Abstract
We report within-host evolution of antibiotic resistance to trimethoprim-sulfamethoxazole and azithromycin in a nontypeable Haemophilus influenzae strain from a patient with common variable immunodeficiency (CVID), who received repeated or prolonged treatment with these antibiotics for recurrent respiratory tract infections. Whole-genome sequencing of three longitudinally collected sputum isolates during the period April 2016 to January 2018 revealed persistence of a strain of sequence type 2386. Reduced susceptibility to trimethoprim-sulfamethoxazole in the first two isolates was associated with mutations in genes encoding dihydrofolate reductase (folA) and its promotor region, dihydropteroate synthase (folP), and thymidylate synthase (thyA), while subsequent substitution of a single amino acid in dihydropteroate synthase (G225A) rendered high-level resistance in the third isolate from 2018. Azithromycin co-resistance in this isolate was associated with amino acid substitutions in 50S ribosomal proteins L4 (W59R) and L22 (G91D), possibly aided by a substitution in AcrB (A604E) of the AcrAB efflux pump. All three isolates were resistant to aminopenicillins and cefotaxime due to TEM-1B beta-lactamase and identical alterations in penicillin-binding protein 3. Further resistance development to trimethoprim-sulfamethoxazole and azithromycin resulted in a multidrug-resistant phenotype. Evolution of multidrug resistance due to horizontal gene transfer and/or spontaneous mutations, along with selection of resistant subpopulations is a particular risk in CVID and other patients requiring repeated and prolonged antibiotic treatment or prophylaxis. Such challenging situations call for careful antibiotic stewardship together with supportive and supplementary treatment. We describe the clinical and microbiological course of events in this case report and address the challenges encountered.
Keywords: Haemophilus influenzae, CVID, multidrug resistance, case report, persistence
Introduction
Nontypeable Haemophilus influenzae (NTHi) frequently colonize the respiratory tract in patients with chronic lung disease or impaired immune system. Bacterial colonization is an independent risk factor for progression to respiratory tract infections, often requiring antibiotic treatment in such patients (Pulvirenti et al., 2018). Recurrent infections and prolonged exposure to antibiotics facilitate development of resistance by increasing the frequency of horizontal acquisition of resistance genes and the rate of adaptive chromosomal resistance mutations and selection of resistant subpopulations (Blazquez et al., 2012; Baquero et al., 2021). With the introduction of whole-genome sequencing (WGS), several examples of within-host resistance development following antibiotic exposure have been described (Didelot et al., 2016; Gatt and Margalit, 2021), but well-documented cases have not been reported in H. influenzae, to our best knowledge.
Common variable immunodeficiency (CVID) is the most prevalent symptomatic primary immune disorder and comprises a heterogenous group of clinical conditions characterized by low levels of circulating immunoglobulins and compromised production of specific antibodies, rendering CVID patients particularly vulnerable to respiratory tract infections (Bonilla et al., 2016). Recurrent lung infections impose a long-term risk of development of bronchiectasis and pulmonary sequelae, further fueling the disposition for bacterial infections and the need for antibiotic treatment (Janssen et al., 2021).
Immunoglobulin substitution therapy is the mainstay of CVID management but is not always sufficient to abate recurring infections. Long-term antimicrobial prophylaxis has been advocated as beneficial in selected patients, but clinical trials supporting this practice are scarce (Bonilla et al., 2016), and the risk of development and selection of resistant strains during treatment is insufficiently investigated. We describe a patient with CVID, where long-term treatment with trimethoprim-sulfamethoxazole and azithromycin led to evolution of resistance towards these antibiotics in an H. influenzae strain persisting in the respiratory tract during April 2016 - January 2018.
Case Report
A 48-year-old Norwegian male with a history of recurring sinopulmonary infections was admitted to Haukeland University Hospital (Bergen, Norway) in 2011 with pneumonia and concurrent agammaglobulinemia. Following a thorough diagnostic evaluation excluding lymphoid and bone marrow malignancy, chronic viral infections, protein loss and drug-induced adverse reactions, the patient was diagnosed with CVID. Immunoglobulin replacement therapy was initiated in 2012, and by the end of 2013 he achieved sustained IgG levels above 6 g/L. The first isolation of H. influenzae from a sputum sample was in November 2013, susceptible to beta-lactam antibiotics and trimethoprim-sulfamethoxazole (Hi-Alpha; Figure 1 ).
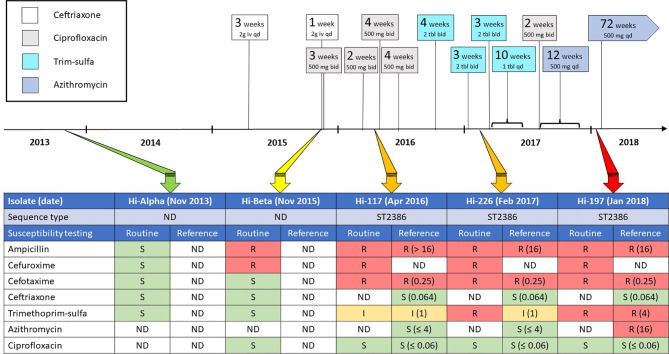
Figure 1.
Time scale annotated with exposure to antibiotics (including dosages), along with results from phenotypic antimicrobial susceptibility testing (AST) to relevant antibiotics for five H. influenzae isolates sampled between November 2013 and January 2018. The Hi-Alpha and Hi-Beta isolates were not available for genomic characterization, and their potential phylogenetic relationship to the Hi-117, Hi226 and Hi-197 isolates could not be explored. Routine AST results for the five isolates represent primary testing with disk diffusion and/or gradient diffusion, whereas reference AST results were produced retrospectively by determination of broth microdilution (BMD) MIC using custom panels. Disk diffusion and BMD were done according to the standards of the EUCAST, while gradient diffusion was performed according to the manufacturer’s recommendations. AST results were interpreted using EUCAST clinical breakpoints (v. 12.0), except azithromycin (no clinical breakpoints), for which susceptibility categorization was based on the epidemiological cut-off value (4 mg/L). bid, twice daily; qd, once daily; Trim-sulfa, trimethoprim-sulfamethoxazole; S, susceptible; I, intermediately susceptible (changed to “susceptible, increased exposure” from 2019); R, resistant; ND, no data.
Over the course of the next two years, the patient experienced few sinopulmonary infections, but in November 2015 he was admitted to hospital with pneumonia. Microbiological samples were procured by bronchoscopy and H. influenzae was identified as sole pathogen (Hi-Beta; Figure 1 ). This isolate displayed resistance to ampicillin and cefuroxime due to beta-lactamase production and mutations in penicillin-binding-protein 3 (PBP3), but was susceptible to cefotaxime, ceftriaxone, trimethoprim-sulfamethoxazole, and ciprofloxacin. The patient received ceftriaxone for five days, followed by three weeks of oral ciprofloxacin.
