Abstract
The clustered, regularly interspaced, short palindromic repeats (CRISPR)-associated endonuclease (Cas) system, referred to as CRISPR/Cas system, has attracted significant interest in scientific community due to its great potential in translating into versatile therapeutic tools in biomedical field. For instance, a myriad of studies has demonstrated that the CRISPR/Cas system is capable of detecting various types of viruses, killing antibiotic-resistant bacteria, treating inherited genetic diseases, and providing new strategies for cancer therapy. Furthermore, CRISPR/Cas systems are also exploited as research tools such as genome engineering tool that allows researchers to interrogate the biological roles of unexplored genes or uncover novel functions of known genes. Additionally, the CRISPR/Cas system has been employed to edit the genome of a wide range of eukaryotic, prokaryotic organisms and experimental models, including but not limited to mammalian cells, mice, zebrafish, plants, yeast, and Escherichia coli. The present review mainly focuses on summarizing recent discoveries regarding the type II CRISPR/Cas9 and type VI CRISPR/Cas13a systems to give researchers a glimpse of their potential applications in the biological and biomedical field.
Keywords: CRISPR/Cas system, Gene therapy, Cancer therapy, Diagnostic tool, Viral detection
Introduction
CRISPR/Cas systems were first found in prokaryotic species and were reported to comprise arrays of short repetitive sequences, referred to as repeats. These repeats were interspersed with non-repetitive nucleotides called spacers. The repeats, with size of 29 base pairs (bp) long, were first inspected by Ishino and his colleagues in Escherichia coli [1]. While the size of the repeats generally ranges from 24 to 47 bp, the number of repeats varies between 2 and 249 [2]. In one CRISPR array, repeats are always the same regarding their size and sequence.
It has also been reported that about 40% of spacers from lactic acid bacteria, namely Streptococcus thermophiles, have a homologue corresponding to either phage or plasmid DNA sequences among the isolated and sequenced phage’s genome [3]. Thus, the spacers are proposed to be derived from foreign DNA including phage and plasmids. Bacteria generally acquire new spacers once they are exposed to new viruses in order to confer resistance to the cognate virus. Therefore, spacers present in a certain bacterium typically reveal the historical record of phages that the bacterium has been exposed to [4]. The leader sequence is commonly located 5’ to the CRISPR locus and is rich in AT nucleotides. However, it has been demonstrated that the leader sequence is incapable of encoding RNAs or proteins, and that it is located next to the short repeats. Apart from the leader sequence, the genes encoding Cas proteins are found to be localized either upstream or downstream of the CRISPR locus.
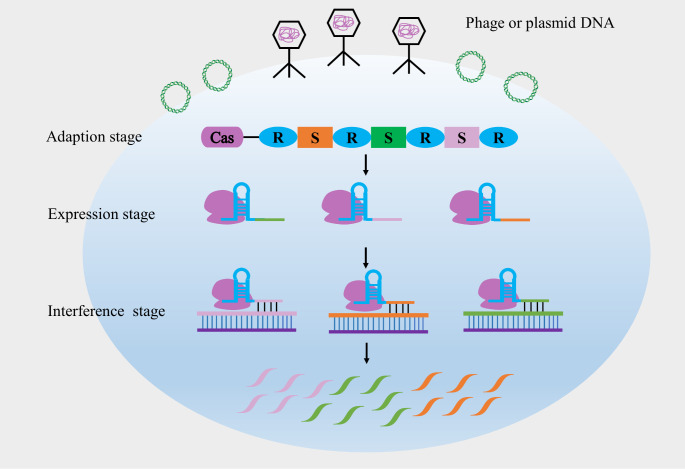
The CRISPR/Cas system is postulated to be formed in three stages. In the adaption stage, bacterial cells acquire sequences derived from foreign DNA such as viral DNA, and then incorporate them into the bacterial CRISPR array. During the expression stage, precursor CRISPR RNA (crRNA) is transcribed and processed into individual mature crRNA comprising a repeat and a spacer region. CRISPR RNA-guided cleavage of the viral DNA via Cas proteins is carried out in the interference stage to protect bacteria against viral infections (Fig. 1) [5].
Fig. 1.
Schematic illustration of bacterial immune response by CRISPR/Cas9 system. Bacteria defend themselves from phage or foreign DNA using CRISPR/Cas9 system through three stages: adaption stage, expression stage and interference stage. In the adaption stage, bacteria acquire phage or plasmid DNA and insert them into the CRISPR array. Cas, CRISPR-associated proteins; R, repeats; S, spacers. In the expression stage, repeat and spacer sequences were transcribed and formed complex with Cas9 protein. In the interference stage, the target DNA matching the spacer sequence were cleaved by Cas9 protein, resulting in degradation of phage or plasmid DNA.
Approximate forty-five Cas proteins adjacent to the CRISPR arrays have been identified and the CRISPR/Cas systems can be further categorized into three types according to the composition of Cas proteins and crRNA. As reported by Makarova et al., Type I CRISPR/Cas system generally harbors a Cas3 protein that displays both helicase and DNase activities [6]. Moreover, the type II CRISPR/Cas system is mainly characterized by the presence of Cas9 protein, which is responsible for generating crRNA and cleaving the target DNA. Type III CRISPR/Cas system is typified by harboring polymerase and repeat-associated mysterious protein (RAMP) modules that are proposed to participate in generating the spacer-repeat transcripts, similar to the Cascade complex [6].
The CRISPR/Cas systems play significant roles in protecting bacteria from invading foreign DNA originated from either bacteriophages or exogenous plasmids [7]. In recent years, the CRISPR/Cas systems, especially CRISPR/Cas9 and CRISPR/Cas13 have been programmed to develop versatile gene-editing tools that could modify, delete or correct precise regions of the target DNA. Moreover, these two systems are also employed to develop highly sensitive and rapid isothermal diagnostic tools that are capable of detecting viruses, bacteria and cancer mutations. The topics on the application of CRISPR in bacteria, cardiovascular disease, gene therapy and metabolic engineering of small molecules has been excellently reviewed [8, 9]. This review aims to focus, consolidate and summarize recent findings on the implementation of CRISPR/Cas systems in the biological and biomedical field, thus giving researchers a comprehensive overview of what can be achieved by the CRISPR/Cas systems.
Working principles of CRISPR/Cas systems
Among these CRISPR/Cas systems, type II CRISPR/Cas9 and type VI CRISPR/Cas13 systems have been widely exploited as genetic modification tool and diagnostic tool, respectively. Cas9 protein recognizes and binds to foreign nucleic acids primarily through Watson-Crick base pairing between its guide RNA and the foreign DNA carrying a short protospacer adjacent motif (PAM) [10]. Once binding to the foreign DNA, the two nuclease domains including HNH and RuvC from Cas9 protein cleave the target DNA site at the complementary and non-complementary DNA strands, respectively.
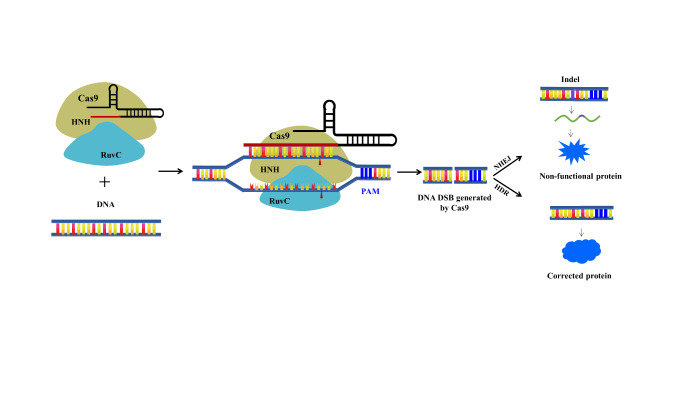
This type of cleavage typically results in a cut from each strand of the foreign DNA, leading to site-specific double strand DNA breaks (DSBs) that can be repaired via either nonhomologous end-joining (NHEJ) or homology-directed repair (HDR) pathway. The former repair pathway frequently contributes to repair errors, which subsequently produce frame-shifting insertions and deletions, also referred to as indels, ultimately inducing gene function disruption. The latter repair pathway can be employed to generate desired alternations of target gene via adding a donor template (Fig. 2) [11]. Therefore, theoretically speaking, any target DNA sites with a PAM motif can be modified by CRISPR/Cas9 system through altering the guide RNA sequence. The easy-to-use property of this unique system makes it an appealing tool for high-throughput sequence-specific genome modification, genome screening, live-cell imaging, as well as other applications [12].
