Abstract
During embryogenesis of eudicots, the apical region of the embryo develops two cotyledon primordia and the shoot meristem. In Arabidopsis thaliana, this process is dependent on the functionally redundant activities of the CUP-SHAPED COTYLEDON (CUC) transcription factors, namely CUC1, CUC2, and CUC3, as well as the phytohormone auxin. However, the relationship between the CUC proteins and auxin has yet to be fully elucidated. In the present study, we examined whether the expression of auxin biosynthetic genes is dependent on CUC gene activities. Comprehensive quantitative RT-PCR analysis of the main auxin biosynthetic gene families of TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1/TRYPTOPHAN AMINOTRANSFERASE RELATED and YUCCA (YUC) showed that YUC1 and YUC4 expression levels were lower in cuc double mutant embryos than the expression levels of these genes in wild type embryos. Reporter analysis also revealed that the expression of YUC1 and YUC4 in the cotyledon boundary region was reduced in cuc double mutant embryos. In contrast, the loss of function mutation in the SHOOT MERISTEMLESS gene, a shoot stem cell regulator that acts downstream of the CUC genes, did not markedly affect YUC1 expression levels. These results demonstrate that CUC genes play an important role in the regulation of auxin biosynthetic gene expression during embryogenesis; furthermore, they raise the possibility that the auxin produced by this regulation contributes to cotyledon boundary development.
Keywords: auxin, CUC, embryogenesis, shoot meristem, YUC
Introduction
Plant development proceeds with a series of patterning and differentiation events in which regulatory transcription factors and signaling molecules, including phytohormones, play important roles. In eudicots, the apical region of the globular-shaped embryo is subdivided into two cotyledon primordia; the shoot meristem is established at the center of the boundary region of these two primordia (Barton 2010; Barton and Poethig 1993). In Arabidopsis thaliana, this developmental process is regulated by the CUP-SHAPED COTYLEDON (CUC) transcription factors (i.e., CUC1, CUC2, and CUC3), which are expressed in the presumptive cotyledon boundary region from the globular stage on (Aida et al. 1999, 1997; Hibara et al. 2006; Takada et al. 2001; Vroemen et al. 2003). By coordinating the expression of regulatory genes including SHOOT MERISTEMLESS (STM), which is involved in stem cell maintenance in the shoot, these transcription factors ensure proper shoot meristem activity in postembryonic development (Aida et al. 1999, 2020; Belles-Boix et al. 2006; Hibara et al. 2006; Scofield et al. 2018; Takada et al. 2001). Because of their functional redundancy, multiple mutations in the CUC genes result in major developmental defects in the apical region of the embryo, whereas CUC genes with single mutations show no defects or those that are mild (Aida et al. 1997; Hibara et al. 2006). Typically, cuc multiple mutants develop a single cup-shaped structure consisting of the two fused cotyledons and they fail to establish the embryonic shoot meristem.
The phytohormone auxin plays important roles in various developmental processes including embryogenesis (Teale et al. 2006). The main pathway of auxin biosynthesis is catalyzed by sequential actions of two kinds of enzymes, each of which is encoded by the 5 TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1/TRYPTOPHAN AMINOTRANSFERASE-RELATED (TAA1/TAR) and 11 YUCCA (YUC) family genes in A. thaliana (Cheng et al. 2006; Mashiguchi et al. 2011; Stepanova et al. 2008). Several multiple mutant combinations of auxin biosynthetic genes, such as yuc1 yuc4 yuc10 yuc11, show the abnormal embryonic phenotype with fused cotyledons and the lack of a root (Cheng et al. 2007; Robert et al. 2013; Stepanova et al. 2008). Moreover, various mutations in other classes of auxin-related genes (e.g., transport, perception, and response genes) cause defects in embryogenesis that include abnormal cotyledon morphology (Möller and Weijers 2009). The phenotype of these mutants suggests that a close relationship exists between the CUC genes and auxin signaling. However, studies on the relationship between CUC genes and auxin signaling in embryogenesis have only rarely been reported (Aida et al. 2002; Furutani et al. 2004). Thus, in the present research, we examined whether the CUC genes regulate the expression of auxin biosynthetic genes.
