Abstract
Eggs contain about 200,000 mitochondria that generate adenosine triphosphate and metabolites essential for oocyte development. Mitochondria also integrate metabolism and transcription via metabolites that regulate epigenetic modifiers, but there is no direct evidence linking oocyte mitochondrial function to the maternal epigenome and subsequent embryo development. Here, we have disrupted oocyte mitochondrial function via deletion of the mitochondrial fission factor Drp1. Fission-deficient oocytes exhibit a high frequency of failure in peri- and postimplantation development. This is associated with altered mitochondrial function, changes in the oocyte transcriptome and proteome, altered subcortical maternal complex, and a decrease in oocyte DNA methylation and H3K27me3. Transplanting pronuclei of fertilized Drp1 knockout oocytes to normal ooplasm fails to rescue embryonic lethality. We conclude that mitochondrial function plays a role in establishing the maternal epigenome, with serious consequences for embryo development.
Mitochondrial dynamics during oogenesis affects embryo development by regulating the maternal epigenome.
INTRODUCTION
Mitochondria are essential organelles that generate cellular energy in the form of adenosine triphosphate (ATP) through oxidative phosphorylation (OXPHOS). Mitochondria also regulate apoptosis through control of caspase activity and have roles in Ca2+ signaling and in maintaining cellular redox balance (1, 2). Mitochondrial quality and function are dependent on mitochondrial fission and fusion events, also known as mitochondrial dynamics (2). These events reestablish mitochondrial function or divert ailing mitochondria to the mitophagy pathway. Mitochondrial fission is driven by dynamin-related protein 1 (DRP1) and its adaptor proteins, while fusion is controlled by mitofusins (MFN1 and MFN2) and optic atrophy 1 (OPA1) (2–5). Knockout (KO) of Drp1 in mice results in embryo death, demonstrating the essential role of mitochondrial fission (3, 5). Furthermore, deletion specifically in neurons results in neuronal cell death (6, 7) and is associated with marked effects on cellular signaling and metabolism (8–12).
Female reproductive capacity is dependent on a finite pool of primordial oocytes (13). The growth of these oocytes from a diameter of approximately 15 to 80 μm is accompanied by a remarkable mitochondrial biogenesis, resulting in the number of mitochondria increasing from about 1000 to 300,000 by the time the oocyte is fully grown (14, 15). This window of mitochondrial biogenesis coincides with remodeling of the maternal epigenome that is critical for embryogenesis (16, 17). Throughout oocyte growth, maturation, and fertilization, mitochondria provide ATP, nucleotide and amino acid precursors, and tricarboxylic acid (TCA) cycle metabolites (18, 19). Unlike most other cell types, oocytes and cleavage stage embryos cannot perform glycolysis and are therefore completely reliant on mitochondria as their source of ATP. Mitochondria undergo substantial changes in localization associated with meiotic progression (20–22) and at fertilization are activated by sperm-induced Ca2+ oscillations for increased ATP production required for fueling the egg-to-embryo transition (23, 24). Thus, mitochondria play key roles in driving and regulating oocyte and early embryo development, while, at the same time, there is a need to minimize any risk to their integrity as they pass from one generation to the next.
Optimal mitochondrial function and quality control in oocytes is therefore essential to ensure the production of a developmentally competent oocyte and viable embryo and for the provision of quality mitochondria to the developing organism. This has been graphically borne out in mice that have a conditional deletion of oocyte Drp1 (25). Infertility of these mice is attributed to the arrest of ovarian follicles early in their growth, although some larger oocytes were recovered. Here, we set out to investigate the question of how inhibition of mitochondrial fission during oocyte growth, when mitochondrial numbers are undergoing rapid expansion, affects subsequent embryo development. We find that Drp1-deleted oocytes undergo fertilization and preimplantation development but that peri- and postimplantation embryonic lethality is markedly increased. This defect is largely assignable to the maternal nuclear genome and is accompanied by changes to the maternal epigenome. Our results reveal a previously unknown role for mitochondrial fission in developmental epigenetic programming in the female germ line.
RESULTS
We generated a conditional Cre/loxP KO of Drp1 (fl) by floxing exons 3 to 5 (fig. S1A). Oocyte-specific deletion of the floxed segment, resulting in the KO allele (∆), was achieved by crossing Drp1fl/fl female mice (3) with transgenic Drp1fl/fl, Gdf9-Cre males in which Cre recombinase is driven by the oocyte-specific Gdf9 (growth differentiation factor 9) promotor (26). Here, Drp1fl/fl, Gdf9-Cre females are referred to as Drp1cKO mice and their oocytes as Drp1Δ/Δ oocytes. We confirmed that DRP1 levels were reduced to background levels in Drp1Δ/Δ oocytes recovered from 4- to 5-week-old Drp1cKO mice (Fig. 1, A to D). These Drp1Δ/Δ oocytes recovered from juvenile mice therefore provide an ideal model to investigate the effect of disrupted mitochondrial fission on oocyte maturation and subsequent embryo development.
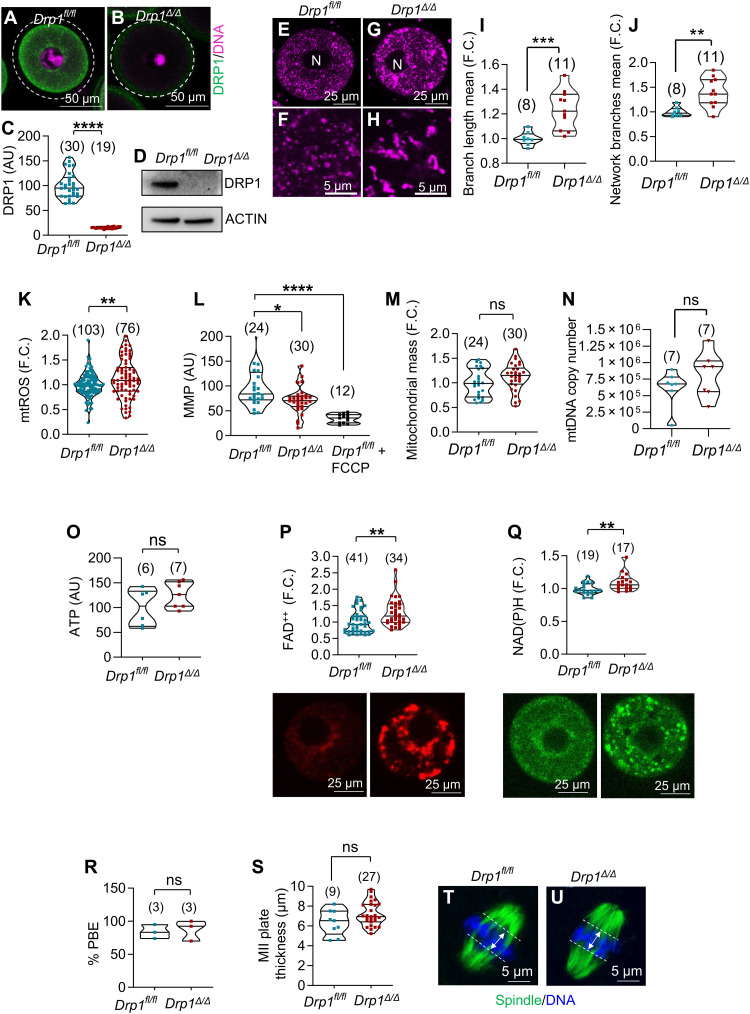
Fig. 1. Oocyte-specific deletion of Drp1 and analysis of oocyte mitochondria.
(A to D) Oocyte-specific deletion of Drp1. DRP1 (green) and DNA (pink) in Drp1fl/fl oocytes (A) but Drp1Δ/Δ oocytes lacked DRP1 (B). Dashed circles represent oocyte margins. (C) DRP1 fluorescence quantification. AU, arbitrary unit. (D) Western blots of DRP1. Each lane contains 50 GV-stage oocytes from 4- to 5-week-old mice. β-Actin was used as a loading control. The experiments were repeated three times, and a representative image is shown. Mitochondrial morphology in GV-stage Drp1fl/fl (E and F) and Drp1Δ/Δ (G and H) oocytes. Branch length mean (I) and network branches mean (J) between Drp1fl/fl and Drp1Δ/Δ oocytes. Oocytes were labeled with mitotracker orange. Number of oocytes analyzed are shown (C, I, and J). (K) Increased mitochondrial ROS (mtROS) levels in Drp1Δ/Δ oocytes. Numbers represent total number of oocytes. (L) Reduced MMP in Drp1Δ/Δ oocytes compared to Drp1fl/fl oocytes. Carbonyl cyanide-4-phenylhydrazone (FCCP) was used to demonstrate assay specificity. Numbers represent total number of oocytes. (M) Similar mitochondrial mass between Drp1fl/fl and Drp1Δ/Δ oocytes. MitoTracker Green signals in (L) were analyzed to calculate mitochondrial mass. ns, not significant. (N) Comparable mtDNA copy number between Drp1fl/fl and Drp1Δ/Δ oocytes. Numbers represent total number of oocytes. (O) Similar ATP levels between Drp1Δ/Δ and Drp1fl/fl oocytes. Numbers represent experimental replicates. Drp1Δ/Δ oocytes have increased levels of FAD++ (P) and NAD(P)H (Q) compared to Drp1fl/fl controls. Total oocyte numbers shown. Representative autofluorescent images are also shown. (R) Comparable PBE rates in vitro. Numbers represent experimental replicates. (S to U) MII oocytes showing spindle (green) and DNA (blue). Chromosome spreads were measured as exemplified in (T) and (U). Total oocyte numbers shown in (S). Results show means ± SD and P values determined by two-tailed Student’s t test (C and I to S). F.C., fold change.
