Abstract
Surface-based morphometry (SBM) is extremely useful for estimating the indices of cortical morphology, such as volume, thickness, area, and gyrification, whereas voxel-based morphometry (VBM) is a typical method of gray matter (GM) volumetry that includes cortex measurement. In cases where SBM is used to estimate cortical morphology, it remains controversial as to whether VBM should be used in addition to estimate GM volume. Therefore, this review has two main goals. First, we summarize the differences between the two methods regarding preprocessing, statistical analysis, and reliability. Second, we review studies that estimate cortical morphological changes using VBM and/or SBM and discuss whether using VBM in conjunction with SBM produces additional values. We found cases in which detection of morphological change in either VBM or SBM was superior, and others that showed equivalent performance between the two methods. Therefore, we concluded that using VBM and SBM together can help researchers and clinicians obtain a better understanding of normal neurobiological processes of the brain. Moreover, the use of both methods may improve the accuracy of the detection of morphological changes when comparing the data of patients and controls.
In addition, we introduce two other recent methods as future directions for estimating cortical morphological changes: a multi-modal parcellation method using structural and functional images, and a synthetic segmentation method using multi-contrast images (such as T1- and proton density-weighted images).
Keywords: confounding covariate, cortex volume, smoothing, surface-based morphometry, voxel-based morphometry
Introduction
Voxel-based morphometry1 (VBM) and surface-based morphometry2,3 (SBM) are effective methods for estimating the indices of cortical morphology, mainly by using T1-weighted MRI. Both methods were introduced almost simultaneously at the end of the last century. While VBM primarily serves to estimate gray matter (GM) volume, including that of the cortex, SBM estimates a variety of features, such as cortex volume, thickness, and area, as well as gyrification.4 These indices are obtained locally and can help differentiate between groups of controls and patients with various conditions such as Alzheimer’s disease.5 These automated analytical methods have significant benefits compared with the method of manual tracing,6 which is much more time-consuming,7 is subject to operator bias, and requires a priori anatomical constraints.
In cases where SBM is used to estimate cortical morphology, it remains controversial as to whether VBM should also be used to estimate GM volume.8–20 Since the two methods were introduced nearly simultaneously, the tools used in both methods have been updated continuously. Therefore, it is essential to consider which tools, such as software versions, have been employed in the existing studies while analyzing advantages and disadvantages of both methods. For example, the tools for spatial normalization,21–23 distortion correction,24,25 and signal intensity non-uniformity correction26–28 have been greatly improved upon for obtaining more accurate results.
Based on reviews of research reports for VBM and SBM from the past 20 years, the authors of this review conclude that both VBM and SBM should be used. The aim of this paper was to discuss the necessity of using VBM in conjunction with SBM, based on the review of various studies regarding the reliability and differences in the analysis of VBM and SBM results.
In the following section, overview of both VBM and SBM processing is described along with their points of difference. Next, factors influencing their reliability are listed, followed by the introduction of several applications for healthy subjects, such as aging. Next, for various types of diseases, SBM-superior examples, cases that are equivalent for both methods, and VBM-superior examples are introduced and reviewed. Then finally, as a future direction, multi-modal parcellation in SBM and synthetic segmentation in VBM are introduced, along with several examples.
Overview of VBM and SBM Processing
VBM and SBM share common processes, which begin with preprocessing and conclude with statistical analysis. An overview of data processing steps for both is described below. In addition, their difference points, factors influencing their reliability, and other details are listed.
Overview of VBM processing
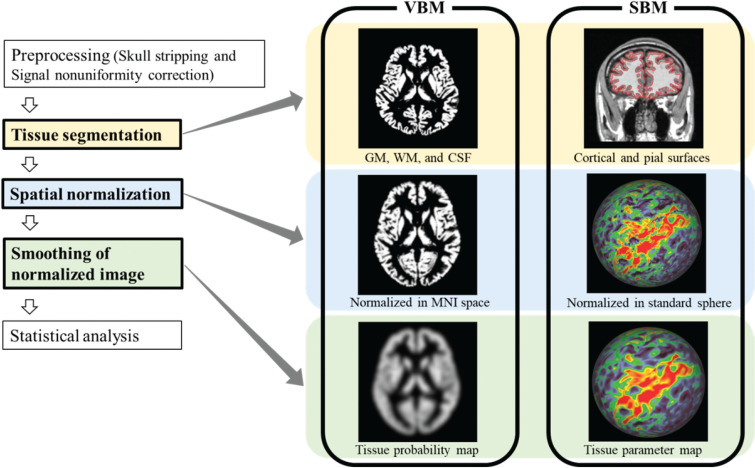
The typical processes involved in VBM include skull stripping29,30 and signal nonuniformity correction27,28 as preprocessing steps, with tissue segmentation, spatial normalization, smoothing of the normalized image, and statistical analysis as the steps of the main process. An overview of the data processing steps for both methods is shown in Fig. 1, and popular software packages used are summarized in Table 1. Two popular software packages: the statistical parametric mapping software (SPM) (The Wellcome Centre for Human Neuroimaging, London, UK) and the FMRIB software library (FSL) (FMRIB, Oxford, UK), are used extensively for VBM. In the following section, each analysis step is described along with corresponding software features.
Fig. 1.
Overview of data processing steps used for VBM and SBM. This figure shows common processes in each method, which are initiated by preprocessing, and finished by statistical analysis. Major points of difference between VBM and SBM processing are highlighted with three colored boxes. SBM, surface-based morphometry; VBM, voxel-based morphometry.
Table 1.
Popular software packages in voxel- and surface-based morphometry
| Package name | Type | Spatial normalization method | Platform | URL |
|---|---|---|---|---|
| SPM | VBM | DARTEL Tool | MATLAB | https://www.fil.ion.ucl.ac.uk/spm/software/ |
| FSL | VBM | FLIRT and FNIRT | Apple, Linux, and Windows (Windows via a Virtual Machine) | https://fmrib.ox.ac.uk/fsl/ |
| FreeSurfer | SBM | Spherical map registration51 | Apple, Linux, and Windows (Windows via a Virtual Machine) | https://surfer.nmr.mgh.harvard.edu/ |
| CIVET | SBM | Robbins’s method49 | Linux and web-based platform | https://mcin.ca/technology/civet/ |
| CAT12 | SBM | Geodesic Shooting50 | MATLAB | www.neuro.uni-jena.de/cat/ |
CAT12, computational anatomy toolbox 12; FSL, FMRIB software library; SBM, surface-based morphometry; SPM, statistical parametric mapping; VBM, voxel-based morphometry.
Tissue segmentation
Both SPM and FSL segmentation have previously been summarized in a report by Tudorascu et al.31 Segmentation in SPM version 12 (SPM12) includes bias-field correction for nonuniformity and registration to a standard anatomic space.31 SPM12 segmentation provides partial volume ratios of six tissue classes: GM, white matter (WM), cerebrospinal fluid (CSF), skull, soft tissue, and air.31 This calculation is based on the registration of the subject data to a prior tissue probability map, which quantifies six partial volume probabilities modeled by a mixture of Gaussians for signal intensity at each voxel in T1-weighted images of the brain.32 That is, for example, GM volume inside each voxel was defined by GM probability and voxel size.
FSL performs similar processes, with minor differences as follows:31 the segmentation in FSL is based on the FMRIB automated segmentation tool algorithm, and consists of bias correction by the N4 (improved nonparametric nonuniformity normalization bias correction) algorithm, skull-stripping, and registration to prior tissue probability maps for three tissue classes (GM, WM, and CSF31).
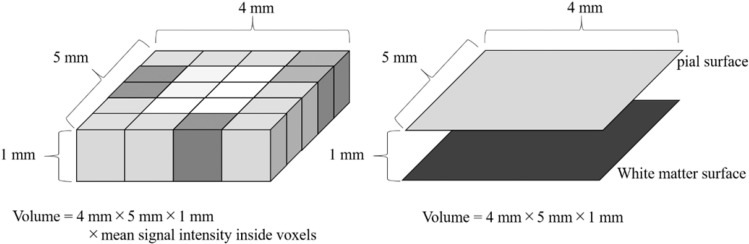
Different segmentation algorithms are used for SPM and FSL segmentation, but the feature that they share is the use of tissue probability maps (segmented tissue images) for measuring tissue volumes. Subsequently, the GM volume inside a voxel of a segmented tissue image is calculated on the segmented GM image using the following equation: signal intensity inside voxel × volume of voxel (Fig. 2).
Fig. 2.
Difference in the measurement of gray matter volume. In VBM (left), volume was calculated from voxel size and signal intensity on a segmented gray matter image used as a tissue probability map. In SBM (right), volume was calculated from area size and gray matter thickness. Gray matter thickness is defined as the distance between the pial and white matter surfaces. SBM, surface-based morphometry; VBM, voxel-based morphometry.
Spatial normalization
3D spatial normalization from the native space to the Montreal Neurological Institute (MNI) space33 is performed on the segmented GM image. To accomplish this task, it is very important to use the most accurate tool of spatial normalization in VBM. In the early phase of VBM application, the registration algorithm for spatial normalization was identical to the one used in the segmentation, which was less flexible. Therefore, there have been discussions on the importance of the algorithm used for spatial normalization by Bookstein34 and Ashburner.35 Recently, diffeomorphic anatomical registration through exponentiated lie algebra (DARTEL) has also attracted attention as a group-wise registration algorithm for building a template for spatial normalization.21 Previous reports have shown that compared with SPM5 unified segmentation, DARTEL significantly improved the accuracy of analysis results for Alzheimer’s disease,36 age-related GM volume change,37 and inter-scanner variances.22 FSL also offers registration tools, which include the FMRIB’s linear image registration tool38,39 in the early versions, and FMRIB’s nonlinear image registration tool40,41 in the updated ones.