Fig. 2.
Working principles of the CRISPR/Cas9 system as genome editing tool. The HNH and RuvC nuclease domains of Cas9 protein cleave complementary and non-complementary DNA strands, respectively. The resulting double DNA strand breaks (DSB) were then repaired through either non-homologous end joining (NHEJ) pathway or homology-directed repair (HDR) pathway with a donor template.
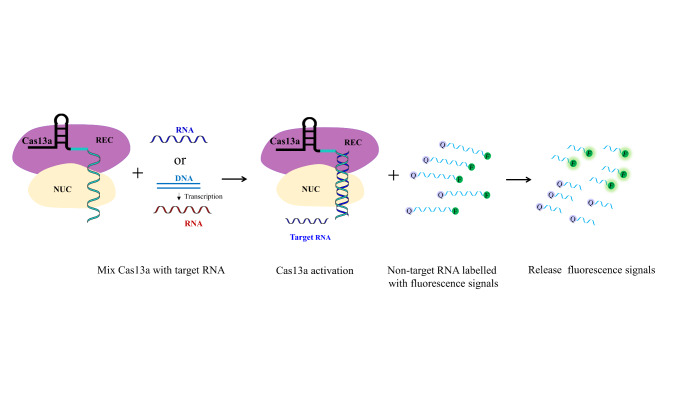
Instead of targeting DNA, CRISPR/Cas13 system has been verified to recognize and bind to RNA. Intriguingly, the RNAase activity of this RNA-guided and RNA-targeting CRISPR effector Cas13a, also termed as C2c2, is activated once guide RNA binds to its target RNA, which then results in cleavage of nearby non-targeted RNAs (Fig. 3)[13]. Thus, the CRISPR/Cas13 system has been employed as molecular diagnostic tools for visualizing, degrading, or binding RNA in a programmable, multiplexed fashion, as well as used in forward transcriptomic pooled screens [14].
Fig. 3.
Adaption of CRISPR /Cas13a system as diagnostic tools. The nuclease activity of Cas13a protein is activated once it binds to the target RNA, leading to cleavage of near-by non-targeting RNA that are labelled with quenched fluorescent dyes. This cleavage subsequently results in release of fluorescence signals.
As mentioned previously, crRNA plays an essential role in recognizing the target sites. It has been reported that the crRNA usually contains a spacer and a repeat region. The spacer region bears a sequence complementary to the target DNA and the repeat sequence bases pair with a trans-activating (tracrRNA) to form a mature crRNA. Thus, both spacer and repeat sequence information are required when designing the crRNA. CRISPR/Cas systems can be delivered into cells through viral vectors, lipid vesicles, gold nanoparticles, carbon nanotubes, microinjection, electroporation, acoustoporation, magnetotransfection and laser optoporation [15]. After transfection, single cells need to be isolated in order to generate clonal lines that can be verified as complete knockouts. Two methods including dilution cloning and Fluorescence-Activated Cell Sorting (FACS) can be employed to obtain single cell-derived clones. FACS is generally more effective in isolating double-positive cells compared with the dilution cloning. However, dilution cloning is cheaper and may result in less cellular stress than sorting [16].
Before the discovery of CRISPR/Cas systems, other types of gene-editing tools have also been programmed to introduce site-specific DNA double strand breaks through fusing a DNA-binding domain to a DNA-cleavage domain. These DNA binding proteins include meganucleases, zinc finger nucleases (ZFNs) and transcription activator-like effectors nucleases (TALENs). However, the widespread application of meganucleases as gene-editing tool is restricted by the low probability of finding a meganuclease cleavage site in a chosen gene [17]. Non-specific DNA-cleavage domain from C-terminus of FokI endonuclease is often linked with ZFNs and TALENs, which can be assembled in desired combinations to generate site-specific double-strand breaks. The ZFNs and TALENs are highly versatile genome-editing tools that allow customizing the DNA-binding domain to target virtually any sequence and precisely edit target gene in a broad spectrum of plant and animal species [18]. However, the design of assembled ZFNs that exhibit high specificity for the targeted sequences can be challenging, time-consuming and labor-intensive. Also, ZF domains display context-dependent binding preference due to crosstalk between adjacent modules when assembled into a larger array [19]. Thus, the adoption of ZFNs is limited by low specificity, which may lead to high levels of off-target effects and the potential for unfavorable mutagenic effects. Although TALENs display greater design flexibility, higher specificity for targeted DNA sequences and less costly compared to the ZNFs, the cloning of repeat TALE arrays can be labor intensive and challenging [20]. To sum up, among these genome-editing tools, the CRISPR/Cas systems have obvious advantages over other types of gene-editing tools in three ways (i) target design simplicity; (ii) high gene-editing efficiency; (iii) capability of altering multiple sites simultaneously. Thus, the CRISPR/Cas systems have attracted significant interest from researchers and are currently being intensively researched worldwide.
Biological applications of CRISPR/Cas systems
Due to the high degree of versatility and easy manipulation properties, CRISPR/Cas systems are programmed to modify genome of almost any desired organism, to edit RNA, to elucidate the biological function of genes, and to image living cells (Fig. 4and Fig. 5).
Fig. 4.
The application of CRISPR/Cas9 or CRISPR/dCas9 systems in biological and biomedical field. CRISPR/Cas9 or CRISPR/dCas9 system has broad biological and biomedical applications including gene therapy, live-cell imaging, genome screening, genome editing, gene therapy and cancer study.
Fig. 5.
The application of CRISPR /Cas13a or CRISPR /dCas13a systems as genome editing and diagnostic tools. CRISPR /Cas13a or CRISPR /dCas13a system is capable of killing antibiotic-resistant bacteria, knocking down RNA, editing RNA, inhibiting RNA virus replication, detecting various types of viruses and bacteria.
Genome editing
Traditional methods utilized for genetic engineering including ZFNs and TALENs are labor intensive and time consuming in comparison with CRISPR/Cas systems [21]. CRISPR/Cas-system-mediated genetic modification has achieved great success in modifying genome of a broad spectrum of bacterial strains and fungus including but not limited to vancomycin-resistant Enterococcus faecium, Mycobacterium tuberculosis, Lactobacillus plantarum, Corynebacterium glutamicum, Lactobacillus casei, Bacillus subtilis, Clostridium beijerinckii Saccharomyces cerevisiae and Streptomyces [22–25]. The genome engineering of microbacteria and fungus enables the modification of specific metabolic pathways, which usually gives rise to production of high value-added metabolites including biochemicals, biofuels, and pharmaceutical products/precursors [26].
Besides its important roles involved in bacterial genome editing, the CRISPR/Cas system has also been demonstrated to harbor critical roles in editing the genome of plants and mammalian cells. For instance, Ming et al. had utilized CRISPR/Cas12b to achieve transcriptional repression and activation in an important crop, rice [27]. In addition, a study manifests that point mutations can be made at specific regions of genome in both rice and tomato by fusing CRISPR/Cas9 with activation-induced cytidine deaminase [28]. Intriguingly, rice containing high amount of carotenoid can be generated via inserting targeted gene using CRISPR/Cas9 with no detectable penalty in morphology or yield of the crop [29]. Another study reveals that CRISPR/Cas9-targeted mutagenesis of a host disease-susceptibility Os8N3 gene endows rice with resistance against Xanthomonas oryzae pv. oryzae (Xoo), which specifically cause bacterial blight in rice [30]. The infection of Xoo in rice can trigger serious production loss worldwide. CRISPR/Cas9 systems are also capable of editing genome of other types of plants or crops including Arabidopsis, wheat, potato, soybean, maize, apple and grapevine [31, 32].