Materials and methods
The A. thaliana accession Columbia (Col) was used as the wild type (WT). The allelic combination of the cuc1 cuc2 double mutant was cuc1-1 cuc2-1, which had originally been isolated from the Landsberg erecta background (Aida et al. 1997) and was backcrossed five times to Col prior to the analyses. The cuc2 cuc3 and stm mutants, which were described previously (cuc2-3 cuc3-105: Hibara et al. 2006; stm-1C: Takano et al. 2010), were in the Col background. The GFP-TAA1, YUC1p-n3xGFP, and YUC4p-n3xGFP reporter lines were also previously reported (Robert et al. 2013; Stepanova et al. 2008). Plants were grown using previously described methods (Takeda et al. 2011).
To conduct quantitative RT-PCR (qRT-PCR) experiments, 20 heart-stage embryos were collected for each sample. The sampled embryos of the cuc1 cuc2 double mutant were cup-shaped progeny of cuc1 cuc2/+ plants. For each genotype, four replicates were obtained from the independent parental plants. Embryos were dissected from seeds in 7% glucose solution as previously described (Imoto et al. 2021). An equal volume of the solution containing embryos and Monarch DNA/RNA Protection Reagent (New England Biolabs, Ipswich, USA) was mixed and then thoroughly homogenized mechanically at room temperature. Total RNA was purified from this lysate using a Monarch Total RNA Miniprep Kit (New England Biolabs). The concentration of total RNA in each sample was measured with a Qubit Fluorometer (Thermo Fisher Scientific, Waltham, USA). Reverse transcription reactions were performed with a LunaScript RT SuperMix Kit (New England Biolabs), and then real-time PCR was performed with a LightCycler system (Roche Diagnostics, Basel, Switzerland) and Luna Universal qPCR Master Mix (New England Biolabs). All RNA samples were adjusted to a final concentration of 25 pg 10 µl−1 for PCR reactions. ACTIN8 (ACT8; AT1G49240) was employed as the internal control gene. The Ct values of each gene relative to that of ACT8 were measured; these were then used to calculate relative expression values. With the exception of YUC11 primers (Song et al. 2019), all primers were designed using QuantPrime (Arvidsson et al. 2008) and are listed in Supplementary Table S1.
In preparation for confocal imaging, embryos were prepared as previously described (Imoto et al. 2021). Images were captured using LSM 5 Live (Carl Zeiss, Oberkochen, Germany) and FV3000 (Olympus, Tokyo, Japan) confocal laser scanning microscopes.
Results
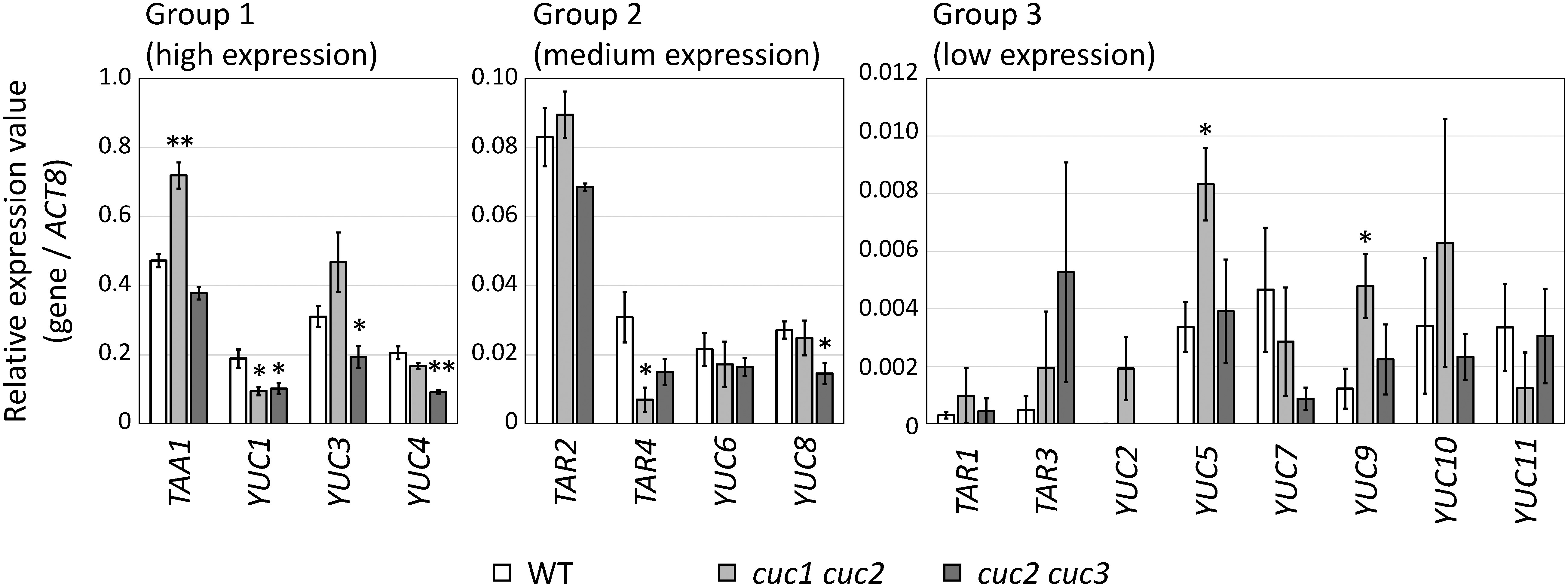