Smoothing of normalized image
The smoothing process for segmented GM images is performed before statistical analysis. Smoothed images are used for statistical analysis in a group comparison and correlation analysis. The repeatability of the measurements of the intracranial tissue volumes in VBM is improved by spatial smoothing.42 Spatial smoothing is performed by the Gaussian smoothing kernel43 as it reduces interindividual variation. The false-positive rate, that is the incorrect significant difference of local GM volume between control and patients, using 12- mm full width at half maximum (FWHM), was reported to be lower than that using 6- mm FWHM.43 By contrast, narrow filters (4-mm FWHM) increased the sensitivity of the correlation procedure, especially when small brain structures were analyzed using VBM.7 Although previous reports have discussed the appropriate filter size (FWHM), this issue remains controversial. We suggest that the appropriate filter size is affected by the size of volume change target (i.e., atrophy), subject number, registration accuracy (i.e., spatial normalization accuracy), and target indices (i.e., volume, area, and thickness). For example, the use of high-dimensional warping tools for registration, such as DARTEL, can reduce the necessity for smoothing to blur out warping errors.43–45
Statistical analysis
Probability density value for the GM tissue is obtained for each voxel in a segmented GM image. Then, statistical analysis is performed voxel by voxel, that is, a comparison between two groups with spatially smoothed GM images in VBM. Conversely, a non-smoothed GM image is used for brain volumetry with the atlas-based method, which can measure the absolute volume of each anatomical region.46
In statistical analysis for VBM, it is important to consider confounding factors: age, sex, handedness, disease duration, and medication use, as well as total intracranial volume (TIV). These factors are the parameters set as confounding covariates in the statistical analysis. For example, patient age is set as a confounding covariate in group comparisons (i.e., control vs. disease group) because cortex volume correlates with age. By contrast, patient age would not serve as a confounding covariate if only age-matched groups were analyzed in the statistical analysis. Therefore, the number of the types of confounding factors can be decreased by using factor-matched groups.
Overview of SBM processing
Similar to VBM, common processes involved in SBM include skull stripping, signal nonuniformity correction as the preprocessing step, and tissue segmentation, spatial normalization, smoothing of the normalized image, and statistical analysis as the steps of the main process. Overview of data processing steps is shown in Fig. 1, and popular software packages are summarized in Table 1. Three popular software packages, the FreeSurfer (Martinos Center for Biomedical Imaging, Boston, MA, USA), CIVET (McConnell Brain Imaging Center, Montreal, Canada), and Computational anatomy toolbox 12 (CAT12) (Structural Brain Mapping Group, Jena, Germany), are used extensively for SBM. Different approaches in cortex segmentation are used in these methods as follows: FreeSurfer uses model-based deformation approach, CIVET uses the skeleton-based reconstruction approach, and CAT12 uses a projection-based thickness approach.47–50 Each step of SBM is described in the following section with examples of FreeSurfer as the most popular package used.
Tissue segmentation
Two surfaces—the WM surface and pial surface—are used to create the segmented tissue images in SBM.2 First, the two borders are determined based on voxel intensity information. As shown in Fig.1, the first border is inside for dividing WM and GM, while the second is outside for dividing GM and CSF. That is, the planar orientation that minimizes within-plane intensity variance is computed for each of these border voxels. Then, WM is divided between the two cortical hemispheres, and surface tessellation is performed for each hemisphere. The tessellation process at each border voxel is a determination of a square consisting of two triangles to separate WM and GM. After the smoothing of these tessellated WM surfaces, the pial surfaces are defined secondarily by expanding the WM surfaces. The vertices of the surfaces are used to estimate indexes of cortical morphology, such as volume, thickness, area, and gyrification. The cortical thickness is defined at each WM vertex as the shortest distance to the pial surface. The cortical area at each vertex can be computed as the average area of all the triangles which include the vertex. Then, the GM volume is defined as the area times the thickness.51,52 The whole brain gyrification index is defined as the ratio of the total pial surface over the total perimeter of the brain. The latter is obtained by shape-smoothing (morphological closing) of the former. The local gyrification index measures the degree of cortical folding at specific points of the cortical surface.4,53,54
Spatial normalization
2D spatial normalization from the native space to a standard surface space was performed on the 2D cortical sheet.3,55 Identifying corresponding points on different cortical surfaces requires the establishment of a uniform surface-based coordinate system. The patient’s WM surface is “inflated” to the shape of a sphere, and the geometric quantification of the WM surface is transferred to the sphere.51 The registration is performed in a spherical space, meaning that the atlas exists as a sphere by the seventh-order subdivision of icosahedron. The folding pattern quantification technique is used to drive a nonlinear, surface-based, inter-subject registration procedure that aligns the cortical folding patterns of each subject to a standard surface space, and this approach is similar to performing a volume-based registration to the MNI space.51
Smoothing of normalized image
The smoothing process is performed before statistical analysis in SBM as it is in VBM. In SBM, spatial smoothing is performed in parameter maps of factors, such as cortical thickness, on the spherical surface by the Gaussian smoothing kernel51 for reduction in inter-individual variation. Larger kernel size yields high repeatability in cortical thickness estimation among scan and rescan, and also produces low repeatability for surface area and volume change.56 The main cause for the former is that the effect of misregistration is decreased by using smoothed image. The latter is mainly based on the fact that small region of surface or volume change is averaged with the surrounding unchanged area by smoothing. Therefore, there exists a trade-off problem in the determination of kernel size (i.e., degree of smoothing), and we should choose the size carefully.
Statistical analysis
Various indices, such as cortical thickness, area, and volume, are obtained at each vertex. Then, statistical analysis is performed vertex by vertex, that is, comparison between two groups with spatially smoothed index images. Conversely, a non-smoothed index image was used for the estimation of morphology with the atlas-based method. The atlas-based method can demonstrate the absolute index values.57
As in the statistical analysis for VBM, SBM considers indispensable factors such as age, sex, handedness, disease duration, and medication use. In the statistical analysis for cortical thickness in SBM, a prominent controversial issue exists such that there is disagreement between two types of studies: studies wherein TIV was used as the confounding covariate,5,11,58,59 and those wherein TIV was not used.8,10,12,14,15,20,60–63 If cortical thickness correlates with TIV, then TIV should be used as the confounding covariate in the statistical analysis. It remains unknown whether results show a correlation between TIV and cortical thickness. The mean female head size is smaller than that of male individuals, but a part of the region within the cortex has a thicker structure in female than in male samples.64 A report by Winkler showed that brain volume is also highly correlated with the surface area of the cortex (R2 = 0.856, P = 1.8 × 10−142) but is not correlated with the average cortical thickness (R2 = 0.046, P = 0.153).65 Therefore, we suggest that TIV should not be used as a confounding covariate in the statistical analysis of cortical thickness; instead, mean cortical thickness of the global brain area may be used for this purpose. Appropriate covariates should be identified in future studies.
Difference points between VBM and SBM processing
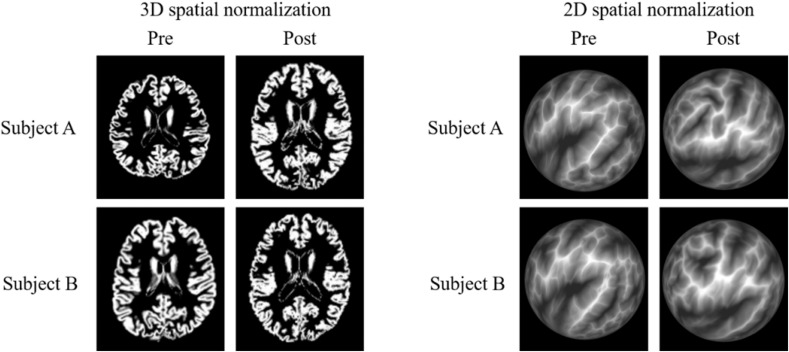
In this paragraph, we describe important points of difference between VBM and SBM. In VBM, the GM volume inside a voxel of a segmented tissue image is calculated on the segmented GM image using the following equation: signal intensity inside voxel × volume of voxel. However, the GM volume in SBM is calculated from GM thickness and area (Fig. 2). Therefore, GM volume in VBM is not equal in meaning to GM volume in SBM. Moreover, previous reports have revealed a difference in accuracy between VBM and SBM. The 3D spatial normalization process was used for VBM, and the 2D spatial normalization process was used for SBM (example images are shown in Fig. 3). A report by Ghosh demonstrated more accurate registration in SBM compared with VBM.66 In contrast, a study by Klein showed no significant difference in registration accuracy between VBM and SBM.67 These contradictory study results from Ghosh and Klein were addressed in Ghosh’s report, noting that the topographic properties used by FreeSurfer (e.g., curvature) may provide better features for matching findings across participants in this age range (i.e., children) than those provided by the intensity-derived properties used by volume-registration algorithms.66 We agree with this discussion point, which is further supported in a report by Wilke,68 showing that the accuracy of spatial normalization with VBM in children is lower than that in older individuals. In addition, Wilke’s report68 suggested that a pediatric template should be used for spatial normalization in VBM for this patient population. No previous report has shown that the spatial normalization performance in VBM is superior to that of SBM. Finally, FWHM in the smoothing process with VBM is smaller than that with SBM (VBM: 4 mm7 to 12 mm12, SBM: 10 mm8 to 25 mm15), which is summarized in Table 2.
Fig. 3.
Difference in spatial normalization. In VBM (figures on the left), 3D spatial normalization was performed on the segmented gray matter image. Signal intensity on the segmented gray matter image represents tissue probability. In SBM (figures on the right), 2D spatial normalization was performed on the 2D cortical sheet that was shown on a sphere surface. The brain sulcal patterns were inflated to the sphere surface space in SBM, and the depths of the sulcus and gyrus were represented by the black and white colors, respectively. SBM, surface-based morphometry; VBM, voxel-based morphometry.