The CRISPR/Cas system was first employed by Feng Zhang’s research group to accomplish precise genome engineering in eukaryotic cells such as mammalian cells [33]. The engineering of a specific gene or multiple genes simultaneously in mammalian cells allows researchers to construct disease-specific models or probe the function of genes involved in particular biochemical pathway. As an example, Zhou et al. have developed a focused CRISPR/Cas-based lentiviral library in human cells and can be used to discover genes that participate in a certain biological process [34]. Moreover, a CRISPR-based genome screen reveals that elongator acetyltransferase complex subunit 5 (ELP5) primarily accounts for the gemcitabine sensitivity in gallbladder cancer [35]. Moreover, Seki and Rutz have optimized the method for Cas9/RNP (Ribonucleoprotein) transfection of primary mouse and human T cells without T cell receptor (TCR) stimulation, which almost causes the entire inhibition of target gene expression. This finding significantly simplify the gene editing process for next-generation immunotherapies [36].CRISPR/Cas systems are also capable of modifying genome in other experimental models including mouse and zebrafish [37].
RNA editing
A plethora of studies indicates that CRISPR/Cas systems also harbor prominent roles in RNA editing [38]. For example, catalytically inactive Cas13 has been used to guide adenosine (A)-to-inosine (I) deaminase, an enzyme essential for programming A to I replacement, to transcripts in mammalian cells, thus allowing the alteration of full-length transcripts consisting of pathogenic mutations [39]. Furthermore, Jing et al. have successfully tethering inactive Cas13a to the catalytic domain of human adenosine deaminase acting on RNA type 2 (hADAR2) to target messenger RNA, ultimately leading to precise modification of specific nucleotide residues [40].
Moreover, the level of reporter or endogenous transcripts can be knocked-down by the CRISPR/Cas13a system with enhanced specificity compared to the RNA interference approach [41]. Likewise, an efficient and specific ribonuclease effector termed as CasRx was discovered and isolated from Ruminococcus flavefaciens XPD3002 by Konermann et al. to target cellular RNA [42]. Furthermore, a novel study has reported a photoactivatable RNA N6-Methyladenosine (m6A) editing approach, which is based on CRISPR/Cas13 system that enables the controlled regulation of m6A editing in response to blue light [43]. This unique system also allows the manipulation of multiple genes robustly and simultaneously. To sum up, the above findings indicate that CRISPR/Cas systems provide a potential novel toolset for RNA editing in a broad spectrum of species and therapeutic development.
Live-cell imaging
Mounting evidence indicates that CRISPR/Cas systems play pivotal roles in live-cell imaging of genomic loci through tagging of either the dead Cas9(dCas9) or the sgRNA with fluorescent protein (FP) molecules for labeling. For instance, Wang et al. have reported a CRISPR-based live-cell fluorescent in situ hybridization (LiveFISH) approach that permits tracking genome loci in a broad spectrum of cell types such as primary cells [44]. Moreover, LiveFISH is capable of accurately detecting Patau syndrome, a chromosomal disorder, and tracking the real-time movement of DSBs induced by CRISPR/Cas9-mediated editing [44]. Additionally, Maass et al. had described an allele-specific CRISPR live-cell DNA imaging technique (SNP-CLING) that enables monitoring of spatiotemporal dynamics of allelic positioning in mouse embryonic stem cells and fibroblasts [45].
Apart from imaging genome loci, CRISPR/Cas9-based system has been engineered to monitor RNA in living cells. For instance, Nelles and his colleagues have proven that targeting RNAs with Cas9 can be achieved through providing the PAM as part of an oligonucleotide (PAMmer), which hybridizes to the target RNA [46]. The resulting system is capable of binding RNA in a nucleic-acid-programmed manner and allowing the detection of endogenous RNA in living cells [46]. Imaging of telomere repeats in live leaf cells of Nicotiana benthamiana has been accomplished through conjugating eGFP/mRuby2 to catalytically inactive Cas9 protein [47].
Furthermore, an all imaging-based pooled-library CRISPR screening approach has been established to image multiple molecular targets, such as investigating modulators of the nuclear speckle localization of a long non-coding RNA (lncRNA) [48]. Likewise, endogenous lncRNA can be visualized and isolated using a CRISPR-mediated endogenous lncRNA tracking and immunoprecipitation system (CERTIS) via attaching a 24-repeat MS2 tag to lncRNA locus. The CERTIS system allows both spatially and temporally tracking of lncRNA modulation in living cells and investigating the lncRNA-protein interactomes [49]. Low-abundance mRNAs in cells can be efficiently monitored by the CRISPR-Sunspot method, which is established using the SunTag signal amplification system [50].
Many efforts have been devoted to improving the CRISPR/Cas-based systems for endogenous imaging of DNA or RNA. For instance, Wu et al. have recently established a novel CRISPR/Cas-based imaging system through attaching the sgRNA with molecular beacons, which are characterized by harboring small molecule dye-based, fluorogenic oligonucleotide probes. The resultant system is referred to as CRISPR/MB that can detect single telomere loci with enhanced sensitivity compared to the conventional approaches employing fluorescent proteins (FP) reports [51]. Similarly, Ma et al. have developed an CRISPR-Sirius method, which is based on octet arrays of aptamers, is more stable in comparison with CRISPR/Cas-based system that are fused with FP [52]. It has been reported that only three unique sgRNAs are required to visualize the non-repetitive genome loci through modifying the CRISPR sgRNA conjugated with two different MBs [53]. Last but not least, genomic DNA and RNA transcripts can be simultaneously visualized through combing CRISPR/Cas9 and CRISPR/Cas13 systems.
Genome screening
The CRISPR/Cas systems have also been employed as high-throughput genomic screening tools to identify regulatory elements, the function of lncRNA and the function of genes that participate in a certain biological process such as protein transport, viral infection, cell migration, cellular reprogramming and oxidative phosphorylation [54, 55]. For example, it has been demonstrated that gene knock-out pooled screening based upon CRISPR/Cas9 system has been employed to reveal essential regulators of immune function. Likewise, a genome-based CRISPR/Cas9 screen is also capable of identifying host factors that modulate killing by Legionella pneumophila, a bacterium that causes a potentially lethal pneumonia termed Legionnaires’ disease [56]. Moreover, one study reveals that CRISPR/Cas9-based genome screen has successfully pinpointed interferon regulatory factor 2 (IRF2), a transcription factor, as an essential factor that regulate caspase-4 in human cells [57]. Additionally, CRISPR/Cas9-based genome screening indicates that a guanine nucleotide exchange factor (RIC8A), harbored a central role in G-alpha protein activation, positively regulates YAP signaling [58].
Several studies have verified that the CRISPR/Cas systems are imperative in elucidating the putative function of lncRNAs. As an example, a CRISPR interference (CRISPRi) screen on 2263 epidermis-expressed lncRNAs reveals that progenitor renewal associated non-coding RNA (PRANCR) plays essential roles in regulating epidermal homeostasis [59]. Furthermore, Zhu and his colleagues have identified 51 lncRNAs as indispensable regulators of human cancer cell growth via a high-throughput screening method based upon CRISPR/Cas system [60].
