Abstract
Proper germ cell sex determination in Caenorhabditis nematodes requires a network of RNA-binding proteins (RBPs) and their target mRNAs. In some species, changes in this network enabled limited XX spermatogenesis, and thus self-fertility. In C. elegans, one of these selfing species, the global sex-determining gene tra-2 is regulated in germ cells by a conserved RBP, GLD-1, via the 3’ untranslated region (3’UTR) of its transcript. A C. elegans-specific GLD-1 cofactor, FOG-2, is also required for hermaphrodite sperm fate, but how it modifies GLD-1 function is unknown. Germline feminization in gld-1 and fog-2 null mutants has been interpreted as due to cell-autonomous elevation of TRA-2 translation. Consistent with the proposed role of FOG-2 in translational control, the abundance of nearly all GLD-1 target mRNAs (including tra-2) is unchanged in fog-2 mutants. Epitope tagging reveals abundant TRA-2 expression in somatic tissues, but an undetectably low level in wild-type germ cells. Loss of gld-1 function elevates germline TRA-2 expression to detectable levels, but loss of fog-2 function does not. A simple quantitative model of tra-2 activity constrained by these results can successfully sort genotypes into normal or feminized groups. Surprisingly, fog-2 and gld-1 activity enable the sperm fate even when GLD-1 cannot bind to the tra-2 3’ UTR. This suggests the GLD-1-FOG-2 complex regulates uncharacterized sites within tra-2, or other mRNA targets. Finally, we quantify the RNA-binding capacities of dominant missense alleles of GLD-1 that act genetically as “hyper-repressors” of tra-2 activity. These variants bind RNA more weakly in vitro than does wild-type GLD-1. These results indicate that gld-1 and fog-2 regulate germline sex via multiple interactions, and that our understanding of the control and evolution of germ cell sex determination in the C. elegans hermaphrodite is far from complete.
Introduction
C. elegans hermaphrodites are essentially modified females, producing first sperm, then oocytes. These self-sperm allow uniparental reproduction, which has evolved at least three times in Caenorhabdtis. Selfing is likely to be adaptive in the patchy, ephemeral environments in which these animals reproduce (Kiontke et al., 2011). How did XX females evolve the ability to produce sperm? Decades of genetic and molecular analyses have identified a network of gene products required to promote XX spermatogenesis in C. elegans. Central to this network is tra-2, a global sex determiner that was first identified by Hodkgin and Brenner (1977) by its Transformer (Tra) null phenotype. In XX tra-2 loss-of-function (tra-2(lf)) mutants, both the soma and germ cells of hermaphrodites adopt the male form. tra-2 gain-of-function alleles also exist that have a feminization of germline (Fog) phenotype: XX animals make oocytes, but no sperm (Doniach, 1986; Goodwin et al., 1993). All such tra-2(gf) alleles map to a direct repeat element (DRE) in the 3’UTR of tra-2, and have little or no effect on male spermatogenesis. These data implicate the presence of a post-transcriptional negative regulator of tra-2 in XX hermaphrodites that is essential for spermatogenesis (Puoti et al., 1997).
Like the tra-2 DREs, gld-1 is required for spermatogenesis in XX hermaphrodites, but not males, although its phenotypes are more pleiotropic (Francis et al., 1995a; Francis et al., 1995b). In addition to loss of XX sperm, gld-1(null) mutants have germ cell tumors of oocyte character, caused by meiotic failure and resumption of mitosis. GLD-1 is a STAR family RBP that is most abundant in early meiotic cells (Jones et al., 1996). It was identified in a molecular screen for its ability to bind the DREs (Jan et al., 1999), indicating it is the germline factor that binds the tra-2 3’UTR. Subsequent studies have confirmed the association of GLD-1 with the tra-2 mRNA (Jungkamp et al., 2011; Wright et al., 2011) and revealed that GLD-1 likely binds its targets as a homodimer (Ryder et al., 2004).
A third component that is essential specifically for XX spermatogenesis is encoded by fog-2. Similar to tra-2 gain-of-function mutants, fog-2 null XX mutants are abnormal only in their lack of sperm, while spermatogenesis of XO male mutants is unaffected (Schedl and Kimble, 1988). This germline-feminizing effect requires tra-2 function, suggesting fog-2 acts upstream of tra-2. The fog-2 gene was eventually cloned by virtue of a physical interaction between FOG-2 and GLD-1 (Clifford et al., 2000). FOG-2 possesses both an F-box and a novel GLD-1-binding domain, the latter distinguishing it from other members of the large F-box family of paralogs (Nayak et al., 2005). F-box proteins often link E3 ubiquitin ligases to their target proteins (Genschik et al., 2013; Ho et al., 2008). Shimada et al. (2006) found that a non-essential proteasome component (RPN-10) is required for XX sperm and tra-2 repression body-wide. These results all are consistent with a model in which FOG-2 and GLD-1 evolved in the C. elegans lineage to cooperate to down-regulate tra-2 activity (Figure 1), and that ubiquitination and/or the proteasome are somehow involved.
Figure 1. Evolutionary history of gld-1 and fog-2 in Caenorhabditis nematodes.

A phylogeny (modified from Kiontke et al., 2011) showing the convergent evolution of selfing in the Elegans group species of Caenorhabditis. Derived aspects of tra-2 regulation in C. elegans are depicted in cartoon form. The GLD-1-tra-2 mRNA regulation was strengthened in XX hermaphrodites by 1) multimerization of GLD-1 binding elements in the tra-2 mRNA’s 3’-untranslated region, or UTR (Beadell et al., 2011), and 2) recruiting the novel cofactor FOG-2 (Nayak et al., 2005). The convergently evolved hermaphrodite C. briggsae lacks these two features, and uses GLD-1 to opposite effect in germline sex determination (Beadell et al., 2011; Nayak et al., 2005).
More recent work has revealed insights into the mechanisms of GLD-1 function, with implications for its role in promoting hermaphrodite spermatogenesis. Ryder et al. (2004) first defined the consensus GLD-1 binding site in RNA targets. The extent of an mRNA’s association with GLD-1 is determined by the strength and number of these (and similar) sites (Galarneau and Richard, 2009; Wright et al., 2011), with most residing in 3’ UTR regions. The PAR-CLIP method, which identifies specific sites bound by RNA-binding proteins in native transcripts (Hafner et al., 2010), has been applied to C. elegans GLD-1 (Jungkamp et al., 2011). This revealed that GLD-1 binds near the start codon of some transcripts, suggesting that it may regulate interaction with the translation initiation machinery. While translation of GLD-1 target mRNAs is often repressed, this repression can also be accompanied by protection from nonsense mediated decay (Lee and Schedl, 2004) and stabilization (Scheckel et al., 2012). Thus, whether loss of gld-1 function has a net positive or negative effect on target gene activity may vary from target to target.
GLD-1 protein is localized to the distal germ line of oogenic XX gonads, with greatest expression concentrated in the region marking entry into meiosis (Jones et al., 1996). GLD-1 expression is repressed in the distal germline stem cell niche by translational control by the FBF PUF family RBPs (Crittenden et al., 2002; Suh et al., 2009) and by glp-1/Notch signaling (Brenner and Schedl, 2016), such that cells upregulate GLD-1 as they leave the niche. The defects seen after loss of gld-1 function (Beadell et al., 2011; Francis et al., 1995a; Francis et al., 1995b; Nayak et al., 2005) and the identity of mRNAs bound by GLD-1 (Beadell et al., 2011; Lee and Schedl, 2001; Ryder et al., 2004; Wright et al., 2011) in multiple Caenorhabditis species suggest that the ancestral role of gld-1 was in the meiotic progression and differentiation of oocytes.
Comparative studies among Caenorhabditis species have revealed that sex determination mechanisms have rapidly evolved. C. briggsae, a convergently evolved hermaphroditic species, has a tra-2 ortholog with a conserved female-promoting function (Kuwabara,1996; Kelleher et al., 2008). However, while gld-1 is present in C. briggsae, its role in self fertility is distinct. RNAi knockdown of gld-1 in C. briggsae results in a sperm-only (Mog) phenotype, unlike the feminization seen in C. elegans (Nayak et al., 2005). These RNAi results were confirmed with Cbr-gld-1 loss-of-function mutants by Beadell et al. (2011). Despite these opposite loss-of-function sex phenotypes, a Cbr-gld-1 transgene can fully rescue the C. elegans gld-1 null mutant. This suggests a pivotal role for changes in target mRNAs in the evolution of GLD-1 function. Consistent with this, the C. briggsae tra-2 3’ UTR lacks the cluster of GLD-1-binding sites that are essential for proper C. elegans tra-2 regulation (Beadell et al., 2011).
The tra-2 ortholog of the male/female C. remanei has a conserved role in female sexual fate, and a GLD-1-like factor binds to the C. remanei tra-2 3’ UTR (Haag and Kimble, 2000). This suggests that key elements of the C. elegans system of germline sex regulation predate the evolution of selfing. However, despite large families of F-box proteins in all Caenorhabditis (Nayak et al., 2005; Thomas, 2006), only C. elegans has a fog-2 paralog. Interestingly, C. briggsae evolved selfing independently (Kiontke et al., 2011; Kiontke et al., 2004), yet requires its own species-restricted F-box protein, SHE-1, for spermatogenesis in hermaphrodites (Guo et al., 2009).
A number of important questions remain unanswered about how XX spermatogenesis is regulated in C. elegans. Many of these relate to tra-2, gld-1, fog-2 and the nature of their interactions. Does FOG-2 impact the steady-state transcript levels of GLD-1 targets, including tra-2? Can the inferred repression of germline TRA-2 levels by the combined action of GLD-1 and FOG-2 be directly visualized? Can the function of fog-2 and gld-1 be ascribed wholly to regulation of tra-2 via its 3’ UTR? What impact do the missense changes in dominant Mog alleles of gld-1, which act like “super repressors,” have on GLD-1 function? Below we describe experiments that address these questions, some using tools not available when the genetic interactions between tra-2, gld-1, and fog-2 were first characterized.
Materials and Methods
C. elegans strains
Strains used in this study:
N2 (reference wild-type)
JK574 fog-2(q71)V
JK992 tra-2(e2020)II; unc-24(e138) fem-3(q95) dpy-20(e1282)IV
JK3107 fbf-1(ok91) fbf-2(704)/mIn1[mIs14 dpy-10(e128)]II
TX189 teIs1[oma-1p::oma-1::GFP + unc-119(+)].
BS3156 unc-13(e51) gld-1(q485)/hT2[dpy-18(h662)]I; +/hT2[bli-4(e937)]III
CP139 and CP141 nm75[tra-2∷HA]II (see below)
CP153 nm75[tra-2∷HA]II; fog-2(q71)V was generated by crossing JK574 males with CP139 or CP141 hermaphrodites. Homozygosity of the HA-tagged endogenous tra-2 allele was confirmed by PCR amplicon band shift; and of the q71 point mutation by PCR and sequencing. In the presence of nm75, the Fog phenotype of q71 is incompletely penetrant.