First, we performed comprehensive expression analyses of auxin biosynthetic genes in WT, cuc1 cuc2, and cuc2 cuc3 double mutant embryos using qRT-PCR. Two cuc double mutant combinations were employed because each shows strong apical defects (i.e., the failure of shoot meristem establishment and cotyledon separation) with high penetrance (Hibara et al. 2006). Five TAA1/TAR family genes and 11 YUC family genes were analyzed; according to their relative expression values (R), these genes were classified into the three groups (Figure 1): Group 1 with high expression levels (R>0.1), Group 2 with medium levels (0.1≥R>0.01), and Group 3 with low levels (R≤0.01). In Group 1, the expression level of TAA1 was significantly higher in the cuc1 cuc2 mutant than that in the WT, whereas YUC3 expression was significantly lower in the cuc2 cuc3 mutant than that in the WT (Figure 1, left panel). On the other hand, YUC1 and YUC4 were expressed at lower levels in both the cuc1 cuc2 and cuc2 cuc3 mutants relative to their expression in the WT, and the differences were significant except for the expression of YUC4 in cuc1 cuc2. In Group 2, TAR4 in cuc1 cuc2 and YUC8 in cuc2 cuc3 mutants showed significantly lower expression levels than those detected in the WT (Figure 1, middle panel). The expression of TAR2, which has redundant functions with TAA1, was not significantly different between the WT and cuc double mutant embryos. Among the eight genes in Group 3, the expression of YUC5 and YUC9 was significantly increased in cuc1 cuc2 relative to their expression in the WT (Figure 1, right panel).
Figure 1. Expression level of auxin biosynthetic genes in the heart-stage embryos of WT, cuc1 cuc2, and cuc2 cuc3. Each gene was classified according to its relative expression value (see main text). Means±SE from four biological replicates are shown. Single and double asterisks indicate p<0.05 and p<0.01, respectively, compared with the WT and cuc double mutants (Welch’s t-test).
Our qRT-PCR results together with the reported spatial expression pattern of each gene are summarized in Table 1. Among the YUC family genes, only YUC1 was consistently downregulated in both the cuc1 cuc2 and cuc2 cuc3 mutants, whereas several other genes, including YUC3, YUC4, and YUC8, were specifically downregulated in the cuc2 cuc3 mutant. Two other YUC genes of Group 3, namely YUC5 and YUC9, were upregulated in the cuc1 cuc2 mutant; however, because their expression levels were very low, the biological significance of this expression is unclear. Two TAA1/TAR family genes, namely TAA1 and TAR4, were significantly upregulated in the cuc1 cuc2 mutant, whereas the expression of the remaining genes in this family did not differ significantly between the WT and the mutants. Notably, mutant combinations that reportedly show the cotyledon fusion phenotype similar to that of the cuc mutants are the taa1 tar1 tar2 triple, yuc1 yuc4 yuc10 yuc11 quadruple, and yuc3 yuc9 double mutants (Cheng et al. 2007; Robert et al. 2013; Stepanova et al. 2008); all of these mutants include at least one gene for which the expression was altered in our qRT-PCR experiments. This raises the possibility that changes in these auxin biosynthetic genes are at least partly responsible for the cuc mutant phenotype.
