Abstract
Human embryonic stem (hES) cells are highly sensitive to apoptotic stimuli such as DNA damage, which allows for the rapid elimination of mutated cells during development. However, the mechanisms that maintain hES cells in the primed apoptotic state are not completely known. Key activators of apoptosis, the BH3-only proteins, are present at low levels in most cell types. In contrast, hES cells have constitutive high levels of the BH3-only protein, NOXA. We examined the importance of NOXA for enabling apoptosis in hES cells. hES cells deleted for NOXA showed remarkable protection against multiple apoptotic stimuli. NOXA was constitutively localized to the mitochondria, where it interacted with MCL1. Strikingly, inhibition of MCL1 in NOXA knockout cells was sufficient to sensitize these cells to DNA damage-induced cell death. Our study demonstrates that an essential function of constitutive high levels of NOXA in hES cells is to effectively antagonize MCL1 to permit rapid apoptosis.
Keywords: human embryonic stem cells, cell death, DNA damage, NOXA, MCL1
Graphical Abstract
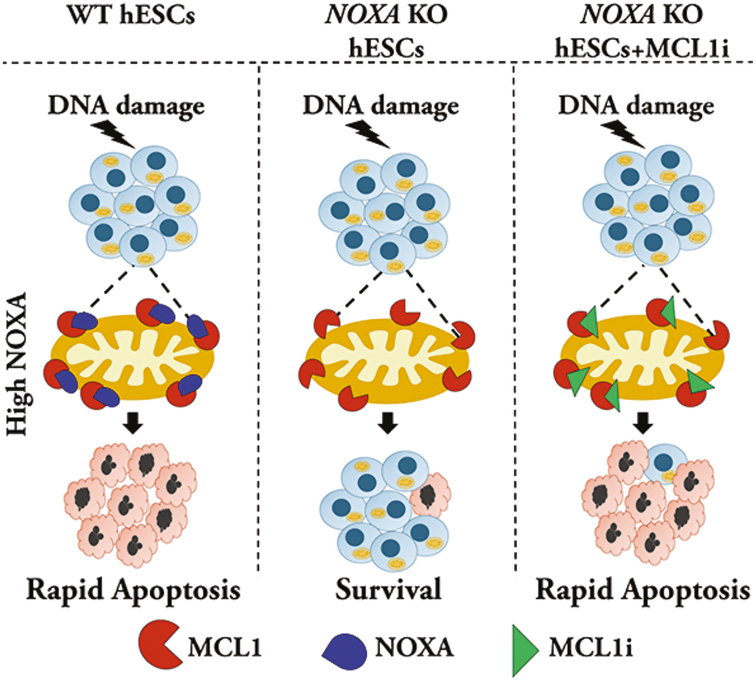
Graphical Abstract.
hES cells express high levels of pro-apoptotic protein NOXA and are highly sensitive to cell death. NOXA-deleted hES cells show remarkable protection against cell death. NOXA is localized to the mitochondria where it interacts with the anti-apoptotic protein, MCL1. Inhibiting MCL1 in NOXA-deleted cells restored apoptotic sensitivity, indicating the importance of the NOXA-MCL1 axis for cell death in hES cells.
Significance Statement.
Human embryonic stem (hES) cells have the ability to differentiate into all cell types, hence understanding how these cells regulate their survival and death is important. These cells undergo rapid death in response to DNA damage thereby removing mutated cells from the developing embryo. We focused on identifying the mechanism underlying the sensitivity of these cells to DNA damage. We discovered that the protein NOXA is essential for cell death in hES cells. Further, the crucial function of NOXA is to neutralize high levels of antiapototic protein, MCL1, thus enabling hES cells to respond rapidly to DNA damage.
Introduction
Cells with varied physiological functions exhibit marked differences in their sensitivity to cell death. However, exactly how these differences in apoptotic thresholds are set in cells remain largely unknown. For example, human embryonic stem (hES) cells are highly sensitive to various genotoxic agents such as etoposide, a topoisomerase II inhibitor, UV exposure, and γ-irradiation.1-6 As these cells can differentiate into all cell types, their ability to undergo rapid apoptosis following DNA damage is crucial to avoid the propagation of DNA mutations during development. hES cells are also widely used to generate different types of cells for modeling human diseases. Therefore, understanding the mechanisms by which the survival and death of hES cells are regulated is significant for both development and disease.
Key initiators of apoptosis in cells are the BH3-only members of the BCL2 family proteins.7 These proteins are activated (eg, transcriptional induction, posttranslational modification) by extracellular or intracellular apoptotic stimuli and initiate the cascade of events that result in cell death. Briefly, the BH3-only proteins fall under 2 categories: The sensitizer BH3-only proteins (eg, NOXA, HRK, BAD) bind to and inactivate the anti-apoptotic BCL2 family proteins such as BCL2, BCL-XL, and MCL1. The activator BH3-only proteins (eg, BIM, BID, PUMA) bind to and directly activate the pro-apoptotic BCL2 family proteins, BAX and BAK.8 Thus, activation of BH3-only proteins results in the activation of BAX and BAK which, by forming pores in the mitochondrial outer membrane, induce the release of cytochrome c (cyt c) into the cytosol to trigger caspases to execute apoptotic cell death.
Interestingly, as the BH3-only proteins consist of many members, they are thought to be functionally redundant. Multiple BH3-only proteins can be engaged in response to a given apoptotic stimuli and several members can act as a sensitizer or direct activators to activate BAX and BAK.9-11 Consistent with the observation that the BH3-only proteins are functionally redundant, the knockout of any single BH3-only protein in mice does not result in complete inhibition of apoptosis. Indeed, with the rare exception in specific contexts, deletion of multiple BH3-only proteins (eg, triple deletion of BIM, BID, and PUMA) is necessary to robustly inhibit apoptosis.12
We previously showed that hES cells undergo apoptosis with DNA damage much more rapidly in comparison to other cell types. For example, while DNA damage typically induces cell death in most cells by 24-48 hours, virtually all hES cells die within 5 hours.1 Multiple factor contribute to this apoptotic priming of hES cells, including increased stabilization of p53 after DNA damage,6 the maintenance of active BAX at the Golgi,1 and the balance between pro- and anti-apoptotic BCL2 family proteins.6 hES cells are known to express high levels of multiple BH3-only proteins with NOXA being expressed at a strikingly 50-fold higher levels as compared to non-hES cells.13
This constitutive high expression of NOXA suggests that it could play a crucial role in hES cells. In this study, we examined the function of NOXA in hES cells by generating hES cells that were CRISPR-deleted for NOXA. Our results identify NOXA as an essential mediator of DNA damage response and emphasize the importance of the NOXA/MCL1 axis in hES cells.
