Abstract
Background:
The ability of the human immunodeficiency virus type 1 (HIV-1) to persist in anatomic compartments and cellular reservoirs is a major obstacle for eradication of replicationcompetent virus in the infected host.
Approach:
We extensively review recent advancements in phylogenetic and phylogeographic techniques that provide a unique opportunity for studies of intra-host HIV-1 compartmentalization and the detection of potential reservoirs.
Conclusion:
We show that infected macrophages in the central nervous system (CNS) harbor viral subpopulations that play a key role in the emergence of escape variants and viral rebound following discontinuation of antiretroviral therapy. An HIV cure, therefore, cannot be achieved without the effective targeting of the virus in the CNS, for which in depth knowledge of viral population dynamics contributing to the development and maintenance of latent reservoirs is critical.
Keywords: CNS, phylogenetics, phylogeography, HIV-1, reservoir, SIV, phyloanatomy
1. EVIDENCE OF CELLULAR AND ANATOMICAL HIV-1 RESERVOIRS
1.1. HIV-1 Reservoirs and the Challenge of Eradication
Thirty years after the discovery of HIV-1 as the causative agent of AIDS, a cure has yet to be found. Highly active anti-retroviral therapy (HAART) has significantly reduced AIDS-related morbidity and mortality, with improved regimens that are more potent, have fewer side effects, and lower medicinal burden. Unfortunately, treatment does not fully restore human health but simply prolongs the onset of this destructive disease and, if interrupted, invariably leads to viral rebound even in patients with low-to-undetectable plasma viremia [1–3]. In addition to lifelong treatment, replacing certain drugs within the HAART regimen has also become eminent owing to the emergence of drug resistant viral variants over the course of infection [4]. The identification and quantification of critical sources of lowlevel viremia and viral rebound, as well as the emergence of drug-resistant variants in the face of treatment has been a well-studied area of research since the development of HAART and has expanded with recent advancements in phylogenetic and sequencing techniques. This research has contributed significantly to our understanding of the complexity of HIV-1 infection and replication and, importantly, the ability of the virus to evolve and adapt to both the host immune response and antiretroviral therapy.
HIV-1 latency was initially attributed to proviral DNA in resting memory CD4+ T cells [5], which have since been shown to contribute to low-level persistent viremia during HAART, emergence of antiviral escape variants, and viral rebound after treatment interruption [6–14]. These cells do not release infectious virus in the resting state but can do so following cellular activation, which can occur under a variety of conditions [5, 6]. Virus production in latently infected cells, occurring in the absence of new rounds of infection of surrounding cells [15–18], is typical of viral reservoirs, defined as “a cell type or anatomical site in association with which a replication-competent form of the virus persists with more stable kinetics than the main pool of actively replicating virus” [19, 20].
Several studies have indicated that in addition to resting naïve and memory CD4+ T cells, viral reservoirs include, but may not be limited to, a recently discovered immature memory T cell population with stem-cell like properties, termed CD4+ T memory stem cells [21–23], as well as monocytes/macrophages [24], tumor-associated macrophages [25], and astrocytes [24]. Follicular dendritic cells are an additional, unique reservoir in that they contribute to viral persistence by trapping virus particles on the cell surface, rather than harboring latent provirus, and this interaction can remain stable for as long as nine months [26, 27]. Another potential reservoir is gut mucosa, although its contribution to viral rebound after treatment interruption seems to be limited [28]. On the other hand, persistence of HIV-1-infection in the spleen [29] has been shown to contribute to viral rebound in patients with incomplete suppression by HAART [9]. Moreover, the ability of the virus to switch from the use of the CCR5 co-receptor for entry to CXCR4 in ~50% of patients [30] has been attributed to increasing replication in resting, naïve CD4+ T cells [31] and subsets of CXCR4-expressing macrophages [32], although higher affinity for the CD4 receptor has specifically been associated with macrophage tropism, as macrophages display a lower CD4 surface density [20]. The timing of the switch from CCR5 to CXCR4 is particularly important for CCR5 antagonist monotherapies used in developing countries, as the re-emergence of pre-treatment CXCR4-using variants has been reported [33].
It has been shown that viral sequences obtained from circulating CD4+ T cells during low-level viremia and viral rebound post-therapy withdrawal are phylogenetically distinct from circulating cell-free virus in plasma [34], as well as viral sequences from monocytes [35]. Such findings not only implicate monocytes and/or additional reservoir cell populations in the low-level production of virus but also indicate that viral reservoirs are characterized by specific evolutionary dynamics due to their ability to remain hidden from both synthetic and host-mediated antiviral response.
1.2. HIV-1 Compartmentalization and Metapopulation Structure
Although difficult to sample in humans, several anatomic “compartments” have been identified that can harbor tissue-specific HIV-1 subpopulations phylogenetically distinct from those circulating in peripheral plasma or other tissues [36]. According to the accepted definition, an anatomic compartment is “a site for which there is limited exchange of viral genetic information with other sites” [20]. Moreover, if the compartment is characterized by suboptimal free drug concentrations, it is referred to as an anatomical sanctuary [20, 37, 38]. Since the first demonstration that genetic differences exist between blood- and brain-derived viral sequences [39], a large proportion of viral compartment studies have focused on the central nervous system (CNS). The CNS is often considered an “immune privileged” site due to its low-density T cell population and weak adaptive immune response attributed to the blood brain barrier (BBB) and blood cerebrospinal fluid (CSF) barrier, which restrict migration of certain cells and other materials [40]. This also renders the CNS a potential sanctuary [41], where poor drug penetration owing to tightly regulated anatomical barriers plays a major role in limiting drug efficacy [42].
CNS compartmentalization is likely the result of founder effects, either during primary or late infection, and tissue-specific selective pressures that shape distinct viral populations, followed by restricted viral trafficking [7, 43–49]. HIV-1 production in the CNS mainly occurs in perivascular macrophages and macrophage-like microglia [50, 51], and has been associated with both the emergence of drug resistance [52, 53] and AIDS-related neuropathology [54, 55], a spectrum of disorders collectively known as HIV-associated neurocognitive disorders (HAND), ranging from asymptomatic to HIV-associated dementia (HAD) [56]. Subjects with and without HAD often harbor viral populations in the CSF that are genetically distinct from virus in the blood [57–62] and exhibit characteristics of macrophage/microglia tropism [63], in which case both latency characterized by low-level replication and anatomical isolation may contribute to the formation of a reservoir during primary infection [64, 65].
The compartmentalization of the virus in the CNS exemplifies HIV-1 metapopulation structure (i.e. division in distinct sub-populations) within different cells and anatomic sites of the infected host. Indeed, tissue-specific viral populations have been detected within female genital tract [44, 66–68], male foreskin [69] and semen [70]. Latent viral forms associated with compartmentalization within the female genital tract and/or semen and foreskin of the male may be important in transmission, since they appear to be preferentially transmitted over more recent variants [71–74]. Limited compartmentalization has also been observed in breast milk [75–77], gut mucosa [78], and lung tissues [79]. The existence of distinct intra-host viral subpopulations has been confirmed by studies using the SIV-macaque model, exhibit compartmentalization in spleen and secondary lymph nodes during primary and early chronic infection [80] as well as in the genital tract [81].
2. PHYLOGENETIC METHODS OF DETERMINING VIRAL COMPARTMENTS AND POTENTIAL RESERVOIRS
2.1. Testing Viral Compartmentalization
The phenomenon of HIV-1 compartmentalization that can ultimately give rise to a reservoir is probably the result of local conditions affecting viral evolution at specific anatomical sites limiting viral trafficking and/or exerting pressures that alter the fitness landscape of the viral population [82, 83]. Compartmentalization can be inferred in a phylogenetic tree of viral sequences amplified from different tissues and/or cell types of an infected subject by the presence of a separate monophyletic clade including all or most of the sequences from a specific site [84]. In addition, sequences sampled from a viral reservoir are expected to lack temporal structure – i.e. sequences sampled at different time points during the infection will intermix since they persist in the reservoir – and show less mean divergence from the most recent common ancestor (MRCA), i.e. the root node of the tree, than other sequences in the tree [36]. Confidence for the monophyletic clades is usually assessed through the use of bootstrapping [85] or Bayesian posterior probabilities estimated by Markov chain Monte Carlo (MCMC) methods [86–88]. Bootstrapping is a technique based on the evaluation of distance based (e.g. neighbor-joining) or maximum likelihood trees inferred from alignment replicates, usually 500 to 1000, built by randomly sampling (with replacement) sites from the original alignment. A bootstrap value represents the proportion of trees in which a particular group of sequences clustered together, with values >80% considered as a significant support for the cluster. Alternatively, Bayesian posterior probabilities are inferred from a posterior distribution of possible phylogenies obtained using an MCMC algorithm that incorporates prior knowledge of evolutionary model parameters [89]. Monophyletic clades present in >90% of the posterior distribution of trees are considered significantly supported.
