Abstract
Texture analysis, as well as its broader category radiomics, describes a variety of techniques for image analysis that quantify the variation in surface intensity or patterns, including some that are imperceptible to the human visual system. Cerebral gliomas have been most rigorously studied in brain tumors using MR-based texture analysis (MRTA) to determine the correlation of various clinical measures with MRTA features. Promising results in cerebral gliomas have been shown in the previous MRTA studies in terms of the correlation with the World Health Organization grades, risk stratification in gliomas, and the differentiation of gliomas from other brain tumors. Multiple MRTA studies in gliomas have repeatedly shown high performance of entropy, a measure of the randomness in image intensity values, of either histogram- or gray-level co-occurrence matrix parameters. Similarly, researchers have applied MRTA to other brain tumors, including meningiomas and pediatric posterior fossa tumors.
However, the value of MRTA in the clinical use remains undetermined, probably because previous studies have shown only limited reproducibility of the result in the real world. The low-to-modest generalizability may be attributed to variations in MRTA methods, sampling bias that originates from single-institution studies, and overfitting problems to a limited number of samples.
To enhance the reliability and reproducibility of MRTA studies, researchers have realized the importance of standardizing methods in the field of radiomics. Another advancement is the recent development of a comprehensive assessment system to ensure the quality of a radiomics study. These two-way approaches will secure the validity of upcoming MRTA studies. The clinical use of texture analysis in brain MRI will be accelerated by these continuous efforts.
Keywords: magnetic resonance imaging, glioblastoma, texture analysis, radiomics, machine learning
Introduction
The occurrence of brain tumor is relatively rare, but it can affect individuals of all ages, from newborn babies to aged people. According to a recent report from the Central Brain Tumor Registry of the United States, the annual U.S. incidence of all primary brain and other central nervous system (CNS) tumors is 23.41 per 100000 population.1 Approximately two-thirds of all cases are nonmalignant, whereas the remainder is malignant.
The World Health Organization (WHO) has adopted histological grades for CNS tumor classification since 2000: from grade I tumors with low proliferative potential and a clinically benign course to grade IV tumors with malignant behaviors and a usually fatal outcome.2 After complete surgical resection, recovery from grade I tumors with no or minimal remaining symptoms can be expected; however, chemoradiotherapy after maximal tumor resection is a choice for the initial treatment of many grade IV tumors. Since treatment strategies considerably vary between tumor grades, estimating the histological grades using noninvasive radiological imaging before surgery is clinically relevant.
The two main components of CNS tissues are neurons and glial cells. Briefly, neurons undertake electrophysiological neural activities, and glial cells build supporting structures for neurons and neural fibers. Within mature CNS tissues, glial cells include astrocytes, oligodendrocytes, and microglial cells. Glioma is a type of brain tumor that is characterized by morphologic and genetic features of glial cells. Glioblastoma is the most aggressive tumor among gliomas and is categorized as grade IV, with a 5-year survival rate of 6.8%.1 Importantly, glioblastoma is the most common malignant primary brain tumor, accounting for approximately 15% of all primary brain tumors.1,2 Although approximately 90% of glioblastomas develop rapidly de novo (i. e., they develop from no preceding lesion), recent evidence in the field of genetics has suggested that, as multiple genetic alterations cumulate, some glioblastomas develop from diffuse astrocytomas (grade II) and anaplastic astrocytomas (grade III).3,4 The mutations of isocitrate dehydrogenase 1 (IDH 1) and less frequently IDH 2 genes, which encode an enzyme that catalyzes oxidative decarboxylation, are key molecular signatures for glioblastomas that develop from lower-grade astrocytic tumors. Consequently, the current edition of the WHO classification published in 2016 divides glioblastomas into two subtypes: glioblastoma, IDH–wildtype and glioblastoma, IDH–mutant.2 Of note, IDH mutation is associated with better outcomes in patients with glioblastomas, with a longer median survival time compared with those with IDH-wild type glioblastomas (27.4 vs. 14 months, respectively).5
Another important genetic signature for gliomas is the combined deletion of the short arm of chromosome 1 (1p) and the long arm of chromosome 19 (19q) (1p/19q co-deletion). In previous research, patients with co-deleted astrocytic tumors lived longer than those with un-co-deleted tumors, irrespective of chemoradiotherapy or radiotherapy alone.6 Depending on the histological evidence of anaplasia, gliomas with 1p/19q co-deletion and IDH mutation are diagnosed with oligodendroglioma (grade II) or anaplastic oligodendroglioma (grade III).2
The current standard treatment is maximal tumor resection followed by radiation therapy and chemotherapy for the initial treatment of glioblastomas. Temozolomide, a deoxyribonucleic acid (DNA)–alkylating agent, is typically chosen for chemotherapy in patients with glioblastomas.7 Humans have a repair mechanism for DNA damage caused by alkylation, in which a protein named O6-methylguanine-DNA methyltransferase (MGMT) removes alkyl groups from the damaged DNAs. Thus, if glioblastomas have a large amount of the MGMT nearby proteins, the damage of the tumor gene due to alkylating agents can be repaired. However, this repair protein synthesis is blocked by epigenetic silencing by the methylation of the MGMT gene promoters. Based on these in vitro findings, a clinical study has shown that the methylation of the MGMT promoter is an independent favorable prognostic factor for patients with glioblastomas. Patients with glioblastomas that contain methylated MGMT promoters demonstrate a longer survival time than those with glioblastomas containing unmethylated MGMT promoters.8
Rationale of Texture Analysis in Brain Tumor MRI
In clinical settings, before making a working diagnosis of a specific disease, radiologists summarize the findings of a brain lesion using several imaging modalities. Unfortunately, however, CT and MRI findings that are recognized and interpreted by a radiologist do not correspond in certain cases to a specific disease on a one-on-one basis. Some brain lesions may resemble each other on MRI, even when they are in totally different disease entities, such as neoplasm and demyelination.
Qualitative imaging characteristics have been, respectively, reported, which may be associated with IDH mutation status, MGMT promoter methylation, and 1p/19q co-deletion status in gliomas;9–12 however, determining the presence or absence of these characteristics in a particular image depends on a subjective judgment based on a radiologist’s experience.
To overcome the uncertainty and limited detectability of human perception, texture analysis (and its broader category, radiomics) has been introduced into the field of neuro-oncology, especially using MRI (Fig. 1). Texture analysis describes a variety of image analysis techniques that quantify the variation in surface intensity or patterns, including some that are imperceptible to the human visual system.13 MR-based texture analysis (MRTA) most often uses hundreds of features of statistical characteristics. In MRTA, intensity values of images are typically discretized into intensity bins in its process. Discretization reduces the burden of computing features and random errors in intensity values by rounding them off to the nearest bin.
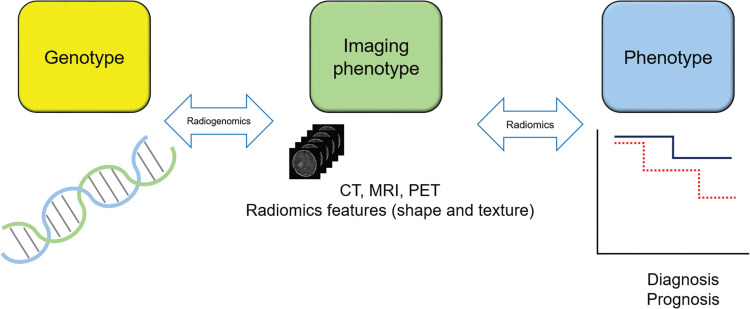
Fig. 1.
Schematic drawing for radiomics bridging medical images to the genotype and phenotype of a disease. Radiomics is a method for medical image analysis that quantifies the variation in the shape and texture of a lesion on medical images. Texture analysis forms the core techniques of radiomics and uses intensity distributions and intensity patterns of the lesion. Radiomics features (i.e., shape and texture) are used to correlate images with clinical measures of disease, including diagnosis, survival time, and histological grades of malignancy. Radiomics features can be linked to genetic and epigenetic alterations of the disease, and this special field of radiomics is often called radiogenomics. Between genotype and phenotype, the information represented by images is called imaging phenotype or image phenotype. PET, positron emission tomography.