In March 2016, saline inhalation therapy was commenced by pulmonologists after bronchiectasis was detected on CT scan. H. influenzae was again isolated from sputum in April 2016 (Hi-117; Figure 1 ). Hi-117 expressed resistance to ampicillin, cefuroxime, and cefotaxime, and was categorized as intermediately susceptible to trimethoprim-sulfamethoxazole. Four weeks of ciprofloxacin was prescribed.
During the period 2016 to 2018 the patient had IgG trough levels above 8 g/L and received intensified inhalation therapy. Despite this, he had frequently recurring infections, requiring repeated and prolonged courses of antimicrobial therapy. Cultivation of sputum in February 2017 revealed H. influenzae (Hi-226) with a resistance pattern similar to Hi-117 from 2016 ( Figure 1 ).
Given the deteriorating clinical course and the short-lived improvement with each antibiotic course, a decision was made to attempt long-term antimicrobial prophylaxis. A ten-week course of low-dose trimethoprim-sulfamethoxazole 80 mg/400 mg daily was initiated in March 2017, followed by two weeks of ciprofloxacin 500 mg bid, immediately succeeded by twelve weeks of azithromycin 500 mg daily from September 2017. Rapid amelioration of respiratory symptoms was observed, and the patient returned to a full working status.
In January 2018, he experienced worsening respiratory symptoms, four weeks after terminating prophylactic azithromycin. H. influenzae cultivated from sputum (Hi-197, Figure 1 ), was resistant to trimethoprim-sulfamethoxazole. Prophylaxis with azithromycin, 500 mg once daily, was resumed on a permanent basis. Retrospective testing using broth microdilution (BMD) revealed increased MICs to both trimethoprim-sulfamethoxazole and azithromycin in Hi-197, compared to Hi-117 and Hi-226. However, as susceptibility to azithromycin was not tested routinely, this information was not available to influence the choice of antibiotic. Although clinical improvement was observed initially, the efficacy of azithromycin gradually declined during 2018.
By January 2019, his disease burden was like the state prior to initiation of prophylaxis. In September 2019, a bronchoalveolar lavage performed at another hospital revealed profuse growth of H. influenzae with a resistance profile identical to that of Hi-197. During 2013-2020, despite comprehensive conventional and molecular investigations, other respiratory pathogens were detected only twice; DNA of Mycoplasma pneumoniae in 2016 and influenza B virus RNA in 2018.
His condition rapidly deteriorated and he developed several CVID-complications, including granulomatous lymphocytic interstitial lung disease, liver cirrhosis, and ultimately a high-grade B-cell lymphoma. He died in July 2020 from a neutropenic sepsis caused by Escherichia coli and Staphylococcus aureus.
Phenotypic and Genotypic Characterization of H. influenzae
Figure 1 shows a time scale with antibiotic exposure, along with results from phenotypic antimicrobial susceptibility testing (AST) to relevant antibiotics for five H. influenzae isolates, identified as the sole pathogen from samples representative for lower respiratory tract, between November 2013 and January 2018. Routine AST results for these five isolates represent primary testing with disk diffusion (Oxoid/Thermo Fisher Scientific, Basingstoke, UK) and/or gradient diffusion (Liofilchem, Roseto degli Abruzzi, Italy), whereas AST for the sequenced strains was also determined by BMD using custom panels (Sensititre NONAG7, Thermo Fisher Scientific). Disk diffusion and BMD were done according to the standards of the European Committee on Antimicrobial Susceptibility Testing (EUCAST), while gradient diffusion was performed according to the manufacturer’s recommendations. AST results were interpreted using EUCAST clinical breakpoints (v. 12.0), except azithromycin, for which susceptibility categorization was based on the epidemiological cut-off value (4 mg/L).
Genetic relationship between Hi-117, Hi-226, and Hi-197 and their molecular basis for resistance development were assessed using WGS (Ion Torrent S5XL, Thermo Fisher Scientific). Trimmed sequencing reads (PHRED score ≥ 20) were analyzed with respect to conventional and core-genome multi-locus sequence typing, ((cg)MLST) with subsequent assignment to Minimum Spanning Tree (MST) clusters (Ridom SeqSphere+ v.8.0). Hi-117, Hi-226, and Hi-197 shared a novel MLST profile (ST2386) comprising a novel adk allele ( Table 1 ) and belonged to the same MST cluster, separated by 1-7 allele differences in 1589 genes. The results confirm that Hi-117, Hi-226, and Hi-197 represent a single strain, persisting during April 2016 to January 2018.
Table 1.
Molecular characteristics of Hi-117, Hi-226, and Hi-197.
