Abstract
Pseudomonas lemoignei is equipped with at least five polyhydroxyalkanoate (PHA) depolymerase structural genes (phaZ1 to phaZ5) which enable the bacterium to utilize extracellular poly(3-hydroxybutyrate) (PHB), poly(3-hydroxyvalerate) (PHV), and related polyesters consisting of short-chain-length hxdroxyalkanoates (PHASCL) as the sole sources of carbon and energy. Four genes (phaZ1, phaZ2, phaZ3, and phaZ5) encode PHB depolymerases C, B, D, and A, respectively. It was speculated that the remaining gene, phaZ4, encodes the PHV depolymerase (D. Jendrossek, A. Frisse, A. Behrends, M. Andermann, H. D. Kratzin, T. Stanislawski, and H. G. Schlegel, J. Bacteriol. 177:596–607, 1995). However, in this study, we show that phaZ4 codes for another PHB depolymeraes (i) by disagreement of 5 out of 41 amino acids that had been determined by Edman degradation of the PHV depolymerase and of four endoproteinase GluC-generated internal peptides with the DNA-deduced sequence of phaZ4, (ii) by the lack of immunological reaction of purified recombinant PhaZ4 with PHV depolymerase-specific antibodies, and (iii) by the low activity of the PhaZ4 depolymerase with PHV as a substrate. The true PHV depolymerase-encoding structural gene, phaZ6, was identified by screening a genomic library of P. lemoignei in Escherichia coli for clearing zone formation on PHV agar. The DNA sequence of phaZ6 contained all 41 amino acids of the GluC-generated peptide fragments of the PHV depolymerase. PhaZ6 was expressed and purified from recombinant E. coli and showed immunological identity to the wild-type PHV depolymerase and had high specific activities with PHB and PHV as substrates. To our knowledge, this is the first report on a PHASCL depolymerase gene that is expressed during growth on PHV or odd-numbered carbon sources and that encodes a protein with high PHV depolymerase activity. Amino acid analysis revealed that PhaZ6 (relative molecular mass [Mr], 43,610 Da) resembles precursors of other extracellular PHASCL depolymerases (28 to 50% identical amino acids). The mature protein (Mr, 41,048) is composed of (i) a large catalytic domain including a catalytic triad of S136, D211, and H269 similar to serine hydrolases; (ii) a linker region highly enriched in threonine residues and other amino acids with hydroxylated or small side chains (Thr-rich region); and (iii) a C-terminal domain similar in sequence to the substrate-binding domain of PHASCL depolymerases. Differences in the codon usage of phaZ6 for some codons from the average codon usage of P. lemoignei indicated that phaZ6 might be derived from other organisms by gene transfer. Multialignment of separate domains of bacterial PHASCL depolymerases suggested that not only complete depolymerase genes but also individual domains might have been exchanged between bacteria during evolution of PHASCL depolymerases.
The ability to degrade extracellular poly(3-hydroxybutyrate) (PHB) and related polyesters is widely distributed among bacteria and depends on the secretion of specific polyester depolymerases which hydrolyze the water-insoluble polyester to water-soluble monomers or oligomers. One of the best-studied polyester-degrading bacteria is Pseudomonas lemoignei (5). It belongs to the beta subclass of Proteobacteria and is related to the Burkholderia-Ralstonia rRNA sublineage. The catabolic abilities of P. lemoignei are restricted to the utilization of a few organic acids (acetate, butyrate, valerate, pyruvate, succinate, and 3-hydroxybutyrate), and polyesters such as PHB, poly(3-hydroxyvalerate) (PHV), and related short-chain-length polyhydroxyalkanoates (PHASCL). Sugars, alcohols, and amino acids do not support growth of P. lemoignei (5, 17). Recently, P. lemoignei (strain A62) was reisolated by application of a specific enrichment procedure with PHV as the sole source of carbon and energy (16).
P. lemoignei is unique among PHA-degrading bacteria because it is able to synthesize at least five different extracellular PHA depolymerases. Three PHA depolymerases are specific for PHB and copolymers of 3-hydroxybutyrate (3-HB) and 3-hydroxyvalerate (3-HV) with low 3-HV content (PHB depolymerases A, B and D). The activity of these enzymes with the homopolyester PHV is below 5% of the activity obtained with PHB as substrate. None of the three PHB depolymerases is able to produce clearing zones on opaque PHV granule-containing agar. The two remaining PHA depolymerases (PHB depolymerases C and PHV depolymerase) also degrade PHB, but are additionally able to hydrolyze PHV, with about 15 and 30% activity compared to PHB hydrolysis. PHB depolymerase C and PHV depolymerase produce clearing zones on opaque PHV agar. Since the latter depolymerase is expressed only during growth on odd-numbered carbon sources (PHV and valerate), this PHA depolymerase was named PHV depolymerase (17).