CP163 unc-13(e51) gld-1(q485)/+I; nm75[tra-2∷HA]II was generated by crossing N2 males with CP141 hermaphrodites, then using F1 males to cross with phenotypically wild-type BS3156 hermaphrodites. Homozygosity of the HA-tagged tra-2 was confirmed by PCR, and of q485 by the gonad tumor and linked Unc phenotypes.
Triple mutant tra-2(e2020) II; fem-3(q95) I; fog-2(q71) V were generated by using mIn1 as a marker on the second chromosome. Dpy mIn1II; fog-2(q71) V males were crossed with JK992 tra-2(e2020) II; unc-24(e138) fem-3(q95) dpy-20(e1282) IV, to yield wild-type like F1s. Non-green, Dpy Unc F2s F2 progeny are tra-2(e2020); fem-3(q95), but of uncertain fog-2 genotype. After phenotyping under a compound microscope, a subset were recovered and a PCR amplicon (produced with primers EH42 5’- CTTCACATGAAGCTTCAAAGC-3’ and EH43 5’-TTTGAGGGGAAAAATGTCCAATT-3’) was sequenced to genotype the point mutation of fog-2(q71).
CP165 unc-24(e138) fem-3(q95) dpy-20(e1282)IV; fog-2(q71) V was created by crossing JK992 hermaphrodites with fog-2(q71) males, and singling many Dpy Unc F2 to create known fem-3(q95) founders. These F2 founders were allowed to lay eggs, and then PCR genotyped to eliminate families carrying tra-2(e2020). Finally, fog-2 genotypes of fem-3(q95); tra-2(+/+) families were determined by PCR and sequencing as described above. Because fog-2(q71) enhances the fertility of fem-3(q95) at permissive temperature (Schedl and Kimble, 1988), q71 homozygotes were over-represented among healthy lines.
Most strains were kept at 20°C on NGM agar plates, except for those carrying fem-3(q95), which is temperature-sensitive. Such strains were held at 15°C for propagation, and assayed for germline feminization by shifting embryos to 25°C and growing them to adulthood at this restrictive temperature.
RNAseq comparisons
Nematodes were synchronized by allowing purified embryos to hatch in food-free M9 salts, and then moving arrested hatchlings to standard NGM plate media with E. coli OP50 bacterial food. Three independent samples of 100 N2 hermaphrodites and fog-2(q71) females were harvested on their first day of adulthood. To avoid potential changes in the oocyte transcriptome related to suppressed ovulation (Jud et al., 2008), only fog-2 XX females that had successfully mated and were gravid were chosen. RNA was extracted in Trizol (Ambion) and purified as previously described (Thomas et al., 2012). cDNA was synthesized using either a 3’-anchored oligo-dT primer or random hexamers via the TotalScript kit (Illumina). As part of ongoing product development work, hexamer-primed libraries were generated using one of two alternative formulations (“standard” or “optimized”) of the reaction buffer for synthesis of the first cDNA strand. The combination of buffer and sequencing run that produced the greatest number of non-rRNA mapped reads was used for subsequent analyses, resulting in use of “optimized” libraries for all but replicate D of the fog-2 samples. Libraries used unique barcodes to allow multiplexing.
Sequencing of the cDNA libraries was performed on an Illumina HiSeq instrument. 30–50 million pairs of 51-nucleotide reads were generated for each library. For the oligo-dT-primed libraries nearly all reads were derived from mRNAs (not shown). For hexamer-primed libraries, roughly 85% of reads were derived from rRNA. After de-multiplexing, reads were mapped to annotated genes of the N2 reference genome (release CE10) using TopHat (Trapnell et al., 2009). Expression levels and statistical tests for significant differences between genotypes was assesses using the program cuffdiff, within the Cufflinks package (Trapnell et al., 2010). Results reported here were obtained using the per-condition dispersion option. Expression measurements, statistical tests, and software details for this study are available from the National Center for Bioinformatics (NCBI)’s Gene Expression Omnibus via accession number GSE110998.
CRISPR genomic modification
Hemagglutinin (HA) epitope tag was knocked into the endogenous tra-2 locus by CRISPR-Cas9 genomic editing. The 9 codons encoding the HA tag was inserted so as to add epitope near the C-terminus of TRA-2, 4 codons before the stop codon. A single guide RNA with the targeting sequence GAAGGCGACCTATCAGACCCAG was in vitro transcribed using the SP6 Megascript system (Invitrogen-Ambion). Oligonucleotide-based transcription templates were designed and synthesized according to the method of Gagnon et al. (2014), with the exception of using Taq DNA polymerase for the annealed oligonucleotide fill-in reactions. The repair oligo sequence (Ultramer, IDT) used for homology-directed repair was 5’ gatgaagcacgggaaggcgacctatcagacTACCCATACGACGTTCCAGACTACGCCccagaggtttaaaatgtctgtttcctttttcag 3’, where uppercase sequences indicate the HA-encoding insertion, and underlined nucleotides indicate the Cas9 guide RNA sequence.
Well-fed young adult worms were injected with the mixture of 800 ng/μl nuclear localization sequence-conjugated Cas9 protein (PNABio), 900 ng/μl sgRNA, and 60nM repair oligo. Edited heterozygous F1 were identified by single worm PCR among the offspring within 8–24 hours time window after injection, using primers flanking the insertion region, looking for a PCR product shift from 203nt to 230nt.
gld-1 RNA interference
An in vitro transcription template for production of gld-1 dsRNA was synthesized by PCR using the forward primer 5’-TAATACGACTCACTATAGGGTGGATGCAAGATTATGGTCCGAGG-3’ and reverse primer 5’-TAATACGACTCACTATAGGGGACCAGGGAATGAGTTGACAAACG-3’ (T7 promoter sequence underlined). The PCR product was purified with a QIAquick PCR purification kit (Qiagen) and 800 ng was used for in vitro transcription using a Megascript T7 kit (Ambion) following the factory protocol. Young adult JK992 and N2 hermaphrodites were injected in the germline or gut at a concentration of 2000 ng/μl, and allowed to recover for 6 hours at 15°C. They were then moved to new plates and allowed to lay for 24 hours, after which embryos were shifted to 25°C or left at 15°C as controls.
Immunoblots and Immunohistochemistry
For immunoblots, 200 worms of each strain were boiled in 2x Laemmli Loading buffer (Bio-Rad), and the entire lysate was fractionated on a 12% SDS-polyacrylamide gel and transferred to nitrocellulose membranes using standard conditions (Harlow and Lane, 1999). Immunoblots used the rat anti-HA monoclonal antibody 3F10 (Roche) 1:100 as the 1st antibody, followed by goat anti-rat IgG-HRP sc-2006 (Santa Cruz) 1:500 as the 2nd antibody. Actin was detected by anti-actin antibody C4 1:1000 (sc-47778), followed by bovine anti-mouse HRP sc-2375 (Santa Cruz) 1:5000. HRP activity was detected on blots using chemiluminescence (SuperSignal West Pico substrate, Thermo Scientific).
For immunohistochemistry, animals were dissected in 0.25 levamisole in PBS, either in 4-well polycarbonate tissue culture trays (for L4 larvae and adults) or on poly-L-lysine-subbed microscope slides (for L3 larvae). Dissected worms were fixed in 3% formaldehyde in 0.1M K2HPO4, followed by dry methanol. Samples were rehydrated and stained with rat anti-HA 3F10 (Roche) 1:100, followed by Alexa Fluor goat anti-rat 555 (Molecular Probes Invitrogen) 1:100, both in PBS with 0.1% Tween. Nuclei were stained with 100 μg/ml Hoechst 33258 just prior to imaging. Images for L4 and adult samples were captured with Zen software on a Zeiss LSM 710 confocal microscope, at 6% laser power and default (1.0) gain, using 10x or 40x oil objectives. Images for L3 larvae were captured with a Leica SP5X confocal microscope, using a 40x oil objective with 3x digital zoom. The 405 nm laser was at 15% power with gain of 648. 555 nm excitation was achieved with a white laser source at 70% power, with the 554nm line at 60% power and 943 gain.
To quantify and compare germline TRA-2:HA expression, a region of at least 2000 pixels from a confocal section that included the distal tip cell was quantified using the histogram tools of Zen (for L4 and adult samples) or ImageJ (for L3 samples) image analysis packages. Samples compared were dissected and stained in parallel using identical reagents on the same day, and imaged in the same session using identical laser and image capture settings.
GLD-1-RNA binding assays
GLD-1 expression plasmids:
The G248R and G250R mutant derivatives of pHMTc-GLD(135–336) (Ryder et al., 2004) were produced by QuikChange mutagenesis (Agilent Genomics, Santa Clara, CA) using primers 5’-CCAGA TTATG GTCCG AAGAA AGGGA TCAAT GC-3’ and 5’-GCATT GATCC CTTTC TTCGG ACCAT AATCT TG-3’ or primers 5’-GGTCC GAGGA AAGAG ATCAA TGCG GG-3’ and 5’-CCCGC ATTGA TCTCT TTCCT CGGAC C-3’, respectively. Sequencing was used to validate the presence of the desired mutations.
Protein purification:
The plasmids encoding pHMTc-GLD(135–336) or mutations thereof were transformed into Escherichia coli strain BL21(DE3). All three maltose-binding protein-GLD-1 fusions were purified as previously described (Ryder et al., 2004). Briefly, bacterial cultures were induced at mid log phase for three hours using 1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG). After harvesting, the cell pellets were lysed, then soluble recombinant protein was purified across an amylose affinity column (New England Biolabs) followed by HiTrap Q and HiTrap S ion exchange columns (GE Healthcare, Wauwatosa, WI) at 4°C. Relative purity of the protein was assessed by SDS-PAGE, and the concentration estimated from the absorption of 280 nm UV light using extinction coefficients calculated from the sequence composition with Expasy Protparam (Wilkins et al., 1999). Purified proteins were dialyzed into storage buffer (50 mM Tris pH 8.0, 20 mM NaCl, 2 mM DTT) and kept at 4°C until use.
Binding assays:
Electrophoretic mobility shift assays using purified recombinant MBP-tagged GLD-1 and mutant variants were performed essentially as described (Farley and Ryder, 2012). The target RNA sequence (RNA: 5’-UUUUU CUUAU UCUAG ACUAA UAUUG UAAGC U-3’-Fl), comprising a fragment of the glp-1 3’UTR previously shown to bind to GLD-1 (Farley and Ryder, 2012), was chemically synthesized and labeled at the 3énd with fluorescein amidite (FAM) by Integrated DNA Technologies (IDT, Coralville, IA). Varying concentrations of purified GLD-1, GLD-1 (G248R) and GLD-1 (G250R) were equilibrated with 3 nM of labeled RNA in equilibration buffer (0.01% IGEPAL, 0.01 mg/ml tRNA, 50 mM Tris, pH 8.0, 100 mM NaCl, 2mM DTT) for 3 hours. After equilibration, bromocresol loading dye was added, the reaction mixture was loaded onto a 5% native polyacrylamide gel in 1X TBE, and allowed to run for 2 hours at 120 volts. The gels were quantified by determining the pixel intensity of the RNA bound species and the total of all RNA species to give the fraction bound of RNA relative to the background using Image Gauge (Fujifilm, Tokyo, Japan). The graph of fraction bound against the protein concentration was then fit to the Hill equation to determine the apparent binding affinity and the Hill coefficient.
