Abstract
Clinical research to achieve antiretroviral therapy-free remission requires quantitative assays of the HIV-1 reservoir. Intact proviral DNA (IPD) measurement has greater throughput than the quantitative viral outgrowth assay (QVOA). In 25 individuals with well-documented long-term viral suppression, IPD levels and infectious units per million CD4+ T cells by QVOA strongly correlated (r = 0.59, P = .002), and IPD correlated with total cell-associated HIV-1 DNA and cell-associated HIV-1 RNA (r = 0.62 and r = 0.59, P ≤ .002). IPD may provide an accessible marker of inducible replication-competent virus, total numbers of infected cells, and cellular expression of HIV-1 RNA.
Keywords: HIV-1 reservoir, intact proviruses, QVOA, HIV-1 expression, HIV-1 cure
Measuring the human immunodeficiency virus-1 (HIV-1) reservoir in persons on suppressive antiretroviral therapy (ART) has long relied on the quantitative viral outgrowth assay (QVOA), which measures the number of infectious units per million CD4+ T cells (IUPM) following maximal ex vivo T-cell stimulation [1]. Although QVOA has been considered the gold-standard measure, a droplet-digital polymerase chain reaction (PCR)-based method (the intact proviral DNA assay [IPDA]) has recently been developed to separately quantify both sequence-intact (presumably replication-competent) and defective HIV-1 proviral DNA [2]. The IPDA is less labor intense and costly than QVOA, facilitating use in larger-scale clinical research studies evaluating experimental interventions designed to reduce the HIV-1 reservoir. In addition, IPDA does not require the large number of cells needed for QVOA, which are often obtained by leukapheresis. Recent reports have shown substantial correlation between IUPM and IPD levels [2, 3]; however, IUPM is on average approximately 100-fold lower than IPD, in part because the single round of ex vivo activation of T cells used in the QVOA does not induce all intact proviruses and because all intact proviruses may not be inducible ex vivo or in vivo [4].
Simpler PCR-based assays can also quantify total proviral DNA, both intact and defective, but these substantially overestimate the size of the reservoir because a large fraction (approximately 90%–95%) of proviruses are defective [1, 4]. In this report, we further evaluate IUPM and IPD levels in a group of individuals with well-documented long-term HIV-1 RNA suppression on ART, and compare IPD levels relative to several other measures of HIV-1 persistence, including 2 assays of total HIV-1 DNA, cell-associated unspliced HIV-1 pol RNA and residual plasma HIV-1 RNA.
METHODS
Study participants were from the AIDS Clinical Trials Group (ACTG) A5321 observational study [5] and its substudy A5341s, and had well-documented long-term viral suppression on ART without exposure to drugs that might affect HIV-1 reservoirs. All participants provided written informed consent. HIV-1 persistence was assessed by measuring (1) IUPM by QVOA; (2) IPD by IPDA; (3) total HIV-1 proviral DNA by IPDA (sum of intact plus 5ʹ defective plus hypermutated/3ʹ defective DNA, copies/million CD4+ T cells); (4) total cell-associated HIV-1 pol (3ʹ integrase) DNA; and (5) cell-associated HIV-1 pol (3ʹ integrase) RNA by quantitative PCR (qPCR, copies/million CD4+ T cells), all from leukapheresis-derived cells. In addition, (6) residual plasma HIV-1 RNA was quantified by reverse transcriptase-initiated qPCR (RT-qPCR) with single-copy sensitivity.
QVOA was performed as described [6], with replicate cultures of 100 000, 300 000, and 1 million CD4+ T cells. CD4+ T cells were obtained by negative selection from leukapheresis product peripheral blood mononuclear cells (PBMCs), serially diluted and plated, and cultured for 21 days. Culture positivity was determined by measuring p24 by enzyme-immunoassay [6]. IUPM was estimated by maximum likelihood from the numbers of positive and negative cultures, and the corresponding number of cells in each culture [7]. Results with all negative cultures were given values less than the result had there been a single positive culture at the lowest cell input. IPDA was performed as described [2, 8]; this droplet digital PCR method distinguishes deleted and/or hypermutated proviruses from intact proviruses using 2 amplicons and hypermutation discrimination probes; intact proviruses per 106 CD4+ T cells were calculated using separate amplification of a cellular gene (RPP30) after correction for DNA shearing. Total cell-associated HIV-1 pol (3ʹ integrase) DNA and cell-associated HIV-1 pol RNA were measured by qPCR/RT-qPCR as reported, using the same PCR primers and probes for HIV-1 DNA and RNA [9]. Plasma HIV-1 RNA by single-copy assay used the same primers and probes targeting 3ʹ integrase (iSCA) as those used for cell-associated HIV-1 DNA and cell-associated HIV-1 RNA [10]. Cell-associated HIV-1 DNA and cell-associated HIV-1 RNA levels per million CD4+ T cells were obtained by dividing the assay-generated copies/million PBMCs by the proximal clinic-reported CD4+ T-cell percentage (× 0.01).