Besides identifying regulatory elements and investigating the function of lncRNAs, the CRISPR/Cas systems have also play momentous roles in screening functional genes. For instance, a genome-based CRISPR screen enables the discovery of human genes required for oxidative phosphorylation (OXPHOS) [61]. Similarly, Bakke et al. have identified that proteasome subunit alpha type-6 (PSMA6) is a key gene required for the survival of pancreatic cancer cells using a negative-selection genome-wide CRISPR screen [62]. Moreover, Bassaganyas et al. have discovered that two new factors including tetratricopeptide repeat protein 17 (TTC17) and coiled-coil domain-containing protein 157 (CCDC157), are involved in protein transport through a pooled genome-wide CRISPRi screen [63]. Intriguingly, Yang et al. have utilized CRISPR activation approach to carry out a genome-scale screen to identify genes that are crucial for cellular reprogramming of mouse primed epiblast stem cells (EpiSCs). The data validated that Sall1, a transcription factor belong to the Spalt-like gene family, harbors key roles in reprogramming EpiSCs [64].
The genome-wide screen based upon CRISPR/Cas systems also have vital roles in probing the functional genes that are participated in viral, bacterial and parasite infection. For instance, several studies have proven that the genome-wide CRISPR-based screen enables the identification of host factors including CD4 and CCR5, which possess a critical role in HIV infection yet are redundant in terms of cell proliferation and viability [65]. Furthermore, a CRISPR activation screen have revealed that a set of interferon-stimulated genes (ISGs) encoding interferon lambda 2 (IFN-λ2) and interferon alpha-inducible protein 6 (IFI6) are required for protecting cells against Zika virus (ZIKV) infection [66]. A recent study has indicated that vascular endothelial growth factor (VEGF) and semaphoring-binding receptor Neuropilin-1 (Nrp-1) are necessary for β-herpesvirus murine cytomegalovirus (MCMV) infection via an antiviral necroptosis-based CRISPR knockout screen [67]. Tian et al. have suggested that semaphorin 6 A (SEMA6A) and 6B (SEMA6B) are crucial receptors for the exotoxin TcsL, a major virulence factor in pathogenic bacterium Paeniclostridium sordellii and responsible for high lethality rate associated with P.sordellii infection [68]. Last but not least, a genome-wide CRISPR screen has also been employed to gauge the significance of each gene from the parasite Toxoplasma gondii, which infects human fibroblasts [69]. In brief, the above evidence has supported the notion that CRISPR/Cas systems are crucial components of genome screening.
Biomedical applications of CRISPR/Cas systems
Viral and bacterial pathogen detection
Over the last few years, studies have validated that CRISPR/Cas13a system is able to detect various types of viruses and bacteria. For instance, a molecular detection system based upon the CRISPR/Cas13a system has been established and referred to as Specific High-Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK), which enables the detection of both Zika and Dengue viruses [13]. In this study, the detection limit of SHERLOCK for Zika virus can be as low as 3.2 aM and the cost of per test can be as low as $0.61, thus opening new avenues for rapid, and sensitive detection of genetic materials in viruses. Moreover, Fozouni et al. have designed an amplification-free CRISPR/Cas13a test, which allows direct detection of the December 2019 outbreak of the novel respiratory virus, SARS-CoV-2, from nasal swab RNA that can be read with a mobile phone microscope [70]. Remarkably, a sensitivity of ~ 100 copies/µL of SARS-CoV-2 RNA can be achieved by this CRISPR/Cas13a-based system within half an hour measurement time with high accuracy and clinical validation [70]. Recently, Liu et al. had established a room temperature influenza A H7N9 detection system using CRISPR/Cas13a system [44]. Additionally, a novel-PCR based CRISPR/Cas13a system with high sensitivity and specificity has been developed to detect hepatitis B virus DNA [71]. One study also indicates that CRISPR/Cas13a-based isothermal diagnostic method enables rapid detection of Ebola virus [72].
Apart from virus detection, emerging evidence suggests that the CRISPR/Cas13a system possesses prominent roles in bacteria detection. For example, Salmonella Enteritidis, one of the causing agents of enterocolitis, can be detected by a combination of fluorescence-labelled nucleic acid probes in conjugation with CRISPR/Cas13a components [73]. Moreover, Zhang et al. has successfully developed a light-up RNA aptamer signaling-CRISPR-Cas13a assay, which enables mix-and-read detection of viable bacteria such as Bacillus cereus and Escherichia coli [74].
Viral inhibition and bacterial killing
Growing evidence has suggested that CRISPR/Cas systems participate in suppressing the replication of viruses and bacteria. For instance, Abbott et al. had established a prophylactic antiviral CRISPR in human cells (PAC-MAN)-based strategy that can efficiently degrade RNA from SARS-CoV-2 sequences and live influenza A virus (IAV) in human lung epithelial cells [75]. These findings demonstrate the feasibility of PAC-MAN in repressing the replication of coronavirus. Moreover, the CRISPR/Cas9 system has also been successfully employed to inhibit the replication of duck hepatitis B virus (DHBV), which is commonly employed to investigate human HBV infection, through designing single-guide RNAs (sgRNAs) specifically targeting HBV genome [76]. Noor and his colleagues have reported a similar finding that employs CRISPR/Cas9-based strategy to inhibit HBV replication [77]. One recent study has exhibited that CRISPR/Cas9 system is capable of efficiently blocking herpes simplex virus type 1 (HSV-1) replication and the occurrence of herpetic stromal keratitis (HSK) in three different infection models including HSK prevention, therapeutic and recurrent models [78]. Additionally, the CRISPR/Cas systems are able to restrain the replication of other types of DNA viruses including African swine fever virus [79], cauliflower mosaic virus [80], Marek’s disease virus [81] and human papilloma virus (HPV) [82].
Besides targeting DNA viruses, RNA virus replication can also be inhibited by the CRISPR/Cas13 system. For example, Li et al. showed that inactivation of dengue virus (DENV) in mammalian cells can be realized through targeting the NS3 region of the DENV genome RNA [83]. The CRISPR/Cas13a system can also be programmed to repress human immunodeficiency virus type 1 (HIV-1), a type of RNA virus, via targeting HIV-1 RNA, thus resulting in decreased viral gene expression [84].
Apart from directly targeting the genome of virus to achieve viral suppression, CRISPR/Cas system also functions to restrict viral infection via targeting the proteins that are essential for viral entry into the cells. For instance, Yang et al. have shown that blocking the N- and O-glycan biosynthesis using CRISPR/Cas9 system remarkably reduces viral entry into HEK293 cells because N- and O-glycans possess a major role in modulating viral entry [49]. Moreover, CRISPR/Cas9-mediated ablation of C-C chemokine receptor type 5 (CCR5), a major coreceptor for HIV-1 entry into human white blood cells, in long-term hematopoietic stem cells (HSCs) protects HSCs against HIV-1 infection [85].
As expected, several studies exhibit that CRISPR/Cas systems not only possess a role in inhibiting viral replication, but also harbor paramount roles in bacterial killing [86, 87]. For instance, Kiga et al. have developed a CRISPR/Cas13a-based antibacterial nucleocapsids, also referred to as CapsidCas13a(s), which enable sequence-specific killing of carbapenem-resistant Escherichia coli and methicillin-resistant Staphylococcus aureus through direct targeting the corresponding antimicrobial resistance genes [87]. Furthermore, a study has utilized conjugative plasmids as effective system to deliver CRISPR nucleases to bacteria, resulting in killing of the targeted bacteria [86]. In brief, these findings verify that CRISPR/Cas systems play key roles in suppressing viral replication and bacterial killing.
Gene therapy
Accumulating evidence proposes that CRISPR/Cas systems hold great potential in treating various types of inherited diseases including Duchenne muscular dystrophy (DMD), sickle cell disease (SCD), β-thalassemia, hemophilia A and hemophilia B, and recessive dystrophic epidermolysis bullosa (RDEB) [88, 89]. As an example, the CRISPR/Cas9-based technique can be applied to fix the exon 44 deletion mutations of the dystrophin gene, of which mutation causes DMD [90]. Furthermore, one recent study has employed he CRISPR/Cas9 system as gene-editing tools to target the enhancer region of B-cell lymphoma/leukemia 11 A (BCL11A) gene in hematopoietic stem and progenitor cells (HSPCs) to treat SCD [91]. Likewise, the CRISPR/Cas9 system is also capable of correcting the human hemoglobin beta (HBB) gene mutations, which are major causing agents of β-thalassemia [92]. Mutation of the factor VIII (F8) gene generally gives rise to monogenic disease Hemophilia A, of which symptoms can be ameliorated in mice through the CRISPR-based therapeutic treatment [93]. Furthermore, Morishige and his colleagues reveal that the CRISPR/Cas system can be employed to repair an in-frame deletion in exon 2 of the factor IX (F9) gene whose mutation is linked with hemophilia B [94]. CRISPR/Cas systems have also been demonstrated to harbor central roles in curing RDEB, a severe inherited skin disorder which is characterized by mutations in the COL7A1 gene that encodes type VII collagen (C7) [95].
