Abstract
Twelve forms of programmed cell death (PCD) have been described in mammalian cells, but which of them occurs during embryonic development and the role played by the p53 transcription factor and tumor suppressor remains enigmatic. Although p53 is not required for mouse embryonic development, some studies conclude that PCD in pluripotent embryonic stem cells from mice (mESCs) or humans (hESCs) is p53-dependent whereas others conclude that it is not. Given the importance of pluripotent stem cells as models of embryonic development and their applications in regenerative medicine, resolving this enigma is essential. This review reconciles contradictory results based on the facts that p53 cannot induce lethality in mice until gastrulation and that experimental conditions could account for differences in results with ESCs. Consequently, activation of the G2-checkpoint in mouse ESCs is p53-independent and generally, if not always, results in noncanonical apoptosis. Once initiated, PCD occurs at equivalent rates and to equivalent extents regardless of the presence or absence of p53. However, depending on experimental conditions, p53 can accelerate initiation of PCD in ESCs and late-stage blastocysts. In contrast, DNA damage following differentiation of ESCs in vitro or formation of embryonic fibroblasts in vivo induces p53-dependent cell cycle arrest and senescence.
Keywords: apoptosis, cell cycle, differentiation, embryo, p53, pluripotent, programmed cell death, stem cells
Graphical Abstract
Neither cell cycle arrest nor programmed cell death requires p53 prior to gastrulation, at which stage DNA damage induces p53-dependent cell cycle arrest and senescence.
Significance Statement.
Programmed cell death (PCD) and survival are inherent components of mammalian development. In addition to its role as “guardian of the genome”, the p53 transcription factor and tumor suppressor has been reported to regulate at least six different forms of PCD. Given the importance of pluripotent stem cells as models of embryonic development and their applications in regenerative medicine, identifying which of the 12 forms of PCD respond to stress imposed at the beginning of embryogenesis and the role of p53 in regulating them is essential.
Introduction
Cell death is a normal event in mammalian development, as well as a cellular response to stressful conditions. The mechanisms that cause cell death are categorized as either necrosis or programmed cell death (PCD). Necrosis results from the progressive degradative action of enzymes and is typically followed by inflammation. Necrosis requires neither energy nor effector proteases1; it is simply a response to physical damage or pathology that does not occur during normal animal development.2 In contrast, PCD is a sequence of genetically programmed events by which a cell provokes its own demise in response to a stimulus. PCD occurs in mammals as early as the blastocyst stage during preimplantation development and as late as tissue homeostasis in adulthood.3 During organogenesis, both PCD and programmed cell senescence are involved in sculpting structures by eliminating interdigital webbings, converting solid structures into hollow tubes, and removing excess cells from nervous, immune, and reproductive systems.4,5 In postimplantation embryos, the proamniotic cavity is formed by PCD of the ectodermal cells in the core of the developing egg cylinder6 (Fig. 1).
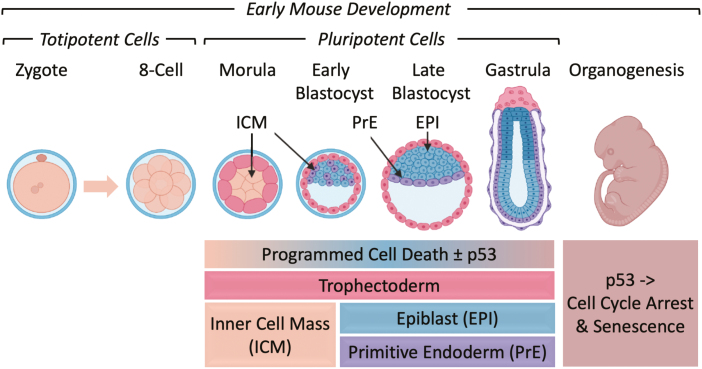
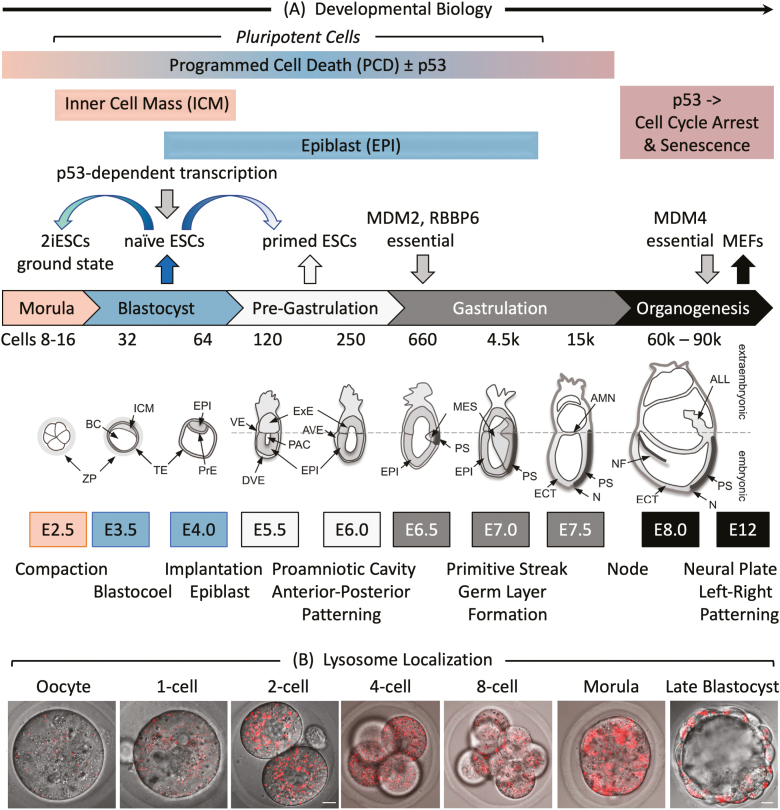
Figure 1.
Early mouse embryonic development. (A) The number of cells, days post-coitum (E2.5-E12), and morphogenetic events are indicated. ALL, allantois; AMN, amnion; AVE, anterior visceral endoderm; BC, blastocyst cavity; DVE, distal visceral endoderm; ECT, ectoderm; EPI, epiblast; ExE, extraembryonic ectoderm; ICM, inner cell mass; MES, mesoderm; N, node; NF, neural fold; PAC, proamniotic cavity; PrE, primitive endoderm; PS, primitive streak; TE, trophectoderm; VE, visceral endoderm; ZP, zona pellucida. Adapted from Ref. 116. Preimplantation development begins with totipotent blastomeres (1-8 cell stage) encapsulated by the zona pellucida. Totipotent cells can give rise to both placental and embryonic cells. When the blastomeres develop cell-to-cell adhesion (compaction), the outer blastomeres differentiate into the trophectoderm while the remaining blastomeres form the inner cell mass. The epithelial trophoblast cells (trophectoderm) are multipotent; they differentiate only into cells required for implantation and placentation. The inner cell mass (recognized upon formation of a blastocoel cavity) differentiates into the epiblast and the primitive endoderm. Postimplantation development begins when the primitive endoderm differentiates into multipotent visceral and parietal endoderm. Mesoderm and ectoderm are derived from the epiblast during gastrulation. Gastrulation begins at the primitive streak, from which mesoderm and endoderm progenitor’s ingress and begin to differentiate.117 Mouse embryonic fibroblasts (MEFs) are derived from E12-E14 embryos. Ablation of the Mdm2, Rbbp6, or Mdm4 gene is lethal in embryos at the indicated times. Mouse embryonic stem cells (mESCs) are derived from the epiblast in blastocysts.49 mESCs cultured in the presence of serum and LIF interleukin-6 are considered “naïve” pluripotent cells, because they can give rise to all the cells of the embryo, but not to the trophectoderm. Naïve mESCs cultured in defined medium (no serum) containing 2 metabolic inhibitors are considered totipotent “ground-state” ESCs (2iESCs), because they give rise to both extraembryonic and embryonic cells. Naïve mESCs cultured in the presence of activin and fibroblast growth factor generate pluripotent “primed” ESCs, because they give rise to the same cells as “naïve mESCs,” but they cannot generate chimeric animals.118 Human embryonic stem cells (hESCs) and mouse epiblast stem cells (EpiSCs) are derived from the epiblast of post-implantation blastocysts.48 (B) Images of LysoTracker Red stained oocytes and preimplantation embryos revealed that the number of lysosomes increased after fertilization.36 Scale bar is 10 µm.