First-order features consider the distribution of discretized intensity values without concern for spatial relationships of voxels.14 In this case, histograms are commonly used to display voxel counts in respective intensity bins, and thus, first-order texture analysis is also termed as histogram analysis. In contrast, second-order statistics provide a feature of the spatial arrangement of the voxel intensities. Higher-order features are obtained by statistical methods after mathematical transforms to the images.14 Gray-level co-occurrence matrix (GLCM) features are categorized in second-order statistics, while higher-order features include gray-level run-length matrix (GLRLM), gray-level size zone matrix (GLSZM), gray-level difference zone matrix, neighborhood gray-tone difference matrix (NGTDM), and neighboring gray-level dependence matrix features. Descriptions of features are summarized in Table 1 (for more details, please refer to standard radiomics features listed in Supplementary file 1).
Table 1.
Feature families used in radiomics
| Feature family | Descriptions of feature family | ||
|---|---|---|---|
| Radiomics features | Nontexture | Morphology | Volume, shape, elongation, compactness, sphericity, etc. |
| Texture | Local intensity | Local intensities within the segmented volume | |
| Intensity-based statistical | Mean, standard deviation, minimum, maximum, kurtosis, skewness, etc. | ||
| Intensity histogram | Bin the intensities of the segmented volume | ||
| Intensity volume histogram | Bin the volume as it relates to intensities | ||
| GLCM | Occurrence of neighboring pixels | ||
| GLSZM | Volume sizes for given intensities | ||
| GLRLM | Length of consecutive pixels for given intensities | ||
| GLDZM | Distance between volumes of varying intensities | ||
| NGTDM | Distance between adjacent gray-tone regions | ||
| NGLDM | Distance between adjacent gray-level regions |
* Adapted from Refs. 19 and 67. GLCM, gray-level co-occurrence matrix; GLDZM, gray-level distance zone matrix; GLRLM, gray-level run-length matrix; GLSZM, gray-level size zone matrix; NGLDM, neighboring gray-level dependence matrix; NGTDM, neighboring gray-tone difference matrix.
Briefly, first-order statistics relate to the likelihood of individual voxels having specific intensity values, whereas second-order statistics relate to the joint likelihood of two random voxels in the image having specific pairs of intensity values.15 Higher-order statistics examine the spatial relationship between three or more voxels.16 Importantly, first-order features do not account for the location of the voxels and lack any reference to the spatial interrelationship between intensities.17 If two different tumors have a similar distribution of intensities but with different spatial interrelationships, first-order features may not be able to differentiate these two tumors; in such a situation, second- or higher-order features may thus be preferable.
Other methods of texture analysis are model-based approaches (e.g., fractal analysis) that analyze a form of pattern and transform-based approaches (e.g., Fourier, Gabor, and wavelet) that use a frequency and a scale domain of images.16
Generic image filters are sometimes used before feature calculation, of which a Laplacian of Gaussian (LoG) filter is most common. The purpose of filtering is to enhance and preserve subtle image features that may be unintentionally lost while analyzing the original images.18 For example, typical LoG filters derive brain textures of different scales or sizes corresponding to filter scale values: fine (emphasizing textures that are approximately two voxels in width), medium (four voxels), and coarse (six voxels), respectively.19 As a result, LoG filtration highlights the gray-level change in fine, medium, and coarse textures depending on the setting. Filtration can be used together with any of the first-, second-, and higher-order features mentioned above.
Studies in the literature have shown that the results of MRTA are promising thus far in brain tumors; however, despite the continued studies, consensus on the clinical role of MRTA remains to be determined.18 In this study, we reviewed the current applications, limitations, and future perspectives of texture analysis in brain tumor MRI.
Technical Considerations
Multiparametric or single-parametric MRI
To elucidate tumor characteristics, MRI for brain tumors typically includes five or more imaging sequences. T1-weighted imaging (T1WI), T2-weighted imaging (T2WI), and fluid-attenuating inversion recovery (FLAIR) imaging can represent anatomical and structural information. Additionally, diffusion-weighted imaging (DWI) and contrast-enhanced T1WI (CE-T1WI) are typically included in brain tumor MRI protocols in many hospitals. The most commonly used index to estimate tumor cellularity is the apparent diffusion coefficient (ADC) derived from DWI.20 CE-T1WI highlights the breakdown of the blood–brain barrier by a tumor; additionally, smaller lesions apart from the main tumor often become evident on CE-T1WI. More advanced MRI techniques include perfusion-weighted imaging (PWI), dynamic contrast-enhanced (DCE)-MRI, and multishell DWI with a complex acquisition scheme, partly for research purposes. Previous MRTA studies for brain tumors have used either multiparametric or single-parametric MRI. Image registration between images with different image acquisition sequences is typically required in multiparametric MRI. In single-parametric MRTA studies on cerebral gliomas, CE-T1WI, followed by T2WI and ADC maps, was most often used to obtain texture features.18
Preprocessing
Noise suppression, skull stripping, nonuniformity correction, and intensity normalization are typically conducted in postacquisition preprocessing (Fig. 2). A recent guideline published by the Image Biomarker Standardization Initiative recommends that researchers report whether and how they perform preprocessing.21
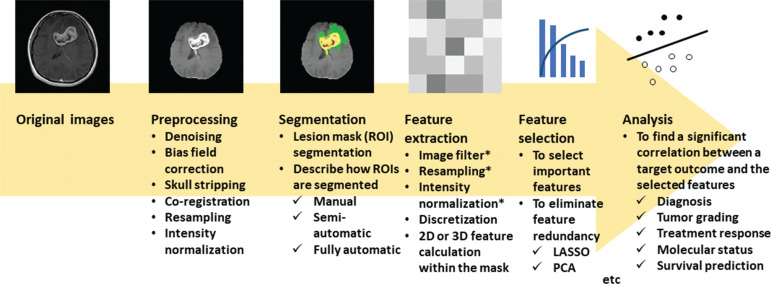
Fig. 2.
Standard flowchart for MRI-based texture analysis. A 35-year-old woman with glioblastoma in the genu of the corpus callosum to bilateral frontal lobes. Legion masks are segmented as described in the current nomenclature by the Multimodal Brain Tumor Segmentation Challenge (https://www.med.upenn.edu/cbica/brats2020/): gadolinium-enhancing tumor (yellow), the peritumoral edema (green), and the necrotic and nonenhancing tumor core (red). The texture analysis comprises several steps, typically in the following order: preprocessing of images, segmentation of the target regions (either diseased lesions or normal-appearing structures), extraction of image features from the regions, selection of important discriminating features, and the subsequent analysis of significant correlations between the selected features and a target outcome, typically using machine learning models. * indicates an optional step. LASSO, least absolute shrinkage and selection operator; PCA, principal component analysis.
Noise suppression and nonuniformity correction are used to mitigate imperceptible bias in MR images that may be specific to the patient or machine. If the image intensity is to be normalized across the head, skull stripping may be required, because fat tissue is a source of a large number of voxels with high signal intensity, which causes bias in the distribution of intensities. Intensity normalization is preferable in images representing relative intensity values that are used in T1WI, T2WI, and CE-T1WI because these sequences have various ranges of intensities for the same image. However, in images with an absolute or quantitative intensity scale, including ADC maps, intensity normalization should not be attempted. Intensity normalization is typically achieved by remapping the brightness to an 8-bit (0–255) scale between the minimum and maximum or within the mean ± 3 standard deviations values of an image.18
Segmentation
Segmenting a tumor is an important but painstaking task. During this process, researchers should decide how to place a ROI on a tumor. There are several choices for ROI placement: (1) ROI on a single or a few representative image slices of a tumor or a volume of interest (VOI; i.e., stacked ROIs) on a whole tumor and (2) fully automated, semiautomated, and manually drawn ROI placement. Presently, no consensus has been reached regarding these technical details, and each has merits and demerits. For example, supposing that the tumor image characteristics are fully represented in the core region of a tumor, nondominant features at the periphery of the tumor may be overstated by a whole-tumor VOI, whereas substantial features might be missed by a single ROI on the image showing the maximal tumor. Regarding tumor-contouring methods, automated segmentation often reports similarity statistics with manual segmentation by experts, and manual segmentation usually requires an interobserver comparison to demonstrate the validity of ROI placement.