Table 2.
Summary of comparison studies of voxel- and surface-based morphometry
| Paper | Year | Patient | Superior | Software (upper: VBM and lower: SBM) | Version | Spat. Norm. | Smoothing | Significant threshold |
|---|---|---|---|---|---|---|---|---|
| Allan8 | 2016 | Tinnitus | SBM | SPM | 8 | DARTEL | FWHM = 10 mm | FWE, P < 0.05 |
| FreeSurfer | 5.3 | FWHM = 10 mm | Corrected, P < 0.05 | |||||
| Juurmaa9 | 2016 | Methcathinone abusers | SBM | FSL | 4.1 | FLIRT | Sigma = 3 mm | FWE, P < 0.05 |
| FreeSurfer | 5.1 | Not specified | Corrected, P < 0.05 | |||||
| Tessitore10 | 2016 | Parkinson’s disease | SBM | SPM | 8 | DARTEL | FWHM = 8 mm | FWE, P < 0.05 |
| FreeSurfer | 4.5 | FWHM = 10 mm | FDR, P < 0.05 | |||||
| Pereira11 | 2012 | Parkinson’s disease | SBM | SPM | 8 | DARTEL | FWHM = 12 mm | FWE, P < 0.05 |
| FreeSurfer | 4.3.1 | FWHM = 15 mm | Corrected, P < 0.05 | |||||
| Hyde12 | 2010 | Autism | SBM | CIVET | Not specified | Not specified | FWHM = 12 mm | FDR, P < 0.05 |
| CIVET | Not specified | FWHM = 20 mm | FDR, P < 0.05 | |||||
| Bär13 | 2015 | Anorexia nervosa | Equivalent | SPM | 8 | DARTEL | FWHM = 8 mm | FWE, P < 0.05 |
| FreeSurfer | 5.3 | FWHM = 10 mm | Corrected, P < 0.05 | |||||
| Baima14 | 2020 | Obstructive sleep apnea syndrome | Equivalent | SPM | 12 | DARTEL | FWHM = 8 mm | Uncrrected, P < 0.001 |
| SPM + CAT | 12 | FWHM = 15 mm | Uncrrected, P < 0.001 | |||||
| Klauser15 | 2015 | ARMS | Equivalent | SPM | 8 | DARTEL | FWHM = 8 mm | Uncrrected, P < 0.001 |
| FreeSurfer | 5.1 | FWHM = 25 mm | FDR, P < 0.05 | |||||
| Grieve16 | 2013 | Major depressive disorder | VBM | SPM | 8 | DARTEL | FWHM = 8 mm | FDR, P < 0.05 |
| FreeSurfer | 4.3 | Not specified | FDR, P < 0.05 | |||||
| Voets17 | 2008 | Schizophrenia | VBM | FSL | Not specified | Not specified | FWHM = 8 mm | Corrected, P < 0.05 |
| FreeSurfer | Not specified | FWHM = 10 mm | FDR, P < 0.05 | |||||
| Palaniyappan18 | 2012 | Schizophrenia | VBM | SPM | 8 | DARTEL | FWHM = 8 mm | FWE, P < 0.01 |
| FreeSurfer | 4.5 | Not specified | Corrected, P < 0.0125 |
DARTEL, diffeomorphic anatomical registration through exponentiated lie algebra; FDR, false-discovery rate; FSL, FMRIB software library; FWE, family-wise error; SBM, surface-based morphometry; SPM, statistical parametric mapping.
Factors Influencing the Measurement Reliability
The reliability of measurements is a very important factor because low reliability obscures changes in brain volume and cortical morphology. Several factors can influence the reliability of measurements. In addition to analysis software,31,69,70 software parameters,71 and quality of the analyzed image24,25,72,73; three major factors are described in the following section: effects of (1) structure and volume, (2) imaging parameters, and (3) statistical analysis parameters.
Effect of structure and volume
In brain morphometry, the goal is to detect small changes. The required level of reliability has been found to change based on the amount (or change rate) of the brain volume change that must be detected. We describe this amount (or change rate) as follows: over time, the GM volume decreases curvilinearly, showing an average volume loss of 0.7 mL/year, whereas the WM volume remains constant during five decades in the normal aging process, as reported in individuals aged 21–70 years.74 The GM volume was found to decrease with age at a rate of 2.37 cm3/year (−0.18%/year) in normal aging (individuals aged 58–95 years).75 Cortical thickness is very thin (overall average of approximately 2.5 mm) for the typical voxel size of an analyzed image, and regional variations in cortical thickness can be quite large (1–4.5 mm).52 Most of the cortical mantle shows thinning rates of ≥ 0.01 mm/decade in normal aging (individuals aged 18–93 years), and the greatest rate (> 0.07 mm/decade) was found in the primary motor cortex.76
Regarding VBM, a report by Jovicich77 showed that: (i) reproducibility errors (= 100 × standard deviation/mean volume) across sessions using the same scanner were < 4.3% in the older group and < 2.3% in the younger group; and (ii) smaller structures (pallidum, amygdala, and inferior lateral ventricles) were associated with higher reproducibility errors (approximately < 10%). Moreover, a report by Ewers78 described repeatability with voxel-wise analysis, and lower repeatability was found in the base of the skull and parietal lobe.
Regarding SBM, a report by Wonderlick79 showed that surface maps of cortical thickness reliability reveal high local intraclass correlation coefficient (ICC) values across most of the cortex, and the areas of relatively low reliability include the entorhinal, medial orbitofrontal, lingual, and right-rostral middle-frontal cortex. Moreover, a report by Han80 also showed that the thickness measurement variability is nonuniform across the cortex. The most variable area is the region surrounding the precentral gyrus. Other reports have shown that the boundaries of the temporal pole and occipital lobe were not defined precisely and that variation in these regions among individuals is often observed.81
Effect of imaging parameters
In VBM, the report by Jovicich77 showed that the choice of imaging sequence (magnetization prepared rapid gradient echo [MPRAGE] or multiecho fast low angle shot [FLASH]) influenced repeatability. There are inconsistent reports regarding static magnetic fields; repeatability in a low-field MRI scanner (0.4 tesla) was similar to that in a 1.5-tesla scanner,82 and bias related to changes in field strength was found in another study.77
In SBM, the report by Wonderlick79 showed that (i) parallel imaging acceleration decreases the ICCs of cortical thickness in a part of the structure (i.e., parietal and cingulate); and (ii) in general, multiecho MR image acquisition increases the ICCs of cortical thickness and volume; and (iii) voxel size has no obvious effect on cortical measurement reliability but produces bias-related effects, such as higher thickness in isotropic voxels compared with anisotropic voxels. Moreover, the report by Han80 also revealed the following three findings on SBM: first, the measurement variability (difference in the average absolute thickness) is < 0.12 mm for the bulk of the cortex when comparing thickness measurements within the same scanner platform; further, comparing these measurements across platforms or field strengths slightly increases measurement variability, but it is still < 0.15 mm for most of the cortex when the platform—but not field strength—differs, and is < 0.2 mm when field strength differs. Second, thickness reliability is poorer when using the non-multiecho FLASH sequence versus MPRAGE. Third, a scanner upgrade improves the thickness measurement reliability. A report by Iscan showed that cortical thickness is more susceptible to scanner differences than cortical surface area and volume.81
In both VBM and SBM processing, signal intensity on MRI is used for tissue segmentation. The signal intensity is affected by imaging parameters, such as field strength, TR, and TE. In addition, MR images show slight differences between initial scans and re-scans even if they are obtained using the same imaging parameters; this is because of random artifacts from motion and flow. These factors decrease the reliability of VBM and SBM. However, we do not yet know the difference in reliability between VBM and SBM. We were not able to find previous reports that show comparisons between the reliability of the methods when the same imaging parameters are used. Moreover, reliability was affected by the software used in VBM and SBM. These differences in imaging parameters and the software used complicate the comparison of reliability.
Effect of statistical analysis parameters
Statistically significant differences were estimated regionally in the brain between control and patient groups in both methods. However, it was found that these statistically significant differences may have included false-positive and -negative results. The major cause of this is the statistical analysis parameters, such as cluster-forming threshold. Otherwise, preprocessing quality (distortion correction and signal intensity non-uniformity) and signal change due to abnormal tissue (iron content) affect analysis in both methods. Therefore, clinicians who use VBM and SBM should not forget the potential for false-positive and -negative results, even if the latest version of analysis software is used for cortex estimation.43,51 As one example, Silver et al.43 assessed 181 patients with mild cognitive impairment from the Alzheimer’s Disease Neuroimaging Initiative database83 who underwent VBM using the SPM software, version 5 (SPM5)1, and created a general linear model for statistical inference. The results showed that the false-positive rates were found to be well controlled (3.9%–5.6%) at a relatively high cluster-forming threshold (αc = 0.001) and that the false-positive rates ranged from 9.8% to 67.6% at a lower cluster-forming threshold (αc = 0.01, 0.05).43 Therefore, it is important to investigate significant positive results in both the VBM and SBM methods, with the hypothesis that VBM or SBM should be used for whole brain testing, and additional methods, such as manual tracing and visual evaluation, should be used to reconfirm the findings.
Review of VBM and SBM Studies on Healthy Subjects
This section discusses two main findings from the literature on VBM and SBM studies: (1) group differences among populations and (2) correlation with aging and indices of cortical morphology.
Group differences among populations
Tang et al.84 have reported on brain morphology differences among populations using both VBM and SBM. On the other hand, Wei’s group85 reported association with brain morphology and second language exposure timing in native English speakers with both methods. Based on the review of both reports, we found differences in brain morphology among populations and differences in statistically significant thresholds between both methods.