Methods and Materials
Cell Culture
Human embryonic stem cell lines H9 (WA09), H14 (WA14), and WA22 were obtained from WiCell Research Institute. hES cells were maintained on hES cell qualified matrigel in mTeSR1 medium. H9 (WA09) cells were used for CRISPR gene editing. Human normal skin fibroblasts CCD-1079Sk were acquired from ATCC and maintained in DMEM with 10% Fetal Bovine Serum and MEM nonessential amino acids. Cells were maintained at 37°C in 5% CO2.
For cell death experiments, cells were seeded at a density of 600 000 cells per well in 6-well plates. Approximately after 24 hours, cells were treated with etoposide or tunicamycin. When MCL1 inhibitors (S63845 and AZD5991) were used, cells were treated with inhibitors in the presence or absence of etoposide. Cells were either stained with Nuclear Blue to label cell nuclei for evaluating cell death or were collected for Western analysis. Images were captured using Leica DMi8 microscope with Leica DFC9000 cMOS camera using the Leica LASX imaging software. Cell death was quantified manually on the basis of nuclear morphology (condensed or fragmented), with an average of more than 800 cells analyzed per condition. For siRNA transfections, cells were seeded at 350 000 cells per well in a 6-well plate. Approximately after 24 hours, control siRNA (MISSION siRNA Fluorescent Universal Negative Control #1, Cyanine 3) and MCL1 siRNA (GGACUUUUAUACCUGUUAUtt) were transfected using Lipofectamine 3000 reagent at 50 nM concentration. Approximately 24 hours post-transfection, cells were treated with DMSO or etoposide and cell death was quantified as described above.
Trilineage differentiation assay was performed using STEMdiff Trilineage differentiation assay kit from STEMCELL Technologies as per the manufacturer\'s protocol. RNA was isolated using high-capacity cDNA reverse transcription kit from Thermofisher and samples were run on TaqMan hPSC score card panel in 384 wells from Thermofisher.
For karyotyping, cells were seeded in a 6-well plate and sent to KaryoLogic for analysis.
Genome Editing of NOXA Locus Using CRISPR/Cas9
Two sgRNAs, TCGAGTGTGCTACTCAACTC and TTCTTGCGCGCCTTCTTCCC (Thermo Fisher Scientific) were delivered into H9 cells along with recombinant True Cut Cas9 v2 using the Neon Transfection system (Thermo Fisher Scientific, Cat. No MPK5000). H9 cells (1 × 105 cells) were electroporated with 33 pmols of each sgRNA and 5 μg/mL Cas9 diluted in 10 μL buffer R. Cells were electroporated using Neon program 17 (850V, 30 seconds, 2 pulses). Seventy-two hours after electroporation, H9 cells were collected. Editing efficiency was assessed using GeneArt Genomic Cleavage Detection Kit. Cell pools with good indel percentage were dissociated into single cells and seeded on 96-well plates coated with matrigel. After 2 weeks, the genomic DNA of single-cell colonies was extracted and the NOXA gene was amplified using primers flanking the sgRNA target sites. Sanger sequencing confirmed the gene knockout. Single-cell clones which were negative for any insertion or deletion at the target site, as per the sequencing data analysis, were selected as negative controls for the study.
Western Blot Analysis
For sample preparation, cells were trypsinized using the TrypLE select enzyme and floating cells were also collected in experiments where apoptosis was induced. Cell pellets were lysed using RIPA buffer for 30 minutes on ice followed by sonication (2×, 10 seconds pulses). Samples were centrifuged at 13 000 rpm for 20 minutes at 4°C. Supernatants were collected and protein quantification was performed using Pierce BCA protein assay kit. Lysates (30-50 μg) were loaded on 12% polyacrylamide gel. After transfer on immobilon FL transfer membrane immunoblotting was performed using specific antibodies (details provided in the Supplementary Information). Membranes were imaged using LICOR imaging system (Odyssey CLx) or chemiluminescence (Amersham Imager 680).
Immunofluorescence
For immunofluorescence study, cells were seeded on glass coverslips coated with matrigel. For staining with pluripotency markers, cells were incubated at 37°C for approximately 24 hours and then fixed with 4% paraformaldehyde at 4°C for 30 minutes or room temperature for 15 minutes. In cell death studies, cells were left untreated or treated with DMSO or etoposide (20 μM) for 5 hours in presence of QVD-OPH (25 mM, SM Biochemicals), thereafter cells were stained for p53, cyt c, TOM20, or pluripotency markers (OCT4, SOX2, NANOG). Images were captured on a Leica DMi8 microscope with Leica DFC9000 cMOS camera using the Leica LASX imaging software. For an unbiased approach for quantification of p53 and cyt c, all images were captured at the same image settings and counted manually for positive or negative signals. For localization study, cells were first transfected with NOXA-GFP plasmid (Vector Builder) using Lipofectamine 3000 reagent as per manufacturers protocol, incubated for 24 hours and treated with etoposide for 5 hours in presence of QVD-OPH followed by fixation and staining for MCL1, TOM20 and NOXA-GFP. Hoechst 33342 was used for nuclear staining at all times. For testing cyt c release in NOXA KO cells, cells were transfected (in presence of QVD-OPH) with either GFP or NOXA-GFP plasmids using Lipofectamine 3000 reagent as per the manufacturer’s protocol. Approximately 24 hours post-transfection, cells were treated with etoposide (20 μM) for 5 hours in presence of QVD-OPH (25 mM, SM Biochemicals). Images were captured on Leica DMi8 microscope and quantified manually as described above.
Cell Fractionation
Cells, seeded at approximately 1.106 cells per well of 6-well plates 24 hours before treatment, were incubated in 100 μL of ice-cold CLAMI (Cell Lysis and Mitochondria Intact) buffer (250 mM sucrose, 70 mM KCl, 137 mM NaCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 200 μg/mL digitonin, pH 7.2) containing complete protease inhibitor for 5 minutes on ice. Samples were pelleted at 1000 g for 5 minutes at 4°C and supernatants (cytosolic fraction) were collected. Pellets were incubated in 100 μL of ice-cold IP buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 0.2% Triton X100, 0.3% Nonidet P-40) containing complete protease inhibitor for 10 minutes on ice. Samples were pelleted at 10 000 g for 10 minutes at 4°C, and supernatants (organelles fraction) were collected. Protein concentrations were estimated using Pierce BCA protein assay kit and samples containing an equal amount of proteins were prepared with SDS-PAGE sample buffer for Western blot analysis.