Cancer Research
Mounting evidence reveals that CRISPR/Cas systems play essential roles in cancer research such as cancer modeling, cancer therapy and interrogation of cancer-related gene function. The following section provides examples of CRISPR/Cas systems involved in cancer investigations.
Cancer modeling
In the past decade, many research studies have indicated that CRISPR/Cas systems harbor significant roles in generating various types of cancer models including breast cancer, colorectal cancer, pancreatic cancer, lung cancer and liver cancer models. For example, a Cre-dependent Cas9 knockin mouse was generated and utilized to produce loss-of-function mutations in tumor suppressor genes as well as gain-of-function mutations in a proto-oncogene [96].This novel biotechnology allows for modeling the dynamics of multiple mutations in carcinogenesis. Similarly, the gene-editing CRISPR/Cas9 technology is also employed to target the tumor suppressor genes phosphatase and tensin homolog deleted on chromosome ten (PTEN) and protein 53 (P53), in order to generate liver cancer models [97]. Additionally, one study reveals that breast cancer organoids can be formed through knocking out four breast cancer-associated tumor suppressor genes (P53, PTEN, RB1, NF1) via the CRISPR/Cas9 system [98]. Furthermore, two independent studies have generated colorectal cancer model using the CRISRP/Cas9 gene editing system [99]. It has also been documented that CRISPR/Cas9 system is capable of generating pancreatic cancer model and Wilms’ tumor model [100].
Cancer therapy
The CRISPR/Cas system has provided a new avenue for cancer treatment. For instance, Dai et al. have established a telomerase-activating gene expression (Tage) system based upon the CRISPR/Cas9 gene-editing technology to effectively kill various types of cancer cells without affecting normal cells [101]. Moreover, knocking out of the programmed death-1 (PD-1) gene from T cells (PD- T cells) using the CRISPR/Cas9 genome editing tool displays higher capability than PD+ T cells to kill liver hepatocellular cells (HepG2), indicating that the CRISPR/Cas9 system possesses a critical role in improving the T-cell immunotherapy [102]. Additionally, suppressing the expression of lncRNA urothelial carcinoma-associated 1 (UCA1) remarkably inhibits cell proliferation, migration and invasion of bladder cancer cells [103].
Interestingly, one study reveals that knockout of macrophage signal regulatory protein-α (SIRP-α) by CRISPR/Cas9 system increases the phagocytic capabilities of the macrophages, which subsequently leads to enhanced phagocytosis of cancer cells [104]. Furthermore, the CRISPR/Cas9 system-mediated targeting of the exons of the human epidermal growth factor receptor 2 (HER2) gene, an extensively investigated oncogene, inhibits the proliferation of HER-2 positive breast cancer cells [105]. Similarly, selective targeting of kirsten rat sarcoma viral oncogene (KRAS) oncogenic alleles, whose mutations accounts for the induction of various kinds of cancers, by CRISRP/Cas9 greatly suppresses the proliferation of different types of cancer cell lines harboring KRAS mutations [106]. Due to the great potential of CRISPR biotechnology in cancer therapy, clinical trials for treating various types of cancer using CRISPR/Cas systems have been conducted in several counties including USA, China, UK and Australia. Some promising results have already been obtained from these clinical studies. For instance, the first-in-human Phase I CRISPR gene-editing trial in China for patients with refractory non-small-cell lung cancer shows that clinical application of CRISPR-Cas9 gene-edited T cells is generally safe and feasible [107]. However, the majority of these clinical trials are still ongoing and whether this novel technology can be widely adopted for cancer treatment in the future remains an open question.
Cancer-related gene function
Accumulating evidence suggests that CRISPR/Cas9 system plays prominent roles in exploring the biological functions of cancer-related genes. For instance, one study has described a novel CRISPR/Cas9-based method to characterize the function of a panel of tumor suppressor genes in well-established autochthonous mouse models of lung cancer. This finding implies that the CRISPR/Cas9-mediated genome editing technique holds great promise in probing the biological function of putative cancer genes [108]. Furthermore, Treuren et al. have indicated that deletions in the migration and invasion enhancer (MIEN1), an oncogene participated in promoting the motility of cancer cells, through the CRISPR/Cas system allow interrogation of the precise function of MIEN1 carried out in breast tumor metastasis [109]. A drug-inducible small guide RNA (sgRNA) CRISPR/Cas9 system has been developed to guide the efficiently and temporally controlled deletion of tumor-essential genes, thus allowing for the recognition of novel gene mutations that drive tumorigenesis [110]. In addition, loss of function mutation in Kelch-like ECH-associated protein 1 (KEAP1), which negatively regulates the nuclear factor erythroid 2-like 2 (NRF2), induced by CRISPR/Cas9 system activates the Nrf2 antioxidant pathway and cooperates with mutant KRAS to facilitate lung adenocarcinoma (LUAD) progression [111]. CRISPR/Cas9-mediated gene knockdown of homeobox protein NANOG (NANOG) and homeobox protein NANOGP8 (NANOGP8) remarkably reduce the malignant capabilities of prostate cancer cell lines. These results reveal that NANOG and NANOGP8 are indispensable factors for increasing the malignant potential of prostate cancer [112]. Last but not least, Phelps and his colleagues have demonstrated that nuclear receptor corepressors (NCORs) and histone deacetylases (HDACs) complex are major suppressors of differentiation in rhabdomyosarcoma using the CRISPR screen [113].
Other applications
Apart from examining cancer gene functions, CRISPR/Cas systems also play significant roles in screening of novel drug resistant genes. For instance, Wei et al. have identified phosphoglycerate dehydrogenase (PHGDH), which plays essential roles in the serine synthesis pathway, as a major cause for Sorafenib resistance in hepatocellular carcinoma [114]. Likewise, Hou et al. have employed a CRISPR-pooled library to discover unknown genes, of which knocked down contribute to the resistance of quizartinib (AC200) drug, which is used to treat acute myeloid leukemia disease [115]. Moreover, variants of tumor suppressor genes in mice can be rapidly mapped through the CRISPR screens using the autochthonous cancer model.
Discussion
As outlined here, the biological and biomedical applications of CRISPR/Cas systems have been intensively explored. In conclusion, it has been demonstrated that CRISPR/Cas systems are exploited as versatile tools that can be utilized to detect virus, kill antibiotic-resistant bacteria, edit genomic DNA, cure genetic diseases and so on. Moreover, it is encouraging that CRISPR/Cas systems also provide new strategies to combat cancer and function as promising therapeutic tools for cancer therapy.
Although CRISPR/Cas systems have displayed great potential in translating into therapeutic tools, the wide application of this novel biotechnology could be hampered by off-target effects, low editing efficiency, the lack of appropriate delivery methods in vivo and ethical issues of editing human genome directly. Thus, much effort has been taken to address these challenges by optimizing sgRNA design, improving the fidelity and specificity of Cas proteins, and improving viral CRISPR delivery and non-viral delivery methods. Continued biochemical and structural investigations to elucidate the mechanism by which Cas proteins interact with and cleave target DNA/RNA will provide invaluable insights to improve the specificity and efficiency of this unique gene-editing system. Furthermore, efforts aimed to overcome the challenges of the CRISPR/Cas system will facilitate its translation into clinic and guide future genetic diseases treatment, cancer therapy and clinical diagnosis.