The report by Tang showed significant group differences in VBM (GM volume) and SBM (cortical thickness, cortical volume, and cortical surface area).84 This study analyzed two comparable samples (45 subjects in each group) from young Chinese and Caucasian populations, well matched for sex, handedness, and education. In VBM, they used the VBM 8 toolbox software (using DARTEL), 8-mm FWHM for image smoothing, and P < 0.001 as the statistically significant threshold without a correction for multiple comparisons. In SBM, they used FreeSurfer (version 5.3.0), 20-mm FWHM for image smoothing, and P < 0.05 as the statistically significant threshold with a false-discovery rate (FDR). They stated that the use of both VBM and SBM for studying group differences was the main strength of their study, the results of which are summarized in Table 3. These results showed that both the VBM and SBM methods were needed for the detection of brain morphological changes. In this report, different statistically significant thresholds were used for group comparisons using VBM (uncorrected, P < 0.001) and SBM (FDR, P < 0.05). It is important to note here that the decision regarding the threshold for statistical significance is a controversial issue for studies using both VBM and SBM. Based on the results of the study by Tang,84 we think that the threshold for statistical significance in VBM is lower than that of SBM, when detectability in VBM is the same as that of SBM. When the statistically significant threshold in VBM was set to the FDR at P < 0.05, the false-negative rate was increased compared with when an uncorrected P < 0.001 was used, but the false-positive rate was decreased.
Table 3.
Summary of result in Tang’s report84 with voxel- and surface-based morphometry
| Type | Index | Result | Middle temporal gyri | Gyrus rectus | Inferior temporal gyri | Parahippocampal gyri | Fusiform gyri | Middle cingulate | Para-cingulate gyri | Middle occipital gyri | Angular gyrus | Olfactory gyri |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VBM | Gray matter volume | Chinese > Caucasian | Bilateral | Bilateral | Bilateral | Bilateral | Right | - | - | - | - | Bilateral |
| SBM | Cortical volume | Chinese > Caucasian | Bilateral | Bilateral | Bilateral | Bilateral | Right | Bilateral | Bilateral | Bilateral | Left | Bilateral |
| SBM | Surface area | Chinese > Caucasian | Left | Left | - | - | - | - | - | Right | Left | - |
| SBM | Cortical thickness | Chinese > Caucasian | Bilateral | Left | Bilateral | Bilateral | Bilateral | Bilateral | Bilateral | Bilateral | - | - |
| Type | Index | Result | Orbitofrontal gyri | Precentral gyrus | Paracentral lobule | Medial prefrontal lobes | Superior frontal gyri | Middle frontal gyri | Post central gyri | Angular gyrus | Motor speech area | |
| VBM | Gray matter volume | Chinese < Caucasian | Bilateral | Bilateral | Left | Bilateral | Bilateral | Bilateral | Bilateral | Bilateral | Bilateral | |
| SBM | Cortical volume | Chinese < Caucasian | Bilateral | Right | Left | Bilateral | Bilateral | - | - | - | - | |
| SBM | Surface area | Chinese < Caucasian | Bilateral | Right | - | Bilateral | Bilateral | - | - | - | - | |
| SBM | Cortical thickness | Chinese < Caucasian | Bilateral | Bilateral | Left | - | - | Bilateral | Bilateral | Right | Bilateral | |
SBM, surface-based morphometry; VBM, voxel-based morphometry.
The statistically significant threshold in VBM and SBM is also discussed in the report by Wei.85 They suggested that VBM results with a lower threshold of uncorrected P < 0.01 still provide useful information. An important point is that VBM and SBM may provide different types of information and thus should be conducted concurrently. The causes of the difference are that the GM volume in VBM is not the same as the cortical volume in SBM and that SBM can provide several kinds of indices. That is, the results strongly depend on various factors such as used indices, smoothing kernel size, and statistically significant threshold. Therefore, it is important to understand the effects of the factors if we interpret the results of the previous studies.
Correlation with aging and indices of cortical morphology
Age and sex differences in brain morphometry were reported in some previous studies on SBM.76,86,87 A study by Zhao examined age differences in SBM measurements of cortical thickness, volume, and surface area in a well-defined sample of 8137 generally healthy participants (age, 45.17–79.37 years) from the United Kingdom Biobank.86 Novel relationships were shown of age-related cortical differences with the following individual factors: sex; cognitive functions of fluid intelligence, reaction time, and prospective memory; cigarette smoking; alcohol consumption; sleep disruption; and genetic markers of apolipoprotein E, brain-derived neurotrophic factor, catechol-O-methyltransferase, and several genome-wide association study loci for Alzheimer’s disease. The results further reveal the combined effects of cognitive functions, lifestyle behaviors, and education on age-related cortical differences.86 In the case of all measurements (cortical volume, area, and thickness), quadratic age effects were observed in the medial orbitofrontal cortex, medial temporal cortex, and cingulate cortex. In addition, quadratic age effects were observed in specific measurements in the following regions: in the lateral temporal cortex and occipital regions, on cortical volume and area; in the dorsolateral prefrontal cortex, on cortical area; in the bilateral temporal poles, on cortical thickness; and in the left-lateral occipital cortex, on cortical volume.86 Negative associations (based on fitted generalized linear regression models) between age and cortical morphology were found across most of the cortical areas, and most pronouncedly, in the prefrontal cortex and lateral temporal cortex in the case of all the measurements.86 Studies of VBM also showed that negative associations between age and GM volume were found across most of the cortical areas and, most pronouncedly, in the lateral temporal cortex.88,89 In addition, the SBM study by Zhao and aforementioned VBM studies showed that the slope of the regression line for the correlation between GM volume and age in male subjects was significantly steeper than that in female subjects.86,88,89
Some previous reports revealed a larger number of clusters of a significant group difference in GM volume on using VBM compared with on using SBM.90–92 Moreover, the report by Winkler showed that cortical surface area is not correlated with cortical thickness (R2 = 0.0003), GM volume is very weakly correlated with cortical thickness (R2 = 0.1815), and GM volume is strongly correlated with surface area (R2 = 0.7881).65 Compared with VBM, SBM can provide more detailed information on brain morphometry—specifically, SBM can provide indices such as volume, thickness, area, and gyrification, whereas VBM can only provide volume measurements. Based on a review of all of the results from these studies on healthy subjects, we suggest that using both VBM (GM volume) and SBM (cortical volume, area, and thickness) would help researchers and clinicians to obtain a better understanding of the neurobiological processes of the brain.
Review of VBM and SBM Patient Studies
In this section, we discuss how VBM compares to SBM in morphological change detection between normal subjects and patients with various diseases. We review them in three classes: (1) superior detection by VBM, (2) superior detection by SBM, and (3) equivalent detection by VBM and SBM.
Superior detection by VBM
In a report by Grieve,16 102 patients with major depressive disorder (MDD) and 34 healthy controls were analyzed using VBM (SPM8) and SBM (FreeSurfer, version 4.3). The VBM analysis showed large areas of decreased GM volume distributed across the brain in MDD, with no regions of increased GM volume. In a comparison of the widespread differences in volume, the measured cortical thickness differences between the MDD and control groups were limited in SBM.16
In a report by Voets,17 25 patients with adolescent-onset schizophrenia and 25 healthy controls were analyzed by VBM (FSL) and SBM (FreeSurfer). Based on their results, nine regions of apparent reduction in GM volume were found in the patients compared with the healthy matched controls using VBM, which were not found with SBM-derived cortical thickness measurements. The study’s discussion made the following points: the measured GM volume with VBM is a mixed result, combining thickness, surface area, and folding differences; and a mixed measurement improves sensitivity relative to that in an analysis that uses each measurement separately.17 In addition, superior detection of brain morphological changes in schizophrenic patients was observed in VBM as compared with in SBM in a study by Palaniyappan.18
In a study by Meyer involving the use of SBM (FreeSurfer, version 5.3.0),19 the researchers reanalyzed the structural MRI data of 257 patients (these data were analyzed in their previous VBM study) with tinnitus and discussed the difference between the results of VBM and SBM. Their discussion expressed that, “even though the architecture of SBM makes it possible to investigate more neuroanatomical traits than just cortical volume, the results that it engenders are statistically less reliable.”19 Moreover, they suggested that SBM should be used complementary to VBM because it allows for the computation of three distinct parameters, namely, cortical volume, area, and thickness.19
In a study by Prins involving the use of SBM (FreeSurfer, version 5.3.0),20 structural MRI data were reanalyzed in 113 participants (34 patients with juvenile macular degeneration, 24 patients with age-related macular degeneration [AMD], and 55 healthy age-matched controls). Their findings suggest that the cortical changes in AMD patients are relatively subtle, and superior detection was observed in VBM compared with SBM in AMD patients. Finally, they suggested that SBM does not always detect changes in cortical features in areas where VBM detects differences in GM volume.20
Superior detection by SBM
In a study by Allan,8 55 controls and 73 patients with tinnitus were analyzed with VBM (SPM8) and SBM (FreeSurfer, version 5.3.0). The results showed that any regions found in the VBM analysis would also appear in the SBM analysis, and a larger proportion and number of significant clusters were observed in SBM compared with VBM.8 Allan noted three reasons for the difference in results between VBM and SBM. First, the nature of the measurement was different because the analytical process is different between VBM and SBM. Second, registration accuracy was different because VBM and SBM used 3D and 2D registration tools, respectively. Third, different types of family-wise error (FWE) corrections and different covariate nuisances were used in the statistical analysis. As we agree with the first and second reasons, we will focus this discussion on the third reason. Allan mentioned in the discussion that it is also likely that the cluster-based FWE correction used in FreeSurfer is more sensitive to large clusters of relatively low significance, which could explain why more clusters were found overall in the SBM analysis.8 As mentioned in a previous subsection of the current review (“Review of VBM and SBM studies on healthy subjects”), many studies used different thresholds of statistical significance between VBM and SBM in the analysis, e.g., uncorrected P < 0.001 in VBM and FDR P < 0.05 in SBM.84 Based on the discussion by Allan8 and result of the Tang study,84 we think that the threshold for significance in the statistical analysis for VBM should be set at a lower level compared with that of SBM. However, a lower statistically significant threshold generates false-positive results corresponding to group differences in brain morphological changes.