Remarkably, the mechanism of PCD during mammalian development, as well as the role of the p53 transcription factor in PCD, remain controversial. Pluripotent embryonic stem cells (ESCs) respond to stressful conditions, such as DNA damage, by arresting cell proliferation and undergoing PCD, but the form of PCD and the role of p53 remain unresolved. Studies not reviewed herein reveal 2 roles of p53 during mouse embryonic development are promoting genomic stability and maintaining a differentiated state by suppressing pluripotent gene expression.7 However, in differentiated cells, the primary role of the p53 transcription factor is regulating cell cycle arrest, senescence, and PCD in response to DNA damage and other stresses. p53 operates primarily through transcriptional activation of the cyclin-dependent kinase inhibitor CDKN1A/p21 to prevent cells from entering S-phase, and the proapoptotic proteins BBC3/PUMA and NOXA/PMAIP18,9 to induce PCD. p53 also regulates transcription of genes involved in DNA repair (DDB2, XPC, GADD45A)10 and cell senescence (p21, PAI1, PML).11,12 Low levels of p53 trigger cell cycle arrest and induction of DNA repair pathways specific to the lesion of concern, but as p53 continues to accumulate with time, it eventually triggers PCD, thereby removing cells.13,14
Remarkably, neither p53, p21, nor PUMA is required for mouse development.15,16 Therefore, either p53 is not required for specific events, or events that require p53 are themselves not required for mammalian development. Alternatively, in the absence of p53, cells utilize a different form of PCD. Therefore, the goal of this review is to summarize the status of PCD at the beginning of mammalian development and to reconcile disparate data based on differences in experimental conditions with mouse ESCs and the fact that p53 cannot induce lethality during mouse embryogenesis until gastrulation.
Programmed Cell Death
Of the 12 forms of PCD characterized in human cells, 4 are associated with mammalian development and 8 with disease states (Table 1). Apoptosis is the most reported PCD and the only one that employs caspases 3, 6, and 7. Apoptosis can be induced by DNA damage, unfolded proteins, reactive oxygen species (ROS), or disruption of cell division using either an intrinsic or an extrinsic pathway. The intrinsic pathway regulates mitochondrial permeability via the Bcl-2 family of cytokines. The extrinsic pathway is triggered by ligand binding to tumor necrosis factor-family receptors in the plasma membrane that activate caspases. Both pathways activate initiator caspase (CASP) 2, 8, 9, or 10, which then activate effector CASP3, 6, and 7, which then degrade cellular proteins indiscriminately. Expression of proapoptotic genes Bax, Puma, Bid, and Bcl-2, as well as Casp6 and Apaf1, a coactivator of Casp9, are upregulated by the p53 transcription factor.
Table 1.
Hallmarks of programmed cell death in mammals.
| Form | p53 | Morphology | Biochemistry |
|---|---|---|---|
| Associated with development | |||
| Apoptosis18,19 | p53 | Cell rounding, nuclear condensation, membrane blebbing, apoptotic bodies | Activates CASP3 and PARP1, DNA fragmentation and loss, ΔΨm dissipation, phosphatidylserine exposure |
| Anoikis20 | p53 | Anchorage-dependent cells detach from the extracellular matrix | Cleaved EMC proteins (laminin, fibronectin, vitronectin) → apoptosis |
| Lysosome dependent21 | Plasma membrane repair, lysosome membrane permeabilization | Release of lysosomal hydrolytic enzymes (cathepsins), lysosomal iron-induced oxidative injury | |
| Autophagy dependent22,23 | p53 | Autophagic vacuolization | LC3-I to LC3-II conversion, increases autophagic flux and lysosomal activity |
| Associated with disease | |||
| Necroptosis24 | p53 | Cell swells, PMR, moderate chromatin condensation | Activates RIPK1, RIPK3, and MLKL, cytosolic necrosome formation |
| Oxeiptosis25 | Apoptosis-like | ROS-dependent, activates KEAP1 and NFE2L2. caspase-independent, no AIFM1 translocation | |
| Ferroptosis26 | p53 | Small mitochondria (mt), reduced mt-crista, elevated mt-membrane densities, mt-membrane rupture | Iron accumulates, lipid peroxidation, ΔΨm dissipation, LC3-I to LC3-II conversion, glutaminolysis, caspase-independent |
| Parthanatos27 | p53 | Chromatin condensation, large DNA fragments, no cell swelling, apoptotic bodies or small DNA fragments, PMR | Oxidative stress (ROS)-induced, PARP1 activation, ΔΨm dissipation, caspase-independent, NAD+ and ATP depletion, accumulates PARP polymers, AIFM1 translocation |
| Alkaliptosis28 | Necrosis-like | Intracellular alkalinization, activates NF-κB, caspase-independent | |
| Pyroptosis29 | No cell swelling, PMR, bubbling, moderate chromatin condensation | Activates CASP1, CASP3, and GSDMD, GSDMD-N-induced pore formation, IL1B released | |
| Entotic30 | One cell invades another | Activates adhesion proteins and actomyosin, LC3-associated phagocytosis | |
| Netotic31 | PMR, nuclear membrane collapse, chromatin fiber release | Forms NETs, release and translocation of granular enzymes, histone citrullination | |
Note: ΔΨm is mitochondrial membrane potential.37 Reactive oxygen species is reactive oxygen species. Neutrophil Extracellular Traps (NETs) are neutrophil extracellular traps. LC3 is MAP1LC3B. Dying cells release small vesicular apoptotic bodies.38 Plasma membrane rupture releases intracellular molecules that propagate inflammatory response.39 Apoptosis-inducing factor 1 (AIFM1) translocates from mitochondria to nucleus.40 EMC is extracellular matrix. Adapted from Ref. 17.
NMRs, neutrophil extracellular traps; PARP, poly(ADP-ribose) polymerase; PMR, plasma membrane rupture; ROS, reactive oxygen species.
Apoptosis is recognized by DNA fragmentation, accumulation of cells containing <2N DNA, binding of annexin-V to detect phosphatidylserine exposure in the plasma membrane, propidium iodide, or trypan blue staining to detect plasma membrane permeability, accumulation of γH2AX to confirm double-strand DNA breaks, and cleavage of poly(ADP-ribose) polymerase (PARP) and CASP3. Apoptosis can be either p53-dependent or p53-independent.
Anoikis is the induction of apoptosis when cells lose their attachment to the extracellular matrix (EMC) and neighboring cells. Anoikis suppresses tumor metastasis and eliminates ectopic proliferation of misplaced progenitor cells during tissue development.
Autophagy-dependent PCD is an unlikely death mechanism because autophagy is primarily a survival mechanism. Autophagy is a process in which cytoplasmic organelles, proteins, and macromolecules are degraded to produce new macromolecules and energy. Thus, starvation activates autophagy to maintain homeostasis and viability, and the autophagy gene ATG7 inhibits p53-dependent cell cycle arrest and PCD.32,33
This paradox could be resolved in 3 ways. First, mild stress might induce autophagy for survival, whereas severe stress induces autophagy for PCD. This hypothesis is analogous to the fact that mild DNA damage induces p53-dependent cell cycle arrest in differentiated human cells, whereas severe DNA damage induces p53-dependent PCD or senescence.34 Alternatively, PCD might disrupt autophagic flux, which results in accumulation of autophagosomes (LC3-I, LC3-II, and p62 proteins),35 a phenomenon that could be misinterpreted as increased autophagy. Finally, lysosome-dependent PCD might be mistaken for autophagy-dependent PCD, because lysosomes are present in zygotes within 2 to 4 h after fertilization and then enriched during reimplantation development36 (Fig. 1B). In autophagy, autophagosomes collect cellular trash and then fuse with lysosomes to degrade the trash within the autolysosome vesicle. In lysosome-dependent PCD, permeabilized lysosomes release their hydrolytic enzymes into the cytoplasm in a process facilitated by p53-upregulation of cathepsin synthesis.17
PCD in Preimplantation Mouse Embryos
During in vitro development of zygotes into blastocysts, the polar bodies in 1-cell and 2-cell embryos and one or 2 of the cells in the morula at the junction between inner cell mass and trophectoderm undergo PCD.41 However, although caspase activity was required for preimplantation development, these early events were not caspase-dependent. Therefore, PCD appeared to occur via a non-canonical form of apoptosis (Fig. 1A).
γH2AX is easily detected throughout preimplantation development, thereby revealing the presence of double-strand DNA breaks even in the absence of any induced DNA damage.42 In contrast, the p53-binding protein “53BP1” that is recruited to sites of double-strand DNA breaks43 was not detected. Thus, p53-dependent PCD does not appear to be induced by low levels of double-strand DNA breaks. In fact, regardless of the presence or absence of p53, induction of double-strand DNA breaks by X-irradiation of two-cell embryos retarded their development to the late blastocyst stage but did not prevent it.44 Wild-type blastocysts exhibited 2 to 3-times more cells with DNA damage than p53−/− blastocysts, thereby revealing that p53 accelerates PCD, but is not required for PCD.