Phylogenetic analysis of HIV/SIV sequences sampled from post mortem brain tissues and longitudinal plasma samples and/or peripheral tissues has shown a clear-cut example of compartmentalization in the CNS, wherein all brain-derived sequences cluster within a distinct and well-supported monophyletic clade (Fig. 1) [48, 54, 55, 90]. However, real data sets may also display partial compartmentalization, characterized by a certain degree of intermixing between sequences obtained from different cell or tissue types, such as, for example, sequences derived from different anatomic sites in the brain (Fig. 1). Therefore, quantitative methods for assessing compartmentalization have been developed that take into account both intracompartmental genetic distances and distances from sequences outside of the proposed compartment (refer to Zárate et al., 2007 [91] for a detailed review). The most commonly used methods can be divided into two main categories: (1) tree-based and (2) distance-based methods. Three well-known tree-based methods are the SlatkinMaddison (SM) test [92], Tree Correlation Coefficients (TCC) [93], and Simmonds Association Index (SAI) [94]. The SM test determines the minimum number of migration events between pre-assigned compartments (i.e. group of viral sequences amplified from different sites) consistent with the topology of the phylogenetic tree and based on the maximum parsimonious reconstruction of the tissue/cell type of origin of ancestral sequences (i.e. the internal nodes of the tree). Statistical support is based on the comparison of observed migrations with a null distribution of randomly intermixed sequences (panmictic population). Significantly less migrations than 99.9% of the null distribution indicate compartmentalization. TCCs (r, and rb) represent correlated distances between two sequences in a phylogenetic tree with the information about whether or not they were isolated from the same compartment. The distance between two sequences can be either the number of tree branches separating the sequences (rb) or the cumulative genetic distance between the sequences (r). Statistical significance is achieved by estimating the distribution of these coefficients by permuting sequences between compartments. The SAI assesses the degree of population structure in the phylogenetic tree by weighting the contribution of each internal node based on its depth in the tree and evaluating the significance of the observed value using a bootstrap sample both over the population structure and tree topology. Distance-based methods include Wright’s measure of population subdivision (FST) [44, 95, 96], nearest neighbor statistic (Snn) [97], and analysis of molecular variance (AMOVA) [98]. FST compares the mean pairwise genetic distance between two sequences sampled from different pre-assigned compartments to the mean distance between sequences sampled from the same compartment, in which statistical significance is derived via population-structure randomization. Snn is simply a measure of how often the nearest neighbors of each sequence are isolated from the same or different assigned compartments. AMOVA calculates an association based on the genetic diversity of the sequences between and within compartments and is an extension of FST, in which the distances are restricted to Euclidean and the variability is calculated from the sum of the squared distances between the sequences.
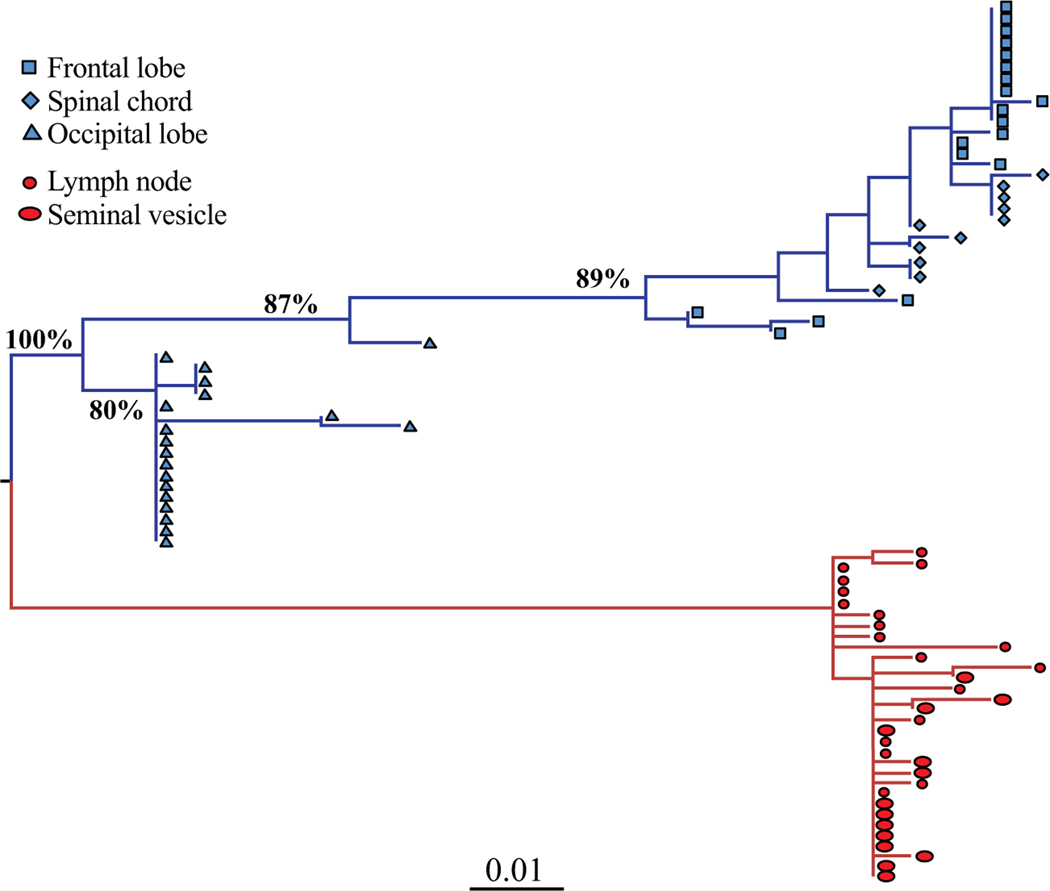
Fig. (1). HIV-1 maximum likelihood (ML) tree of brain and peripheral tissue derived sequences from a patient with HIV-associated dementia.
HIV-1 gp120 sequences were amplified from tissues sampled at necropsy and are a subset of the sequences described in Salemi et al. (2005) [54]. The ML tree was inferred with the HKY+G model. Values along the branches represent bootstrap support (500 replicates); only values >80% are shown. Branch lengths are scaled in nucleotide substitutions per site according to the scale bar at the bottom. Sequences from different brain (frontal lobe, spinal chord, occipital lobe) and peripheral (lymph node, seminal vesicle) tissues are indicated in blue and red, respectively, and by different symbols according to the legend in the figure. Brain sequences are completely compartmentalized (bootstrap support 100%). Sub-compartmentalization of individual CNS as well as peripheral tissues is also evident, although not all supported by bootstrapping.
Comparison of different compartmentalization tests based on 92 separate HIV-1 data sets and 1,500 simulated data sets not surprisingly resulted in disagreement between tree-based and distance-based methods [91]. Tree-based methods are expected to be more robust, since they include phylogenetic information otherwise ignored. However, they can also be misleading if branches are unresolved and have low support, since in such cases they lead to an overestimation of topological distance, or degree of segregation, between sequences. Furthermore, each method can be affected by sampling bias, for example underrepresentation of sequences from a particular compartment, which is often the case for difficult-to-sample anatomical locations prior to autopsy, such as the brain, or sites/cell populations with low-level replication, such as peripheral monocytes. The largest change (>70%) in the proportion of sequences classified as compartmentalized with skewed sampled size was observed for the TCC and SM tests, with SAI next in line (56%), indicating an increased effect of sampling underrepresentation on the power of tree-based over distance-based compartmentalization detection methods. Also, as with other phylogenetic analyses, recombination can greatly bias the results. Introduction of recombinant sequences results in reduced power, particularly for tree-based methods, to detect compartmentalization due to increased intermixing between clades of the tree, thereby increasing phylogenetic uncertainty. Finally, since the development and improvement of sequencing methods using single genome amplification (SGA) [99–103], comparisons of SGA with sequencing methods that rely on earlier PCR/cloning techniques have shown that the latter can introduce significant errors that influence the degree of diversity and thus compartmentalization [100, 104], although the general validity of this conclusion has been questioned [105]. In summary, given the potential limitations of each method, compartmentalization should always be tested using more than one algorithm and discrepancies should be evaluated by taking into account the specific assumptions of each test in relation with the actual data set under investigation.