Drp1Δ/Δ oocytes have altered mitochondrial function
Drp1Δ/Δ oocytes contained elongated and aggregated mitochondria (Fig. 1, G to J) compared to the discrete and evenly distributed mitochondria in the ooplasm of Drp1fl/fl oocytes (Fig. 1, E, F, I, and J). In fully grown germinal vesicle (GV)–stage oocytes, we observed a moderate but significant increase in mitochondrial reactive oxygen species (ROS) levels in Drp1Δ/Δ oocytes compared to Drp1fl/fl controls (Fig. 1K). On measurement of mitochondrial membrane potential (MMP), Drp1Δ/Δ oocytes had a reduced MMP compared to Drp1fl/fl oocytes (Fig. 1L). These changes were not associated with any detectable effects on mitochondrial mass (Fig. 1M) or mitochondrial DNA (mtDNA) copy number between Drp1fl/fl and Drp1Δ/Δ oocytes (Fig. 1N). The differences observed in mitochondrial activity also did not translate into consistent differences in oocyte ATP levels (Fig. 1O).
Mitochondrial FADH2 [hydroquinone form of FAD (flavin adenine dinucleotide)] and NADH (reduced form of nicotinamide adenine dinucleotide) are generated as a result of reduction in the TCA cycle and are then oxidized by the mitochondrial OXPHOS pathway to provide electrons for the electron transport chain (ETC). FAD++ and NAD(P)H are autofluorescent and can be measured to provide an indication of the metabolic state of the cell. Drp1Δ/Δ oocytes were found to have increased levels of FAD++ (Fig. 1P) and NAD(P)H (Fig. 1Q) compared to Drp1fl/fl controls. NAD(P)H can be cytosolic or mitochondrial in origin; the increase in NAD(P)H fluorescence appears to be primarily in the mitochondria rather than in the cytosol (Fig. 1Q). These changes provide evidence that mitochondrial metabolism and/or ETC activity is altered in Drp1Δ/Δ oocytes.
Drp1Δ/Δ oocytes undergo normal meiotic maturation yet Drp1cKO females are severely subfertile
To investigate whether DRP1 is necessary to maintain ploidy and normal spindle function, we monitored meiotic maturation and assessed the first and second meiotic spindles. The maturation rates, as indicated by first polar body extrusion (PBE), were comparable between control (84 ± 10.6%) and KO oocytes (87 ± 15.6%; Fig. 1R). Consistent with this, mature Drp1Δ/Δ oocytes formed normal metaphase II (MII) spindles with aligned chromosomes (Fig. 1, S and U) similar to control oocytes (Fig. 1, S and T). Histological analysis of ovaries of 5-week-old Drp1cKO mice revealed the presence of follicles at all developmental stages (fig. S1C), similar to Drp1fl/fl controls (fig. S1B). Furthermore, superovulation of 4- to 5-week-old Drp1cKO females and Drp1fl/fl controls resulted in similar numbers of ovulated oocytes (fig. S1D). Together, these results indicate that DRP1 is dispensable for the production of oocytes with intact spindles and normal chromosome organization.
While Drp1cKO mice were able to produce oocytes and exhibited normal mating behavior, six females produced only one litter over 10 weeks of being housed with wild-type (WT) males. By contrast, control Drp1fl/fl females regularly produced litters, with an average size of 6 ± 1.37 pups/litter (fig. S1, E and F). Histological analysis of ovarian sections showed 10-week-old Drp1cKO females contained only primary and secondary stage follicles, with no evidence of more advanced antral stages (fig. S1, G and H). These findings are consistent with a previous study that identified the development of this ovarian phenotype in 10-week-old Drp1cKO mice (25).
DRP1 is essential for preimplantation development
To investigate the developmental capacity of zygotes derived from Drp1Δ/Δ oocytes, we mated 5-week-old control Drp1fl/fl and Drp1cKO females with WT (+/+) males (Fig. 2A). Despite disrupted mitochondrial organization in Drp1Δ/Δ oocytes, Drp1matΔ/+ two-cell stage embryos displayed equal mitochondrial inheritance between blastomeres (Fig. 2, B to D), and their rate of preimplantation development was similar to Drp1fl/+ controls (Fig. 2, E to G). Drp1matΔ/+ blastocysts had comparable TMRM (tetramethylrhodamine, methyl ester, perchlorate) intensity to Drp1fl/+ controls, indicating similar MMP (fig. S2A). Drp1matΔ/+ blastocysts had comparable levels of NAD(P)H (fig. S2B) but slightly lower levels of FAD++ (fig. S2C) compared to Drp1fl/+ controls. These results indicate that during preimplantation, development paternal expression of Drp1 normalizes some of the mitochondrial differences observed in Drp1Δ/Δ oocytes. Moreover, preferential immunolabeling of Octamer-binding transcription factor 4 (OCT-4) and CDX2 revealed no detectable difference in the number of inner cell mass (ICM) cells in Drp1matΔ/+ blastocysts compared to Drp1fl/+ controls, although a 25% reduction in the number of trophectoderm (TE) cells was observed (Fig. 2, H to J). Thus, loss of the maternal Drp1 allele appears to preferentially affect the proliferation or survival of TE cells in blastocysts.
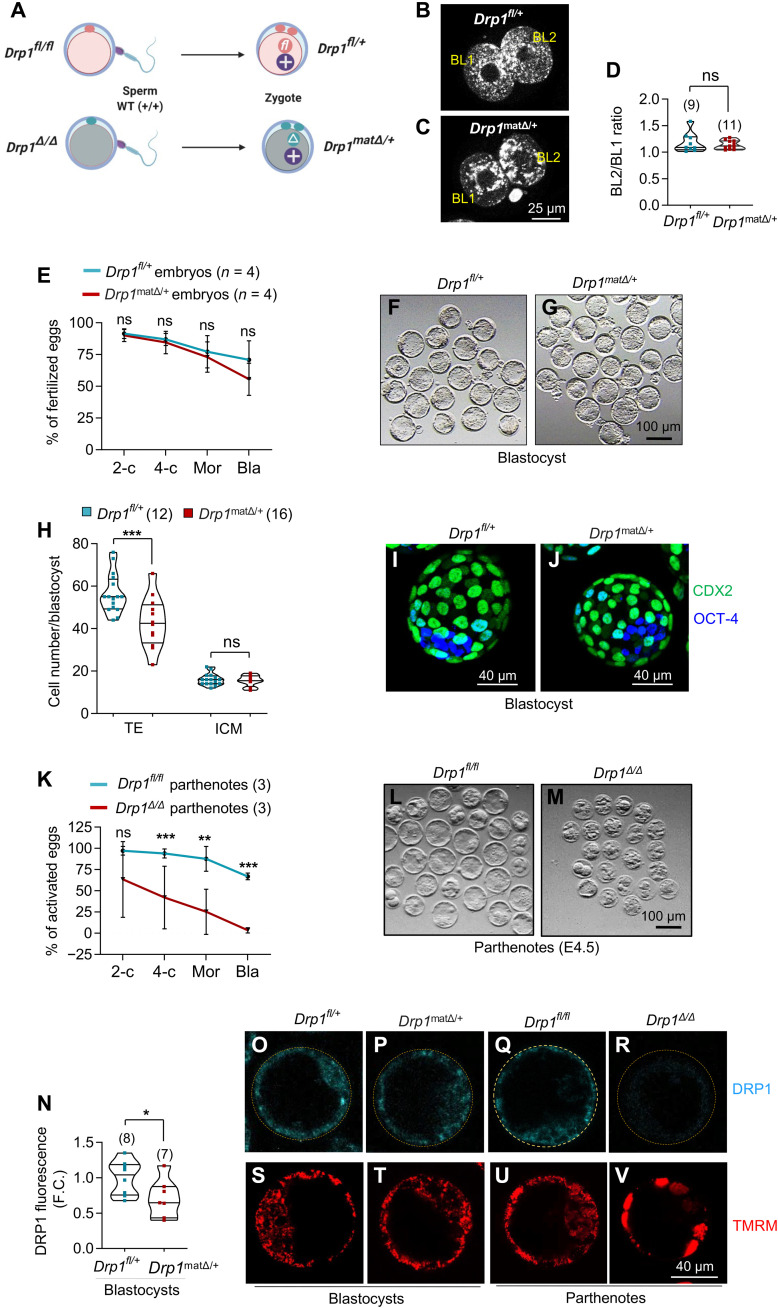
Fig. 2. Preimplantation development of Drp1matΔ/+ embryos.