Results
Impact of fog-2 on the transcriptome
If GLD-1 regulates protein expression at the translational level, and FOG-2 functions via its association with GLD-1, then loss of FOG-2 may not affect steady-state levels of mRNAs targeted by GLD-1. Alternatively, the demonstrated impact of gld-1 on target stability noted above would predict that some targets may show altered transcript levels. We therefore used RNAseq to compare the abundance of mRNAs extracted from gravid young adult N2 and fog-2(q71) XX worms, which are genetic nulls for fog-2 function (Schedl and Kimble, 1988). Because GLD-1 binding sites can regulate the extent of tra-2 mRNA polyadenylation (Thompson et al., 2000), and because deadenylation can enhance mRNA turnover (Alonso, 2012), separate sequencing libraries produced from cDNA synthesized with oligo-dT and random hexamer primers were generated for each sample.
In the oligo-dT-primed experiment, we found 235 of 19,291 transcripts (1.2%) were significantly differentially expressed (FDR = 0.05) between the wild-type and fog-2(q71) samples (Figure 2, Suppl. Table 1). 14 of these encode paralogous major sperm proteins (MSPs), sperm-specific cytoskeletal and signaling proteins (Burke and Ward, 1983; Ward et al., 1988). All 14 showed roughly four-fold reduced expression in fog-2(q71), as did the sperm-specific family S gene sss-1 and the msp-related msd-4. The differentially expressed msp genes represent half of the total (Smith, 2014), far more than the one case expected by chance. Absent from the set of differentially expressed transcripts was tra-2 (mean FPKM values in wild-type and fog-2(q71) of 48.9 and 49.6, respectively). Furthermore, no known regulators of sex-determination were differentially expressed, with the exception of fog-2 itself. The four-fold reduction of fog-2 mRNA abundance in fog-2(q71) homozygotes indicates that the q71 mutation, a premature stop (Clifford et al. 2000), may induce nonsense-mediated decay.
Figure 2. Steady-state mRNA levels in wild-type vs fog-2(q71) mutants.
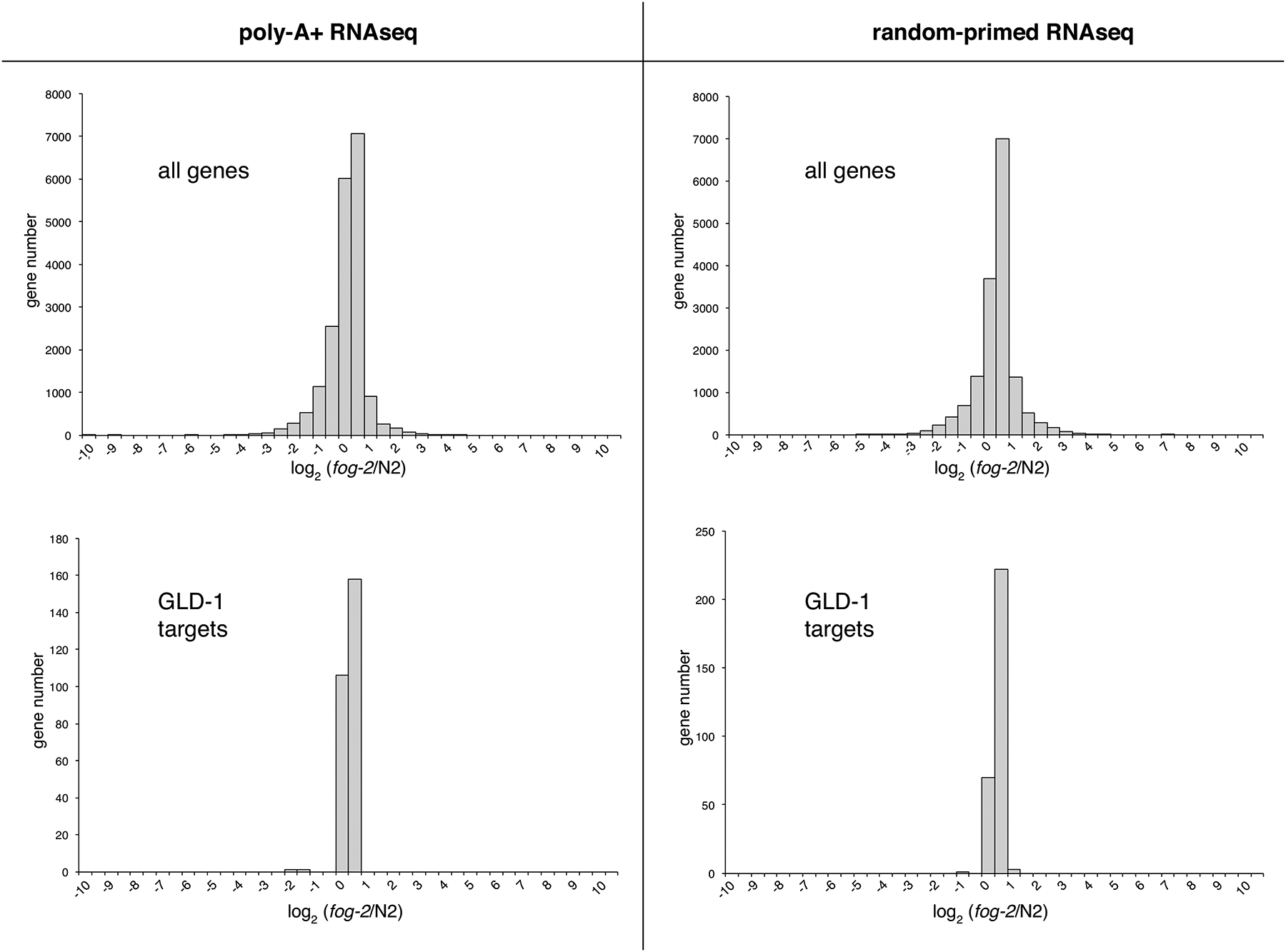
Top panels: Distributions of the fog-2/wild-type (N2) expression ratios for all transcripts consistently detected by RNAseq in young adult XX transcriptomes. Sequencing libraries were primed with oligo-dT (left) or random hexamers (right). Lower panels: Distributions of the fog-2/wild-type (N2) expression ratios for the 302 GLD-1 targets identified by both Wright et al. (2011) and Jungkamp et al. (2011). Data summarized from Supplemental Dataset 1.
In the RNAseq libraries produced using random hexamer primers (Figure 2, Supplemental Table 1), the burden of rRNA fragments reduced the number of genes whose expression could be consistently quantified to less than 10,000. Using the same significance threshold of FDR=0.05, 81 transcripts were judged as differentially expressed (0.08%). Reminiscent of the dT-primed data, the most significantly differentially expressed transcript was fog-2. In contrast with the oligo-dT-primed data, however, no msp paralogs were judged to significantly differentially expressed. However, for all 14 of the msp genes whose expression was detected in both genotypes in the random-primed libraries, mean expression was lower in the fog-2 samples. This is highly unlikely to occur by chance given the overall symmetrical distribution of fog-2/N2 expression ratios (sign test P=.00006), and is therefore not a clear contradiction of the result from the dT-primed libraries.
Because FOG-2 lacks an RNA-binding domain, any direct role it may have in tra-2 regulation likely relies on its association with GLD-1 (Clifford et al., 2000). We therefore examined the abundance of a set of 304 GLD-1-associated transcripts that were identified in two independent studies (Jungkamp et al., 2011; Wright et al., 2011). In both dT-primed and random-primed datasets, GLD-1 targets appear typical of the transcriptome as a whole, and are not enriched in differentially expressed transcripts (Figure 2). For the dT-primed case, two GLD-1 targets were judged to be differentially expressed (FDR=0.05), gipc-1 and R09E10.6. This small number is similar to the number expected by chance (304 GLD-1 targets × 1.2% overall differential expression = 3.6). gipc-1 encodes a PDZ domain-containing protein implicated in clearing of protein aggregates (Hamamichi et al., 2008), but with no known germ cell function. R09E10.6 is a nematode-specific protein enriched in germ cells that may function in mitochondria (Sakamoto et al., 2012). In the random-primed dataset, none of the GLD-1 targets were judged to be differentially expressed. Overall, it appears that loss of fog-2 has only subtle impacts on the transcriptome that are not biased towards GLD-1 targets or sex-determination genes. The similar results of the two libraries also indicate that FOG-2 does not regulate GLD-1 target polyadenylation.
Expression of TRA-2 in wild-type and mutant contexts
To examine the regulation of TRA-2, we created an epitope-tagged allele. The two predicted TRA-2 isoforms are a full-length (~170 kDa) membrane protein (TRA-2A), transcribed from a 4.7kb mRNA, and a shorter TRA-2B protein (Fig. 3a). TRA-2B is translated directly in germ cells from a 1.8kb mRNA (Kuwabara et al., 1998), resulting in an ~55 kDa cellular protein. TRA-2B also corresponds to the intracellular domain of TRA-2A (i.e. “TRA-2ic”) after TRA-3 cleavage (Sokol and Kuwabara, 2000). To detect both TRA-2 isoforms, we added the small (9 amino acid) hemagluttinin (HA) epitope tag to endogenous TRA-2 using CRISPR genomic editing, inserting HA four amino acids before the stop codon. The resulting tra-2:HA(nm75) hermaphrodites are self-fertile and appear normal. TRA-2:HA is detected in larval and adult hermaphrodites (Fig. 3b), but not in males, consistent with their greatly reduced transcript levels (Okkema and Kimble, 1991). In anti-HA immunoblots, only the 55 kDa TRA-2B/ic isoform was detected.
Figure 3. Impact of tra-2 repressors on overall TRA-2B expression.