Acknowledgements
Not applicable.
Author contributions
All authors contributed to the study conception and design. The manuscript was written by Y.Y. W and W.Q.Z. Literature search and figure preparation was performed by Y.Y. W and C. H. All authors read and approved the final manuscript.
Funding
The work in this study was supported by the China Postdoctoral Science Foundation (2020M683751), National Science Basic Research Plan in Shaanxi Province of China (2021JQ-559) and start-up funds from Xi’an University of Science and Technology (6310118015 and 2050217109).
Data Availability
Not applicable.
Declarations
Conflict of interest
The authors declare no competing or financial interests.
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Consent to participate
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ishino Y, Shinagawa H, Makino. K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorek R, Kunin V, Hugenholtz P. CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6(3):181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 3.Bolotin A, Quinquis B, Sorokin. A et al (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading). 151:2551–2561Pt 8 [DOI] [PubMed]
- 4.Shipman SL, Nivala J, Macklis JD, et al. Molecular recordings by directed CRISPR spacer acquisition. Science. 2016;353(6298):aaf1175. doi: 10.1126/science.aaf1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 6.Makarova KS, Haft DH, Barrangou. R, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sampson TR, Saroj SD, Llewellyn. AC, et al. Author Correction: A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2019;570(7760):E30–e31. doi: 10.1038/s41586-019-1253-9. [DOI] [PubMed] [Google Scholar]
- 8.Bhagwat AC, Patil AM, Saroj SD. CRISPR/Cas 9-Based Editing in the Production of Bioactive Molecules. Mol Biotechnol. 2022;64(3):245–251. doi: 10.1007/s12033-021-00418-4. [DOI] [PubMed] [Google Scholar]
- 9.Cring MR, Sheffield VC. Gene therapy and gene correction: targets, progress, and challenges for treating human diseases. Gene Ther. 2022;29(1–2):3–12. doi: 10.1038/s41434-020-00197-8. [DOI] [PubMed] [Google Scholar]
- 10.Kweon J, Yoon JK, Jang AH, et al. Engineered prime editors with PAM flexibility. Mol Ther. 2021;29(6):2001–2007. doi: 10.1016/j.ymthe.2021.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu YW, Dai XY, Wang. WT, et al. Dynamics and competition of CRISPR-Cas9 ribonucleoproteins and AAV donor-mediated NHEJ, MMEJ and HDR editing. Nucleic Acids Res. 2021;49(2):969–985. doi: 10.1093/nar/gkaa1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barman A, Deb B, Chakraborty S. A glance at genome editing with CRISPR-Cas9 technology. Curr Genet. 2020;66(3):447–462. doi: 10.1007/s00294-019-01040-3. [DOI] [PubMed] [Google Scholar]
- 13.Gootenberg JS, Abudayyeh OO, Lee JW et al (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 356:438–4426336 [DOI] [PMC free article] [PubMed]
- 14.Wessels HH, Méndez-Mancilla A, Guo. X, et al. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat Biotechnol. 2020;38(6):722–727. doi: 10.1038/s41587-020-0456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fajrial AK, He QQ, Wirusanti. NI et al (2020) A review of emerging physical transfection methods for CRISPR/Cas9-mediated gene editing. Theranostics. 10:5532–554912 [DOI] [PMC free article] [PubMed]
- 16.Giuliano CJ, Lin A, Girish. V, et al. Generating Single Cell-Derived Knockout Clones in Mammalian Cells with CRISPR/Cas9. Curr Protoc Mol Biol. 2019;128(1):e100. doi: 10.1002/cpmb.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith J, Grizot S, Arnould. S, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34(22):e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaj T, Sirk SJ, Shui. SL et al (2016) Genome-Editing Technologies: Principles and Applications. Cold Spring Harb Perspect Biol. 8(12) [DOI] [PMC free article] [PubMed]
- 19.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JC, Tan S, Qiao. G, et al. ).A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 21.Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung CY, McNeil MB, Cook GM. Utilization of CRISPR interference to investigate the contribution of genes to pathogenesis in a macrophage model of Mycobacterium tuberculosis infection. J Antimicrob Chemother. 2022;77(3):615–619. doi: 10.1093/jac/dkab437. [DOI] [PubMed] [Google Scholar]
- 23.Chua MJ, Collins J (2022) Rapid, Efficient, and Cost-Effective Gene Editing of Enterococcus faecium with CRISPR-Cas12a. Microbiol Spectr. 10:e02427211 [DOI] [PMC free article] [PubMed]
- 24.Huang H, Zheng G, Jiang. W, et al. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin (Shanghai) 2015;47(4):231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 25.Sachla AJ, Alfonso AJ, Helmann JD. A Simplified Method for CRISPR-Cas9 Engineering of Bacillus subtilis. Microbiol Spectr. 2021;9(2):e0075421. doi: 10.1128/Spectrum.00754-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh R, Chandel S, Ghosh. A, et al. Application of CRISPR/Cas System in the Metabolic Engineering of Small Molecules. Mol Biotechnol. 2021;63(6):459–476. doi: 10.1007/s12033-021-00310-1. [DOI] [PubMed] [Google Scholar]
- 27.Ming M, Ren Q, Pan. C, et al. CRISPR-Cas12b enables efficient plant genome engineering. Nat Plants. 2020;6(3):202–208. doi: 10.1038/s41477-020-0614-6. [DOI] [PubMed] [Google Scholar]
- 28.Shimatani Z, Kashojiya S, Takayama. M, et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35(5):441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- 29.Dong OX, Yu S, Jain. R, et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat Commun. 2020;11(1):1178. doi: 10.1038/s41467-020-14981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim YA, Moon H, Park CJ. CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae Rice (N Y) 2019;12(1):67. doi: 10.1186/s12284-019-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaillochet C, Develtere W, Jacobs TB. CRISPR screens in plants: approaches, guidelines, and future prospects. Plant Cell. 2021;33(4):794–813. doi: 10.1093/plcell/koab099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rana S, Aggarwal PR, Shukla. V, et al. Genome Editing and Designer Crops for the Future. Methods Mol Biol. 2022;2408:37–69. doi: 10.1007/978-1-0716-1875-2_3. [DOI] [PubMed] [Google Scholar]
- 33.Ran FA, Hsu PD, Wright. J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Zhu S, Cai. C, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509(7501):487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 35.Xu S, Zhan M, Jiang. C, et al. Genome-wide CRISPR screen identifies ELP5 as a determinant of gemcitabine sensitivity in gallbladder cancer. Nat Commun. 2019;10(1):5492. doi: 10.1038/s41467-019-13420-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seki A, Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J Exp Med. 2018;215(3):985–997. doi: 10.1084/jem.20171626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hart-Johnson S, Mankelow K. Archiving genetically altered animals: a review of cryopreservation and recovery methods for genome edited animals. Lab Anim. 2022;56(1):26–34. doi: 10.1177/00236772211007306. [DOI] [PubMed] [Google Scholar]
- 38.Terns MP. CRISPR-Based Technologies: Impact of RNA-Targeting Systems. Mol Cell. 2018;72(3):404–412. doi: 10.1016/j.molcel.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13. Science. 2017;358(6366):1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jing X, Xie B, Chen. L, et al. Implementation of the CRISPR-Cas13a system in fission yeast and its repurposing for precise RNA editing. Nucleic Acids Res. 2018;46(15):e90. doi: 10.1093/nar/gky433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abudayyeh OO, Gootenberg JS, Essletzbichler. P et al (2017) ).RNA targeting with CRISPR-Cas13. Nature. 550:280–2847675 [DOI] [PMC free article] [PubMed]
