Abstract
Cerebral malaria (CM) is the severest form of Plasmodium falciparum infection. Children under 5 years old are those most vulnerable to CM, and they consequently have the highest risk of malaria-related death. Parasite-associated factors leading to CM are not yet fully elucidated. We therefore sought to characterize the gene expression profile associated with CM, using RNA sequencing data from 15 CM and 15 uncomplicated malaria isolates from Benin. Cerebral malaria parasites displayed reduced circulation times, possibly related to higher cytoadherence capacity. Consistent with the latter, we detected increased var genes abundance in CM isolates. Differential expression analyses showed that distinct transcriptome profiles are signatures of malaria severity. Genes involved in adhesion, excluding variant surface antigens, were dysregulated, supporting the idea of increased cytoadhesion capacity of CM parasites. Finally, we found dysregulated expression of genes in the entry into host pathway that may reflect greater erythrocyte invasion capacity of CM parasites.
Keywords: cerebral malaria, cytoadherence, Plasmodium falciparum, transcriptomics, var genes
Transcriptome analysis of P falciparumisolates from Benin revealed that CM parasites have reduced circulation time and an overall increase in vargenes expression. These results may be related to a higher cytoadherence capacity of CM isolates.
Malaria is a vector-borne disease that affected 241 million people in 2020 [1]. Plasmodium falciparum is the species responsible for most cases and deaths in sub-Saharan Africa. Cerebral malaria (CM) is the most severe form of P falciparum infection and mainly affects children under 5 years old, who account for approximately 80% of all malaria-related deaths in the World Health Organization (WHO) African Region [1]. Cerebral malaria pathophysiology is linked to the ability of P falciparum to adhere to specific receptors in host brain capillaries [2, 3] during the asexual intraerythrocytic development cycle (IDC). Throughout this cycle, the parasite evolves from ring, young to late trophozoite, schizont, and finally merozoite form. Erythrocytes infected with P falciparum mature forms (ie, trophozoites and schizonts) can adhere to the host endothelium, through the expression of variant surface antigens (VSAs) exported to the erythrocyte membrane [4].
Recent studies have shown that infected erythrocytes’ (iE) circulation time is decreased in severe malaria attacks compared to nonsevere cases, reflecting the greater adherence to endothelial cells of iE containing mature parasite forms [5–8]. Higher adherence capacity confers a real advantage to the parasite by protecting iE from splenic clearance, thus promoting increased parasitemia [7]. This phenomenon is modulated by the host’s immune system [9] and by iE adherence capacity that is mainly mediated by expression of the Plasmodium falciparum Erythrocyte Membrane Protein 1 (PfEMP1) encoded by the multigenic var genes family [10]. Specific PfEMP1 domains [11], among which domain cassette 8 (DC8) and DC13, are specifically associated with CM [12, 13]. The ability of these DCs to bind endothelial protein C receptor (EPCR) and intercellular adhesion molecule 1 (ICAM-1) in host brain capillaries is a major virulence factor of CM [2, 14, 15].
The present work is part of the NeuroCM study, in which host and parasite factors that lead to neuroinflammation in CM are investigated. Children presenting with CM and uncomplicated malaria (UM) were enrolled in a cross-sectional study in Benin, West Africa during the malaria transmission season [16]. A first comparative study highlighted that CM isolates display greater adherence to a human brain endothelial cell line (Hbec-5i) than UM isolates, and a specific var genes expression profile in CM was confirmed [17].
Although var genes domains expression associated with CM has been extensively studied [13, 17, 18], the involvement of other parasite factors is not yet fully understood. Previous studies have shown that parasites display distinct gene expression profiles associated with severe malaria (SM) [5, 6]. A more metabolically quiescent phenotype associated with SM was demonstrated [5]. In addition, deregulated genes implicated in var genes expression or PfEMP1 presentation to the iE membrane were identified [5, 6]. In this study, we aimed to characterize P falciparum isolates’ gene expression in the context of pediatric malaria, by comparing the whole transcriptome of isolates from Beninese children enrolled in the NeuroCM study.
MATERIALS AND METHODS
Participant Recruitment
Ethical clearance was obtained from Comité National d’Ethique pour la Recherche en Santé au Bénin (N°67/MS/DC/SGM/DRFMT/CNERS/SA; October 17, 2017) and by Comité Consultative de Déontologie et d’éthique of Institut de Recherche pour le Développement (October 24, 2017). Children under 6 years old presenting with CM or UM were recruited in 2018 in Southern Benin. Malaria infection was diagnosed with positive P falciparum thin or thick blood smear. For CM, a Blantyre score ≤2 was required. UM was defined by a parasitemia between 1000 and 5.105 parasites/μL and no clinical or biological signs of severe malaria. The recruitment of participants has been described previously [16, 17].
Sample Preparation and RNA Sequencing
Plasmodium falciparum RNA was extracted from whole blood, preserved at −80°C in TRIzol LS reagent (Life Technologies), with a phenol-chloroform protocol, and purified using RNeasy minikit (QIAGEN). The RNA quality, purity, and concentration were assessed with a NanoDrop 2000c (Thermo Fisher Scientific) and Agilent RNA 6000 Pico Kit (Agilent Biotechnologies) (Supplemental Data 1). Samples with both RNA Integrity Number ≥6 and at least 100 ng of total RNA available were retained for sequencing. Libraries were prepared using the TruSeq Stranded mRNA protocol (Illumina) and sequenced after qPCR quantification on a NovaSeq 6000 (Illumina) system, using 2 × 100 cycles (paired-end reads, 100 nucleotides) and aiming to obtain approximately 20M clusters per sample (ICGex, next-generation sequencing [NGS] platform; Institut Curie).