Spearman correlations assessed associations between the different assays, analyzing results below the assay limit as the lowest rank. To quantify differences between the HIV-1 persistence measures, the median ratio between assays is presented. For this ratio, when below-limit results were in the numerator those ratios were assigned the lowest rank; when below-limit results were in the denominator those ratios were assigned the highest rank; participants with below-limit (undetectable) results for both numerator and denominator were excluded. Confidence intervals for medians were obtained using nonparametric methods based on order statistics. Analyses were performed using SAS Statistical Package, version 9.4.
RESULTS
Twenty-five participants were analyzed (92% male, sex corresponding with gender; 16% black; median 8.9 (minimum–maximum, 3.7–17.4) years on ART at sampling; all enrolled at sites in the United States). The majority (19, 76%) initiated ART during chronic infection; 4 (16%) initiated ART within 45 days of acute HIV-1 infection and 2 (8%) initiated ART as an HIV-1 controller (pre-ART HIV-1 RNA levels of 516 and 670 copies/mL, respectively). Three additional participants (3/28, 11%) were excluded due to amplification or detection failure of the IPD assay [8, 11].
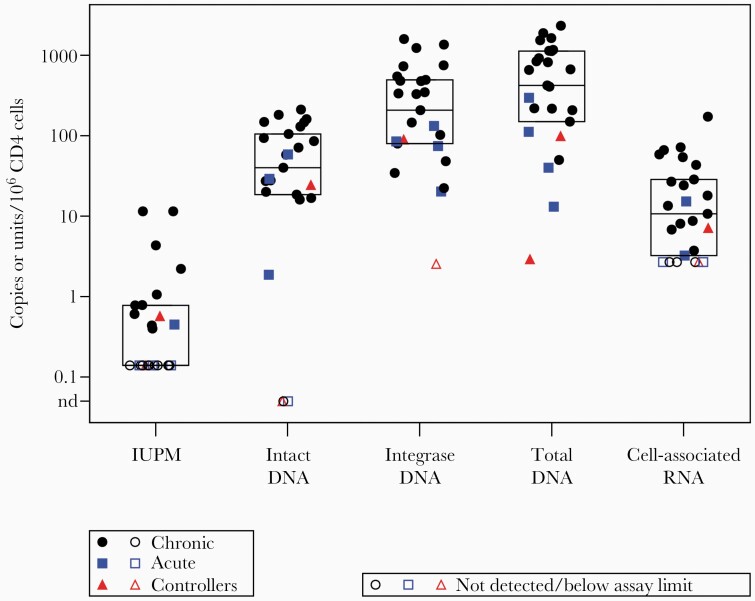
The median IUPM was < 0.14 per 106 CD4+ T cells, in comparison to median IPD of 40.1 copies per 106 CD4+ T cells. Total HIV-1 DNA levels were median 208 and 421 copies per 106 CD4+ T cells for cell-associated HIV-1 DNA (by integrase DNA qPCR) and total DNA by IPDA (sum of intact and defective DNA), respectively. Median cell-associated HIV-1 RNA was 11 copies per 106 CD4+ T cells (Figure 1). The majority (68%, 17/23) of these long-term suppressed individuals had plasma HIV-1 RNA < 0.3 copies/mL; the maximum level was 3.6 copies/mL.
Figure 1.
Cell-associated HIV-1 measurements in people with HIV-1 on long-term ART. Boxes display median, quartile 1, and quartile 3 levels. Total DNA is the sum of intact, 5ʹ defective, and hypermutated/3ʹ defective HIV-1 proviruses from the intact proviral DNA assay. Total HIV-1 DNA was also estimated by measuring integrase DNA. Circles represent participants who had initiated ART in chronic HIV-1 infection, squares represent ART initiation in acute HIV-1 infection, and triangles represent participants who had initiated ART as HIV-1 controllers. Abbreviations: ART, antiretroviral therapy; HIV-1, human immunodeficiency virus-1; IUPM, infectious units per million CD4+ T cells by the quantitative viral outgrowth assay; nd, not detected.