2.2. Phyloanatomy
As discussed above, restricted exchange of genetic information with other sites is the defining property of a viral compartment. Recently, Bayesian phylogeography methods, originally developed to study the spread of viral epidemics [106, 107], have been successfully applied to the study of viral trafficking between different anatomical sites and cell populations, also referred to as viral gene flow or phyloanatomy [49, 108a]. Phyloanatomy can provide significant insights into the establishment and maintenance of potential viral reservoirs, and it is based on the incorporation of spatial information (sampled tissue) into the evolutionary analysis of an organism that is evolving according to a population structure, or coalescent, model. Earlier implementations of phyloanatomy based on maximum parsimony have also been utilized for the investigation of HIV-1 intra-host evolution [48], and have been described elsewhere [108b-110], but are not the focus of this review.
By using the Bayesian phylogeography framework implemented in the BEAST software package [111, 112], information on intra-host migration rates, origin and timing of specific migration events, as well as the number of migrations (“Markov jumps”) and time spent in a given location (“Markov rewards”) can be inferred from an alignment of heterochronous sequences from various anatomical locations [113]. Such migrations, supported in the form of Bayesian posterior probabilities, are interpreted as the probability that sequences from one anatomical location gave rise to, or shared ancestral history with, sequences in another location at a given point in time. Although seemingly complex, the use of the continuous-time Markov Chain (CTMC) model to trace transitions between discrete (or continuous) geographical traits along branches of a phylogenetic tree is analogous to the use of the general time-reversible nucleotide substitution model of transitions between nucleotide character traits along the same tree, both of which are inherent viral evolutionary processes naturally embedded in the genealogy.
Unfortunately, like other epidemiological studies, even for a moderately small number of spatial locations, most migration events might not even be sampled. Therefore, recent improvements of the Bayesian phylogeographic methodology, using hierarchical modeling, have incorporated joint analysis of paralleled migration processes, such as similar data sets from multiple patients, for a more statistical approach to gene flow analysis. Referred to as the Bayesian hierarchical phylogenetic model (HPM), this framework allows for a more accurate interpretation of summary data [114] than would be possible for consensus [115, 116] or combined [117, 118] approaches. Essentially, viral sequence data from multiple subjects can be used in a single analysis, akin to a strict combined approach, while simultaneously allowing for different phylogenetic parameters in the individual paralleled partitions, e.g. patients, as in the consensus approach. Results from the individual partitions are combined to provide overall summaries of each parameter, such as migration rates. The overall model and individual partition models are fitted simultaneously, enabling the overall model to feed back information, in the form of a prior, into the estimation of the individual paralleled partitions. This feedback mechanism results in a more precise estimate of each evolutionary parameter. Details of the application of HPM methodology to HIV-1 phylogeography and phyloanatomy are outlined in Cybis et al. (2013) [107]. Although the study assessed viral gene flow only between CD8+ and CD4+ T cell compartments, the application of this statistical method to phyloanatomy has the potential to greatly impact future investigations of HIV-1 compartmentalization and reservoir dynamics.
2.3. Analyzing the Temporal Structure of Longitudinally Sampled Viral Sequences
Compartmentalization and restricted viral migration are necessary but not sufficient conditions to infer the presence of a viral reservoir. The flow of virus and/or infected cells between compartments can mask low-level viral persistence, as is the case for the memory CD4+ T-cell population in lymphoid tissues during chronic infection [11, 80]. Mannioui et al. (2009) used molecular biology techniques to detect in vivo viral replication during low-level viremia, represented by the presence of episomal DNA, indicative of recent infection [119] and, thus, persistent viral replication within a population. However, evidence is also needed of the contribution of a potential reservoir to the reemergence of archival variants [120] and/or viral rebound that could be determined based on shared ancestry between sequences during rebound and sequences obtained prior to treatment withdrawal. From a phylogenetic perspective, the clustering pattern of a viral reservoir would be characterized by lack of significant cumulative genetic divergence between sequences sampled at different time points [36]. To which extent real phylogenies differ from perfectly temporally structured trees – representing the progressive increase in viral diversity and divergence from the MRCA during the course of the infection [121, 122] – can be accurately quantified by the temporal structure statistic, defined between 0 (no temporal structure) and 1(perfect temporal structure) [123]. In particular, the clustering of sequences sampled late in infection with earlier ones would reduce temporal structure and indicate the reemergence of latent viruses (Fig. 2). Similar to the previously mentioned SM test of compartmentalization, topological temporal structure can be evaluated by measuring the extent of clustering whereby sequences are categorized as discrete states corresponding to sampling time, rather than anatomical location [123]. The number of state changes within the inferred phylogeny is then compared to that of a null distribution to assess statistical significance.
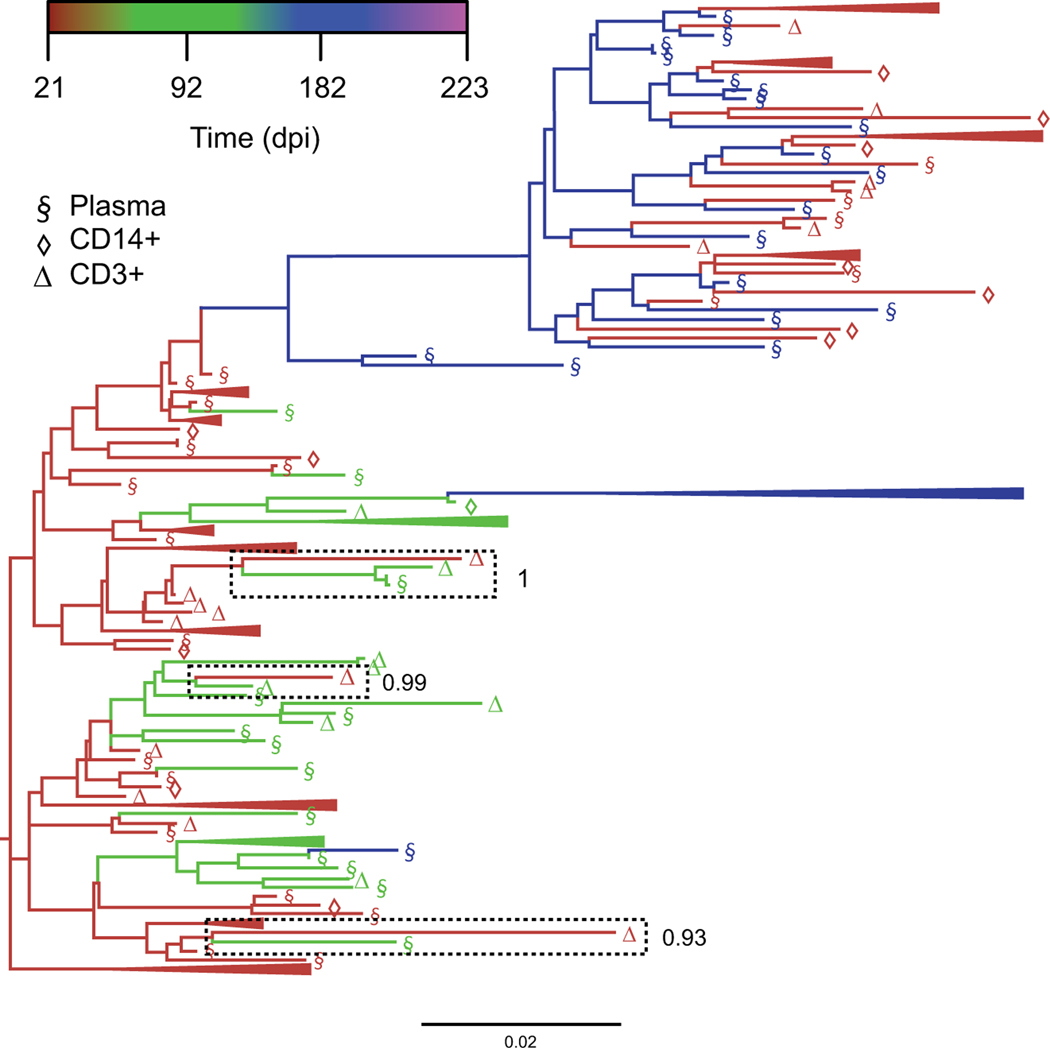
Fig. (2). Bayesian maximum clade credibility (MCC) tree highlighting reduced intra-host temporal structure.