Table 1. Summary of qRT-PCR results.
| Group | Gene | cuc1 cuc2 | cuc2 cuc3 | Previously reported expression pattern* |
|---|---|---|---|---|
| 1 | TAA1 | Up | — | AP1, CB2 |
| YUC1 | Down | Down | AP1, CB3 | |
| YUC3 | — | Down | S1 | |
| YUC4 | — | Down | AP1, CB3, CT3, R1, S1 | |
| 2 | TAR2 | — | — | n.a. |
| TAR4 | Down | — | n.a. | |
| YUC6 | — | — | n.a. | |
| YUC8 | — | Down | P1 | |
| 3 | TAR1 | — | — | n.a. |
| TAR3 | — | — | n.a. | |
| YUC2 | — | — | n.a. | |
| YUC5 | Up | — | n.a. | |
| YUC7 | — | — | n.a. | |
| YUC9 | Up | — | S3 | |
| YUC10 | — | — | overlaps YUC1 and YUC43 | |
| YUC11 | — | — | overlaps YUC1 and YUC43 |
1Robert et al. 2013; 2Stepanova et al. 2008; 3Chen et al. 2007; *Domains of expression are abbreviated as follows: AP, apical protoderm; CB, cotyledon boundary; CT, cotyledon tip; P, provascular; R, root pole; S, suspensor. n. a., not applicable.
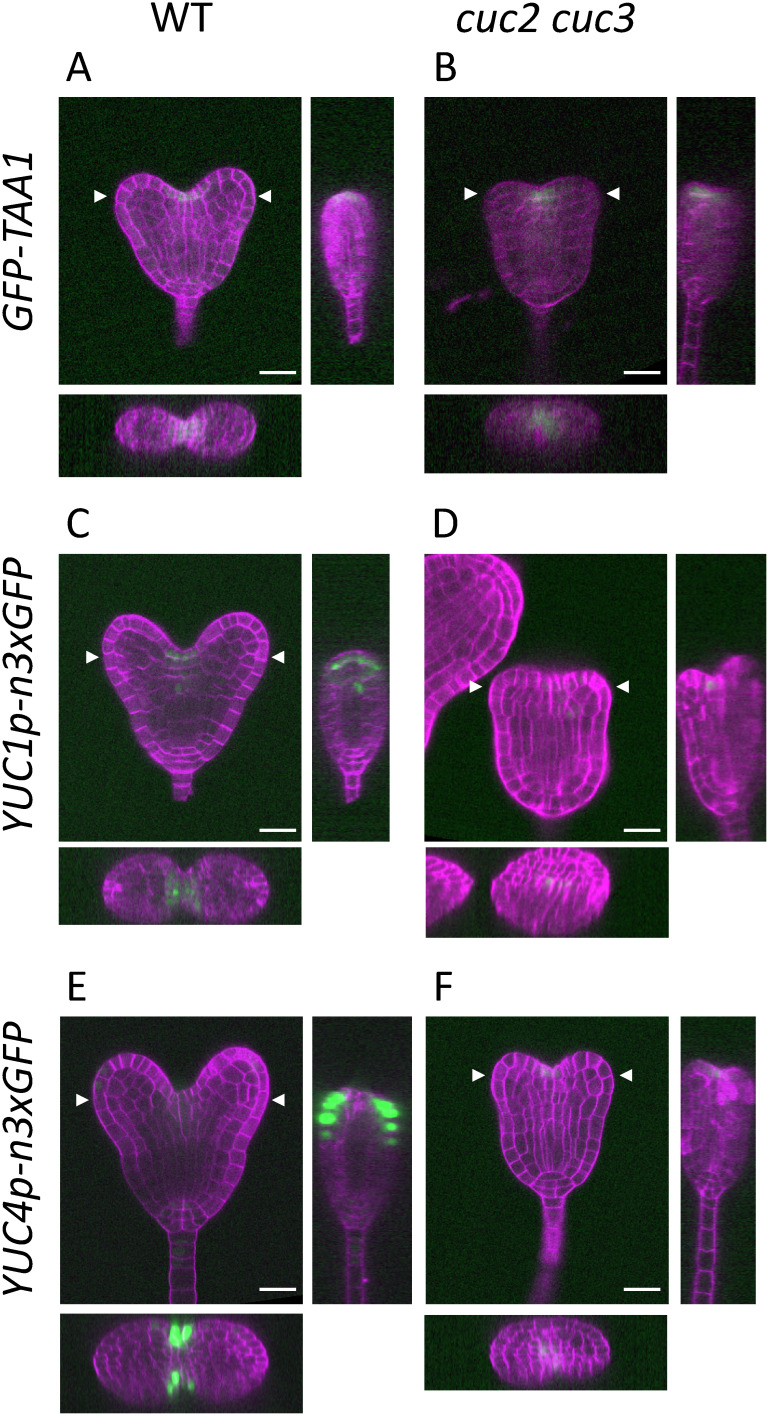
We also performed reporter analysis of TAA1, YUC1, and YUC4, the expression of which spatially overlaps that of the CUC genes in the cotyledon boundary region (Table 1; Cheng et al. 2007; Robert et al. 2013; Stepanova et al. 2008), and we compared their expression patterns in the WT and cuc2 cuc3 double mutant embryos at the heart stage. The expression of TAA1 was detected in the protodermal cells that lay along the cotyledon boundary in the WT (Figure 2A). Within the boundary region, TAA1 expression was localized at the center, as revealed by transverse and sagittal longitudinal sections reconstructed from serial, frontal longitudinal sections (Figure 2A, left and bottom panels). In the cuc2 cuc3 mutant, TAA1 expression was detected in the protodermal cells of the embryo apex corresponding to the cotyledon boundary (Figure 2B). Consistent with the results of the qRT-PCR experiments, the GFP signals of the TAA1 reporter were essentially the same in the WT and cuc2 cuc3 embryos, both in terms of their