In a report by Juurmaa,9 12 methcathinone abusers and 12 controls were analyzed using VBM (FSL, version 4.1) and SBM (FreeSurfer, version 5.1). For the statistically significant threshold, the P value with a multiple-comparison correction was set to 0.05 in both methods. In SBM, clusters of significant cerebral cortical thinning were observed in a large area of the cortex in the methcathinone abusers compared with in the controls. By contrast, in VBM, significant GM loss was limited to small areas (i.e., putamen, caudate, and left-temporal lobe) in the patients compared with the controls.
In a report by Tessitore,10 30 patients with Parkinson’s disease (PD) and 24 controls were analyzed using VBM (SPM8) and SBM (FreeSurfer, version 4.5). For the statistically significant threshold, the P value with a multiple-comparison correction was set to 0.05 in both methods. In SBM, statistically significant results indicating a thicker cortex were shown in the anterior cingulate and orbitofrontal cortices in PD patients. In contrast, in VBM, no significant GM loss was seen in the patients compared with in the controls. In addition, the study’s discussion expressed that cortical thickness in SBM may be more sensitive than the findings in VBM in revealing PD-related structural changes. Pereira11 also reported the superior detection in SBM for PD and highlighted the following reasons: this difference in sensitivity between the two methods has been related to limitations of VBM in cortical GM assessments, as it merges information regarding morphology, size, and position; and consequently, the final measurements were comprised of a combination of thickness and cortical folding, therefore producing less specific results.11 In reports by Tessitore10 and Pereira,11 superior detection by SBM compared with that by VBM was shown in PD patients.
In a report by Hyde,12 superior detectability was found in SBM compared with in VBM by group comparison. They used CIVET software for cortical thickness analysis and VBM analysis.12 The study participants were 15 young adult men with autism and 13 controls. The VBM results converged with the cortical thickness findings of an increased orbital frontal gyrus and decreased pre- and post-central gyri. Moreover, the cortical thickness analysis also revealed GM differences in various other cortical areas, consistent with previous findings in autistic individuals. The researchers expressed that an estimation of cortical thickness was more sensitive in detecting subtle cortical differences in autism.12
Cases finding VBM and SBM equivalent
In a report by Bär,13 26 patients with anorexia nervosa and 26 healthy controls were analyzed using VBM (SPM8) and SBM (FreeSurfer, version 5.3.0). In both VBM and SBM, significant GM reductions were observed in the midcingulate cortex and posterior cingulate cortex in the patients with anorexia nervosa compared with the healthy controls. In addition, in the patients, significant GM reductions were observed in the supplementary motor area by VBM, whereas significant cortical thickness reductions were observed in some gyri (i.e., middle frontal, superior frontal, and superior temporal gyrus) by SBM.
In a report by Baima,14 18 patients with obstructive sleep apnea syndrome and 32 healthy controls were analyzed using VBM (SPM12) and SBM (CAT12). Significant GM reductions were observed in the superior temporal gyrus, temporal pole, cerebellum, and thalamus in the patients with obstructive sleep apnea syndrome compared with the healthy controls in VBM. By contrast, significant cortical thickness reductions were observed in the precentral gyrus in the patients by SBM.
In a report by Klauser,15 69 patients with an at-risk mental state (ARMS) for psychosis and 32 healthy controls were analyzed using VBM (SPM8) and SBM (FreeSurfer, version 5.1.0). Using VBM, a significant GM volume increase in the precentral gyrus and decrease in the frontal inferior gyrus were observed in the patients with ARMS compared with the healthy controls. In contrast, using SBM, increased cortical thickness was observed in the frontal pole of the patients with ARMS.
Based on these reviews of three classes of VBM and SBM studies, a summary on the use of VBM and SBM follows. In some diseases such as MDD, schizophrenia, and AMD, VBM outperformed SBM in the detection of cortical morphological changes, while SBM showed superior detection power over VBM in other diseases, including methcathinone abuse, PD, and autism. That is, the appropriate method appears to depend on the target disease with further reports for each disease expected. However, we should remember that even when SBM (as the first choice) detected group differences, VBM may contribute to the detection of other regions of significant difference, and vice versa. In summary, VBM and SBM can serve as complimentary methods for the detection of morphological changes in the cortex.
Future Directions for the Estimation of Cortical Morphological Changes
Based on our review, we posit that both VBM and SBM are useful for the detection of the brain morphological change. In the future, if a new method that offers the advantages of both VBM and SBM is developed, it will be the first choice for the brain morphometry. In the following section, we describe two recent methods that are candidates for becoming the preferred new method. The first method is multi-modal parcellation, in which several types of images—T1-weighted, T2-weighted, functional, and diffusion-weighted MR images—are captured using several types of MRI pulse sequences and used for the analysis of cortical morphological changes; however, the effect of spatial misregistration exists in each type of contrast image, thereby complicating the results of the analysis. In the present report, parcellation means the division of the cortex into a larger number of small areas, and segmentation means the division of tissue into GM and WM. The second method is multi-contrast segmentation using synthetic MRI data, and is called synthetic segmentation. In synthetic MRI, several types of contrast—for example T1-, T2-, and proton density-weighted MRI—are obtained for one pulse sequence. In this process of segmentation with multi-contrast images of synthetic MRI, no effect of spatial misregistration occurs between contrast images, thereby not complicating the results of the analysis.
Multi-modal parcellation
Multi-modal parcellation is a new analysis method based on the definition of ROIs with fine division, which is used for the evaluation of the brain morphological and functional changes. In comparison with conventional VBM and SBM, more than double number of ROIs are used in multi-modal parcellation. In addition, unlike geometric registration of pre-determined ROIs, multi-modal information such as T1-weighted, T2-weighted, functional, and diffusion-weighted MRI provides more reliable ROI configuration. Consequently, fine and reliable ROIs enable us to obtain new findings. For example, Glasser et al. showed 180 areas (97 new areas in addition to 83 previously reported areas) in one hemisphere with multi-modal parcellation.93 These new areas, especially the language-related area (known as Area 55b), provide new knowledge.94–96
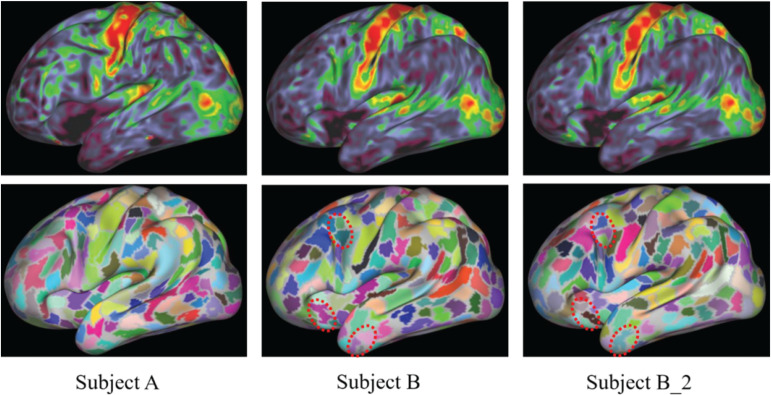
The Human Connectome Project (HCP) developed a method and released a software tool for a typical multi-modal parcellation, which uses T1-weighted, T2-weighted, functional, and diffusion-weighted MRI.97 The minimal preprocessing of the HCP pipelines is comprised of three parts: HCP Structural Pipelines, HCP Functional Pipelines, and Diffusion Preprocessing Pipelines. Based on the pipeline, the HCP atlas of multi-modal parcellation, version 1.0 (HCP_MMP1.0), was released to the public by Washington University (USA).93 Sample images of areal parcellation captured using structural and resting-state functional MRI are shown in Fig. 4. Fig. 4A and 4B shows parcellation results in different subjects, which indicate individual-level differences. Fig. 4B and 4B2 shows repeatability of analysis and reanalysis. The parcellation results changed slightly between analysis and reanalysis, and the myelin map did not change (Fig. 4B and 4B2). The slight differences were caused by randomness introduced in the algorithm. Parcellation analyses (Fig. 4) were conducted with the following methods that are the same as those used by Osada et al. and Suda et al..98,99 Each image data type in a subject was transferred to normalized space in the single-modal analysis, and several types of image data in a subject were transferred to normalized space in the multi-modal analysis. In general, this normalization scheme may yield registration error modality by modality. Therefore, accurate spatial normalization is more difficult in multi-modal than single-modal analysis. However, we suggest that the accuracy of spatial normalization in HCP pipelines is at a sufficient level in clinical settings, as previous reports have shown its usefulness in patients.94–96,98,99
Fig. 4.
Sample images of the myelin map (upper) and parcels (lower) displayed on an inflated cortical surface in the left hemisphere. Individual differences between subject A and subject B were visually found in both the myelin map and parcels. B_2 shows the results of the reanalysis of subject B. Slight differences between the results in B and B_2 were visually found in the parcels inside the circle with the dotted line but were not found in the myelin maps.