The coalescent framework implemented in BEAST [111, 112] was used for a serially sampled SIV-infected macaque dataset to obtain a posterior distribution of trees scaled in substitutions/site, from which the MCC tree was derived using Tree Annotator [112]. Branches are colored according to sampling time point in days post-infection (dpi), with temporally clustered sequences (>3) collapsed for illustrative purposes. Three separate instances of shared ancestry between sequences sampled relatively far apart (92 and 223 dpi) have been highlighted, with corresponding clade posterior probabilities reported to the right for which the temporal states have been randomized. The final result is referred to as the temporal clustering statistic, ranging from 0 (absence of temporal structure) and 1 (perfect temporal structure).
2.4. Intra-Host Evolutionary Rate Variation
In addition to temporal structure, analyzing evolutionary rate variation among phylogenetic lineages can be used to investigate temporal signal. In the case of latent virus reactivation, shared direct ancestry (clustering) between sequences sampled during early and late infection will significantly reduce the evolutionary rate during that period of time [84, 124]. In contrast, migration of virus between anatomical sanctuaries may increase the evolutionary rate. Both incidences act to reduce the ‘clock-like’ signal associated with the molecular clock hypothesis, which assumes a linear increase in the number of substitutions with time along all phylogenetic lineages [125]. Temporal signal can be quantified using a simple regression of root-to-tip genetic distances in the phylogeny against sampling time (Fig. 3) with the program Path-O-Gen (v1.3; available from http://tree.bio.ed.ac.uk/). Path-O-Gen-derived R2 values indicate ‘clock-likeness’ of the sequence data, wherein high R2 values (>0.5) are indicative of sequences that evolve under a strict or relaxed molecular clock.
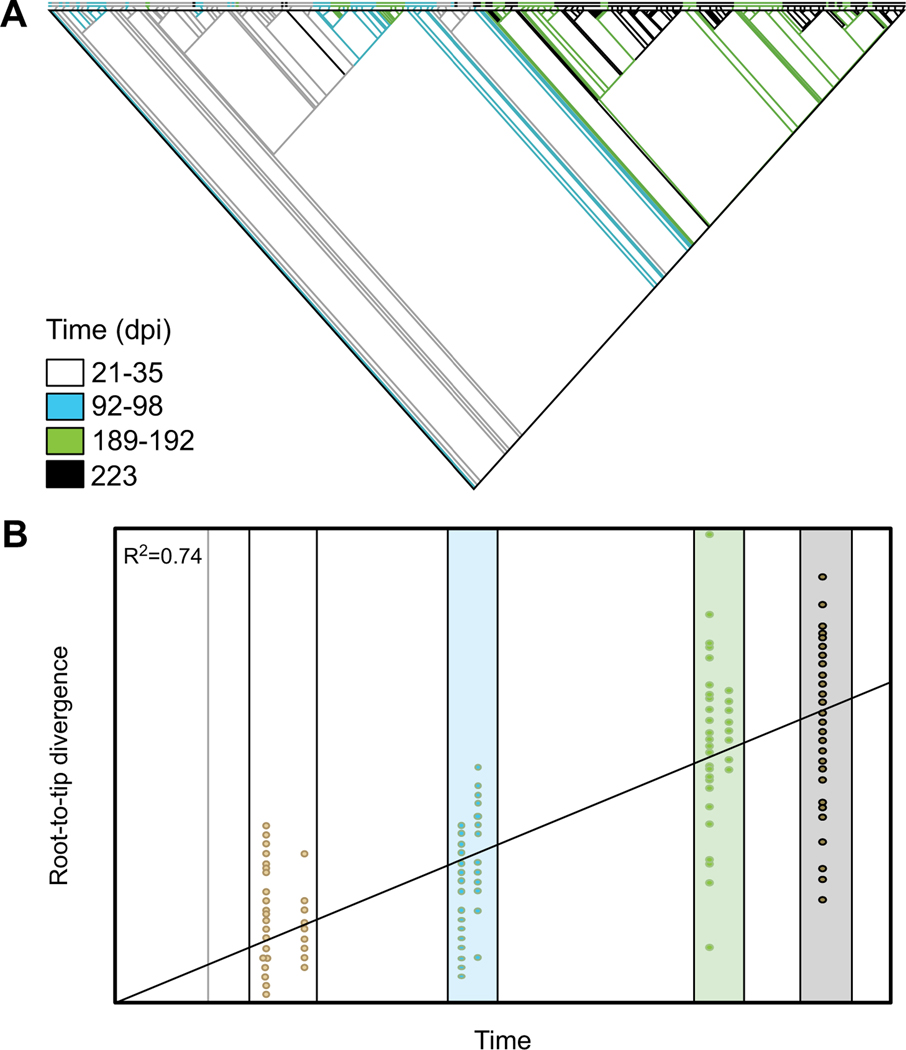
Fig. (3). Result of temporal signal analysis in Path-O-Gen.
(A) A maximum likelihood tree was inferred using RaxML for serially sampled sequences from a SIV-infected macaque dataset. Branches are colored according to sampling time, with interior branches treated as temporal ancestral states inferred using maximum parsimony in MacClade (available from http://macclade.org). (B) A linear regression model was used to measure the temporal signal, or “clocklikeness,” in Path-O-Gen for root-to-tip genetic divergence (y-axis) for each sequence (yellow dot) in the phylogeny above against sampling time (x-axis). Colored boxes again correspond to sampling time for visual comparison.
Immomen & Leitner (2014) have recently developed a method to quantify the impact of potential reservoirs on viral evolutionary rates by identifying lineages within a serially sampled phylogeny that evolve at a significantly lower rate than at least one other lineage in the tree [126]. The method assumes that HIV mutates according to a Poisson process with a single molecular clock rate when replicating but does not mutate when latent. For sequence pairs that share a common ancestry, latency is calculated based on the Poisson probability of accumulating the number of mutations observed on the shorter lineage dShort, given the evolutionary rate of the longer lineage dLong. Thus, latency is defined when the shorter lineage provides a Pr[(λk)/(k!e-λ)] ≤ 0.05, where λ=dLong*L and k<=dShort*L for sequence length L. However, because hypermutation, recombination, and selection can have substantial effect on evolutionary rates, sequences experiencing these evolutionary events should be removed prior to analysis and only 3rd codon positions analyzed.
3. RECENT EVIDENCE OF CNS AS A KEY VIRAL RESERVOIR
Each of the phylogenetic methods described above is not sufficient alone to identify with certainty a viral reservoir. Additionally, over- or under-sampling of sequences from specific cell types or tissues, as well as the number and time intervals of available longitudinal sequences, can greatly influence the results. Experimental design and optimization of sampling schemes is crucial for any phylogeny-based investigation of viral reservoirs [124]. However, when properly designed phylogenetic analyses are used in combination to and complemented with studies of viral replication kinetics within specific tissues and/or cell populations, they can be powerful tools for the advancement of reservoir investigation and drug treatment strategies.