Autophagy-dependent PCD is unlikely because autophagy is essential for preimplantation development.45 Autophagy-defective oocytes derived from oocyte-specific Atg5 (autophagy-related 5) knockout mice failed to develop beyond the 4- to 8-cell stages if fertilized by Atg5−/− sperm, but they did develop if fertilized by wild-type sperm. However, lysosomes rapidly accumulate after fertilization (Fig. 1B) and lysosome accumulation is required for development,36 suggesting that induced stress might activate lysosome-dependent PCD. In fact, PIKfyve, a phosphoinositide kinase essential for maintaining lysosome homeostasis and autophagic flux,35 is essential for preimplantation mouse development.46
PCD in ESCs
Embryonic stem cells can exhibit multiple physiological states (Fig. 1 legend). Naïve mESCs correspond to transient populations in pre- or peri-implantation embryonic epiblast whereas primed mESCs (termed EpiSCs) isolated from post-implantation blastocysts model the postimplantation epiblast.47 hESCs are similar to primed mESCs.48 Ground state mESCs are produced by culturing naïve mESCs with inhibitors of MAP2K1/MEK1 and FRAT2/GSK-3, and therefore termed 2iESCs.49 An intermediate state between naïve and primed ESCs has recently been described.50
Doxorubicin/Adriamycin is an anticancer drug commonly used to induce PCD in mammalian cells by causing double-strand DNA breaks. Doxorubicin induces PCD equally well in either p53+/+ or p53−/− naïve ESCs, as evident from visual inspection of cultured cells, DNA loss, annexin-V binding, propidium iodide, and trypan blue staining, cleaved PARP, and CASP3.51 Robust PCD did not require p53 or its primary targets, the CDK2 inhibitor p21 and pro-apoptotic protein PUMA, to cleave PARP and CASP3 (Fig. 2A), arrest cell proliferation (cells accumulate with 4N DNA content, Fig. 2C) and complete apoptosis (cells accumulate with <2N DNA content) (Fig. 2C and D). Thus, DNA damage in naïve ESCs induced a p53-independent form of noncanonical apoptosis. This conclusion was confirmed by the translocation of AIFM from mitochondria to the nucleus in p53−/− as well as p53+/+ ESCs (Fig. 2B).
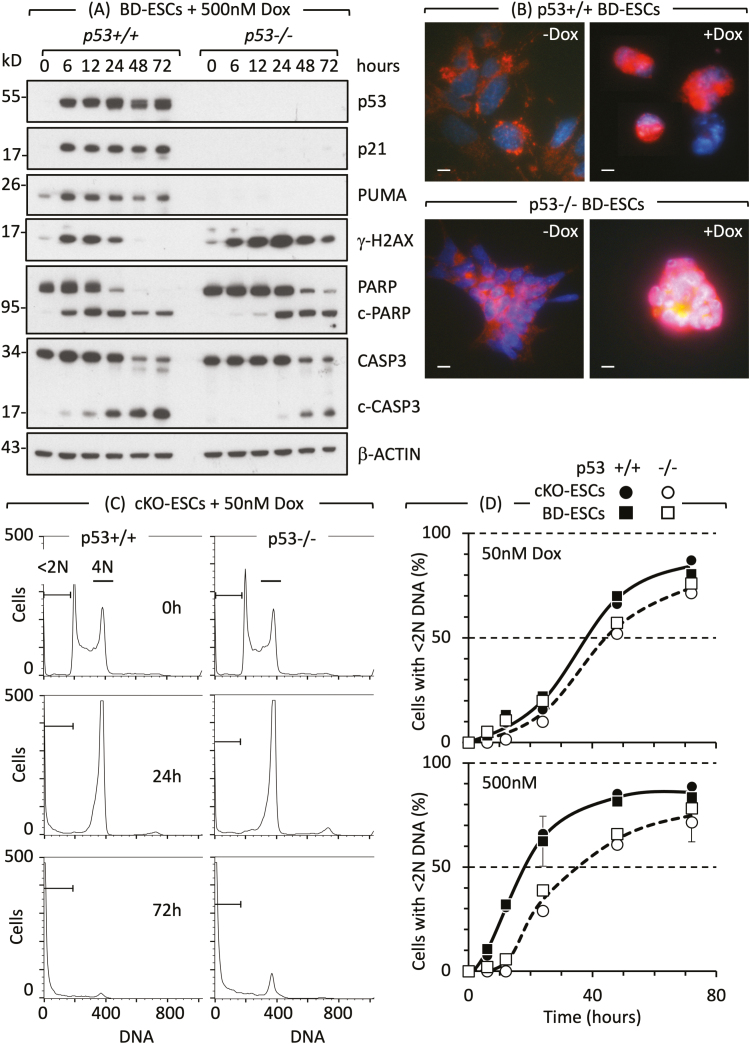
Figure 2.
Cell cycle arrest and apoptosis in naïve ESCs are not dependent on p53. (A) Doxorubicin/Adriamycin (Dox) induced DNA damage (γH2AX expression), DNA damage response (PARP to c-PARP cleavage) and apoptosis (CASP3 to c-CASP3 cleavage) in ESCs derived from p53+/+ and p53−/− mouse blastocysts (BD-ESCs, “chronic phenotype”). ESCs were cultured with or without 500 nM Dox. At the times indicated, attached and unattached cells were combined, and total cellular proteins analyzed by immunoblotting. (B) PCD was detected by translocation of AIFM (red) from cytoplasm to nuclei (blue) in BD-ESCs cultured with 500 nM Dox for 16 h. Scale bar is 15 µm. (C) A transient accumulation of cells with 4N DNA content is characteristic of a DNA damage-induced G2-arrest. The G2-checkpoint was activated within 24 h and apoptosis within 72 h by 50 nM Dox in both conditional knockout p53−/− ESCs and their p53+/+ parent (cKO-ESCs, “acute phenotype”). Attached and unattached cells were combined, and their DNA content quantified by fluorescence-activated cell sorting. Cells with <2N DNA content (apoptotic cells) and cells with 4N DNA content (G2/M phase cells) are indicated. Equivalent results were obtained with BD-ESCs. (D) Cells with <2N DNA content were quantified as a function of time cultured with Dox and normalized to 0% at zero hours. Error bars indicate ±SEM. Panels A and B are from Fig. 3B and C in Ref. 51 and panels C and D are from Figs. 2 and S2 in Ref. 51.
Similar results occurred using etoposide to cause double-strand DNA breaks.52 DNA fragmentation was accompanied by annexin-V binding, plasma membrane permeabilization, and cleavage of PARP, but not caspase-3. Instead, PCD was accompanied by increased levels of cathepsins. Pifithrin-α reduced PCD, suggesting these effects were p53-dependent, but pifithrin-α also suppresses ESC self-renewal53 via mechanisms unrelated to p53.54 These results are consistent with a p53-independent form of lysosome-dependent PCD.
PIKfyve inhibitors induce non-canonical apoptosis (no caspase-3 cleavage) in mouse and human ESCs,51,55 as well as in autophagy-dependent human cells,35,56 thereby confirming the dependence of pluripotent stem cells on either lysosome homeostasis or autophagic flux. Because PIKfyve inhibitors alter lysosome permeability and cathepsin maturation,35,57 these results suggest lysosome-dependent PCD.
Necroptosis initiates cell death in the absence of caspase cleavage by activating death receptors in the plasma membrane that trigger assembly of a “necrosome complex” followed by permeabilization of the plasma membrane and an inflammatory response. High levels of autophagosome-associated proteins ATG5, ATG8/LC3, or SQSTM1/p62 promote necrosome assembly and activation in human cancer cells,58-62 suggesting that disruption of autophagy by inhibition of PIKfyve, which causes accumulation of LC3 and p62, might trigger necroptosis in ESCs. Thus, different cellular stresses appear to trigger different forms of PCD.
PCD in Postimplantation Mouse Embryos
Leukemia inhibitory factor (LIF) deprivation of naïve ESCs induces differentiation, and comparison of LIF deprived p53+/+ with p53−/− ESCs revealed that the roles of p53 in cell cycle regulation, apoptosis, and senescence are acquired during pluripotent stem cell differentiation.51 Senescence prevents cell proliferation permanently, while retaining cell function. Wild-type mouse embryonic fibroblasts (MEFs, Fig. 1) treated with doxorubicin undergo G1-arrest and senescence, but p53−/− MEFs fail to do so and ultimately undergo noncanonical apoptosis.51,63-66 As little as 50 nM doxorubicin induces p53-independent apoptosis in ESCs (Fig. 2D), but MEFs require excessive concentrations of doxorubicin (Fig. 3B).51,67 Only MEFs expressing an oncogene exhibit p53-dependent apoptosis.68-70 Thus, senescence rather than apoptosis is the normal response to DNA damage in differentiated cells.