| Parameter | Characteristics | |||
|---|---|---|---|---|
| Strain ID (accession) | Hi-117 (GCA_923276745) | Hi-226 (GCA_923282765) | Hi-197 (GCA_923283335) | |
| Sample type (date) | Sputum (April 2016) | Sputum (February 2017) | Sputum (January 2018) | |
| MLST a | ST2386 (254-11-18-18-62-1-5) | ST2386 (254-11-18-18-62-1-5) | ST2386 (254-11-18-18-62-1-5) | |
| cgMLST b | MST cluster 9 | MST cluster 9 | MST cluster 9 | |
| Capsular serotype c | Nontypeable | Nontypeable | Nontypeable | |
| Other virulence determinants d | hmw, hap, igaA1 | hmw, hap, igaA1 | hmw, hap, igaA1 | |
| Transferable resistance genes e | bla TEM-1B | bla TEM-1B | bla TEM-1B | |
| Chromosomal resistance f | ||||
| - Beta-lactams g | PBP3 (ftsI) | D350N, S357N, M377I, S385T, R517H, T532S, V547I | D350N, S357N, M377I, S385T, R517H, T532S, V547I | D350N, S357N, M377I, S385T, R517H, T532S, V547I |
| PBP3 group | High III-like(-) | High III-like(-) | High III-like(-) | |
| - Quinolones h | GyrA (gyrA) | – | – | – |
| ParC (parC) | – | – | – | |
| - Azithromycin i | L4 (rpl4) | – | – | W59R |
| L22 (rpl22) | – | – | G91D | |
| 23S rRNA | – | – | – | |
| - Trimethoprim-sulfamethoxazole j | DHFR (folA) | N13S, W31R, L67P, E69K, I74V, F79L, I95L, K107Q, E135K | N13S, W31R, L67P, E69K, I74V, F79L, I95L, K107Q, E135K | N13S, W31R, L67P, E69K, I74V, F79L, I95L, K107Q, E135K |
| DHFR (promoter) | A(-32)C, T(-24)C, G(-4)A | A(-32)C, T(-24)C, G(-4)A | A(-32)C, T(-24)C, G(-4)A | |
| DHPS (folP) | N87S, V95A, V101I, N108S, A150V, I177V, G189C, I210N, I236V, A240V, V268I, A273E | N87S, V95A, V101I, N108S, A150V, I177V, G189C, I210N, I236V, A240V, V268I, A273E | N87S, V95A, V101I, N108S, A150V, I177V, G189C, I210N, G225A, I236V, A240V, V268I, A273E | |
| TS (thyA) | H26R, V107I, E238K, T253S | H26R, V107I, E238K, T253S | H26R, V107I, E238K, T253S | |
| - Efflux k | AcrR (acrR) | S14L, R22K, N26D, Q27R, L31H, L33I, T77S, I121V, H131D, Q134K | S14L, R22K, N26D, Q27R, L31H, L33I, T77S, I121V, H131D, Q134K | S14L, R22K, N26D, Q27R, L31H, L33I, T77S, I121V, H131D, Q134K, S181F |
| AcrA (acrA) | M20I, G32E, M67L, A75T, V76I, V147L, S149N, A156V, D253N, V345A, D369G, I473V | M20I, G32E, M67L, A75T, V76I, V147L, S149N, A156V, D253N, V345A, D369G, I473V | M20I, G32E, M67L, A75T, V76I, V147L, S149N, A156V, D253N, V345A, D369G, I473V | |
| AcrB (acrB) | P660A, T828N, F837Y, A854T, V855T, A858I, I862V, H942Y, V1015I | P660A, T828N, F837Y, A854T, V855T, A858I, I862V, H942Y, V1015I | A604E, P660A, T828N, F837Y, A854T, V855T, A858I, I862V, H942Y, V1015I | |
Multi-locus sequence typing (MLST) with assignment to sequence types (ST) based on allelic profiles of seven housekeeping genes (adk, atpG, frdB, fucK, mdh, pgi, and recA). ST2386 is a single-locus variant (SLV) of ST836 with the novel allele adk-254 (Jolley et al., 2018).
Core genome MLST (cgMLST) with assignment to Minimum Spanning Tree (MST) cluster was performed with Ridom SeqSphere+ v. 8.0 (Münster, Germany) on a collection of 222 clinical isolates of H. influenzae from Norway or Sweden (BioProject PRJEB49398).
Capsular serotyping was performed with Hicap v.1.0.3 (Watts and Holt, 2019).
Virulence determinants were called using a locally installed version of MyDbFinder v.2.0 with a custom database comprising genes from the virulence factor database (VFDB) (Liu et al., 2022) and the following additional sequences, database downloaded 2021-08-24: hmwA1 (first 1269 bp) (NZ_LN831035.1), hap (U11024.1), hia (U38617.2), igaA2 (NDZN01000054.1), igaB1 (DQ423203), and igaB2 (KC607498). Thresholds of 60% were used for identity and coverage.
Transferable resistance genes were called with ResFinder v.4.1 (Bortolaia et al., 2020), using thresholds of 60% for identity and coverage. blaTEM-1B, 100% identity and coverage (AY458016).
Alterations in chromosomally encoded proteins, genes (in brackets) or promoter regions were called by multiple sequence alignment of translated coding genes using the msa package for R (Bodenhofer et al., 2015) and H. influenzae Rd KW20 (GCA_000027305.1) as reference. Amino acid substitutions were confirmed by mapping of quality-trimmed sequencing reads (PHRED score ≥ 20) against the reference sequence using BWA (Li, 2013), with subsequent variant calling and annotation using FreeBayes (Garrison and Marth, 2012) and SnpEFF (Cingolani et al., 2012).
Substitutions in penicillin-binding protein 3 (PBP3) (transpeptidase region, aa 327-610) and grouping according to Skaare et al., 2014 (Skaare et al., 2014).
Substitutions in DNA gyrase (GyrA, subunit A) or DNA topoisomerase IV (ParC, subunit A) (quinolone-resistance determining regions, QRDR; aa 80-92) (Georgiou et al., 1996).
Substitutions in 50S ribosomal proteins L4 or L22, or single nucleotide polymorphisms (SNPs) in the six copies of the 23S rRNA gene (rrnA23S-rrnR23S) (peptidyl transferase center, nt 1900-2520) (Fyfe et al., 2016).
Substitutions in dihydrofolate reductase (DHFR) (or SNPs in promoter region), dihydropteroate synthase (DHPS), or thymidylate synthase (TS) (Fernandez-Villa et al., 2019).
Alterations in the operon encoding and regulating the AcrAB efflux pump.
Differences between strains in bold.
Table 1 summarizes the molecular basis of resistance to the different antibiotic groups, and depicts virulence determinants that might influence persistence, despite antibiotic therapy. Hi-117, Hi-226, and Hi-197 had bla TEM-1B and identical amino acid substitution patterns in PBP3, including the S385T substitution associated with high-level beta-lactam resistance (Skaare et al., 2014). They also shared multiple amino acid substitutions in enzymes involved in the folate pathway; dihydrofolate reductase (DHFR), dihydropteroate synthase (DHPS), and thymidylate synthase (TS) (Fernandez-Villa et al., 2019), in addition to single nucleotide polymorphisms (SNPs) in the DHFR promoter region. Variant calling (see Table 1 for methodology) showed that Hi-197, which expressed significantly higher MICs to trimethoprim-sulfamethoxazole and azithromycin compared to Hi-117 and Hi-226 ( Figure 1 ), possessed additional substitutions in DHPS (G225A), 50S ribosomal proteins L4 (W59R) and L22 (G91D), and the components AcrR (S181F) and AcrB (A604E) of the AcrAB efflux pump. All these mutations had 100% allelic uniformity and convincing depths of coverage (range 41-74). The macrolide binding site of 23S rRNA (Fyfe et al., 2016) and the quinolone-resistance determining regions of GyrA and ParC (Georgiou et al., 1996) lacked resistance-conferring mutations. bla TEM-1B was the only horizontally acquired resistance gene.
Discussion
This study describes persistent colonization of the respiratory tract with an NTHi strain despite antibiotic treatment in line with contemporary CVID guidelines, complicated by within-host evolution of resistance-conferring mutations and selection of subpopulations resistant to trimethoprim-sulfamethoxazole and azithromycin, during long-term prophylaxis with these antibiotics.