Synthetic segmentation: multi-contrast segmentation using synthetic MRI
Recently, 3D quantification involving an interleaved Look–Locker acquisition sequence along with a T2 preparation pulse (3D-QALAS) sequence was developed for the simultaneous quantification of relaxation times (longitudinal T1 and transverse T2 relaxation times) and proton density100; and this method has been applied in the brain.101,102 Using information obtained through T1, T2, and proton density maps, the synthetic MRI method using SyMRI software (SyntheticMR, Linköping, Sweden) can capture any type of contrast image, including T1-weighted, T2-weighted, proton density-weighted, and inversion recovery images.103 Therefore, VBM with synthetic MR images overcomes problems, such as additional image scan time and misalignment between different images, incurred during multi-contrast segmentation. Moreover, in image analysis with VBM and SBM, synthetic MR images have fewer drawbacks, such as signal intensity nonuniformity and low-contrast-to-noise ratio, compared with conventional MR images because synthetic images are generated by quantitative values, namely, T1, T2, and proton density maps.104
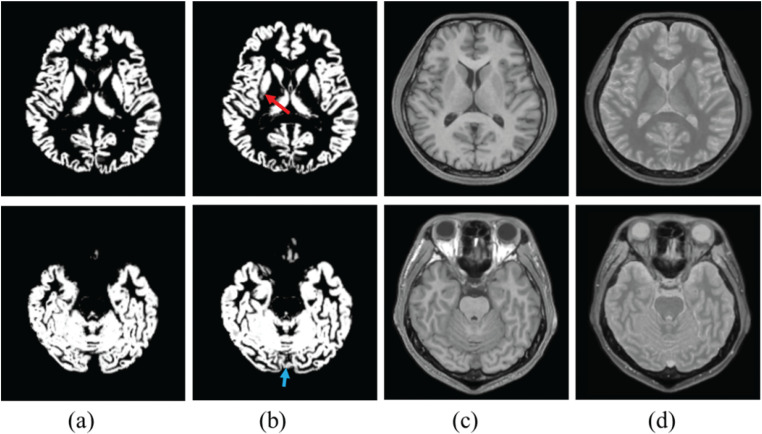
A previous study demonstrated improved accuracy of the brain extraction by using synthetic segmentation.30 We additionally note that the accuracy of GM segmentation was also improved by using the synthetic segmentation.105 Sample images are shown in Fig. 5. Sample images were processed with the following methods: based on the 3D-QALAS data, SyMRI software (version 0.45.14) was used to create images with two contrast mechanisms (T1-weighted and proton density-weighted). Multi-contrast (T1-weighted + proton density-weighted) segmentations were performed using the SPM 12 software. In the putamen and dural sinus, mis-segmentation was decreased by using synthetic segmentation (Fig. 5). It is easy for mis-segmentation to arise in the putamen and dural sinus when using the single-contrast method.106 Reasons for mis-segmentation are that signal intensity differences between anterior and posterior parts in putamen, and signal intensity is similar in the dural sinus and cortex. Proton density-weighted images have a lower signal intensity variation in the putamen as well as dissimilar signal intensities in the dural sinus and cortex. Then, multi-contrast segmentation with proton density-weighted images has lower mis-segmentation in the dural sinus and cortex, compared with single-contrast (T1-weighted) segmentation.
Fig. 5.
Representative image of segmented gray matter image in (a) synthetic (T1- and proton density-weighted) segmentation and (b) single contrast (T1-weighted) segmentation. (c) is a T1-weighted image and (d) is a proton density-weighted image. Red and blue arrows show mis-segmentation in single-contrast segmentation.
Conclusion
Based on our reviews, we suggest that using both VBM (GM volume) and SBM (cortical volume, area, and thickness) can help researchers and clinicians to obtain a better understanding of the neurobiological processes of the brain. Moreover, VBM can contribute to the detection of group differences in morphological changes in the cortex, even when SBM showed superior detection power in some diseases (i.e., methcathinone abuse, PD, and autism). In addition, VBM has superior detection abilities for the brain morphological changes in other diseases (i.e., MDD, schizophrenia, and AMD). That is, VBM and SBM can serve as complimentary methods for the detection of morphological changes in the cortex. Therefore, the use of both VBM and SBM has the potential to improve the accuracy of the detection of morphological changes in the cortex. Finally, we described two recent methods, multi-modal parcellation and synthetic segmentation, which may open new horizons for the brain morphometry. Recent reports on these methods provide new knowledge of the estimation of cortical morphology.
Funding Statement
This work was supported by JSPS KAKENHI: Grant Number JP20K08057.
Footnotes
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Ashburner J, Friston KJ. Voxel-based morphometry—the methods. Neuroimage 2000; 11:805–821. 10.1006/nimg.2000.0582 [DOI] [PubMed] [Google Scholar]
- 2.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 1999; 9:179–194. 10.1006/nimg.1998.0395 [DOI] [PubMed] [Google Scholar]
- 3.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage 1999; 9:195–207. 10.1006/nimg.1998.0396 [DOI] [PubMed] [Google Scholar]
- 4.Mills KL, Tamnes CK. Methods and considerations for longitudinal structural brain imaging analysis across development. Dev Cogn Neurosci 2014; 9:172–190. 10.1016/j.dcn.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehmann M, Crutch SJ, Ridgway GR, et al. Cortical thickness and voxel-based morphometry in posterior cortical atrophy and typical Alzheimer’s disease. Neurobiol Aging 2011; 32:1466–1476. 10.1016/j.neurobiolaging.2009.08.017 [DOI] [PubMed] [Google Scholar]
- 6.Giuliani NR, Calhoun VD, Pearlson GD, Francis A, Buchanan RW. Voxel-based morphometry versus region of interest: a comparison of two methods for analyzing gray matter differences in schizophrenia. Schizophr Res 2005; 74:135–147. 10.1016/j.schres.2004.08.019 [DOI] [PubMed] [Google Scholar]
- 7.Uchida RR, Del-Ben CM, Araujo D, et al. Correlation between voxel based morphometry and manual volumetry in magnetic resonance images of the human brain. An Acad Bras Cienc 2008; 80:149–156. 10.1590/S0001-37652008000100010 [DOI] [PubMed] [Google Scholar]
- 8.Allan TW, Besle J, Langers DR, et al. Neuroanatomical alterations in tinnitus assessed with magnetic resonance imaging. Front aging neurosci 2016; 8:221. 10.3389/fnagi.2016.00221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juurmaa J, Menke RA, Vila P, et al. Grey matter abnormalities in methcathinone abusers with a Parkinsonian syndrome. Brain Behav 2016; 6:e00539. 10.1002/brb3.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tessitore A, Santangelo G, De Micco R, et al. Cortical thickness changes in patients with Parkinson’s disease and impulse control disorders. Parkinsonism Relat Disord 2016; 24:119–125. 10.1016/j.parkreldis.2015.10.013 [DOI] [PubMed] [Google Scholar]
- 11.Pereira JB, Ibarretxe-Bilbao N, Marti MJ, et al. Assessment of cortical degeneration in patients with Parkinson’s disease by voxel-based morphometry, cortical folding, and cortical thickness. Hum Brain Mapp 2012; 33:2521–2534. 10.1002/hbm.21378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyde KL, Samson F, Evans AC, Mottron L. Neuroanatomical differences in brain areas implicated in perceptual and other core features of autism revealed by cortical thickness analysis and voxel-based morphometry. Hum Brain Mapp 2010; 31:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bär KJ, de la Cruz F, Berger S, Schultz CC, Wagner G. Structural and functional differences in the cingulate cortex relate to disease severity in anorexia nervosa. J Psychiatry Neurosci 2015; 40:269–279. 10.1503/jpn.140193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baima CB, Fim NC, Alves KF, Resende LAL, Fonseca RG, Betting LE. Analysis of patients with obstructive sleep apnea with and without pharyngeal myopathy using brain neuroimaging. Sleep (Basel) 2020; 43: zsz216. [DOI] [PubMed] [Google Scholar]
- 15.Klauser P, Zhou J, Lim JK, et al. Lack of evidence for regional brain volume or cortical thickness abnormalities in youths at clinical high risk for psychosis: Findings from the longitudinal youth at risk study. Schizophr bull 2015; 41:1285–1293. 10.1093/schbul/sbv012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grieve SM, Korgaonkar MS, Koslow SH, Gordon E, Williams LM. Widespread reductions in gray matter volume in depression. Neuroimage Clin 2013; 3:332–339. 10.1016/j.nicl.2013.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voets NL, Hough MG, Douaud G, et al. Evidence for abnormalities of cortical development in adolescent-onset schizophrenia. Neuroimage 2008; 43:665–675. 10.1016/j.neuroimage.2008.08.013 [DOI] [PubMed] [Google Scholar]
- 18.Palaniyappan L, Liddle PF. Differential effects of surface area, gyrification and cortical thickness on voxel based morphometric deficits in schizophrenia. Neuroimage 2012; 60:693–699. 10.1016/j.neuroimage.2011.12.058 [DOI] [PubMed] [Google Scholar]
- 19.Meyer M, Neff P, Liem F, et al. Differential tinnitus-related neuroplastic alterations of cortical thickness and surface area. Hear Res 2016; 342:1–12. 10.1016/j.heares.2016.08.016 [DOI] [PubMed] [Google Scholar]
- 20.Prins D, Plank T, Baseler HA, et al. Surface-based analyses of anatomical properties of the visual cortex in macular degeneration. PLoS One 2016; 11:e0146684. 10.1371/journal.pone.0146684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage 2007; 38:95–113. 10.1016/j.neuroimage.2007.07.007 [DOI] [PubMed] [Google Scholar]
- 22.Goto M, Abe O, Aoki S, et al. Diffeomorphic anatomical registration through exponentiated lie algebra provides reduced effect of scanner for cortex volumetry with atlas-based method in healthy subjects. Neuroradiology 2013; 55:869–875. 10.1007/s00234-013-1193-2 [DOI] [PubMed] [Google Scholar]