distribution and intensity. In contrast to TAA1, both YUC1 and YUC4 showed significantly decreased expression in cuc2 cuc3 mutants compared with their expression in the WT (Figure 2C, D, E, F). A belt-shaped expression pattern was observed in the WT for the YUC1 reporter in the protodermal layer along the cotyledon boundary; YUC1 was also expressed in the inner cells that lay just above the provascular cells (Figure 2C). In WT embryos, YUC4 expression was observed in the peripheral region of the cotyledon boundary; additionally, weak YUC4 expression was detected in the cotyledon tips, root primordium, and suspensor (Figure 2E). Contrastingly, YUC1 and YUC4 expression in the cuc2 cuc3 mutant was substantially weakened and often difficult to detect (Figure 2D, F); when expression was observed, it was detected in the apical and central region corresponding to the cotyledon boundary. The reduction of YUC1 and YUC4 expression in the cuc2 cuc3 mutant was not restricted to the boundary region; their expression was reduced in other regions such as in the inner cells for YUC1 and in the cotyledon tips, root primordium, and suspensor for YUC4 (Figure 2D, F). These results indicate that CUC2 and CUC3 are required to promote expression of YUC1 and YUC4 but not the expression of TAA1.
Figure 2. Spatial expression patterns of auxin biosynthetic genes in heart-stage embryos. Confocal images of the embryos carrying the GFP reporters of TAA1 (A and B), YUC1 (C and D), and YUC4 (E and F) in the WT (A, C, F) and cuc2 cuc3 (B, D, F) backgrounds. Images of the reconstructed cross and sagittal sections are shown in the bottom and right panels, respectively. Arrowheads indicate the y-axis position of the cross sections. The sagittal sections were reconstructed from the medial lines of each embryo. The signals of Calcofluor White staining of the cell wall and GFP are represented in magenta and green, respectively. Scale bars: 20 µm.