Anti-Plasmodium falciparum Antibody Quantification in Patient Plasma
Detection and quantification of total antibodies against P falciparum was performed to evaluate previous exposure. Immunoglobulin (Ig)G/A/M antiplasmodial antibodies were quantified by indirect immunofluorescence assay using whole schizonts of the 3D7 P falciparum strain as crude antigens and fluorescein-linked antihuman IgG/A/M (BioRad; Hercules, CA) as conjugate. Plasma antibody levels were estimated by serologic titers using 4-fold dilution series (1:16 to 1:4096), by recording the last positive dilution. Two blinded readers quantified the intensity of fluorescence by microscopy. For statistical analysis, antibody titers were classified into 3 groups: negative, (or 0) corresponding to titers <1:64; positive, corresponding to titers 1:64, 1:256, and 1:1024; and strongly positive, corresponding to titers ≥1:4096 [19].
IgG against VSAs expressed on 3D7 P falciparum iE were also detected and quantified by flow cytometry. Patient plasma was brought into contact with the iE suspension and binding revealed, after washing, with goat IgG antihuman IgG Fc labeled with phycoerythrin. SYTO 61 label (Thermo Fisher Scientific) was also used for parasite DNA labeling in iE (Supplementary Method). Control of unspecific fluorescence signal was performed for goat IgG antihuman on infected and uninfected erythrocyte (uE) and for each patient plasma on uE. Mean fluorescence intensity (MFI) was defined as follows: MFI = (MFIiE—MFIiE goat IgG) – (MFIuE—MFIuE goat IgG).
Gene Expression Analysis
Bioinformatical analysis was performed following a previously published script: https://github.com/gtonkinhill/falciparum_transcriptome_manuscript [5]. Raw reads were mapped on P falciparum (3D7 strain, PlasmoDB v49) and human (hg38, UCSC genome browser) reference genomes with Hisat2 v2.2. Samples below a minimum cutoff of 4 million reads mapped to the parasite genome were excluded. Parasite stages proportions were evaluated, referring to published RNA-seq dataset [20] (Supplementary Method). Unwanted variations factors were estimated using control genes that are not expected to vary with disease severity (from a previous study [21] comparing pregnancy-associated and childhood malaria) with the Remove Unwanted Variation algorithm (R package ruv 0.9.7 [22]), adjusting for systematic errors. Differential expression (DE) analysis was performed including proportions of parasites at the ring-stage and unwanted variations factors as input of the Limma/Voom pipeline [23]. A gene was considered as DE for P adjusted values ≤.05 (Benjamini-Hochberg) and a fold-change ≥1.5. The Gene Ontology (GO) [24] and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway [25] enrichment analyses were performed by applying a hypergeometric test (P ≤ .05) on the GO and KEGG annotations downloaded from PlasmoDB (version 49). The protein-protein interaction (PPI) network was generated from the STRING online tool (version 11.5) [26]. Settings were set to high confidence interaction score (0.7) and coexpression interactions source. Then, cluster analysis was performed in Cytoscape 3.8.2 [27] with MCode application version 2.0 [28]. The RNA-seq data are uploaded in Gene Expression Omnibus (GEO) repository (GSE186820).
var Genes Transcripts
var transcripts were assembled de novo from RNA-seq reads using a published script (https://github.com/PapenfussLab/assemble_var) [5]. The nature of each subdomain (eg, DBLα1.1-2-3/2) was assessed by performing an alignment with MAFFT [29] against the Vardom database [11]. The FPKM (fragments per kilobase million) values were evaluated with edgeR. To evaluate var genes assembly quality, reads were aligned with Hisat2 v2.2.1 [30] to a database composed of ATS domains from Vardom’s 7 reference genomes [11], varDB [31], and RNA-seq assembly and to assembled exon 1 sequences in each sample. To evaluate var genes abundances, reads mapped to ATS were counted and were normalized accounting for both the library size and the parasite’s stage proportions, based on synchronized 3D7 strain RNA-seq data [20].
RT-qPCR Validations
Expression of 8 genes was measured to confirm the RNA-seq results on the Rotor-Gene qPCR system (QIAGEN). Gene levels transcripts were evaluated with serial dilutions of plasmids (GenScript). Results were normalized with a housekeeping gene expression and accounting for the parasite’s developmental stage (Supplementary Method). var genes expression was assessed by 3 independent methods. Published DBLα primers [12] and newly developed primers targeting the ATS domain (ATS-2 primers based on varDB dataset [31], Supplemental Data 2) were used on the Rotor-Gene qPCR system (QIAGEN) following the same methodology. The qPCR-TaqMan developed by Hofmann et al [32] targeting the ATS domain (ATS-1) was performed on a Viia7 qPCR system (Applied Biosystems) (Supplementary Method).
Statistical Analysis
Quantitative variables are presented as medians (10th–90th percentiles). Statistical differences were assessed using the Mann-Whitney U test. Qualitative variables are presented as proportions and statistical differences were measured with the χ2 test.
RESULTS
Samples Analysis
Parasite RNA was extracted from a total of 73 CM and 98 UM cases. Based on RNA quantification and quality criteria, 21 UM and 16 CM samples were subsequently selected for high-throughput sequencing (Supplemental Data 1). From RNA-seq mapping statistics, 30 samples (15 CM and 15 UM) were retained for transcriptome analysis (Supplemental Data 3). Selection criteria are detailed in the methods.
Clinical and biological features are displayed in Table 1. Children with CM presented a higher parasitemia (P < .001, Mann-Whitney U test), and children with UM were older (P = .004, Mann-Whitney U test). Hemoglobinemia and blood glucose were lower in CM (P < .001, Mann-Whitney U test). Total antibodies titers, reflecting previous exposure to Plasmodium sp, did not differ between CM and UM (P = .57, χ2 test). Moreover, the number of patients having IgG against VSA was not different between clinical presentations, with 3 of 15 CM (20%) and 7 of 15 UM (47%) (P = .12, χ2 test) (Supplemental Data 4).
Table 1.