(A) Schematic representation of mating schemes and the genotype of resultant embryos. (B to D) Comparison of mitochondrial mass between blastomeres of 2-cell embryos. One-cell zygotes harvested from gonadotropin-stimulated females and mated with WT males were cultured for 24 hours and labelled with MitoTracker Orange. Numbers represent total number of zygotes. (E to G) After gonadotropin stimulations, 4- to 5-week-old Drp1fl/fl and Drp1cKO mice were mated with WT males to harvest Drp1fl/+ and Drp1matΔ/+ one-cell zygotes. One-cell zygotes were cultured in vitro for 96 hours. Number of replicates are shown. Bla, blastocyst. (H to J) Blastocysts were labelled with CDX2 and OCT-4 to count the trophectoderm (TE) and inner cell mass (ICM), respectively. Representative maximum projection images of Drp1fl/+ (I) and Drp1matΔ/+ (J) blastocysts are shown. Number of blastocysts analyzed in (H) are shown. (K to M) Absolute requirement of DRP1 for preimplantation embryo development. The rate of parthenogenetic activation of Drp1Δ/Δ oocytes was significantly reduced compared to Drp1fl/fl controls (K). Numbers of experimental repeats are shown. Bla, blastocyst. Representative images of Drp1fl/fl control (L) and Drp1Δ/Δ (M) parthenotes at the end of culture. (N to V) Recovery of DRP1 and mitochondrial morphology in fertilized Drp1matΔ/+ embryos. DRP1 expression was largely recovered in fertilized Drp1matΔ/+ embryos (N). Number of blastocysts analyzed are shown. Immunofluorescence of DRP1 (cyan) was evident in Drp1fl/+ (O) and Drp1matΔ/+ (P) blastocysts. Unfertilized Drp1fl/fl control parthenote expressed DRP1 (Q) but Drp1Δ/Δ parthenote blastocysts completely lacked DRP1 immunofluorescence (R). Reflecting the DRP1 expression profiles, TMRM labelling revealed normal singular mitochondria in control fertilized Drp1fl/+ (S) and Drp1matΔ/+ (T) blastocysts. Unfertilized Drp1fl/fl control parthenote also contained dispersed mitochondria (U) but Drp1Δ/Δ parthenogenetic blastocysts had mitochondria aggregated into large clumps (V). Results show mean ± SD and P values determined by two-tailed Student’s t test (E, H, K, N).
To examine the developmental competence of Drp1Δ/Δ oocytes directly, i.e., in the absence of the paternal Drp1 allele, parthenogenetic activation was used to generate diploid Drp1Δ/Δ zygotes. The rate of two-cell formation after parthenogenetic activation of Drp1Δ/Δ oocytes was significantly reduced compared to Drp1fl/fl controls (60% versus 90%, respectively; Fig. 2K) and, remarkably, less than 4% of Drp1Δ/Δ two-cell parthenotes developed to the blastocyst stage (Fig. 2, K and M) compared to 68% of Drp1fl/fl controls (Fig. 2, K and L). Immunofluorescence confirmed that fertilized Drp1matΔ/+ blastocysts recovered DRP1 protein, while it was not detectable in the few Drp1Δ/Δ parthenogenetic blastocysts produced (Fig. 2, N to R). In addition, TMRM labeling showed that fertilized Drp1matΔ/+ blastocysts (Fig. 2T) had a mitochondrial distribution indistinguishable from control fertilized Drp1fl/+ blastocysts (Fig. 2S) and Drp1fl/fl control parthenogenetic blastocysts (Fig. 2U), while in Drp1Δ/Δ parthenogenetic blastocysts, mitochondria were highly aggregated into large clumps (Fig. 2V). The catastrophic failure of Drp1Δ/Δ embryos to undergo preimplantation development is not due to parthenogenesis per se but likely mediated by impacts on mitochondrial function, although we cannot rule out other DRP1-dependent factors playing a role. These findings demonstrate an absolute reliance on oocyte DRP1 for preimplantation development and that the paternal allele is sufficient to support development in vitro, albeit with a reduced number of TE cells.
Embryos from Drp1cKO females exhibit developmental arrest
Previous studies show Drp1 to be haplosufficient because viable Drp1 heterozygous progeny (Drp1+/−) are produced at normal frequency in Drp1+/− intercrosses (3, 5, 27). However, because of heterozygous parents, these matings are unable to determine whether there is a maternal requirement for DRP1. Therefore, to investigate the role of DRP1 in oocytes, we mated 5-week-old Drp1cKO and control Drp1fl/fl females with WT males (as in Fig. 2A). We focused on 5-week-old mice as we knew they were capable of generating oocytes at similar rates to controls (fig. S1D). Plugged females were assessed between embryonic day 10.5 (E10.5) to E18.5. A confirmed pregnancy was defined if the uteri of plugged females contained decidua, resorptions, or live embryos. By this criterion, pregnancies were significantly reduced in Drp1cKO females (60%; n = 25) compared to Drp1fl/fl females (94%; n = 17; Fig. 3A and table S1). Of confirmed pregnancies, Drp1cKO females had a significantly reduced number of implanted embryos per mating compared to Drp1fl/fl females (4.2 ± 2.3 versus 8.0 ± 1.1, respectively; Fig. 3B and table S1). Notably, in Drp1cKO females, most of the implanted embryos had died and were resorbing (76%, compared to 3% in controls; Fig. 3, C to E, and table S1). In addition, when live fetuses were present, most of these were severely growth retarded (Fig. 3E, arrowheads).
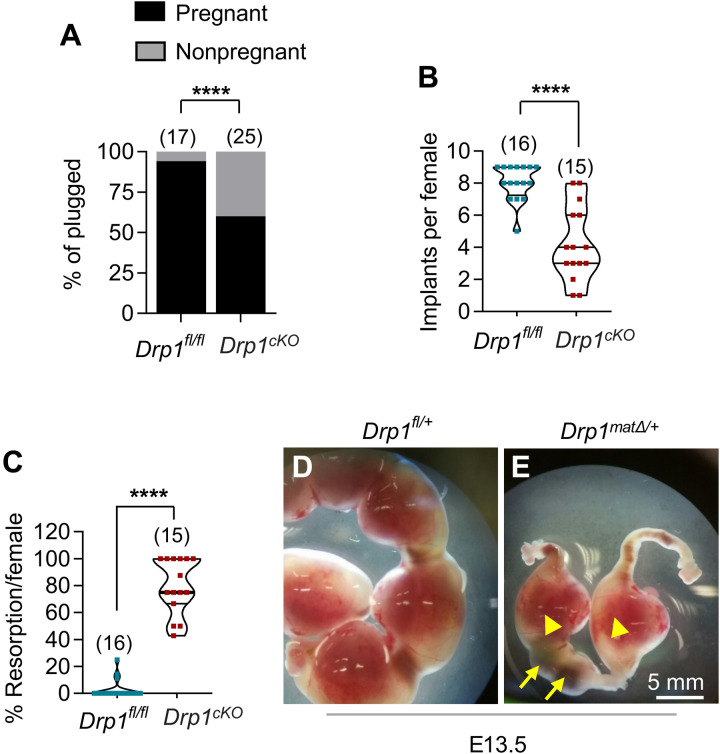
Fig. 3. Postimplantation death of Drp1matΔ/+ embryos derived from Drp1Δ/Δ oocytes.
Five-week-old Drp1cKO and control Drp1fl/fl females were mated with WT (+/+) males and assessed fetal development between E10.5 to E18.5 of pregnancy. (A) Uteri of 94% of plugged control Drp1fl/fl females showed embryos as compared to only 60% of plugged Drp1cKO females contained any embryo. Number of mice analyzed are shown. (B) Control females had an average of eight implants per pregnancy, whereas Drp1cKO females had only four implants per pregnancy. Number of mice analyzed are shown. (C) Within confirmed pregnancies, there was a higher percentage of embryo resorption in Drp1cKO females (76%) compared to only 3% in Drp1fl/fl females. Number of mice analyzed are shown. While the uteri of E13.5 Drp1fl/fl females showed normal developing embryos (D), the embryos in uteri of Drp1cKO females were either dead (E, arrows) or severely growth retarded (E, arrowheads). Results show means ± SD (B and C). P value determined by chi-square test (A), two-tailed Student’s t test (B and C).