Immunoprecipitation
Cells (~5.106 cells) were lysed with 500 µL CHAPS buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1 mM DTT, 1% CHAPS) containing complete protease inhibitor. Lysates were sonicated and centrifuged at 16 000 g for 5 minutes at 4°C. Supernatants were precleared with 40 µL protein A/G beads for 30 minutes at 4°C. Precleared lysates were incubated with 40 µL protein A/G beads preconjugated and crosslinked (5 mM BS3) with 4 µg normal mouse IgG or 4 µg mouse anti-MCL1 IgG1 mAb for 4 hours at 4°C. Beads were washed 3 times with CHAPS lysis buffer before being resuspended with Laemmli buffer for Western blot analysis.
Gene Copy Number Variation (CNV) Assay
qRT PCR-based gene copy number variation assay for 20q11.21 was performed as per the manufacturer’s protocol. The TaqMan assays used were Rnase P and POFUT1 from Thermo Fisher.
Statistical Analysis
For all experiments, the value “n” indicated in the figure legend represents the number of independent replicates. Data visualization and statistical analyses were performed using GraphPad Prism v.8.3.1. Error bars represent SEM. Before conducting statistical analysis, normality was tested using the Shapiro-Wilk normality test. No data were excluded for the statistical analysis. For all assays comparing 2 groups with normal distributions, the Student’s t-test (unpaired, 2-tailed) with a 95% confidence interval (CI) was performed. For multiple comparison tests, a 2-way analysis of variance (ANOVA) with post-hoc Dunnett’s multiple comparisons test was performed.
Additional details are provided in the Supplementary Materials section.
Results
hES Cells Express High Level of NOXA
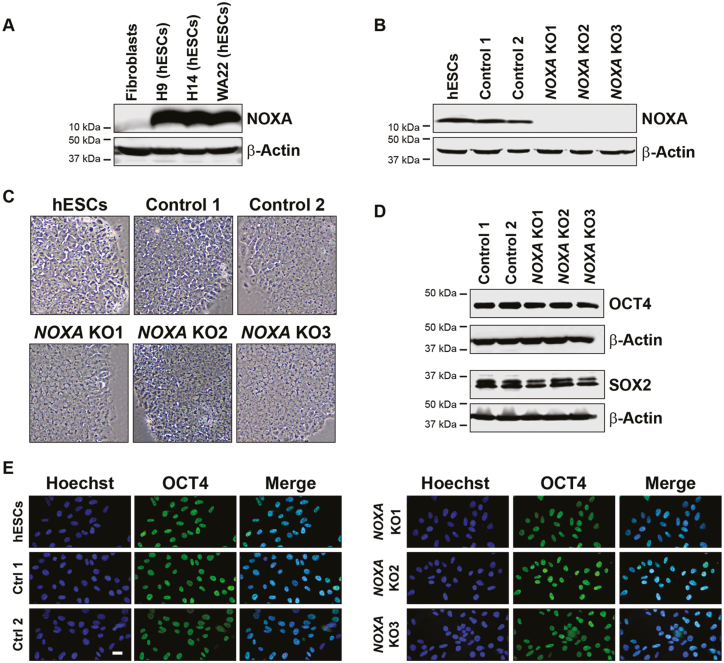
NOXA (also called PMAIP1) is a BH3-only gene that is known to be induced in response to DNA damage in cells.11,14,15 We examined the basal expression of NOXA, as well as other BH3-only proteins (eg, PUMA, BIM, BID, BIK) in hES cells compared to fibroblasts. Consistent with previous reports,13,16 NOXA was present at very high levels in all 3 hES cell lines (H9, H14, and WA22) as compared to human dermal fibroblasts (Fig. 1A, Supplementary Fig. S1A).13,17 Since hES cells are known to be primed for apoptosis in response to DNA damage1 and NOXA is a key mediator of DNA damage-induced apoptosis, we considered the possibility that NOXA could be functionally important for the rapid DNA damage response in hES cells.
Figure 1.
hES cells express high levels of NOXA. (A) Western blot for NOXA expression in human normal skin fibroblasts (CCD-1079Sk), H9 hES cells (WA09), H14 hES cells (WA14), and WA22 hES cells. (B) Western blot for NOXA expression in hES cells (H9), control cells (cells negative for indels), and NOXA knockout cells generated by CRISPR/Cas9. (C) Bright field images for hES cells (H9), control and NOXA knockout cells. Scale bar 100 μm (representative for all the figures in the panel). (D) Western blot for pluripotency markers OCT4 and SOX2 in control and NOXA knockout cells. (E) Immunofluorescence staining for the pluripotency marker OCT4 in hES cells (H9), control and NOXA knockout cells. Scale bar 20 μm (representative for all the figures in the panel).
To examine the function of NOXA in hES cells, we utilized CRISPR-Cas9 technology to generate NOXA knockout (NOXA KO) hES cells. Single-cell clone screening was done by sequencing and Western blot to identify hES cells deleted for NOXA. Two independent control hES cells (single-cell clones which were negative for any insertion or deletion at the target site as confirmed by sequencing analysis were selected as a negative control for the study) and 3 NOXA knockout hES cells were selected for further analysis (Fig. 1B).
NOXA knockout hES cells maintained their stem cell characteristics. They exhibited stem cell morphology (Fig. 1C). Cytogenetic analysis of 2 control and 2 NOXA knockout cell lines (with 20 metaphase spreads analyzed for each clone) confirmed normal karyotype (Supplementary Fig. S2). Also, the expression of pluripotency markers (eg, OCT4, SOX2, NANOG) (Fig. 1D,E, Supplementary Fig. S3A,B) and Trilineage-differentiation assay (Supplementary Fig. S4A-C) confirmed their pluripotent potential. Thus, NOXA deletion alone did not alter the pluripotent status of hES cells. We also examined and confirmed that the expression of other BH3-only proteins was unaffected in the NOXA-deleted cells (Supplementary Fig. S1B).