Five PHA depolymerase structural genes (phaZ1 to PhaZ5) of P. lemoignei were cloned in earlier studies (3, 10, 11). Four genes, namely phaZ1, phaZ2, phaZ3, and phaZ5, were identified as encoding PHB depolymerases C, B, D, and A, respectively, by agreement of the DNA-deduced amino acid sequences with the sequences derived from Edman degradation of the purified proteins. When the N-terminal amino acid sequence of the PHV depolymerase was compared with the DNA-deduced amino acid sequence of the remaining gene, phaZ4, 23 of 24 amino acids were identical. A threonine codon had been determined at the position of the seventh codon of the deduced mature depolymerase, but an alanine had been identified in the N-terminal amino acid of the purified PHV depolymerase (10). From these results, it was hypothesized that phaZ4 might encode a different, yet unknown, PHA depolymerase and that the true PHV depolymerase structural gene had not been cloned so far. In that case, a sixth PHA depolymerase gene should be present and prompted us to undertake the present study.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. P. lemoignei was grown in Nakayama mineral salts medium (MM) with 0.4% (wt/vol) (R,S)-3-hydroxybutyrate as described recently (26). Recombinant E. coli strains harboring PHA depolymerase genes were grown in nutrient broth (NB) medium supplemented with ampicillin (Ap; 100 μg/ml) and IPTG (isopropyl-β-d-thiogalactopyranoside [0.2 g/liter]) at 37°C. Identification of recombinant Escherichia coli strains expressing PHA depolymerase genes was performed by clearing zone formation on NB-PHB agar (NB-Ap-IPTG underlay [25 ml], NB-Ap-IPTG agar supplemented with 0.2% [wt/vol] denatured PHA granule overlay [7 ml]) and on M9-Ap-PHB mineral salts medium supplemented with ampicillin, IPTG, proline (50 mg/liter), thiamine (1 mg/liter), glucose (1 g/liter), and glycerol (5 ml/liter) underlay (25 ml) plus a 7-ml overlay agar of the same composition, but with addition of PHA granules as described above.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference(s) |
|---|---|---|
| Strains | ||
| Ralstonia eutropha H16 | Source of PHB | DSM 428, ATCC 17699 |
| Chromobacterium violaceum | Source of PHV | DSM 30191 |
| Pseudomonas lemoignei | Growth on PHB and PHV | LMG 2207 |
| Escherichia coli | ||
| JM83 | PHB−ara Δ(lac-proAB) rpsL (Smr) thi-1 φ8 lacZΔ15 | 27 |
| XL1 Blue | PHB−recA1 endA1 gyrA96 thi hsdR17 (rK− mK+) supE44 relA1 λ− lac [F′ proAB lacIq ZΔM15 Tn10(Tcr)] | 4 |
| JM83(pSN654) | PHB+ PHV+ pUC9-2::1.7-kbp HindIII::phaZ1 | 11 |
| XL-1-Blue(pSN1260-39) | PHB+ PHV+ pUC9-::4.1-kbp MboI::phaZ1 | This study |
| XL1-Blue(pSN1270-49) | PHB+ PHV+ pUC9::2.7-kbp MboI::phaZ1 | This study |
| XL1-Blue(pSN1280-109) | PHB+ PHV+ pUC9-1::2.1-kbp MboI::phaZ1 | This study |
| XL1-Blue(pSN1268-46) | PHB+ PHV± pUC19-1::2.6-kbp MboI::phaZ2 | This study |
| XL1-Blue(pSN1258-62) | PHB+ PHV− pCU9::2.3-kbp MboI::phaZ3 | This study |
| XL1-Blue(pSN612-TF21) | PHB+ PHV− pUC9-2::3.1-kbp BamHI::phaZ4 | 10, 11 |
| XL1-Blue(pSN874) | PHB+ pBluescriptKS−::2.5-kbp HindIII-BamHI::phaZ4 | This study |
| XL1-Blue(pSN1292-110) | PHB+ PHV− pUC9-2::4.5-kbp MboI::phaZ5 | This study |
| XL1-Blue(pSN1262-66) | PHB+ PHV+ pUC9::2.6-kbp MboI::phaZ6 | This study |
| JM83(pSN1324) | PHB+ PHV+ pBluescript KS−::2.1-kbp EcoRI::phaZ6 | This study |
| Plasmids | ||
| pUC9, pUC9-1, pUC9-2 | AprlacPOZ′ | 7, 27 |
| pBluescript KS− | AprlacPOZ′; T7 and T3 promoters | Stratagene, San Diego, Calif. |
+ and − indicate ability and inability to form a clearing zone on opaque polymer agar plates.
Preparation of polymer suspension and assay of PHA depolymerases.
The polymers PHB, PHV, poly(3-hydroxyoctanoate) (PHO), and poly(3-hydroxyoctanoate-co-3-hydroxydecanoate) (PHO/HD) were isolated from sodium gluconate-grown cells of Ralstonia eutropha H16 (PHB), sodium valerate-grown cells of C. violaceum (PHV), sodium octanoate-grown cells of Pseudomonas oleovorans (PHO), and sodium gluconate-grown cells of Pseudomonas putida KT2440 (PHO/HD) as described previously (10, 11). PHA depolymerase activity was assayed by measuring the decrease in the optical density (650 nm) of the respective polymer suspension at 37°C. The reaction mixture contained 50 mM Tris-HCl (pH 8) with 1 mM CaCl2 and 60 μl of a 0.3% (wt/vol) suspension of denatured PHB granules (total volume, 1 ml). The reaction was started by the addition of 5 to 50 μl of depolymerase preparation. Alternatively, clearing zone formation of opaque agar media containing PHA granules in 100 mM Tris-HCl (pH 8) and 1 mM CaCl2 upon the addition of 1 to 2 μl of PHA depolymerase solution and incubation at 37°C semiquantitatively indicated the activity of the depolymerase.
Purification of PHA depolymerases.
PHV depolymerase from P. lemoignei was purified from 5 liters of cell-free culture fluid of 0.2% (wt/vol) sodium valerate-grown (30°C, 24 to 36 h) cells by (i) ammonium sulfate precipitation (35 to 75%), (ii) chromatography on DEAE-Sephacel, and (iii) chromatography on CM-Sepharose-CL6B as described earlier (17), except that one additional purification step (iv) was required (size exclusion chromatography on Sephadex G-75S).
