Abstract
The electrophile N-ethylmaleimide (NEM) elicits rapid K+ efflux from Escherichia coli cells consequent upon reaction with cytoplasmic glutathione to form an adduct, N-ethylsuccinimido-S-glutathione (ESG) that is a strong activator of the KefB and KefC glutathione-gated K+ efflux systems. The fate of the ESG has not previously been investigated. In this report we demonstrate that NEM and N-phenylmaleimide (NPM) are rapidly detoxified by E. coli. The detoxification occurs through the formation of the glutathione adduct of NEM or NPM, followed by the hydrolysis of the imide bond after which N-substituted maleamic acids are released. N-Ethylmaleamic acid is not toxic to E. coli cells even at high concentrations. The glutathione adducts are not released from cells, and this allows glutathione to be recycled in the cytoplasm. The detoxification is independent of new protein synthesis and NAD+-dependent dehydrogenase activity and entirely dependent upon glutathione. The time course of the detoxification of low concentrations of NEM parallels the transient activation of the KefB and KefC glutathione-gated K+ efflux systems.
Electrophiles can severely damage biological nucleophiles, such as the bases of DNA and the sulfydryl groups of proteins (21). Consequently, some electrophiles are potent bacteriocidal agents that exert their effects through the modification of cellular macromolecules. Bacterial cells, especially gram-negative bacteria, have evolved specific efflux systems, DNA protective proteins, metabolic pathways, and ion channels, the combined function of which is to protect the cell during exposure to toxic chemicals (5, 9–14). The sensitivity of cells to electrophiles is affected by inducible stress responses (e.g., the RpoS and Dps regulons [1, 13, 22]) that lead to the expression of proteins that either protect or repair DNA. However, new protein synthesis is not required for the survival of exposure to electrophiles, and thus it is the preexisting mechanisms that are central to survival (10, 12). Exposure of cells to N-ethylmaleimide (NEM) or methylglyoxal (MG) leads to rapid sequestration of the electrophile by glutathione (GSH), leading to the formation of GSH adducts that activate the KefB and KefC K+ efflux systems (5, 10). Activation of the efflux systems causes a rapid decline of the cytoplasmic pH, which effects protection against electrophiles (9, 11, 12). For MG, a further component of the protection arises from the rapid metabolism of the GSH adduct, which regenerates free GSH that can form further adducts and thus continue the detoxification process (4, 14, 20). Although there are multiple detoxification pathways for MG in Escherichia coli, the dominant one is the GSH-dependent glyoxalase I-II pathway (20) that catalyzes an internal oxidation-reduction reaction (ketone to alcohol coupled with aldehyde to carboxylic acid). Interference with any of these processes greatly sensitizes cells to the electrophile (10–14, 20).
NEM is a strong activator of the KefC system via the formation of the GSH adduct, N-ethylsuccinimido-S-GSH (ESG), and survival of exposure to this compound is enhanced by the activation of KefB and KefC (5, 12, 13). Cells rapidly recover from exposure to low concentrations of NEM, and metabolism of low concentrations of NEM by E. coli cells has been reported, but the product was not identified (32). NEM is a man-made chemical and itself should not have figured in the evolution of E. coli. Consequently, the fate of this GSH adduct is of interest since its breakdown would indicate that E. coli cells have been exposed to compounds similar to NEM, forcing the evolution of systems for their metabolism. Work with other electrophiles has shown that they may be metabolized in the cytoplasm (20), exported to the periplasm or growth medium (16), or degraded in the periplasm to yield glutamate, glycine, and nontoxic cysteine adducts (34). Thus, we have investigated the fate of NEM and its analogue N-phenylmaleimide (NPM) as convenient model compounds and because of the linkage of their metabolism to the activation of the KefB and KefC systems. We demonstrate that both compounds are rapidly metabolized to their corresponding maleamic acids that are then excreted into the medium. Throughout this process the GSH adducts are retained in the cytoplasm.
MATERIALS AND METHODS
Reagents.
All chemicals were of analytical grade and supplied by Sigma or BDH. The radiochemicals were supplied by New England Nuclear.
Media.
The media used is Kx minimal medium (pH 7), where x is the K+ concentration; thus, K120 contains 120 mM K+, K10 contains 10 mM K+, etc. (7). K120 medium consists of the following: K2HPO4, 46 mM; KH2PO4, 23 mM; (NH4)2SO4, 8 mM; MgSO4, 0.4 mM; FeSO4, 6 mM; sodium citrate, 1 mM; thiamine hydrochloride, 1 mg · liter−1; and glucose, 2 g · liter−1. K0 medium is similar, with equimolar sodium salts replacing the potassium phosphate. K1 was prepared by adding 1 M KCl to K0; medium with higher K concentrations was made by mixing suitable proportions of K0 and K120. LNa plates contain (per liter) 10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl. Chloramphenicol was added to incubations at a final concentration of 25 μg · ml−1.
Bacterial strains.
The E. coli strains used were Frag5 (F− thi gal rha lacZ kdpABC5), Frag56 (Frag5, gshA::Tn10Kan), MJF274 (Frag5, lacI trkD1), MJF276 (MJF274, kefB157 kefC::Tn10) (16), MJF374 [MJF274, Δ(yabF-kefC) Δ(yheR-kefB)] (26), NK6033 (Hfr PO1 Δ(gpt-lac)5 nadA50::Tn10 relA1 spoT1 thi-1 λ−), MJF342 [MJF274, gshA::Tn10 (Kan)], and MJF407 (MJF342, ggt-2 zhg::Tn10). Strain MJF407 was constructed by P1 transduction of strain MJF274 to Tetr with SH682 (F− ggt-2 zhg::Tn10) (31) as donor. Transductants derived from SH682 were screened for the acquisition of the ggt-2 mutant allele by their resistance to a toxic peptide, gamma-glutamylvaline (30).