Such an approach, applied to the studies of HIV/SIV brain infection and neuroAIDS, has recently provided several lines of evidence that the CNS, along with its cellular components, are a key viral reservoir, actively contributing to both the re-seeding of peripheral virus and emergence of drug resistance. For example, parenchymal-derived sequences from patients diagnosed with HAD display reduced evolutionary rates [55], which may be explained by the low-level persistent expression of unintegrated viral DNA in CNS-resident macrophages for prolonged periods of time (at least 30 days) [127]. Levels of unintegrated viral DNA relative to the integrated form are 10-fold elevated in brain of patents with HAD [128, 129], and CNS-resident macrophage populations that harbor latent, integrated forms of viral DNA (perivascular macrophages and microglia) have relatively low turnover rates, ranging from months to years. As previously mentioned, anatomical barriers such as the BBB and blood-CSF barrier inhibit viral gene flow and drug penetration, resulting in isolated, or compartmentalized, viral replication. Viral compartmentalization in the CNS may be a critical phenomenon from the standpoint of neurological impairment, but migration of the virus, although limited, in and out of this compartment is of more importance when considering the reservoir potential of the CNS. Clear evidence of reseeding peripheral tissues by CNS-derived viral variants was found for HIV-1-infected patients, as it can be inferred from the emergence of peripheral sequences from CNS-associated ancestral nodes in Bayesian genealogical reconstructions (Fig. 4) [55]. Furthermore, use of the well-characterized SIVmac251-infected rhesus macaque model of neuroAIDS demonstrated that this recirculation was not simply limited to terminal illness but can also happen during early stages of infection (Fig. 4) [49]. Evolution of drug resistance has also been identified in the CNS compartment of treated patients [51, 53], which poses a significant challenge to treatment optimization, since specific antiretroviral regimens are based on resistance testing of circulating plasma virus.
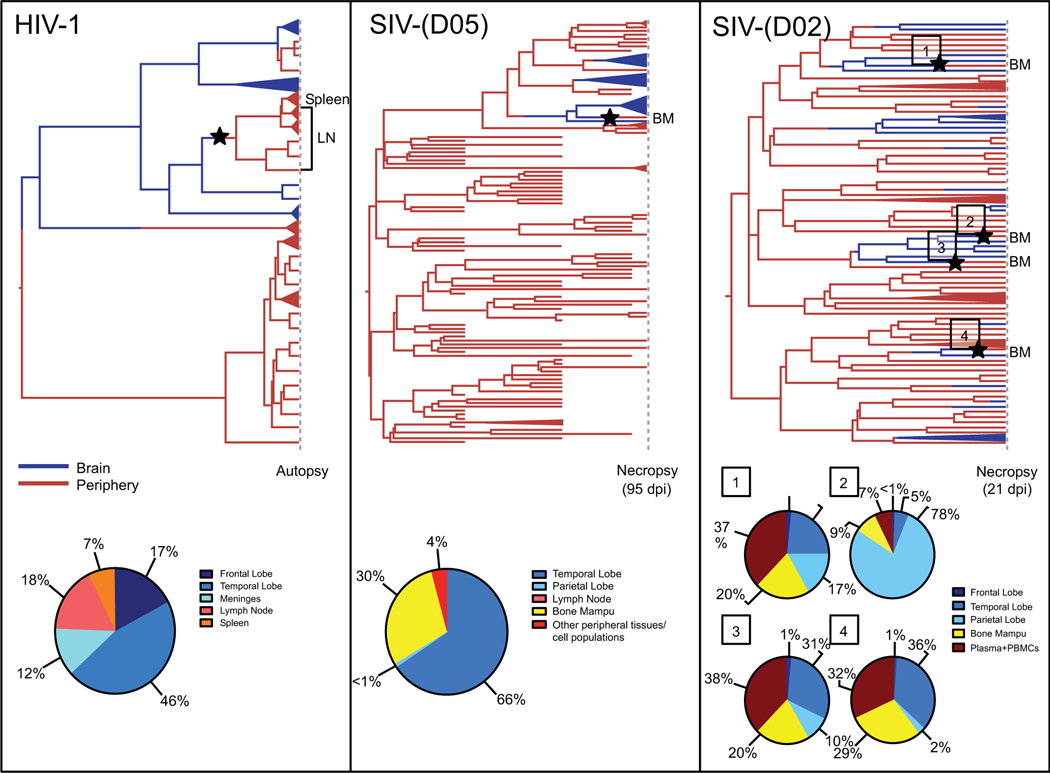
Fig. (4). Bayesian maximum clade credibility (MCC) trees with highlighted viral migration events from brain into circulation and estimated ancestral state posterior probabilities using phyloanatomy algorithms.
The phylogeographic framework implemented in the BEAST package [111, 112] was used to obtain a posterior distribution of trees from which MCC trees were derived using Tree Annotator [112]. The trees in the figure include sequences from an HIV-1-infected patient (analyzed in Lamers et al. (2010) [55] and reanalyzed here with the Bayesian phylogeography framework (left panel)) and sequences from two SIV-infected CD8+ lymphocyte-depleted macaques (middle and right panels) [49]. Branches are colored according to the sampled tissue/cell location (exterior) or highest posterior probability location of the reconstructed ancestral sequence (interior), with blue representing brain-derived sequences and red representing sequences derived from peripheral tissues or blood. Peripheral blood mononuclear cells (PBMCs) and cell populations in the SIV trees refer to flow cytometry-sorted CD3+ lymphocyte and CD14+ monocyte cell populations. Collapsed branches represent monophyletic clades consisting of sequences from the same tissue/cell type and sampling time point. Cross-sectional sequences from the HIV-1-infected patient (left panel) and one of the SIV-infected, CD8+ lymphocyte-depleted macaques (right panel) were taken at autopsy/necropsy, the time in days post-infection (dpi) depicted for the macaque. Longitudinal sequences were derived from the macaque depicted in the middle panel prior to necropsy following euthanization due to the onset of SAIDS [49]. Stars represent inferred viral entry into the periphery from the brain and are located at the midpoint of the branches along which these events occur. The posterior probability, expressed as a percent of total posterior probability, of the ancestral state at the preceding node is reported at the bottom for each migration event. Ancestral state posterior probabilities can be observed using FigTree (available from http://tree.bio.ed.ac.uk/software/figtree/).
Based on the accessibility of CSF relative to parenchymal brain tissue, researchers have proposed that sampling of viral sequences from the CSF could provide genetic information on viral populations in the brain reservoir as early as primary infection [64, 65]. However, other studies have indicated that CSF viral sequences are not representative of the viral population dynamics observed in the parenchyma [130, 131], wherein individual parenchymal lobe tissue may harbor distinct viral sub-populations [48, 54]. Moreover, extensive sequence analyses in SIV-infected macaques have identified heterogeneous populations within the CNS, defined also by genetically distinct subcompartments, such as meninges and parenchyma [49], basal ganglia, cerebellum, and hippocampus [132] (Fig. 1). Although subpopulations of virus found in the CSF are likely derived from regions within the brain, trafficking of virus between brain parenchyma compartments and peripheral blood is not well understood. This lack of knowledge, together with evidence of the complexity of viral genetic composition within the brain not accurately mirrored in the CSF underlines the significant limitation of using sequences from the CSF to characterize the CNS reservoir to a sufficient depth or pre-screen for drug-resistant variants within tissue sanctuaries. Therefore, although phylogenetic methods have identified the CNS as a key viral reservoir and potentially crucial barrier to treatment and eradication, more studies are urgently needed to improve our understanding of the contribution of individual CNS components in HIV reservoir establishment and maintenance.
4. FUTURE DIRECTIONS
The past few years have witnessed significant advances in the development of novel and powerful phylogenetic methods applicable to the study of intra-host viral compartments and reservoirs of fast evolving viruses, such as SIV/HIV [25, 48, 49, 54] and HCV [133, 134], and made clear the central role played by CNS infection in HIV persistence and rebound. The use of such analysis tools in AIDS research, however, is limited by obvious ethical restraints. HIV evolutionary dynamics of deep tissues are extremely difficult to investigate in vivo since the required sampling often impractical, when not unethical. In addition, most of the findings mentioned in this review highlight the importance of investigating the establishment of viral reservoirs during primary infection, which is difficult to detect in humans. On the other hand, non-human primate models provide a viable alternative. SIV infection of rhesus macaques (Macaca mulatta) is one of the most widely used models for HIV-related pathogenesis studies [135]. Infected macaques exhibit similar clinical manifestations as HIV-1 infection, albeit on a shorter time scale of approximately one to three years [136]. More importantly, they offer the unique opportunity for a wide range of infected tissue and cell population sampling that is critical to HIV-1 reservoir studies and can be performed in the presence or absence of treatment. Much can, and still needs to be, learned from this model with respect to viral latency and the contribution of viral evolutionary and population dynamics to reservoir formation.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health funding grants R01 NS063897.