PHA depolymerases from recombinant E. coli harboring pSN874 (phaZ4) or pSN1324 (phaZ6) were purified from periplasm extracts of cells that had been grown in NB medium (800 ml) supplemented with ampicillin (50 μg/ml) for 12 to 16 h at 37°C. All subsequent steps were performed at 0 to 6°C. Cells were harvested by centrifugation and washed three times with 0.9% (wt/vol) NaCl. Cells were resuspended in 1 volume (800 ml) of 30 mM Tris-HCl (pH 8), containing 20% (wt/vol) sucrose and 1/4,000 volume (0.2 ml) of 250 mM EDTA and shaken for 10 min at room temperature. Cells were collected by centrifugation, and the proteins of the periplasm were released by resuspension and shaking the cells in 1 volume of 5 mM MgSO4 for about 7 min. After centrifugation, PHA depolymerase was purified from the supernatant as described above for the wild-type PHV depolymerase. Purified PHA depolymerase proteins were stored in 10 to 50 mM Tris-HCl (pH 7 to 7.5) containing 1 mM CaCl2 at −20°C.
Proteolytic digestion by endoproteinase GluC.
Purified wild-type PHV depolymerase (340 μg) in 50 mM K2HPO4 (pH 7.5), containing 0.05% (wt/vol) sodium dodecyl sulfate (SDS), was incubated at 100°C for 3 min. After cooling to room temperature, 1/30 (11 μg) endoproteinase GluC from Staphylococcus aureus V8 was added, and the complete reaction mixture was incubated at 37°C for 28 h. The cleavage products were separated by denaturing SDS-polyacrylamide gel electrophoresis (PAGE) and subsequently blotted onto a polyvinylidene difluoride membrane at 5 mA/cm2 for 50 min and stained with Coomassie blue.
Immunological techniques.
Rabbit polyclonal antisera were raised against (i) PHV depolymerase purified from P. lemoignei and (ii) PHB depolymerase purified from Comamonas sp. The immunoglobulin G (IgG) fractions were purified from the antisera (three injections of 100 to 500 μg of purified protein in 50% [vol/vol] complete Freund's adjuvant [first injection], 50% (vol/vol) incomplete Freund's adjuvant [second injection], and without adjuvant [third injection]) by standard chromatography on protein A Sepharose and stored at −20°C. Purified PHA depolymerase proteins were tested for immunological relatedness by Ouchterlony double-immune-diffusion testing in 1% (wt/vol) agarose in 100 mM Tris-HCl (pH 7.6) (19). Precipitation bands were visualized by staining with Coomassie blue.
Molecular biology techniques.
Standard molecular biology techniques were used throughout this study, including double-stranded DNA sequencing by dideoxy chain termination and primer walking.
Cloning of the PHV depolymerase structural gene phaZ6.
Agarose gel-separated and size-fractioned (2 to 10 kbp) partially MboI-digested genomic DNA of P. lemoignei was ligated to a mixture of BamHI-linearized pUC expression vectors (pUC9, pUC9-1, and pUC9-2) and transformed in E. coli XL1-Blue. Colonies harboring recombinant plasmids were screened for halo formation on PHB- and PHV-containing NB and M9 agar (described above).
Nucleotide sequence accession number.
The DNA sequence of phaZ6 is available under accession no. AF222999.
RESULTS
Biochemical and immunological characterization of the PhaZ4 depolymerase from recombinant E. coli and the wild-type PHV depolymerase from P. lemoignei.
In order to elucidate whether phaZ4 or another gene encoded the PHV depolymerase of P. lemoignei, the recombinant PhaZ4 depolymerase and the wild-type PHV depolymerase were purified, and the relevant characteristics of both depolymerases were compared. Recombinant E. coli cells, which harbored phaZ4 (pSN874), were grown on NB-PHB and M9-PHB as well as on NB-PHV and M9-PHV agar plates at 37°C, respectively. Large clearing zones appeared on NB-PHB and M9-PHB medium after 2 days and indicated the functional expression of PhaZ4. However, no clearing zone formation was observed on PHV agar even after 1 week of incubation. Apparently, the activity of PhaZ4 with PHV as a substrate was very low. The recombinant PhaZ4 depolymerase and the wild-type PHV depolymerase were purified as described in Materials and Methods. The purified proteins were homogeneous, as judged by denaturing SDS-PAGE and silver staining. The ratios between the specific PHB and PHV depolymerase activity remained almost constant throughout purification and amounted to 25:1 for PhaZ4 and 3:1 for the wild-type PHV depolymerase (Table 2). Therefore, it appears unlikely that PhaZ4 represents the PHV depolymerase.
TABLE 2.
Relative PHV depolymerase activities of PHASCL depolymerases of P. lemoignei
| Gene | Protein | PHV hydrolysis (% of PHB hydrolysis)a |
|---|---|---|
| phaZ1 | PHB depolymeraseC | 15–20b |
| phaZ2 | PHB depolymerase B | <5 |
| phaZ3 | PHB depolymerase D | <5 |
| phaZ4 | PHB depolymerase E | <5 |
| phaZ5 | PHB depolymerase A | <5 |
| phaZ6 | PHV depolymerase | 28–35b |
Hydrolysis obtained with denatured PHB granules was taken as 100%.
Values varied depending on the particular batch of PHA granules and purified depolymerase.
Both purified depolymerase proteins were subjected to an Ouchterlony double-immune-diffusion test against a polyclonal antiserum that had been raised against the wild-type PHV depolymerase. A strong cross-reaction was found for the positive control with wild-type PHV depolymerase, but no reaction occurred with different amounts of recombinant PhaZ4. Eleven N-terminal amino acids of the purified recombinant PhaZ4 depolymerase were determined by Edman degradation (LTPGSGT7WVKE). The sequence obtained matched exactly the phaZ4-deduced sequence, including amino acid 7 (threonine), whereas an alanine was determined at position 7 for the PHV depolymerase purified from P. lemoignei (LTPGSGA7WVKESATYGTPNLQ). The purified PHV depolymerase of P. lemoignei was subjected to proteolytic cleavage by endoproteinase GluC, as described in Materials and Methods. The cleavage products were separated by denaturing SDS-PAGE and Western blotted. Four of the endopeptidase GluC cleavage products were isolated, and the following sequences were determined by Edman degradation: SATYGTPNLQ, peptide 1; AYVYV, peptide 2; QYGMVVIAP, peptide 3; and YFAGVV, peptide 4. The sequences of peptides 1 and 4 were the same as amino acids 12 to 21 and 177 to 182 of the DNA-deduced sequence of mature PhaZ4, respectively. However, the amino acid sequences of peptides 2 and 3 were not present in the DNA-deduced sequence of PhaZ4, although similar sequences, each having one or two mismatches (underlined), were found in PhaZ4 (A23YLYY and K70YGMVVVAP) (Fig. 1). These data proved that phaZ4 did not code for the PHV depolymerase, but encoded a PHA depolymerase which is very similar to the PHV depolymerase with respect to its amino acid sequence. As a consequence, another depolymerase gene (phaZ6) had to be postulated that encoded the true PHV depolymerase of P. lemoignei.