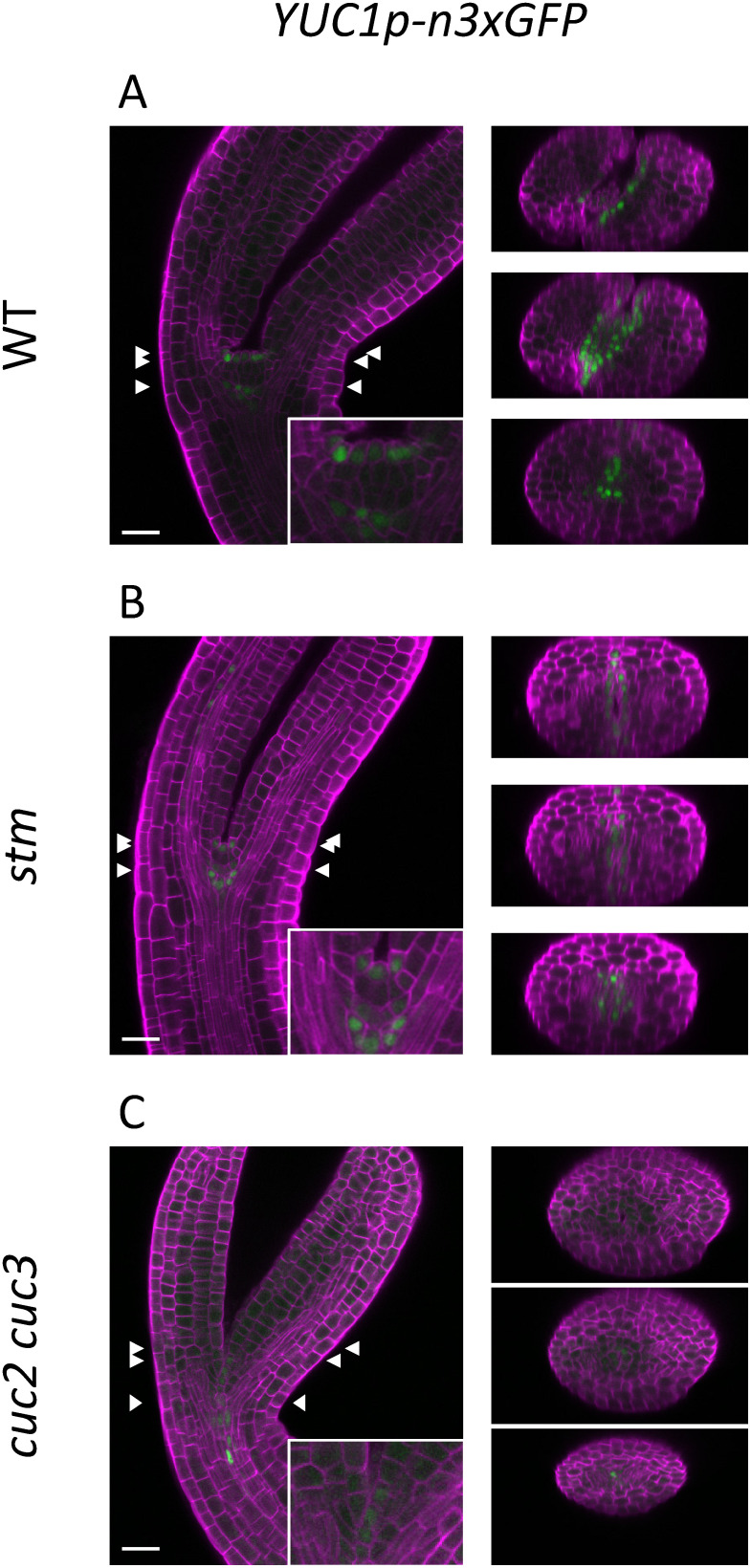
CUC genes are reportedly required for the expression of several downstream genes, among which STM plays a major role in promoting cotyledon separation and shoot meristem establishment (Aida et al. 1999, 2020; Scofield et al. 2018). Thus, it is possible that STM is required to promote YUC1 and YUC4 expression, and the observed reduction in expression in cuc2 cuc3 embryos may be due to the reduced expression of STM (Hibara et al. 2006). To test this possibility, we performed reporter analysis of YUC1 in the stm mutant background. Because the homozygous stm mutant is sterile, we selected homozygous progeny from heterozygous parental plants based on their morphology; the homozygous stm embryos are initially recognizable at the bending-cotyledon stage as those lacking a dome-shaped shoot meristem between the cotyledon primordia (Barton and Poethig 1993; compare Figure 3A, B, insets). In the WT, YUC1 expression was detected in the protoderm cells of the shoot meristem, inner cells underneath the meristem, and the boundary region between the cotyledon primordia and the meristem (Figure 3A). In stm, despite the lack of a shoot meristem, YUC1 expression was detected in the boundary region of the cotyledon primordia and inner cells positioned a few cells away from the protoderm; a similar intensity to that of the signals in the WT was shown (Figure 3B). The cuc2 cuc3 double mutant was also examined at the same stage; only a weak signal at the base of the cotyledon primordia was detected (Figure 3C). Taken together, these results indicate that the expression of YUC1 is dependent on CUC2 and CUC3 but not markedly dependent on STM.
Figure 3. Spatial expression patterns of the auxin biosynthetic gene YUC1 in bending-cotyledon-stage embryos. Confocal images of the embryos carrying the GFP reporter of YUC1 in the WT (A), stm (B), and cuc2 cuc3 (C) backgrounds. The three pairs of arrowheads indicate the y-axis positions of the reconstructed cross sections shown in the right panels. The signals of Calcofluor White staining of the cell wall and GFP are represented in magenta and green, respectively. Scale bars: 20 µm.