Clinical and Biological Features of Participants
| Clinical and Biological Characteristics | CM (n = 15) | UM (n = 15) | P Value |
|---|---|---|---|
| Age (months) | 42.4 [30.2–55.5] | 55 [44.5–70.1] | .004 |
| Gender: female no./total no. (% female) | 6/15 (40%) | 4/15 (27%) | NS |
| Parasitemia (parasites/μL) | 459 000 [281 600–1 152 286] | 252 000 [148 542–325 626] | <.001 |
| Hemoglobinemia (g/dL) | 6.5 [3.78–9.18] | 9.8 [8.34–10.68] | <.001 |
| Blood glucose (g/L) |
0.14 [0.1–0.71] | 1.05 [0.82–1.36] | <.001 |
| Malarial retinopathy | Yes: 5 | NA | NA |
| No: 4 | |||
| Not evaluated: 6 | |||
| Blantyre score | 0: 1 (6.7%) | NA | NA |
| 1: 5 (33.3%) | |||
| 2: 9 (60%) | |||
| Mortality (%) | 57% | NA | NA |
Abbreviations: CM, cerebral malaria; NA, not applicable; NS, not significant; UM, uncomplicated malaria.
Circulating Parasites’ Gene Expression
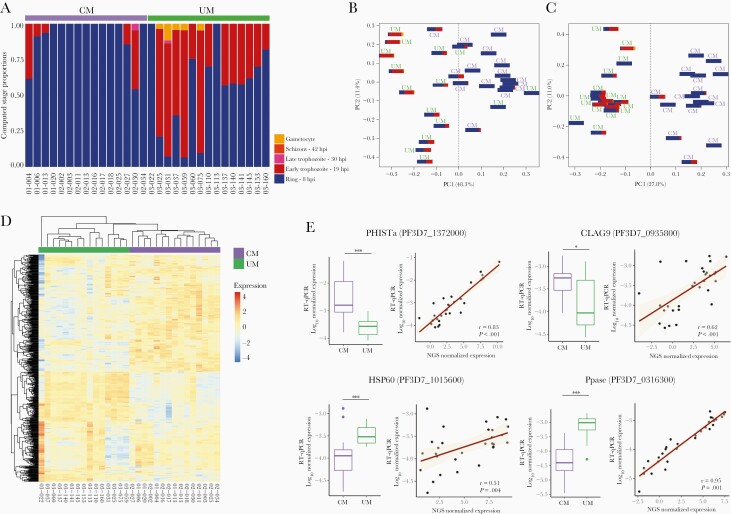
The all-gene analysis was focused on the parasite core genome (VSAs were excluded). Parasite developmental stage proportions were evaluated in each sample based on total gene expression measured by RNA-seq [5] (Figure 1A, Supplemental Data 5). The mean parasite age expressed in hours post infection (hpi) was statistically lower in CM cases, with a median age of 8 hpi (8–11.2) compared with 12.6 hpi (8.8–16.7) in UM (P < .001, Mann-Whitney U test). The difference was confirmed by the microscopic reading of thin blood smears (P < .001, Mann-Whitney U test) (Supplementary Method, Supplemental Data 5). The all-gene expression principal component analysis (PCA) showed that both clinical presentation and parasite developmental stage affect the separation by the first component (Figure 1B). After correcting gene expression for both life-cycle stage effect and unwanted variations, CM and UM isolates were perfectly separated based on clinical presentation (Figure 1C). Differential expression analysis was performed considering the gene expression represented in Figure 1C and revealed 551 DE genes (Supplemental Table 2), 284 of which were upregulated and 267 were downregulated in CM compared with UM. Further hierarchical clustering of the DE genes showed a separation of CM and UM isolates, reflecting a common gene expression pattern within each clinical group (Figure 1D).
Figure 1.
Cerebral malaria (CM) and uncomplicated malaria (UM) parasites gene expression. (A) Proportions of ring, early trophozoite, late trophozoite, schizont, and gametocyte stage parasites in each sample. (B) Principal component analysis (PCA) plots of the all-gene expression normalized for library size (read counts available in Supplemental Table 1); (C) PCA plot of gene expression normalized for library size, life-cycle stage effect, and unwanted variations (read counts available in Supplemental Table 1). (D) Heatmap of differentially expressed genes. An upregulated expression is represented in red and the downregulation is in blue. (E) Distribution of normalized expression values measured by RT-qPCR in CM and UM is represented (boxplot). The P values were calculated with the Mann-Whitney U test and are represented by as follows: *** <.001, ** <.01, and * <.05. The correlation between the normalized expression values measured by RT-qPCR and RNA-seq was represented through the regression line and tested by the Spearman rank test (Spearman rank correlation coefficient ρ and P values are shown).
The differential gene expression measured by RNA-seq was validated by RT-qPCR with 8 genes (Figure 1D, Supplemental Data 6) for which the corresponding proteins have variable functions and locations. The RT-qPCR expression was strongly correlated with gene expression measured by RNA-seq, with all Spearman rank correlation coefficients ρ greater than 0.5 (all P < .01, Spearman rank test) (Figure 1E).