FIG. 1.
Amino acid alignment of PHA depolymerases PhaZ6 (PHV depolymerase) and PhaZ4 (PHB depolymerase E). The first 300 amino acids of the mature proteins are aligned (catalytic domain). The N-terminal amino acid sequences of purified PHV depolymerase and of four endoproteinase GluC-generated internal peptides are indicated by italic letters and by underlining, respectively. Mismatches between amino acids determined by Edman degradation and the DNA-deduced sequence of phaZ4 are indicated by asterisks (∗).
Cloning of the PHV depolymerase structural gene (phaZ6).
Due to the apparent high amino acid sequence similarity between PhaZ4 and the PHV depolymerase, the identification of a recombinant PHV depolymerase-containing clone by colony hybridization with a PHV depolymerase-specific oligonucleotide or by a specific PCR-based amplification of the PHV depolymerase structural gene appeared not to be promising. A less selective approach was chosen for the detection of PHA depolymerase activity-expressing clones: about 6,500 recombinant strains of a genomic library of P. lemoignei in E. coli XL1-Blue were screened for haloformation on opaque M9-PHB and NB-PHB agar. Eight positive clones (no. 26, 39, 46, 49, 62, 66, 109, and 110) were identified that produced large clearing zones on PHB agar and indicated the functional expression of a PHA depolymerase. After purification, these clones were tested for halo formation on M9-PHV and NB-PHV agar. Except for no. 62 and 110, all other six clones produced clearing zones on both PHV agar types. The diameters of the clearing zones were significantly smaller on PHV agar and appeared later than those on PHB agar. The plasmid DNA of all eight clones was isolated, and the DNA sequences of the corresponding insert ends were determined. Seven clones harbored already known PHB depolymerase genes, namely phaZ1 (clones 39, 49, and 109), phaZ2 (clones 26 and 46), phaZ3 (clone 62), and phaZ5 (clone 110). Clone 66, which showed the highest clearing zone on PHV agar, was different from all other clones, and the complete DNA sequence (2,595 bp) of this clone was determined for both strands. One open reading frame (ORF1) of 1,254 bp was identified that encoded the PHV depolymerase structural gene phaZ6 (see below).
Characterization of phaZ6.
A potential ribosome binding site (5′-GGAGA-3′) and a sequence with homology to ς70-dependent promoters (5′-TTGACT-N17-AAATCT-3′) were present 7 and 172 bp upstream of phaZ6, respectively. Another potential ATG start codon 18 bp upstream of phaZ6 was unlikely to represent the initiation site of translation due to the absence of a ribosome binding motif. A potential hairpin structure followed by a 7-bp poly(T) sequence was identified 88 bp downstream of phaZ6 and might represent a factor-independent termination site of transcription. The G+C contents of phaZ6 and of the total sequenced DNA amounted to 54.5 and 52%, respectively, which is a slightly lower value than that of total genomic DNA (58%) (5). The codon usage of phaZ6 was partially different from that of other genes sequenced from P. lemoignei. This difference was most evident for the His, Asp, Asn, and Lys codons: all eight His codons had the sequence CAT (100%), compared to only 63% for CAT and 37% for CAC as the average of other sequenced genes of P. lemoignei. The codons for Asp, Asn, and Lys with A or T in the wobble position were preferentially used in phaZ6 (63, 68, and 75%) compared to only 40, 42, and 25% in other genes of P. lemoignei, respectively. Codons with a G or C in the wobble position were preferentially used for most of the remaining amino acids and resulted in an average G+C content of the total gene above 50%. Inspection of the G+C content of the sequenced DNA revealed a local minimum of 35 to 40% in the region up to 400 bp upstream of phaZ6.
Characterization of PhaZ6.
phaZ6 coded for a protein of 417 amino acids (43,610 Da). The N-terminal deduced amino acid sequence had the characteristics of signal peptides of secretory precursors: two positively charged amino acids (K5 and R9) were followed by several hydrophobic residues, and a putative signal peptidase cleavage site was predicted between A25 and L26, resulting in a molecular mass of 41,048 Da for the mature protein. The DNA-deduced amino acid sequence was in complete agreement with the 21 amino acids that had been determined by Edman degradation of the purified PHV depolymerase of P. lemoignei from amino acid 26 onward and sustained the predicted signal peptidase cleavage site. The sequence included an alanine at position 7 of the mature protein and contained the complete sequences of the four endoproteinase GluC-derived peptide fragments of the PHV depolymerase (Fig. 1). These results confirmed that phaZ6 encoded the true PHV depolymerase. The homology of the PhaZ6 amino acid sequence (mature protein) to other extracellular PHB depolymerases varied between 25% (Streptomyces exfoliatus PHB depolymerase) and 50% identical amino acids for the PhaZ4 depolymerase of P. lemoignei. The homology of PhaZ6 to PhaZ4 was particularly high within the first 200 amino acids (72.5%, Fig. 1).