Feature extraction
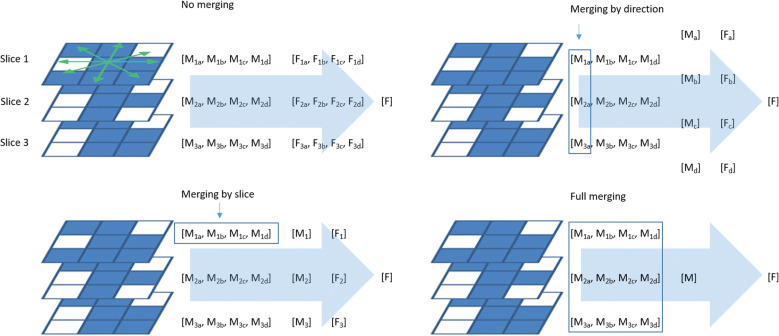
Feature extraction also raises brainstorming concerns. Both 2D and 3D feature calculation methods are available in the state-of-the-art software programs for radiomics. The 3D feature calculation is typically accompanied by isotropic interpolation of the original images and the segmented mask images for the lesions (i.e., ROI masks), whereas the 2D texture calculation is also available for a tumor volume, and the calculation is conducted for every single image slice and then averaged for the whole tumor. In this setting, there are four options for averaging: no merging, merging by slice, merging by direction, and full merging (Fig. 3).
Fig. 3.
Variety in 2D texture feature computation. In gray-level co-occurrence and run-length matrices, connections between neighboring voxels on an image slice have four different directions (green double arrows). Depending on the presence or absence of merging by slice or direction, texture feature computation has four options, resulting in different values of the same feature. The subscript characters indicate slice numbers (1–3) and directions (a–d). F, feature; M, matrix.
Another important concern is discretization in image intensities. Because many texture features are calculated based on histogram-style discretization, fixed bin size and fixed bin number (typically 32 or 64) are available options for discretization.22
By applying generic image filters to the original images before feature extraction, the number of texture features can be easily doubled or tripled. Several current software programs offer a dozen kinds of these generic filters, and hundreds of features can be calculated depending on the parameter settings. However, a geometric increase in texture features results in feature redundancy.23,24
Feature selection
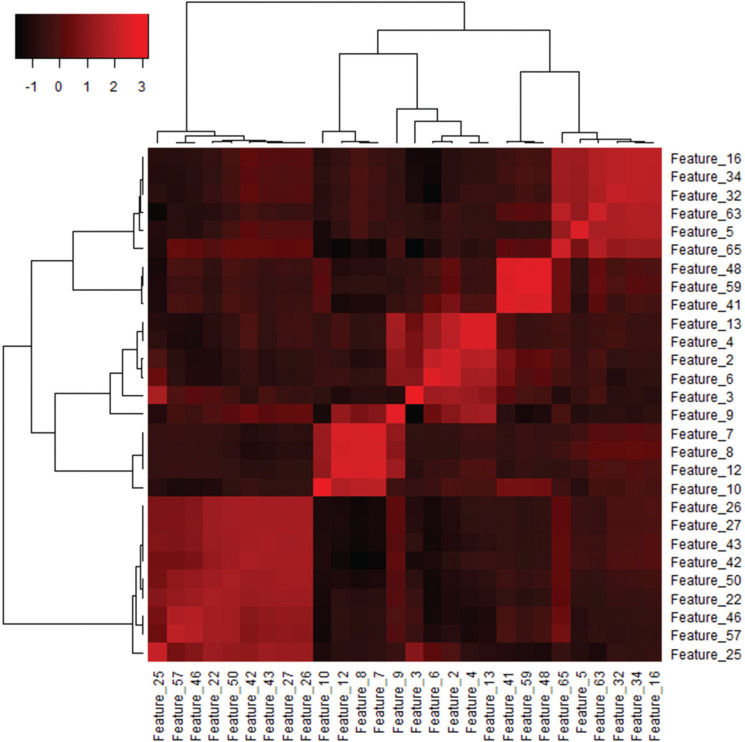
It is well known that texture features demonstrate considerable collinearities and redundancies (Fig. 4). Feature selection is the process whereby important discriminating features are chosen to describe a response (e.g., diagnosis or prognosis). The main reasons for using feature selection are to make the model easier to interpret, to remove features with multicollinearities, to reduce computation costs, and to reduce overfitting. To select a feature subset, machine learning techniques are commonly used, where important discriminating features are selected based on the model’s performance for different subsets of features. These supervised methods include Pearson’s correlation-based feature ranking, a least absolute shrinkage and selection operator technique, support vector machines (SVMs) with recursive feature elimination, and artificial neural networks. Unsupervised methods are also available, and principal component analysis is an example.24
Fig. 4.
Heatmap presentation of the variance–covariance matrix of texture features. A heatmap of the variance–covariance matrix can be used to find collinearities among features. Data from our previous study50 were reused. Unsupervised clustering was performed with Ward’s method using a free software for statistical computing (R: A language and environment for statistical computing; R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org/.) and its heatmap3 package (https://github.com/cran/heatmap3). Features within the same branch of the dendrograms have substantial collinearities showing highly similar patterns of the covariance across the features. This heatmap indicates that three to five features are enough and optimal to describe a response.
Analysis with discriminating features
After feature selection in typical MRTA studies, supervised or unsupervised machine learning is conducted to detect any significant correlations between discriminating features and a target clinical outcome. In this process, regression or classification models, incorporating texture features, are typically developed with one or more machine learning algorithms (however, discussion on machine learning is beyond the aim of this review, and please refer to nice reviews on machine learning in the field of radiology).25,26 In some earlier MRTA studies, especially those using a relatively limited number of texture features, pairwise comparisons were used; however, this strategy is now deprecated because of the continuous rise in computing capability and the exploding number of texture features used in recent studies.
Application of Texture Analysis to Brain Tumor MRI
Glioma grading
Survival rates for gliomas with WHO grades I–IV show a decreasing trend with increasing grades.27 Other than these four grades, gliomas are commonly classified into two broader categories: low grade and high grade. A standard scheme categorizes gliomas with WHO grades I and II as low-grade gliomas (LGGs) and those with WHO grades III and IV as high-grade gliomas (HGGs).2
Differentiation of LGGs and HGGs is mandatory for risk stratification and to tailor the best management strategies for patients.28 In a study by Ryu et al., enrolling 40 patients with gliomas (eight grade II, 10 grade III, and 22 grade IV tumors), they found that GLCM entropy and fifth percentile values of the ADC histogram differed significantly between HGGs and LGGs (hereafter, see Supplemental file 2 for feature definitions); however, only entropy was proven to show significant differences between grade III and IV tumors.29 The ADC entropy showed a sensitivity of 78.1%, a specificity of 87.5%, an accuracy of 80.0%, and an area under the curve (AUC) value of 0.830 in a receiver-operating characteristic analysis in differentiating HGGs from LGGs. Skogen et al. examined 95 patients with gliomas (27 grade II, 34 grade III, and 34 grade IV tumors) with an LoG filtration-based histogram analysis on CE-T1WI.30 According to their results, HGGs were best discriminated from LGGs using the standard deviation values at a fine texture scale (sensitivity and specificity of 93% and 81%, respectively, and an AUC value of 0.910); however, they also reported that the diagnostic ability to differentiate grades II–IV was lower than that in differentiating HGGs and LGGs. Similar results were replicated by Ditmar et al., who examined 94 patients with gliomas (14 LGGs and 80 HGGs).31
Xie et al. used texture analysis for kinetic DCE-MRI parameters in 42 patients with gliomas (15 grade II, 13 grade III, and 14 grade IV tumors).32 They showed that the entropy and inverse difference moment (IDM) of the DCE-MRI parameters were able to differentiate glioma grade II from grade III and grade III from grade IV. By contrast, in a study involving 42 patients with gliomas (10 LGGs and 30 HGGs), Su et al. applied texture analysis to DCE-MRI and found that uniformity of permeability images had the highest AUC value of 0.917 with 93% sensitivity and 90% specificity.33
Regarding glioma grading using multiparametric MRTA, Vamvakas et al. reported that the higher-ranked discriminating parameters were skewness and variance of CE-T1WI, mean of diffusion anisotropy, and IDM of mean diffusivity images.34 An SVM classifier achieved an accuracy of 95.5%, sensitivity of 95%, specificity of 96%, and an AUC value of 0.955 at cross-validation. In their recent study, Alis et al. enrolled 181 patients with gliomas (84 LGGs and 97 HGGs) and evaluated the classification ability of an artificial neural network model with 10 hidden layers.28 Models using texture features obtained from FLAIR and CE-T1WI images achieved AUC values of 0.87 and 0.86, respectively. The combined artificial neural network model with eight selected texture features achieved the highest diagnostic accuracy of 88.3%, with an AUC value of 0.92. The results of these previous studies are summarized in Table 2.
Table 2.