Growth and assay of K+ pools.
The standard cell inoculum was obtained by initial growth of overnight cultures at 37°C in Kx plus limiting glucose (0.04% [wt/vol]). The overnight culture was then supplemented with glucose (0.2% [wt/vol]), and cells were permitted to double once prior to dilution 15-fold into prewarmed Kx medium for growth of cells prior to experimentation. Addition of electrophiles was made from fresh solutions. NEM was dissolved in 50% (vol/vol) ethanol, and NPM was dissolved in ethanol. For growth experiments with NEM and NPM, exponential-phase cells were prepared as described above. At an optical density at 650 nm (OD650) of 0.6, aliquots were taken and diluted to an OD650 of 0.1 and either NEM or NPM (5 to 15 μM) was added. Samples were taken at intervals, and the growth was monitored. A similar protocol was utilized for analysis of the growth in the presence of N-phenylmaleamic acid (NPMA), except that cells were diluted to an OD650 of 0.04 prior to the addition of the acid (0.1 to 20 mM). K+ pools were assayed as described previously (5, 11). An equivalence of 2 OD650 units = 1 mg of dry cell weight · ml−1 and a cytoplasmic volume of 1.6 μl · mg of dry cell weight−1 have been assumed throughout.
Strain NK6033 was grown in the presence of proline (50 μg · ml−1), thiamine (1 μg · ml−1), and nicotinamide (1 μg · ml−1; 8.1 μM). For starvation experiments, overnight cultures were grown to exponential phase in K120 medium with the above supplements and glucose (0.2% [wt/vol]) as sole carbon source. When the OD650 reached approximately 0.3, the culture was filtered, washed, and resuspended in prewarmed medium lacking nicotinamide and reincubated at 37°C in a shaking incubator. Samples were removed for analysis of respiration in a Clarke oxygen electrode, for measurement of NEM-elicited K+ efflux, and for analysis of metabolism of 14C-NEM. In parallel incubations, samples were labeled with 14C-nicotinamide (final concentration, 8.1 μM; specific activity, 62 μCi · μmol−1). These cultures were starved as described above, and the pools of pyridine nucleotides were determined from the retained radioactivity measured by scintillation counting.
NEM and NPM detoxification.
The metabolism of NEM and NPM was monitored by measuring the disappearance of the absorption at 305 nm (A305) due to loss of the unsaturated imide bond of NEM and NPM. The formation of the breakdown product was followed by the increase in A260. UV spectra were obtained with a Shimadzu UV 2101 PC spectrometer. The product of detoxification was recovered from the culture supernatant. The cells were harvested by centrifugation (4,500 × g at 4°C for 10 min), followed by filtration (Whatman HA; pore size, 0.45 μm) of the supernatant. The supernatant was desalted by passage through a Water Oasis HLB solid-phase extraction cartridge conditioned with methanol and equilibrated with water. The NEM-NPM detoxification product present in the supernatant was eluted with a small amount of water. This fraction was freeze-dried, and the residue was dissolved in 2D2O for 1H nuclear magnetic resonance (NMR) analyses. Authentic NPMA was synthesized by the method of Patel and Balasubramaniyan (29).
NMR spectroscopy.
All NMR spectra were acquired on a Varian Unity INOVA 400-MHz spectrometer. 1H NMR spectra were acquired with water presaturation in 5-mm NMR tubes. Chemical shifts were standardized to an HDO resonance of 4.61 ppm.
Measurement of intracellular 14C-NEM and 3H-GSH.
Harvested cells were washed with K10 medium prior to resuspension in K0 medium. 14C-NEM was added either from a solution prepared by diluting the original pentane with 50% (vol/vol) ethanol and sparging the solution with N2 gas to remove the pentane or from a secondary stock made by mixing this solution with nonradioactive NEM. Cell suspensions were filtered (Whatman; 0.45 μm [pore size], 2.5-cm diameter) and washed on the filter with several drops of K0 medium at 37°C. The filter was transferred to a scintillation vial and dried, and 3 ml of scintillation fluid was added and then counted for radioactivity. Osmotic downshock was performed as described previously (23). Measurement of intracellular 3H-GSH was by a similar protocol. Harvested cells were resuspended in K10 medium containing glucose (0.2% [wt/vol]), and 3H-GSH (500 μM; 2 Ci · mol−1) was added. Samples were treated as described above. After 20 min, GSH was removed from the medium by harvesting the cells by centrifugation and resuspension in the same volume of K10 medium, prewarmed to 37°C, containing glucose. The culture was divided equally between two water-jacketed vessels held at 37°C, and 500 μM NEM was added to one of them. Samples were taken for the analysis of the radioactivity in the cells as described above.
RESULTS
Rapid GSH-dependent detoxification of NEM by E. coli cells.