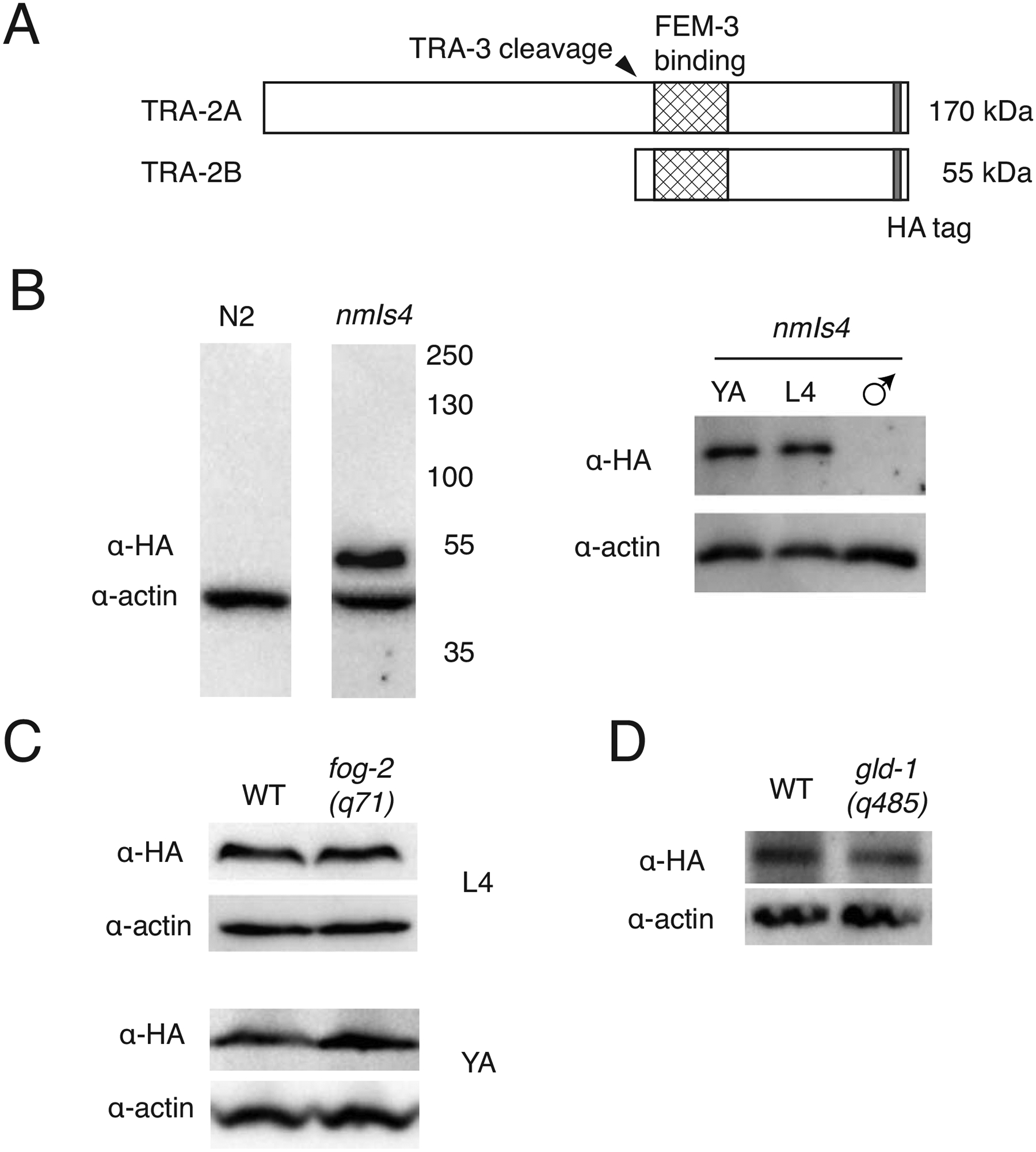
A. Schematic of the two TRA-2 protein products, TRA-2A (a membrane protein) and TRA-2B (which lacks transmembrane domains). The HA epitope marks both forms near the carboxy terminus. B. Anti-HA antibodies detect TRA-2B:HA in lysates from XX tra-2(nm75) animals, but not in control N2 XX (left) nor XO nm75 males (right). The full-length membrane protein TRA-2A:HA was not detected in either sex. C. TRA-2B:HA expression compared between wild-type XX hermaphrodites and fog-2(q71) XX mutant females at L4 (top) and young adult (bottom) stages. D. Comparison of TRA-2B:HA expression in lysates from adult XX wild-type (WT) and gld-1(q485) null mutants.
To reveal impacts of germline repressors of tra-2 on TRA-2:HA expression, we first crossed the tra-2:HA(nm75) allele into the null fog-2(q71) allele (Schedl and Kimble, 1988). Hermaphrodite fog-2(q71) mutants do not produce sperm (the Fog, or feminization of germ line, phenotype), while males are unaffected. Surprisingly, 35% (N=40) of tra-2:HA(nm75); fog-2(q71) double mutants were self-fertile, and the strain can be propagated without any males present. Thus, the HA-tagged TRA-2 is a partial suppressor of fog-2(q71), indicating that insertion of even the small (9-residue) HA tag encoded by nm75 produces a mild reduction-of-function variant.
Because fog-2 and gld-1 are implicated in the suppression of tra-2 translation, we hypothesized that TRA-2:HA expression would be increased in both fog-2 and gld-1 null mutant backgrounds. However, anti-HA immunoblots (Fig. 3c, d) revealed no elevation of TRA-2:HA signal in animals homozygous for null mutations in either fog-2 (q71) or gld-1(q485). Thus, at the whole-worm level, TRA-2 expression was not detectably increased when fog-2 or gld-1 activities are absent.
A quantitative model of tra-2 regulation in the hermaphrodite germ line
TRA-2 is a dose-sensitive protein (Kuwabara and Kimble, 1995), and both gain-of-function and loss-of-function tra-2 alleles have defects in reproduction (Doniach, 1986; Hodgkin and Brenner, 1977). The precise level of tra-2 activity thus appears to be crucial for germline sex determination (Puoti et al., 1997). In keeping with this, the tra-2(nm75) HA-tagged allele suppresses the fog-2(q71) Fog phenotype, yet does not reduce tra-2 function enough to overtly masculinize an otherwise wild-type hermaphrodite. Similarly, one copy of the tra-2(e1095) strong loss-of-function allele also suppresses fog-2(q71) (Schedl and Kimble, 1988). tra-2 null mutations are completely recessive (Hodgkin and Brenner, 1977), indicating that 50% of usual activity is adequate for development of a hermaphrodite germline. These observations suggest there is a range of tra-2 activities that can allow the sperm-then-oocyte pattern. The multiple factors involved justify formalization of a quantitative model.
If the haploid dose of unmodified germline tra-2 activity is defined as T, then the operational tra-2 activity could be determined by functional copy number (0, 1, or 2) reduced by repressive factors (i.e. between 0 and 1), imparted by fog-2 (F), gld-1 (G), or the HA epitope tag (H). Similarly, the tra-2 3’ UTR deletion allele, e2020 (Doniach, 1986; Goodwin et al., 1993), augments T by a factor E (where E>1). Table 1 presents expressions for various situations using these terms, assuming that a single copy of a negative regulator is sufficient for full function (i.e. they are dominant), that the activity of the two tra-2 alleles is regulated independently and additively, and that E, F, G, and H are independent of each other. Justification for this last assumption comes from data presented in Tables 2 and 3, but is at best an approximation. Despite its limitations, the above model supports some inferences. For example, the inferred tra-2 activity in the wild-type case, 2TFG, yields a hermaphrodite, while the tra-2(e2020)/tra-2(null) expression, TEFG, is Fog. Since tra-2 activity must be greater in the latter, it implies that E>2, or that the effect of the e2020 3’ UTR deletion is to at least double the basal level of tra-2 activity. Similarly, though the repressive parameters F, G, and H are unknown, evaluating each the expression of tra-2 activity for each genotype in Table 1 with E>2 and F=G=H=1/2 (right column) produces a clean separation between hermaphroditic and Fog genotypes, with hermaphrodite activity of up to T/2 (for wild-type) and Fog activity as low as 3T/4 (for tra-2:HA/+; fog-2(null)). This discrimination between the two phenotypes is seen for any value between 1/3 and 1 when F = G = H. Allowing the values of F, G, or H to diverge can either increase or decrease the extent to which estimates of tra-2 activity in the two phenotypic classes overlap. Given the data presented in Figure 6 below (which indicate that gld-1 repression is greater than that of fog-2), the effect of lowering G below F and H is worth noting (Fig. 4). Setting G=0.1, complete separation is seen when F and H are 0.5 or greater. The model is therefore robust to observed differences in repressor activity. More generally, it also highlights the bistable nature of germline sex determination, in which a continuous range of activity states produces only two discreet phenotypes.
Table 1. Quantitative model of germline tra-2 activity.
T represents the activity of one wild-type tra-2 locus without trans-acting repression. H, F, and G represent factors (ranging from 0–1) that adjust T downward in response to the HA epitope tag of tra-2(nm75), activity of fog-2(+), or activity of gld-1(+), respectively. E represents a factor (>1) by which T is increased in the tra-2(e2020) dominant Fog allele. Because reduction of gld-1 and fog-2 function impact sexual fate independently and in e2020 mutants (see Tables 2 and 3), F and G are retained as factors even when combined with tra-2(e2020) alleles.
| Genotype | Germline phenotype | tra-2 activity | value for F=G=H=0.5 E>2 |
|---|---|---|---|
| Wild-type | Hermaphrodite | 2TFG | T/2 |
| tra-2(null)/+ | Hermaphroditea | TFG | T/4 |
| tra-2:HA | Hermaphroditeb | 2TFGH | T/4 |
| tra-2(null)/+; fog-2(null) | Hermaphroditec | TG | T/2 |
| tra-2:HA; fog-2(null) | Hermaphroditeb | 2TGH | T/2 |
| tra-2:HA/+; fog-2(null) | Fogb | TG+TGH | 3T/4 |
| fog-2(null) | Fogc | 2TG | T |
| gld-1(null) | Fog (Tum)d | 2TF | T |
| tra-2(e2020)/+ | Foge | TEFG+TFG | >3T/4 |
| tra-2(e2020)/tra-2(null) | Foge | TEFG | >T/2 |
| tra-2(null) | Moga | 0 | 0 |
This study
Table 2. Evidence for a tra-2 3’ UTR-independent role for fog-2.
Families of tra-2 (e2020); fem-3 (q95) mutants segregating for fog-2(q71) were grown at 25° C and scored for the presence of oocytes with a compound microscope (“total scored”). A subset of animals thus scored were recovered and genotyped at fog-2 by single-worm PCR and sequencing.
| Mog | Fog (includes self-fertile) | |
|---|---|---|
| Total scored: | 85 (78%) | 24 (22%) |
| Number sequenced: | 9 | 12 |
| fog-2 genotypes | Mog | Fog (includes self-fertile) |
| +/+ | 4 | 0 |
| q71/+ | 5 | 1 |
| q71 | 0 | 11 |
Table 3. Genetic interactions between gld-1(lf) and fog-2(lf), tra-2 (gf), and fem-3 (gf).
Progeny of control and gld-1(RNAi) hermaphrodite injectees from the indicated strains were raised at either 15° or 25° C until young adulthood, and then scored with DIC microscopy. JK992 is tra-2(e2020gf), fem-3(q95gf), and CP165 is fem-3(q95); fog-2(q71), both in the N2 background.
| Tum | |||||
|---|---|---|---|---|---|
| N2 | control | 15° C | 23 | 0 | 0 |
| N2 | control | 25° C | 1300 | 0 | 0 |
| N2 | gld-1(RNAi) | 15° C | 11 | 0 | 230 |
| N2 | gld-1(RNAi) | 25° C | 34 | 0 | 925 |
| JK992 | control | 15° C | 32 | 0 | 0 |
| JK992 | control | 25° C | 0 | 250 | 0 |
| JK992 | gld-1(RNAi) | 15° C | 14 | 0 | 70 |
| JK992 | gld-1(RNAi) | 25° C | 0 | 160 | 19 |
| CP165 | control | 25° C | 0 | 40 | 0 |
| CP165 | gld-1 (RNAi) | 25° C | 0 | 43 | 32 |
Figure 6. TRA-2 expression in fog-2 and gld-1 null mutants.