Glioma Grading
| Author (Year) | Number of Subjects | MRI Sequence | Texture Software | Type of Texture Features | Best Discriminating Feature | Prediction Model | Main Findings |
|---|---|---|---|---|---|---|---|
| Differentiation between HGGs and LGGs | |||||||
| Ryu et al. (2014)29 | 40 (8 grade II, 10 grade III, and 22 grade IV gliomas) | ADC | In-house | First-order, GLCM | GLCM entropy | No (pairwise, ROC) | AUC= 0.830 Accuracy = 80.0% |
| Skogen et al. (2016)30 | 95 (27 grade II, 34 grade III, and 34 grade IV gliomas) | CE-T1WI | TexRAD*1 | First-order with LoG filtration | SD at fine scale | No (pairwise, ROC) | AUC = 0.910 |
| Ditmar et al. (2018)31 | 94 (14 LGGs and80 HGGs) | ADC, FLAIR, CE-T1WI | TexRAD*1 | First-order with LoG filtration | Mean at fine scale (CE-T1WI) | No (pairwise, ROC) | AUC = 0.900 |
| Su et al. (2019)33 | 42 (10 LGGs and30 HGGs) | DCE-MRI | OmniKinetics*2 | First-order | Uniformity of Ktrans | No (pairwise, ROC) | AUC = 0.917 |
| Vamvakas et al. (2019)34 | 40 (20 LGGs and20 HGGs) | T1WI, T2WI, FLAIR, CE-T1WI, DTI, PWI, MRS | MaZda*3 | First-order, GLCM, GLRLM | 21 top-ranked features | SVM | AUC = 0.955 Accuracy = 95.5% |
| Alis et al. (2020)28 | 181 (84 LGGs and 97 HGGs) | FLAIR, CE-T1WI | MaZda*3 | First-order, HoG, gradient-map-based features, GLCM, GLRLM, autoregressive model, Haar wavelet features, Gabor transform features, and local binary patterns | 8 features (5 GLCM and 3 GLRLM features) | ANN (MLP with 10 hidden layers) | AUC = 0.92 |
| Differentiating grade III from grade IV | |||||||
| Xie et al. (2018)32 | 42 (15 grade II, 13 grade III, and 14 grade IV gliomas) | DCE-MRI | OmniKinetics | GLCM | Entropy and IDM (Vp images) | No (pairwise, ROC) | AUC = 0.885 (Entropy) AUC = 0.901 (IDM) |
*1 https://fbkmed.com/texrad-landing-2/. *2 GE Healthcare, Waukesha, WI, USA. *3 http://www.eletel.p.lodz.pl/programy/mazda/. ADC, apparent diffusion coefficient; ANN, artificial neural network; AUC, area under the curve; CE-T1WI, contrast-enhanced T1-weighted imaging; DCE-MRI, dynamic contrast-enhanced MR imaging; DTI, diffusion tensor imaging; FLAIR, fluid-attenuating inversion recovery; GLCM, gray-level co-occurrence matrix; GLRLM, gray-level run-length matrix; HGG, high-grade glioma; HoG, histogram of oriented gradients; IDM, inverse difference moment; LGG, low-grade glioma; LoG, Laplacian of Gaussian; MLP, multi-layer perceptron; MRS, MR spectroscopy; PWI, perfusion-weighted imaging; ROC, receiver-operating characteristic; SD, standard deviation; SVM, support vector machine; T1WI, T1-weighted imaging; T2WI, T2-weighted imaging; Vp, blood plasma volume.
Risk stratification in gliomas
Molecular status
Another important topic in oncology is tumor risk stratification. As mentioned above, IDH mutation, 1p/19q co-deletion, and MGMT promoter methylation are significant prognostic factors for gliomas.5,6,8
Regarding IDH mutation, Zhou et al., in their study involving 165 patients with grade II or III gliomas, found that a logistic regression model incorporating skewness, run-length variance, and short-run low gray-level emphasis of GLRLM on T2WI reached an AUC value of 0.86, sensitivity of 0.75, and specificity of 0.78 for the presence of IDH1 mutation (a major subtype of IDH mutation).35 For 1p/19q co-deletion status, the set of features comprised GLRLM low gray-level run emphasis on CE-T1WI, short-zone low gray-level emphasis of GLSZM on T2WI, and GLRLM long-run high gray-level emphasis on T2WI images reached an AUC value of 0.96, sensitivity of 0.90, and specificity of 0.89. Each of the aforementioned models outperformed an established human visual scoring system in terms of diagnostic ability. Another study conducted by Bahrami et al. involving 61 patients with grade II or III gliomas showed that compared with patients with IDH–mutant tumors, patients with IDH–wild-type tumors showed greater GLCM-based heterogeneity and lower gradient magnitude at the lesion edges within the tumor on FLAIR images.36 Among patients with IDH–mutant tumors, 1p/19q co-deleted tumors had greater signal heterogeneity and lower edge contrast than 1p/19q intact tumors. Tumors with methylated MGMT promoters showed lower edge contrasts than those with unmethylated MGMT promoters (Table 3).
Table 3.
Risk Stratification (Molecular Status)
| Author (Year) | Number of Subjects | MRI Sequence | Texture Software | Type of Texture Features | Best Discriminating Features | Prediction Model | Main Findings |
|---|---|---|---|---|---|---|---|
| IDH mutation | |||||||
| Zhou et al. (2017)35 | 165 (grades II and III gliomas, TCIA/TCGA dataset) | T1WI, T2WI, FLAIR, CE-T1WI | In-house | First-order, GLCM, GLRLM, GLSZM, NGTDM | Skewness, run-length variance, and short-run low gray-level emphasis of GLRLM (T2WI) | Logistic regression | AUC = 0.86 |
| Bahrami et al. (2018)36 | 61 (grades II and III gliomas) | T1WI, FLAIR, CE-T1WI | Not described | GLCM, edge contrast (gradient magnitude of lesion edges) | Correlation, edge contrast (FLAIR) | Logistic regression | Greater heterogeneity and lower edge contrast in wildtype tumors |
| Lewis et al. (2019)38 | 97 (54 grade II, 20 grade III, 23 grade IV gliomas) | T2WI, CE-T1WI, ADC | TexRAD*1 | First-order | Kurtosis (CE-T1WI, at all scales) | No (pairwise, ROC) | IDH mutation in glioblastoma AUC = 0.945 |
| 1p/19q co-deletion | |||||||
| Zhou et al. (2017)35 | 165 (grades II and III gliomas, TCIA/TCGA dataset) | T1WI, T2WI, FLAIR, CE-T1WI | In-house | First-order, GLCM, GLRLM, GLSZM, NGTDM | GLRLM low gray-level run emphasis (CE-T1WI)GLRLM long-run high gray-level emphasis, GLSZM short-zone low gray-level emphasis (T2WI) | Logistic regression | AUC = 0.86 |
| Bahrami et al. (2018)36 | 61 (grades II and III gliomas) | T1WI, FLAIR, CE-T1WI | Not described | GLCM, edge contrast (gradient magnitude of lesion edges) | Correlation, edge contrast (FLAIR) | Logistic regression | Greater heterogeneity and lower edge contrast in co-deleted tumors |
| Lewis et al. (2019)38 | 97 (54 grade II, 20 grade III, 23 grade IV gliomas) | T2WI, CE-T1WI, ADC | TexRAD*1 | First-order | Skewness (ADC, no LoG filtration) | No (pairwise, ROC) | 1p/19q co-deletion in grades II and III AUC = 0.811 |
| MGMT methylation | |||||||
| Bahrami et al. (2018)36 | 61 (grades II and III gliomas) | T1WI, FLAIR, CE-T1WI | Not provided | GLCM, edge contrast (gradient magnitude of lesion edges) | Edge contrast (FLAIR) | Logistic regression | Lower edge contrast in methylated tumors |
| Kanazawa et al. (2019)37 | 48 glioblastomas | ADC | Synapse Vincent*2 | First-order | Mean, entropy | No (pairwise, ROC) | The combination of mean ADC value and ADC entropy predicted MGMT promoter methylation, with a positive predictive value of 81.2% and specificity of 88.9% |
*1 https://fbkmed.com/texrad-landing-2/. *2 Fujifilm, Tokyo, Japan. ADC, apparent diffusion coefficient; AUC, area under the curve; CE-T1WI, contrast-enhanced T1-weighted imaging; FLAIR, fluid-attenuating inversion recovery; GLCM, gray-level co-occurrence matrix; GLRLM, gray-level run-length matrix; GLSZM, gray-level size zone matrix; LoG, Laplacian of Gaussian; MGMT, O6-methylguanine-DNA methyltransferase; NGTDM, neighborhood gray-tone difference matrix; ROC, receiver-operating characteristic; T1WI, T1-weighted imaging; T2WI, T2-weighted imaging; TCIA, The Cancer Imaging Archive; TCGA, The Cancer Genome Atlas.