Discussion
Our results indicate that the expression of the auxin biosynthetic genes YUC1 and YUC4 are positively regulated by CUC2 and CUC3; at least for YUC1, this regulation does not require STM, which is a major downstream gene of the CUC genes. Expression of these two YUC family genes overlaps that of the CUC genes in the cotyledon boundary region; additionally, their expression was found to be reduced in cuc2 cuc3 double mutant embryos. Our qRT-PCR experiments indicate that CUC1 also positively regulates YUC1 expression. Conversely, among the TAA1/TAR family genes, only TAA1 is reportedly expressed in the cotyledon boundary region and its expression did not require any of the CUC genes. Because the YUC family proteins catalyze a rate-limiting step of indole-3-acetic acid biosynthesis (Mashiguchi et al. 2011), it is possible that the regulation of YUC1 and YUC4 expression by the CUC genes plays a role in auxin biosynthesis in the apical embryo.
The biological relevance of CUC gene-regulated expression of YUC1 and YUC4 is currently unclear. The yuc1 yuc4 double mutant does not show any prominent phenotype of embryogenesis (Cheng et al. 2006); thus, the cotyledon fusion phenotype observed in the cuc2 cuc3 mutant cannot be explained by the mere reduction of YUC1 and YUC4 expression levels. Studies of the cellular localization and activity of the auxin efflux protein PIN1 indicate that auxin molecules that are synthesized in the apical region of the embryo are transported to the tips of the cotyledon primordia (Petrášek and Friml 2009). Previous researchers have suggested that the auxin that accumulates in the primordium tips contributes to restricting the expression domain of the CUC genes to the cotyledon boundary region (Aida et al. 2002; Furutani et al. 2004). One possibility is that CUC genes may fine-tune their own expression domain via this negative feedback regulation involving (YUC gene-dependent) auxin biosynthesis.
Our results also indicate the cell-nonautonomous effect of the CUC genes on YUC1 and YUC4, as well as on other YUC genes that are expressed outside the cotyledon boundary region. The expression of YUC1 and YUC4 is not only restricted to the boundary region but also occurs in the inner cells above the provascular cells (YUC1) and in the tips of the cotyledon primordia and the root (YUC4), where a reduction in expression was observed in the cuc2 cuc3 mutant embryos (Figure 2E, F). Moreover, the cuc2 cuc3 double mutation also significantly reduced the expression levels of YUC3 and YUC8, which are reportedly expressed in the suspensor and provascular cells, respectively (Figure 1; Table 1; Robert et al. 2013); this further supports the cell-nonautonomous actions of the CUC genes on auxin biosynthetic genes. The expression of several YUC family genes, including YUC1, YUC4, and YUC8, is negatively regulated by active auxin in seedlings or in roots (Suzuki et al. 2015; Takato et al. 2017). Because a reduction in local auxin biosynthesis can systemically affect auxin distribution in the embryo (Robert et al. 2013), it is possible that the reduced expression of the YUC1 and YUC4 in the cotyledon boundary region of the cuc2 cuc3 mutant would affect overall distribution of auxin, which would in turn alter YUC gene expression in other regions of the embryo. Therefore, the regulation of YUC1 and YUC4 expression by CUC genes may contribute to the fine-tuning of systemic auxin biosynthesis in the whole embryo rather than specifically affecting the differentiation of the apical region of the embryo.
It is also possible that interactions occur between auxin signaling and other CUC downstream factors such as STM. Auxin is known to negatively regulate expression of STM, which in turn positively regulates cytokinin biosynthesis (Heisler et al. 2005; Jasinski et al. 2005; Yanai et al. 2005). The antagonistic crosstalk of auxin and cytokinin signaling plays an important role in establishing and regulating the meristems (Kurepa et al. 2019; Su et al. 2011). Moving forward, further research should aim to clarify the role of CUC genes in balancing auxin and cytokinin signaling through the regulation of YUC1, YUC4, and STM expression.
Acknowledgments
We thank Jiří Friml for providing seeds of the YUC1 and YUC4 reporters, Arabidopsis Biological Resource Center for seeds of the TAA1 reporter (Stock Number CS16432), Kazuko Onga and Shoko Nagame for technical assistance, and ENAGO (www.enago.jp) for the English language review. This work was supported by MEXT KAKENHI (Grant No. 18H04842, 20H04889 to MA); JSPS KAKENHI (Grant No. 16K07401 to MA); the Yamada Science Foundation to MA; IROAST operational funds to MA.