A. Comparison of TRA-2B:HA expression in the distal region (typical of gonad as a whole) of otherwise wild-type nm75 gonads (“WT”) and in fog-2(q71) mutants. Confocal micrographs of L4 larvae and young adults are shown for each, along with non-tagged N2 as a negative control (far left). The arrowheads indicate staining of the distal tip cell nucleus. Bright punctae in the fog-2(q71) adult sample (far right) were not consistently associated with a particular genotype from experiment to experiment. B. Differential interference contrast (top) and epifluorescence (bottom) micrographs of the oma-1:GFP reporter teIs1 in live, anaesthetized wild-type (left) or fog-2(q71) homozygotes (right). GFP fluorescence is abundant in mature oocytes (mo), but lacking in both cases in the distal germline region (d, dashed line) where GLD-1 is expressed (Jones et al., 1996). Scale bar = 100 μm. C. Confocal micrographs of dissected gonads of tra-2(nm75) animals either heterozygous (left) or homozygous (right) for the gld-1(q485) null mutation, stained for TRA-2B:HA and imaged under identical conditions in the same session. Each image represents maximum intensity projections of 12–14 μm, or about half the depth of each gonad. As in Fig. 4 and panel 5A above, arrowheads, arrows, and wedges indicate the HA-positive distal tips cell, sheath cells, and gut nuclei, respectively; sheath cell nuclei in gld-1(q485) gonad are not visible in this focal plane. The far right column is an enlarged version of the region boxed in the tumorous q485 gonad, a single focal plane with the red channel pixel intensity rescaled uniformly to highlight nuclear accumulation in a subset of pachytene nuclei (dashed circles). The scale bars in the left and middle columns are 100 μm, and 20 μm in the right column.
Figure 4. Modeling tra-2 activity with strong gld-1 repression.

Each line corresponds to a hermaphroditic (solid) or feminized (dashed) genotype in Table 1. In calculating the expression for tra-2 activity, the strength of tra-2(e2020) activation, E, is fixed at 2.1, and the parameter for gld-1 repression, G, is fixed at 0.1 (i.e. very strong repression). The parameters for repression by fog-2 and the tra-2(nm75) HA tag (F and H, respectively) are equal but allowed to vary along the X-axis as indicated. Note the separation of phenotypic classes by inferred tra-2 activity when F = H > 0.5.
TRA-2B:HA is detected in somatic nuclei, but not germ cells
The TRA-2:HA tagged allele described above (nm75) was next used to determine in which tissues TRA-2B is expressed, with particular emphasis on gonads. TRA-2B:HA was detected exclusively in the nuclei of dissected tissues of larvae and adults, including those of the intestine and somatic gonad (sheath and distal tip cells (Fig. 5). Nuclear localization of TRA-2B in somatic cells is consistent with previous studies using antibodies (Shimada et al., 2006) and reporter transgenes (Lum et al., 2000; Mapes et al., 2010). As judged by northern blotting (Kuwabara et al., 1998) and in situ hybridization (Kohara, 2005), tra-2 transcripts are especially abundant in the germ line of adult hermaphrodites. Surprisingly, however, germ cell expression of TRA-2:HA was not consistently above background levels. Similar lack of germline expression was also reported with a polyclonal antibody raised against TRA-2B (Shimada et al., 2006). The TRA-2B expression observed in total worm lysates (Fig. 3) is therefore contributed almost completely from somatic tissues.
Figure 5. TRA-2:HA expression in wild-type animals.
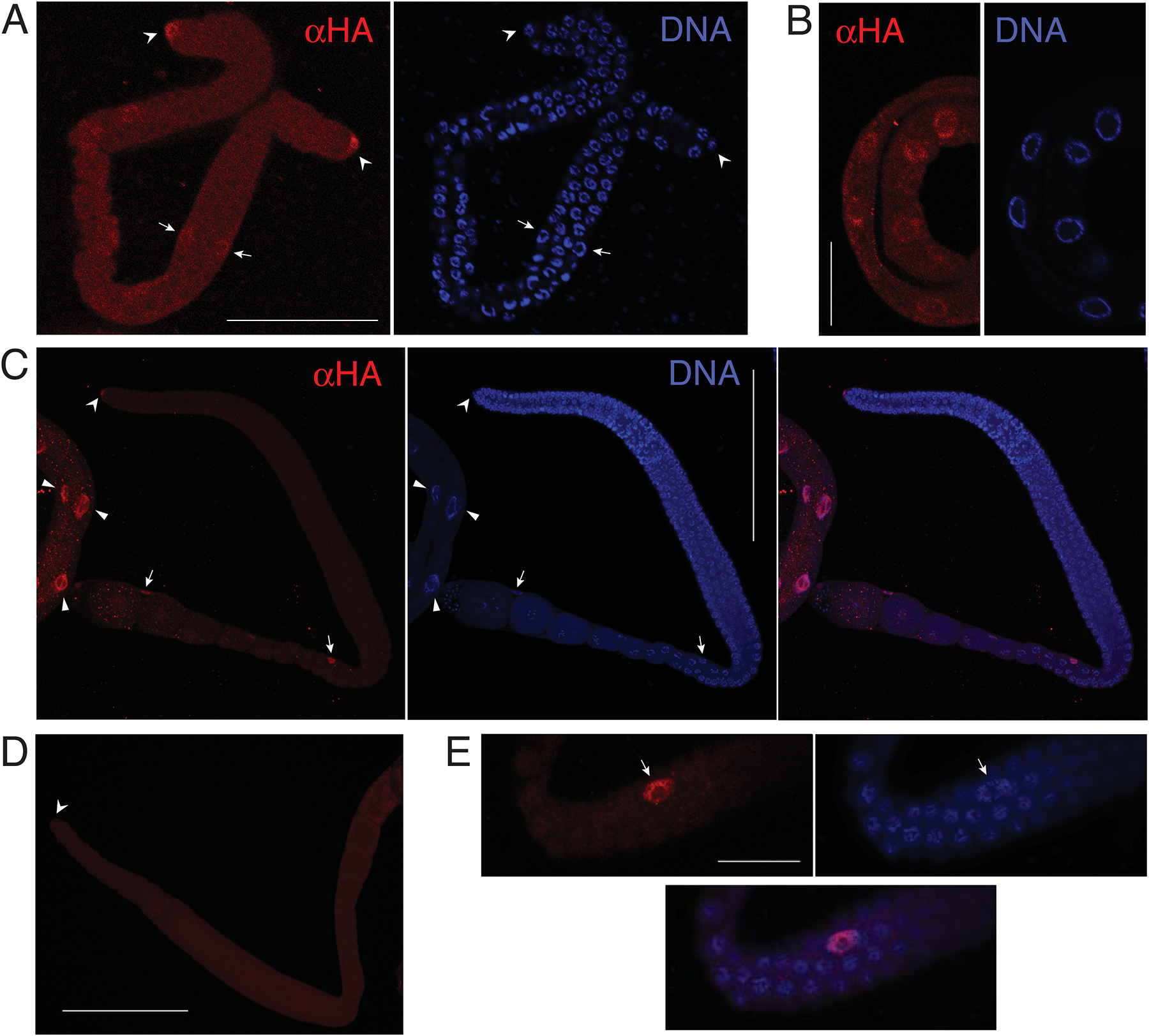
A, B. Confocal micrographs of dissected L3 larval hermaphrodite gonad (A) and intestine (B) showing TRA-2:HA localization (α-HA, left), Hoechst 33258 dye (DNA, right). TRA-2B:HA is detected in the nuclei of the distal tip cells (arrow heads) and more weakly in more proximal somatic cells (arrows). Scale bar is μm in A, 25 μm in B. C. Confocal micrograph of dissected adult hermaphrodite, showing TRA-2:HA localization (α-HA, left), Hoechst 33258 dye (DNA, center), and the two channels merged (right). TRA-2B:HA is detected in the somatic gonad, including the nuclei of the distal tip cell (arrow head), gonad sheath cells (arrows), and large intestinal nuclei (wedges). Several other gonadal sheath nuclei are not captured in the 8 μm-thick slice (see panel E). Scale bar: 100 μm. D. Negative control gonad preparation of the same TRA-2:HA strain as in C, but omitting the anti-HA primary antibody. Arrowhead marks the distal tip cell. Scale bar: 100 μm. Faint background in the germ line is comparable to that in A. E. Higher-magnification, 3 μm-thick slice of an HA-positive sheath cell nucleus (arrowhead) and adjacent germ cell nuclei from the same gonad in A (alternative focal plane, near the bend, displayed rotated 90 degrees).
We next examined the TRA-2B:HA gonad expression in the fog-2 null and gld-1 null backgrounds. No clear elevation of TRA-2∷HA signal was observed in the germ line of adult or L4 larval fog-2(q71) animals relative to wild type (Fig. 6A). Meiosis of XX spermatocytes begins in L3 (Hansen et al., 2004), it is likely sexual fate is also specified at this time. We therefore also compared wild-type and fog-2(q71) backgrounds at this stage. Anti-HA germline fluorescence was quantified by confocal microscopy and image analysis for 12 L3 gonads of each genotype. Again, no difference in expression was observed (mean pixel intensity of 41.17 vs. 41.2, two-tailed t Test, P=0.99). Thus, the repression of TRA-2 expression provided by fog-2 is insufficient to raise the germ cell immunofluorescence signal above background levels when eliminated. Loss of fog-2 does not alter the spatial expression of a GFP reporter for oma-1 (Lin, 2003), a conserved direct GLD-1 target (Beadell et al., 2011; Lee and Schedl, 2004) easily detected in germ cells (Fig. 6B). We conclude that expression of mRNAs targeted by GLD-1 is not systematically up-regulated in the absence of FOG-2.
For gld-1(q485) mutants, immunofluorescence in the red (anti-HA) channel was elevated (roughly two-fold) compared to non-tumorous gld-1(485/+) siblings (Fig. 6C). This signal is mostly non-localized, but above this uniform background pachytene nuclei often show localized fluorescence. This suggests that there may be some up-regulation of TRA-2B in gld-1(q485) mutant germ cells. However, gld-1(q485) gonads are a qualitatively different tissue than wild-type or q485/+ gonads, being composed of a vast excess of mitotically proliferating germ cells. We therefore considered the possibility that their elevated fluorescence was due to intrinsically higher nonspecific background. To address this, we quantified the mean pixel intensity in sheath cell-free regions of gld-1(q485) gonads that were either processed normally, or without the anti-HA primary antibody to reveal nonspecific background. Processed and imaged in parallel on the same day, fluorescence of samples with anti-HA primary antibody (mean pixel intensity 12.69, S.E.M. 0.32, N=7) was significantly higher (unpaired two-tailed t test, P<0.0001) than of those that did not (mean pixel intensity 8.34, S.E.M. 0.47, N=4). The magnitude of the difference (1.5-fold) is similar to that seen between gld-1(q485/+) and gld-1(q485) samples stained with both primary and secondary antibodies. We conclude that loss of gld-1, unlike loss of fog-2, is sufficient to elevate TRA-2B expression to detectable levels in germ cells. The magnitude of the change is not large enough to be revealed in immunoblots of whole-worm lysates.