NOXA-deleted hES Cells Are Resistant to Cell Death
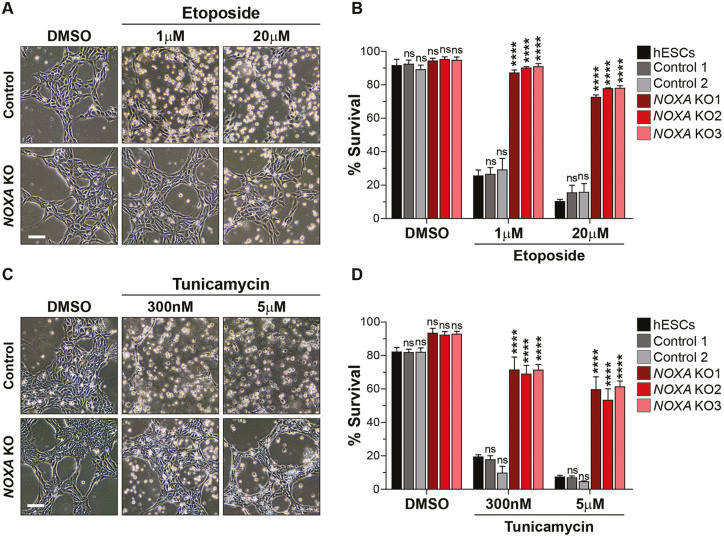
To examine the functional importance of NOXA in mediating the DNA damage-induced apoptosis in hES cells, we exposed control and NOXA knockout hES cells to the DNA damage drug, etoposide. hES cells are very sensitive to DNA damage with even a low dose of 1 μM etoposide resulting in the death of 75% of cells within 5 hours of treatment (Fig. 2A,B). Strikingly, NOXA knockout cells were nearly completely protected under these conditions. Additionally, even at a much higher dose of etoposide (20 μM), NOXA knockout cells were markedly protected with greater than 70% cell survival, whereas less than 15% of control hES cells were alive at this concentration (Fig. 2A,B).
Figure 2.
NOXA-deleted hES cells are resistant to cell death. (A) Representative bright field images of control and NOXA knockout hES cells treated with DMSO or etoposide (1 μM and 20 μM) for 5 hours. Scale bar 100 μm (representative for all the figures in the panel). (B) Quantification of the survival of hES, control and NOXA knockout cells treated with DMSO or etoposide (1 μM and 20 μM) for 5 hours. Cells were stained with nuclear blue and cell death was counted on the basis of nuclear morphology (condensed or fragmented). (n = 3 independent replicates; error bars represent SEM; Dunnett’s multiple comparison test compared to hESCs, ∗∗∗∗ p < 0.0001, ns: p ≥ 0.05). (C) Representative bright field images of control and NOXA knockout hES cells treated with DMSO or tunicamycin (300 nM and 5 μM) for 20 hours. Scale bar 100 μm (representative for all the figures in the panel). (D) Quantification of the survival of hES, control and NOXA knockout cells treated with DMSO or tunicamycin (300 nM and 5 μM) for 20 hours (n = 3 independent replicates; error bars represent SEM; Dunnett’s multiple comparison test compared to hESCs, ∗∗∗∗P < .0001, ns: P ≥ .05).
To verify that DMSO used as a solvent for etoposide in this experiment did not have any influence on stem cell pluripotency or differentiation, we checked for expression of pluripotency factors (OCT4, SOX2, and NANOG) in the presence or absence of DMSO and found DMSO alone did not have any effect on the expression of pluripotency factors (Supplementary Fig. S5A-C), hence the results we see on etoposide treatment are not due to differentiation of the cells.
To determine if the importance of NOXA in hES cells was specific to DNA damage mediated apoptosis, we examined whether NOXA knockout hES cells were also protected in response to endoplasmic reticulum (ER) stress. We focused on ER stress since NOXA has been linked to ER stress-induced apoptosis in various cell lines.18,19 Control and NOXA knockout hES cells were treated with the ER stress-inducing drug tunicamycin. We found that NOXA deletion conferred significant protection at both low (300 nM) and high (5 μM) doses of tunicamycin in hES cells (Fig. 2C,D). Similarly, NOXA deletion also protected hES cells treated with the broad-spectrum kinase inhibitor staurosporine (data not shown). Together, these results identify NOXA as an essential regulator of apoptosis in hES cells.
NOXA-deficient hES Cells Induce p53 but do not Release cyt c After DNA Damage
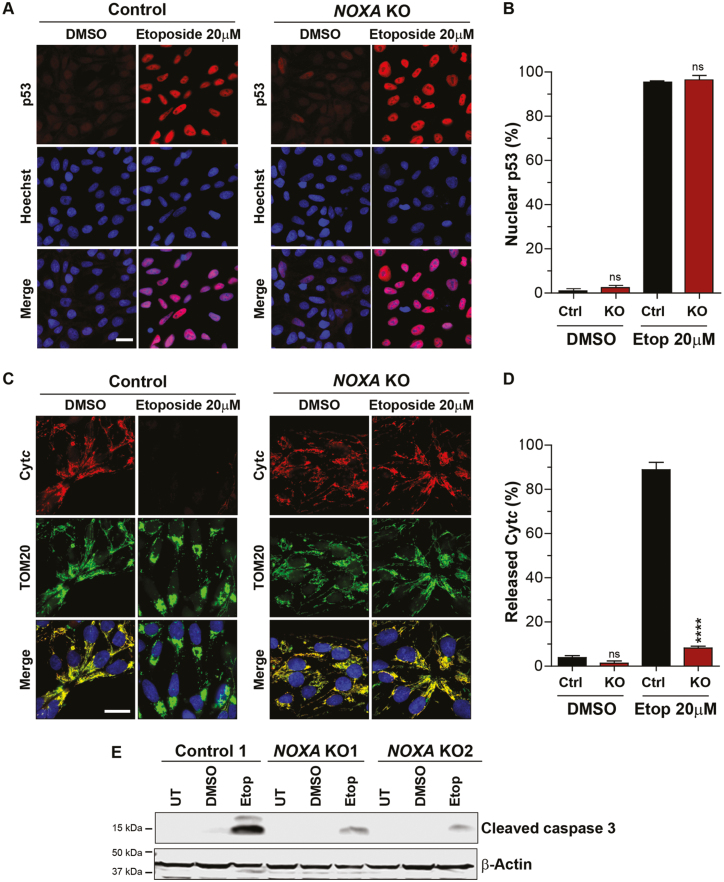
To confirm that NOXA deletion protected against mitochondrial permeabilization in response to apoptotic stimuli in hES cells, we specifically probed for 3 events that are known to occur in response to DNA damage: (1) p53 induction; (2) cyt c release from mitochondria; and (3) caspase-3 activation. Control and NOXA knockout hES cells were treated with 20 μM etoposide for 5 hours and probed for these events. We found that p53 expression was comparably induced in control and NOXA knockout cells (Fig. 3A,B, Supplementary Fig. S6). This suggests that the upstream pathway is unaffected in the NOXA knockout cells. However, while cyt c was released in approximately 90% of control hES cells, only 8% of NOXA knockout cells showed release of cyt c (Fig. 3C,D). Consistent with the cyt c results, while etoposide-induced robust cleavage of caspase-3 in control hES cells, only minimal caspase-3 cleavage could be observed in NOXA knockout hES cells (Fig. 3E). These results confirm that the p53 pathway is functional in NOXA knockout hES cells but these cells are unable to release cyt c and activate caspase-3 activation after DNA damage.