Abbreviations
- Col
Columbia
- CUC
CUP-SHAPED COTYLEDON
- qRT-PCR
quantitative RT-PCR
- STM
SHOOT MERISTEMLESS
- TAA1
TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1
- TAR
TRYPTOPHAN AMINOTRANSFERASE RELATED
- WT
wild type
- YUC
YUCCA
Supplementary Data
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Tasaka M (1999) Shoot apical meristem and cotyledon formation during Arabidopsis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126: 1563–1570 [DOI] [PubMed] [Google Scholar]
- Aida M, Tsubakimoto Y, Shimizu S, Ogisu H, Kamiya M, Iwamoto R, Takeda S, Karim MR, Mizutani M, Lenhard M, et al. (2020) Establishment of the embryonic shoot meristem involves activation of two classes of genes with opposing functions for meristem activities. Int J Mol Sci 21: 5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Vernoux T, Furutani M, Traas J, Tasaka M (2002) Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129: 3965–3974 [DOI] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B (2008) QuantPrime—a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK (2010) Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev Biol 341: 95–113 [DOI] [PubMed] [Google Scholar]
- Barton MK, Poethig RS (1993) Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119: 823–831 [Google Scholar]
- Belles-Boix E, Hamant O, Witiak SM, Morin H, Traas J, Pautot V (2006) KNAT6: An Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18: 1900–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M (2004) PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131: 5021–5030 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hibara K, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M (2006) Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto A, Yamada M, Sakamoto T, Okuyama A, Ishida T, Sawa S, Aida M (2021) A ClearSee-based clearing protocol for 3D visualization of Arabidopsis thaliana embryos. Plants 10: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Shull TE, Smalle JA (2019) Antagonistic activity of auxin and cytokinin in shoot and root organs. Plant Direct 3: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller B, Weijers D (2009) Auxin control of embryo patterning. Cold Spring Harb Perspect Biol 1: a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J, Friml J (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Robert HS, Grones P, Stepanova AN, Robles LM, Lokerse AS, Alonso JM, Weijers D, Friml J (2013) Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr Biol 23: 2506–2512 [DOI] [PubMed] [Google Scholar]
- Scofield S, Murison A, Jones A, Fozard J, Aida M, Band LR, Bennett M, Murray JAH (2018) Coordination of meristem and boundary functions by transcription factors in the SHOOT MERISTEMLESS regulatory network. Development 145: dev157081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Sun S, Ren H, Grison M, Boutté Y, Bai W, Men S (2019) The SMO1 family of sterol 4α-methyl oxidases is essential for auxin- and cytokinin-regulated embryogenesis. Plant Physiol 181: 578–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Su YH, Liu YB, Zhang XS (2011) Auxin-cytokinin interaction regulates meristem development. Mol Plant 4: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Yamazaki C, Mitsui M, Kakei Y, Mitani Y, Nakamura A, Ishii T, Soeno K, Shimada Y (2015) Transcriptional feedback regulation of YUCCA genes in response to auxin levels in Arabidopsis. Plant Cell Rep 34: 1343–1352 [DOI] [PubMed] [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128: 1127–1135 [DOI] [PubMed] [Google Scholar]
- Takano S, Niihama M, Smith HMS, Tasaka M, Aida M (2010) Gorgon, a novel missense mutation in the SHOOT MERISTEMLESS gene, impairs shoot meristem homeostasis in Arabidopsis. Plant Cell Physiol 51: 621–634 [DOI] [PubMed] [Google Scholar]
- Takato S, Kakei Y, Mitsui M, Ishida Y, Suzuki M, Yamazaki C, Hayashi K-I, Ishii T, Nakamura A, Soeno K, et al. (2017) Auxin signaling through SCFTIR1/AFBs mediates feedback regulation of IAA biosynthesis. Biosci Biotechnol Biochem 81: 1320–1326 [DOI] [PubMed] [Google Scholar]
- Takeda S, Hanano K, Kariya A, Shimizu S, Zhao L, Matsui M, Tasaka M, Aida M (2011) CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J 66: 1066–1077 [DOI] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K (2006) Auxin in action: Signaling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7: 847–859 [DOI] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N (2005) Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.