MRTA with ADC and CE-T1WI images may be useful for risk stratification in grade IV glioblastomas. Kanazawa et al. conducted ADC histogram analyses on 48 patients with newly diagnosed glioblastomas and found that the combination of mean ADC value and ADC entropy predicted MGMT promoter methylation, with a positive predictive value of 81.2% and a specificity of 88.9%.37 In another study conducted by Lewis et al., CE-T1WI had the best performance for IDH typing of glioblastoma (sensitivity of 91.9%, specificity of 100%, and AUC of 0.945) (Table 3).38
Prognosis
In the clinical course of patients with LGGs, malignant transformation to HGGs can be a fatal problem. A recent study by Zhang et al. demonstrated that the most discriminating features based on linear discriminant analysis resulted in AUC values of 0.90 (85% sensitivity and 84% specificity) for FLAIR, 0.92 (86% sensitivity and 94% specificity) for ADC, 0.96 (97% sensitivity and 84% specificity) for T1WI, and 0.82 (78% sensitivity and 75% specificity) for CE-T1WI images.39 Their models correctly discriminated LGGs with early signs of malignant transformation from those without in 93%, 100%, 93%, and 92%, respectively. Longer survival times often indicate favorable outcomes of treated brain tumors. Chaddad et al. studied the association between the overall survival and texture features of FLAIR and CE-T1WI images obtained from 39 patients with glioblastomas.40 The results of their study showed that four features (energy, correlation, variance, and inverse of variance) obtained from contrast-enhancing regions and homogeneity from edema regions on FLAIR images were shown to be associated with survival times. Similarly, on CE-T1WI images, three features (energy, correlation, and variance) from contrast-enhancing regions were found to be useful for the prediction of patient overall survival. In another study, Kickingereder et al. demonstrated that supervised principal component analysis with standard deviations (between wavelet-transformed FLAIR and CE-T1WI) of six GLCM features and standard deviations or mean values of five GLRLM features allowed for the stratification into a low- or high-risk group for progression-free survival and overall survival.41 Prasanna et al. reported that MRTA features in the peritumoral brain zone were most predictive of survival time across T1WI, T2WI, FLAIR, and multiparametric combination of these methods.42 Interestingly, in their prediction models, the authors found that Laws features, which identify textural patterns corresponding to spots, level, waves, ripples, or edges in an image, were the most predictive (Table 4).
Table 4.
Risk Stratification (Prognosis)
| Author (Year) | Number of Subjects | MRI Sequence | Texture Software | Type of Texture Features | Best Discriminating Features | Prediction Model | Main Findings |
|---|---|---|---|---|---|---|---|
| Risk of malignant transformation | |||||||
| Zhang et al. (2019)39 | 68 LGGs | T1WI, FLAIR, CE-T1WI, ADC | MaZda*1 | Not provided (279 features) | 30 top-ranked features | Linear discriminant analysis | Accuracy = 93% (FLAIR), 100% (ADC), 93% (T1WI), 92% (CE-T1WI) |
| Survival | |||||||
| Chaddad et al. (2016)40 | 39 glioblastomas (TCIA/TCGA dataset) | FLAIR, CE-T1WI | Matlab*2 | GLCM | Energy, correlation, variance, inverse variance, homogeneity (FLAIR) Energy, correlation, variance (CE-T1WI) | No (Kaplan-Meier) | Longer survival time was associated with:Higher energy, higher correlation, lower variance, lower inverse variance (FLAIR)Higher energy, higher correlation, lower variance (CE-T1WI) |
| Kickingereder et al. (2016)41 | 119 glioblastomas | FLAIR, CE-T1WI | Medical Imaging Toolkit*3 | First-order, GLCM, GLRLM | SD of 6 GLCM features (FLAIR) Mean or SD of 5 GLRLM features (FLAIR) | Cox regression | Significant association with both PFS (HR, 2.28; P = 0.032) and OS (HR, 3.45; P = 0.004) |
| Prasanna et al. (2017)42 | 65 glioblastomas (TCIA/TCGA dataset) | T2WI, FLAIR, CE-T1WI | In-house | GLCM, laws features, HoG, Laplacian pyramids | 10 most predictive features | Random forest | Intensity heterogeneity and textural patterns were found to be predictive of survival (P = 1.47 × 10-5) |
*1 http://www.eletel.p.lodz.pl/programy/mazda/. *2 MathWorks, Natick, MA, USA. *3 https://www.mitk.org/wiki/The_Medical_Imaging_ Interaction_Toolkit_(MITK). ADC, apparent diffusion coefficient; CE-T1WI, contrast-enhanced T1-weighted imaging; FLAIR, fluid-attenuating inversion recovery; GLCM, gray-level co-occurrence matrix; GLRLM, gray-level run-length matrix; HoG, histogram of gradient orientations; HR, hazard ratio; LGG, low-grade glioma; OS, overall survival; PFS, progression-free survival; SD, standard deviation; T1WI, T1-weighted imaging; T2WI, T2-weighted imaging; TCIA, The Cancer Imaging Archive; TCGA, The Cancer Genome Atlas.
Differentiation from other brain tumors and tumor mimics
Metastatic brain tumor
Distant metastasis to the brain tissue typically occurs in advanced cancers, especially in lung and breast cancers. Because breast or lung cancer occurs more frequently than primary brain tumors, the incidence of metastatic brain tumors is comparable with the total incidence of all primary brain tumors in the adult population.43 However, the therapeutic strategy for a metastatic brain tumor is considerably different from that for a primary brain tumor, because patients with metastatic brain tumors typically suffer from local recurrent tumors and other metastatic lesions in parts of the body other than the brain.
Regarding the differentiation of glioblastomas or HGGs from brain metastases, breast and lung cancers are the primary sites of metastases in the previous studies. An early study conducted by Mouthuy et al. showed a significant difference between metastatic tumors and glioblastomas in GLCM parameters (energy, entropy, homogeneity, correlation, inverse differential moment, and sum average) calculated on PWI.44 Using T2WI and CE-T1WI images, Petrujkić et al. demonstrated that IDM yielded the highest sensitivity and specificity in the differentiation of brain metastases from glioblastomas.45 Zhang et al. found that homogeneity and IDM calculated on ADC images were significantly higher in glioblastomas than in metastases, with AUC values of up to 0.886, sensitivity of 83.3%, and specificity of 76.9%.46 Using 12 texture features obtained on T2WI, CE-T1WI, and ADC images, the prediction performance with SVM was calculated at an AUC value of 0.92, which may be comparable with the performance of radiologists.47 Other than the metastatic tumors themselves, peritumoral edema in glioblastoma may show higher heterogeneity than the edema surrounding metastatic tumors, differentiating them with a sensitivity of 80% and specificity of 90% (Table 5).48
Table 5.