- 42.Konermann S, Lotfy P, Brideau. NJ et al (2018) Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. 173:665–676e6143 [DOI] [PMC free article] [PubMed]
- 43.Zhao J, Li B, Ma. J et al (2020) ).Photoactivatable RNA N(6) -Methyladenosine Editing with CRISPR-Cas13. Small. 16:e190730130 [DOI] [PubMed]
- 44.Wang H, Nakamura M, Abbott. TR, et al. CRISPR-mediated live imaging of genome editing and transcription. Science. 2019;365(6459):1301–1305. doi: 10.1126/science.aax7852. [DOI] [PubMed] [Google Scholar]
- 45.Maass PG, Barutcu AR, Shechner. DM, et al. Spatiotemporal allele organization by allele-specific CRISPR live-cell imaging (SNP-CLING) Nat Struct Mol Biol. 2018;25(2):176–184. doi: 10.1038/s41594-017-0015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelles DA, Fang MY, O’Connell. MR et al (2016) Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell. 165:488–4962 [DOI] [PMC free article] [PubMed]
- 47.Dreissig S, Schiml S, Schindele. P, et al. Live-cell CRISPR imaging in plants reveals dynamic telomere movements. Plant J. 2017;91(4):565–573. doi: 10.1111/tpj.13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C, Lu T, Emanuel. G, et al. Imaging-based pooled CRISPR screening reveals regulators of lncRNA localization. Proc Natl Acad Sci U S A. 2019;116(22):10842–10851. doi: 10.1073/pnas.1903808116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Q, Hughes TA, Kelkar. A et al (2020) Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. Elife. 9. [DOI] [PMC free article] [PubMed]
- 50.Sun NH, Chen DY, Ye. LP, et al. CRISPR-Sunspot: Imaging of endogenous low-abundance RNA at the single-molecule level in live cells. Theranostics. 2020;10(24):10993–11012. doi: 10.7150/thno.43094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu X, Ying Y, Mao. S, et al. Live-Cell Imaging of Genomic Loci Using CRISPR/Molecular Beacon Hybrid Systems. Methods Mol Biol. 2020;2166:357–372. doi: 10.1007/978-1-0716-0712-1_21. [DOI] [PubMed] [Google Scholar]
- 52.Ma H, Tu LC, Naseri. A, et al. CRISPR-Sirius: RNA scaffolds for signal amplification in genome imaging. Nat Methods. 2018;15(11):928–931. doi: 10.1038/s41592-018-0174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mao S, Ying Y, Wu. X, et al. CRISPR/dual-FRET molecular beacon for sensitive live-cell imaging of non-repetitive genomic loci. Nucleic Acids Res. 2019;47(20):e131. doi: 10.1093/nar/gkz752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta D, Bhattacharjee O, Mandal. D, et al. CRISPR-Cas9 system: A new-fangled dawn in gene editing. Life Sci. 2019;232:116636. doi: 10.1016/j.lfs.2019.116636. [DOI] [PubMed] [Google Scholar]
- 55.Yu JSL, Yusa K (2019) Genome-wide CRISPR-Cas9 screening in mammalian cells. Methods. 164–165:29–35 [DOI] [PubMed]
- 56.Jeng EE, Bhadkamkar V, Ibe NU, et al. Systematic Identification of Host Cell Regulators of Legionella pneumophila Pathogenesis Using a Genome-wide CRISPR Screen. Cell Host Microbe. 2019;26(4):551–563e556. doi: 10.1016/j.chom.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benaoudia S, Martin A, Puig M, Gamez, et al. A genome-wide screen identifies IRF2 as a key regulator of caspase-4 in human cells. EMBO Rep. 2019;20(9):e48235. doi: 10.15252/embr.201948235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng H, Castillo-Cabrera J, Manser. M et al (2019) Genome-wide CRISPR screening reveals genetic modifiers of mutant EGFR dependence in human NSCLC. Elife. 8 [DOI] [PMC free article] [PubMed]
- 59.Cai P, Otten ABC, Cheng. B, et al. A genome-wide long noncoding RNA CRISPRi screen identifies PRANCR as a novel regulator of epidermal homeostasis. Genome Res. 2020;30(1):22–34. doi: 10.1101/gr.251561.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu S, Li W, Liu. J et al (2016) Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 34:1279–128612 [DOI] [PMC free article] [PubMed]
- 61.Arroyo JD, Jourdain AA, Calvo. SE, et al. A Genome-wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation. Cell Metab. 2016;24(6):875–885. doi: 10.1016/j.cmet.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bakke J, Wright WC, Zamora. AE, et al. Genome-wide CRISPR screen reveals PSMA6 to be an essential gene in pancreatic cancer cells. BMC Cancer. 2019;19(1):253. doi: 10.1186/s12885-019-5455-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bassaganyas L, Popa SJ, Horlbeck. M, et al. New factors for protein transport identified by a genome-wide CRISPRi screen in mammalian cells. J Cell Biol. 2019;218(11):3861–3879. doi: 10.1083/jcb.201902028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J, Rajan SS, Friedrich. MJ, et al. Genome-Scale CRISPRa Screen Identifies Novel Factors for Cellular Reprogramming. Stem Cell Reports. 2019;12(4):757–771. doi: 10.1016/j.stemcr.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park RJ, Wang T, Koundakjian. D, et al. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat Genet. 2017;49(2):193–203. doi: 10.1038/ng.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dukhovny A, Lamkiewicz K, Chen. Q et al (2019) A CRISPR Activation Screen Identifies Genes That Protect against Zika Virus Infection. J Virol. 93(16) [DOI] [PMC free article] [PubMed]
- 67.Lane RK, Guo H, Fisher. AD, et al. Necroptosis-based CRISPR knockout screen reveals Neuropilin-1 as a critical host factor for early stages of murine cytomegalovirus infection. Proc Natl Acad Sci U S A. 2020;117(33):20109–20116. doi: 10.1073/pnas.1921315117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tian S, Liu Y, Wu. H, et al. Genome-Wide CRISPR Screen Identifies Semaphorin 6A and 6B as Receptors for Paeniclostridium sordellii Toxin TcsL. Cell Host Microbe. 2020;27(5):782–792e787. doi: 10.1016/j.chom.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sidik SM, Huet D, Ganesan. SM, et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell. 2016;166(6):1423–1435e1412. doi: 10.1016/j.cell.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fozouni P, Son S, M. Díaz de León Derby. et al (2021) Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 184(2): 323–333.e329 [DOI] [PMC free article] [PubMed]
- 71.Wang S, Li H, Kou. Z et al (2020) Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the PCR-based CRISPR-Cas13a system. Clin Microbiol Infect [DOI] [PubMed]
- 72.Barnes KG, Lachenauer AE, Nitido. A, et al. Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time. Nat Commun. 2020;11(1):4131. doi: 10.1038/s41467-020-17994-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen J, Zhou X, Shan. Y, et al. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat Commun. 2020;11(1):267. doi: 10.1038/s41467-019-14135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang T, Zhou W, Lin. X, et al. Light-up RNA aptamer signaling-CRISPR-Cas13a-based mix-and-read assays for profiling viable pathogenic bacteria. Biosens Bioelectron. 2021;176:112906. doi: 10.1016/j.bios.2020.112906. [DOI] [PubMed] [Google Scholar]
- 75.Abbott TR, Dhamdhere G, Liu. Y, et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell. 2020;181(4):865–876e812. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng Q, Bai L, Zheng. S, et al. Efficient inhibition of duck hepatitis B virus DNA by the CRISPR/Cas9 system. Mol Med Rep. 2017;16(5):7199–7204. doi: 10.3892/mmr.2017.7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Noor S, Rasul A, Iqbal. MS, et al. Inhibition of Hepatitis B Virus with the Help of CRISPR/Cas9 Technology. Crit Rev Eukaryot Gene Expr. 2020;30(3):273–278. doi: 10.1615/CritRevEukaryotGeneExpr.2020028453. [DOI] [PubMed] [Google Scholar]
- 78.Yin D, Ling S, Wang. D et al (2021) Targeting herpes simplex virus with CRISPR-Cas9 cures herpetic stromal keratitis in mice. Nat Biotechnol [DOI] [PMC free article] [PubMed]