With such a marked effect of Drp1 deletion in oocytes on subsequent embryonic development, we investigated a number of potential contributory causes. First, we confirmed that the fetuses generated were of the correct heterozygous Drp1matΔ/+ genotype (fig. S2D) and that they were expressing DRP1 from the paternal allele (fig. S2E). Furthermore, to test whether the heterozygote level of DRP1 was sufficient to support normal mitochondrial morphology, we derived fetal fibroblasts from E13.5 Drp1matΔ/+ and control Drp1fl/+ embryos. The fetal fibroblasts from both genotypes exhibited comparable mitochondrial morphology (fig. S2, F and G), demonstrating that the DRP1 level in Drp1matΔ/+ fetuses is sufficient to support overtly normal mitochondrial organization. An increase in aneuploidy may be a factor contributing to embryonic failure, but no significant differences were seen in chromosome counts of blastomeres of Drp1matΔ/+ and control Drp1fl/+ four-cell embryos (fig. S2, H to K). Another source of embryonic failure could potentially be related to the timing of onset of zygotic genome activation (ZGA) at the two-cell stage, but no significant difference in 5-ethynyl uridine (EU) incorporation was found between Drp1fl/+ and Drp1matΔ/+ embryos (fig. S2, L, N, O, Q, and R). In addition, phosphorylated RNA polymerase II at serine-2 (pS2, a marker of RNA polymerase II activation) levels were also similar between Drp1fl/+ and Drp1matΔ/+ embryos (fig. S2, M, N, P, Q, and S). These results indicate that there is no marked difference in transcriptional activation between Drp1fl/+ and Drp1matΔ/+ embryos. Another common feature of embryonic lethal mouse models is a defect in placental development (28), a possibility consistent with the observed decrease in TE cells in Drp1matΔ/+ blastocysts (Fig. 2H). However, a detailed histological analysis of placental structure in surviving E13.5 embryos of Drp1matΔ/+ and control embryos uncovered no obvious differences in placental or embryonic structures (fig. S2, T to W). The developmental failure of Drp1matΔ/+ embryos appears not to be caused by a failure to express sufficient DRP1 in development, or to any obvious abnormalities in chromosome number, ZGA, or even placental organization. We therefore returned to a deeper analysis of Drp1Δ/Δ oocytes.
Altered transcriptome and proteome in Drp1Δ/Δ oocytes
To understand the effects of DRP1 deletion on oocytes, we used single-cell RNA sequencing (RNA-seq) to compare the transcriptome of Drp1fl/fl and Drp1Δ/Δ GV-stage oocytes. This analysis revealed 717 differentially expressed genes (DEGs; adjusted P < 0.05) between Drp1fl/fl and Drp1Δ/Δ oocytes, of which 126 (17.5%) were up-regulated and 591 (82.5%) were down-regulated in Drp1Δ/Δ oocytes (Fig. 4, A and B). Several DEGs between Drp1fl/fl and Drp1Δ/Δ oocytes are known epigenetic regulators, such as Kdm5a, Kdm5b, Tet2, Dppa4, and Dnmt3a (Fig. 4C and fig. S3, A and B), indicating that loss of DRP1 during oocyte growth may lead to alterations in the maternal epigenome. In addition, maternal-effect genes of the subcortical maternal complex (SCMC), Tle6 and Khdc3, were significantly down-regulated in Drp1Δ/Δ oocytes as compared to Drp1fl/fl oocytes (fig. S3C). In contrast, there were no significant changes in the expression of oocyte-specific genes (fig. S3D), consistent with the minimal impact of Drp1 deletion on oogenesis, oocyte maturation, and morphology. Perhaps unexpectedly, we also found no evidence for changes in mtDNA-encoded gene products (mt-Nd1, mt-Nd2, mt-Nd3, mt-Nd4, mt-Nd5, mt-Nd6, mt-Cytb, and mt-Co1; fig. S3E), suggesting that the nuclear genome provides the main mechanism for adapting to mitochondrial dysfunction due to compromised DRP1 expression.
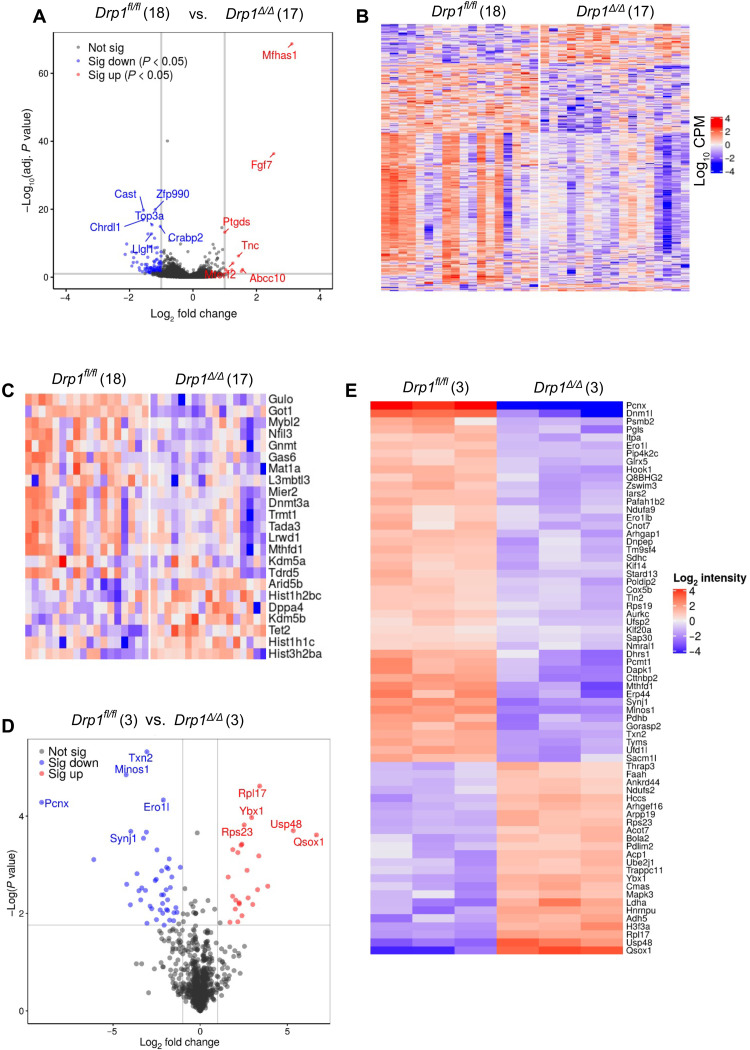
Fig. 4. Altered gene expression profile of Drp1Δ/Δ oocytes.
(A) Volcano plot showing DEGs (down-regulated, blue; up-regulated, red; and not different, black) between Drp1fl/fl and Drp1Δ/Δ oocytes. Highly dysregulated genes are labeled. (B) Heatmap of differential gene expression in Drp1fl/fl and Drp1Δ/Δ oocytes. CPM, counts per million. (C) Heatmap showing differential expression of epigenetic regulators in Drp1fl/fl and Drp1Δ/Δ oocytes. Number of single GV-stage oocytes per genotype used for RNA-seq analysis are shown (A to C). (D) Volcano plot showing differentially abundant proteins (down-regulated, blue; up-regulated, red; and not different, black) between MII-stage Drp1fl/fl and Drp1Δ/Δ oocytes. Highly dysregulated proteins are labeled. (E) Hierarchically clustered heatmap of relative abundancies of proteins in Drp1fl/fl and Drp1Δ/Δ oocytes. Number of replicates per genotype used for proteomic analysis are shown (D and E).
Proteomic analysis of Drp1Δ/Δ and Drp1fl/fl MII oocytes demonstrated that 69 proteins were differentially accumulated between Drp1fl/fl and Drp1Δ/Δ oocytes, of which 24 proteins were increased and 45 proteins were decreased in Drp1Δ/Δ oocytes (Fig. 4, D and E, and table S2). Enrichment analyses indicate that proteins regulating cellular metabolism (LDHA, CMAS, NDUFA9, SYNJ1, ADH5, COX5B, PDHB, MTHFD1, TYMS, PGLS, ITPA, SDHC, PAFAH1B2, and NDUFS2) are significantly affected in Drp1Δ/Δ oocytes compared to Drp1fl/fl oocytes (fig. S3F and table S2). Many of the affected proteins in Drp1Δ/Δ oocytes are known regulators of epigenetic modifications. For example, SAP30 is a regulator of histone acetylation (29), SDHC is a known regulator of histone modifications in mammalian cells (30) and MTHFD1 plays an essential role in one-carbon metabolism (31) and maternal Mthfd1 disruption is known to impair fetal growth (32).
Together, these omics studies show that disrupting mitochondrial fission in oocytes leads to marked changes to the transcriptomic and proteomic profile of Drp1Δ/Δ oocytes and that epigenetic and metabolic pathways are substantially altered. Given the importance of metabolites in the regulation of gene expression via modulation of epigenetic enzyme activity in somatic cells (33–35), we examined the effects of Drp1 deletion during oocyte growth on the oocyte epigenome.