Diagnosis (Differentiation from non-Glioma)
| Author (Year) | Number of Subjects | MRI Sequence | Texture Software | Type of Texture Features | Best Discriminating Features | Prediction Model | Main Findings |
|---|---|---|---|---|---|---|---|
| Differentiation from metastatic brain tumor | |||||||
| Mouthuy et al. (2012)44 | 41 glioblastomas and 14 metastases | PWI | MaZda*1 | GLCM | Energy, entropy, homogeneity, correlation, inverse differential moment, sum average | No (pairwise, ROC) | Glioblastomas showed higher energy, higher homogeneity, higher inverse differential moment, and lower entropy. Highest AUC = 0.75 (correlation) |
| Petrujkić et al. (2019)45 | 30 glioblastomas and 25 solitary brain metastases | T2WI, CE-T1WI, SWI | ImageJ*2 | GLCM | Angular second moment, inverse difference moment, contrast, correlation, entropy | No (pairwise, ROC) | All five GLCM parameters obtained from T2WI showed significant difference between glioblastomas and solitary metastases. Highest AUC = 0.795 (inverse difference moment, CE-T1WI) |
| Zhang et al. (2019)46 | 36 glioblastomas and 26 solitary brain metastases | ADC | In-house | First-order, GLCM | Homogeneity, inverse difference moment | No (pairwise, ROC) | AUC = 0.886 (homogeneity) AUC = 0.732 (inverse difference moment) |
| Tateishi et al. (2020)47 | 73 glioblastomas and 53 metastases | T2WI, CE-T1WI, ADC | LIFEx*3 | First-order, GLCM | 12 a-priori texture features | Logistic regression, SVM | Highest AUC = 0.92 (SVM model) |
| Skogen et al. (2019)48 | 22 glioblastomas and 21solitary brain metastases | ADC, FA | TexRAD*4 | First-order | Entropy | No (pairwise, ROC) | Texture features were derived from peritumoral edema Highest AUC = 0.911 (combined ADC and FA, no LoG filtration) |
| Differentiation from PCNSL | |||||||
| Alcaide-Leon et al. (2017)49 | 71 glioblastomas and 35 PCNSL | CE-T1WI | In-house | First-order, GLCM, GLRLM, GLSZM, NGTDM | Not provided | SVM | Mean AUC = 0.877 (at cross-validation) |
| Kunimatsu et al. (2018, 2019)50, 51 | Training: 44 glioblastomas/16 PCNSL) Test: 11 glioblastomas/5 PCNSL | CE-T1WI | R*5 | First-order, GLCM, GLRLM, GLSZM | Entropy, median (first-order)Run-length nonuniformity, run percentage (GLRLM) | SVM | Highest AUC = 0.99 (at cross-validation) Accuracy = 75% (in test dataset) |
*1 http://www.eletel.p.lodz.pl/programy/mazda/. *2 https://imagej.nih.gov/ij/. *3 http://www.lifexsoft.org/. *4 https://fbkmed.com/texrad-landing-2/. *5 https://cran.r-project.org/. ADC, apparent diffusion coefficient; AUC, area under the curve; CE-T1WI, contrast-enhanced T1-weighted imaging; FA, fractional anisotropy; GLCM, gray-level co-occurrence matrix; GLRLM, gray-level run-length matrix; GLSZM, gray-level size zone matrix; LoG, Laplacian of Gaussian; NGTDM, neighborhood gray-tone difference matrix; PCNSL, primary central nervous system lymphoma; PWI, perfusion-weighted imaging; ROC, receiver operating characteristic; SVM, support vector machine; SWI, susceptibility-weighted imaging; T2WI, T2-weighted imaging.
CNS lymphoma
Lymphoma can arise throughout the body, and primary CNS lymphoma is a subtype of lymphoma that is prone to be confined to the CNS tissues. Primary CNS lymphomas most commonly affect older adults and elderly people, as do glioblastomas, and notably, imaging findings often resemble each other; however, chemotherapy is the first choice of treatment for primary CNS lymphomas, whereas surgery and subsequent chemoradiotherapy are typically chosen for glioblastoma treatment. Thus, differentiation by imaging before surgery is of high clinical relevance.
Researchers have reported an MRTA-based classification between primary CNS lymphomas and glioblastomas using CE-T1WI images. Alcaide-Leon et al. demonstrated that the classification performance between primary CNS lymphomas and glioblastomas by SVM classifiers was comparable with that by radiologists, incorporating first-order, GLCM, GLRLM, GLSZM, and NGTDM features, with a mean AUC value of 0.877 for the SVM classifier in cross-validation.49 In another study conducted by Kunimatsu et al., the first-order median and entropy, and GLRLM run-length nonuniformity and run percentage were shown to be the most efficient in differentiating primary CNS lymphomas from glioblastomas using principal component analysis.50 They also demonstrated that the AUC values of SVM classifiers were as high as 0.99 in cross-validation and that the prediction accuracy of 75% was achieved by the classifiers with individual test images (Table 5).51
Radiation necrosis
When tumor cell death induced by chemoradiation therapy or radiation therapy alone surpasses tumor cell proliferation, the tumor will stabilize in size or even become smaller. In such a condition, the dead region of a brain tumor, as well as typically the surrounding brain tissues, sometimes mimics tumor recurrence after treatment, showing a ringlike contrast enhancement on CE-T1WI. This phenomenon is referred to as radiation necrosis. Because radiation necrosis means that the therapy is effective, differentiation from tumor recurrence by noninvasive imaging has gained clinical attention. Regarding the differentiation of radiation necrosis from treated brain metastases, only one study by Larroza et al. is available.52 They investigated the performance of SVM classifiers using the top-ranked features for two different datasets. The GLCM sum entropy and first-order variance were selected repeatedly for both classifiers: however, the remaining discriminating features were different between the classifiers. Despite the substantial difference in the included features between the classifiers, both classifiers showed excellent performance with the AUC values of greater than 0.9.
Texture analysis in other brain tumors
Meningiomas are more common than gliomas, and they account for approximately one-third of all primary CNS tumors.1 Although the vast majority of meningiomas are benign and categorized as WHO grade I, more aggressive grades II and III meningiomas comprise 10% of all meningiomas.53 Conventional MRI remains the standard radiologic technique for the provisional diagnosis and surveillance of meningioma; however, recent progress in radiomics research in cerebral gliomas has encouraged researchers to apply radiomics methodologies to meningiomas as well.
As in cerebral gliomas, the differentiation of meningioma from clinically pertinent entities is of high clinical relevance. In an early study, Georgiadis et al. reported 100% overall classification accuracy in the differentiation of gliomas and meningiomas using GLCM and GLRLM features and SVMs.54 Similarly, several previous machine learning studies with a similar study purpose used MRI datasets, including intra-axial brain tumors (e.g., glioblastomas and metastases) other than extra-axial meningiomas.55–57 In this regard, however, the real radiological challenge in the differential diagnosis of meningioma would be its distinction from dura-based tumors, such as dural metastases and solitary fibrous tumors/hemangiopericytomas.58,59
Meningioma grading is another attractive topic. Previous studies have reported AUC values of 0.63–0.97 for the prediction of WHO grade or for the differentiation of low- and high-grade meningiomas, with the reflection of various types of machine learning algorithms and either single-parametric or multiparametric MRI, and the types of images they used.60–64 A few studies have addressed the prognosis of meningiomas.65,66 Morin et al. enrolled 314 patients with meningiomas and reasonably included many cases of higher grades of meningiomas (57% grade I, 35% grade II, and 8% grade III).67 They demonstrated an accuracy of up to 77% for overall survival using a random forest classifier incorporating both MRTA and non-MRTA features. Common limitations in the previous MRTA studies in meningioma are the retrospective design and the fact that meningioma datasets predominantly comprised grade I lesions (i.e., there was an imbalance in categories).59
Other than meningioma, the classification performance with texture analysis was reported in pediatric patients with brain tumors arising in the posterior cranial fossa.68–70 In an earlier study using first-order and GLCM features, Rodriguez Gutierrez et al. suggested that first-order features in ADC yielded the best tumor classification accuracy (78.9%–91.4%).71 Differentiation between pituitary adenomas and craniopharyngiomas is another interesting topic in MRTA. A recent study by Zhang et al. suggested that first-order skewness and GLCM contrast in T2WI, and GLCM energy in CE-T1WI may be significant predictors between these tumors.72
Challenges and Perspectives
As shown above, MRTA studies have shown promising results; however, several criticisms currently hinder the clinical use of MRTA. One major criticism of MRTA is that it is not hypothesis-driven.18 MRTA collects numerous mathematical computations and clinical parameters to determine what are significantly correlated using valid statistical methods; nevertheless, whether this particular significant correlation indicates a true causal relationship or a chance finding is impossible to know.
Researchers should note that the MRI acquisition process has considerable effects on MRTA measurements. Previous studies have shown that MRTA can be affected by many acquisition parameters of MRI, including scanner platforms, magnetic strength, number of coil elements and coil arrangements, spin-echo or gradient recalled-echo acquisition, TE, TR, flip angle, number of excitations, noise level, and image reconstruction algorithms.73–75
Another important criticism is the fact that MRTA requires explicit engineering of features (e.g., ROI selection, feature calculation, and selection of features). In contrast to conventional MRTA, the extraction of machine-learned features is becoming widely applicable with deep learning technologies.24,76 However, higher processing powers and a huge number of images accompanied by high-quality ground truth data are required for deep learning methods. Compared with deep learning, MRTA is less data-hungry. Additionally, the internal algorithms in deep learning may not always be apparent (i.e., black box), whereas MRTA features can be explained more easily.18
Finally, the common weaknesses of previous MRTA studies are their poor reproducibility and external validity (or generalizability) of the results. The previous MRTA studies have typically been conducted in a single institution with fewer than 100 cases, using a single-vendor MRI and unified MRI protocols. Homogeneity in subjects and analysis methods is of high importance in most of the scientific studies; however, because the MRI acquisition process has an impact on MRTA, such homogeneity unintentionally becomes a source of bias that might result in overfitting to the present study samples in MRTA. The risk for overfitting problems also increases when the models are developed especially with limited (and thus highly biased) data. Therefore, when conducting MRTA, sufficiently large datasets with unbiased images are desirable. In this context, multi-institutional image data using multivendor MRI machines will be advantageous in MRTA.77 Conversely, because of the variation in image processing, feature extraction, and feature selection, MRTA methods should be standardized and systematically reported. When developing machine learning models using MRTA features to correlate with some clinical outcomes, researchers should compare the performance in multiple models with different algorithms.78
To improve the robustness and validity of texture analysis, researchers in this field have proposed standardized schemes to conduct and evaluate texture analysis. One example is provided by the Image Biomarker Standardization Initiative (https://theibsi.github.io/); the purpose of which is to standardize the extraction of image biomarkers from acquired images for high-throughput quantitative image analysis (i.e., radiomics).21 Another example is the radiomics quality score, which has been introduced to evaluate the quality of radiomics studies.79 These two-way attempts will contribute to the further improvement of MRTA reliability.