DISCLAIMER: The above article has been published in Epub (ahead of print) on the basis of the materials provided by the author. The Editorial Department reserves the right to make minor modifications for further improvement of the manuscript.
Footnotes
CONFLICT OF INTEREST
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
REFERENCES
- [1].Davey R T Jr, Bhat N, Yoder C, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci USA 1999; 96: 15109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Garcia F, Plana M, Vidal C, et al. Dynamics of viral load rebound and immunological changes after stopping effective antiretroviral therapy. AIDS 1999; 13: F79–86. [DOI] [PubMed] [Google Scholar]
- [3].Mata RC, Viciana P, de Alarcon A, et al. Discontinuation of antiretroviral therapy in patients with chronic HIV infection: clinical, virologic, and immunologic consequences. AIDS Patient Care Stds 2005; 19: 550–62. [DOI] [PubMed] [Google Scholar]
- [4].International AIDS Society Scientific Working Group on HIV Cure, Deeks SG, Autran B et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 2012; 12: 607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med 1995; 1: 1284–90. [DOI] [PubMed] [Google Scholar]
- [6].Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997; 387: 183–8. [DOI] [PubMed] [Google Scholar]
- [7].Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997; 278: 1295–300. [DOI] [PubMed] [Google Scholar]
- [8].Wong JK, Ignacio CC, Torriani F, Havlir D, Fitch NJ, Richman DD. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol 1997; 71: 2059–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Siliciano JD, Kajdas J, Finzi D, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 2003; 9: 727–8. [DOI] [PubMed] [Google Scholar]
- [10].Zhang L, Chung C, Hu B, et al. Genetic characterization of rebounding HIV-1 after cessation of highly active anti-retroviral therapy. J Clin Invest 2000; 106: 839–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Furtado MR, Callaway DS, Phair JP, et al. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N Engl J Med 1999; 340: 1614–22. [DOI] [PubMed] [Google Scholar]
- [12].Josefsson L, Von Stockenstrom S, Faria NR, et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci USA 2013; 110: E4987–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bacchus C, Cheret A, Avettand-Fenoël V, et al. A single HIV-1 cluster and a skewed immune homeostasis drive the early spread of HIV among resting CD4+ cell subsets within one month post-infection. PLoS ONE 2013; 8: e64219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schrager LK & D’Souza MP. Cellular and anatomical reservoirs of HIV-1 in patients receiving potent antiretroviral combination therapy. JAMA 1998; 1: 67–71. [DOI] [PubMed] [Google Scholar]
- [15].Perelson AS, Essunger P, Cao Y, et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 1997; 8: 387: 188–91. [DOI] [PubMed] [Google Scholar]
- [16].Redel L, Le Douce V, Cherrier T, et al. HIV-1 regulation of latency in the monocyte-macrophage lineage and in CD4+ T lymphocytes. J Leukoc Biol 2010; 87: 575–88. [DOI] [PubMed] [Google Scholar]
- [17].Battistini A, Sgarbanti M. HIV-1 latency: An update of molecular mechanisms and therapeutic strategies. Viruses 2014; 6: 1715–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Traylen CM, Patel HR, Fondaw W, et al. Virus reactivation: a panoramic view in human infections. Future Virol 2011; 6: 451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Blankson JN, Persaud D, Siliciano RF. The challenge of viral reservoirs in HIV-1 infection. Annu Rev Med 2002; 53: 557–93. [DOI] [PubMed] [Google Scholar]
- [20].Eisele E, Siliciano RF. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 2012; 37: 377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gattinoni L, Lugli E, Ji Y, et al. A human memory T-cell subset with stem cell-like properties. Nat Med 2011; 17: 1290–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cieri N, Camisa B, Cocchiarella F, et al. IL-7 and IL-15 instruct the generation of human memory stem T cells from naive precursors. Blood 2013; 121: 573–84. [DOI] [PubMed] [Google Scholar]
- [23].Buzon MJ, Sun H, Li C, et al. HIV-1 persistence in CD4+-T cells with stem-like properties. Nat Med 2014; 20: 139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kumar A, Abbas W, Herbein G. HIV-1 latency in monocytes/macrophages. Viruses 2014; 6: 1837–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Salemi M, Lamers SL, Huysentruyt LC, et al. Distinct patterns of HIV-1 evolution within metastatic tissues in patients with NonHodgkins Lymphoma. PLoS ONE 2009; 4: e8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Keele BF, Tazi L, Gartner S, et al. Characterization of the follicular dendritic cell reservoir of human immunodeficiency virus type 1. J Virol 2008; 82: 5548–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Smith BA, Gartner S, Liu Y, et al. Persistence of infectious HIV on follicular dendritic cells. J Immunol 2001; 166: 90–6. [DOI] [PubMed] [Google Scholar]
- [28].Lerner P, Guadalupe M, Donovan R, et al. The gut mucosal viral reservoir in HIV-infected patients is not the major source of rebound plasma viremia following interruption of highly active antiretroviral therapy. J Virol 2011; 85: 4772–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Delassus S, Cheynier R, Wain-Hobson S. Nonhomogeneous distribution of human immunodeficiency virus type 1 proviruses in the spleen. J Virol 1992; 66: 5642–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol 1999; 17: 657–700. [DOI] [PubMed] [Google Scholar]
- [31].Zamarchi R, Allavena P, Borsetti A, et al. Expression and functional activity of CXCR-4 and CCR-5 chemokine receptors in human thymocytes. Clin Exp Immunol 2002; 127: 321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA 1999; 96: 5215–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Westby M, Lewis M, Whitcomb J, et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol 2006; 80: 4909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sahu GK, Paar D, Frost SDW, Smith MM, Weaver S, Cloyd MW. Low-level plasma HIVs in patients on prolonged suppressive highly active antiretroviral therapy are produced mostly by cells other than CD4 T-cells. J Med Virol 2009; 81:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fulcher JA, Hwangbo Y, Zioni R, et al. Compartmentalization of human immunodeficiency virus type 1 between blood monocytes and CD4+ T cells during infection. J Virol 2004; 78: 7883–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nickle DC, Jensen MA, Shriner D, et al. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol 2003; 77: 5540–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ramratnam B, Ribeiro R, Chung C, et al. Intensification of antiretroviral therapy accelerates the decay of the HIV-1 latent reservoir and decreases, but does not eliminate, ongoing virus replication. J Acquir Immune Defic Syndr 2004; 35: 33–7. [DOI] [PubMed] [Google Scholar]
- [38].Chun TW, Nickle DC, Justement JS, et al. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J Clin Invest 2005; 115: 3250–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Korber BT, Kunstman KJ, Patterson BK, et al. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol 1994; 68: 7467–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ousman SS, Kubes P. Immune surveillance in the central nervous system. Nature Neuro 2012; 15: 1096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Varatharajan L, Thomas SA. The transport of anti-HIV drugs across blood-CNS interfaces: Summary of current knowledge and recommendations for further research. Antiviral Res 2009; 82: A99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Katlama C, Deeks SG, Autran B, et al. Barriers to a cure: New concepts in targeting and eradicating HIV-1 reservoirs. Lancet 2014; 381: 10.1016/S0140-6736(13)601040X. [DOI] [Google Scholar]
- [43].Zhang L, Rowe L, He T, et al. Compartmentalization of surface envelope glycoprotein of human immunodeficiency virus type 1 during acute and chronic infection. J Virol 2002; 76: 9465–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kemal KS, Foley B, Burger H, et al. HIV-1 in genital tract and plasma of women: compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc Natl Acad Sci USA 2003; 100: 12972–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Crotty S, Lohman BL, Lu FX, Tang S, Miller CJ, Andino R. Mucosal immunization of cynomolgus macaques with two serotypes of live poliovirus vectors expressing simian immunodeficiency virus antigens: Stimulation of humoral, mucosal, and cellular immunity. J Virol 1999; 73: 9485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gallichan WS, Rosenthal KL. Long-lived cytotoxic T lymphocyte memory in mucosal tissues after mucosal but not systemic immunization. J Exp Med 1996; 184: 1879–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shacklett B, Cu-Uvin S, Beadle T, et al. Quantification of HIV-1-specific T-cell responses at the mucosal cervicovaginal surface. AIDS 2000; 14: 1911–5. [DOI] [PubMed] [Google Scholar]