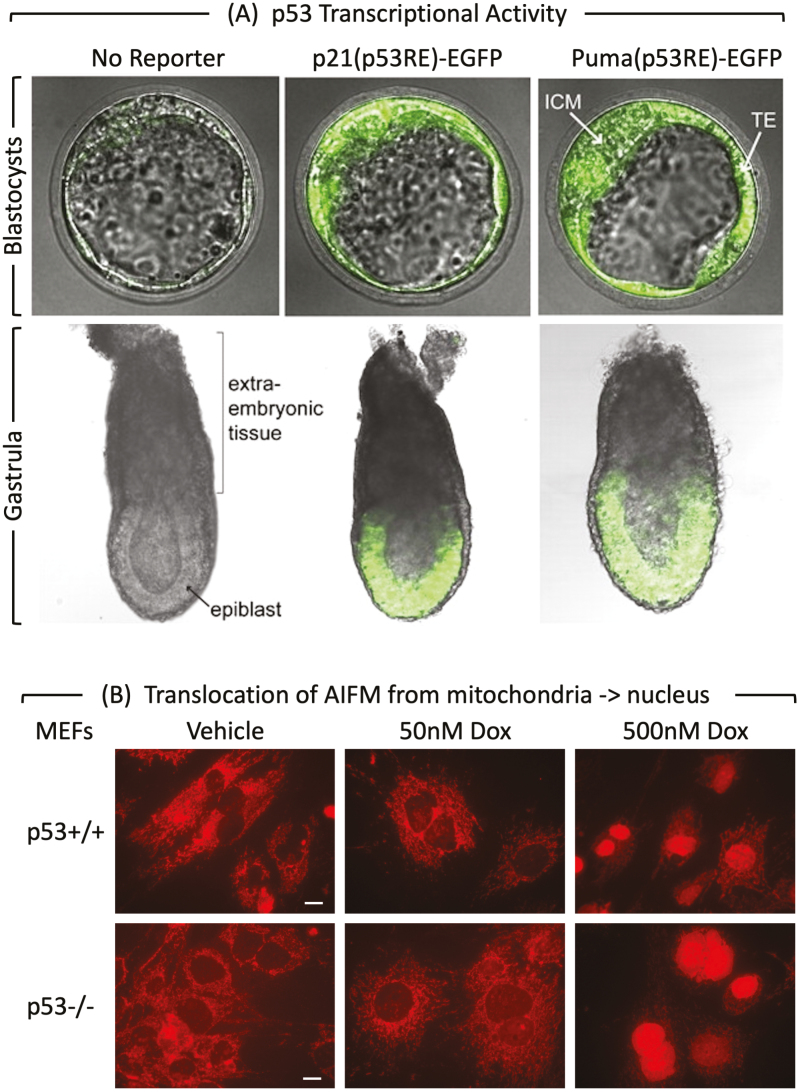
Figure 3.
p53 activity and MEF PCD response at the beginning of mouse development. (A) p53 activity assayed in embryos isolated from mice homozygous for reporter genes expressing enhanced green fluorescence protein driven by either the Cdkn1a/p21 or the Bbc3/Puma gene’s p53 response element.75 At embryonic day E3.5, fluorescence was detected in the inner cell mass (ICM), and trophectoderm (TE) of blastocysts. The large blastocoel cavity identifies these examples as late-stage blastocysts containing early epiblast (Fig. 1). At embryonic day E6.5, fluorescence was detected in the epiblast but not in the extraembryonic tissue of gastrula. (B) MEFs cultured for 24 h with doxorubicin and then stained for “apoptosis-inducing factor” AIFM.51 Scale bar is 15 μm. Translocation of AIFM from mitochondria to nucleus occurred in both p53+/+ and p53−/− cells, thereby confirming non-canonical apoptosis in MEFs treated with 500 nM doxorubicin, but not in MEFs with 50 nM doxorubicin.
In the absence of pro-apoptotic genes Bax and Bak, MEFs appear to undergo autophagy-dependent PCD in response to either etoposide or Staurosporine.71,72p53−/− MEFs were not characterized. Alternatively, DNA damage in MEFs lacking both Bax and Bak induced “parthanatos” (programable necrosis), a mechanism largely controlled by p53-mediated transcription of cathepsin Q in cooperation with DNA damage-induced ROS.73,74Parthanatos is a PARP1-dependent form of PCD that relies on the AIFM1-macrophage migration inhibitory factor (MIF) pathway. MIF is an AIFM1-binding protein with nuclease activity that produces large DNA fragments. Thus, alternative forms of PCD can be activated in different types of cells.
p53 Regulation of Cell Proliferation, PCD, and Senescence
p53-dependent transcription is first detected during mouse development at the late blastocyst stage, but p53 levels are not great enough to induce embryonic lethality until gastrulation. Therefore, a role for p53 in arresting cell proliferation or activating PCD or senescence begins with cell differentiation.
p53 Activity
Expression of an ectopic EGFP reporter gene driven by a p53-dependent response element demonstrated that p53-dependent transcriptional activity exists as early as late-stage blastocysts and is confined to the epiblast in post-implantation embryos75 (Figure 3A). Double-strand DNA breaks introduced by X-irradiation of embryos at either E3.5 (blastocysts) or E9.5 (organogenesis) revealed that p53+/+ embryos die more frequently than p53−/− embryos, whereas p53−/− embryos exhibit more developmental anomalies.76 X-irradiated p53+/+ embryos undergoing organogenesis contain a greater number of apoptotic cells than p53−/− embryos. p53 facilitates apoptosis in X-irradiated embryos only after preimplantation embryos developed into late-stage blastocysts (E5).44 No significant change in cell proliferation was observed following X-irradiation, but late-stage p53+/+ blastocysts exhibited 2 to 3-times more apoptotic cells than p53−/− blastocysts. Thus, p53-dependent transcriptional activity and apoptosis are first evident in late-stage blastocysts and increase during organogenesis.
p53 Regulation
p53 activity is tightly regulated posttranslationally. Under normal conditions, p53 expression is very low; it is a short-lived protein whose stability and activity are regulated by phosphorylation, methylation, and acetylation events, and by association with specific p53 regulatory proteins such as MDM2 and MDM4/MDMX,77-79 RBBP6/PACT,80-82 and PRKRA/RAX/PACT.83,84 MDM2, MDM4, and RBBP6 are essential for cell viability and embryonic development. Mice lacking p53 and mice lacking both p53 and MDM2 display the same incidence and spectrum of spontaneous tumor formation,65 thereby revealing that, in the absence of p53, MDM2 has no effect on cell proliferation, cell cycle regulation, or tumorigenesis. Thus, ablation of the Mdm2, Mdm4, or Rbbp6 gene in mouse embryos is lethal, but only in the presence of p53 protein. Thus, unregulated p53 expression during embryonic development is lethal.
p53-Dependent Lethality
Unregulated p53 activity does not induce embryonic lethality until the onset of gastrulation (Fig. 1). For Mdm2−/− embryos, demise occurs after implantation of the embryo in the wall of the uterus but before day 7.5 of gestation (≈E5.5).85,86 Deletion of the Mdm2 gene has no additional effect on cell proliferation, cell cycle control, or tumorigenesis when p53 gene is absent.65,87 Therefore, lethality in the absence of MDM2 is due solely to p53 activity. For RBBP6/PACT−/− embryos, lethality occurs after implantation but before E7.5.81 For Mdm4/Mdmx−/− embryos, lethality occurs between E7.5 and E12 from the p53-dependent arrest of cell proliferation (presumably senescence).88-90 All 3 phenotypes could be rescued by transferring the mutated p53 negative regulator gene (Mdm2, RBBP6, or Mdm4) to a p53-nullizygous background, in which case mice develop normally. Thus, embryonic death in the absence of a p53 regulator resulted from activation of p53 protein.
p53-Dependent Senescence
Given that p53 activity is first detectable during the late blastocyst stage and confined to the epiblast in early gastrula, p53 expression is too low to induce either cell cycle arrest or cell death upon release from post-translational regulation until after the blastocyst has implanted (E4.5) and gastrulation has begun (E6.25-E7.5). However, DNA damage begins to accumulate in Mdm2−/− blastocysts, but less so if embryos also lack the p53-dependent proapoptotic gene Bax, suggesting that unregulated p53 initiates apoptosis in blastocysts.91 Nevertheless, embryonic lethality still occurs at E6.5-E7.5 due to the arrest of cell proliferation (cell senescence) rather than PCD.
p53 and Naïve ESCs
Cell cycle checkpoints are p53-independent. The G1 checkpoint is a response to cell stress that retards entrance into the S phase.92 Naïve mESCs lack a G1 DNA damage checkpoint.51,93-97 The G2 checkpoint is a transient accumulation of cells with 4N DNA content in response to DNA damage prior to induction of apoptosis.98 Double-strand DNA breaks induced by culturing cells with doxorubicin activated the G2 checkpoint in naïve mESCs regardless of the presence or absence of p53,51,95,97,99 p21, or PUMA51 (Fig. 2A). In contrast, both hESCs and mouse EpiSCs derived from the epiblast in postimplantation blastocysts exhibit a G1 checkpoint.48
PCD is not dependent on p53. Of the 9 studies that investigated the role of p53 in PCD, 3 concluded that p53 is not required51,93,97 and 6 concluded that p53 is required.95,96,100-102 To resolve this paradox, Jaiswal et al51 quantified the effects of doxorubicin on p53+/+ and p53−/− ESCs derived by 2 different methods. To eliminate the possibility that conclusions depended on either the source or derivation of p53−/− ESCs, both wild-type and p53−/− ESCs derived directly from blastocysts were characterized in parallel with ESCs in which the p53 genes were ablated in vitro. ESCs isolated from p53−/− blastocysts exhibit the effects of p53 loss through multiple generations in vivo (chronic phenotype), whereas p53−/− ESCs engineered in vitro from p53+/+ ESCs exhibit the effects of immediate p53 loss (acute phenotype).