Summary
Studies have shown promising results for the use of MRTA in brain tumors. Entropy, a feature that describes the randomness in intensity values of an image, has been shown to highlight the differences among multiple MRTA studies of brain tumors. Discussions for methodological standardization and quality checks for MRTA studies are maturing. Continuous efforts to improve the reliability and reproducibility of this method will accelerate the clinical use of texture analysis in brain MRI.
Funding Statement
This work was partially supported by JSPS KAKENHI Grant Number JP18K07629.
Footnotes
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the united states in 2012-2016. Neuro Oncol 2019; 21:v1–v100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol, 2016; 131:803–820. [DOI] [PubMed] [Google Scholar]
- 3.Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet 2012; 205:613–621. [DOI] [PubMed] [Google Scholar]
- 4.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res 2013; 19:764–772. [DOI] [PubMed] [Google Scholar]
- 5.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 2009; 27:4150–4154. [DOI] [PubMed] [Google Scholar]
- 6.Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol 2013; 31:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005; 352:987–996. [DOI] [PubMed] [Google Scholar]
- 8.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005; 352:997–1003. [DOI] [PubMed] [Google Scholar]
- 9.Pope WB, Sayre J, Perlina A, et al. MR imaging correlates of survival in patients with high-grade gliomas. AJNR Am J Neuroradiol 2005; 26:2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 10.Pope WB, Chen JH, Dong J, et al. Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology 2008; 249:268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moon WJ, Choi JW, Roh HG, et al. Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology 2012; 54:555–563. [DOI] [PubMed] [Google Scholar]
- 12.Smits M. Imaging of oligodendroglioma. Br J Radiol 2016; 89:20150857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kassner A, Thornhill RE. Texture analysis: a review of neurologic MR imaging applications. AJNR Am J Neuroradiol 2010; 31:809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Ma XL, Pu LT, et al. Prediction of overall survival and progression-free survival by the 18F-FDG PET/CT radiomic features in patients with primary gastric diffuse large B-cell lymphoma. Contrast Media Mol Imaging 2019; 2019:5963607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Cataldo S, Ficarra E. Mining textural knowledge in biological images: Applications, methods and trends. Comput Struct Biotechnol J 2017; 15:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davnall F, Yip CS, Ljungqvist G, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging 2012; 3:573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lubner MG, Smith AD, Sandrasegaran K, et al. CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics 2017; 37:1483–1503. [DOI] [PubMed] [Google Scholar]
- 18.Soni N, Priya S, Bathla G. Texture analysis in cerebral gliomas: a review of the literature. AJNR Am J Neuroradiol 2019; 40:928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganeshan B, Miles KA, Young RC, et al. Three-dimensional selective-scale texture analysis of computed tomography pulmonary angiograms. Invest Radiol 2008; 43:382–394. [DOI] [PubMed] [Google Scholar]
- 20.Sugahara T, Korogi Y, Kochi M, et al. Usefulness of diffusion-weighted MRI with echo-planar technique in the evaluation of cellularity in gliomas. J Magn Reson Imaging 1999; 9:53–60. [DOI] [PubMed] [Google Scholar]
- 21.Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020; 295:328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carré A, Klausner G, Edjlali M, et al. Standardization of brain MR images across machines and protocols: bridging the gap for MRI-based radiomics. Sci Rep 2020; 10:12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology 2018; 288:407–415. [DOI] [PubMed] [Google Scholar]
- 24.Avanzo M, Wei L, Stancanello J, et al. Machine and deep learning methods for radiomics. Med Phys 2020; 47:e185–e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erickson BJ, Korfiatis P, Akkus Z, et al. Machine learning for medical imaging. Radiographics 2017; 37:505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohli M, Prevedello LM, Filice RW, et al. Implementing machine learning in radiology practice and research. AJR Am J Roentgenol 2017; 208:754–760. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen BK, Hansen S, Laursen RJ, et al. Epidemiology of glioma: clinical characteristics, symptoms, and predictors of glioma patients grade I-IV in the the Danish Neuro-Oncology Registry. J Neurooncol 2017; 135:571–579. [DOI] [PubMed] [Google Scholar]
- 28.Alis D, Bagcilar O, Senli YD, et al. The diagnostic value of quantitative texture analysis of conventional MRI sequences using artificial neural networks in grading gliomas. Clin Radiol 2020; 75:351–357. [DOI] [PubMed] [Google Scholar]
- 29.Ryu YJ, Choi SH, Park SJ, et al. Glioma: application of whole-tumor texture analysis of diffusion-weighted imaging for the evaluation of tumor heterogeneity. PLoS One 2014; 9:e108335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skogen K, Schulz A, Dormagen JB, et al. Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol 2016; 85:824–829. [DOI] [PubMed] [Google Scholar]
- 31.Ditmer A, Zhang B, Shujaat T, et al. Diagnostic accuracy of MRI texture analysis for grading gliomas. J Neurooncol 2018; 140:583–589. [DOI] [PubMed] [Google Scholar]
- 32.Xie T, Chen X, Fang J, et al. Textural features of dynamic contrast-enhanced MRI derived model-free and model-based parameter maps in glioma grading. J Magn Reson Imaging 2018; 47:1099–1111. [DOI] [PubMed] [Google Scholar]
- 33.Su CQ, Lu SS, Han QY, et al. Intergrating conventional MRI, texture analysis of dynamic contrast-enhanced MRI, and susceptibility weighted imaging for glioma grading. Acta Radiol 2019; 60:777–787. [DOI] [PubMed] [Google Scholar]
- 34.Vamvakas A, Williams SC, Theodorou K, et al. Imaging biomarker analysis of advanced multiparametric MRI for glioma grading. Phys Med 2019; 60:188–198. [DOI] [PubMed] [Google Scholar]
- 35.Zhou H, Vallières M, Bai HX, Su C, et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol 2017; 19:862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahrami N, Hartman SJ, Chang YH, et al. Molecular classification of patients with grade II/III glioma using quantitative MRI characteristics. J Neurooncol 2018; 139:633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanazawa T, Minami Y, Jinzaki M, et al. Predictive markers for MGMT promoter methylation in glioblastomas. Neurosurg Rev 2019; 42:867–876. [DOI] [PubMed] [Google Scholar]
- 38.Lewis MA, Ganeshan B, Barnes A, et al. Filtration-histogram based magnetic resonance texture analysis (MRTA) for glioma IDH and 1p19q genotyping. Eur J Radiol 2019; 113:116–123. [DOI] [PubMed] [Google Scholar]
- 39.Zhang S, Chiang GC, Magge RS, et al. Texture analysis on conventional MRI images accurately predicts early malignant transformation of low-grade gliomas. Eur Radiol 2019; 29:2751–2759. [DOI] [PubMed] [Google Scholar]
- 40.Chaddad A, Desrosiers C, Toews M. Radiomic analysis of multi-contrast brain MRI for the prediction of survival in patients with glioblastoma multiforme. Annu Int Conf IEEE Eng Med Biol Soc 2016; 2016:4035–4038. [DOI] [PubMed] [Google Scholar]
- 41.Kickingereder P, Burth S, Wick A, et al. Radiomic Profiling of Glioblastoma: Identifying an Imaging Predictor of Patient Survival with Improved Performance over Established Clinical and Radiologic Risk Models. Radiology 2016; 280:880–889. [DOI] [PubMed] [Google Scholar]
- 42.Prasanna P, Patel J, Partovi S, et al. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur Radiol 2017; 27:4188–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brain Tumor Registry of Japan (2005-2008). Neurol Med Chir (Tokyo) 2017; 57(Suppl 1):9–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mouthuy N, Cosnard G, Abarca-Quinones J, et al. Multiparametric magnetic resonance imaging to differentiate high-grade gliomas and brain metastases. J Neuroradiol 2012; 39:301–307. [DOI] [PubMed] [Google Scholar]