Altered DNA and histone modifications in Drp1Δ/Δ oocytes and Drp1matΔ/+ zygotes
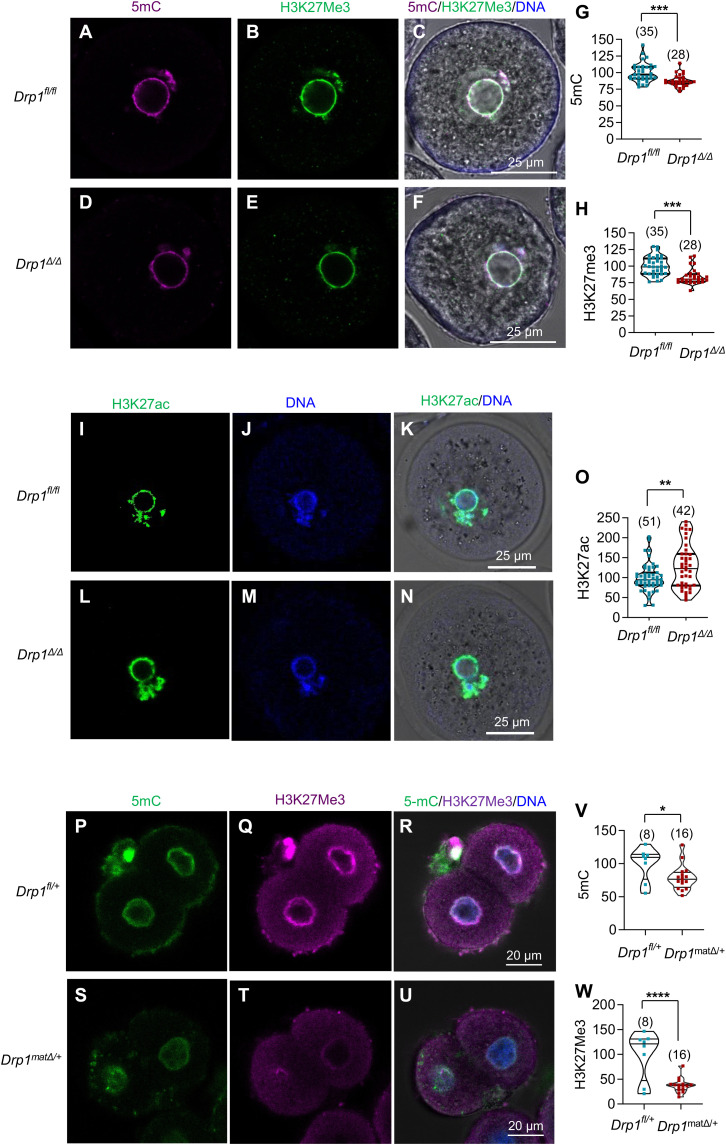
Given the dysregulation of metabolic and epigenetic gene products in Drp1Δ/Δ oocytes, we tested the hypothesis that deletion of Drp1 during oocyte growth is sufficient to disrupt the establishment of the oocyte epigenome. We examined DNA methylation and H3K27me3 in oocytes as they are known to regulate embryo development (36, 37). Immunofluorescence showed that the levels of 5mC and H3K27me3 were significantly decreased in Drp1Δ/Δ oocyte nuclei compared to Drp1fl/fl oocytes (Fig. 5, A to H) while the level of H3K27ac was significantly higher in the nuclei of Drp1Δ/Δ than in Drp1fl/fl oocytes (Fig. 5, I to O). We found that 5mC and H3K27me3 were also significantly decreased in Drp1matΔ/+ two-cell embryos compared to Drp1fl/+ embryos (Fig. 5, P to W). Other epigenetic marks, H3K4me3 (fig. S4, A to G) and H3K9me3 (fig. S4, H to N), were not altered. We matched the DEGs between Drp1fl/fl and Drp1Δ/Δ oocytes with previously published genes known to be marked with H3K27me3 in mouse oocytes (38, 39). About 18% of DEGs between Drp1fl/fl and Drp1Δ/Δ oocytes are among the known H3K27me3-marked genes (fig. S4O and table S3), suggesting that altered H3K27me3 level could be a determinant, at least in part, of differential gene expression in Drp1Δ/Δ oocytes.
Fig. 5. Altered epigenetic marks in the nuclei of Drp1Δ/Δ oocytes and Drp1matΔ/+ 2-cell zygotes.
Immunofluorescence (IF) showing 5mC (A, magenta), H3K27me3 (B, green), and merged (C) in Drp1fl/fl oocytes. IF showing 5mC (D), H3K27me3 (E), and merged (F) in Drp1Δ/Δ oocytes. (G) Quantification of 5mC fluorescence. (H) Quantification of H3K27me3 fluorescence. IF showing H3K27ac (I, green), DNA (J, blue), and merged (K) in Drp1fl/fl oocytes. IF showing H3K27ac (L, green), DNA (M, blue), and merged (N) in Drp1Δ/Δ oocytes. (O) Quantification of H3K27ac fluorescence. IF showing 5mC (P, green), H3K27me3 (Q, magenta) and merged (R) in Drp1fl/+ 2-cell zygotes. IF showing 5mC (S), H3K27me3 (T) and merged (U) in Drp1matΔ/+ 2-cell zygotes. (V) Quantification of 5mC fluorescence. (W) Quantification of H3K27me3 fluorescence. Results show mean ± SD and P values determined by two-tailed Student’s t test (G, H, O, V and W). Number of oocytes/zygotes analyzed are shown (G, H, O, V, and W).
Developmental defects of Drp1matΔ/+ embryos associated with the maternal nuclear genome
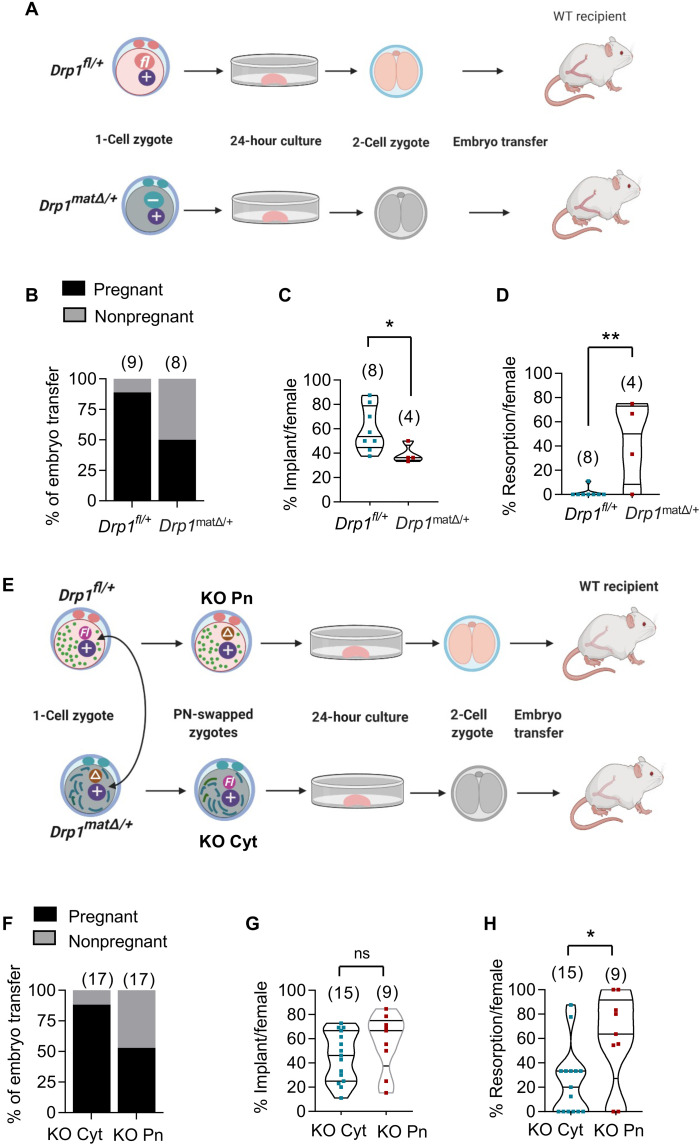
The epigenetic changes seen in Drp1Δ/Δ oocytes suggest a potential explanation for subsequent embryonic failure. To investigate this hypothesis, we have undertaken two series of embryo transfer experiments. First, Drp1fl/+ and Drp1matΔ/+ two-cell embryos were transferred into identical surrogates for gestation to obtain a baseline rate of development after embryo transfer (Fig. 6A). Seventy-four Drp1fl/+ (8.2 ± 1.6 per transfer, n = 9) and 65 Drp1matΔ/+ (8.1 ± 2.3 per transfer, n = 8) two-cell embryos were transferred to the oviducts of WT pseudo-pregnant recipients (table S4). Recipients culled between E15.5 to E20.5 of pregnancy revealed that eight of nine recipients receiving Drp1fl/+ embryos were confirmed pregnant compared to only four of eight recipients receiving Drp1matΔ/+ embryos (Fig. 6B and table S4). Of Drp1fl/+ embryos transferred, 40 of 74 (54%) implanted and 39 (53%) formed viable embryos (Fig. 6, C and D, and table S4). In contrast, 14 of 65 (22%) of Drp1matΔ/+ embryos implanted and only 8 (12%) formed morphologically normal fetuses (P < 0.0001; table S4). These embryo transfer results confirm that effects on fetal development are independent of the maternal environment in Drp1cKO mice and demonstrate that the absence of DRP1 specifically during oocyte growth markedly compromises subsequent embryo development.
Fig. 6. Development after embryo transfer.