- [48].Lamers SL, Gray RR, Salemi M, Huysentruyt LC, McGrath M. HIV-1 phylogenetic analysis shows HIV-1 transits through the meninges to brain and peripheral tissues. Infect Genet Evol 2011; 11: 31–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Strickland SL, Rife BD, Lamers SL, et al. Spatiotemporal dynamics of simian immunodeficiency virus brain infection in CD8+ lymphocyte-depleted rhesus macaques with neuro AIDS. J Gen Virol 2014; 95: 2784–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Burdo T, Lackner A, Williams KC. Monocyte/macrophages and their role in HIV neuropathogenesis. Immunol Rev 2013; 254: 102–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Williams KC, Corey S, Westmoreland SV, et al. Perivascular macrophages are the primary cell type productively infected by simian immunodeficiency virus in the brains of macaques: implications for the neuropathogenesis of AIDS. J Exp Med 2001; 193: 905–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Strain MC, Letendre S, Pillai SK, et al. Genetic composition of human immunodeficiency virus type 1 in cerebrospinal fluid and blood without treatment and during failing antiretroviral therapy. J Virol 2005; 79:1772–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Smit TK, Brew BJ, Tourtellotte W, Morgello S, Gelman BB, Saksena NK. Independent evolution of human immunodeficiency virus (HIV) drug resistance mutations in diverse areas of the brain in HIV-infected patients, with and without dementia, on antiretroviral treatment. J Virol 2004; 78: 10133–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Salemi M, Lamers SL, Yu S, de Oliveira T, Fitch WM, McGrath MS. Phylodynamic analysis of human immunodeficiency virus type 1 in distinct brain compartments provides a model for the neuropathogenesis of AIDS. J Virol 2005; 79: 11343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lamers SL, Salemi M, Galligan DC, et al. Human immunodeficiency virus-1 evolutionary patterns associated with pathogenic processes in the brain. J Neurovirol 2010; 16: 230–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Clifford DB, Ances BM. HIV-associated neurocognitive disorder. Lancet Inf Dis 2013; 13: 976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Schnell G, Spudich S, Harrington P, Price RW, Swanstrom R. Compartmentalized human immunodeficiency virus type 1 originates from long-lived cells in some subjects with HIV-1associated dementia. PLoS Pathog 2009; 5: e1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Schnell G, Price RW, Swanstrom R, Spudich S. Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J Virol 2010; 84: 2395–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pillai SK, Kosakovsky-Pond SL, Good BM, et al. Genetic attributes of cerebrospinal fluid-derived HIV-1 env. Brain 2006; 129: 1872–83. [DOI] [PubMed] [Google Scholar]
- [60].Dunfee RL, Thomas ER, Gorry PR, et al. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc Natl Acad Sci USA 2006; 41: 15160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Falcone EL, Adegbulugbe AA, Sheikh V, et al. Cerebrospinal fluid HIV-1 compartmentalization in a patient with AIDS and acute varicella-zoster virus meningomyeloradiculitis. Clin Infect Dis 2013; 57: e135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ohagen A, Devitt A, Kunstman KJ, et al. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J Virol 2003; 77: 12336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog 2011; 7: e1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Harrington PR, Schnell G, Letendre SL, et al. Cross-sectional characterization of HIV-1 env compartmentalization in cerebrospinal fluid over the full disease course. AIDS 2009; 23: 907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sturdevant CB, Joseph SB, Schnell G, Price RW, Swanstrom R, Spudich S. Compartmentalized replication of R5 T cell-tropic HIV1 in the central nervous system early in the course of infection. PLoS Pathog 2015; 11: e1004720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Philpott S, Burger H, Tsoukas C, Foley B, Anastos K, Kitchen C, Weiser B. Human immunodeficiency virus type 1 genomic RNA sequences in the female genital tract and blood: compartmentalization and intrapatient recombination J Virol 2004; 79: 353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Andreoletti L, Skrabal K, Perrin V, et al. Genetic and phenotypic features of blood and genital viral populations of clinically asymptomatic and antiretroviral-treatment-naive clade A human immunodeficiency virus type 1-infected women. J Clin Microbiol 2007; 45: 1838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Poss M, Rodrigo A, Gosink JJ, et al. Evolution of envelope sequences fro the genital tract and peripheral blood of women infected with Clade A human immunodeficiency virus type 1. J Virol 1998; 72: 8240–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Galiwango RM, Lamers SL, Redd AD, et al. HIV type 1 genetic variation in foreskin and blood from subjects in Rakai Uganda. AIDS Res Hum Retrov 2012; 28: 729–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Delwart EL, Mullins JI, Gupta P, et al. Human immunodeficiency virus type 1 populations in blood and semen J Virol 1998; 72: 617–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Vrancken B, Rambaut A, Suchard MA, et al. The genealogical population dynamics of HIV-1 in a large transmission chain: bridging within and among host evolutionary rates. PLoS Comput Biol 2014; 10: e1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lythgoe KA, Fraser C. New insights into the evolutionary rate of HIV-1 at the within-host and epidemiological levels. Proc Biol Sci 2012; 279: 3367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Redd AD, Collinson-Streng AN, Chatziandreou N, et al. Previously transmitted HIV-1 strains are preferentially selected during subsequent sexual transmissions. J Infect Dis 2012; 206: 1433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Alizon S, Fraser C. Within-host and between-host evolutionary rates across the HIV-1 genome. Retrovirol 2013; 10: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Gantt S, Carlsson J, Heath L, et al. Genetic analysis of HIV-1 env sequences demonstrate limited compartmentalization in breast milk and suggest viral replication within the breast that increases with mastitis. J Virol 2010; 84: 10812–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Salazar-Gonzalez JF, Salazar MG, Learn GH, et al. Center for HIV/AIDS Vaccine Immunology A0167854. Origin and evolution of HIV-1 in breast milk determined by single-genome amplification and sequencing. J Virol 2011; 85: 2751–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Gray RR, Salemi M, Lowe A, et al. Goodenow MM Multiple independent lineages of HIV-1 persist in breast milk and plasma. AIDS 2011; 25: 143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].van Marle G, Gill MJ, Kolodka D, McManus L, Grant T, Church DL. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirol 2007; 4: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Heath L, Fox A, McClure J, et al. Evidence for limited genetic compartmentalization of HIV-1 between lung and blood. PLoS One 2009; 4: e6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Manniou A, Bourry O, Sellier P, et al. Dynamics of viral replication in blood and lymphoid tissues during SIVmac251 infection of macaques. Retrovirol 2009; 6: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Whitney JB, Hraber PT, Luedemann C, et al. Genital tract sequestration of SIV following acute infection. PLoS Pathog 2011; 7: e1001293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kovacs A, Wasserman SS, Burns D, et al. Determinants of HIV-1 shedding in the genital tract of women. Lancet 2001; 358: 1593601. [DOI] [PubMed] [Google Scholar]
- [83].Patterson B, Landay A, Andersson J, et al. Repertoire of chemokine receptor expression in the female genital tract: implications for human immunodeficiency virus transmission. Am J Pathol 1998; 153: 481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Salemi M. The intra-host evolutionary and population dynamics of human immunodeficiency virus type 1: a phylogenetic perspective. Inf Dis Rep 2013; 5: s1e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39: 783–91. [DOI] [PubMed] [Google Scholar]
- [86].Li S, Pearl DK, Doss H. Phylogenetic tree construction using Markov chain Monte Carlo. J Am Stat Assoc 2000; 95: 493. [Google Scholar]
- [87].Mau B, Newton MA, Larget B. Bayesian phylogenetic inference via Markov chain Monte Carlo methods. Biometrics 1999; 55: 112. [DOI] [PubMed] [Google Scholar]