- 45.Petrujkić K, Milošević N, Rajković N, et al. Computational quantitative MR image features - a potential useful tool in differentiating glioblastoma from solitary brain metastasis. Eur J Radiol 2019; 119:108634. [DOI] [PubMed] [Google Scholar]
- 46.Zhang G, Chen X, Zhang S, et al. Discrimination between solitary brain metastasis and glioblastoma multiforme by using ADC-based texture analysis: a comparison of two different ROI placements. Acad Radiol 2019; 26:1466–1472. [DOI] [PubMed] [Google Scholar]
- 47.Tateishi M, Nakaura T, Kitajima M, et al. An initial experience of machine learning based on multi-sequence texture parameters in magnetic resonance imaging to differentiate glioblastoma from brain metastases. J Neurol Sci 2020; 410:116514. [DOI] [PubMed] [Google Scholar]
- 48.Skogen K, Schulz A, Helseth E, et al. Texture analysis on diffusion tensor imaging: discriminating glioblastoma from single brain metastasis. Acta Radiol 2019; 60:356–366. [DOI] [PubMed] [Google Scholar]
- 49.Alcaide-Leon P, Dufort P, Geraldo AF, et al. Differentiation of enhancing glioma and primary central nervous system lymphoma by texture-based machine learning. AJNR Am J Neuroradiol 2017; 38:1145–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunimatsu A, Kunimatsu N, Kamiya K, et al. Comparison between glioblastoma and primary central nervous system lymphoma using MR image-based texture analysis. Magn Reson Med Sci 2018; 17:50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kunimatsu A, Kunimatsu N, Yasaka K, et al. Machine learning-based texture analysis of contrast-enhanced MR imaging to differentiate between glioblastoma and primary central nervous system lymphoma. Magn Reson Med Sci 2019; 18:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larroza A, Moratal D, Paredes-Sánchez A, et al. Support vector machine classification of brain metastasis and radiation necrosis based on texture analysis in MRI. J Magn Reson Imaging 2015; 42:1362–1368. [DOI] [PubMed] [Google Scholar]
- 53.Bhat AR, Wani MA, Kirmani AR, et al. Histological-subtypes and anatomical location correlated in meningeal brain tumors (meningiomas). J Neurosci Rural Pract 2014; 5:244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Georgiadis P, Cavouras D, Kalatzis I, et al. Enhancing the discrimination accuracy between metastases, gliomas and meningiomas on brain MRI by volumetric textural features and ensemble pattern recognition methods. Magn Reson Imaging 2009; 27:120–130. [DOI] [PubMed] [Google Scholar]
- 55.Shrot S, Salhov M, Dvorski N, et al. Application of MR morphologic, diffusion tensor, and perfusion imaging in the classification of brain tumors using machine learning scheme. Neuroradiology 2019; 61:757–765. [DOI] [PubMed] [Google Scholar]
- 56.Huang M, Yang W, Wu Y, et al. Content-based image retrieval using spatial layout information in brain tumor T1-weighted contrast-enhanced MR images. PLoS One 2014; 9:e102754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Svolos P, Tsolaki E, Kapsalaki E, et al. Investigating brain tumor differentiation with diffusion and perfusion metrics at 3T MRI using pattern recognition techniques. Magn Reson Imaging 2013; 31:1567–1577. [DOI] [PubMed] [Google Scholar]
- 58.Wei J, Li L, Han Y, et al. Accurate preoperative distinction of intracranial hemangiopericytoma from meningioma using a multihabitat and multisequence-based radiomics diagnostic technique. Front Oncol 2020; 10:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neromyliotis E, Kalamatianos T, Paschalis A, et al. Machine learning in meningioma MRI: Past to present. A narrative review. J Magn Reson Imaging 2022; 55:48–60. [DOI] [PubMed] [Google Scholar]
- 60.Yan PF, Yan L, Hu TT, et al. the potential value of preoperative MRI texture and shape analysis in grading meningiomas: a preliminary investigation. Transl Oncol 2017; 10:570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park YW, Oh J, You SC, et al. Radiomics and machine learning may accurately predict the grade and histological subtype in meningiomas using conventional and diffusion tensor imaging. Eur Radiol 2019; 29:4068–4076. [DOI] [PubMed] [Google Scholar]
- 62.Laukamp KR, Shakirin G, Baeßler B, et al. Accuracy of radiomics-based feature analysis on multiparametric magnetic resonance images for noninvasive meningioma grading. World Neurosurg 2019; 132:e366–e390. [DOI] [PubMed] [Google Scholar]
- 63.Lu Y, Liu L, Luan S, et al. The diagnostic value of texture analysis in predicting WHO grades of meningiomas based on ADC maps: an attempt using decision tree and decision forest. Eur Radiol 2019; 29:1318–1328. [DOI] [PubMed] [Google Scholar]
- 64.Chen C, Guo X, Wang J, et al. The diagnostic value of radiomics-based machine learning in predicting the grade of meningiomas using conventional magnetic resonance imaging: a preliminary study. Front Oncol 2019; 9:1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Yao K, Liu P, et al. A radiomics model for preoperative prediction of brain invasion in meningioma non-invasively based on MRI: A multicentre study. EBioMedicine 2020; 58:102933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gennatas ED, Wu A, Braunstein SE, et al. Preoperative and postoperative prediction of long-term meningioma outcomes. PLoS One 2018; 13:e0204161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morin O, Chen WC, Nassiri F, et al. Integrated models incorporating radiologic and radiomic features predict meningioma grade, local failure, and overall survival. Neurooncol Adv 2019; 1:vdz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orphanidou-Vlachou E, Vlachos N, Davies NP, et al. Texture analysis of T1 - and T2 -weighted MR images and use of probabilistic neural network to discriminate posterior fossa tumours in children. NMR Biomed 2014; 27:632–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fetit AE, Novak J, Peet AC, et al. Three-dimensional textural features of conventional MRI improve diagnostic classification of childhood brain tumours. NMR Biomed 2015; 28:1174–1184. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Chen C, Tian Z, et al. The diagnostic value of MRI-based texture analysis in discrimination of tumors located in posterior fossa: a preliminary study. Front Neurosci 2019; 13:1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodriguez Gutierrez D, Awwad A, Meijer L, et al. Metrics and textural features of MRI diffusion to improve classification of pediatric posterior fossa tumors. AJNR Am J Neuroradiol 2014; 35:1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, Chen C, Tian Z, et al. Discrimination between pituitary adenoma and craniopharyngioma using MRI-based image features and texture features. Jpn J Radiol 2020; 38:1125–1134. [DOI] [PubMed] [Google Scholar]
- 73.Yang F, Dogan N, Stoyanova R, et al. Evaluation of radiomic texture feature error due to MRI acquisition and reconstruction: A simulation study utilizing ground truth. Phys Med 2018; 50:26–36. [DOI] [PubMed] [Google Scholar]
- 74.Ford J, Dogan N, Young L, et al. Quantitative radiomics: impact of pulse sequence parameter selection on MRI-based textural features of the brain. Contrast Media Mol Imaging 2018; 2018:1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buch K, Kuno H, Qureshi MM, et al. Quantitative variations in texture analysis features dependent on MRI scanning parameters: a phantom model. J Appl Clin Med Phys 2018; 19:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yasaka K, Akai H, Kunimatsu A, et al. Deep learning with convolutional neural network in radiology. Jpn J Radiol 2018; 36:257–272. [DOI] [PubMed] [Google Scholar]
- 77.Bluemke DA, Moy L, Bredella MA, et al. Assessing radiology research on artificial intelligence: a brief guide for authors, reviewers, and readers-from the radiology editorial board. Radiology 2020; 294:487–489. [DOI] [PubMed] [Google Scholar]
- 78.Gregory J, Welliver S, Chong J. Top 10 reviewer critiques of radiology Artificial Intelligence (AI) articles: qualitative thematic analysis of reviewer critiques of machine learning/deep learning manuscripts submitted to JMRI. J Magn Reson Imaging 2020; 52:248–254. [DOI] [PubMed] [Google Scholar]
- 79.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14:749–762. [DOI] [PubMed] [Google Scholar]