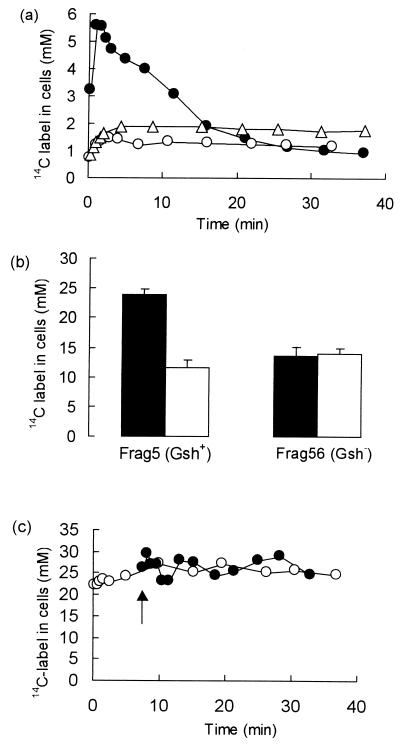
We have previously shown that the formation of the GSH adduct, ESG, is central to the ability of E. coli cells to survive exposure to low concentrations of NEM (9). However, the fate of this adduct has not been established. Addition of low concentrations of 14C-NEM (11 μM) to Frag5 cells resulted in the rapid influx of radiolabel, which peaked after 1 min at approximately 5 mM and was followed by a decline in the pool of radiolabeled material to approximately 1 mM over the next 25 min (Fig. 1a). Preincubating E. coli cells with chloramphenicol for 30 min did not affect the rate of breakdown of ESG, (t1/2 = 17 ± 0.8 min; n = 3, in the presence or absence of chloramphenicol), which suggests that de novo protein synthesis is not required for detoxification to take place. Strain Frag56 (GshA−), which lacks GSH synthesis, did not display the rapid influx of NEM seen in the GSH+ parent; rather, accumulation was progressive for the first 4 min, and the pool was then essentially unchanged over a 30-min incubation period. As a consequence, the maximum labeling of cells with 14C-NEM was much less than the parent and was equivalent to that seen after 25 min in the parent (Fig. 1a). Similarly, accumulation of 14C-NEM was significantly reduced when the GSH pool was sequestered by preincubation of Frag5 (GshA+) with excess iodoacetate, which we have previously shown forms carboxymethyl-S-GSH (5). Thus, the rapid accumulation and release of 14C-NEM is dependent upon the possession of a significant GSH pool.
FIG. 1.
Metabolism of NEM by E. coli cells. Cells were grown to exponential phase in K120 medium and prepared for the determination of 14C-NEM metabolism as described in Materials and Methods. (a) Rapid metabolism of NEM takes place in E. coli cells. 14C-NEM (specific activity, 6.5 Ci · mol−1) was added to cells suspended in K0 (OD650 = 1), and samples were taken at intervals, filtered, and washed dropwise with 1 ml of prewarmed K0 medium. The filters were then dried, and the radioactivity was measured by scintillation counting. A standard aliquot of 14C-NEM was dried onto a filter for the estimation of counting efficiency. Symbols: ●, Frag5 (11 μM NEM); ○, Frag56 (GshA−) (12 μM NEM); ▵, Frag5 preincubated with 2 mM iodoacetate for 5 min with 16 μM NEM added. (b) Incubation with high concentrations of NEM forms a pool of osmotically active metabolites. Cells of Frag5 (GshA+) and Frag56 (GshA−) were grown in double-strength K120 medium to facilitate osmotic downshock and then incubated with 500 μM 14C-NEM (64 μC · μmol−1, final specific activity) for up to 30 min. At intervals the samples were filtered and washed with prewarmed medium, and then either the radioactivity was determined or the cells were subjected to osmotic downshock with ice-cold water (23) prior to determination of the radioactivity in the cells. Filled bars, without osmotic downshock; open bars, after osmotic downshock. (c) Rapid turnover of the ESG pool in cells incubated with high concentrations of NEM. Cells were grown in K120, washed, and suspended in K0 medium (OD650 = 0.9). Symbols: ○, 500 μM 14C-NEM (130 mCi · mol−1) added (t = 0); ●, 500 μM nonradioactive NEM added (t = 0) and 20 μM 14C-NEM added after 7 min (arrow) to a final specific activity of 204 mCi · mol−1. Samples were filtered, washed, and dried, and the radioactivity was determined.
We have previously shown that the cells accumulate NEM as the GSH adduct, ESG (5). After 1 to 2 min of incubation with 10 to 15 μM NEM, approximately 50% of the GSH pool, which is 10 mM (8, 15, 29), was converted to ESG. Higher concentrations of 14C-NEM (200 to 500 μM) resulted in the immediate accumulation of a much larger intracellular pool of radiolabel, approximately 24 mM, which was steady for the next 30 min (Fig. 1b). The pool of 14C-NEM comprises both NEM attached to macromolecules and free ESG (Fig. 1b). Thus, when Frag5 (GshA+) cells incubated with 500 μM 14C-NEM were subjected to a hypo-osmotic shock with ice-cold water, which releases all free solutes from the cytoplasm (23), approximately 60% of the radiolabel was released by hypo-osmotic shock (Fig. 1b). This pool released by hypo-osmotic shock was equivalent in size to that expected for free GSH in E. coli cells (i.e., approximately 10 mM [8, 15, 28]). The bound pool represents between 2% (NEM concentration, 200 to 500 μM) and 10% (NEM concentration, 10 to 20 μM) of the total NEM present in the incubation. By spectrophotometric analysis (A305), approximately 90 to 95% of the NEM is metabolized via ESG (data not shown). The amount of label retained in the cells was found to be similar to that observed in the GSH-deficient mutant, Frag56 (GshA−) (Fig. 1b). Hypo-osmotic shock did not release radiolabeled material from Frag56 (GshA−), suggesting that the 14C-NEM accumulating in such cells was bound to protein or nucleic acids and that in the parent GSH is the major low-molecular-weight solute that binds NEM.
Rapid turnover of the ESG pool was demonstrated by preincubating Frag5 (GshA+) cells with unlabeled NEM and adding 20 μM 14C-NEM after the steady state had been established. If the ESG pool was slowly metabolized there should be no significant accumulation of 14C-NEM since the GSH pool would already be sequestered by unlabeled NEM. However, rapid influx of the radioactivity occurred with the steady state established within 30 s and generated a pool similar to that seen in the control incubation that had 500 μM 14C-NEM added at time zero (Fig. 1c). These data suggest that GSH remains available for the rapid reaction with the added 14C-NEM, which can only be generated from the breakdown of ESG formed with unlabeled NEM.