(A to D) Embryonic lethality is due to defective Drp1Δ/Δ oocytes. (A) Schematic representation of embryo transfer experiment. Superovulated 5-week-old Drp1fl/fl and Drp1CKO females were mated with WT (+/+) males to generate WT Drp1fl/+embryos and heterozygous Drp1matΔ/+ zygotes, respectively. One-cell zygotes harvested from the oviducts of plugged donor females were cultured in vitro for 24 hours until they reached the two-cell stage. Two-cell stage embryos were transferred to the oviducts of different WT pseudo-pregnant recipients. (B) Recipients culled between E15.5 and E20.5 of pregnancy revealed that eight of nine recipients receiving Drp1fl/+ embryos became pregnant compared to only four of eight recipients receiving Drp1Drp1matΔ/+ embryos. (C) Per-pregnancy implantation rate of Drp1matΔ/+ embryos was markedly reduced than Drp1fl/+ embryos. (D) Within confirmed pregnancies, there was a higher percentage of embryo resorption in Drp1cKO females compared to Drp1fl/fl females. (E to H) Embryo development after pronuclear swapping and embryo transfer. (E) Schematic representation of reciprocal pronuclear swapping between one-cell zygotes, in vitro culture, and embryo transfer. (F) Comparison of pregnancy rates after implanting KO Cyt and KO Pn zygotes. (G) Implantation rates of KO Pn and KO Cyt embryos. Within positive pregnancies, there was a higher percentage of resorption of KO Pn embryos compared to KO Cyt embryos (H). P values determined by chi-square test (B and F) and two-tailed Student’s t test (C, D, G, and H). Results show means ± SD (C, D, G, and H). Number of mice analyzed are shown (B to D and F to H).
Next, we performed nuclear transfer at the pronuclear stage followed by embryo transfer to test the relative contributions of the nuclear genome (and its epigenetic modifications) and cytoplasm (with fission defective mitochondria) to the increased rate of embryonic lethality seen in Drp1matΔ/+ embryos. Nuclear transfer was used to swap the pronuclei between Drp1matΔ/+ and Drp1fl/+ zygotes to generate Drp1matΔ/+ pronuclei with WT cytoplasm (KO Pn) and Drp1fl/+ pronuclei with Drp1matΔ/+ cytoplasm (KO Cyt; Fig. 6E). Similar numbers of KO Pn embryos (10.5 ± 2.7 per transfer) and KO Cyt embryos (10.7 ± 2.2 per transfer) were transferred to individual oviducts of recipients. The pregnancy rate of dams receiving KO Pn embryos was significantly reduced compared to KO Cyt embryos (53%, n = 17 versus 88%, n = 17, respectively; Fig. 6F and table S5). Of the dams that became pregnant, implantation rates were variable but not significantly different (Fig. 6G and table S5). However, the proportion of resorption sites and dying embryos was markedly increased for KO Pn embryos with a mean of 60% resorptions compared to just over 20% in KO Cyt embryos (Fig. 6H and table S5). As a result, only half as many KO Pn embryos survived to midgestation [24 of 178 (13%)] compared to KO Cyt embryos [49 of 182 (27%); P < 0.001, chi-square test; table S5]. These data support the hypothesis that alterations to the maternal genome during growth of Drp1Δ/Δ oocytes is the primary reason for the high frequency of developmental failures in Drp1matΔ/+ fetuses.
DISCUSSION
In this study, we reveal a marked maternal effect resulting from the loss of Drp1 during oocyte development. The large majority of peri- and postimplantation-stage embryos and fetuses derived from Drp1 KO oocytes fail in utero, despite having sufficient DRP1 derived from the paternal allele for normal survival. Overall, our findings support the conclusion that DRP1 in oocytes is essential for normal mitochondrial function, which, in turn, is required for establishing a maternal epigenome compatible with viable embryo development.
A number of lines of evidence support this conclusion. Using pronuclear transplantation, we demonstrate that WT ooplasm is insufficient to completely rescue the developmental lethality seen in Drp1matΔ/+ embryos and, conversely, that the cytoplasm of Drp1matΔ/+ zygotes is compatible with higher rates of postimplantation development. The fact that the embryonic lethality more consistently follows the Drp1 KO maternal genome rather than the cytoplasm is strong evidence that epigenetic programming of the maternal genome is irreversibly disrupted during oocyte growth. This is further supported by the observed reduction in DNA methylation and H3K27me3 in the nucleus of Drp1 KO oocytes. These findings directly demonstrate the maternal epigenome is altered because of the loss of DRP1 during oocyte development. DNA methylation and H3K27me3 in the female germ line are responsible for regulating gene expression in resultant embryos (40, 41) and, consistent with our findings, others have shown that disruption of these epigenetic marks predominantly affects postimplantation development with little effect on oocytes or preimplantation embryos (41–43).
Single-cell oocyte RNA-seq and proteomics revealed genes involved with epigenetic regulation to be among the most disrupted in the absence of DRP1, including Got1, Dnmt3a, Mthfd1, Kdm5a, Kdm5b, Tet2, Hist1h1c, and Hist3h2ba, providing a potential mechanism for the observed marked recalibration of the oocyte transcriptome and proteome. This analysis also revealed that Drp1 deletion leads to disruption of the SCMC. Loss of this complex has been shown to be associated with maternal effects leading to embryonic arrest in mice [reviewed in (44)]. A number of SCMC components are disrupted including Tle6, which primarily affects preimplantation embryo development in mice (45), while TLE6 mutations are linked with human embryonic lethality (46, 47). KHDC3L mutation causes early embryonic failure, gestational abnormalities, and recurrent pregnancy loss in human and causes widespread DNA methylation deficiency (48), and NLRP5 mutations are associated with zygotic imprinting (49). Nlrp2 in mouse oocyte is essential for embryo development and postzygotic DNA methylation maintenance (50). How loss of DRP1 causes changes in SCMC genes remains unknown; however, given the decreased global DNA and histone methylation in Drp1 KO oocytes, SCMC genes may be differentially expressed because of the associated epigenetic modifications.
Drp1 deletion via Gdf9-Cre occurs during oocyte growth and is coincident with the period during which the oocyte epigenome is being established. The parental epigenetic profile needs to be erased each generation before being reestablished largely during oocyte growth (16, 17, 36). This is well established for classical genomic imprinting (17, 42) but less well established for noncanonical imprinting mediated by heritable histone modifications, including H3K27me3. Nevertheless, this temporal association of Drp1 deletion and establishing the oocyte epigenetic landscape provides additional support for the link between mitochondrial function and the oocyte epigenome. This conclusion is entirely consistent with the increasingly prevalent link between mitochondrial metabolism and epigenetic regulation in cancer, immunity, and cell differentiation (51–54). Cell culture models show that mitochondrial dysfunction can alter DNA methylation (55) and histone modifications (56) via disrupted metabolic pathways. Furthermore, a recent study demonstrates that metabolites can indeed alter DNA and histone methylation in oocytes and embryos (57).
How the deletion of Drp1 in oocytes causes the changes in the epigenome is likely to be a complex integration of impacts on mitochondrial metabolism and downstream modification of the activity of epigenetic regulators. Metabolic and epigenetic genes were the most disrupted in single-oocyte transcriptomics and proteomics. Mitochondrial structure is affected in Drp1 KO oocytes, consistent with the known role of DRP1 in mitochondrial fission (3, 5), and this is accompanied by a marked increase in levels of mitochondrial FAD++ and NAD(P)H as well as small but significant changes in MMP and mitochondrial ROS. It is unclear exactly how these mitochondrial changes come about and whether they are all coupled, but an increase in mitochondrial NADH and FAD++ is consistent with a decrease in Complex I activity and a switch to Complex II. The accumulation of NADH and FAD++ may affect normal TCA cycle activity, in part because of the failure of the ETC to generate NAD+ needed for oxidation of mitochondrial metabolites, as well as its availability for NAD+-dependent epigenetic regulators (52). Thus, the loss of DRP1 may ultimately disrupt the TCA cycle, the source of epigenetic modifying metabolites such as S-adenosyl methionine, succinate, and fumarate (11, 12, 33), thereby providing a potential link between DRP1 loss and epigenetic modifications.
Despite the marked impact on postimplantation development, Drp1 KO oocytes used in this study underwent growth, maturation, fertilization, and preimplantation development. This minimal impact on oocyte maturation and preimplantation development is consistent with our observation that aneuploidy was not increased because of the absence of DRP1. Mitochondrial mass has also been shown to influence histone modifications and gene expression (58), although we did not find any evidence for Drp1 deletion in oocytes causing a decrease in mitochondria or mtDNA copy number. In contrast, the decrease in MMP in Drp1 KO oocytes may contribute to changes in gene expression as it is a known mediator of mitochondria-directed nuclear gene expression (51). It remains uncertain whether the relatively modest decrease in MMP and increase in ROS seen in Drp1 KO oocytes is contributing to these effects, particularly given that oocyte meiosis and subsequent preimplantation development is not affected.
While Drp1matΔ/+ zygotes carrying a paternal Drp1 allele develop to the blastocyst stage with only a minor change in TE cell number, Drp1Δ/Δ zygotes generated by parthenogenetic activation undergo developmental arrest in the early cleavage stages. Given that the development of control parthenotes and fertilized embryos is similar, this demonstrates that at least one Drp1 allele is essential for preimplantation development. The precise explanation for the catastrophic failure of preimplantation development in the complete absence of DRP1 is not known but may reflect cumulative mitochondrial damage, building on compromised oocyte mitochondria. Alternatively, DRP1 may be essential to support the developmental transition of mitochondrial function as they undergo marked changes in structure and function from the eight-cell stage, particularly in cells destined to become the TE (59).