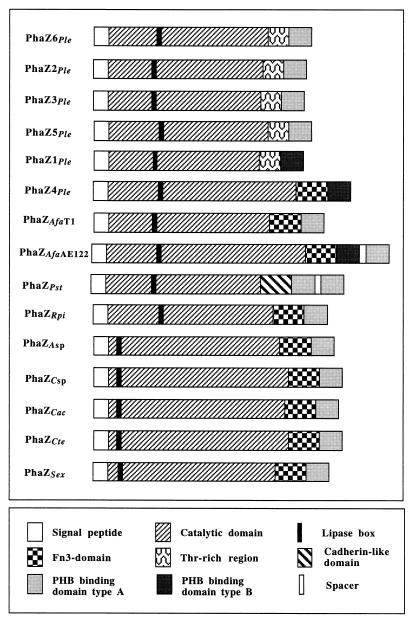
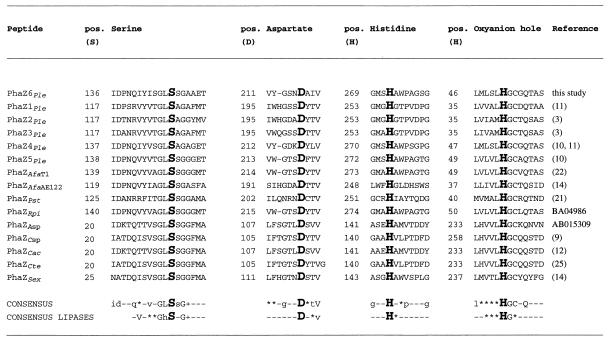
Inspection of the primary sequence of PhaZ6 revealed the presence of three domains, namely, a catalytic domain, a linking domain, and a substrate binding domain, as has been found for all other PHB depolymerases (Fig. 2). The catalytic domain (residues 1 to 306) contains a pentapeptide sequence, G-X-S136-X-G (lipase box), which is a characteristic of serine esterases such as lipases, esterases, and all PHA depolymerases analyzed so far. The central serine of the lipase box has been shown to be essential for activity in many lipases and several PHA depolymerases (8, 23, 24). Two histidine residues (H46 and H269) and one aspartate residue (D211) are strictly conserved in PhaZ6 and all PHA depolymerases (Fig. 3). The serine, histidine, and aspartate residues are known to constitute a catalytic triad including an oxyanion pocket (H46), as is known for serine esterases (8). The amino acid sequence of PhaZ6 between G307 and T334 is extremely enriched in threonine and other residues with small side chains (threonine-rich region). Similar threonine-rich regions have been identified in the PHB depolymerases PhaZ1, PhaZ2, PhaZ3, and PhaZ5 of P. lemoignei and are assumed to have the function of a linking domain. The 60 amino acids of the C terminus of PhaZ6 have high homologies to the substrate-binding domains of other PHB depolymerases (Fig. 4) and apparently are responsible for binding of the depolymerase to the hydrophobic water-insoluble polyester.
FIG. 2.
Domain structure of PHASCL depolymerases. An interpretation of the DNA-deduced amino acid sequences is shown. The last three letters of each depolymerase refer to the bacterial origin: P. lemoignei (Ple), A. faecalis strains T1 and AE122 (Afa), P. stutzeri (Pst), R. pickettii (Rpi), Acidovorax sp. (Asp), Comamonas sp. (Csp), C. acidovorans (Cac), C. testosteroni (Cte), and S. exfoliatus (Sex). For references describing the PHA depolymerase genes, see Fig. 3.
FIG. 3.
Alignment of catalytic amino acids of PHASCL depolymerases. An alignment of the regions around the catalytic triad amino acids serine (S), aspartate (D), and histidine (H) and around the putative oxyanion histidine (H) (indicated by boldface letters) is shown. The numbering refers to the mature polypeptides. Positions (pos.) of amino acids with hydrophobic (∗) or small (+) side chains are indicated in the consensus sequence.
FIG. 4.
Alignment of PHASCL depolymerase C-terminal substrate-binding domains. An alignment of the C-terminal 61 amino acids is shown (numbering refers to PhaZ6 depolymerase). The substrate-binding domains of types A and B are aligned separately under A and B, respectively. Residues that are conserved in almost all sequences are indicated by boldface letters. For abbreviations of depolymerases and for definitions of abbreviations, see the legends to Fig. 2 and 3.
Purification and characterization of PhaZ6 from recombinant E. coli.
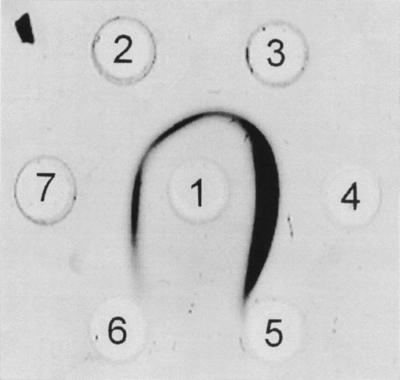
PhaZ6 was purified from the periplasm of IPTG-induced NB-grown cells. The elution behavior of PhaZ6 was very similar to that of the wild-type PHV depolymerase during all stages of purification. The purified PhaZ6 depolymerase hydrolyzed PHASCL, such as PHB and PHV, at high rates, but was unable to degrade medium-chain-length PHA (PHAMCL) such as PHO or P(HO/HD). The ratio between the specific PHB and PHV depolymerase activity remained constant at about 3:1 throughout all stages of purification and is in agreement with the same ratio that had been determined for the PHV depolymerase purified from P. lemoignei (Table 2). The purified PhaZ6 depolymerase was subjected to an Ouchterlony double-immune-diffusion test against a polyclonal antiserum that had been raised against the purified wild-type PHV depolymerase. The PHV depolymerase and the recombinant PhaZ6 depolymerase both cross-reacted with the antibodies, and the precipitation bands perfectly mingled without the formation of a spur (Fig. 5). This indicated that both proteins are immunologically identical. On the contrary, no cross-reaction occurred with purified PHB depolymerases A and B (Fig. 5). PhaZ6 depolymerase did not cross-react with antibodies raised against PHB depolymerase of Comamonas sp., and antibodies raised against the wild-type PHV depolymerase did not cross-react with PHB depolymerase A (PhaZ5) of P. lemoignei (data not shown).