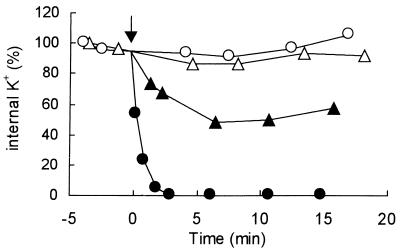
The kinetics of detoxification of ESG paralleled the activation of the KefB and KefC systems. Thus, 10 μM NEM provoked an initial K+ efflux that was slower than that observed with 500 μM NEM (Fig. 2), which saturates the GSH pool (5). Potassium efflux ceased after 7 min of incubation with 10 μM NEM, which corresponded to the decay of the ESG pool to approximately 2 mM (Fig. 1a and 2). Thus, the transient nature of the ESG pool in cells treated with low concentrations of NEM (10 μM) was paralleled by the degree of activation of KefB and KefC. High concentrations of NEM (>100 μM) give maximum rates of K+ efflux with no recovery within 30 min (Fig. 2), which matches the sustained ESG pool over this time period (Fig. 1b and c).
FIG. 2.
Potassium efflux consequent upon activation of KefB and KefC by NEM. K+ efflux was assayed in K0 medium with strain MJF274 as described previously (5, 11) using either 15 μM (▴) or 500 μM (●) NEM (arrow); the control incubations, with no addition, is indicated by the open symbols. The experiments have been replicated at least three times.
Determination of the identity of the detoxification product of NEM.
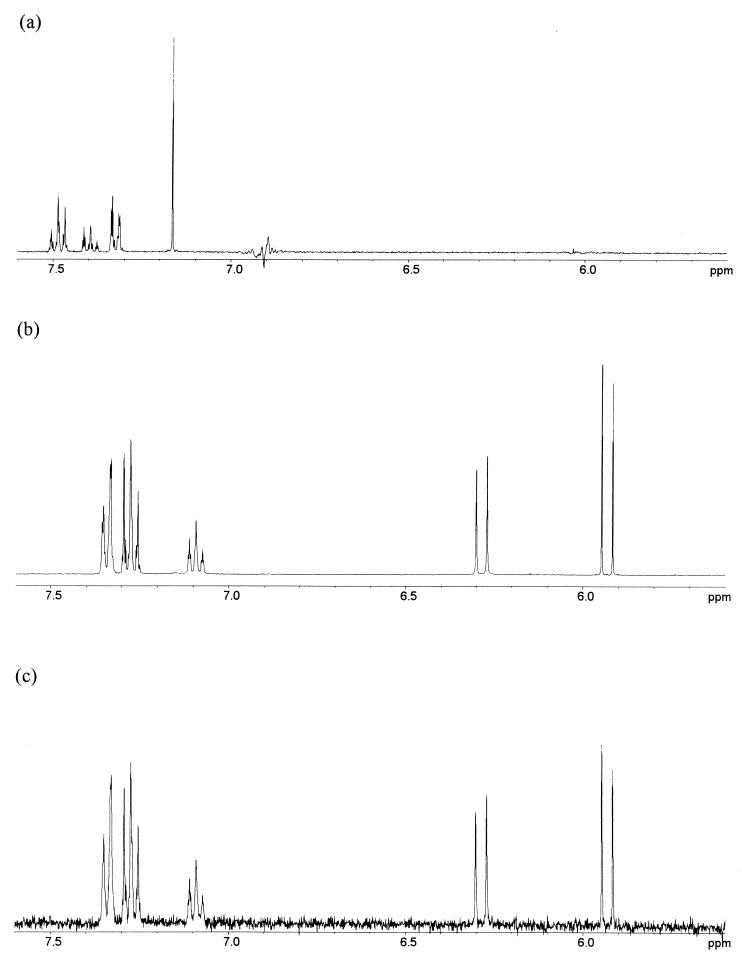
The analysis described above suggested a rapid turnover of ESG consistent with the detoxification of this compound. Thus, we analyzed the supernatant of cells treated with NEM and NPM to identify the degradation product. The supernatant of cells (either Frag5 or MJF274) treated with 300 μM NEM or NPM exhibited a characteristic A305 due to the conjugated alkene bond of the maleimide ring. This absorbance disappeared over the time course of the incubation (2 h) and was paralleled by an increase of A260 (24), which was subsequently shown to be characteristic of the product of detoxification (see below). NPM, which has a similar effect on cell growth and KefB and KefC activity, was chosen for the identification of the detoxification product because its phenyl group allowed easier detection by NMR and UV spectroscopy and its lower polarity made isolation easier. Analysis of the proton spectrum in D2O of the detoxification product from NPM, extracted from the supernatant of E. coli MJF274 cells incubated for 2 h with 300 μM NPM, revealed the presence of a single compound, NPMA (Fig. 3c). The structural assignment of the detoxification products of NEM and NPM (Table 1) was achieved by comparing the chemical shifts, multiplicities, and integration ratios in the proton spectra to those of a synthetic sample of NPMA (Fig. 3b). The 1H NMR spectrum of this compound exhibited resonances for a phenyl moiety, and a pair of doublets in the ratio of 1:1 arising from a cis alkene moiety with protons in different environments (Fig. 3b and c, coupling constant characteristic for the cis double bond, 12.4 Hz). The chemical shifts and coupling constants are listed in Table 1. Similarly, the 1H NMR spectrum in D2O of the extract of E. coli MJF274 cells after incubation with NEM revealed the product of the detoxification metabolism as N-ethylmaleamic acid (NEMA). The resonances for the ethyl moiety are maintained, and the signals for an asymmetrically substituted alkene are again evident (Table 1). Similarly, the UV spectrum of NPMA matched that observed in supernatants of cells detoxifying NPM (i.e., an absorption maximum at 260 nm and no absorbance at 305 nm) (D. McLaggan, H. Ruffino, M. Jaspars, and I. R. Booth, unpublished data). Quantification of the disappearance of NEM and NPM and the appearance of NEMA and NPMA by NMR would be unreliable since an isolation procedure was involved to obtain the maleamic acids. NMR work on the crude mixture was initially attempted but proved difficult since the signals of the maleimides and maleamic acids were obscured by other components of the medium.