- 23.Matsuda H, Mizumura S, Nemoto K, et al. Automatic voxel-based morphometry of structural MRI by SPM8 plus diffeomorphic anatomic registration through exponentiated lie algebra improves the diagnosis of probable Alzheimer Disease. AJNR Am J Neuroradiol 2012; 33:1109–1114. 10.3174/ajnr.A2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jovicich J, Czanner S, Greve D, et al. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage 2006; 30:436–443. 10.1016/j.neuroimage.2005.09.046 [DOI] [PubMed] [Google Scholar]
- 25.Goto M, Abe O, Kabasawa H, et al. Effects of image distortion correction on voxel-based morphometry. Magn Reson Med Sci 2012; 11:27–34. 10.2463/mrms.11.27 [DOI] [PubMed] [Google Scholar]
- 26.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010; 29:1310–1320. 10.1109/TMI.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 1998; 17:87–97. 10.1109/42.668698 [DOI] [PubMed] [Google Scholar]
- 28.Goto M, Abe O, Miyati T, et al. Influence of signal intensity non-uniformity on brain volumetry using an atlas-based method. Korean J Radiol 2012; 13:391–402. 10.3348/kjr.2012.13.4.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp 2002; 17:143–155. 10.1002/hbm.10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goto M, Hagiwara A, Kato A, et al. Effect of changing the analyzed image contrast on the accuracy of intracranial volume extraction using Brain Extraction Tool 2. Radiol Phys Technol 2020; 13:76–82. 10.1007/s12194-019-00551-5 [DOI] [PubMed] [Google Scholar]
- 31.Tudorascu DL, Karim HT, Maronge JM, et al. Reproducibility and bias in healthy brain segmentation: Comparison of two popular neuroimaging platforms. Front Neurosci 2016; 10:503. 10.3389/fnins.2016.00503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner J, Friston KJ. Unified segmentation. Neuroimage 2005; 26:839–851. 10.1016/j.neuroimage.2005.02.018 [DOI] [PubMed] [Google Scholar]
- 33.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002; 15:273–289. 10.1006/nimg.2001.0978 [DOI] [PubMed] [Google Scholar]
- 34.Bookstein FL. “Voxel-based morphometry” should not be used with imperfectly registered images. Neuroimage 2001; 14:1454–1462. 10.1006/nimg.2001.0770 [DOI] [PubMed] [Google Scholar]
- 35.Ashburner J, Friston KJ. Why voxel-based morphometry should be used. Neuroimage 2001; 14:1238–1243. 10.1006/nimg.2001.0961 [DOI] [PubMed] [Google Scholar]
- 36.Cuingnet R, Gerardin E, Tessieras J, et al. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage 2011; 56:766–781. 10.1016/j.neuroimage.2010.06.013 [DOI] [PubMed] [Google Scholar]
- 37.Peelle JE, Cusack R, Henson RN. Adjusting for global effects in voxel-based morphometry: gray matter decline in normal aging. Neuroimage 2012; 60:1503–1516. 10.1016/j.neuroimage.2011.12.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal 2001; 5:143–156. 10.1016/S1361-8415(01)00036-6 [DOI] [PubMed] [Google Scholar]
- 39.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 2002; 17:825–841. 10.1006/nimg.2002.1132 [DOI] [PubMed] [Google Scholar]
- 40.Woolrich MW, Jbabdi S, Patenaude B, et al. Bayesian analysis of neuroimaging data in FSL. Neuroimage 2009; 45(Suppl):S173–S186. 10.1016/j.neuroimage.2008.10.055 [DOI] [PubMed] [Google Scholar]
- 41.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. Neuroimage 2012; 62:782–790. 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 42.Goto M, Miyati T, Abe O, et al. Repeatability of measured brain volume by atlas-based method using T1-weighted image. J Digit Imaging 2012; 25:173–178. 10.1007/s10278-011-9412-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silver M, Montana G, Nichols TE. Alzheimer’s Disease Neuroimaging I. False positives in neuroimaging genetics using voxel-based morphometry data. Neuroimage 2011; 54:992–1000. 10.1016/j.neuroimage.2010.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergouignan L, Chupin M, Czechowska Y, et al. Can voxel based morphometry, manual segmentation and automated segmentation equally detect hippocampal volume differences in acute depression?. Neuroimage 2009; 45:29–37. 10.1016/j.neuroimage.2008.11.006 [DOI] [PubMed] [Google Scholar]
- 45.McLaren DG, Kosmatka KJ, Kastman EK, Bendlin BB, Johnson SC. Rhesus macaque brain morphometry: a methodological comparison of voxel-wise approaches. Methods 2010; 50:157–165. 10.1016/j.ymeth.2009.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goto M, Abe O, Miyati T, et al. Accelerated hippocampal volume reduction in post-menopausal women: an additional study with Atlas-based method. Radiol Phys Technol 2011; 4:185–188. 10.1007/s12194-011-0120-7 [DOI] [PubMed] [Google Scholar]
- 47.Redolfi A, Manset D, Barkhof F, et al. Head-to-head comparison of two popular cortical thickness extraction algorithms: a cross-sectional and longitudinal study. PLoS One 2015; 10:e0117692. 10.1371/journal.pone.0117692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Righart R, Schmidt P, Dahnke R, et al. Volume versus surface-based cortical thickness measurements: A comparative study with healthy controls and multiple sclerosis patients. PLoS One 2017; 12:e0179590. 10.1371/journal.pone.0179590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robbins S, Evans AC, Collins DL, Whitesides S. Tuning and comparing spatial normalization methods. Med Image Anal 2004; 8:311–323. 10.1016/j.media.2004.06.009 [DOI] [PubMed] [Google Scholar]
- 50.Ashburner J, Friston KJ. Diffeomorphic registration using geodesic shooting and Gauss-Newton optimisation. Neuroimage 2011; 55:954–967. 10.1016/j.neuroimage.2010.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greve DN, Fischl B. False positive rates in surface-based anatomical analysis. Neuroimage 2018; 171:6–14. 10.1016/j.neuroimage.2017.12.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA 2000; 97:11050–11055. 10.1073/pnas.200033797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zilles K, Armstrong E, Schleicher A, Kretschmann HJ. The human pattern of gyrification in the cerebral cortex. Anat Embryol (Berl) 1988; 179:173–179. 10.1007/BF00304699 [DOI] [PubMed] [Google Scholar]
- 54.Schaer M, Cuadra MB, Tamarit L, Lazeyras F, Eliez S, Thiran JP. A surface-based approach to quantify local cortical gyrification. IEEE Trans Med Imaging 2008; 27:161–170. 10.1109/TMI.2007.903576 [DOI] [PubMed] [Google Scholar]
- 55.Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp 1999; 8:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liem F, Merillat S, Bezzola L, et al. Reliability and statistical power analysis of cortical and subcortical FreeSurfer metrics in a large sample of healthy elderly. Neuroimage 2015; 108:95–109. 10.1016/j.neuroimage.2014.12.035 [DOI] [PubMed] [Google Scholar]
- 57.Zheng F, Liu Y, Yuan Z, et al. Age-related changes in cortical and subcortical structures of healthy adult brains: A surface-based morphometry study. J Magn Reson Imaging 2019; 49:152–163. 10.1002/jmri.26037 [DOI] [PubMed] [Google Scholar]
- 58.Liu C, Li C, Gui L, et al. The pattern of brain gray matter impairments in patients with subcortical vascular dementia. J Neurol Sci 2014; 341:110–118. 10.1016/j.jns.2014.04.017 [DOI] [PubMed] [Google Scholar]
- 59.Braga AM, Fujisao EK, Verdade RC, et al. Investigation of the cingulate cortex in idiopathic generalized epilepsy. Epilepsia 2015; 56:1803–1811. 10.1111/epi.13205 [DOI] [PubMed] [Google Scholar]
- 60.Pappaianni E, Siugzdaite R, Vettori S, Venuti P, Job R, Grecucci A. Three shades of grey: detecting brain abnormalities in children with autism using source-, voxel- and surface-based morphometry. Eur J Neurosci 2018; 47:690–700. 10.1111/ejn.13704 [DOI] [PubMed] [Google Scholar]
- 61.Li T, Liu C, Lyu H, et al. Alterations of sub-cortical gray matter volume and their associations with disease duration in patients with restless legs syndrome. Front Neurol 2018; 9:1098. 10.3389/fneur.2018.01098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Madeira N, Duarte JV, Martins R, Costa GN, Macedo A, Castelo-Branco M. Morphometry and gyrification in bipolar disorder and schizophrenia: A comparative MRI study. Neuroimage Clin 2020; 26:102220. 10.1016/j.nicl.2020.102220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cerasa A, Morelli M, Augimeri A, et al. Prefrontal thickening in PD with levodopa-induced dyskinesias: new evidence from cortical thickness measurement. Parkinsonism Relat Disord 2013; 19:123–125. 10.1016/j.parkreldis.2012.06.003 [DOI] [PubMed] [Google Scholar]
- 64.Luders E, Narr KL, Thompson PM, et al. Gender effects on cortical thickness and the influence of scaling. Hum Brain Mapp 2006; 27:314–324. 10.1002/hbm.20187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winkler AM, Kochunov P, Blangero J, et al. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage 2010; 53:1135–1146. 10.1016/j.neuroimage.2009.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghosh SS, Kakunoori S, Augustinack J, et al. Evaluating the validity of volume-based and surface-based brain image registration for developmental cognitive neuroscience studies in children 4 to 11 years of age. Neuroimage 2010; 53:85–93. 10.1016/j.neuroimage.2010.05.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klein A, Ghosh SS, Avants B, et al. Evaluation of volume-based and surface-based brain image registration methods. Neuroimage 2010; 51:214–220. 10.1016/j.neuroimage.2010.01.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilke M, Schmithorst VJ, Holland SK. Assessment of spatial normalization of whole-brain magnetic resonance images in children. Hum Brain Mapp 2002; 17:48–60. 10.1002/hbm.10053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajagopalan V, Pioro EP. Disparate voxel based morphometry (VBM) results between SPM and FSL softwares in ALS patients with frontotemporal dementia: which VBM results to consider?. BMC Neurol 2015; 15:32. 10.1186/s12883-015-0274-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fellhauer I, Zollner FG, Schroder J, et al. Comparison of automated brain segmentation using a brain phantom and patients with early Alzheimer’s dementia or mild cognitive impairment. Psychiatry Res 2015; 233:299–305. 10.1016/j.pscychresns.2015.07.011 [DOI] [PubMed] [Google Scholar]