- 79.Hübner A, Petersen B, Keil. GM, et al. Efficient inhibition of African swine fever virus replication by CRISPR/Cas9 targeting of the viral p30 gene (CP204L) Sci Rep. 2018;8(1):1449. doi: 10.1038/s41598-018-19626-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu H, Soyars CL, Li. J, et al. CRISPR/Cas9-mediated resistance to cauliflower mosaic virus. Plant Direct. 2018;2(3):e00047. doi: 10.1002/pld3.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hagag IT, Wight DJ, Bartsch. D, et al. Abrogation of Marek’s disease virus replication using CRISPR/Cas9. Sci Rep. 2020;10(1):10919. doi: 10.1038/s41598-020-67951-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ling K, Yang L, Yang. N et al (2020) Gene Targeting of HPV18 E6 and E7 Synchronously by Nonviral Transfection of CRISPR/Cas9 System in Cervical Cancer. Hum Gene Ther. 31:297–3085–6 [DOI] [PubMed]
- 83.Li H, Wang S, Dong. X, et al. CRISPR-Cas13a Cleavage of Dengue Virus NS3 Gene Efficiently Inhibits Viral Replication. Mol Ther Nucleic Acids. 2020;19:1460–1469. doi: 10.1016/j.omtn.2020.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yin L, Zhao F, Sun. H et al (2020) CRISPR-Cas13a Inhibits HIV-1 Infection. Mol Ther Nucleic Acids. 21:147–155 [DOI] [PMC free article] [PubMed]
- 85.Xu L, Yang H, Gao. Y, et al. CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo. Mol Ther. 2017;25(8):1782–1789. doi: 10.1016/j.ymthe.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hamilton TA, Pellegrino GM, Therrien. JA, et al. Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat Commun. 2019;10(1):4544. doi: 10.1038/s41467-019-12448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiga K, Tan XE, Ibarra-Chávez. R, et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat Commun. 2020;11(1):2934. doi: 10.1038/s41467-020-16731-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han JP, Kim M, Choi. BS et al (2022) In vivo delivery of CRISPR-Cas9 using lipid nanoparticles enables antithrombin gene editing for sustainable hemophilia A and B therapy. Sci Adv. 8:eabj69013 [DOI] [PMC free article] [PubMed]
- 89.Wu SS, Li QC, Yin. CQ, et al. Advances in CRISPR/Cas-based Gene Therapy in Human Genetic. Dis Theranostics. 2020;10(10):4374–4382. doi: 10.7150/thno.43360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Y, Li H, Min. YL, et al. Enhanced CRISPR-Cas9 correction of Duchenne muscular dystrophy in mice by a self-complementary AAV delivery system. Sci Adv. 2020;6(8):eaay6812. doi: 10.1126/sciadv.aay6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frangoul H, Altshuler D, Cappellini MD, et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N Engl J Med. 2021;384(3):252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 92.Gabr H, El Ghamrawy MK, Almaeen. AH, et al. CRISPR-mediated gene modification of hematopoietic stem cells with beta-thalassemia IVS-1-110 mutation. Stem Cell Res Ther. 2020;11(1):390. doi: 10.1186/s13287-020-01876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen H, Shi M, Gilam. A, et al. Hemophilia A ameliorated in mice by CRISPR-based in vivo genome editing of human Factor VIII. Sci Rep. 2019;9(1):16838. doi: 10.1038/s41598-019-53198-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morishige S, Mizuno S, Ozawa. H, et al. CRISPR/Cas9-mediated gene correction in hemophilia B patient-derived iPSCs. Int J Hematol. 2020;111(2):225–233. doi: 10.1007/s12185-019-02765-0. [DOI] [PubMed] [Google Scholar]
- 95.Jacków J, Guo Z, Hansen. C, et al. CRISPR/Cas9-based targeted genome editing for correction of recessive dystrophic epidermolysis bullosa using iPS cells. Proc Natl Acad Sci U S A. 2019;116(52):26846–26852. doi: 10.1073/pnas.1907081116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Platt RJ, Chen S, Zhou. Y, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xue W, Chen S, Yin. H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514(7522):380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dekkers JF, Whittle JR, Vaillant. F, et al. Modeling Breast Cancer Using CRISPR-Cas9-Mediated Engineering of Human Breast Organoids. J Natl Cancer Inst. 2020;112(5):540–544. doi: 10.1093/jnci/djz196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roper J, Tammela T, Cetinbas. NM, et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol. 2017;35(6):569–576. doi: 10.1038/nbt.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jo N, Sogabe Y, Yamada. Y, et al. Platforms of in vivo genome editing with inducible Cas9 for advanced cancer modeling. Cancer Sci. 2019;110(3):926–938. doi: 10.1111/cas.13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dai W, Xu X, Wang. D, et al. Cancer therapy with a CRISPR-assisted telomerase-activating gene expression system. Oncogene. 2019;38(21):4110–4124. doi: 10.1038/s41388-019-0707-8. [DOI] [PubMed] [Google Scholar]
- 102.Lu S, Yang N, He. J, et al. Generation of Cancer-Specific Cytotoxic PD-1(-) T Cells Using Liposome-Encapsulated CRISPR/Cas System with Dendritic/Tumor Fusion Cells. J Biomed Nanotechnol. 2019;15(3):593–601. doi: 10.1166/jbn.2019.2712. [DOI] [PubMed] [Google Scholar]
- 103.Zhen S, Hua L, Liu. YH et al (2017) Inhibition of long non-coding RNA UCA1 by CRISPR/Cas9 attenuated malignant phenotypes of bladder cancer. Oncotarget. 8:9634–96466 [DOI] [PMC free article] [PubMed]
- 104.Ray M, Lee YW, Hardie. J, et al. ).CRISPRed Macrophages for Cell-Based Cancer Immunotherapy. Bioconjug Chem. 2018;29(2):445–450. doi: 10.1021/acs.bioconjchem.7b00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang H, Sun W. CRISPR-mediated targeting of HER2 inhibits cell proliferation through a dominant negative mutation. Cancer Lett. 2017;385:137–143. doi: 10.1016/j.canlet.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 106.Lee W, Lee JH, Jun. S, et al. Selective targeting of KRAS oncogenic alleles by CRISPR/Cas9 inhibits proliferation of cancer cells. Sci Rep. 2018;8(1):11879. doi: 10.1038/s41598-018-30205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lu Y, Xue J, Deng. T, et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat Med. 2020;26(5):732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 108.Sánchez-Rivera FJ, Papagiannakopoulos T, Romero. R, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516(7531):428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Treuren T, Vishwanatha JK. CRISPR deletion of MIEN1 in breast cancer cells. PLoS One. 2018;13(10):e0204976. doi: 10.1371/journal.pone.0204976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aubrey BJ, Kelly GL, Kueh. AJ, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10(8):1422–1432. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 111.Romero R, Sayin VI, Davidson. SM, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med. 2017;23(11):1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kawamura N, Nimura K, Nagano. H, et al. CRISPR/Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget. 2015;6(26):22361–22374. doi: 10.18632/oncotarget.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Phelps MP, Bailey JN, Vleeshouwer-Neumann T, et al. CRISPR screen identifies the NCOR/HDAC3 complex as a major suppressor of differentiation in rhabdomyosarcoma. Proc Natl Acad Sci U S A. 2016;113(52):15090–15095. doi: 10.1073/pnas.1610270114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wei L, Lee D, Law. CT, et al. Genome-wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for Sorafenib resistance in HCC. Nat Commun. 2019;10(1):4681. doi: 10.1038/s41467-019-12606-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hou P, Wu C, Wang. Y, et al. A Genome-Wide CRISPR Screen Identifies Genes Critical for Resistance to FLT3 Inhibitor AC220. Cancer Res. 2017;77(16):4402–4413. doi: 10.1158/0008-5472.CAN-16-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.