Figure 3.
NOXA-deficient hES cells induce p53 but do not release cyt c after DNA damage. (A) Representative immunofluorescence images of p53 in control and NOXA knockout cells treated with DMSO or etoposide (20 μM) for 5 hours. p53 (red) and Hoechst (blue). Scale bar 20 μm (representative for all the figures in the panel). (B) Quantification of the cells presenting p53 in the nucleus in Fig. 3A (n = 3 independent replicates; error bars represent SEM; unpaired, 2-tailed Student’s t-test compared to Ctrl, ∗∗∗∗P < .0001, ns: P ≥ .05). (C) Representative immunofluorescence images of cyt c in control and NOXA knockout cells treated with DMSO or etoposide (20 μM) for 5 hours. Cyt c (red), TOM20, mitochondrial marker (green) and Hoechst (blue). Scale bar 20 μm (representative for all the figures in the panel). (D) Quantification of the cells showing cyt c released in Fig. 3C (n = 3 independent replicates; error bars represent SEM; unpaired, 2-tailed Student’s t-test compared to Ctrl, ∗∗∗∗ P < .0001, ns: P ≥ .05). (E) Western blot for cleaved caspase 3 in control and NOXA knockout cells untreated or treated with DMSO and etoposide (20 μM) for 5 hours (n = 3).
It is known that stem cells can acquire genomic aberration during culture. For example, 20q11.21 is known to amplify in stem cells and provide resistance to apoptosis due to overexpression of the anti-apoptotic protein BCL-XL located in this chromosome region.20,21 To confirm that the NOXA KO cells did not exhibit resistance to cell death due to the gain of chromosome region 20q11.21 or increased expression of BCL-XL, we performed a qRT PCR based CNV assay to assess the number of copies of the chromosome region 20q11.21 (Supplementary Fig. S7A). CNV results show no gain of chromosome region 20q11.21 in the NOXA KO cells. Additionally, NOXA KO cells did not show increased expression of BCL-XL (Supplementary Fig. S7B). Thus, the increased resistance to cell death observed in NOXA KO cells is not due to the overexpression of anti-apoptotic protein BCL-XL or gain of 20q11.21 region of the chromosome.
To confirm that the resistance to cell death exhibited by NOXA-deleted cells was due to specific deletion of NOXA and not an off-target effect, we overexpressed GFP or NOXA-GFP in NOXA-deleted cells and treated these cells with etoposide. As expected, cells overexpressing NOXA-GFP restored the release of cyt c (Supplementary Fig. S8A,B), confirming that the resistance to cell death observed in NOXA-deleted cells was specific to the lack of NOXA expression.
NOXA Colocalizes and Interacts with MCL1 at the Mitochondria in hES Cells
Individual BH3-only proteins regulate cell death by either neutralizing the pro-survival proteins (eg, BCL2, BCL-XL, MCL1) or by directly activating pro-apoptotic proteins (eg, BAX, BAK), or both.9 NOXA is an outlier among the BH3-only proteins because it has limited capacity to interact with multiple BCL2 family proteins and its best-defined association is with the pro-survival protein MCL1.22 Thus, we examined whether NOXA colocalizes and interacts with MCL1 in hES cells, under normal and apoptotic conditions.
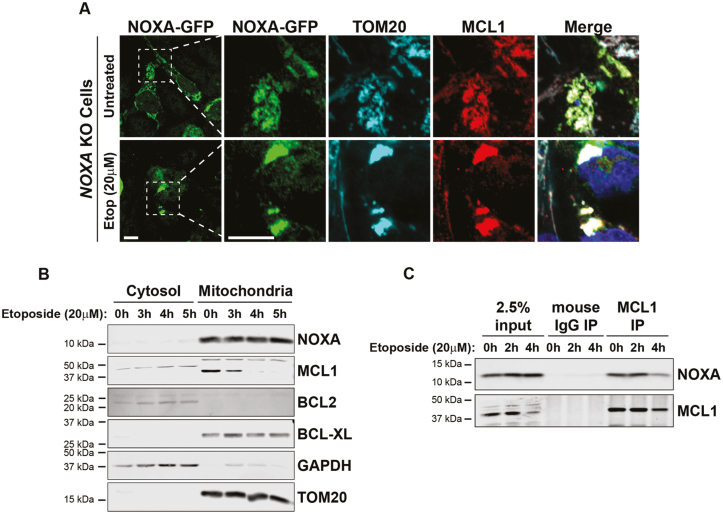
We first tested the localization of NOXA and MCL1 by immunofluorescence. Since we were unable to identify an antibody that specifically detected endogenous NOXA by immunofluorescence in hES cells, we examined the localization of exogenously expressed NOXA-GFP in NOXA knockout hES cells. We found NOXA-GFP to be localized to the mitochondria in untreated hES cells as its signal overlapped with the mitochondrial marker TOM20 (Fig. 4A). MCL1 staining also overlapped with NOXA and TOM20 staining indicating that both NOXA and MCL1 were present at the mitochondria in untreated hES cells (Fig. 4A). Importantly, upon etoposide treatment, the mitochondria appeared aggregated, but both NOXA and MCL1 remained colocalized to the aggregated mitochondria under these conditions (Fig. 4A).