To eliminate methodology-dependent biases, the rate and extent of cell cycle arrest and cell death were quantified by time-dependent changes in DNA content (Fig. 2C and D), by staining with annexin-V and propidium iodide to distinguish apoptosis from necrosis, by exclusion of trypan blue to distinguish live cells from dead cells, by Western immunoblotting of p53, p21, PUMA, γH2AX, PARP, and CASP3 to confirm genotypes, DNA damage, and caspase cleavage (Fig. 2A), and by cellular localization of AIFM to confirm caspase-independent apoptosis (Fig. 2B).
The results revealed that, regardless of their derivation, naïve mouse ESCs do not require p53, p21, or PUMA either to activate the G2-checkpoint (Fig. 2C) or to undergo robust apoptosis (Fig. 2D). Depending on conditions such as seeding density and doxorubicin concentration, p53 can accelerate initiation of apoptosis in ESCs in response to DNA damage by 8.4 ± 0.5 h, but the rate and extent of apoptosis in ESCs are equivalent and complete PCD within 72 h, regardless of the presence or absence of p53. The inhibitory effect of only 50 nM doxorubicin is evident from visual inspection of cultured cells, and the lethal effect is evident from the accumulation of cells with <2N DNA content. Short exposure (24 h) to a low concentration (50 nM) of doxorubicin to ESCs, then allowing them to recover for 96 h proved that even minimal DNA damage is enough to induce apoptosis in ESCs regardless of presence or absence of p53.
p53 and Ground-State 2iESCs
Naïve ESCs are characterized by hyper-phosphorylated RB1 protein, lack of G1 control, and rapid progression through the cell cycle. In contrast, ground-state 2iESCs, which are derived from naïve ESCs (Fig. 1), have a longer G1-phase with hypo-phosphorylated RB1, implying that they have a functional G1 checkpoint. The RB1-dependent G1 restriction point is active in 2iESCs but abrogated when cultured in serum.103 Moreover, the p53-p21 pathway appears active in 2iESCs, and its role in the G1-checkpoint is abolished in naïve ESCs.104
DNA damage in 2iESCs caused by doxorubicin-induced p53-dependent cell death.104 DNA damage in 2iESCs caused either by doxorubicin or by aphidicolin inhibition of DNA polymerase-α activated expression of DUX transcription factors that are involved in zygotic gene activation in mouse 2-cell to 4-cell embryos.105,106 Both studies concluded that this phenomenon is mediated by an ATR and CHK1 response to double-strand DNA breaks. Critical experiments in which p53+/+ and p53−/− 2iESCs were compared were carried out in both studies. However, one study concluded that this phenomenon required p53 expression,106 whereas the other study concluded that it did not.105 Ironically, even if p53 is essential for DUX expression, loss of p53 would still not affect embryonic development, because DUX is not required for mouse development.107
Experimental Conditions Could Account for Contradictory Conclusions
Cell Culture
Culture conditions are critical to maintaining the pluripotent state.49 Suboptimal conditions promote DNA damage108 and ESC differentiation, thereby selecting for p53 dependence.109 ESCs under stress characteristically undergo either differentiation or apoptosis.110,111 In fact, the culture conditions used to convert naïve ESCs into 2iESCs enforce self-renewal and a dramatic loss of spontaneously differentiating cells; neither primed ESCs nor differentiated somatic cells survive these conditions.112 Remarkably, 2 studies used blastocyst derived-ESCs from the same source (Rudolf Jaenisch, MIT, Cambridge, MA) but reported contradictory results. Culturing ESCs to “sub-confluence” before adding doxorubicin101 might have created conditions in which excessively high concentrations of doxorubicin-induced apoptosis in p53+/+ cells more rapidly than in p53−/− cells.51,93
Time
The only effect of p53 on apoptosis in naïve ESCs was to accelerate its initiation. Once initiated, apoptosis continued at the same rate and to the same extent as in the absence of p53. Experiments with a single time point and a single-drug concentration cannot reveal the relationship between DNA damage and the significance of p53.
DNA Damage
As little as 0.05 µM doxorubicin is sufficient to induce apoptosis in either p53+/+ and p53−/− naïve ESCs. Yet most studies used from 0.5 µM to 1.8 µM. With naïve ESCs, high doxorubicin concentrations-initiated apoptosis more quickly in p53+/+ ESCs than in p53−/− ESCs, but once initiated, PCD occurred at equivalent rates and to equivalent extents (Fig. 2D). With 2iESCs, one study used 1 µM doxorubicin for 6 h and concluded that the DNA damage response was p53-dependent.106 A second study used 1 µg/mL (1.84 µM) doxorubicin for 48 h and concluded that the DNA damage response was p53-independent.105 Still a third study cultured 2iESCs with 1 μM doxorubicin for 16 h and observed that p53+/+ cells underwent apoptosis more quickly than p53−/− cells (63% p53+/+ cells vs. 13% p53−/− cells).104 These results might be reconciled if both studies avoided excessively high concentrations of doxorubicin and monitored the effects of doxorubicin over time.
Viability
Two studies concluded that DNA damage-induced PCD was p53-dependent in naïve mESCs95 and hESCs,102 because cells in which p53 was suppressed constitutively by shRNA did not exhibit doxorubicin-induced apoptosis. However, this technology raises 2 caveats. First, isolation of viable clones also selects for “off-target” mutations that promote cell proliferation or prevent cell death, as evidenced by the fact that constitutive suppression of p53-expression ESCs and embryos promotes clonal heterogeneity by disrupting DNA methylation homeostasis.113 Furthermore, since ESCs under stress characteristically undergo either differentiation or apoptosis,110,111 changes observed in gene expression and relocalization of p53 from the cytoplasm to the nucleus are characteristics of ESC differentiation as well as apoptosis. The same caveats apply to the application of CRISPR-Cas9 technology to ablate p53 in 2iESCs.104-106
p53 Null Mutation
Comparing p53+/+ with p53−/− ESCs is essential to establish a role for p53. Two studies with contradictory conclusions relied on inadequately characterized ESCs.97,100 Another study on hESCs based its conclusion solely on changes in p53 expression in response to apoptotic stimuli.114 Studies that rely upon changes in p53 protein in response to stress and p53 inhibitors ignore the fact that the p53 transcription factor regulates at least 343 target genes involved in maintaining genomic stability, cell differentiation, cell senescence, cell cycle regulation, and PCD.7 The fact that ectopic over-expression of certain p53 mutations also suppressed doxorubicin-induced apoptosis101 simply reflects the fact that p53 affects expression of hundreds of different genes, some of which affect apoptosis. Many naturally occurring p53 mutations have the opposite effect; they gain additional oncogenic functions that endow cells with growth and survival advantages.115
Reproducibility
Two studies using the same source of ESCs (Yang Xu, Univ. California, San Diego) concluded that p53 is not required for cell cycle arrest51,96 and their results with p53+/+ ESCs are indistinguishable.51,96 However, one study concluded that p53 is essential for doxorubicin-induced apoptosis96 whereas the other study concluded that it is not.51 The first study relied on caspase-3 cleavage to confirm apoptosis, which they detected with a monoclonal antibody specific for the cleaved form. Thus, the fact that the extent of CASP3 cleavage was insignificant was not recognized. Moreover, the time delay for initiation of apoptosis exhibited by p53−/− ESCs cultured with excess doxorubicin delayed the appearance of cleaved caspase-3, thereby allowing cleaved-caspase-3 to be detected in p53+/+ cells under conditions where it appeared to be absent in p53−/− cells. Apoptosis is also delayed in p53−/− ESCs cultured under stress, such as the extremely high seeding density (260 000 cells/cm2) used in the first study.96
Conclusions
Of the 12 forms of PCD described in human cells, only noncanonical apoptosis, autophagy-dependent, and lysosome-dependent PCD have been reported in ESCs, preimplantation, or gastrulating embryos. However, autophagy-dependent PCD might be confused with autophagy disruption which could activate non-canonical apoptosis, lysosome-dependent PCD, or necroptosis. Another candidate is parthanatos.
The importance of p53 in PCD has been characterized extensively, but conclusions are often enigmatic. Based solely on studies comparing wild-type with p53−/− ESCs, MEFs, or mice, 3 conclusions appear uncontested; p53 is not required for activation of the G2-checkpoint, for embryonic lethality prior to gastrulation, or for embryonic development. The form of PCD and the role of p53 might change as preimplantation embryos develop from totipotent (2iESCs) to pluripotent (naïve ESCs) to primed pluripotent cells in post-implantation embryos (hESCs, mEpiSCs). However, contradictory conclusions concerning the role of p53 during PCD in ESCs can be reconciled by differences in experimental conditions, such as the amount of stress and the length of time stress was induced, culture conditions, and assay conditions.