FIG. 5.
Ouchterlony double-immune-diffusion assay. The immunological reaction of a polyclonal PHV depolymerase antiserum against PHASCL depolymerases in 1% agarose is shown. The central well (well 1) contained 10 μg of a PHV depolymerase antiserum (IgG fraction). The other wells contained 8 to 10 μg of purified recombinant PhaZ6 depolymerase (wells 7, 2, and 3), purified wild-type PHV depolymerase (well 4), purified PHB depolymerase A (30 μg, well 5), or purified PHB depolymerase B (30 μg, well 6). The reaction was run to completion for 18 h at room temperature, before the agarose was stained with Coomassie blue.
DISCUSSION
P. lemoignei is unique among PHA-degrading bacteria because of its high number of extracellular PHA depolymerases (at least six) and its ability to degrade the homopolyester PHV in addition to PHB. Most other PHB-degrading bacteria are not able to degrade PHV and usually have only one PHB depolymerase. Five PHB depolymerase genes of P. lemoignei (phaZ1 to phaZ5) and the corresponding proteins have been described in earlier studies (3, 5, 10, 11, 18; for a recent review, see reference 9). In this report, we studied a novel PHA depolymerase of P. lemoignei which has high activities with PHV (and PHB) as a substrate and which is specifically synthesized during growth on PHV or odd-numbered carbon sources, such as valerate (PHV depolymerase). The structural gene of the PHV depolymerase, phaZ6, was undoubtedly identified by (i) complete agreement of the N-terminal amino acid sequence of the purified wild-type PHV depolymerase and the N-terminal amino acid sequences of four internal endoproteinase GluC-generated peptides with the DNA-deduced amino acid sequence of phaZ6 (Fig. 2), (ii) by cross-reaction of purified recombinant PhaZ6 protein with PHV depolymerase-specific antibodies (Fig. 5), and (iii) by the same ratio of PHV and PHB depolymerase activity of the purified recombinant enzyme and the wild-type PHV depolymerase (Table 2). PhaZ6 showed significant amino acid sequence homologies to all 14 PHB depolymerases known from databases (25 to 50% identity). PhaZ1 of P. lemoignei is the only PHB depolymerase, except PhaZ6, that has any significant activity with PHV (Table 2). However, the degree of amino acid homology of PhaZ6 to PHB depolymerase C (PhaZ1) was lower than to any other depolymerase of P. lemoignei. Therefore, it is impossible to correlate particular amino acids or amino acid sequences of PhaZ1 and PhaZ6 with the ability to utilize PHV as a substrate.
Comparison of the primary amino acid sequences of all available PHASCL depolymerases revealed that mature PHASCL depolymerases generally consist of three domains in the following sequential order: (i) a catalytic domain including a catalytic triad of serine, aspartate, histidine, and the oxyanion histidine; (ii) a linking domain between the catalytic domain and the C-terminal domain; and (iii) a C-terminal substrate-binding domain (Fig. 2). Each domain can be present in different isoforms: two types of catalytic domains are differentiated by the primary sequence and by a different sequential order of the lipase box and the other catalytic triad amino acids. The sequential order of the catalytic amino acids of all six P. lemoignei PHA depolymerases and of the depolymerases of Alcaligenes faecalis AE122, A. faecalis T1, Ralstonia pickettii, and Pseudomonas stutzeri is (i) histidine (oxyanion), (ii) lipase box serine, (iii) aspartate, and (iv) histidine (Fig. 3). These depolymerases degrade PHB to a mixture of 3-HB and oligomers of 3-HB. The sequential order in the other PHB depolymerases (Acidovorax sp., Comamonas sp., C. acidovorans, C. testosteroni, and S. exfoliatus) is lipase box serine, aspartate, histidine, and oxyanion hole histidine (Fig. 3). The catalytic domains of these depolymerases share relatively high amino acid homologies to each other, but have rather low similarities to the catalytic domains of all other PHB depolymerases. As far as has been analyzed, these depolymerases have high internal 3-HB dimer hydrolase activity in addition to the depolymerase activity. As a consequence, monomeric 3-HB is the major end product of PHB hydrolysis in the presence of an excess of these PHB depolymerases, and the amount of oligomeric end products is relatively low. At present, it is unclear whether the composition of the PHB hydrolysis end products can be generally predicted from the sequential order of the catalytic triad amino acids.
Three types of linking domains have been identified in bacterial PHA depolymerases, namely (i) threonine-rich region, (ii) fibronectin type 3 domain (Fn3), and (iii) a cadherin-like domain (Cad). Threonine-rich regions, which consist of an amino acid sequence approximately 40 amino acids in length highly enriched in threonine and other amino acids with hydroxylated and/or small side chains, have been found only in P. lemoignei PHA depolymerases PhaZ1, PhaZ2, PhaZ3, PhaZ5, and PhaZ6. Similar domains with either a high degree of serine residues or with a high proportion of glycine, alanine, serine, threonine, and proline are known to occur in many polymer hydrolases (6). These domains are assumed to constitute a highly flexible linker between adjacent domains and might facilitate the hydrolysis of neighboring polymer chains, while the enzyme remains bound to the substrate via its substrate-binding domain. Fn3 domains have been found in the PHB depolymerases of Acidovorax sp., A. faecalis T1 and AE122, Comamonas sp., C. acidovorans, C. testosteroni, and S. exfoliatus and in PhaZ4 of P. lemoignei. They also occur in other polymer hydrolases, such as chitinases, cellulases, pullulanases, and amylases (15). Recently, a depolymerase with a novel type of linking domain with high amino acid homology to a cadherin domain has been described for the P. stutzeri PHB depolymerase (21). Since the Fn3 domain of the A. faecalis T1 PHB depolymerase was successfully replaced by a threonine rich region without any significant effect on activity, whether the linking region has any additional sequence-specific function is questionable (19).