Figure 4.
NOXA colocalizes and interacts with MCL1 at the mitochondria in hES cells. (A) Representative images of immunofluorescence staining of NOXA, MCL1, TOM20 in NOXA knockout cells transfected with NOXA-GFP plasmid in presence of caspase inhibitor QVD. Cells were either left untreated or treated with etoposide (20 μM) for 5 hours. Cells were immunostained with antibodies to GFP (NOXA; green), TOM20, (cyan), MCL1 (red), and Hoechst (blue). Scale bar 10 μm (representative for all the figures in the panel). (B) Cytosolic and mitochondrial fractions of cells treated with etoposide (20 μM) for indicated period of time. Fractions were probed for NOXA, MCL1, BCL2, and BCL-XL. GAPDH was used as marker for cytosolic fractions and TOM20 as a marker for mitochondrial fraction. (C) Immunoprecipitation of endogenous MCL1 in hES cells treated with etoposide (20 μM) for indicated period of time. Membranes were probed for both NOXA and MCL1.
To confirm the localization of endogenous NOXA and MCL1, we conducted cell fractionation studies in untreated and etoposide-treated hES cells. Analysis of cytosolic and mitochondrial fractions confirmed that both NOXA and MCL1 were present in the mitochondrial fractions of untreated and etoposide-treated hES cells (Fig. 4B). Interestingly, MCL1 levels decreased with etoposide treatment, an observation that is consistent with the fact that MCL1 is targeted for degradation in cells undergoing apoptosis.23,24
To directly examine whether NOXA and MCL1 interact in hES cells, we conducted immunoprecipitation experiments. Our results show that NOXA is associated with MCL1 in untreated hES cells (Fig. 4C). This interaction is initially maintained but subsequently reduced with etoposide treatment, as MCL1 is targeted for degradation (Fig. 4B,C). Together, these results show that in hES cells, NOXA colocalizes and interacts with MCL1 at the mitochondria under normal conditions, and this interaction is disrupted upon etoposide treatment as MCL1 levels decrease to permit apoptosis.
MCL1 Inhibition Permits Apoptosis in NOXA Knockout hES Cells
Our results with the NOXA knockout cells show that NOXA is essential for apoptosis in hES cells (Fig. 2). We considered 2 possible mechanisms by which NOXA could promote apoptosis in hES cells. While it is generally considered to act as an inhibitor of MCL1, NOXA can also bind to BAX and function as a direct activator of BAX.25 Thus, the function of NOXA in hES cells could be inhibition of MCL1 or activation of BAX.
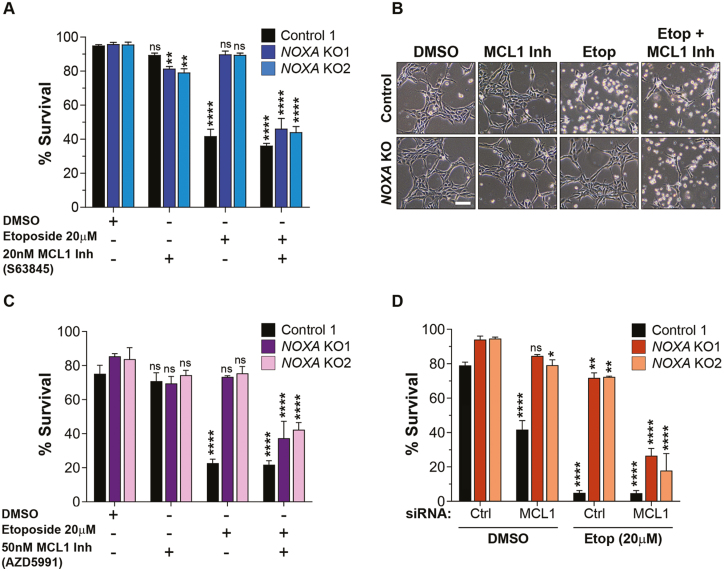
To evaluate the specific function of NOXA in hES cells, we examined whether inhibition of MCL1 can restore apoptosis in NOXA-deficient hES cells. Control and NOXA knockout hES cells were treated with the MCL1 inhibitor S6384526 either in the absence or presence of etoposide. Our results show that MCL1 inhibition in NOXA knockout cells strikingly enabled these cells to undergo apoptosis with DNA damage (Fig. 5A,B). To confirm these results, we conducted similar experiments with a second, more recently described, MCL1 inhibitor (AZD599127). Here too, MCL1 inhibition permitted the NOXA-deficient hES cells to undergo apoptosis with DNA damage (Fig. 5C). We also evaluated the expression of MCL1 in presence of MCL1 inhibitors or etoposide and observed no significant difference in the level of MCL1 in presence of MCL1 inhibitors (Supplementary Fig. S9A,B). Lastly, as an independent approach to inhibit MCL1, we utilized siRNAs to knockdown MCL1. Our results show that treatment with MCL1 siRNA, but not control siRNA, restored the capability of NOXA knockout cells to undergo apoptosis with DNA damage (Fig. 5D, Supplementary Fig. S10). Together, these results indicate that the important function of NOXA in hES cells is to inhibit MCL1 and inactivation of MCL1 is necessary for apoptosis to proceed in hES cells.
Figure 5.
MCL1 inhibition permits apoptosis in NOXA knockout hES cells. (A) Quantification of the survival of control and NOXA knockout cells in presence of DMSO or MCL1 inhibitor (S63845). Cells were either left untreated or treated with etoposide (20 μM) for 3 hours, as shown. Cells were stained with nuclear blue and cell death was counted on the basis of nuclear morphology (condensed or fragmented). (n = 3 independent replicates; error bars represent SEM; Dunnett’s multiple comparison test compared to DMSO, NOXA KO1 MCL1 Inh ∗∗ P = .0037, NOXA KO2 MCL1 Inh ∗∗ P = .0011, ∗∗∗∗ P < .0001, ns P ≥ .05). (B) Representative bright field images for Fig. 5A. Scale bar 100 μm (representative for all the figures in the panel). (C) Quantification of the survival of control and NOXA knockout cells in presence of DMSO or MCL1 inhibitor (AZD5991). Cells were either left untreated or treated with 20 μM etoposide for 5 hours, as shown (n = 3 independent replicates; error bars represent SEM; Dunnett’s multiple comparison test compared to DMSO, ∗∗∗∗ P < .0001, ns P ≥ .05). (D) Quantification of the survival of control and NOXA knockout cells transfected with control siRNA or MCL1 siRNA in presence of DMSO or etoposide (20 μM) for 7 hours, as shown (n = 3 independent replicates; error bars represent SEM; Dunnett’s multiple comparison test compared to Ctrl siRNA DMSO; NOXA KO2, DMSO, MCL1 siRNA ∗ P = .0229; NOXA KO1, Etop, Ctrl siRNA ∗∗ P = .0011; NOXA KO2, Etop, Ctrl siRNA ∗∗ P = .0011; ∗∗∗∗ P < .0001, ns P ≥ .05).