Differential Expression Gene Characterization
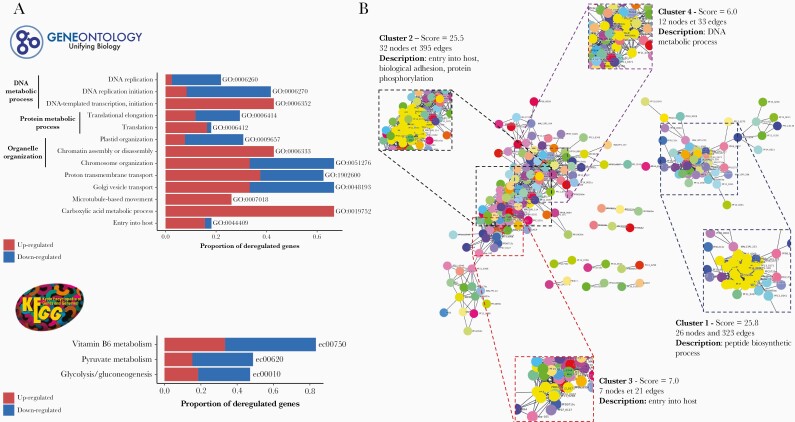
The GO and KEGG pathways gene set enrichment analysis (GSEA) were performed including all the DE genes (Figure 2A, Supplemental Data 7). Thirteen GO pathways were deregulated, with a minimum of 18% deregulated genes identified in each (P ≤ .05, hypergeometric test). Three of 13 deregulated pathways concerned DNA metabolic processes, 2 of 13 were involved in protein metabolic processes, and 3 of 13 were implicated in organelle organization. The entry into host pathway was deregulated, including 12 upregulated genes in CM, among which were apical membrane antigen 1 (ama-1, PF3D7_1133400), plasmepsin IX and plasmepsin X (pmIX, PF3D7_1430200 and pmX, PF3D7_0808200), and erythrocyte binding antigen-175 (eba175, PF3D7_0731500). Concerning KEGG pathways results, we found 3 associated pathways, with at least 40% deregulated genes.
Figure 2.
Gene set enrichment analysis (GSEA) and protein-protein interaction (PPI) network of differential expression (DE) genes analysis. (A) Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis. Barplots show the proportion of deregulated genes in each pathway. Upregulated genes in cerebral malaria (CM) are in red and downregulated in CM are in blue. (B) String’s PPI network of DE genes. Cluster found with MCode and associated pathways are shown.
A PPI network analysis was performed on 546 of 551 DE genes that were annotated in the STRING database. Results showed a very dense network with 212 nodes and 2976 edges with a significant PPI enrichment (P < .001), reflecting biologically connected proteins (Figure 2B). Four highly interconnected clusters were found (Figure 2B, Supplemental Data 8). Cluster 1, containing 26 nodes and 323 edges, was composed exclusively of ribosomal proteins (all downregulated), reflecting peptide biosynthesis process deregulation. In cluster 2, there were 32 nodes and 395 edges, and the main pathways represented in this cluster were entry into host (rhoptry-associated protein 2 [PF3D7_0501600], AMA-1 (PF3D7_1133400) and 6-cysteine protein (PF3D7_0508000), biological adhesion (CLAG proteins), and protein phosphorylation. In cluster 3, composed of 7 nodes and 21 edges, proteins were implicated in entry into host through erythrocyte binding antigen-181 (EBA-181, PF3D7_0102500) and PMX (PF3D7_0808200). Cluster 4 contained 12 nodes and 33 edges representing activities related to DNA metabolic processes.
Upregulated genes involved in PfEMP1 expression regulation were manually identified, including PHISTa and PHISTb family genes (PF3D7_1372000/MAL13P1.470, PF3D7_0424600), set9 and set10 (PF3D7_0508100, PF3D7_1221000), apiAP2 (PF3D7_1350900), fikk9.6 (PF3D7_0902500), and semp1 (PF3D7_0702400). Downregulated genes, including exported protein 2 and exported protein 3 (exp2 PF3D7_1471100; exp3 PF3D7_1024800), were also identified.
var Genes Expression
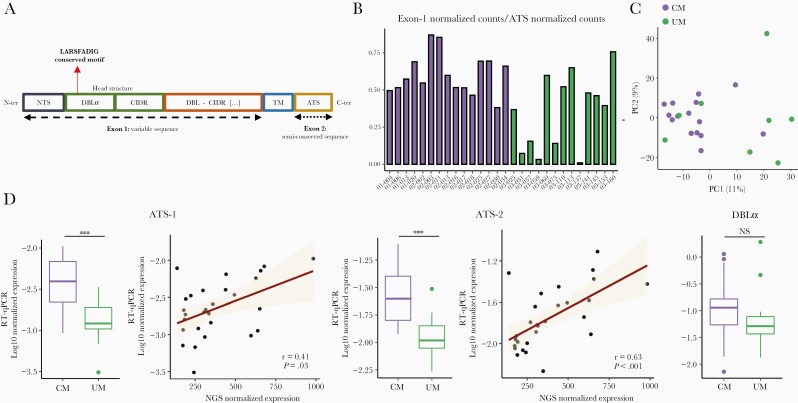
var genes from each isolate were assembled from RNA-seq data (Supplemental Tables 3 and 4). Fifteen CM and 13 UM samples were analyzed with at least 1 var genes domain identified. As we obtained more P falciparum mapped reads in CM isolates compared with UM (P = .016, Mann-Whitney U test), we sought to assess the quality of the assemblies by comparing the number of mapped reads to exon 1 and ATS semiconserved sequence (Figures 3A and B). With a 0.3 exon1/ATS counts proportion threshold, we considered 15 CM and 8 UM for downstream analysis.
Figure 3.
var genes expression. (A) Plasmodium falciparum Erythrocyte Membrane Protein 1 (PfEMP1) schematic organization. (B) Proportion of assembled var genes in each sample. We compared the number of reads mapped to exon 1 and ATS to assess the quality of reconstructions. (C) Principal component analysis (PCA) based on var genes domains expression. (D) Var genes expression based on DBLα and ATS domains. Distribution of normalized expression values measured by RT-qPCR in cerebral malaria (CM) and uncomplicated malaria (UM) is represented (boxplot). The P values were calculated with the Mann-Whitney U test and are represented as follows: *** <.001, ** <.01, and * <.05. The correlation between the normalized expression values measured by RT-qPCR and RNA-seq was represented through the regression line and tested by the Spearman rank test (Spearman rank correlation coefficient ρ and P values are shown).