- 71.Goto M, Abe O, Aoki S, et al. Influence of parameter settings in voxel-based morphometry 8. Using DARTEL and region-of-interest on reproducibility in gray matter volumetry. Methods Inf Med 2015; 54:171–178. 10.3414/ME14-01-0049 [DOI] [PubMed] [Google Scholar]
- 72.Shuter B, Yeh IB, Graham S, Au C, Wang SC. Reproducibility of brain tissue volumes in longitudinal studies: effects of changes in signal-to-noise ratio and scanner software. Neuroimage 2008; 41:371–379. 10.1016/j.neuroimage.2008.02.003 [DOI] [PubMed] [Google Scholar]
- 73.Goto M, Abe O, Aoki S, et al. Combining segmented grey and white matter images improves voxel-based morphometry for the case of dilated lateral ventricles. Magn Reson Med Sci 2018; 17:293–300. 10.2463/mrms.mp.2017-0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pfefferbaum A, Mathalon DH, Sullivan EV, Rawles JM, Zipursky RB, Lim KO. A quantitative magnetic resonance imaging study of changes in brain morphology from infancy to late adulthood. Arch Neurol 1994; 51:874–887. 10.1001/archneur.1994.00540210046012 [DOI] [PubMed] [Google Scholar]
- 75.Smith CD, Chebrolu H, Wekstein DR, Schmitt FA, Markesbery WR. Age and gender effects on human brain anatomy: a voxel-based morphometric study in healthy elderly. Neurobiol Aging 2007; 28:1075–1087. 10.1016/j.neurobiolaging.2006.05.018 [DOI] [PubMed] [Google Scholar]
- 76.Salat DH, Buckner RL, Snyder AZ, et al. Thinning of the cerebral cortex in aging. Cereb Cortex 2004; 14:721–730. 10.1093/cercor/bhh032 [DOI] [PubMed] [Google Scholar]
- 77.Jovicich J, Czanner S, Han X, et al. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. Neuroimage 2009; 46:177–192. 10.1016/j.neuroimage.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ewers M, Teipel SJ, Dietrich O, et al. Multicenter assessment of reliability of cranial MRI. Neurobiol Aging 2006; 27:1051–1059. 10.1016/j.neurobiolaging.2005.05.032 [DOI] [PubMed] [Google Scholar]
- 79.Wonderlick JS, Ziegler DA, Hosseini-Varnamkhasti P, et al. Reliability of MRI-derived cortical and subcortical morphometric measures: effects of pulse sequence, voxel geometry, and parallel imaging. Neuroimage 2009; 44:1324–1333. 10.1016/j.neuroimage.2008.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han X, Jovicich J, Salat D, et al. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. Neuroimage 2006; 32:180–194. 10.1016/j.neuroimage.2006.02.051 [DOI] [PubMed] [Google Scholar]
- 81.Iscan Z, Jin TB, Kendrick A, et al. Test-retest reliability of freesurfer measurements within and between sites: Effects of visual approval process. Hum Brain Mapp 2015; 36:3472–3485. 10.1002/hbm.22856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goto M, Suzuki M, Mizukami S, et al. Repeatability of brain volume measurements made with the atlas-based method from T1-weighted images acquired using a 0.4 tesla low field MR scanner. Magn Reson Med Sci 2016; 15:365–370. 10.2463/mrms.mp.2015-0107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jack CR, Jr., Bernstein MA, Fox NC, et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging 2008; 27:685–691. 10.1002/jmri.21049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang Y, Zhao L, Lou Y, et al. Brain structure differences between Chinese and Caucasian cohorts: A comprehensive morphometry study. Hum Brain Mapp 2018; 39:2147–2155. 10.1002/hbm.23994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wei M, Joshi AA, Zhang M, et al. How age of acquisition influences brain architecture in bilinguals. J Neurolinguistics 2015; 36:35–55. 10.1016/j.jneuroling.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao L, Matloff W, Ning K, Kim H, Dinov ID, Toga AW. Age-related differences in brain morphology and the modifiers in middle-aged and older adults. Cereb Cortex 2019; 29:4169–4193. 10.1093/cercor/bhy300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lv B, Li J, He H, et al. Gender consistency and difference in healthy adults revealed by cortical thickness. Neuroimage 2010; 53:373–382. 10.1016/j.neuroimage.2010.05.020 [DOI] [PubMed] [Google Scholar]
- 88.Sato K, Taki Y, Fukuda H, Kawashima R. Neuroanatomical database of normal Japanese brains. Neural Netw 2003; 16:1301–1310. 10.1016/j.neunet.2003.06.004 [DOI] [PubMed] [Google Scholar]
- 89.Farokhian F, Yang C, Beheshti I, Matsuda H, Wu S. Age-related gray and white matter changes in normal adult brains. Aging Dis 2017; 8:899–909. 10.14336/AD.2017.0502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang J, Zhang H, Li J, et al. Adaptive modulation of adult brain gray and white matter to high altitude: structural MRI studies. PLoS One 2013; 8:e68621. 10.1371/journal.pone.0068621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bailey JA, Zatorre RJ, Penhune VB. Early musical training is linked to gray matter structure in the ventral premotor cortex and auditory-motor rhythm synchronization performance. J Cogn Neurosci 2014; 26:755–767. 10.1162/jocn_a_00527 [DOI] [PubMed] [Google Scholar]
- 92.Thayer RE, Hagerty SL, Sabbineni A, Claus ED, Hutchison KE, Weiland BJ. Negative and interactive effects of sex, aging, and alcohol abuse on gray matter morphometry. Hum Brain Mapp 2016; 37:2276–2292. 10.1002/hbm.23172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Glasser MF, Coalson TS, Robinson EC, et al. A multi-modal parcellation of human cerebral cortex. Nature 2016; 536:171–178. 10.1038/nature18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang EF, Kurteff G, Andrews JP, et al. Pure apraxia of speech after resection based in the posterior middle frontal gyrus. Neurosurgery 2020; 87:E383–E389. 10.1093/neuros/nyaa002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Milton CK, Dhanaraj V, Young IM, et al. Parcellation-based anatomic model of the semantic network. Brain Behav 2021; 11:e02065. 10.1002/brb3.2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hazem SR, Awan M, Lavrador JP, et al. Middle frontal gyrus and area 55b: Perioperative mapping and language outcomes. Front Neurol 2021; 12:646075. 10.3389/fneur.2021.646075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Glasser MF, Smith SM, Marcus DS, et al. The human connectome project’s neuroimaging approach. Nat Neurosci 2016; 19:1175–1187. 10.1038/nn.4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Osada T, Ohta S, Ogawa A, et al. An essential role of the intraparietal sulcus in response inhibition predicted by parcellation-based network. J Neurosci 2019; 39:2509–2521. 10.1523/JNEUROSCI.2244-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Suda A, Osada T, Ogawa A, et al. Functional organization for response inhibition in the right inferior frontal cortex of individual human brains. Cereb Cortex 2020; 30:6325–6335. 10.1093/cercor/bhaa188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kvernby S, Warntjes M, Engvall J, Carlhall CJ, Ebbers T. Clinical feasibility of 3D-QALAS - Single breath-hold 3D myocardial T1- and T2-mapping. Magn Reson Imaging 2017; 38:13–20. 10.1016/j.mri.2016.12.014 [DOI] [PubMed] [Google Scholar]
- 101.Fujita S, Hagiwara A, Hori M, et al. 3D quantitative synthetic MRI-derived cortical thickness and subcortical brain volumes: Scan-rescan repeatability and comparison with conventional T1 -weighted images. J Magn Reson Imaging 2019; 50:1834–1842. 10.1002/jmri.26744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fujita S, Hagiwara A, Hori M, et al. Three-dimensional high-resolution simultaneous quantitative mapping of the whole brain with 3D-QALAS: An accuracy and repeatability study. Magn Reson Imaging 2019; 63:235–243. 10.1016/j.mri.2019.08.031 [DOI] [PubMed] [Google Scholar]
- 103.Hagiwara A, Warntjes M, Hori M, et al. SyMRI of the brain: Rapid quantification of relaxation rates and proton density, with synthetic MRI, automatic brain segmentation, and myelin measurement. Invest Radiol 2017; 52:647–657. 10.1097/RLI.0000000000000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deoni SC, Rutt BK, Peters TM. Synthetic T1-weighted brain image generation with incorporated coil intensity correction using DESPOT1. Magn Reson Imaging 2006; 24:1241–1248. 10.1016/j.mri.2006.03.015 [DOI] [PubMed] [Google Scholar]
- 105.Goto M, Fukunaga I, Hagiwara A, et al. Decreased mis-segmentation in SPM 12: multi-channel analysis of synthetic magnetic resonance imaging. Proceedings of the 22nd Congress of Japan Human Brain Mapping Society 2020. [Google Scholar]
- 106.Goto M, Abe O, Miyati T, Aoki S, Gomi T, Takeda T. Mis-segmentation in voxel-based morphometry due to a signal intensity change in the putamen. Radiol Phys Technol 2017; 10:515–524. 10.1007/s12194-017-0424-3 [DOI] [PubMed] [Google Scholar]