eckhardt SG, Fleming RYD, Kim S, Park S, Koh H, Yoo TK, Kim HJ. 2020. Three-dimensional regeneration of patient-derived intestinal organoid epithelium in a physiodynamic mucosal interface-on-a-chip. Micromachines 11:663. 10.3390/mi11070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cherne MD, Sidar B, Sebrell TA, Sanchez HS, Heaton K, Kassama FJ, Roe MM, Gentry AB, Chang CB, Walk ST, Jutila M, Wilking JN, Bimczok D. 2021. A synthetic hydrogel, VitroGel ORGANOID-3, improves immune cell-epithelial interactions in a tissue chip co-culture model of human gastric organoids and dendritic cells. Front Pharmacol 12:707891. 10.3389/fphar.2021.707891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sidar B, Jenkins BR, Huang S, Spence JR, Walk ST, Wilking JN. 2019. Long-term flow through human intestinal organoids with the gut organoid flow chip (GOFlowChip). Lab Chip 19:3552–3562. 10.1039/C9LC00653B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Beaurivage C, Kanapeckaite A, Loomans C, Erdmann KS, Stallen J, Janssen RAJ. 2020. Development of a human primary gut-on-a-chip to model inflammatory processes. Sci Rep 10:21475. 10.1038/s41598-020-78359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gijzen L, Marescotti D, Raineri E, Nicolas A, Lanz HL, Guerrera D, van Vught R, Joore J, Vulto P, Peitsch MC, Hoeng J, Lo Sasso G, Kurek D. 2020. An Intestine-on-a-chip model of plug-and-play modularity to study inflammatory processes. SLAS Technol 25:585–597. 10.1177/2472630320924999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kasendra M, Luc R, Yin J, Manatakis DV, Kulkarni G, Lucchesi C, Sliz J, Apostolou A, Sunuwar L, Obrigewitch J, Jang K-J, Hamilton GA, Donowitz M, Karalis K. 2020. Duodenum Intestine-Chip for preclinical drug assessment in a human relevant model. Elife 9:e50135. 10.7554/eLife.50135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Achberger K, Probst C, Haderspeck J, Bolz S, Rogal J, Chuchuy J, Nikolova M, Cora V, Antkowiak L, Haq W, Shen N, Schenke-Layland K, Ueffing M, Liebau S, Loskill P. 2019. Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform. Elife 8:e46188. 10.7554/eLife.46188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xue Y, Seiler MJ, Tang WC, Wang JY, Delgado J, McLelland BT, Nistor G, Keirstead HS, Browne AW. 2021. Retinal organoids on-a-chip: a micro-millifluidic bioreactor for long-term organoid maintenance. Lab Chip 21:3361–3377. 10.1039/d1lc00011j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Manafi N, Shokri F, Achberger K, Hirayama M, Mohammadi MH, Noorizadeh F, Hong J, Liebau S, Tsuji T, Quinn PMJ, Mashaghi A. 2021. Organoids and organ chips in ophthalmology. Ocul Surf 19:1–15. 10.1016/j.jtos.2020.11.004. [DOI] [PubMed] [Google Scholar]
- 195.Karzbrun E, Kshirsagar A, Cohen SR, Hanna JH, Reiner O. 2018. Human brain organoids on a chip reveal the physics of folding. Nat Phys 14:515–522. 10.1038/s41567-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang Y, Wang L, Zhu Y, Qin J. 2018. Human brain organoid-on-a-chip to model prenatal nicotine exposure. Lab Chip 18:851–860. 10.1039/c7lc01084b. [DOI] [PubMed] [Google Scholar]
- 197.Ndyabawe K, Cipriano M, Zhao W, Haidekker M, Yao K, Mao L, Kisaalita WS. 2021. Brain-on-a-chip device for modeling multiregional networks. ACS Biomater Sci Eng 7:350–359. 10.1021/acsbiomaterials.0c00895. [DOI] [PubMed] [Google Scholar]