To our best knowledge, this is the first well-documented example of within-host evolution of antibiotic resistance in H. influenzae. Pfeifer et al. reported the emergence of a multidrug-resistant H. influenzae strain in a CVID patient but were unable to demonstrate persistence because earlier strains were not available for molecular characterization (Pfeifer et al., 2013). A study conducting partial genomic analyses on serially collected NTHi from a bronchiectasis patient revealed genetic changes associated with resistance to antimicrobial peptides in a persistent strain (Garmendia et al., 2014). Two later studies applied WGS for investigation of genetic changes associated with host adaption during persistent respiratory tract infection with NTHi, but within-host evolution of antibiotic resistance was not demonstrated (Moleres et al., 2018; Pettigrew et al., 2018).
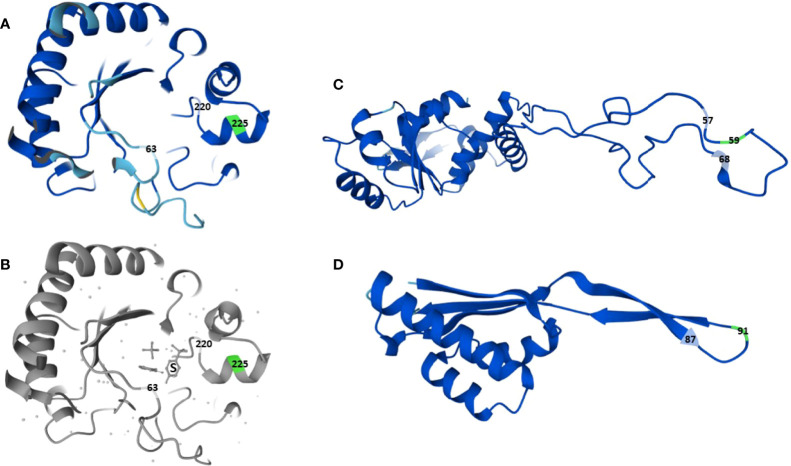
Trimethoprim interferes with the folate pathway by inhibiting the enzyme DHFR (encoded by folA), and sulfamethoxazole by inhibiting DHPS (encoded by folP) (Fernandez-Villa et al., 2019). Resistance to trimethoprim-sulfamethoxazole in H. influenzae is usually due to target alterations caused by folA or folP mutations, DHFR overexpression due to mutations in the folA promoter region, thymidine auxothrophy because of loss-of-function mutations in thyA, or horizontally acquired sul genes encoding sulfonamide-resistant isoforms of DHPS (Enne et al., 2002; Rodriguez-Arce et al., 2017; Sierra et al., 2020). The low-level resistant Hi-117 and Hi-226 harbored several substitutions in DHFR and DHPS, including three SNPs in the DHFR promoter region ( Table1 ), previously described in resistant H. influenzae (Enne et al., 2002; Sierra et al., 2020). The additional DHPS substitution G225A in Hi-197 is to our best knowledge novel. Crystallography shows that sulfonamides are sandwiched between amino acids 63 and 220 in DHPS in E. coli (Achari et al., 1997). The proximity of position 225 to the sulfonamide binding site ( Figure 2 ) and the lack of other changes in relevant genes make it plausible that G225A caused the significant increase in trimethoprim-sulfamethoxazole MIC in Hi-197.
Figure 2.
Three-dimensional structure of dihydropteroate synthase (DHPS) and 50S ribosomal proteins L4 and L22 in H. influenzae Rd KW20, with positions of newly acquired substitutions in strain Hi-197 highlighted (green). (A top left) DHPS in Rd KW20 (P43776), with focus on the binding site for sulfonamides. (B bottom left), DHPS in E. coli K12 (P0AC13) with the sulfonamide molecule (S) sandwiched between amino acid positions 63 and 220. (C, D right), 50S ribosomal proteins L4 [P44345, (C top)] and L22 [P44360, (D bottom)] in Rd KW20, with focus on the highly conserved regions 57KPWRQKGTGRAR68 (L4) and 87PRAKG91 (L22) of the extended hairpin loops. Screenshots from UniProt (UniProt, 2021), licenced under CC BY 4.0.
Azithromycin inhibits protein synthesis in H. influenzae through dual inhibitory effects on synthesis and function of the 50S ribosomal subunit (Champney and Miller, 2002). The drug binds to 23S rRNA (nt 2058-2059) downstream of the peptidyl transferase center, blocking the peptide exit tunnel close to a constriction formed by hairpin loops of ribosomal proteins L4 and L22 (Fyfe et al., 2016). Azithromycin resistance in H. influenzae is predominantly caused by chromosomal mechanism such as target alterations due to 23s rRNA mutations or altered ribosome-macrolide interaction due to L4 and/or L22 substitutions; however, rare strains with efflux-mediated resistance encoded by horizontally acquired mel and mef genes have been reported (Atkinson et al., 2017). Hi-197 contained substitutions G91D in L22 and W59R in L4. While G91D has been described in macrolide-resistant H. influenzae, W59R seems novel and is located within the highly conserved region 57KPWRQKGTGRAR68 of the extended hairpin loop of L4 ( Figure 2 ), where alterations are associated with macrolide resistance (Fyfe et al., 2016). This strongly suggests that both substitutions contributed to the azithromycin-resistant phenotype of Hi-197. In addition, while Hi-117, Hi-226 and Hi-197 shared several substitutions in AcrA, AcrB, and the repressor AcrR of the AcrAB efflux pump, the azithromycin resistant Hi-197 had additional substitutions in AcrB (A604E) and AcrR (S181F). Stepwise resistance to azithromycin has been reported with truncated AcrR and a subsequent substitution in AcrB (R327S) (Seyama et al., 2017). Truncated AcrR was not found in our strain, but like R327S, the A604E substitution is in the periplasmic domain where ligand binding occurs (Daley et al., 2005). Accordingly, we cannot exclude that the AcrB substitution might have contributed to azithromycin resistance; however, as Hi-197 expressed wild-type susceptibility to quinolones, chloramphenicol, tetracyclines, gentamicin, and rifampicin (data not shown), the effect of A604E might be discrete and/or azithromycin selective.
Whole-genome phylogenetic analysis revealed persistence of an ST2386 H. influenzae strain during April 2016 - January 2018, despite repeated and prolonged exposure to antibiotics.