IPD levels were approximately 100- to 300-fold higher than IUPM. Ratios were quantified between the HIV-1 DNA assays, which had minimal assay censoring. Total HIV-1 DNA by IPDA (sum of intact and defective proviruses) were median 11-fold higher than intact proviruses (95% confidence interval [CI], 8- to 20-fold). Note that this 11-fold ratio corresponds to median 9% intact proviruses (95% CI, 5%–13%). Cell-associated HIV-1 DNA copies (by integrase DNA qPCR) were median 5-fold higher than intact proviruses (95% CI, 3- to 10-fold). Total HIV-1 DNA levels by IPDA were median 1.9-fold higher than cell-associated HIV-1 DNA by integrase DNA qPCR (95% CI, 1.2- to 2.3-fold).
Levels of latent intact HIV-1 estimated by IUPM and IPD were positively correlated (r = 0.59; P = .002), though there were 5 participant samples with IPD levels > 50 copies per 106 CD4+ T cells but IUPM below the limit of quantification. IPD levels also strongly correlated with measures of total HIV-1 DNA (r = 0.62, r = 0.76; P = .001, P < .001) and levels of cell-associated HIV-1 RNA (r = 0.59; P = .002; Figure 2). In contrast, IUPM only correlated with IPD; significant correlations of IUPM were not identified with the 2 total HIV-1 DNA measures (r = 0.23, r = 0.16; P = .28, P = .45) and cell-associated RNA (r = 0.16; P = .45). No apparent associations were observed between iSCA and any of the other virology measures (r values 0.04 to 0.21; P ≥ .33).
Figure 2.
Spearman correlations between IPD levels and the other cell-associated HIV-1 measurements. Total DNA is the sum of intact, 5ʹ defective, and hypermutated/3ʹ defective HIV-1 proviruses from the intact proviral DNA assay. Total HIV-1 DNA was also estimated by measuring integrase DNA. Abbreviations: HIV-1, human immunodeficiency virus-1; IPD, intact proviral DNA; IUPM, infectious units per million CD4+ T cells by the quantitative viral outgrowth assay; nd, not detected by the intact proviral DNA assay.
DISCUSSION
Our results add to the growing body of literature on the IPDA and other measures of HIV-1 persistence on ART. Consistent with recent reports [2, 3], inducible replication-competent proviruses by IUPM strongly correlated with PCR-based IPD levels. IUPM levels are typically 100 times lower than IPD, in part because the single round of activation in QVOA does not induce all proviruses [4]. In addition, proviruses determined as intact by IPDA might have inactivating mutations, insertions, or deletions elsewhere in the proviral genome rendering them defective and unable to produce infectious virus. Thus, IPD levels may overestimate the intact reservoir [3]. Intact proviruses by IPDA may also be in deep latency in regions of the human genome that are not activated despite strong ex vivo stimulation [12]. In this study we observed 5 cases of IPD levels > 50 copies per 106 CD4+ T cells but IUPM below the limit of quantification (Figure 2), which could be explained by intact proviruses that are noninducible by QVOA activation.
While IPD levels correlated well with measures of total DNA, these latter measurements are mostly quantifying defective proviruses, which cannot lead to production of infectious virus. IPD levels also correlated with cell-associated HIV-1 RNA, which may be expressed from intact or defective proviruses. While plasma HIV-1 RNA levels by iSCA did not correlate substantively with the other cell-based assays, we were limited by our modest sample size and the large fraction of iSCA levels below assay limit (<0.3 copies/mL). The advent of the IPD assay is a substantive advance to the cure field [2], as a scalable assay to increase our understanding of HIV-1 persistence on ART and to evaluate the effects of interventions on the intact subset of proviruses.
The percentage of intact proviruses (9%) in HIV-1–infected CD4+ T cells in our participants agrees with other reports [3, 5, 8]. Efforts are underway to build a larger IPD database with relevant participant characteristics to better understand factors that may influence the frequency of intact proviruses. These include timing of ART initiation, CD4+ T-cell count nadir, and duration of suppressive ART [5]. Participants in our study who started ART during acute infection or as an HIV-1 controller often had lower measurements of HIV-1 persistence, but comparisons would need to address potentially differing durations of ART, because measures of HIV-1 persistence decline over years of suppressive ART [1, 3, 5]. The modest sample size of the present study did not support robust assessment of correlates.
Viral diversity including subtype differences and multiple founder viruses can affect the ability to quantify HIV-1 reservoirs by IPDA or other nucleic acid amplification and detection methods [8, 11]. Although only 3 participants in our study could not be analyzed due to IPDA amplification or detection failure, current early-phase research studies might involve a small number of participants who receive intensive interventions designed to reduce the viral reservoir. Obtaining pre- versus postintervention measurements for every participant may require additional sequencing and tailored primers/probes for such cases to quantify effects on intact proviral levels and other measures of viral nucleic acids [11, 13].