Sensitivity of tra-2(e2020) to loss of fog-2 and gld-1 activity
The tra-2 gain-of-function (gf) alleles eliminate some or all GLD-1 binding sites in the 3’ UTR (Goodwin et al., 1993; Jan et al., 1999), and also eliminate XX spermatogenesis (Doniach, 1986). Such mutants can be restored to self-fertility by combination with a suitable fem-3(gf) allele, which on its own fully masculinizes the germ line (Barton et al., 1987). Schedl and Kimble (1988) found that in this context, seven different tra-2(gf) alleles tested were sensitive to fog-2 loss. However, none of these alleles eliminate all known GLD-1 3’ UTR binding sites, suggesting continued sensitivity may be due to residual repression mediated by remaining sites. The tra-2 gain-of-function (gf) allele e2020 has a 108 nt deletion of its 3’ UTR that eliminates all GLD-1 binding in vitro (Goodwin et al., 1993; Jan et al., 1999). If FOG-2 regulates germline sex solely through its regulation of the tra-2 mRNA via GLD-1 and its 3’ UTR binding sites, then tra-2(e2020) may be uniquely insensitive to fog-2 loss. Conversely, feminization of the e2020 allele by fog-2(lf) would suggest a role for fog-2 that does not rely on the known tra-2 3’ UTR binding sites for GLD-1.
JK992 (tra-2(e2020); fem-3(q95)) animals are normally Mog at 25° C, thus creating a situation in which abundant sperm (and no oocytes) are made without the known GLD-1-binding sites in the tra-2 3’ UTR. To assess whether fog-2 has an impact on germline sex in this situation, we produced a tra-2(e2020), fem-3(q95) population that was also segregating for the fog-2(q71) null allele. Similar to results using weaker tra-2(gf) alleles with residual GLD-1-binding capacity (Schedl and Kimble, 1988), tra-2(e2020gf); fem-3(q95gf); fog-2(q71) triple mutants were all Fog (Table 2). However, their fog-2(q71/+) or fog-2(+/+) siblings remained Mog, with the exception of one heterozygote. Thus, lack of all fog-2 function feminizes the germ line even when the tra-2(gf) allele used eliminates all known binding sites of GLD-1 in the tra-2 3’ UTR.
The above result could indicate that the FOG-2-GLD-1 complex has previously unrecognized targets, or that FOG-2 has GLD-1-independent functions. If loss of gld-1 function in the same tra-2(e2020gf); fem-3(q95gf) context also feminizes, it would support the former explanation. We therefore performed an RNAi knock-down of gld-1 in JK992 via injection (Table 3). This produces a highly penetrant oocyte tumor (Tum) phenotype in wild-type animals at both 15° and 25° C, and in JK992 at the permissive (oocyte-producing) 15° C. At 25°, gld-1(RNAi) shifted 12% of adults from Mog to Tum, indicating partial feminization. This is qualitatively similar to the effect of fog-2(q71), but less penetrant. Whether this difference is due to incomplete knockdown or a GLD-1-independent function of FOG-2 remains unclear.
As another way to examine the interaction of gld-1 and fog-2 function, we examined the impact of reducing gld-1 function in fem-3(q95); fog-2(q71) double mutants (Table 3). At the non-permissive temperature (25° C), this strain is 100% Mog. However, when gld-1 function was reduced by RNA interference, over ten percent of the progeny of injected mothers were Tum. A similar enhancement was observed at permissive temperature. This indicates that GLD-1 promotes male fate even in the absence of its dimeric partner FOG-2.
Dominant GLD-1 gain-of-function mutant exhibits weaker RNA-binding capacity
GLD-1 mutant alleles are categorized into 6 phenotypic classes (Francis et al., 1995a). The subclass C1 alleles, q93, oz17, oz34, oz35, and q62 change Gly-248 into Arg (G248R); subclass C2 alleles, oz30, oz16, oz29, oz33, and oz70 have a point mutation of G250R. The glycine residues altered by these missense mutations are highly conserved in the GSG domain (Jones and Schedl, 1995). XX animals from both sub-classes over-produce sperm and fail to switch to oogenesis (the masculinization of germline, or Mog phenotype). This has been interpreted as a gain-of-function phenotype, and alleles from both subclasses dominantly suppress the Fog phenotype of fog-2(q71) (Francis et al., 1995b).
One possible mechanism of a dominant gain-of-function GLD-1 mutant would be tighter binding to its target mRNA, which could suppress the translation to a greater degree. To test this possibility, we examined the GLD-1 wild type, G250R and G248R mutants for their binding affinity to a synthetic fragment encoding the 3’ UTR of glp-1, a well-characterized GLD-1 target mRNA (Farley and Ryder, 2012). Unlike the multimerized GLD-1-binding motifs (GBMs) in the C. elegans tra-2 3’ UTR, glp-1 contains a single GLD-1 binding site (Marin and Evans, 2003; Ryder et al., 2004) that makes interpretation of affinity straightforward. We found that while wild type GLD-1 bound to its target efficiently, the two mutant proteins had reduced binding (Figure 7). The G248R mutant had particularly poor binding. The C1 alleles bearing this mutation show a more severe Mog phenotype than the C2 alleles (G250R). Thus, contrary to one simple hypothesis, lower binding capacity of GLD-1 to its target mRNA is associated with a stronger dominant gain-of-function phenotype.
Figure 7. Binding analysis of GLD-1 WT, GLD-1 G248R, and GLD-1 G250R.
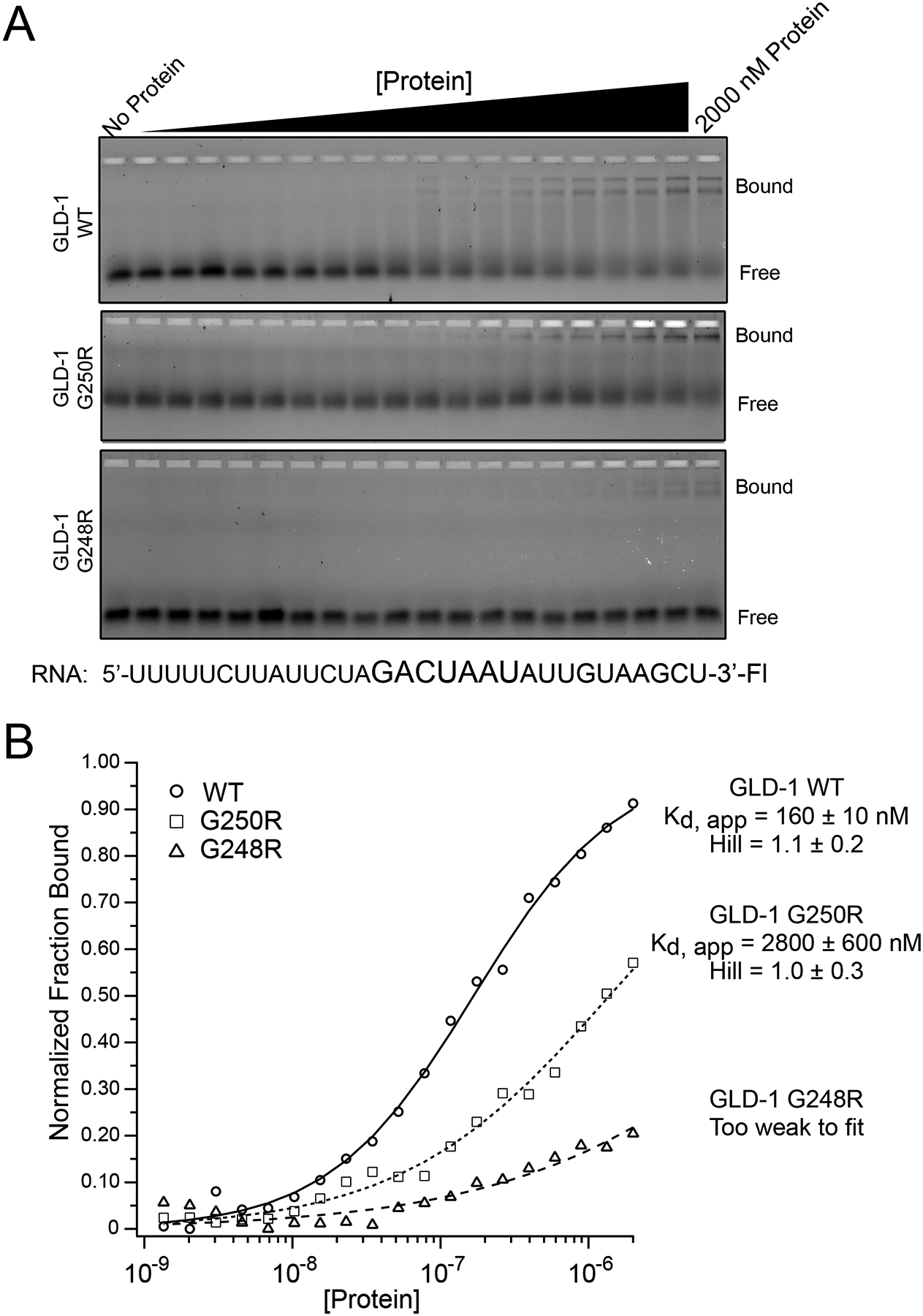
A. An increasing concentration of purified, recombinant MBP-tagged GLD-1 or mutant variants was incubated with a fluorescein labeled RNA oligonucleotide comprising a fragment from the glp-1 3ÚTR that contains a GLD-1 binding motif, shown in larger font. Bound RNA was resolved from free using a slab polyacrylamide gel, and visualized using a FUJI FLA9000 Imaging System. B. The fraction of bound RNA was quantified using MultiGauge software. These data were plotted as a function of the protein concentration and fit to the Hill Equation in order to determine the apparent equilibrium dissociation constant (Kd, app) and the Hill coefficient. The average and standard deviation of the Kd, app and the Hill equation for at least three experimental replicates are presented to the right of the graph. Data were normalized to the local minimum and the fitted window of GLD-1 WT binding to facilitate comparison of the three mutants on the same set of axes. Un-normalized data were used to obtain the fitted parameters. For GLD-1 G248R, the data were not fit because little to no binding was observed across three replicates.