In mice, p53 dependent transcription is first evident in late-stage blastocysts, and the ability of p53 to induce embryonic lethality is first evident during gastrulation. Depending on experimental conditions, p53 can accelerate initiation of PCD in mESCs and late-stage blastocysts, but once initiated, PCD occurs at equivalent rates and to equivalent extents regardless of the presence or absence of p53. Following either mESC differentiation in vitro or the formation of MEFs in vivo, DNA damage induces p53-dependent cell cycle arrest and senescence. Given the sensitivity of MEFs to p53-dependent senescence, failure of embryonic development likely results from cell senescence rather than PCD, although excessive DNA damage induces PCD.
Funding
The work was funded by the National Institute of Child Health and Human Development (NICHD) Intramural Research Program, Grant/Award Numbers: ZIA HD000506, ZIA HD000507.
Conflict of Interest
The authors declare no potential conflicts of interest.
Author Contributions
All the authors wrote and revised the manuscript. All the authors have read and agreed to the published version of the manuscript.
Data Availability
No new data were generated or analyzed in support of this research.
References
- 1. Galluzzi L, Bravo-San Pedro JM, Vitale I, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ. 2015;22(1):58-73. https://doi.org/10.1038/cdd.2014.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wyllie AH, Kerr JF, Currie AR.. Cell death: the significance of apoptosis. Int Rev Cytol. 1980;68:251-306. https://doi.org/10.1016/s0074-7696(08)62312-8 [DOI] [PubMed] [Google Scholar]
- 3. Hardy K, Handyside AH, Winston RM.. The human blastocyst: cell number, death and allocation during late preimplantation development in vitro. Development. 1989;107(3):597-604. [DOI] [PubMed] [Google Scholar]
- 4. Fuchs Y, Steller H.. Programmed cell death in animal development and disease. Cell. 2011;147(4):742-758. https://doi.org/10.1016/j.cell.2011.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Munoz-Espin D, Canamero M, Maraver A, et al. Programmed cell senescence during mammalian embryonic development. Cell. 2013;155(5):1104-1118. https://doi.org/10.1016/j.cell.2013.10.019 [DOI] [PubMed] [Google Scholar]
- 6. Coucouvanis E, Martin GR.. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell. 1995;83(2):279-287. https://doi.org/10.1016/0092-8674(95)90169-8 [DOI] [PubMed] [Google Scholar]
- 7. Jaiswal SK, Raj S, DePamphilis ML.. Developmental acquisition of p53 functions. Genes. 2021;12(11):1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aubrey BJ, Kelly GL, Janic A, et al. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25(1):104-113. https://doi.org/10.1038/cdd.2017.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levine AJ, Berger SL.. The interplay between epigenetic changes and the p53 protein in stem cells. Genes Dev. 2017;31(12):1195-1201. https://doi.org/10.1101/gad.298984.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bieging KT, Mello SS, Attardi LD.. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359-370. https://doi.org/10.1038/nrc3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riley T, Sontag E, Chen P, et al. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402-412. https://doi.org/10.1038/nrm2395 [DOI] [PubMed] [Google Scholar]
- 12. Shiloh Y, Y Z.. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14(4):197-210. [PubMed] [Google Scholar]
- 13. Sengupta S, Harris CC.. p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6(1):44-55. https://doi.org/10.1038/nrm1546 [DOI] [PubMed] [Google Scholar]
- 14. Tilgner K, Neganova I, Moreno-Gimeno I, et al. A human iPSC model of Ligase IV deficiency reveals an important role for NHEJ-mediated-DSB repair in the survival and genomic stability of induced pluripotent stem cells and emerging haematopoietic progenitors. Cell Death Differ. 2013;20(8):1089-1100. https://doi.org/10.1038/cdd.2013.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lozano G. Mouse models of p53 functions. Cold Spring Harb Perspect Biol. 2010;2(4):a001115. https://doi.org/10.1101/cshperspect.a001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu J, Zhang L.. PUMA, a potent killer with or without p53. Oncogene. 2008;27(suppl 1):S71-S83. https://doi.org/10.1038/onc.2009.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang D, Kang R, Berghe TV, et al. The molecular machinery of regulated cell death. Cell Res. 2019;29(5):347-364. https://doi.org/10.1038/s41422-019-0164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doherty J, Baehrecke EH.. Life, death and autophagy. Nat Cell Biol. 2018;20(10):1110-1117. https://doi.org/10.1038/s41556-018-0201-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singh R, Letai A, Sarosiek K.. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol. 2019;20(3):175-193. https://doi.org/10.1038/s41580-018-0089-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laszlo ZI, Lele Z, Zoldi M, et al. ABHD4-dependent developmental anoikis safeguards the embryonic brain. Nat Commun. 2020;11(1):4363. https://doi.org/10.1038/s41467-020-18175-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stahl-Meyer J, Stahl-Meyer K, Jaattela M.. Control of mitosis, inflammation, and cell motility by limited leakage of lysosomes. Curr Opin Cell Biol. 2021;71:29-37. https://doi.org/10.1016/j.ceb.2021.02.003 [DOI] [PubMed] [Google Scholar]
- 22. Denton D, Kumar S.. Autophagy-dependent cell death. Cell Death Differ. 2019;26(4):605-616. https://doi.org/10.1038/s41418-018-0252-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. White E. Autophagy and p53. Cold Spring Harb Perspect Med. 2016;6(4):a026120. https://doi.org/10.1101/cshperspect.a026120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weinlich R, Oberst A, Beere HM, et al. Necroptosis in development, inflammation and disease. Nat Rev Mol Cell Biol. 2017;18(2):127-136. https://doi.org/10.1038/nrm.2016.149 [DOI] [PubMed] [Google Scholar]
- 25. Holze C, Michaudel C, Mackowiak C, et al. Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway. Nat Immunol. 2018;19(2):130-140. https://doi.org/10.1038/s41590-017-0013-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang X, P G.. Parthanatos in the pathogenesis of nervous system diseases. Neuroscience. 2020;449:241-250. [DOI] [PubMed] [Google Scholar]
- 28. J L, Kuang F, Kang R, et al. Alkaliptosis: a new weapon for cancer therapy. Cancer Gene Ther. 2020;27(5):267-269. [DOI] [PubMed] [Google Scholar]
- 29. Y H, Wang B, S L, et al. Pyroptosis, and its role in central nervous system disease. J Mol Biol. 2021;167379. [DOI] [PubMed] [Google Scholar]
- 30. Hamann JC, Kim SE, Overholtzer M.. Methods for the study of entotic cell death. Methods Mol Biol. 2019;1880:447-454. https://doi.org/10.1007/978-1-4939-8873-0_28 [DOI] [PubMed] [Google Scholar]
- 31. Inoue M, Enomoto M, Yoshimura M, et al. Pharmacological inhibition of sodium-calcium exchange activates NADPH oxidase and induces infection-independent NETotic cell death. Redox Biol. 2021;43:101983. https://doi.org/10.1016/j.redox.2021.101983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee IH, Kawai Y, Fergusson MM, et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336(6078):225-228. https://doi.org/10.1126/science.1218395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang Y, White E.. Autophagy suppresses TRP53/p53 and oxidative stress to enable mammalian survival. Autophagy. 2020;16(7):1355-1357. https://doi.org/10.1080/15548627.2020.1765522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb Perspect Med. 2016;6(3):a026104. https://doi.org/10.1101/cshperspect.a026104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharma G, Guardia CM, Roy A, et al. A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis. Autophagy. 2019;15(10):1694-1718. https://doi.org/10.1080/15548627.2019.1586257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsukamoto S, Hara T, Yamamoto A, et al. Functional analysis of lysosomes during mouse preimplantation embryo development. J Reprod Dev. 2013;59(1):33-39. https://doi.org/10.1262/jrd.2012-096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Charlot JF, Pretet JL, Haughey C, et al. Mitochondrial translocation of p53 and mitochondrial membrane potential (Delta Psi m) dissipation are early events in staurosporine-induced apoptosis of wild type and mutated p53 epithelial cells. Apoptosis. 2004;9(3):333-343. https://doi.org/10.1023/b:appt.0000025810.58981.4c [DOI] [PubMed] [Google Scholar]
- 38. Battistelli M, Falcieri E.. Apoptotic bodies: particular extracellular vesicles involved in intercellular communication. Biology (Basel). 2020;9(1). https://doi.org/10.3390/biology9010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kayagaki N, Kornfeld OS, Lee BL, et al. NINJ1 mediates plasma membrane rupture during lytic cell death. Nature. 2021;591(7848):131-136. https://doi.org/10.1038/s41586-021-03218-7 [DOI] [PubMed] [Google Scholar]