Ultimately, the field of HIV-1 cure research will need to fully evaluate which on-ART measures of persistence best predict the clinical end points of time to viral rebound [14] or viral control following ART withdrawal. Such clinically validated biomarkers [15] are likely to accelerate progress towards identifying therapies that can achieve ART-free remission.
Notes
Acknowledgments. We gratefully acknowledge all the members of the A5321 and A5341s teams, and commend all the staff at AIDS Clinical Trials Group sites for their invaluable contributions to enroll and evaluate the study population. Particular thanks go to P. Nathan Enick, Michele Sobolewski, and Jana Jacobs for their work and expertise on the laboratory assays. In addition, we thank Greg Laird and Accelevir Diagnostics for performing the intact proviral DNA assays. We especially thank and acknowledge the study participants for their ongoing contributions without whom this research would not be possible.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grant numbers UM1AI068636, UM1AI106701, and UM1AI068634). R.T.G. receives grant funding from the Harvard University Center for AIDS Research (National Institute of Allergy and Infectious Diseases P30 AI060354) and the AIDS Clinical Trials Group (National Institute of Allergy and Infectious Diseases 2 UMAI069412-09).
Potential conflicts of interest. R. T. G. has served on a scientific advisory board for Merck. J. W. M. has been a consultant to Gilead Sciences, has received research grants from Gilead, and owns share options in Co-Crystal Pharmac*euticals, Infectious Diseases Connect, and Abound Bio, which have no relationship to the current work. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed
Contributor Information
Ronald J Bosch, Center for Biostatistics in AIDS Research, Harvard TH Chan School of Public Health, Boston, Massachusetts, USA.
Rajesh T Gandhi, Infectious Diseases Division, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Hanna Mar, Center for Biostatistics in AIDS Research, Harvard TH Chan School of Public Health, Boston, Massachusetts, USA.
Joseph J Eron, Department of Medicine, University of North Carolina, Chapel Hill, North Carolina, USA.
Joshua C Cyktor, Division of Infectious Diseases, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Deborah K McMahon, Division of Infectious Diseases, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
John W Mellors, Division of Infectious Diseases, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
References
- 1. Bruner KM, Hosmane NN, Siliciano RF.. Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol 2015; 23:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bruner KM, Wang Z, Simonetti FR, et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019; 566:120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Falcinelli SD, Kilpatrick KW, Read J, et al. Longitudinal dynamics of intact HIV proviral DNA and outgrowth virus frequencies in a cohort of ART-treated individuals. J Infect Dis 2021; 224:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ho YC, Shan L, Hosmane NN, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gandhi RT, Cyktor JC, Bosch RJ, et al. Selective decay of intact HIV-1 proviral DNA on antiretroviral therapy. J Infect Dis 2021; 223:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Enick PN, Brooker JP, Tumiotto CM, et al. Comparison of methods to quantify inducible HIV-1 outgrowth. J Virus Erad 2021; 7:100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rosenbloom DI, Elliott O, Hill AL, et al. Designing and interpreting limiting dilution assays: general principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect Dis 2015; 2:ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simonetti FR, White JA, Tumiotto C, et al. Intact proviral DNA assay analysis of large cohorts of people with HIV provides a benchmark for the frequency and composition of persistent proviral DNA. Proc Natl Acad Sci USA 2020; 117:18692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hong F, Aga E, Cillo AR, et al. Novel assays for measurement of total cell-associated HIV-1 DNA and RNA. J Clin Microbiol 2016; 54:902–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cillo AR, Vagratian D, Bedison MA, et al. Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. J Clin Microbiol 2014; 52:3944–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kinloch NN, Ren Y, Conce Alberto WD, et al. HIV-1 diversity considerations in the application of the intact proviral DNA assay (IPDA). Nat Commun 2021; 12:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Einkauf KB, Lee GQ, Gao C, et al. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J Clin Invest 2019; 129:988–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaebler C, Falcinelli SD, Stoffel E, et al. Sequence evaluation and comparative analysis of novel assays for intact proviral HIV-1 DNA. J Virol 2021; 95:e01986-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li JZ, Etemad B, Ahmed H, et al. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS 2016; 30:343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mildvan D, Landay A, De Gruttola V, et al. An approach to the validation of markers for use in AIDS clinical trials. Clin Infect Dis 1997; 24:764–74. [DOI] [PubMed] [Google Scholar]