Discussion
In this study, we report that the BH3-only protein NOXA is essential for rapid apoptosis in hES cells. We find that NOXA knockout hES cells are resistant to multiple apoptotic stimuli including DNA damage and ER stress. NOXA was also recently identified in a genome-wide CRISPR screen where its deletion was found to improve hES cell survival in response to targeted double-stranded breaks.17 Interestingly, while BH3-only proteins are typically induced with apoptotic stimuli, NOXA is constitutively present at high levels and exists in a complex with MCL1 at the mitochondria even in healthy, untreated, hES cells. Our results show that NOXA-mediated inactivation of MCL1 is a crucial event for apoptosis to occur in hES cells, as MCL1 inhibition permitted apoptosis to occur in NOXA-deficient hES cells. Although the BH3-only proteins are generally considered to function redundantly in cells, our results highlight the importance of a single BH3-only protein, NOXA, for apoptosis in hES cells.
NOXA is well recognized as a p53-induced gene upon DNA damage in most cells.15 In hES cells, p53 is induced with DNA damage and required for apoptosis.2 Strikingly, however, its transcriptional activity is not required.3 This is consistent with the observation that NOXA shows constitutive high expression, and its levels do not substantially increase with DNA damage in hES cells. Interestingly, we have shown previously that p53 is required for the translocation of active BAX from the Golgi to mitochondria.1 Thus, p53 function in hES cells appears atypical, where it is not required to transcriptionally induce NOXA and activate BAX.
In contrast to other BH3-only proteins that interact with multiple BCL2 family proteins, NOXA primarily binds to only MCL1 with high affinity.14,22 This ability of NOXA to interact with and inhibit MCL1 is important in contexts where the inactivation of MCL1 is necessary for apoptosis to proceed. For example, NOXA is induced in response to multiple apoptotic stimuli including DNA damage, ER stress, hypoxia, and a viral infection where it is important for apoptosis.15,19,28-30 hES cells not only express high levels of NOXA but also MCL1,6,31 both of which co-exist at the mitochondria and interact with each other under normal and apoptotic conditions (Fig. 4). Importantly, however, MCL1 is targeted for degradation with apoptotic stimuli,24 including in ES cells.30 Exactly how the apoptotic stimulus promotes MCL1 degradation in hES cells remains unknown. In hematopoietic cells, where NOXA is constitutively expressed,32 or in gastric epithelial cells infected with Helicobacter pylori, where NOXA is induced, NOXA is found to be phosphorylated and localized to the cytoplasm.33 The model proposed is that dephosphorylation of NOXA results in its localization to mitochondria, where it interacts with MCL1 and promotes its degradation. It is unclear if a similar mechanism is engaged in hES cells since NOXA and MCL1 are constitutively at the mitochondria and interact even in untreated conditions. Previous studies have shown that apoptotic stimuli can also cause the phosphorylation of MCL1 that results in its degradation.34 Thus, we hypothesize that DNA damage likely induces either a posttranslational modification in NOXA or MCL1 or could activate an interacting protein that promotes the degradation of MCL1 in hES cells. Our finding that MCL1 inhibition is sufficient to permit apoptosis in NOXA-deficient hES cells indicates that the key function of NOXA in hES cells in the context of cell death is the inhibition of MCL1. These results also indicate that NOXA does not function as a direct activator of the proapoptotic proteins in these cells, since DNA damage-induced apoptosis can proceed in the absence of NOXA with MCL1 inhibition alone.
Examples of cells that constitutively express elevated levels of NOXA are rare. hES cells express NOXA at nearly 50-fold higher levels as compared to other cells.13 An interesting question here is why are NOXA levels constitutively high in hES cells? In hematopoietic cells where NOXA is also strongly expressed, it functions to increase glucose flux via the pentose phosphate pathway.32 As ES cells are known to be highly glycolytic,35 NOXA could be important for maintaining their high glycolytic state. Additionally, the levels of MCL1 are also high in hES cells where it was found to have non-apoptotic activity for the maintenance of pluripotency.31 Even though maintaining high levels of MCL1 is physiologically important for hES cells, the ability of hES cells to undergo rapid apoptosis in response to DNA damage is equally crucial to prevent the propagation of mutations during early embryonic development. Thus, the constitutive high levels of NOXA in hES cells may serve to effectively counteract the anti-apoptotic activity of MCL1 and enable rapid apoptosis. Together, these results highlight the finding that levels and activities of apoptotic proteins are fine-tuned not only to enable their specific non-apoptotic functions in different cell types but also to set the precise apoptotic thresholds that are physiologically appropriate for those cells.
Supplementary Material
Acknowledgments
We thank the Deshmukh lab members for their discussions and critical review of this work. We also thank Dr. Anirban Kar, Dr. Ayumi Nakamura and Dr. Vijay Swahari for reviewing the manuscript.
Funding
The Neuroscience Microscopy Core Facility, supported, in part, by funding from the National Institutes of Health - National Institute of Neurological Disease and Stroke Neuroscience Center Support Grant P30 NS045892 and the National Institutes of Health - National Institute of Child Health and Human Development Intellectual and Developmental Disabilities Research Center Support Grant U54 HD079124. This work was supported by National Institutes of Health grant GM118331 to M.D.
Conflict of Interest
M.D. declared a leadership position with Lucidiun Bio and patent holder for the use of miR-29 for neuroprotection. This patent is owned by the author’s university. The other authors declared no potential conflicts of interest.
Author Contributions
R.B.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; S.K., E.H.: collection and assembly of data, data analysis and interpretation; N.K.: data analysis; A.B.: contributed in CRISPR knockout generation; N.M.M.: contributed in Western blot experiments; M.D.: conception and design, financial support, administrative support, manuscript writing, and final approval of the manuscript.