The number of var genes assemblies with at least 1 predicted domain did not differ between CM and UM (96.0 [70.4–141.2] vs 64 [32.7–114.2]; P = .11, Mann-Whitney U test). Sixteen full-length PfEMP1 sequences were obtained (8 CM and 2 UM). A search for the ICAM-1 binding motif showed the pattern in 12 of 15 CM and 5 of 8 UM isolates. Regarding PfEMP1 protein structure, the DC8 domain (DBLα2-CIDRα1.1/8-DBLβ12) was found in 7 of 15 CM and 2 of 8 UM, and DC13 (DBLα1.7-CIDRα1.4) was found in 7 of 15 CM and 3 of 8 UM, respectively. We considered that we have too few isolates in each clinical group to be able to compare the expression values. However, PCA-based analysis of domains expression revealed a partial grouping of CM isolates (Figure 3C).
Global var gene expression was first assessed by evaluating the ATS domain abundance, measured by RNA-seq. Expression was normalized accounting for library size and IDC stage proportions. We found a 1.8-fold-increase in the CM group (P = .02, Mann-Whitney U test). The increased var genes expression measured in CM by RNA-seq was validated by 3 independent RT-qPCR experiments, targeting both DBLα (2.2-fold increase, P = .1; Mann-Whitney U test) and ATS domain (3.2- and 2.4-fold increase for ATS-1 and ATS-2 primers, respectively; P < .001, Mann-Whitney U test) (Figure 3D, Supplemental Data 9). A moderate to strong correlation (ρ > 0.4, P ≤ .05, Spearman rank test) was found between the RT-qPCR and NGS expression (Figure 3D). Next, we focused on the expression associated with assemblies’ sequences. The comparison of var gene expression data (reads per kilobase of transcript per million reads mapped [RPKM] normalized for developmental stage) of the most expressed assembly for each patient (at least NTS-DBLα-CIDRα + 1 domain) revealed higher expression in CM (2.2-fold increase; P = .007, Mann-Whitney U test).
DISCUSSION
Although recent studies have focused on host transcriptome signatures associated with CM [33–35], characterization of the gene expression profile of P falciparum isolated from CM needs further improvement. In this context, we analyzed the gene expression of 15 CM and 15 UM P falciparum isolates from a Beninese pediatric population, to better understand the parasite factors associated with CM.
A greater proportion of ring stage parasites was measured in CM, suggesting a decrease in parasites circulation time. Recent studies that included datasets with various disease severity also demonstrated that lower circulating parasites hpi was correlated with increased severity [5–8].
Andrade et al [7] predicted that cytoadhesion alone may explain the parasites age difference observed in febrile compared with asymptomatic malaria isolates in a cohort of Malian individuals. Previous in vitro cytoadherence experiments, using fresh parasite isolates from our cohort, showed that CM isolates exhibited a higher cytoadherence level on a human brain endothelial cell line (Hbec-5i) [17], a frequently used cellular model of CM [36, 37]. Consistent with this hypothesis, we found an overall upregulated expression of var genes in CM. For ATS domain-based measure, an upregulated expression of a factor 1.8 to 3.2 was established by both RNA-seq and RT-qPCR assay. For DBLα domain-based var gene expression, a nonsignificant 2.2-fold increase (P = .1) in CM isolates was measured by RT-qPCR. Because DBLα sequences are more diverse [11], the nature of the corresponding primers (including degenerated bases) [12] may impact the quality of detection and subsequent quantification. By comparing the most expressed var genes in each sample, a trend of increased expression was reported in febrile malaria compared with asymptomatic infections [7]. Tonkin-Hill et al [5] did not measure a difference in var genes abundance in SM compared with UM, on the basis of de novo assemblies.
The var genes assemblies’ quality in UM samples was not sufficient to perform domain expression comparison. These decreased reconstruction qualities were directly linked to the patients’ lower parasitemia thus affecting the depth of the sequencing. However, the PCA based on var genes domains expression showed a grouping of CM, which may reflect a more restricted var genes repertoire. Indeed, CM isolates express var genes domains allowing specific adhesion to EPCR, which we previously confirmed in our cohort, and ICAM-1 [12, 13, 17, 18]. Regarding the expression of the DBLβ1/3 ICAM-1 binding motif [14], results were concordant with previous RT-qPCR results [17], with expression in both CM and UM, still questioning the implication of this motif in PfEMP1 binding to ICAM-1 receptor.
In addition to iE increased adherence capacity mainly mediated by PfEMP1, an efficient immune response against iE surface proteins may also affect adhesion [9], but was not confirmed in our study through total antibody titers determination on crude whole P falciparum antigen and through detection and quantification of IgG against VSA expressed on iE.
Further experiments including more recombinant proteins from specific domains of PfEMP1 could increase the accuracy of the measurement [38]. Binding inhibition assays could help to understand more precisely the influence of the immune system in iE cytoadhesion mechanisms in further studies [39–41]. However, humoral immunity is not the only host protection against malaria infection and the role of complement component 1s, modulating iE adherence, has recently been demonstrated [42].
The decreased circulation time observed in CM may result in iE avoiding splenic clearance, leading to higher parasitemia [7], as we found in our dataset. A correlation between decreased circulation time and increased parasitemia was retrospectively demonstrated from different studies, by Thomson-Luque et al [8].
Because adhesion capacity does not fully explain CM pathophysiology, we then focused on all-gene expression to decipher CM specificities. After normalization accounting for parasite developmental stage, gene expression profile was a signature of malaria severity. More than 500 genes were deregulated in the CM group compared with the UM isolates.