FIG. 3.
NMR spectra for NPM and NPMA. (a) The 1H NMR spectrum of NPM in d6-dimethyl sulfoxide (DMSO) at 400 MHz (DMSO reference at 2.50 ppm). (b) The 1H NMR spectrum of synthetic NPMA in K10 medium-D2O at 400 MHz. (c) 1H NMR of the product of detoxification of NPM by E. coli cells in D2O at 400 MHz.
TABLE 1.
1H NMR data of NPM, NEM, NPMA, and NEMA in D2O at 400 MHz
| Assignment | No. of protons, chemical shifts (δ, ppm), multiplicity and coupling constant (J, Hz)a for:
|
|||
|---|---|---|---|---|
| NPM | NEM | NPMA | NEMA | |
| Ha | 2H, 7.10, s | 2H, 6.64, s | 1H, 6.28, d, 12.4 Hz | 1H, 6.13, d, 12.4 Hz |
| Hb | 1H, 5.93, d, 12.4 Hz | 1H, 5.75, d, 12.4 Hz | ||
| ArH | 5H, 7.25–7.45, m | 1H, 7.07–7.10, m 4H, 7.25–7.35, m | ||
| -CH2 | 2H, 3.54, q, 7.2 Hz | 2H, 3.06, q, 7.2 Hz | ||
| -CH3 | 3H, 1.15, t, 7.2 Hz | 3H, 0.95, t, 7.2 Hz | ||
Spectrum in DMSO-d6: nH = number of protons, where n is a number between 1 and 5; the chemical shift (δ) is shown in ppm; s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet of peaks on the NMR spectrum, and J (Hz) is the coupling constant between these peaks.
We have previously shown that E. coli cells recover from exposure to 10 μM NEM and reestablish exponential growth at the same rate as nontreated cultures (12). To confirm that the product of NPM detoxification was nontoxic, NPMA was synthesized (29) and added to growing cells of MJF274. No growth inhibition was observed with up to 20 mM NPMA; in contrast, 10 μM NPM inhibited growth for at least 3 h (McLaggan et al., unpublished). Thus, the detoxification products are essentially inactive as growth inhibitors for E. coli cells.
NAD+ is not required for detoxification.
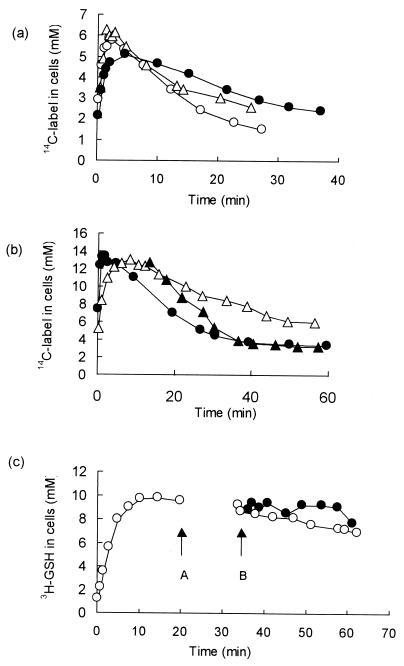
The mechanism of breakdown of ESG to GSH and NEMA is not known. ESG is stable when incubated in aqueous solution (17). An NEM reductase activity has been reported in E. coli (24), and a GSH conjugate-specific dehydrogenase has been characterized in Rhodococcus spp. (33). Since such enzymes were potential candidates for the detoxification of ESG, the rate of detoxification in E. coli cells depleted of NAD+ was investigated. An E. coli mutant (NK6033, nadA::Tn10) auxotrophic for nicotinic acid was starved for nicotinamide for either 6 or 24 h to deplete the pools of NAD+ and NADP (19), and the metabolism of ESG was determined. The nicotinamide pool fell from approximately 1.6 ± 0.3 mM during steady-state growth to approximately 30 to 70 μM after 24 h of starvation. The initial decline of the pool was rapid, after 8 h the pool had fallen to approximately 0.1 mM (McLaggan et al., unpublished), and our data reproduced earlier studies (19) both quantitatively and qualitatively. The initial decrease in the pool has previously been shown to preferentially affect NAD(H) (19), and this was supported in our experiments by the observed decline in the rate of glucose-stimulated respiration, which fell rapidly after the removal of nicotinamide from the medium and ceased after 4 h of starvation for the vitamin. These data are consistent with a major loss of the NAD(H) pool, which is required for glyceraldehyde-3-phosphate dehydrogenase and for enzymes of the tricarboxylic acid cycle. The formation of ESG was unaffected by starvation for nicotinamide. Similarly, ESG continued to be broken down, although there was a slightly reduced rate in cells starved for 24 h, but this was equivalent to the effect seen with cells depleted of glucose for 3 h (Fig. 4a). Thus, it is unlikely that an NAD+-dependent dehydrogenase is required for the metabolism of ESG. The activation of KefB and KefC by NEM was also unaffected by starvation of the nadA mutant (McLaggan et al., unpublished). The only other factor that was found to significantly affect the rate of detoxification was the presence of glucose. The rate of breakdown of the ESG was reduced if cells were incubated for 3 h in the absence of glucose (Fig. 4b). Addition of glucose after the formation of the ESG pool restored rapid efflux of the labeled material (Fig. 4b). Thus, energy is either required for the breakdown of ESG or for the expulsion of the product NEMA from the cell.