The C-terminal part of all PHASCL depolymerases consists of a PHA-specific substrate-binding domain. This has been shown experimentally for the PHB depolymerases of P. lemoignei (PhaZ4), A. faecalis, C. acidovorans, C. testosteroni, and P. stutzeri (1, 2, 21, 25). Two types of substrate-binding domains can be differentiated by sequence alignment (Fig. 4). However, some amino acids, including the motif HxxAGR* (with x and * representing an amino acid with a variable or hydrophobic side chain, respectively) are conserved in both types of binding domains. Recently, two PHB depolymerases with two substrate-binding domains have been described for A. faecalis AE122 and P. stutzeri (13, 20). Interestingly, both subtypes of substrate-binding domains were found in the A. faecalis AE122 depolymerase (Fig. 4). The selective advantage of two substrate-binding domains is not known.
The highest degrees of amino acid homology between (mature) PHASCL depolymerases were found for the PHB depolymerase of Comamonas sp. and C. testosteroni (98% identical amino acid); for the depolymerases of A. faecalis T1 and R. pickettii (88%); and for the depolymerases PhaZ1, PhaZ2, and PhaZ3 of P. lemoignei (69 to 74% identity). The high degrees of similarity could be explained by the close relatedness of both Comamonas strains and of A. faecalis T1 and R. pickettii or by gene duplication in the case of P. lemoignei. For other depolymerases, high degrees of amino acid identity were found only for selected domains. For example, the catalytic domains and substrate-binding domains of the A. faecalis T1 and the PhaZ5 PHB depolymerase of P. lemoignei shared 72 and 68% identity, respectively, but both depolymerases had different types of linking domains (Fn3 domains versus threonine-rich region). A comparable result was obtained by alignment of the PHV depolymerase PhaZ6 with the PHB depolymerase PhaZ4. In this case, only the first 200 amino acids of the catalytic domains were highly identical (73%). In particular, 26 of the 28 N-terminal amino acids of the mature proteins were identical (93% identity; Fig. 1). A similar result was found even for depolymerases from not related bacteria, e.g., S. exfoliatus and PhaZ5 of P. lemoignei: relatively high homologies were found for the substrate-binding domains (60%), but the catalytic domain and the linking domain belonged to different types (Fig. 2). It appears as if the bacteria have exchanged and randomly combined selected domains of PHASCL depolymerases during evolution. This assumption is supported by experimental data in at least one case: the codon usage of phaZ6 was slightly different for the codons Asp, Asn, and His from those of other genes that have been sequenced from P. lemoignei. The AAT and AAC codons (Asn) occurred 21 and 10 times in phaZ6, corresponding to 68 and 32%, respectively. The average values for all other (eight) genes of P. lemoignei that have been sequenced amounted to 42% (AAT) and 58% (AAC). This apparent difference in the frequency of both codons was mainly caused by the high frequency of AAT within the DNA sequence of the catalytic domain (17 codons [corresponding to 74%], compared to only 6 AAC codons [26%]). The distributions of both codons within the linker and substrate-binding domain were different (four AAT and four AAC codons, respectively, each corresponding to 50%). Apparently, the levels of codon usage can be different in different domains of multidomain peptides and might indicate domain exchange.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft and the Graduiertenkolleg “Chemische Aktivitäten von Mikroorganismen.”
REFERENCES
- 1.Behrends A, Klingbeil B, Jendrossek D. Poly(3-hydroxybutyrate) depolymerases bind to their substrate by a C-terminal located substrate binding site. FEMS Microbiol Lett. 1996;143:191–194. doi: 10.1111/j.1574-6968.1996.tb08479.x. [DOI] [PubMed] [Google Scholar]
- 2.Briese B-H, Jendrossek D. Biological basis of enzyme-catalyzed polyester degradation: 59 C-terminal amino acids of poly(3-hydroxybutyrate) (PHB) depolymerase A from Pseudomonas lemoignei are sufficient for PHB binding. Macromol Sympos. 1998;130:205–216. [Google Scholar]
- 3.Briese B-H, Schmidt B, Jendrossek D. Pseudomonas lemoignei has five poly(hydroxyalkanoic acid) (PHA) depolymerase genes: a comparative study of bacterial and eukaryotic depolymerases. J Environ Polym Degrad. 1994;2:75–87. [Google Scholar]
- 4.Bullock W O, Fernandez J M, Stuart J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 5.Delafield F P, Doudoroff M, Palleroni N J, Lusty C J, Contopoulos R. Decomposition of poly-β-hydroxybutyrate by pseudomonads. J Bacteriol. 1965;90:1455–1466. doi: 10.1128/jb.90.5.1455-1466.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilkes N R, Henrissat B, Kilburn D G, Miller R C, Jr, Warren R A J. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanna Z, Fregeau C, Prefontaine G, Brousseau R. Construction of a family of universal expression plasmid vectors. Gene. 1984;30:247–250. doi: 10.1016/0378-1119(84)90128-8. [DOI] [PubMed] [Google Scholar]