Data Availability
No new data were generated or analyzed in support of this research.
References
- 1. Dumitru R, Gama V, Fagan BM, et al. Human embryonic stem cells have constitutively active Bax at the Golgi and are primed to undergo rapid apoptosis. Mol Cell 2012;46:573-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grandela C, Pera MF, Grimmond SM, et al. p53 is required for etoposide-induced apoptosis of human embryonic stem cells. Stem Cell Res 2007;1:116-128. [DOI] [PubMed] [Google Scholar]
- 3. Qin H, Yu T, Qing T, et al. Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J Biol Chem 2007;282:5842-5852. [DOI] [PubMed] [Google Scholar]
- 4. Setoguchi K, TeSlaa T, Koehler CM, et al. P53 regulates rapid apoptosis in human pluripotent stem cells. J Mol Biol 2016;428:1465-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sokolov MV, Neumann RD.. Human embryonic stem cell responses to ionizing radiation exposures: current state of knowledge and future challenges. Stem Cells Int 2012;2012:579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu JC, Guan X, Ryan JA, et al. High mitochondrial priming sensitizes hESCs to DNA-damage-induced apoptosis. Cell Stem Cell 2013;13:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Willis SN, Adams JM.. Life in the balance: how BH3-only proteins induce apoptosis. Curr Opin Cell Biol 2005;17:617-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kale J, Osterlund EJ, Andrews DW.. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ 2018;25:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Happo L, Strasser A, Cory S.. BH3-only proteins in apoptosis at a glance. J Cell Sci 2012;125(Pt 5):1081-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Topham C, Tighe A, Ly P, et al. MYC is a major determinant of mitotic cell fate. Cancer Cell 2015;28:129-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Villunger A, Michalak EM, Coultas L, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003;302:1036-1038. [DOI] [PubMed] [Google Scholar]
- 12. Ren D, Tu HC, Kim H, et al. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science 2010;330:1390-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Madden DT, Davila-Kruger D, Melov S, et al. Human embryonic stem cells express elevated levels of multiple pro-apoptotic BCL-2 family members. PLoS One 2011;6:e28530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ploner C, Kofler R, Villunger A.. Noxa: at the tip of the balance between life and death. Oncogene 2008;27 Suppl 1:S84-S92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shibue T, Takeda K, Oda E, et al. Integral role of Noxa in p53-mediated apoptotic response. Genes Dev 2003;17:2233-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J, Hirst AJ, Duan F, et al. Anti-apoptotic mutations desensitize human pluripotent stem cells to mitotic stress and enable aneuploid cell survival. Stem Cell Reports 2019;12:557-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ihry RJ, Salick MR, Ho DJ, et al. Genome-scale CRISPR screens identify human pluripotency-specific genes. Cell Rep 2019;27:616-630.e6. [DOI] [PubMed] [Google Scholar]
- 18. Gupta S, Giricz Z, Natoni A, et al. NOXA contributes to the sensitivity of PERK-deficient cells to ER stress. FEBS Lett 2012;586:4023-4030. [DOI] [PubMed] [Google Scholar]
- 19. Iurlaro R, Muñoz-Pinedo C.. Cell death induced by endoplasmic reticulum stress. FEBS J 2016;283:2640-2652. [DOI] [PubMed] [Google Scholar]
- 20. Avery S, Hirst AJ, Baker D, et al. BCL-XL mediates the strong selective advantage of a 20q11.21 amplification commonly found in human embryonic stem cell cultures. Stem Cell Reports 2013;1:379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nguyen HT, Geens M, Mertzanidou A, et al. Gain of 20q11.21 in human embryonic stem cells improves cell survival by increased expression of Bcl-xL. Mol Hum Reprod 2014;20:168-177. [DOI] [PubMed] [Google Scholar]
- 22. Chen L, Willis SN, Wei A, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 2005;17:393-403. [DOI] [PubMed] [Google Scholar]
- 23. Nakajima W, Sharma K, Lee JY, et al. DNA damaging agent-induced apoptosis is regulated by MCL-1 phosphorylation and degradation mediated by the Noxa/MCL-1/CDK2 complex. Oncotarget 2016;7:36353-36365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Senichkin VV, Streletskaia AY, Gorbunova AS, et al. Saga of Mcl-1: regulation from transcription to degradation. Cell Death Differ 2020;27:405-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Du H, Wolf J, Schafer B, et al. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J Biol Chem 2011;286:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kotschy A, Szlavik Z, Murray J, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 2016;538:477-482. [DOI] [PubMed] [Google Scholar]
- 27. Tron AE, Belmonte MA, Adam A, et al. Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia. Nat Commun 2018;9:5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim JY, Ahn HJ, Ryu JH, et al. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1alpha. J Exp Med 2004;199:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun Y, Leaman DW.. Involvement of Noxa in cellular apoptotic responses to interferon, double-stranded RNA, and virus infection. J Biol Chem 2005;280:15561-15568. [DOI] [PubMed] [Google Scholar]
- 30. Huskey NE, Guo T, Evason KJ, et al. CDK1 inhibition targets the p53-NOXA-MCL1 axis, selectively kills embryonic stem cells, and prevents teratoma formation. Stem Cell Reports 2015;4:374-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rasmussen ML, Kline LA, Park KP, et al. A non-apoptotic function of MCL-1 in promoting pluripotency and modulating mitochondrial dynamics in stem cells. Stem Cell Reports 2018;10:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lowman XH, McDonnell MA, Kosloske A, et al. The proapoptotic function of Noxa in human leukemia cells is regulated by the kinase Cdk5 and by glucose. Mol Cell 2010;40:823-833. [DOI] [PubMed] [Google Scholar]
- 33. Rath S, Das L, Kokate SB, et al. Regulation of Noxa-mediated apoptosis in Helicobacter pylori-infected gastric epithelial cells. FASEB J 2015;29:796-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gomez-Bougie P, Ménoret E, Juin P, et al. Noxa controls Mule-dependent Mcl-1 ubiquitination through the regulation of the Mcl-1/USP9X interaction. Biochem Biophys Res Commun 2011;413:460-464. [DOI] [PubMed] [Google Scholar]
- 35. Yu L, Ji KY, Zhang J, et al. Core pluripotency factors promote glycolysis of human embryonic stem cells by activating GLUT1 enhancer. Protein Cell 2019;10:668-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were generated or analyzed in support of this research.