FIG. 4.
Requirements for metabolism of NEM by E. coli cells. (a) Strain NK6033 (NadA−) was grown and starved for nicotinamide as described in Materials and Methods. Samples were taken and incubated with 10 μM 14C-NEM (specific activity, 6.5 Ci · mol−1). Symbols: ▵, control, no starvation; ○, after 6 h of starvation for nicotinamide; ● NK6033 starved of nicotinamide for 24 h. (b) Cells of strain MJF274 were grown to mid-exponential phase in K10 medium, filtered, and washed with glucose-free medium and suspended to a final OD650 of 0.1. One aliquot was incubated with glucose (0.2% [wt/vol]) and assayed immediately at 37°C for 14C-NEM metabolism (●) as described for Fig. 1a. An identical aliquot was incubated without glucose for 3 h at 37°C and then assayed for NEM metabolism (▵). After 15 min the culture was split into two identical portions, and glucose was added to one culture (▴), and the incubation was continued. (c) Cells of MJF407 (GshA− Ggt−) were prepared as described in Materials and Methods and incubated with 500 μM 3H-GSH (2 Ci · mol−1) (○). After 20 min (arrow A), the cells were harvested by centrifugation and resuspended in prewarmed medium lacking 3H-GSH; the culture was then split, and 500 μM NEM was added to one aliquot of cells (●) (arrow B) while the other was used as a control (○). The radioactivity in the cells determined as described for Fig. 1a. All experiments were repeated at least three times, and the data shown are representative of the phenomena observed on each occasion.
ESG is not exported from E. coli cells.
GSH adducts can be pumped from E. coli cells as a component of the detoxification process (16). In addition, a number of multisubstrate efflux pumps are also found in great variety in bacterial cells (27), and it is known that the conjugate formed between chlorodinitrobenzene (CDNB) and GSH can be exported to the periplasm in some bacteria (16, 34). Extracellular GSH can be either transported into cells or metabolized in the periplasm by gamma-glutamyl peptidase (Ggt), followed by the transport of the constituent amino acids into the cytoplasm (30). To enable the GSH pool to be labeled for analysis of the efflux of GSH conjugates, a GshA− Ggt− strain (MJF407) lacking significant periplasmic Ggt activity was constructed. MJF407 (GshA− Ggt−) is unable to degrade GSH and, thus, incubation of such cells with 500 μM 3H-GSH created a stable cytoplasmic pool of approximately 10 mM GSH (Fig. 4c). The addition of 500 μM NEM did not stimulate the efflux of the GSH pool even upon extended incubation (15 min) (Fig. 4c). Under these conditions the GSH is converted almost stoichiometrically to ESG (see above), and thus any efflux should be evident as a decrease of the 3H-GSH pool. These data show that the GSH pool is retained in the cytoplasm during the detoxification of NEM and thus suggest that export of the conjugate is unlikely to be significant. Further, detoxification was also independent of the Ggt, since a Ggt- strain exhibited NEM detoxification essentially identical to that shown by MJF274 when incubated with 15 μM NEM (McLaggan et al., unpublished).
DISCUSSION
We have demonstrated that E. coli cells can detoxify NEM and NPM to the equivalent maleamic acids. Detoxification occurs via a GSH adduct, and the final step leads to regeneration of GSH via hydrolysis of the C-S bond formed between the electrophile and the cysteine of GSH (Fig. 5). Thus, E. coli cells are able to break down the GSH S conjugate to recycle GSH and release a nonreactive form of the electrophile from the cell. Formation of the adduct is rapid (Fig. 1a); at 10 to 15 μM NEM, approximately t1/2 = 30 s (n = 6) was required for formation, whereas the breakdown was relatively slow (t1/2 = 15 ± 3 min [n = 6]). Although NEM is a man-made compound, there are a number of natural products with structures related to the maleimide ring, e.g., showdomycin, pencolide, and maleimycin (3, 6), that are produced by bacteria and fungi. Such compounds may be detoxified by the formation of GSH adducts. Resistance to fosfomycin, (1,2-epoxipropyl)phosphonic acid, in E. coli is dependent upon formation of a GSH adduct catalyzed by a specific GSH S-transferase (18). While this compound is not a maleimide, it illustrates the general principle that natural antibacterial compounds can be detoxified by the formation of GSH conjugates. The fate of the fosfomycin-GSH adduct is not clear; however, the high levels of resistance conferred by this mechanism (18) suggest that this could only be achieved if the adduct is further metabolized, leading to recycling of free GSH.
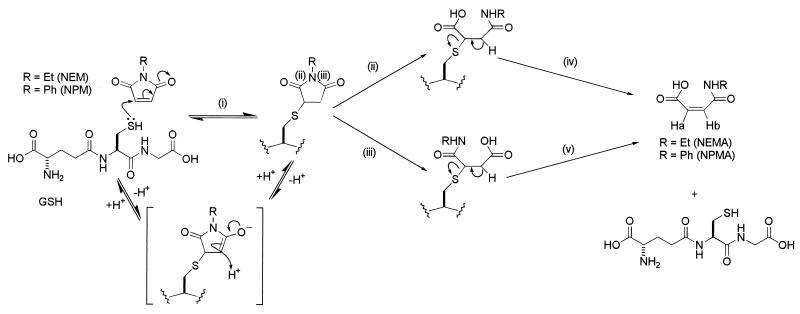
FIG. 5.