Focusing on differentially expressed genes, DNA and protein metabolism pathways were dysregulated in CM samples, reflecting the overall gene expression dysregulation. plasmepsin IX (pmIX, PF3D7_1430200) and plasmepsin X (pmX, PF3D7_0808200), involved in the GO pathway entry into host, were upregulated in CM. These 2 proteins are druggable mediators, implicated in both entry and exit from host [43]. eba175 (PF3D7_0731500), encoding for a receptor having a key role in the binding between merozoite and erythrocyte [44], was also upregulated. ama-1 (PF3D7_1133400) was upregulated in CM. The corresponding protein is implicated in erythrocyte invasion [45] and was already identified as a vaccine target [46]. The numerous upregulated genes in CM could reflect an increased invasion capacity of such isolates’ parasites, which may also promote the greater parasitemia measured in CM.
Regarding pathways implicated in cytoadhesion and in PfEMP1 regulation, we also found numerous upregulated genes in CM. Genes from the PHISTa and PHISTb family (PF3D7_1372000, PF3D7_0424600) were upregulated. Several members of the PHIST family have been described to act as an anchor for PfEMP1 by binding to the ATS domain [47]. The PHISTa protein has already been characterized as overexpressed in pregnancy-associated malaria [21, 48]. clag9 (PF3D7_0935800) was upregulated in CM and the corresponding protein was identified as essential for VAR2CSA presentation, which means that it plays a major role in addressing PfEMP1 to the membrane [49]. Other genes implicated in PfEMP1 regulation were dysregulated as set9, set10 and exp2, exp3 which are involved in the regulation of var genes expression and the export of PfEMP1 through the parasitophorous vacuole membrane [50].
CONCLUSIONS
To conclude, parasite and host factors leading to CM should be further investigated and particularly factors influencing parasite decreased circulation time. Difference in terms of parasite adhesion capacity linked to var genes overexpression and increased invasion of erythrocytes should be explored in detail.
In summary, an increased ring stage transcriptomic signature was identified in CM circulating parasites compared with UM, which may result from an increased parasite adherence capacity. Consistent with this result, var genes were overexpressed in CM. We also found numerous upregulated genes involved in entry into host pathway, which could reflect an increased invasion capacity of CM isolates. Deregulated genes involved in adhesion, except for VSAs, support the hypothesis of a stronger CM adhesion.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We are grateful to all patients and their parents for participating in this study. We thank the nurses of the CHU-MEL hospital (CHU-Mère et Enfant de la Lagune) and Hôpital de Zone de Calavi for help in the collection of samples. We also thank the health center of So-Ava. We thank Adrian Luty for help in reviewing the manuscript. We acknowledge Antoine Claessens and all of his team (Prince Nyarko, Marc-Antoine Guéry, and Camille Cohen) for help and counseling regarding data analysis.
Author contributions. E. G. designed the experiments, performed the experiments, performed bioinformatic methodology, performed data analysis, and wrote the article. J. F. designed the experiments, performed the experiments, and revised the manuscript. V. J. designed the experiments, performed the experiments, and revised the manuscript. C. K. designed experiments and revised the manuscript. B. V. performed the experiments. L. H. performed the experiments. L. G. B. performed the experiments. J. F. F. supervised patients’ recruitment, clinical analysis. A. A. supervised patients’ recruitment. S. H. supervised clinical analysis. M. C. revised the manuscript. N. A. supervised the experiments, performed the experiments, and revised the manuscript. O. T. supervised data analysis and revised the manuscript. G. I. B. designed the study, supervised the experiments, supervised data analysis, and wrote the article.
Disclaimer. The funding organization did not play any role in the trial design, data collection, analysis of results, or writing of the manuscript.
Financial support. High-throughput sequencing was performed by the ICGex next-generation sequencing platform of the Institut Curie supported by the grants ANR-10-EQPX-03 (Equipex) and ANR-10-INBS-09-08 (France Génomique Consortium) from the Agence Nationale de la Recherche (“Investissements d’Avenir” program), by the ITMO-Cancer Aviesan (Plan Cancer III) and by the SiRIC-Curie program (SiRIC Grant INCa-DGOS- 465 and INCa-DGOS-Inserm_12554). E. G. benefits from a French Minister for the Research PhD scholarship through Ecole Doctorale 393 Pierre Louis de Santé Publique from the Sorbonne University. This work was funded by the French National Research Agency (ANR-17-CE17-0001).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
NeuroCM Group
Jules Alao (Pediatric Department, Calavi Hospital, Calavi, Benin); Dissou Affolabi (Pediatric Department, Calavi Hospital, Calavi, Benin); Bibiane Biokou (Pediatric Department, Mother and Child University and Hospital Center [CHUMEL], Cotonou, Benin); Jean-Eudes Degbelo (Institut de Recherche Clinique du Bénin [IRCB], Calavi, Benin); Philippe Deloron (MERIT UMR 261, IRD, Université Paris Cité, Paris, France); Latifou Dramane (Institut de Recherche Clinique du Bénin [IRCB], Abomey Calavi, Benin); Sayeh Jafari-Guemouri (MERIT UMR 261, IRD, Université Paris Cité, Paris, France); Anaïs Labrunie (NET, INSERM, Université de Limoges, Limoges, France); Yélé Ladipo (Pediatric Department, Mother and Child University and Hospital Center [CHUMEL], Cotonou, Benin); Thomas Lathiere (Ophthalmology Department, Limoges University Hospital, Limoges, France); Achille Massougbodji (Institut de Recherche Clinique du Bénin [IRCB], Abomey Calavi, Benin); Audrey Mowendabeka (Paediatric Department, Hopital de la Mère et de l’Enfant, Limoges, France); Jade Papin (MERIT UMR 261, IRD, Université Paris Cité, Paris, France); Bernard Pipy (PHARMADEV, Université de Toulouse, IRD, UPS, France); Pierre-Marie Preux (NET, INSERM, Université de Limoges, Limoges, France); Marie Raymondeau (NET, INSERM, Université de Limoges, Limoges, France); Jade Royo (PHARMADEV, Université de Toulouse, IRD, UPS, France); Darius Sossou (Institut de Recherche Clinique du Bénin [IRCB], Abomey Calavi, Benin); Brigitte Techer (MERIT UMR 261, IRD, Université Paris Cité, Paris, France).