Evading antibiotic activity by biofilm formation and intracellular invasion of host cells appear to be pivotal mechanisms promoting bacterial persistence (Clementi and Murphy, 2011). Beta-lactams and trimethoprim-sulfamethoxazole penetrate NTHi biofilm poorly. In an in vitro study, amoxicillin/clavulanic acid eliminated 100% of planktonic NTHi isolates, but only 3.6% of isolates in biofilm (Slinger et al., 2006). Trimethoprim-sulfamethoxazole eliminated 68% of planktonic but not biofilm isolates. Ciprofloxacin and azithromycin eliminated 100% of planktonic isolates and had biofilm elimination rates of 68% and 57%, respectively.
A notable observation in the present study was that repeated and prolonged courses with ciprofloxacin did not eradicate the strain, despite wild-type susceptibility to ciprofloxacin. This is consistent with the suboptimal abilities of ciprofloxacin to eliminate NTHi in biofilm (Slinger et al., 2006; Cavaliere et al., 2014). As quinolones increase the genome-wide spontaneous mutation rate (Long et al., 2016; Song et al., 2016) and may promote mutational resistance to other drugs (Tanimoto et al., 2008; Didier et al., 2011; Song et al., 2016), an intriguing yet unresolved question is whether ciprofloxacin exposure immediately before azithromycin prophylaxis from September 2017 contributed to the emergence of L4, L22, and AcrR substitutions in Hi-197.
Azithromycin is an appealing choice in the treatment of NTHi infections based on high intracellular activity, a biofilm-penetrating ability superior to most other drugs (albeit far from 100%), and a proven inhibitory effect against biofilm formation (Lode et al., 1996; Slinger et al., 2006; Starner et al., 2008). Moreover, several experimental studies suggest an immunomodulatory effect of azithromycin on host inflammatory response, reducing mucus production and ameliorating chronic inflammation (Parnham et al., 2014). Consequently, azithromycin prophylaxis has gained momentum in the management of chronic and recurrent lung infections.
Contemporary CVID guidelines advocate the use of long-term antibiotics in selected patients, although criteria for identifying such patients are lacking (Bonilla et al., 2016; Polverino et al., 2017; Hanitsch et al., 2020). The use of antimicrobial prophylaxis is nevertheless widespread, administered to 20% - 65% of CVID patients in different temporal and geographic settings, and azithromycin is by far the most frequently prescribed antibiotic (Kuruvilla and de la Morena, 2013; Sperlich et al., 2018). The recommendations and clinical practice predominantly lend support from three randomized controlled trials exploring macrolide prophylaxis for six to twelve months in non-cystic fibrosis bronchiectasis patients (Wong et al., 2012; Altenburg et al., 2013; Serisier et al., 2013). All demonstrated reduced number of infectious exacerbations in the macrolide group.
However, the risk of macrolide resistance is not negligible. In one study, macrolide resistance in pathogens isolated from the respiratory tract increased from 35% to 88% in the azithromycin group during the twelve-month study period, compared to 28% and 25% in the placebo group (Altenburg et al., 2013). Interestingly, emergence of macrolide resistance was not associated with loss of efficacy in the subsequent months, suggesting a pivotal role of the anti-inflammatory properties of azithromycin. This may also have been the case in our patient; however, azithromycin gradually lost activity after twelve months, highlighting the need for publications with long-term follow up.
EUCAST state that there is conflicting clinical evidence for the efficacy of macrolides in H. influenzae respiratory infections and have removed the clinical breakpoints (EUCAST, 2017). The Clinical and Laboratory Standards Institute (CLSI) state that susceptibility testing of azithromycin is often not necessary for management of individual patients and categorize the drug as Group C (alternative or supplemental agents) (CLSI, 2021). Accordingly, azithromycin susceptibility was not investigated routinely in the presented case. Retrospective testing revealed that azithromycin prophylaxis was continued for 18 months after the emergence of high-level resistance, exemplifying that discrepancy between clinical and laboratory guidelines might lead to non-beneficial or even potentially harmful treatment. Considering the widespread use of macrolide prophylaxis in CVID and bronchiectasis patients, EUCAST and CLSI recommendations for azithromycin susceptibility testing of H. influenzae from these patient groups should be revisited, with more emphasis on harmonization of clinical and laboratory guidelines. Moreover, this case report underlines the importance of maintaining close collaboration between clinicians and microbiology laboratories.
The presented case illustrates that the immunologic impairment in CVID patients extends beyond reduced IgG levels, and they remain at risk of infections despite aggressive IgG substitution therapy. Highly variable titers of antibodies specific to Streptococcus pneumoniae and H. influenzae in immunoglobulin replacement products, reduced half-life of infused immunoglobulins, and lack of mucosal immunity all likely contribute to the persistent susceptibility to these pathogens (Mooney et al., 2017). Restoration of mucosal immunity is likely not feasible through traditional immunoglobulin substitution therapy. Recent studies with nebulized administration of immunoglobulins or local immunotherapy by sublingual administration of inactivated bacterial pathogens report promising effects on mucosal immunity (Vonarburg et al., 2019; Guevara-Hoyer et al., 2020). However, their clinical efficacy is yet to be evaluated in larger trials. Importantly, the repertoire of virulence determinants in H. influenzae includes IgA proteases, as illustrated by the presence of an igaA1 gene in our strain ( Table 1 ).
Reduced vaccine response is one of the diagnostic criteria for CVID and the therapeutic potential of active immunization in this patient group remains questionable. Moreover, no vaccine is available for NTHi, which currently constitute the major concern in CVID patients (Slack et al., 2021).
Biofilm inhibitors have attracted interest as possible supplements to antibiotic therapy or prophylaxis in patients with chronic NTHi infections. DNase I, a mucolytic approved for human use often combined with inhaled antibiotics in cystic fibrosis patients (Manos, 2021), destabilizes biofilm matrix and enhances the efficacy of antibiotic treatment of NTHi biofilms in vitro (Izano et al., 2009; Cavaliere et al., 2014). Clarification is needed as to whether DNase I, or other biofilm inhibitors or mucolytic supplements represent useful adjuvants in bronchiectasis and CVID patients.
A weakness of this study is that isolates from 2013 (Hi-Alpha) and 2015 (Hi-Beta) were not available for retrospective characterization, and we had incomplete information about antibiotics prescribed by the patient’s general practitioner during this period. Assessment with respect to development of beta-lactam resistance and duration of persistence prior to 2016 was therefore not possible. Moreover, it is difficult to determine the relative contribution of the persistent H. influenzae strain to the patient's deteriorating clinical course after January 2018, especially in the late stages with progressive interstitial lung disease (Granulomatous and Lymphocytic Interstitial Lung Disease (GLILD)). However, we did not detect any other likely causative pathogens in respiratory samples in this period.