- [88].Yang Z, Ranna B. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Mol Biol Evol 1997; 14: 717–24. [DOI] [PubMed] [Google Scholar]
- [89].Lemey P, Salemi M, Vandamme A. The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing. 2nd ed. Cambridge University Press; 2009. [Google Scholar]
- [90].Shapshak P, Segal DM, Crandall KA, et al. Independent evolution of HIV type 1 in different brain regions. AIDS Res Hum Retroviruses 1999; 15: 811–20. [DOI] [PubMed] [Google Scholar]
- [91].Zárate S, Kosakovsky-Pond SL, Shapshak P, Frost SDW. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J Virol 2007; 81: 6643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 1989; 123: 603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Critchlow DE, Li S, Nourijelyani K, Pearl DK. Some statistical methods for phylogenetic trees with application to HIV disease. Math Comput Model 200; 32: 69–81. [Google Scholar]
- [94].Wang TH, Donaldson YK, Brettle RP, Bell JE, Simmonds P. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J Virol 2001; 75: 11686–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Hudson R, Boos D, Kaplan N. A statistical test for detecting geographic subdivision. Mol Biol Evol 1992a; 9: 138–51. [DOI] [PubMed] [Google Scholar]
- [96].Hudson R, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics 1992b; 132: 583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hudson RR. A new statistic for detecting genetic differentiation. Genetics 2000; 155: 2011–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 1992; 131: 479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Butler DM, Pacold ME, Jordan PS, Richman DD, Smith DM. The efficiency of single genome amplification and sequencing is improved by quantitation and use of a bioinformatics tool. J Virol Methods 2009; 162: 280–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Palmer S, Kearney M, Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol 2005; 43: 406–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Shriner D, Rodrigo AG, Nickle DC, Mullins JI. Pervasive genomic recombination of HIV-1 in vivo. Genetics 2004; 167: 1573–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Simmonds P, Balfe P, Ludlam CA, Bishop JO, Brown AJ. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol 1990; 64: 5840–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Simmonds P, Balfe P, Peutherer JF, Ludlam CA, Bishop JO, Brown AJ. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol 1990; 64: 864–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom, R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci USA 2011; 108: 20166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Jordan MR, Kearney M, Palmer S, et al. Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. J Virol Methods 2010; 168: 114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comp Biol 2009; 5: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Cybis GB, Sinsheimer JS, Lemey P, Suchard MA. Graph hierarchies for phylogeography. Philos Trans R Soc Lond B Biol Sci 2013; 368: 20120206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].(a) Shirreff G, Garcia V, Vanderford TH, Silvestri G, Regoes RR. The phyloanatomy of early SIV infection. In: Mathematical and Computational Evolutionary Biology; 2013. May 27-31; Saint Martin de Londres, France. [Google Scholar]; (b) Knowles L & Maddison W. Statistical phylogeography. Mol Ecol 2002; 11: 2623–35. [DOI] [PubMed] [Google Scholar]
- [109].Slatkin M. Detecting small amounts of gene flow from phylogenies of alleles. Genetics 1989; 121: 609–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Templeton A. Statistical phylogeography: methods for evaluating and minimizing inference errors. Mol Ecol 2004; 13: 789–809. [DOI] [PubMed] [Google Scholar]
- [111].Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007; 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 2012; 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Faria NR, Suchard MA, Rambaut A, Lemey P. Towards a quantitative understanding of viral phylogeography. Curr Opin Virol 2012; 1: 423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Suchard MA, Kitchen CMR, Sinsheimer JS, Weiss RE. Hierarchical phylogenetic models for analyzing multipartite sequence data. Syst Biol 2003; 52: 649:64. [DOI] [PubMed] [Google Scholar]
- [115].Penny D, Hendy M. Estimating the reliability of evolutionary trees. Mol Biol Evol 1986; 3: 403–17. [DOI] [PubMed] [Google Scholar]
- [116].Miyamoto M, Fitch W. Testing species phylogenies and phylogenetic methods with congruence. Syst Biol 1995; 44: 64–76. [Google Scholar]
- [117].Kluge AG. A concern for evidence and a phylogenetic hypothesis of relationships among Epicrates (Boidae, Serpentes). Syst Zool 1989; 38: 7–25. [Google Scholar]
- [118].Miyamoto M. Consensus cladograms and general classification. Cladistics 1985; 1: 186–9. [DOI] [PubMed] [Google Scholar]
- [119].Sharkey M, Triques K, Kuritzkes DR, Stevenson M. In vivo evidence for instability of episomal human immunodeficiency virus type 1 cDNA. J Virol 2005; 79: 5203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sharkey M, Babic DZ, Greenough T, Gulick R, Kuritzkes DR, Stevenson M. Episomal viral cDNAs identify a reservoir that fuels viral rebound after treatment interruption and that contributes to treatment failure. PLoS Pathog 2011; 7: e1001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Shankarappa R, Margolick JB, Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. Virol 1999; 73: 10489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Ruff CT, Ray SC, Kwon P, et al. Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J Virol 2002; 76: 9481–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Gray RR, Pybus OG, Salemi M. Measuring the temporal structure in serially sampled phylogenies. Methods Ecol Evol 2011; 5: 437–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Norström MM, Karlsson AC, Salemi M. Toward a new paradigm linking virus molecular evolution and pathogenesis: experimental design and phylodynamic inference. New Microbiologica 2012; 35: 101–11. [PubMed] [Google Scholar]
- [125].Zuckerkandl E, Pauling L. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. Evolutionary divergence and convergence in proteins. New York: Academic Press; 1965; 97–166. [Google Scholar]
- [126].Immomen TT, Leitner T. Reduced evolutionary rates in HIV-1 reveal extensive latency periods among replicating lineages. Retrovirol 2014; 11: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kelly J, Beddall MH, Yu D, Iyer SR, Marsh JW, Wu Y. Human macrophages support persistent transcription from unintegrated HIV-1 DNA. Virology 2008; 372: 300–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Pang S, Koyanagi Y, Miles S, Wiley C, Vinters HV, Chen IS. High levels of unintegrated HIV-1 DNA in brain tissue of AIDS dementia patients. Nature 1990; 343: 85–9. [DOI] [PubMed] [Google Scholar]
- [129].Teo I, Veryard C, Barnes H, et al. Circular forms of unintegrated human immunodeficiency virus type 1 DNA and high levels of viral protein expression: Association with dementia and multinucleated giant cells in the brains of patients with AIDS. J Virol 1997; 71: 2928–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Matsuda K, Brown CR, Foley B, et al. Laser capture microdissection assessment of virus compartmentalization in the central nervous systems of macaques infected with neurovirulent simian immunodeficiency virus. J Virol 2013; 87: 8896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Demuth M, Czub S, Sauer U, et al. Relationship between viral load in blood, cerebrospinal fluid, brain tissue and isolated microglia with neurological disease in macaques infected with different strains of SIV. J Neurovirol 2000; 6: 187–201. [DOI] [PubMed] [Google Scholar]
- [132].Reeve AB, Patel K, Pearce NC, et al. Reduced genetic diversity in lymphoid and central nervous system tissues and selection-induced tissue-specific compartmentalization of neuropathogenic SIVsmmFGb during acute infection. AIDS Res Hum Retroviruses 2009; 25: 583–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Gray RR, Salemi M, Klenerman P, Pybus OG. A new evolutionary model for hepatitis C virus chronic infection. PLoS Pathogens 2012; 8(5), e1002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Gray RR, Strickland SL, Veras NMC, et al. Unexpected Maintenance of Hepatitis C Viral Diversity Following Liver Transplantation. J Virol 2012; 86(16), 8432–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Evans DT, Silvestri G. Non-human primate models in AIDS research. Curr Opin HIV AIDS 2013; 8: 255–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Hatziioannou T, Evans DT. Animal models for HIV/AIDS research. Nat Rev Microbiol 2012; 10: 852–67. [DOI] [PMC free article] [PubMed] [Google Scholar]