Our findings have important implications for understanding the role of oocyte mitochondrial function in embryo development and offspring health. We and others have shown that oocyte mitochondrial function is disrupted in obesity (60, 61), metabolic disease (62, 63), and maternal aging (64, 65). These conditions are also often associated with heritable and even transgenerational changes in offspring health (63, 66), but a direct link between oocyte mitochondrial function and heritable epigenetic changes has not been established. Our demonstration that a direct mitochondrial intervention, deletion of Drp1, leads to changes to the oocyte epigenome provides strong support that the heritable phenotypic changes are mediated via perturbation of mitochondrial function.
Our findings are consistent with recent developments in other systems, whereby mitochondria act as hubs, sensing environmental changes and directing nuclear responses through epigenetic modulation. The fact that mitochondria perform this function in oocytes elevates them from mere providers of ATP to pivotal modulators of embryonic development and offspring health.
MATERIALS AND METHODS
Mice
Drp1loxP/loxP mice (3) were crossed with transgenic mice that carried Gdf-9 promoter–mediated Cre recombinase (26). After multiple rounds of crossing, homozygous Drp1cKO female mice lacking Drp1 in oocytes (Drp1loxP/loxP; Gdf9-Cre) or Drp1Δ/Δ oocytes were obtained. Mice that do not carry the Cre transgene are referred to as Drp1loxP/loxP and were used as controls. The mice were housed under controlled environmental conditions with free access to water and food. All animal experimentation was approved by Monash University Animal Experimentation Ethics Committee and was performed in accordance with Australian National Health and Medical Research Council Guidelines on Ethics in Animal Experimentation.
Collection of oocytes and early embryos
To obtain GV oocytes, 4 to 5-week-old female mice were intraperitoneally injected with 8-IU pregnant mare’s serum gonadotropin (PMSG; Intervet), and 44 to 48 hours later, the mice were euthanized. Fully grown GV-stage oocytes surrounded by cumulus cells were released by puncturing the ovaries in M2 medium (Sigma-Aldrich) containing 200 μM 3-isobutyly-1-methylx-anthine (Sigma-Aldrich). Oocytes were freed from the attached cumulus cells by repetitive pipetting through a narrow-bore glass pipette. Oocytes were in vitro cultured in drops of M16 medium (Sigma-Aldrich) under mineral oil (Sigma-Aldrich) at 37°C in a humidified atmosphere of 5% CO2 in air or in M2 medium at 37°C. For collection of ovulated MII-stage oocytes, mice were superovulated by sequential intraperitoneal injections of 8-IU PMSG and 8-IU human chorionic gonadotropin (hCG; Intervet) at an interval of 48 hours, and the mice were culled 14 to 16 hours after hCG injection to harvest eggs from oviducts. For collection of embryos, females were mated with stud C57BL/6J males after hCG injection, and the mice were culled 14 to 16 hours after hCG injection to harvest one-cell zygotes from oviducts. Zygotes were further cultured in SAGE 1-Step medium (Cooper Surgical) up to blastocyst stage at 37°C in a humidified atmosphere of 5% CO2 in air.
Detection of RNA synthesis
Two-cell stage zygotes were incubated with 2 mM EU (Thermo Fisher Scientific) for 2 hours in SAGE 1-Step medium at 37°C in a humidified atmosphere of 5% CO2 in air. Zona pellucida was removed by Tyrode’s solution (Sigma-Aldrich) and then fixed in 4% paraformaldehyde (PFA) for 30 min at room temperature (RT) and permeabilized with 0.5% Triton X-100 in phosphate-buffered saline (PBS) for 20 min at RT. Zygotes were stained using the Click-iT RNA imaging kit for nascent RNA (Thermo Fisher Scientific) and then incubated with rabbit polyclonal RNA PolII (phospho-Ser2) antibodies (1:400) overnight at 4°C. This was followed by an incubation with specific Alexa Fluor 647 (1:1000 diluted in blocking buffer; Invitrogen Molecular Probes) for 1 hour at RT. DNA was labeled using 10-min incubation in Hoechst 33342 (10 μg/ml; Sigma-Aldrich).
mtDNA copy number assay
Individual oocytes were transferred into 10 μl of lysis buffer [50 mM tris-HCl (pH 8.0), 1 mM EDTA, 0.5% Tween 20, and proteinase K (200 μg/ml)]. Sample was incubated at 55°C for 2 hours, followed by inactivation of proteinase K at 95°C for 10 min. Genomic DNA was diluted five times for quantitative polymerase chain reaction (PCR). A plasmid containing 12S ribosomal (r)RNA region of mtDNA was used as quantification standards. The standard stock was serially diluted to prepare the standard curve. Real-time fluorescence–monitored quantitative PCR using the primer pair 5′-CGT TAG GTC AAG GTG TAG CC-3′ and 5′-CCA AGC ACA CTT TCC AGT ATG-3′ was performed in Stratagene Mx3000P (Agilent Technologies). Each reaction of 20 μl consisted of 10 μl of SYBR Green, 100 nM each of the forward and reverse primers and 2 μl of diluted DNA sample. Standard curves were created for each run, and sample copy number was generated from the equation of Ct value against copy number for the corresponding standard curve.
Parthenogenesis and culture
Ovulated Drp1fl/fl and Drp1Δ/Δ eggs were collected from oviduct 14 to 16 hours after hCG in M2 medium, and cumulus cells were removed with hyaluronidase (0.3 mg/ml; Sigma-Aldrich) in M2 medium. After two quick washes in calcium-free CZB medium [81.62 mM NaCl, 4.83 mM KCl, 1.18 mM MgSO4, 0.11 mM EDTA, 5.6 mM glucose, 37 mM sodium lactate, 1.2 mM KH2PO4, 25.1 mM NaHCO3, 0.27 mM pyruvate, 1 mM l-glutamine, and bovine serum albumin (BSA; 5 mg/ml), pH 7.4] containing cytochalasin B (5 μg/ml; Sigma-Aldrich) and 10 mM SrCl2 (Sigma-Aldrich), oocytes were cultured in this medium for 2 hours. Oocytes were further cultured for 3 hours in M2 medium containing cytochalasin B (5 μg/ml). Activated eggs were cultured in SAGE 1-Step medium (CooperSurgical) up to blastocyst stage at 37°C in a humidified atmosphere of 5% CO2 in air.
Timed mating and harvesting postimplantation embryos and histology
Female mice at 5 weeks of age were individually housed with C57BL/6J WT male (age, 3 to 4 months) and checked every morning until vaginal plugs were detected. The day on which a vaginal plug was detected was designated as E0.5. Pregnant females were euthanized by cervical dislocation, and the uteri were collected in ice-chilled PBS kept on ice. Embryos and placentae either were snap-frozen in liquid nitrogen and stored at −80°C until further analysis or were fixed in Bouin’s solution (Sigma-Aldrich) for histological analysis.
Pronuclear swapping and embryo transfer
The protocol was previously described (67). Briefly, one-cell stage Drp1fl/+ and Drp1matΔ/+ embryos were collected (24 hours after hCG injection) in M2 medium containing hyaluronidase (0.3 mg/ml) to remove cumulus cells. Embryos were then incubated in M16 medium containing nocodazole (0.3 μg/ml; Sigma-Aldrich) and cytochalasin B (5 μg/ml) in a humidified incubator with 5% CO2 in air for 15 to 45 min at 37°C. Both the pronuclei from a zygote were removed by micropipette and drawn a small volume of inactivated Sendai virus (Cosmo Bio, USA; ~3000 hemagglutinating units/ml) into the enucleation/injection pipette. The virus and karyoplast were injected into the perivitelline space of a second enucleated one-cell stage embryo. The reconstructed embryos were cultured in inhibitor-free M16 medium overnight to allow their cleavage. Two-cell embryos were transferred into the oviducts of pseudo-pregnant C57BL6J/MARP x BALBC F1 recipient females for further embryo development.
Determination of mitochondrial ROS
Mitochondrial ROS were measured using Mitosox Red (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Oocytes were incubated with 5 μM Mitosox in M2 media at 37°C for 30 min. Epifluorescence and bright-field images were acquired using an SP8 confocal microscope (Leica, Germany), and fluorescence was measured at an excitation wavelength of 488-nm and 520-nm long-pass filter. Images were acquired using a 40× water immersion 1.2–numerical aperture (NA) objective with line averaging set to 2 to 4. Confocal sections obtained from the LAS AF Lite software were analyzed using ImageJ [National Institutes of Health (NIH)]. Gray values were derived from the “analyze” function of ImageJ to evaluate the fluorescent intensity of oocyte mitochondria.
Measurement of MMP
Oocytes were labeled with 50 nM TMRM (Sigma-Aldrich) and MitoTracker Green (Thermo Fisher Scientific) in M2 medium for 30 min at 37°C. TMRM was excited using the 552-nm laser, and fluorescence collected using a 560- to 630-nm band-pass filter. MitoTracker Green was excited using the 488-nm laser and fluorescence collected using a 495- to 540-nm band-pass filter. Confocal sections obtained from the LASX software (Leica) were analyzed using ImageJ for measurements of MMP. Gray values were derived from the analyze function of ImageJ to evaluate the fluorescent intensity of oocyte mitochondria. Ratio of TMRM and MitoTracker Green was calculated to measure the MMP (21). Values were normalized to the mean of control values in each experimental replicate to avoid the impact of day-to-day variations in the fluorescence ratio.