Conclusion
We describe chromosomal mutations responsible for within-host evolution of resistance to trimethoprim-sulfamethoxazole and azithromycin in a persisting NTHi strain in the respiratory tract of a CVID patient, after prolonged exposure to these antibiotics. The presented case illustrates the precariousness of long-term antimicrobial prophylaxis in immunocompromised patients, and the current clinical management and guidelines for CVID should be scrutinized. We highlight the need for a multi-targeted approach for treatment of respiratory tract infections and measures to hinder pathogen persistence and development of antibiotic resistance in this challenging patient group.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, accession numbers: GCA_923276745, GCA_923282765, GCA_923283335.
Ethics Statement
The use of personal data in this study was approved by the Regional Committees for Medical and Health Research Ethics in Norway (reference number 2018/1558) and the Norwegian Data Protection Services (reference number 232381). The patient’s spouse provided written informed consent to the use of patient data in this study.
Author Contributions
The idea of the case report was conceived by DS, PL, and HM. PL, HM, OO, and DS wrote the paper. OO is the infectious disease specialist involved in treatment of the patient. The bacterial isolates were obtained and primarily investigated at the laboratory of PL and HM. IA, NZ, and DS performed the reference analyses. All the authors contributed to the revision of the manuscript and approved the final version.
Funding
This work is funded by a research grant to DS from the South-Eastern Norway Regional Health Authority (project# 2016132).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Kanita Pimpaporn and Anita Bratfoss Andreassen at Vestfold Hospital Trust for technical assistance.
References
- Achari A., Somers D. O., Champness J. N., Bryant P. K., Rosemond J., Stammers D. K. (1997). Crystal Structure of the Anti-Bacterial Sulfonamide Drug Target Dihydropteroate Synthase. Nat. Struct. Biol. 4 (6), 490–497. doi: 10.1038/nsb0697-490 [DOI] [PubMed] [Google Scholar]
- Altenburg J., de Graaff C. S., Stienstra Y., Sloos J. H., van Haren E. H., Koppers R. J., et al. (2013). Effect of Azithromycin Maintenance Treatment on Infectious Exacerbations Among Patients With Non-Cystic Fibrosis Bronchiectasis: The BAT Randomized Controlled Trial. JAMA 309 (12), 1251–1259. doi: 10.1001/jama.2013.1937 [DOI] [PubMed] [Google Scholar]
- Atkinson C. T., Kunde D. A., Tristram S. G. (2017). Expression of Acquired Macrolide Resistance Genes in Haemophilus Influenzae. J. Antimicrob. Chemother. 72 (12), 3298–3301. doi: 10.1093/jac/dkx290 [DOI] [PubMed] [Google Scholar]
- Baquero F., Martinez J. L., Lanza V. F., Rodriguez-Beltran J., Galan J. C., San Millan A., et al. (2021). Evolutionary Pathways and Trajectories in Antibiotic Resistance. Clin. Microbiol. Rev. 34 (4), e0005019. doi: 10.1128/CMR.00050-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez J., Couce A., Rodriguez-Beltran J., Rodriguez-Rojas A. (2012). Antimicrobials as Promoters of Genetic Variation. Curr. Opin. Microbiol. 15 (5), 561–569. doi: 10.1016/j.mib.2012.07.007 [DOI] [PubMed] [Google Scholar]
- Bodenhofer U., Bonatesta E., Horejs-Kainrath C., Hochreiter S. (2015). Msa: An R Package for Multiple Sequence Alignment. Bioinformatics 31 (24), 3997–3999. doi: 10.1093/bioinformatics/btv494 [DOI] [PubMed] [Google Scholar]
- Bonilla F. A., Barlan I., Chapel H., Costa-Carvalho B. T., Cunningham-Rundles C., de la Morena M. T., et al. (2016). International Consensus Document (ICON): Common Variable Immunodeficiency Disorders. J. Allergy Clin. Immunol. Pract. 4 (1), 38–59. doi: 10.1016/j.jaip.2015.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolaia V., Kaas R. S., Ruppe E., Roberts M. C., Schwarz S., Cattoir V., et al. (2020). ResFinder 4.0 for Predictions of Phenotypes From Genotypes. J. Antimicrob. Chemother. 75 (12), 3491–3500. doi: 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaliere R., Ball J. L., Turnbull L., Whitchurch C. B. (2014). The Biofilm Matrix Destabilizers, EDTA and DNaseI, Enhance the Susceptibility of Nontypeable Hemophilus Influenzae Biofilms to Treatment With Ampicillin and Ciprofloxacin. Microbiologyopen 3 (4), 557–567. doi: 10.1002/mbo3.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champney W. S., Miller M. (2002). Inhibition of 50S Ribosomal Subunit Assembly in Haemophilus Influenzae Cells by Azithromycin and Erythromycin. Curr. Microbiol. 44 (6), 418–424. doi: 10.1007/s00284-001-0016-6 [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., et al. (2012). A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain W1118; Iso-2; Iso-3. Fly (Austin) 6 (2), 80–92. doi: 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi C. F., Murphy T. F. (2011). Non-Typeable Haemophilus Influenzae Invasion and Persistence in the Human Respiratory Tract. Front. Cell Infect. Microbiol. 1. doi: 10.3389/fcimb.2011.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI . (2021). “"Performance Standards for Antimicrobial Susceptibility Testing,” in CLSI Supplement M100", 31st Edition (Wayne, PA: Clinical and Laboratory Standards Institute; ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley D. O., Rapp M., Granseth E., Melen K., Drew D., von Heijne G. (2005). Global Topology Analysis of the Escherichia Coli Inner Membrane Proteome. Science 308 (5726), 1321–1323. doi: 10.1126/science.1109730 [DOI] [PubMed] [Google Scholar]
- Didelot X., Walker A. S., Peto T. E., Crook D. W., Wilson D. J. (2016). Within-Host Evolution of Bacterial Pathogens. Nat. Rev. Microbiol. 14 (3), 150–162. doi: 10.1038/nrmicro.2015.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier J. P., Villet R., Huggler E., Lew D. P., Hooper D. C., Kelley W. L., et al. (2011). Impact of Ciprofloxacin Exposure on Staphylococcus Aureus Genomic Alterations Linked With Emergence of Rifampin Resistance. Antimicrob. Agents Chemother. 55 (5), 1946–1952. doi: 10.1128/AAC.01407-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enne V. I., King A., Livermore D. M., Hall L. M. (2002). Sulfonamide Resistance in Haemophilus Influenzae Mediated by Acquisition of Sul2 or a Short Insertion in Chromosomal folP. Antimicrob. Agents Chemother. 46 (6), 1934–1939. doi: 10.1128/AAC.46.6.1934-1939.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST (2017) EUCAST General Consultation: Proposal to Remove Macrolide Breakpoints for Haemophilus Influenzae But Include a Note on Possible Clinical Efficacy and ECOFFs to Distinguish Wild Type From Isolates With Acquired Resistance. Available at: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Consultation/2017/Haem_and_macrolides/H_influenzae_macrolides_consultation_document_2017.pdf (Accessed 2022.02.17 2022).