- 40. Hevler JF, Zenezeni Chiozzi R, Cabrera-Orefice A, et al. Molecular characterization of a complex of apoptosis-inducing factor 1 with cytochrome c oxidase of the mitochondrial respiratory chain. Proc Natl Acad Sci USA. 2021;118(39). https://doi.org/10.1073/pnas.2106950118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zakeri Z, Lockshin RA, Criado-Rodriguez LM, et al. A generalized caspase inhibitor disrupts early mammalian development. Int J Dev Biol. 2005;49(1):43-47. https://doi.org/10.1387/ijdb.041920zz [DOI] [PubMed] [Google Scholar]
- 42. Ziegler-Birling C, Helmrich A, Tora L, et al. Distribution of p53 binding protein 1 (53BP1) and phosphorylated H2A.X during mouse preimplantation development in the absence of DNA damage. Int J Dev Biol. 2009;53(7):1003-1011. https://doi.org/10.1387/ijdb.082707cz [DOI] [PubMed] [Google Scholar]
- 43. J L, Priest DG, Solano A, et al. Spatiotemporal dynamics of 53BP1 dimer recruitment to a DNA double strand break. Nat Commun. 2020;11(1):5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilson Y, Morris ID, Kimber SJ, et al. The role of Trp53 in the mouse embryonic response to DNA damage. Mol Hum Reprod. 2019;25(7):397-407. https://doi.org/10.1093/molehr/gaz029 [DOI] [PubMed] [Google Scholar]
- 45. Tsukamoto S, Kuma A, Murakami M, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321(5885):117-120. https://doi.org/10.1126/science.1154822 [DOI] [PubMed] [Google Scholar]
- 46. Ikonomov OC, Sbrissa D, Delvecchio K, et al. The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve−/− embryos but normality of PIKfyve+/- mice. J Biol Chem. 2011;286(15):13404-13413. https://doi.org/10.1074/jbc.M111.222364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith A. Formative pluripotency: the executive phase in a developmental continuum. Development. 2017;144(3):365-373. https://doi.org/10.1242/dev.142679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zaveri L, Dhawan J.. Cycling to meet fate: connecting pluripotency to the cell cycle. Front Cell Dev Biol. 2018;6:57. https://doi.org/10.3389/fcell.2018.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mulas C, Kalkan T, von Meyenn F, et al. Defined conditions for propagation and manipulation of mouse embryonic stem cells. Development. 2019;146(6):dev178970. https://doi.org/10.1242/dev.178970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kinoshita M, Barber M, Mansfield W, et al. Capture of mouse and human stem cells with features of formative pluripotency. Cell Stem Cell. 2021;28(3):453-471.e8. https://doi.org/10.1016/j.stem.2020.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jaiswal SK, Oh JJ, DePamphilis ML.. Cell cycle arrest and apoptosis are not dependent on p53 prior to p53-dependent embryonic stem cell differentiation. Stem Cells. 2020;38(9):1091-1106. https://doi.org/10.1002/stem.3199 [DOI] [PubMed] [Google Scholar]
- 52. Tichy ED, Stephan ZA, Osterburg A, et al. Mouse embryonic stem cells undergo charontosis, a novel programmed cell death pathway dependent upon cathepsins, p53, and EndoG, in response to etoposide treatment. Stem Cell Res. 2013;10(3):428-441. https://doi.org/10.1016/j.scr.2013.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Abdelalim EM, Tooyama I.. The p53 inhibitor, pifithrin-alpha, suppresses self-renewal of embryonic stem cells. Biochem Biophys Res Commun. 2012;420(3):605-610. https://doi.org/10.1016/j.bbrc.2012.03.041 [DOI] [PubMed] [Google Scholar]
- 54. J Z, Singh M, Selivanova G, et al. Pifithrin-alpha alters p53 post-translational modifications pattern and differentially inhibits p53 target genes. Sci Rep. 2020;10(1):1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chakraborty AR, Vassilev A, Jaiswal SKet al. Selective elimination of pluripotent stem cells by PIKfyve specific inhibitors. Stem Cell Rep. 2021;17(2):397-412. https://doi.org/10.1016/j.stemcr.2021.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gayle S, Landrette S, Beeharry N, et al. Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood. 2017;129(13):1768-1778. https://doi.org/10.1182/blood-2016-09-736892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. O’Connell CE, Vassilev A.. Combined inhibition of p38MAPK and PIKfyve synergistically disrupts autophagy to selectively target cancer cells. Cancer Res. 2021;81(11):2903-2917. https://doi.org/10.1158/0008-5472.CAN-20-3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Basit F, van Oppen LM, Schockel L, et al. Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells. Cell Death Dis. 2017;8(3):e2716. https://doi.org/10.1038/cddis.2017.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arya BD, Mittal S, Joshi P, et al. Graphene oxide-chloroquine nanoconjugate induce necroptotic death in A549 cancer cells through autophagy modulation. Nanomedicine (Lond). 2018;13(18):2261-2282. https://doi.org/10.2217/nnm-2018-0086 [DOI] [PubMed] [Google Scholar]
- 60. Goodall ML, Fitzwalter BE, Zahedi S, et al. The autophagy machinery controls cell death switching between apoptosis and necroptosis. Dev Cell. 2016;37(4):337-349. https://doi.org/10.1016/j.devcel.2016.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kharaziha P, Chioureas D, Baltatzis G, et al. Sorafenib-induced defective autophagy promotes cell death by necroptosis. Oncotarget. 2015;6(35):37066-37082. https://doi.org/10.18632/oncotarget.5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sulkshane P, Teni T.. BH3 mimetic Obatoclax (GX15-070) mediates mitochondrial stress predominantly via MCL-1 inhibition and induces autophagy-dependent necroptosis in human oral cancer cells. Oncotarget. 2017;8(36):60060-60079. https://doi.org/10.18632/oncotarget.11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Micco R D, Cicalese A, Fumagalli M, et al. DNA damage response activation in mouse embryonic fibroblasts undergoing replicative senescence and following spontaneous immortalization. Cell Cycle. 2008;7(22):3601-3606. [DOI] [PubMed] [Google Scholar]
- 64. Harvey M, Sands AT, Weiss RS, et al. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993;8(9):2457-2467. [PubMed] [Google Scholar]
- 65. Jones SN, Sands AT, Hancock AR, et al. The tumorigenic potential and cell growth characteristics of p53-deficient cells are equivalent in the presence or absence of Mdm2. Proc Natl Acad Sci USA. 1996;93(24):14106-14111. https://doi.org/10.1073/pnas.93.24.14106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brown JM, Attardi LD.. The role of apoptosis in cancer development and treatment response. Nat Rev Cancer. 2005;5(3):231-237. https://doi.org/10.1038/nrc1560 [DOI] [PubMed] [Google Scholar]
- 67. Lowe SW, Ruley HE, Jacks T, et al. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74(6):957-967. https://doi.org/10.1016/0092-8674(93)90719-7 [DOI] [PubMed] [Google Scholar]
- 68. Lowe SW, Bodis S, McClatchey A, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266(5186):807-810. https://doi.org/10.1126/science.7973635 [DOI] [PubMed] [Google Scholar]
- 69. Lowe SW, Jacks T, Housman DE, et al. Abrogation of oncogene-associated apoptosis allows transformation of p53-deficient cells. Proc Natl Acad Sci USA. 1994;91(6):2026-2030. https://doi.org/10.1073/pnas.91.6.2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vater CA, Bartle LM, Dionne CA, et al. Induction of apoptosis by tamoxifen-activation of a p53-estrogen receptor fusion protein expressed in E1A and T24 H-ras transformed p53−/− mouse embryo fibroblasts. Oncogene. 1996;13(4):739-748. [PubMed] [Google Scholar]
- 71. Shimizu S, Kanaseki T, Mizushima N, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6(12):1221-1228. https://doi.org/10.1038/ncb1192 [DOI] [PubMed] [Google Scholar]
- 72. Arakawa S, Tsujioka M, Yoshida T, et al. Role of Atg5-dependent cell death in the embryonic development of Bax/Bak double-knockout mice. Cell Death Differ. 2017;24(9):1598-1608. https://doi.org/10.1038/cdd.2017.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tu HC, D R, Wang GX, et al. The p53-cathepsin axis cooperates with ROS to activate programmed necrotic death upon DNA damage. Proc Natl Acad Sci USA. 2009;106(4):1093-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vaseva AV, Marchenko ND, K J, et al. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149(7):1536-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Goh AM, Lim CY, Chiam PC, et al. Using targeted transgenic reporter mice to study promoter-specific p53 transcriptional activity. Proc Natl Acad Sci USA. 2012;109(5):1685-1690. https://doi.org/10.1073/pnas.1114173109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Norimura T, Nomoto S, Katsuki M, et al. p53-dependent apoptosis suppresses radiation-induced teratogenesis. Nat Med. 1996;2(5):577-580. https://doi.org/10.1038/nm0596-577 [DOI] [PubMed] [Google Scholar]