Discussion
fog-2 and tra-2 regulation
The fog-2 gene was formed in the C. elegans lineage by duplication and modification of one of many F-box proteins (Nayak et al., 2005). In contrast, tra-2 and gld-1 are ancient regulators of overall sex determination and oocyte development (Beadell et al., 2011; Haag and Kimble, 2000; Kuwabara, 1996). FOG-2 is thus a new player in an old system, and its recent origin and its essential role in the newly acquired self-fertility of C. elegans are likely not coincidental. By what mechanism does FOG-2 contribute to the regulation of germline sex in the C. elegans hermaphrodite? An important first step is to examine potential impacts on mRNA abundance. The transcriptome of fog-2(q71) XX gonads was previously characterized, and compared to those of fem-3(q96) mutants, which express only sperm (Ortiz et al., 2014). This comparison highlighted the extreme differences between the male and female germ cell transcriptomes, and revealed reductions in tra-2 and gld-1 expression in q96 germ cells consistent with a negative feedback between sperm fate and their expression. However, to understand the role of FOG-2 in an otherwise female germ line, the comparison of fog-2(q71) with wild-type hermaphrodites is more appropriate.
Using both oligo-dT primed and random-primed libraries, we found that the expression of 99% of detected transcripts was unchanged in fog-2 mutants relative to wild-type hermaphrodites. Similarly, nearly all GLD-1 target mRNAs, including tra-2, were also unchanged in their abundance. This finding is consistent with the hypothesis that FOG-2 regulates tra-2 activity post-transcriptionally. It should be noted, however, that in our adult samples the XX spermatocytes that rely upon fog-2 for their formation are not present. L4 samples would include such cells, but they would also likely include many spermatogenesis-related transcripts in the wild-type sample that would dominate the set of differentially expressed genes. The choice to analyze adults is somewhat justified because FOG-2 is expressed in all stages of development (Clifford et al., 2000).
Although neither the wild-type nor fog-2(q71) animals compared with RNAseq were undergoing active spermatogenesis, we nevertheless found that expression of 14 major sperm protein (msp) paralogs was reduced in fog-2 mutants. One possible explanation is that cryptic misexpression of sperm genes occurs in oocytes, and this is reduced (feminized) in fog-2(q71). Alternatively, because the fog-2 mutant hermaphrodites were inseminated by fog-2(q71) males, the difference may reflect residual transcripts in spermatozoa. It would not be surprising if fog-2 male sperm were cryptically feminized, and thus transcribed msp genes at lower levels. This could be tested in the future.
Assuming that FOG-2 does indeed regulate translation, the mechanism by which this would occur remains elusive. Many F-box proteins have roles in marking target proteins for ubiquitination and subsequent proteasomal degradation, and FOG-2 interacts with the C. elegans Skp1 homolog SKR1 (Nayak et al., 2005). What might the target of a putative FOG-2-mediated ubiquitination be? It is unlikely to be GLD-1, as both are required for the same process. One interesting possibility is that, by virtue of being tethered to the tra-2 3’ UTR via GLD-1, FOG-2 targets the nascent TRA-2 peptide itself. Such co-translational modification via a 3’ UTR scaffold has been reported for the human protein CD47 (Berkovits and Mayr, 2015).
Tissue- and sex-specificity of TRA-2 expression
Three other groups have examined the subcellular, tissue-level, and sex-biased expression of TRA-2. Lum et al. (2000) detected a transgene fusion of TRA-2B to GFP in somatic nuclei. The multi-copy array used would not be expected to express in germ cells due to silencing. However, using a polyclonal antibody, endogenous TRA-2B was similarly reported in intestinal nuclei of XX hermaphrodites, but not detected in male intestine or germ cells from both sexes (Shimada 2006). Finally, an integrated 3xFlag-tagged TRA-2 transgene was expressed in nuclei of the head (Mapes 2010), and could be detected in males as long as it was heterozygous (reporter homozygotes were feminized). Our immunoblots and immunohistochemistry of HA-tagged endogenous TRA-2 are broadly consistent with these earlier studies. We observed expression of TRA-2B in XX hermaphrodites, but could not detect it in germ cells, nor in males. The full-length 170 kDa TRA-2A isoform did not appear on our blots. This could be because it does not readily solubilize in sample buffer, but the absence of plasma membrane expression in stained tissues is more consistent with rapid cleavage by the TRA-3 protease in XX animals.
Weak germline expression of TRA-2B is surprising, given available in situ data (Kohara, 2005) and extensive evidence suggesting cell-autonomous function in germ cells. This includes a 1.8 kb transcript encoding TRA-2B that is abundant in oocytes (Kuwabara et al., 1998). It is consistent, however, with extensive translational repression of the tra-2 mRNAs in germ cells, which is well documented (Goodwin et al., 1993; Jan et al., 1999). The modest upregulation of TRA-2B signal in the germ line of gld-1 mutants (Fig. 6C) indicates that complete loss of GLD-1 does have a measurable effect (though other, uncharacterized negative regulators likely remain). In contrast, fog-2 either exerts a more subtle influence on TRA-2 expression than does gld-1, or it regulates tra-2 independently of its steady-state protein product abundance. The formal model of tra-2 regulation developed here (Table 1, Fig. 4) suggests that a scenario in which fog-2 exerts a weaker effect on tra-2 activity than gld-1 is consistent with current genetic data. However, a full assessment of the processes that regulate tra-2 activity awaits more sensitive methods.
The low levels of TRA-2 in the germ line might be essential to hermaphrodite development. TRA-2 and FEM-3 levels are kept in approximate balance by a battery of regulators (Puoti et al., 2001). If the absolute expression levels of both were low, then very small changes in either would suffice to push the germ line towards either spermatogenesis or oogenesis. fog-2 appears to represent one such small change.
Though undetectable in wild-type germ cells via immunohistochemistry, TRA-2B:HA is highly expressed in distal tip cells and gonad sheath cells, somatic cells tightly directly attached to the germ line. This raises the possibility that the somatic TRA-2 expression may play a role in determining the sex of germ cells. Ablations of sheath cells can feminize C. elegans hermaphrodites (McCarter et al., 1997). Because sheath cell TRA-2 expression would be expected to promote oocyte fate, the possible connection between this and the sheath cells’ role in supporting XX spermatogenesis remains mysterious. Preliminary experiments in which the male-promoting protein FEM-3 was overexpressed in the distal tip cell of hermaphrodites led to abnormal somatic gonad morphology, but failed to masculinize germ cells (S.H. and E.S.H., unpublished data).
Uncharacterized roles of fog-2 and gld-1 in germline sex determination
We present two surprising findings about the roles of fog-2 and gld-1 in the regulation of germline sex determination. First, the tra-2(e2020); fem-3(q95); fog-2(q71) and tra-2(e2020); fem-3(q95); gld-1(RNAi) triple mutants (Tables 2 and 3) indicate that loss of either fog-2 or gld-1 function can feminize germ cells in a background that eliminates all known GLD-1-interacting motifs of the tra-2 3’UTR. This result reveals an unexpected aspect of their function. One possibility is that the 108 nucleotide deletion in the tra-2(e2020) 3’UTR does not abolish in vivo GLD-1 binding completely. For example, GLD-1, along with FOG-2, may bind the tra-2 mRNA outside of the 3’ UTR. Some GLD-1 targets are repressed by binding in 5’ UTRs (Jungkamp et al., 2011), and a GLD-1 site in the tra-2 open reading frame was predicted computationally (Brummer et al., 2013). Such an association may be enough to render sexual fate of e2020 mutants sensitive to fog-2 loss. Another possibility is that fog-2 may be able to regulate TRA-2 expression in a GLD-1-independent manner, perhaps via its F-box domain. Finally, fog-2 may promote sperm fate in an entirely TRA-2-independent manner, with or without interacting with GLD-1.
Second, the results in Table 3 suggest that the sperm-promoting effects of gld-1 and fog-2 are additive. One issue our data cannot resolve, however, is whether fog-2 has any gld-1-independent functions. However, comparison of our results with the literature suggests that it might. In the context of the fem-3(q95) dominant Mog mutation, gld-1(lf) suppresses masculinization less than 10% of the time (Francis et al., 1995b). However, gld-1(RNAi); fem-3(q95); fog-2(q71) animals are feminized (Tum) over half of the time, even though RNAi is unlikely to impart a loss of gld-1 function that is as strong as the q485 mutation. This is unexpected, as recruitment of FOG-2 to the tra-2 mRNA would seem to be dependent upon interaction with the GLD-1 RBP. Known FOG-2 protein interactors are GLD-1 (Jan et al., 1999) and the Skp1 homolog SKR-1 (Nayak et al., 2005), but these results suggest there may be others. Characterization of novel FOG-2-associated macromolecules is thus a priority for future research.
Mechanism of translational repression by GLD-1
Transcriptome-wide examination of mRNA translation via ribosomal profiling found that many GLD-1 targets are translationally repressed (Scheckel et al., 2012). The feminized germline phenotypes of both gld-1(lf) and tra-2(gf) mutants are consistent with such negative regulation of TRA-2 expression by GLD-1. Our observation of enhanced immunofluorescence of TRA-2B:HA in gld-1(q485) gonads (Fig. 6B) provides further support for this. Dominant gld-1(Mog) alleles with the opposite phenotype as gld-1 null mutants might therefore be interpreted as “hyper-repressors” of tra-2. However, we find that the missense mutant proteins these alleles encode actually bound a representative GLD-1 binding motif more weakly in vitro than wild-type GLD-1. It is possible, however, that the tandem GBMs in the tra-2 3’ UTR interact with the G248R and G250R mutant forms of GLD-1 in ways that are distinct from that seen in glp-1.
How could poor binding lead to a phenotype that appears to represent a gain of function? Previous studies have suggested that GLD-1 not only represses translation, but also protects its targets from nonsense-mediate decay and perhaps other forms of degradation (Lee and Schedl, 2004; Scheckel et al., 2012). Thus, a mutation that weakens binding might have the counter-intuitive effect of actually hyper-repressing tra-2 expression, due to destabilization of the mRNA. Consistent with this, tra-2 mRNA is slightly less abundant in gld-1 mutants (Scheckel et al., 2012). However, other observations suggest the situation is not so simple. First, both gld-1 null mutations and deletion of the GBMs of tra-2 in the e2020 allele have the opposite (Fog) phenotype of these gld-1(gf) alleles (Doniach, 1986; Francis et al., 1995b; Goodwin et al., 1993). Disruption of GLD-1-mRNA interaction via the C-class Mog alleles of GLD-1 is therefore not equivalent to a general loss of gld-1 function. Second, the dominant Mog phenotype of gld-1(q93) is suppressed to self-fertility by one copy of the tra-2(gf) allele, q122 (Francis et al., 1995b). This is consistent with the existence of a mode of tra-2 repression for GLD-1 that does not require (and may be blocked by) wild-type levels of RNA-binding activity. Alternatively, the gld-1(gf) alleles tested here may confer novel protein-protein interactions that more than compensate for reduced RNA-binding capacity. Which, if either, of these scenarios is correct awaits further characterization of GLD-1’s many roles in germ cell gene regulation and development.
fog-2 and the evolution of self-fertility
fog-2 evolved in the C. elegans lineage via a tandem duplication, and its protein product is distinguished from those of closely related paralogs by a C-terminal GLD-1-binding domain (Clifford et al., 2000; Nayak et al., 2005). GLD-1 has roughly 1000 target mRNAs, most of which appear to be translationally repressed as a result (Jungkamp et al., 2011; Scheckel et al., 2012; Wright et al., 2010). The model and experiments presented here indicate that very little TRA-2 protein is normally ever made in germ cells, and that GLD-1 contributes the bulk of tra-2 repression. We therefore suggest that the evolution of the FOG-2 cofactor contributed a relatively modest, but nevertheless crucial, enhancement of tra-2 repression. The weakly feminized phenotypes of tra-2(q122) mutants, which that lack some, but not all GBMs (Goodwin et al., 1993), further suggests that amplification of the GBMs was another important step in the evolution of reliable XX spermatogenesis.