- Fernandez-Villa D., Aguilar M. R., Rojo L. (2019). Folic Acid Antagonists: Antimicrobial and Immunomodulating Mechanisms and Applications. Int. J. Mol. Sci. 20 (20), 4996. doi: 10.3390/ijms20204996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe C., Grossman T. H., Kerstein K., Sutcliffe J. (2016). Resistance to Macrolide Antibiotics in Public Health Pathogens. Cold Spring Harb. Perspect. Med. 6 (10), a025395. doi: 10.1101/cshperspect.a025395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmendia J., Viadas C., Calatayud L., Mell J. C., Marti-Lliteras P., Euba B., et al. (2014). Characterization of Nontypable Haemophilus Influenzae Isolates Recovered From Adult Patients With Underlying Chronic Lung Disease Reveals Genotypic and Phenotypic Traits Associated With Persistent Infection. PloS One 9 (5), e97020. doi: 10.1371/journal.pone.0097020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E., Marth G. (2012) Haplotype-Based Variant Detection From Short-Read Sequencing. Available at: https://arxiv.org/abs/1207.3907v2.
- Gatt Y. E., Margalit H. (2021). Common Adaptive Strategies Underlie Within-Host Evolution of Bacterial Pathogens. Mol. Biol. Evol. 38 (3), 1101–1121. doi: 10.1093/molbev/msaa278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou M., Munoz R., Roman F., Canton R., Gomez-Lus R., Campos J., et al. (1996). Ciprofloxacin-Resistant Haemophilus Influenzae Strains Possess Mutations in Analogous Positions of GyrA and ParC. Antimicrob. Agents Chemother. 40 (7), 1741–1744. doi: 10.1128/AAC.40.7.1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-Hoyer K., Saz-Leal P., Diez-Rivero C. M., Ochoa-Grullon J., Fernandez-Arquero M., Perez de Diego R., et al. (2020). Trained Immunity Based-Vaccines as a Prophylactic Strategy in Common Variable Immunodeficiency. A Proof of Concept Study. Biomedicines 8 (7), 203. doi: 10.3390/biomedicines8070203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanitsch L., Baumann U., Boztug K., Burkhard-Meier U., Fasshauer M., Habermehl P., et al. (2020). Treatment and Management of Primary Antibody Deficiency: German Interdisciplinary Evidence-Based Consensus Guideline. Eur. J. Immunol. 50 (10), 1432–1446. doi: 10.1002/eji.202048713 [DOI] [PubMed] [Google Scholar]
- Izano E. A., Shah S. M., Kaplan J. B. (2009). Intercellular Adhesion and Biocide Resistance in Nontypeable Haemophilus Influenzae Biofilms. Microb. Pathog. 46 (4), 207–213. doi: 10.1016/j.micpath.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen L. M. A., van der Flier M., de Vries E. (2021). Lessons Learned From the Clinical Presentation of Common Variable Immunodeficiency Disorders: A Systematic Review and Meta-Analysis. Front. Immunol. 12. doi: 10.3389/fimmu.2021.620709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley K. A., Bray J. E., Maiden M. C. J. (2018). Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.org Website and Their Applications. Wellcome Open Res. 3, 124. doi: 10.12688/wellcomeopenres.14826.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruvilla M., de la Morena M. T. (2013). Antibiotic Prophylaxis in Primary Immune Deficiency Disorders. J. Allergy Clin. Immunol. Pract. 1 (6), 573–582. doi: 10.1016/j.jaip.2013.09.013 [DOI] [PubMed] [Google Scholar]
- Li H. (2013) Aligning Sequence Reads, Clone Sequences and Assembly Contigs With BWA-MEM. Available at: http://arxiv.org/abs/1303.3997.
- Liu B., Zheng D., Zhou S., Chen L., Yang J. (2022). VFDB 2022: A General Classification Scheme for Bacterial Virulence Factors. Nucleic Acids Res. 50 (D1), D912–D917. doi: 10.1093/nar/gkab1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lode H., Borner K., Koeppe P., Schaberg T. (1996). Azithromycin–review of Key Chemical, Pharmacokinetic and Microbiological Features. J. Antimicrob. Chemother. 37 Suppl C, 1–8. doi: 10.1093/jac/37.suppl_c.1 [DOI] [PubMed] [Google Scholar]
- Long H., Miller S. F., Strauss C., Zhao C., Cheng L., Ye Z., et al. (2016). Antibiotic Treatment Enhances the Genome-Wide Mutation Rate of Target Cells. Proc. Natl. Acad. Sci. U.S.A. 113 (18), E2498–E2505. doi: 10.1073/pnas.1601208113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manos J. (2021). Current and Emerging Therapies to Combat Cystic Fibrosis Lung Infections. Microorganisms 9 (9), 1874. doi: 10.3390/microorganisms9091874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moleres J., Fernandez-Calvet A., Ehrlich R. L., Marti S., Perez-Regidor L., Euba B., et al. (2018). Antagonistic Pleiotropy in the Bifunctional Surface Protein FadL (OmpP1) During Adaptation of Haemophilus Influenzae to Chronic Lung Infection Associated With Chronic Obstructive Pulmonary Disease. mBio 9 (5), e01176-18. doi: 10.1128/mBio.01176-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney D., Edgar D., Einarsson G., Downey D., Elborn S., Tunney M. (2017). Chronic Lung Disease in Common Variable Immune Deficiency (CVID): A Pathophysiological Role for Microbial and non-B Cell Immune Factors. Crit. Rev. Microbiol. 43 (4), 508–519. doi: 10.1080/1040841X.2016.1268568 [DOI] [PubMed] [Google Scholar]
- Parnham M. J., Erakovic Haber V., Giamarellos-Bourboulis E. J., Perletti G., Verleden G. M., Vos R. (2014). Azithromycin: Mechanisms of Action and Their Relevance for Clinical Applications. Pharmacol. Ther. 143 (2), 225–245. doi: 10.1016/j.pharmthera.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Pettigrew M. M., Ahearn C. P., Gent J. F., Kong Y., Gallo M. C., Munro J. B., et al. (2018). Haemophilus Influenzae Genome Evolution During Persistence in the Human Airways in Chronic Obstructive Pulmonary Disease. Proc. Natl. Acad. Sci. U.S.A. 115 (14), E3256–E3265. doi: 10.1073/pnas.1719654115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer Y., Meisinger I., Brechtel K., Grobner S. (2013). Emergence of a Multidrug-Resistant Haemophilus Influenzae Strain Causing Chronic Pneumonia in a Patient With Common Variable Immunodeficiency. Microb. Drug Resist. 19 (1), 1–5. doi: 10.1089/mdr.2012.0060 [DOI] [PubMed] [Google Scholar]