- 8.Jaeger K E, Ransac S, Dijkstra B W, Colson C, van Heuvel M, Misset O. Bacterial lipases. FEMS Microbiol Rev. 1994;15:29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 9.Jendrossek D. Microbial degradation of polyesters: a review on extracellular poly(hydroxyalkanoic acid) depolymerases. Polym Degrad Stab. 1998;59:317–325. [Google Scholar]
- 10.Jendrossek D, Frisse A, Behrends A, Andermann M, Kratzin H D, Stanislawski T, Schlegel H G. Biochemical and molecular characterization of the Pseudomonas lemoignei polyhydroxyalkanoate depolymerase system. J Bacteriol. 1995;177:596–607. doi: 10.1128/jb.177.3.596-607.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jendrossek D, Müller B, Schlegel H G. Cloning and characterization of the poly(hydroxyalkanoic acid)-depolymerase gene locus, phaZ1, of Pseudomonas lemoignei and its gene product. Eur J Biochem. 1993;218:701–710. doi: 10.1111/j.1432-1033.1993.tb18424.x. [DOI] [PubMed] [Google Scholar]
- 12.Kasuya K-I, Inoue Y, Tanaka T, Akehata T, Iwata T, Fukui T, Doi Y. Biochemical and molecular characterization of the polyhydroxybutyrate depolymerase of Comamonas acidovorans YM1609, isolated from freshwater. Appl Environ Microbiol. 1997;63:4844–4852. doi: 10.1128/aem.63.12.4844-4852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kita K, Mashiba S, Nagita M, Ishimaru K, Okamoto K, Yanase H, Kato N. Cloning of poly(3-hydroxybutyrate) depolymerase from a marine bacterium, Alcaligenes faecalis AE122, and characterization of its gene product. Biochim Biophys Acta. 1997;1352:113–122. doi: 10.1016/s0167-4781(97)00011-0. [DOI] [PubMed] [Google Scholar]
- 14.Klingbeil B, Kroppenstedt R, Jendrossek D. Taxonomical identification of Streptomyces exfoliatus K10 and characterization of its poly(3-hydroxybutyrate) depolymerase gene. FEMS Microbiol Lett. 1996;142:215–221. doi: 10.1111/j.1574-6968.1996.tb08433.x. [DOI] [PubMed] [Google Scholar]
- 15.Little E, Bork P, Doolittle R F. Tracing the spread of fibronectin type III domains in bacterial glycohydrolases. J Mol Evol. 1994;39:631–643. doi: 10.1007/BF00160409. [DOI] [PubMed] [Google Scholar]
- 16.Mergaert J, Schirmer A, Hauben L, Mau M, Hoste B, Kersters K, Jendrossek D, Swings J. Isolation and identification of poly(3-hydroxyvalerate)-degrading strains of Pseudomonas lemoignei. Int J Syst Bacteriol. 1996;46:769–773. doi: 10.1099/00207713-46-3-769. [DOI] [PubMed] [Google Scholar]
- 17.Müller B, Jendrossek D. Purification and properties of poly(3-hydroxyvaleric acid) depolymerase from Pseudomonas lemoignei. Appl Microbiol Biotechnol. 1993;38:487–492. [Google Scholar]
- 18.Nakayama K, Saito T, Fukui T, Shirakura Y, Tomita K. Purification and properties of extracellular poly(3-hydroxybutyrate) depolymerases from Pseudomonas lemoignei. Biochim Biophys Acta. 1985;827:63–72. doi: 10.1016/0167-4838(85)90101-3. [DOI] [PubMed] [Google Scholar]
- 19.Nojiri M, Saito T. Structure and function of poly(3-hydroxybutyrate) depolymerase from Alcaligenes faecalis T1. J Bacteriol. 1997;179:6965–6970. doi: 10.1128/jb.179.22.6965-6970.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oakley C L. Antigen-antibody reactions in microbiology. Methods Microbiol. 1971;5A:173–218. [Google Scholar]
- 21.Ohura T, Kasuya K-I, Doi Y. Cloning and characterization of the polyhydroxybutyrate depolymerase gene of Pseudomonas stutzeri and analysis of the function of substrate-binding domains. Appl Environ Microbiol. 1999;65:189–197. doi: 10.1128/aem.65.1.189-197.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito T, Suzuki K, Yamamoto J, Fukui T, Miwa K, Tomita K, Nakanishi S, Odani S, Suzuki J-I, Ishikawa K. Cloning, nucleotide sequence, and expression in Escherichia coli of the gene for poly(3-hydroxybutyrate) depolymerase from Alcaligenes faecalis. J Bacteriol. 1989;171:184–189. doi: 10.1128/jb.171.1.184-189.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schirmer A, Matz C, Jendrossek D. Substrate specificities of PHA-degrading bacteria and active site studies on the extracellular poly(3-hydroxyoctanoic acid) [P(3HO)] depolymerase of Pseudomonas fluorescens GK13. Can J Microbiol. 1995;41(Suppl. 1):170–179. doi: 10.1139/m95-184. [DOI] [PubMed] [Google Scholar]
- 24.Shinohe T, Nojiri M, Saito T, Stanislawski T, Jendrossek D. Determination of the active sites serine of the poly(3-hydroxybutyrate) depolymerases of Pseudomonas lemoignei (PhaZ5) and of Alcaligenes faecalis. FEMS Microbiol Lett. 1996;141:103–109. doi: 10.1111/j.1574-6968.1996.tb08370.x. [DOI] [PubMed] [Google Scholar]
- 25.Shinomiya M, Iwata T, Kasuya K, Doi Y. Cloning of the gene for poly(3-hydroxybutyric acid) depolymerase of Comamonas testosteroni and functional analysis of its substrate-binding domain. FEMS Microbiol Lett. 1997;154:89–94. doi: 10.1111/j.1574-6968.1997.tb12628.x. [DOI] [PubMed] [Google Scholar]
- 26.Terpe K, Kerkhoff K, Pluta E, Jendrossek D. Relationship between succinate transport and production of extracellular poly(3-hydroxybutyrate) depolymerase in Pseudomonas lemoignei. Appl Environ Microbiol. 1999;65:1703–1709. doi: 10.1128/aem.65.4.1703-1709.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]