Possible mechanism for the detoxification of N-substituted maleimides in E. coli. (i) Spontaneous nucleophilic Michael attack of the cysteine lone pair in GSH on the NEM to give the hemithioacetal ESG. This reaction is reversible. (ii and iii) Hydrolysis of the ESG (at the positions marked) by a putative imidase. The reaction can occur at both sites to give the intermediates shown, both giving the same final product. (iv and v) Elimination of GSH from both intermediates to form the double bond in NEMA and NPMA. The NMR chemical shifts for Ha and Hb are given in Table 1.
For NEM the formation of maleamic acid probably occurs via an enzymatic mechanism, and a reaction pathway can be predicted that involves a broad-specificity imidase (Fig. 5). None of the steps in the breakdown of the ESG to yield NEMA requires a redox reaction but rather arise by hydrolysis of the adduct (Fig. 5, ii and iii) and the elimination of the GSH (iv and v). This contrasts with the internal redox reaction required for the detoxification of MG. An NEM reductase has been reported in E. coli cells, but this does not appear to be involved in the detoxification of ESG since depletion of the NAD+ pool did not inhibit the rate of ESG breakdown. Induction of the putative ESG imidase is not required since chloramphenicol did not inhibit either ESG formation or destruction. Preliminary data suggest that the enzyme catalyzing hydrolysis of the ESG adduct is not a component of the OxyR and RpoS regulons, which are the major stress regulons required for survival of exposure to NEM (13). Thus, mutants lacking these regulons continue to detoxify NEM at the same rate as the parent (McLaggan et al., unpublished). Excretion of the maleamic acid rather than the expulsion of the GSH conjugate, as favored by plant cells, animal cells, and some bacterial cells, is an economical solution for E. coli since it leads to retention of GSH that can then participate in further rounds of detoxification. Only in this respect does the detoxification system for NEM resemble that for MG, which also leads to recycling of the GSH. In contrast, GSH conjugates of CDNB are exported from members of the Enterobacteriaciae (16, 34), but this may only take place due to the slow rate of hydrolysis of these compounds, which leads to their extended presence in the cytoplasm. Clearly, the cell needs alternative strategies for dealing with those GSH adducts that are rapidly metabolized and those that are more recalcitrant.
ACKNOWLEDGMENTS
We thank H. Suzuki (Kyoto University, Kyoto, Japan) for his kind donation of strain SH682 and G. Storz (National Institutes of Health, Bethesda, Md.) for the oxyR::Tn10Kan. We thank the E. coli Genetic Stock Centre for strain NK6033. We also thank Conor O'Byrne for his careful reading and criticism of the manuscript.
This work was supported by a Wellcome Trust Programme grant to I.R.B. (040174) and a supplement grant to I.R.B. and M.J.
REFERENCES
- 1.Almirón M A, Link A J, Furlong D, Kolter R D. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli cells. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 2.Arca P, Rico M, Brana A F, Villar C J, Hardisson C, Suarez J E. Formation of an adduct between fosfomycin and glutathione: a new mechanism of antibiotic resistance in bacteria. Antimicrob Agents Chemother. 1988;32:1552–1556. doi: 10.1128/aac.32.10.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birkinshaw J H, Kalyanpur M G, Stickings C E. Studies on the biochemistry of microorganisms. 113. Pencolide, a nitrogen-containing metabolite of Penicillium multicolor Grigorievna-Manilova and poradievova. Biochem J. 1963;86:237–243. doi: 10.1042/bj0860237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper R A. Metabolism of methylglyoxal in microorganisms. Annu Rev Microbiol. 1984;38:49–68. doi: 10.1146/annurev.mi.38.100184.000405. [DOI] [PubMed] [Google Scholar]
- 5.Elmore M J, Lamb A J, Ritchie G Y, Douglas R M, Munro A, Gajewska A, Booth I R. Activation of potassium efflux from Escherichia coli by glutathione metabolites. Mol Microbiol. 1990;4:405–412. doi: 10.1111/j.1365-2958.1990.tb00607.x. [DOI] [PubMed] [Google Scholar]
- 6.Elstner E F, Carnes D M, Suhadolnik R J, Kreishman G P, Schweizer M P, Robbins R K. Isolation, structural elucidation, biological properties and biosynthesis of maleimycin, a new bicyclic maleimide antibiotic isolated from culture filtrates of Streptomyces showdoensis. Biochemistry. 1973;12:4992–4997. doi: 10.1021/bi00748a027. [DOI] [PubMed] [Google Scholar]
- 7.Epstein W, Kim B S. Potassium transport loci in Escherichia coli K-12. J Bacteriol. 1971;108:639–644. doi: 10.1128/jb.108.2.639-644.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahey R C, Brown W C, Adams W B, Worsham M B. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson G P, Booth I R. Importance of glutathione for growth and survival of Escherichia coli cells: detoxification of methylglyoxal and maintenance of intracellular K+ J Bacteriol. 1998;180:4314–4318. doi: 10.1128/jb.180.16.4314-4318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson G P, Munro A W, Douglas R M, McLaggan D, Booth I R. Activation of potassium channels during metabolite detoxification in Escherichia coli. Mol Microbiol. 1993;9:1297–1303. doi: 10.1111/j.1365-2958.1993.tb01259.x. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson G P, McLaggan D, Booth I R. Potassium channel activation by glutathione-S-conjugates in Escherichia coli: protection against methylglyoxal is mediated by cytoplasmic acidification. Mol Microbiol. 