Presented in part: BioMalPar XVII (virtual session) EMBL Conference, 25 May 2021.
Contributor Information
E Guillochon, Université Paris Cité, MERIT, IRD, Paris, France; Université Paris Cité, INSERM U1133, CNRS UMR 8251, Paris, France.
J Fraering, Université Paris Cité, MERIT, IRD, Paris, France.
V Joste, Université Paris Cité, MERIT, IRD, Paris, France; Parasitology Laboratory, Hôpital Bichat - Claude-Bernard, APHP, Paris, France; French Malaria Reference Center, Hôpital Bichat, APHP, Paris, France.
C Kamaliddin, Cumming School of Medicine, The University of Calgary, Calgary, Alberta, Canada.
B Vianou, Université Paris Cité, MERIT, IRD, Paris, France; Institut de Recherche Clinique du Bénin, Cotonou, Bénin.
L Houzé, Université Paris Cité, MERIT, IRD, Paris, France.
L G Baudrin, Institut Curie Genomics of Excellence Platform, PSL Research University, Research Center, Institut Curie, Paris, France.
J F Faucher, INSERM, Univ. Limoges, CHU Limoges, IRD, U1094 Tropical Neuroepidemiology, Institute of Epidemiology and Tropical Neurology, GEIST, Limoges, France.
A Aubouy, Université de Toulouse, PHARMADEV, IRD, UPS, Toulouse, France.
S Houzé, Université Paris Cité, MERIT, IRD, Paris, France; Parasitology Laboratory, Hôpital Bichat - Claude-Bernard, APHP, Paris, France; French Malaria Reference Center, Hôpital Bichat, APHP, Paris, France.
M Cot, Université Paris Cité, MERIT, IRD, Paris, France.
N Argy, Université Paris Cité, MERIT, IRD, Paris, France; Parasitology Laboratory, Hôpital Bichat - Claude-Bernard, APHP, Paris, France; French Malaria Reference Center, Hôpital Bichat, APHP, Paris, France.
O Taboureau, Université Paris Cité, INSERM U1133, CNRS UMR 8251, Paris, France.
G I Bertin, Université Paris Cité, MERIT, IRD, Paris, France.
NeuroCM group:
Jules Alao, Dissou Affolabi, Bibiane Biokou, Jean-Eudes Degbelo, Philippe Deloron, Latifou Dramane, Sayeh Jafari-Guemouri, Anaïs Labrunie, Yélé Ladipo, Thomas Lathiere, Achille Massougbodji, Audrey Mowendabeka, Jade Papin, Bernard Pipy, Pierre-Marie Preux, Marie Raymondeau, Jade Royo, Darius Sossou, and Brigitte Techer
References
- 1. World malaria report 2021. Available at: https://www.who.int/publications-detail-redirect/9789240040496. Accessed 18 January 2022.
- 2. Turner L, Lavstsen T, Berger SS, et al. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature 2013; 498:502–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Silamut K, Phu NH, Whitty C, et al. A quantitative analysis of the microvascular sequestration of malaria parasites in the human brain. Am J Pathol 1999; 155:395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller LH, Baruch DI, Marsh K, Doumbo OK.. The pathogenic basis of malaria. Nature 2002; 415:673–9. [DOI] [PubMed] [Google Scholar]
- 5. Tonkin-Hill GQ, Trianty L, Noviyanti R, et al. The Plasmodium falciparum transcriptome in severe malaria reveals altered expression of genes involved in important processes including surface antigen-encoding var genes. PLoS Biol 2018; 16:e2004328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wichers JS, Tonkin-Hill G, Thye T, et al. Common virulence gene expression in adult first-time infected malaria patients and severe cases. eLife 2021; 10. doi: 10.7554/eLife.69040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Andrade CM, Fleckenstein H, Thomson-Luque R, et al. Increased circulation time of Plasmodium falciparum underlies persistent asymptomatic infection in the dry season. Nat Med 2020; 26:1929–40. [DOI] [PubMed] [Google Scholar]
- 8. Thomson-Luque R, Votborg-Novél L, Ndovie W, et al. Plasmodium falciparum transcription in different clinical presentations of malaria associates with circulation time of infected erythrocytes. Nat Commun 2021; 12:4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duffy MF, Noviyanti R, Tsuboi T, et al. Differences in PfEMP1s recognized by antibodies from patients with uncomplicated or severe malaria. Malar J 2016; 15:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saul A. The role of variant surface antigens on malaria-infected red blood cells. Parasitol Today Pers Ed 1999; 15:455–7. [DOI] [PubMed] [Google Scholar]
- 11. Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T.. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes--divide and conquer. PLoS Comput Biol 2010; 6:e1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lavstsen T, Turner L, Saguti F, et al. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci USA 2012; 109:E1791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bertin GI, Lavstsen T, Guillonneau F, et al. Expression of the domain cassette 8 Plasmodium falciparum erythrocyte membrane protein 1 is associated with cerebral malaria in Benin. PLoS One 2013; 8:e68368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lennartz F, Adams Y, Bengtsson A, et al. Structure-guided identification of a family of dual receptor-binding PfEMP1 that is associated with cerebral malaria. Cell Host Microbe 2017; 21:403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jensen AR, Adams Y, Hviid L.. Cerebral Plasmodium falciparum malaria: the role of PfEMP1 in its pathogenesis and immunity, and PfEMP1-based vaccines to prevent it. Immunol Rev 2020; 293:230–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Joste V, Maurice L, Bertin GI, et al. Identification of Plasmodium falciparum and host factors associated with cerebral malaria: description of the protocol for a prospective, case-control study in Benin (NeuroCM). BMJ Open 2019; 9:e027378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Joste V, Guillochon E, Fraering J, et al. PfEMP1 a-type ICAM-1-binding domains are not associated with cerebral malaria in Beninese children. mBio 2020; 11:e02103–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Storm J, Jespersen JS, Seydel KB, et al. Cerebral malaria is associated with differential cytoadherence to brain endothelial cells. EMBO Mol Med 2019; 11:e9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Argy N, Kendjo E, Augé-Courtoi C, et al. Influence of host factors and parasite biomass on the severity of imported Plasmodium falciparum malaria. PLoS One 2017; 12:e0175328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. López-Barragán MJ, Lemieux J, Quiñones M, et al. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 2011; 12:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vignali M, Armour CD, Chen J, et al. NSR-seq transcriptional profiling enables identification of a gene signature of Plasmodium falciparum parasites infecting children. J Clin Invest 2011; 121:1119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gagnon-Bartsch JA, Speed TP.. Using control genes to correct for unwanted variation in microarray data. Biostat Oxf Engl 2012; 13:539–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Law CW, Chen Y, Shi W, Smyth GK.. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014; 15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M.. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 2016; 44:D457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res 2017; 45:D362–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bader GD, Hogue CWV.. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf 2003; 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katoh K, Standley DM.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013; 30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL.. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019; 37:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Otto TD, Assefa SA, Böhme U, et al. Evolutionary analysis of the most polymorphic gene family in falciparum malaria. Wellcome Open Res 2019; 4:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hofmann N, Mwingira F, Shekalaghe S, Robinson LJ, Mueller I, Felger I.. Ultra-sensitive detection of Plasmodium falciparum by amplification of multi-copy subtelomeric targets. PLoS Med 2015; 12:e1001788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li K, Wang H, Zhang H-F, Zhao X-X, Lai Y-J, Liu F-F.. Genomic analysis of host gene responses to cerebral Plasmodium falciparum malaria. Immun Inflamm Dis 2021; 9:819–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cabantous S, Poudiougou B, Bergon A, et al. Understanding human cerebral malaria through a blood transcriptomic signature: evidences for erythrocyte alteration, immune/inflammatory dysregulation, and brain dysfunction. Mediators Inflamm 2020; 2020:3280689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thiam A, Sanka M, Ndiaye Diallo R, et al. Gene expression profiling in blood from cerebral malaria patients and mild malaria patients living in Senegal. BMC Med Genomics 2019; 12:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Claessens A, Adams Y, Ghumra A, et al. A subset of group A-like var genes encodes the malaria parasite ligands for binding to human brain endothelial cells. Proc Natl Acad Sci U S A 2012; 109:E1772–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Avril M, Tripathi AK, Brazier AJ, et al. A restricted subset of var genes mediates adherence of Plasmodium falciparum-infected erythrocytes to brain endothelial cells. Proc Natl Acad Sci U S A 2012; 109:E1782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rambhatla JS, Turner L, Manning L, et al. Acquisition of antibodies against endothelial protein C receptor-binding domains of Plasmodium falciparum erythrocyte membrane protein 1 in children with severe malaria. J Infect Dis 2019; 219:808–18. [DOI] [PubMed] [Google Scholar]
- 39. Doritchamou J, Teo A, Morrison R, et al. Functional antibodies against placental malaria parasites are variant dependent and differ by geographic region. Infect Immun 2019; 87:e00865–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Olsen RW, Ecklu-Mensah G, Bengtsson A, et al. Natural and vaccine-induced acquisition of cross-reactive IgG-inhibiting ICAM-1-specific binding of a Plasmodium falciparum PfEMP1 subtype associated specifically with cerebral malaria. Infect Immun 2018; 86:e00622–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Avril M, Cartwright MM, Hathaway MJ, Smith JD.. Induction of strain-transcendent antibodies to placental-type isolates with VAR2CSA DBL3 or DBL5 recombinant proteins. Malar J 2011; 10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Azasi Y, Low LM, Just AN, et al. Complement C1s cleaves PfEMP1 at interdomain conserved sites inhibiting Plasmodium falciparum cytoadherence. Proc Natl Acad Sci U S A 2021; 118:e2104166118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nasamu AS, Glushakova S, Russo I, et al. Plasmepsins IX and X are essential and druggable mediators of malaria parasite egress and invasion. Science 2017; 358:518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klotz FW, Orlandi PA, Reuter G, et al. Binding of Plasmodium falciparum 175-kilodalton erythrocyte binding antigen and invasion of murine erythrocytes requires N-acetylneuraminic acid but not its O-acetylated form. Mol Biochem Parasitol 1992; 51:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Silvie O, Franetich J-F, Charrin S, et al. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J Biol Chem 2004; 279:9490–6. [DOI] [PubMed] [Google Scholar]
- 46. Sirima SB, Durier C, Kara L, et al. Safety and immunogenicity of a recombinant Plasmodium falciparum AMA1-DiCo malaria vaccine adjuvanted with GLA-SE or Alhydrogel® in European and African adults: a phase 1a/1b, randomized, double-blind multi-centre trial. Vaccine 2017; 35:6218–27. [DOI] [PubMed] [Google Scholar]
- 47. Yang B, Wang X, Jiang N, et al. Interaction analysis of a Plasmodium falciparum PHISTa-like protein and PfEMP1 proteins. Front Microbiol 2020; 11:2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Francis SE, Malkov VA, Oleinikov AV, et al. Six genes are preferentially transcribed by the circulating and sequestered forms of Plasmodium falciparum parasites that infect pregnant women. Infect Immun 2007; 75:4838–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goel S, Valiyaveettil M, Achur RN, et al. Dual stage synthesis and crucial role of cytoadherence-linked asexual gene 9 in the surface expression of malaria parasite var proteins. Proc Natl Acad Sci U S A 2010; 107:16643–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. LaCount DJ, Vignali M, Chettier R, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 2005; 438:103–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.