- Polverino E., Goeminne P. C., McDonnell M. J., Aliberti S., Marshall S. E., Loebinger M. R., et al. (2017). European Respiratory Society Guidelines for the Management of Adult Bronchiectasis. Eur. Respir. J. 50 (3), 1700629. doi: 10.1183/13993003.00629-2017 [DOI] [PubMed] [Google Scholar]
- Pulvirenti F., Camilli R., Giufre M., Milito C., Pimentel de Araujo F., Mancini F., et al. (2018). Risk Factors for Haemophilus Influenzae and Pneumococcal Respiratory Tract Colonization in CVID. J. Allergy Clin. Immunol. 142 (6), 1999–2002 e1993. doi: 10.1016/j.jaci.2018.08.014 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Arce I., Marti S., Euba B., Fernandez-Calvet A., Moleres J., Lopez-Lopez N., et al. (2017). Inactivation of the Thymidylate Synthase thyA in Non-Typeable Haemophilus Influenzae Modulates Antibiotic Resistance and Has a Strong Impact on Its Interplay With the Host Airways. Front. Cell Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serisier D. J., Martin M. L., McGuckin M. A., Lourie R., Chen A. C., Brain B., et al. (2013). Effect of Long-Term, Low-Dose Erythromycin on Pulmonary Exacerbations Among Patients With non-Cystic Fibrosis Bronchiectasis: The BLESS Randomized Controlled Trial. JAMA 309 (12), 1260–1267. doi: 10.1001/jama.2013.2290 [DOI] [PubMed] [Google Scholar]
- Seyama S., Wajima T., Nakaminami H., Noguchi N. (2017). Amino Acid Substitution in the Major Multidrug Efflux Transporter Protein AcrB Contributes to Low Susceptibility to Azithromycin in Haemophilus Influenzae. Antimicrob. Agents Chemother. 61 (11), e01337-17. doi: 10.1128/AAC.01337-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra Y., Tubau F., Gonzalez-Diaz A., Carrera-Salinas A., Moleres J., Bajanca-Lavado P., et al. (2020). Assessment of Trimethoprim-Sulfamethoxazole Susceptibility Testing Methods for Fastidious Haemophilus Spp. Clin. Microbiol. Infect. 26 (7), 944 e941–944 e947. doi: 10.1016/j.cmi.2019.11.022 [DOI] [PubMed] [Google Scholar]
- Skaare D., Anthonisen I. L., Kahlmeter G., Matuschek E., Natas O. B., Steinbakk M., et al. (2014). Emergence of Clonally Related Multidrug Resistant Haemophilus Influenzae With Penicillin-Binding Protein 3-Mediated Resistance to Extended-Spectrum Cephalosporins, Norway 2006 to 2013. Euro. Surveill. 19 (49), 20986. doi: 10.2807/1560-7917.es2014.19.49.20986 [DOI] [PubMed] [Google Scholar]
- Slack M. P. E., Cripps A. W., Grimwood K., Mackenzie G. A., Ulanova M. (2021). Invasive Haemophilus Influenzae Infections After 3 Decades of Hib Protein Conjugate Vaccine Use. Clin. Microbiol. Rev. 34 (3), e0002821. doi: 10.1128/CMR.00028-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slinger R., Chan F., Ferris W., Yeung S. W., St Denis M., Gaboury I., et al. (2006). Multiple Combination Antibiotic Susceptibility Testing of Nontypeable Haemophilus Influenzae Biofilms. Diagn. Microbiol. Infect. Dis. 56 (3), 247–253. doi: 10.1016/j.diagmicrobio.2006.04.012 [DOI] [PubMed] [Google Scholar]
- Song L. Y., Goff M., Davidian C., Mao Z., London M., Lam K., et al. (2016). Mutational Consequences of Ciprofloxacin in Escherichia Coli. Antimicrob. Agents Chemother. 60 (10), 6165–6172. doi: 10.1128/AAC.01415-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperlich J. M., Grimbacher B., Workman S., Haque T., Seneviratne S. L., Burns S. O., et al. (2018). Respiratory Infections and Antibiotic Usage in Common Variable Immunodeficiency. J. Allergy Clin. Immunol. Pract. 6 (1), 159–168 e153. doi: 10.1016/j.jaip.2017.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starner T. D., Shrout J. D., Parsek M. R., Appelbaum P. C., Kim G. (2008). Subinhibitory Concentrations of Azithromycin Decrease Nontypeable Haemophilus Influenzae Biofilm Formation and Diminish Established Biofilms. Antimicrob. Agents Chemother. 52 (1), 137–145. doi: 10.1128/AAC.00607-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K., Tomita H., Fujimoto S., Okuzumi K., Ike Y. (2008). Fluoroquinolone Enhances the Mutation Frequency for Meropenem-Selected Carbapenem Resistance in Pseudomonas Aeruginosa, But Use of the High-Potency Drug Doripenem Inhibits Mutant Formation. Antimicrob. Agents Chemother. 52 (10), 3795–3800. doi: 10.1128/AAC.00464-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C. (2021). UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 49 (D1), D480–D489. doi: 10.1093/nar/gkaa1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarburg C., Loetscher M., Spycher M. O., Kropf A., Illi M., Salmon S., et al. (2019). Topical Application of Nebulized Human IgG, IgA and IgAM in the Lungs of Rats and non-Human Primates. Respir. Res. 20 (1), 99. doi: 10.1186/s12931-019-1057-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts S. C., Holt K. E. (2019). Hicap: In Silico Serotyping of the Haemophilus Influenzae Capsule Locus. J. Clin. Microbiol. 57 (6), e00190-19. doi: 10.1128/JCM.00190-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C., Jayaram L., Karalus N., Eaton T., Tong C., Hockey H., et al. (2012). Azithromycin for Prevention of Exacerbations in non-Cystic Fibrosis Bronchiectasis (EMBRACE): A Randomised, Double-Blind, Placebo-Controlled Trial. Lancet 380 (9842), 660–667. doi: 10.1016/S0140-6736(12)60953-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, accession numbers: GCA_923276745, GCA_923282765, GCA_923283335.