1995;17:1025–1033. doi: 10.1111/j.1365-2958.1995.mmi_17061025.x. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson G P, Nikolaev Y, McLaggan D, Maclean M J, Booth I R. Survival during exposure to the electrophilic reagent N-ethylmaleimide in Escherichia coli: role of KefB and KefC potassium channels. J Bacteriol. 1997;179:1007–1012. doi: 10.1128/jb.179.4.1007-1012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson G P, Creighton R I, Nikolaev Y, Booth I R. The importance of RpoS and Dps in the survival of both exponential- and stationary-phase Escherichia coli cells against the electrophile, N-ethylmaleimide. J Bacteriol. 1998;180:1030–1036. doi: 10.1128/jb.180.5.1030-1036.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson G P, Tötemeyer S, MacLean M J, Booth I R. Methylglyoxal production in bacteria: suicide or survival? Arch Microbiol. 1998;170:209–219. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg J T, Demple B. Glutathione in Escherichia coli is dispensable for resistance to H2O2 and gamma radiation. J Bacteriol. 1986;168:1026–1029. doi: 10.1128/jb.168.2.1026-1029.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaluzna A, Bartosz G. Transport of glutathione-S-conjugates in Escherichia coli. Biochem Mol Biol Int. 1997;43:161–171. doi: 10.1080/15216549700203931. [DOI] [PubMed] [Google Scholar]
- 17.Kermack W O, Matheson N A. The synthesis of some analogues of glutathione. Biochem J. 1957;65:45–48. doi: 10.1042/bj0650045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llaneza J, Villar C J, Salas J A, Suarez J E, Mendoza M C, Hardisson C. Plasmid-mediated fosfomycin resistance is due to enzymatic modification of the antibiotic. Antimicrob Agents Chemother. 1985;28:163–164. doi: 10.1128/aac.28.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundquist R, Olivera B M. Pyridine nucleotide metabolism in Escherichia coli. II. Niacin starvation. J Biol Chem. 1973;248:5137–5143. [PubMed] [Google Scholar]
- 20.MacLean M J, Ness L S, Ferguson G P, Booth I R. The role of glyoxalase I in the detoxification of methylglyoxal and in the activation of the KefB K+ efflux system in Escherichia coli. Mol Microbiol. 1998;27:563–571. doi: 10.1046/j.1365-2958.1998.00701.x. [DOI] [PubMed] [Google Scholar]
- 21.Mannervik B, Danielson U H. Glutathione-S-transferases—structure and catalytic activity. Crit Rev Biochem. 1988;23:283–337. doi: 10.3109/10409238809088226. [DOI] [PubMed] [Google Scholar]
- 22.Matin A. Physiology, molecular biology and the applications of the bacterial starvation response. J Appl Bacteriol. 1992;73:49S–57S. doi: 10.1111/j.1365-2672.1992.tb03624.x. [DOI] [PubMed] [Google Scholar]
- 23.McLaggan D, Naprstek J, Buurman E T, Epstein W. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J Biol Chem. 1994;269:1911–1917. [PubMed] [Google Scholar]
- 24.Miura K, Tomioka Y, Hoshi Y, Suzuki H, Yonezawa M, Hishinuma T, Mizugaki M. The effects of unsaturated fatty acids, oxidizing agents and Michael reaction acceptors on the induction of N-ethylmaleimide reductase in Escherichia coli: possible application for drug design of chemoprotectors. Methods Find Exp Clin Pharmacol. 1997;19:147–151. [PubMed] [Google Scholar]
- 25.Ness L S, Booth I R. Different foci for the regulation of the activity of the KefB and KefC glutathione-gated K+ efflux systems. J Biol Chem. 1999;274:9524–9530. doi: 10.1074/jbc.274.14.9524. [DOI] [PubMed] [Google Scholar]
- 26.Ness L S, Ferguson G P, Nikolaev Y, Booth I R. Survival of Escherichia coli cells exposed to iodoacetate and chlorodinitrobenzene is independent of the glutathione-gated K+ efflux systems KefB and KefC. Appl Environ Microbiol. 1997;63:4083–4086. doi: 10.1128/aem.63.10.4083-4086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido H. Multidrug efflux systems of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oktyabrsky O N, Smirnova G V. Changes in intracellular potassium and thiol levels in Escherichia coli K-12 under various stresses. Biochem Mol Biol Int. 1993;30:377–383. [PubMed] [Google Scholar]
- 29.Patel M V, Balasubramaniyan V. Maleamic acids from maleic anhydride and aromatic amines. Indian J Chem. 1977;15B:1142–1143. [Google Scholar]
- 30.Suzuki H, Hashimoto W, Kumagai H. Escherichia coli K-12 can utilize an exogenous gamma-glutamyl peptide as an amino acid source, for which gamma-glutamyl peptidase is essential. J Bacteriol. 1993;175:6038–6040. doi: 10.1128/jb.175.18.6038-6040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki H, Kumagai H, Tochikura T. Isolation, genetic mapping and characterization of Escherichia coli K-12 mutants lacking γ-glutamyl peptidase. J Bacteriol. 1987;169:3926–3931. doi: 10.1128/jb.169.9.3926-3931.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tötemeyer S, Barrett-Bee K, Booth I R. Generation of a transient nonculturable state in Pseudomonas putida during detoxification of N-ethylmaleimide. Microbiology. 1996;142:2857–2862. [Google Scholar]
- 33.Van Hylckama Vlieg J E T, Kingma J, Kruizinga W, Janssen D B. Purification of a glutathione S-transferase and a glutathione conjugate-specific dehydrogenase involved in isoprene metabolism in Rhodococcus sp. strain AD45. J Bacteriol. 1999;181:2094–2101. doi: 10.1128/jb.181.7.2094-2101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zablotowicz R M, Hoagland R E, Locke M A, Hickey W J. Glutathione S-transferase activity and metabolism of glutathione conjugates by rhizosphere bacteria. Appl Environ Microbiol. 1995;61:1054–1060. doi: 10.1128/aem.61.3.1054-1060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]