Supplementary Material
Acknowledgements
We thank Tim Schedl for advice and for sharing strain JK992, Amy Beaven for assistance with confocal microscopy, and two anonymous reviewers for perceptive suggestions. This research was supported by NSF award IOS-1355119 to E.S.H. and an Ithaca College H&S STEM Summer Scholar award to L.E.R. Some strains were provided by the Caenorhabditis Genetics Center at the University of Minnesota, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Declarations of interest: none
References
- Alonso CR, 2012. A complex ‘mRNA degradation code’ controls gene expression during animal development. Trends Genet 28, 78–88. [DOI] [PubMed] [Google Scholar]
- Barton MK, Schedl TB, Kimble J, 1987. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadell AV, Liu Q, Johnson DM, Haag ES, 2011. Independent recruitments of a translational regulator in the evolution of self-fertile nematodes. Proc Natl Acad Sci U S A 108, 19672–19677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, Mayr C, 2015. Alternative 3’ UTRs act as scaffolds to regulate membrane protein localization. Nature 522, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner JL, Schedl T, 2016. Germline Stem Cell Differentiation Entails Regional Control of Cell Fate Regulator GLD-1 in Caenorhabditis elegans. Genetics 202, 1085–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummer A, Kishore S, Subasic D, Hengartner M, Zavolan M, 2013. Modeling the binding specificity of the RNA-binding protein GLD-1 suggests a function of coding region-located sites in translational repression. Rna 19, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke DJ, Ward S, 1983. Identification of a large multigene family encoding the major sperm protein of Caenorhabditis elegans. J Mol Biol 171, 1–29. [DOI] [PubMed] [Google Scholar]
- Clifford R, Lee M, Nayak S, Ohmachi M, Giorgini F, Schedl T, 2000. FOG-2, a novel F-box-containing protein, associates with the GLD-1 RNA-binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development 127, 5265–5276. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J, 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417, 660–663. [DOI] [PubMed] [Google Scholar]
- Doniach T, 1986. Activity of the sex-determining gene tra-2 is modulated to allow spermatogenesis in the C. elegans hermaphrodite. Genetics 114, 53–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley BM, Ryder SP, 2012. POS-1 and GLD-1 repress glp-1 translation through a conserved binding-site cluster. Mol Biol Cell 23, 4473–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Barton MK, Kimble J, Schedl T, 1995a. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139, 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Maine E, Schedl T, 1995b. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139, 607–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF, 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9, e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau A, Richard S, 2009. The STAR RNA binding proteins GLD-1, QKI, SAM68 and SLM-2 bind bipartite RNA motifs. BMC Mol Biol 10, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Sumara I, Lechner E, 2013. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J 32, 2307–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EB, Okkema PG, Evans TC, Kimble J, 1993. Translational regulation of tra-2 by its 3’ untranslated region controls sexual identity in C. elegans. Cell 75, 329–339. [DOI] [PubMed] [Google Scholar]
- Guo Y, Lang S, Ellis R, 2009. Independent recruitment of F-box genes to regulate hermaphrodite development during nematode evolution. Curr Biol 19, 1853–1860. [DOI] [PubMed] [Google Scholar]
- Haag ES, Kimble J, 2000. Regulatory elements required for development of Caenorhabditis elegans hermaphrodites are conserved in the tra-2 homologue of C. remanei, a male/female sister species. Genetics 155, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr., Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T, 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamichi S, Rivas RN, Knight AL, Cao S, Caldwell KA, Caldwell GA, 2008. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model. Proc Natl Acad Sci U S A 105, 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D, Hubbard EJ, Schedl T, 2004. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev Biol 268, 342–357. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D, 1999. Using antibodies: A laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbory, NY. [Google Scholar]
- Ho MS, Ou C, Chan YR, Chien CT, Pi H, 2008. The utility F-box for protein destruction. Cellular and molecular life sciences : CMLS 65, 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin JA, Brenner S, 1977. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics 86, 275–287. [PMC free article] [PubMed] [Google Scholar]
- Jan E, Motzny CK, Graves LE, Goodwin EB, 1999. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 18, 258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T, 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol 180, 165–183. [DOI] [PubMed] [Google Scholar]
- Jones AR, Schedl T, 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev 9, 1491–1504. [DOI] [PubMed] [Google Scholar]
- Jud MC, Czerwinski MJ, Wood MP, Young RA, Gallo CM, Bickel JS, Petty EL, Mason JM, Little BA, Padilla PA, Schisa JA, 2008. Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev Biol 318, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungkamp AC, Stoeckius M, Mecenas D, Grun D, Mastrobuoni G, Kempa S, Rajewsky N, 2011. In vivo and transcriptome-wide identification of RNA binding protein target sites. Molecular cell 44, 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K, Félix M-A, Ailion M, Rockman M, Braendle C, Penigault J-B, Fitch D, 2011. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol 11, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K, Gavin NP, Raynes Y, Roehrig C, Piano F, Fitch DH, 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc Natl Acad Sci U S A 101, 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y, 2005. NEXTDB: The Nematode Expression Pattern DataBase. National Institute of Genetics, Japan. [Google Scholar]
- Kuwabara PE, 1996. Interspecies comparison reveals evolution of control regions in the nematode sex-determining gene tra-2. Genetics 144, 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara PE, Kimble J, 1995. A predicted membrane protein, TRA-2A, directs hermaphrodite development in Caenorhabditis elegans. Development 121, 2995–3004. [DOI] [PubMed] [Google Scholar]
- Kuwabara PE, Okkema PG, Kimble J, 1998. Germ-line regulation of the Caenorhabditis elegans sex-determining gene tra-2. Dev. Biol 204, 251–262. [DOI] [PubMed] [Google Scholar]
- Lee MH, Schedl T, 2001. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev 15, 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Schedl T, 2004. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev 18, 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, 2003. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev Biol 258, 226–239. [DOI] [PubMed] [Google Scholar]
- Lum D, Kuwabara P, Zarkower D, Spence A, 2000. Direct protein-protein interaction between the intracellular domain of TRA-2 and the transcription factor TRA-1A modulates feminizing activity in C. elegans. Genes Dev 14, 3153–3165. [PMC free article] [PubMed] [Google Scholar]
- Mapes J, Chen JT, Yu JS, Xue D, 2010. Somatic sex determination in Caenorhabditis elegans is modulated by SUP-26 repression of tra-2 translation. Proc Natl Acad Sci U S A 107, 18022–18027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin VA, Evans TC, 2003. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 130, 2623–2632. [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T, 1997. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol 181, 121–143. [DOI] [PubMed] [Google Scholar]
- Nayak S, Goree J, Schedl T, 2005. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biology 3, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema PG, Kimble J, 1991. Molecular analysis of tra-2, a sex determining gene in Caenorhabditis elegans. EMBO J. 10, 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz MA, Noble D, Sorokin EP, Kimble J, 2014. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda) 4, 1765–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A, Gallegos M, Zhang B, Wickens MP, Kimble J, 1997. Controls of cell fate and pattern by 3’ untranslated regions: the Caenorhabditis elegans sperm/oocyte decision. Cold Spring Harb Symp Quant Biol 62, 19–24. [PubMed] [Google Scholar]
- Puoti A, Pugnale P, Belfiore M, Schlappi AC, Saudan Z, 2001. RNA and sex determination in Caenorhabditis elegans. Post-transcriptional regulation of the sex-determining tra-2 and fem-3 mRNAs in the Caenorhabditis elegans hermaphrodite. EMBO reports 2, 899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder SP, Frater L, Abramovitz D, Goodwin E, Williamson J, 2004. RNA target specificity of the STAR/GSG domain posttranscriptional regulatory protein GLD-1. Nat Struct Mol Biol 11, 20–28. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Inoue T, Otomo Y, Yokomori N, Ohno M, Arai H, Nakagawa Y, 2012. Deficiency of cardiolipin synthase causes abnormal mitochondrial function and morphology in germ cells of Caenorhabditis elegans. J Biol Chem 287, 4590–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheckel C, Gaidatzis D, Wright JE, Ciosk R, 2012. Genome-Wide Analysis of GLD-1-Mediated mRNA Regulation Suggests a Role in mRNA Storage. PLoS Genet 8, e1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T, Kimble J, 1988. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119, 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Kanematsu K, Tanaka K, Yokosawa H, Kawahara H, 2006. Proteasomal ubiquitin receptor RPN-10 controls sex determination in Caenorhabditis elegans. Mol Biol Cell 17, 5356–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HE, 2014. Nematode sperm motility. WormBook, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol S, Kuwabara P, 2000. Proteolysis in Caenorhabditis elegans sex determination: cleavage of TRA-2A by TRA-3. Genes Dev 14, 901–906. [PMC free article] [PubMed] [Google Scholar]
- Suh N, Crittenden SL, Goldstrohm A, Hook B, Thompson B, Wickens M, Kimble J, 2009. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics 181, 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CG, Li R, Smith HE, Woodruff GC, Oliver B, Haag ES, 2012. Simplification and desexualization of gene expression in self-fertile nematodes. Curr Biol 22, 2167–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, 2006. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res 16, 1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR, Goodwin EB, Wickens M, 2000. Rapid deadenylation and Poly(A)-dependent translational repression mediated by the Caenorhabditis elegans tra-2 3’ untranslated region in Xenopus embryos. Mol Cell Biol 20, 2129–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL, 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L, 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Burke DJ, Sulston JE, Coulson AR, Albertson DG, Ammons D, Klass M, Hogan E, 1988. Genomic organization of major sperm protein genes and pseudogenes in the nematode Caenorhabditis elegans. J Mol Biol 199, 1–13. [DOI] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF, 1999. Protein identification and analysis tools in the ExPASy server. Methods in molecular biology (Clifton, N.J.) 112, 531–552. [DOI] [PubMed] [Google Scholar]
- Wright JE, Gaidatzis D, Senften M, Farley BM, Westhof E, Ryder SP, Ciosk R, 2010. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J 30, 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JE, Gaidatzis D, Senften M, Farley BM, Westhof E, Ryder SP, Ciosk R, 2011. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J 30, 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