- 77. Klein AM, de Queiroz R, M, Venkatesh D, et al. The roles and regulation of MDM2 and MDMX: it is not just about p53. Genes Dev. 2021;35(9-10):575-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ivanov GS, Ivanova T, Kurash J, et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol. 2007;27(19):6756-6769. https://doi.org/10.1128/MCB.00460-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kruse JP, W G.. Modes of p53 regulation. Cell. 2009;137(4):609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hull R, Oosthuysen B, Cajee UF, et al. The Drosophila retinoblastoma binding protein 6 family member has two isoforms and is potentially involved in embryonic patterning. Int J Mol Sci. 2015;16(5):10242-10266. https://doi.org/10.3390/ijms160510242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li L, Deng B, Xing G, et al. PACT is a negative regulator of p53 and essential for cell growth and embryonic development. Proc Natl Acad Sci USA. 2007;104(19):7951-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yang X, Zhang Q, Yang X, et al. PACT cessation overcomes ovarian cancer cell chemoresistance to cisplatin by enhancing p53-mediated apoptotic pathway. Biochem Biophys Res Commun. 2019;511(4):719-724. https://doi.org/10.1016/j.bbrc.2019.02.089 [DOI] [PubMed] [Google Scholar]
- 83. Bennett RL, Pan Y, Christian J, et al. The RAX/PACT-PKR stress response pathway promotes p53 sumoylation and activation, leading to G(1) arrest. Cell Cycle. 2012;11(2):407-417. https://doi.org/10.4161/cc.11.2.18999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fujitani K, Otomo A, Nagayama Y, et al. PACT/PRKRA and p53 regulate transcriptional activity of DMRT1. Genet Mol Biol. 2020;43(2):e20190017. https://doi.org/10.1590/1678-4685-GMB-2019-0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Montes de Oca Luna R, Wagner DS, Lozano G.. Rescue of early embryonic lethality in Mdm2-deficient mice by deletion of p53. Nature. 1995;378(6553):203-206. https://doi.org/10.1038/378203a0 [DOI] [PubMed] [Google Scholar]
- 86. Jones SN, Roe AE, Donehower LA, et al. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378(6553):206-208. https://doi.org/10.1038/378206a0 [DOI] [PubMed] [Google Scholar]
- 87. McMasters KM, Montes de Oca Luna R, Pena JR, et al. Mdm2 deletion does not alter growth characteristics of p53-deficient embryo fibroblasts. Oncogene. 1996;13(8):1731-1736. [PubMed] [Google Scholar]
- 88. Migliorini D, Lazzerini Denchi E, Danovi D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22(15):5527-5538. https://doi.org/10.1128/MCB.22.15.5527-5538.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Finch RA, Donoviel DB, Potter D, et al. mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 2002;62(11):3221-3225. [PubMed] [Google Scholar]
- 90. Parant J, Chavez-Reyes A, Little NA, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29(1):92-95. https://doi.org/10.1038/ng714 [DOI] [PubMed] [Google Scholar]
- 91. Chavez-Reyes A, Parant JM, Amelse LL, et al. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res. 2003;63(24):8664-8669. [PubMed] [Google Scholar]
- 92. Hume S, Dianov GL, Ramadan K.. A unified model for the G1/S cell cycle transition. Nucleic Acids Res. 2020;48(22):12483-12501. https://doi.org/10.1093/nar/gkaa1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Aladjem MI, Spike BT, Rodewald LW, et al. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 1998;8(3):145-155. https://doi.org/10.1016/s0960-9822(98)70061-2 [DOI] [PubMed] [Google Scholar]
- 94. Hong Y, Stambrook PJ.. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc Natl Acad Sci USA. 2004;101(40):14443-14448. https://doi.org/10.1073/pnas.0401346101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. He H, Wang C, Dai Q, et al. p53 and p73 regulate apoptosis but not cell-cycle progression in mouse embryonic stem cells upon DNA damage and differentiation Stem Cell Rep. 2016;7(6):1087-1098. https://doi.org/10.1016/j.stemcr.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. M L, H G, Tripathi BK, et al. An Apela RNA-containing negative feedback loop regulates p53-mediated apoptosis in embryonic stem cells. Cell Stem Cell. 2015;16(6):669-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Prost S, Bellamy CO, Clarke AR, et al. p53-independent DNA repair and cell cycle arrest in embryonic stem cells. FEBS Lett. 1998;425(3):499-504. https://doi.org/10.1016/s0014-5793(98)00296-8 [DOI] [PubMed] [Google Scholar]
- 98. Stark GR, Taylor WR.. Analyzing the G2/M checkpoint. Methods Mol Biol. 2004;280:51-82. https://doi.org/10.1385/1-59259-788-2:051 [DOI] [PubMed] [Google Scholar]
- 99. M L, Y H, Dubois W, et al. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell. 2012;46(1):30-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Corbet SW, Clarke AR, Gledhill S, et al. P53-dependent and -independent links between DNA-damage, apoptosis and mutation frequency in ES cells. Oncogene. 1999;18(8):1537-1544. [DOI] [PubMed] [Google Scholar]
- 101. de Vries A, Flores ER, Miranda B, et al. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proc Natl Acad Sci USA. 2002;99(5):2948-2953. https://doi.org/10.1073/pnas.052713099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Grandela C, Pera MF, Grimmond SM, et al. p53 is required for etoposide-induced apoptosis of human embryonic stem cells. Stem Cell Res. 2007;1(2):116-128. https://doi.org/10.1016/j.scr.2007.10.003 [DOI] [PubMed] [Google Scholar]
- 103. Ter Huurne M, Chappell J, Dalton S, et al. Distinct cell-cycle control in two different states of mouse pluripotency. Cell Stem Cell. 2017;21(4):449-455.e4. https://doi.org/10.1016/j.stem.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ter Huurne M, Peng T, G Y, et al. Critical role for P53 in regulating the cell cycle of ground state embryonic stem cells. Stem Cell Rep. 2020;14(2):175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Atashpaz S, Samadi Shams S, Gonzalez JM, et al. ATR expands embryonic stem cell fate potential in response to replication stress. Elife. 2020;9:e54756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Grow EJ, Weaver BD, Smith CM, et al. p53 convergently activates Dux/DUX4 in embryonic stem cells and in facioscapulohumeral muscular dystrophy cell models. Nat Genet. 2021;53:1207-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Chen Z, Zhang Y.. Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat Genet. 2019;51(6):947-951. https://doi.org/10.1038/s41588-019-0418-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jacobs K, Zambelli F, Mertzanidou A, et al. Higher-density culture in human embryonic stem cells results in DNA damage and genome instability. Stem Cell Rep. 2016;6(3):330-341. https://doi.org/10.1016/j.stemcr.2016.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mansouri A, Fukumitsu H, Schindehuette J, et al. Differentiation of embryonic stem cells. Curr Protoc Neurosci. 2009;Chapter 3:Unit3 6. [DOI] [PubMed] [Google Scholar]
- 110. Jain AK, Y X, McCarthy R, et al. LncPRESS1 is a p53-regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol Cell. 2016;64(5):967-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Maimets T, Neganova I, Armstrong L, et al. Activation of p53 by nutlin leads to rapid differentiation of human embryonic stem cells. Oncogene. 2008;27(40):5277-5287. https://doi.org/10.1038/onc.2008.166 [DOI] [PubMed] [Google Scholar]
- 112. Navarro P. 2i, or not 2i: the soliloquy of nanog-negative mouse embryonic stem cells. Stem Cell Rep. 2018;11(1):1-3. https://doi.org/10.1016/j.stemcr.2018.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Tovy A, Spiro A, McCarthy R, et al. p53 is essential for DNA methylation homeostasis in naive embryonic stem cells, and its loss promotes clonal heterogeneity. Genes Dev. 2017;31(10):959-972. https://doi.org/10.1101/gad.299198.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Setoguchi K, TeSlaa T, Koehler CM, et al. P53 regulates rapid apoptosis in human pluripotent stem cells. J Mol Biol. 2016;428(7):1465-1475. https://doi.org/10.1016/j.jmb.2015.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Rivlin N, Brosh R, Oren M, et al. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2(4):466-474. https://doi.org/10.1177/1947601911408889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kojima Y, Tam OH, Tam PP.. Timing of developmental events in the early mouse embryo. Semin Cell Dev Biol. 2014;34:65-75. https://doi.org/10.1016/j.semcdb.2014.06.010 [DOI] [PubMed] [Google Scholar]
- 117. Bardot ES, Hadjantonakis AK.. Mouse gastrulation: coordination of tissue patterning, specification and diversification of cell fate. Mech Dev. 2020;163:103617. https://doi.org/10.1016/j.mod.2020.103617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Samanta M, Kalantry S.. Generating primed pluripotent epiblast stem cells: a methodology chapter. Curr Top Dev Biol. 2020;138:139-174. https://doi.org/10.1016/bs.ctdb.2020.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analyzed in support of this research.