Measurement of oocyte ATP levels
Denuded GV oocytes (10 pooled per group) collected from PMSG-primed mice were collected in 50 μl of filtered ultrapure water and stored at −80°C until use. To prepare standards, 10−7 M ATP standard stock was obtained from ENLITEN ATP Assay System Bioluminescence Detection Kit (Promega) and was diluted with filtered ultrapure water. ATP levels were assayed using the ATP bioluminescent somatic cell assay kit (FLASC, Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, 100 μl of ATP Assay Mix Working Solution was added to a 96-well plate (reaction vial, M0187, Greiner) and allowed to stand at RT for 3 min. Somatic Cell ATP Releasing Reagent (100 μl), 50 μl of filtered ultrapure water, and 50 μl of sample were added to a new tube, and 100 μl was transferred to the reaction vial. ATP concentration was measured immediately using a luminometer (BMG, Clariostar, 76G58). Data are normalized to the mean ATP levels of control group.
Autofluorescence imaging of NAD(P)H and FAD++
Autofluorescent signals of NAD(P)H and FAD++ were detected as described previously (19). Briefly, the reduced nucleotides NADH and NADPH were excited by ultraviolet light (405 nm), and emission was collected using a 435- to 485-nm band-pass filter. NAD(P)H autofluorescence is both detected in mitochondria and cytosol. The oxidized flavoproteins (FAD++) autofluorescence was collected with a 505- to 550-nm band-pass filter after exciting with 488-nm lasers. FAD++ autofluorescence is exclusively localized into the mitochondria.
Western blot
Oocytes were collected in SDS sample buffer and heated for 10 min at 70°C. Proteins were fractionated at 200 V for 50 min using a 10% NuPAGE bis-tris precast gel (Invitrogen) and Mops running buffer. Proteins were blotted onto polyvinylidene fluoride membranes (Invitrogen) for 1.5 hours at 30 V, and then the membrane was blocked in tris-buffered saline-Tween 20 containing 5% skimmed milk for 2 hours, followed by incubation overnight at 4°C with primary antibodies, β-actin (Abcam no. ab3280) and DRP1 (Cell Signaling Technology no. 8570). For primary antibody detection, we used goat horseradish peroxidase–conjugated anti-mouse or anti-rabbit secondary antibodies (1:1000, 1 hour at RT, Dako). Last, the membrane was processed using standard enhanced chemiluminesence techniques (GE Healthcare) and processed using the ChemiDoc MP imaging system (Bio-Rad).
Immunofluorescence and imaging
Oocytes or embryos were fixed in a solution of 4% PFA (Sigma-Aldrich) and 2% Triton X-100 in PBS for 20 min at RT. Blocking was performed in PBS with 10% BSA and 2% Tween 20 for 60 min at RT. For 5mC and H3K27me3 labeling, oocytes or embryos were collected in M2 medium, and zona pellucida was removed by Tyrode’s solution (Sigma-Aldrich) and then fixed in 4% PFA for 30 min at RT and permeabilized with 0.5% Triton X-100 in PBS for 20 min at RT. They were denatured with 4 N hydrochloric acid for 10 min, neutralized with 100 mM tris-HCl (pH 8.5) for 15 min, blocked in PBS with 10% BSA and 2% Tween for 60 min at RT, and incubated with specific primary antibodies overnight at 4°C. The primary antibodies used were DRP1 (Cell Signaling Technology no. 8570), 5mC (Bio-Rad no. MCA2201), H3K27ac (Cell Signaling Technology no. 8173), H3K27me3 (Merck Millipore no. 07-449), H3K4me3 (Active Motif no. 39159), CDX2 (Novus Biologicals no. NB100-2136), OCT-4 (Abcam no. ab184665), H3K9me3 (Cell Signaling Technology no. 13969), TOM20 (Santa Cruz Biotechnology no. sc-11415), and RNA Pol II (phospho S2) (Abcam no. ab5095). This was followed by an incubation with specific Alexa Fluors (1:1000 diluted in blocking buffer; Invitrogen Molecular Probes) for 1 hour at RT. DNA was labeled using 10-min incubation in Hoechst 33342 (10 μg/ml; Sigma-Aldrich). Serial Z sections of fixed oocytes/eggs in PBS were acquired at RT in a glass-bottomed dish (World Precision Instruments Inc., Sarasota, FL) using a laser-scanning confocal microscope imaging system (SP8, Leica).
Images were analyzed using ImageJ software (NIH). The midsection of each nucleus was identified by Hoechst staining and determined by maximal area. Total intensities of DNA modifications in the midsections were measured, and background signals corresponding to the equal cytoplasmic area were subtracted. Within each experiment, oocytes/zygotes were imaged using the same setting on the microscope.
Analysis of oocyte mitochondrial morphology
Germinal vesicle stage oocytes were labeled with MitoTracker Orange (25 nM) in M2 medium for 30 min at 37°C. Images acquired by an SP8 confocal microscope were analyzed using MiNA (Mitochondrial Network Analysis, https://github.com/StuartLab/MiNA), an ImageJ (NIH)–based utility to aid in quantitatively describe the appearance of mitochondrial morphology in fluorescence micrographs. The mean number of attached lines used to represent each structure (network branches mean) and the mean length of all the lines used to represent the mitochondrial structures (branch length mean) were calculated.
Chromosome spreads
For collection of four-cell embryos, females were mated with stud C57BL/6J males after hCG injection, and the mice were culled 14 to 16 hours after hCG injection to harvest one-cell zygotes from oviducts. Zygotes were further cultured in SAGE 1-Step medium (Cooper Surgical) up to four-cell stage at 37°C in a humidified atmosphere of 5% CO2 in air. Four-cell embryos were transferred into SAGE 1-Step medium containing Colcemid (100 ng/ml; Gibco) and were cultured overnight at 37°C to arrest at metaphase. Embryos were exposed to Acid Tyrodes solution (Sigma-Aldrich) for 1 min at 25°C to remove the zona pellucida. After a 2-min recovery in M2 medium, the embryos were transferred into a 0.5% sodium citrate (Sigma-Aldrich) drop for 2 min and then moved onto a clean microscope slide that had been previously dipped in a solution of 1% PFA in MilliQ water (pH 9.2), containing 0.15% Triton X-100 and 3 mM dithiothreitol (68). The slides were allowed to dry slowly in a humid chamber overnight. DNA on the slides was stained with Hoechst 33342 (10 μg/ml; Sigma-Aldrich) for 10 min, and slides were mounted for observation using a Zeiss Imager M2 fluorescence microscope linked to an AxioCam MRm charge-coupled device camera system and processed using the Zen software 2011 (Carl Zeiss Microscopy).
Oocyte meiotic spindle analysis
MII oocytes were fixed in 4% PFA (Sigma-Aldrich) and 2% Triton X-100 (Sigma-Aldrich) in PBS for 30 min at RT. Oocytes were blocked in PBS containing 10% BSA and 2% Tween 20 for 1 hour at RT and incubated with Alexa Fluor 488–conjugated mouse anti–α-tubulin (1:200; Thermo Fisher Scientific no. 322588) for 1 hour at RT. After three washes, chromosomes were labeled using Hoechst 33342 (10 μg/ml; Sigma-Aldrich) for 10 min, and oocytes were mounted in PBS drops on glass-bottomed dish (World Precision Instruments, Sarasota, FL, USA) for imaging. Serial Z sections of labeled oocytes were acquired using a laser-scanning confocal microscope imaging system (SP8; Leica) and 40× water immersion objective (1.2 NA) at 25°C. Fluorescein isothiocyanate was excited using the 488-nm laser, and a band-pass of green-fluorescent emission was 495 to 540 nm. Hoechst 33342 was excited with a 405-nm laser, and emission was collected with a band-pass filter of 438 to 458 nm. Images were analyzed using Las X software (Leica).
Histological analysis of ovary
Histological analysis of ovarian tissue was performed as previously described (69). Briefly, ovaries were fixed in 4% PFA, dehydrated, and embedded in paraffin. Paraffin-embedded ovaries were serially sectioned at 5-μm thickness and stained with hematoxylin for morphological observation.
Single-oocyte RNA-seq
To perform single-cell RNA-seq analyses, GV-stage oocytes (18 oocytes/genotype) were individually placed in an Eppendorf tube containing 10 μl of nuclease-free water. Tubes were snap frozen in liquid nitrogen and stored at −80°C until further experiment. The whole lysate was supplied for complementary DNA synthesis and RNA-seq library preparation with, respectively, a SMARTer Ultra Low Input RNA Kit (Clontech, CA USA) and Nextera XT library prep kit (Illumina, CA, USA), each according to the manufacturer’s instructions. Indexed libraries were pooled (10 nM each) and sequenced using an Illumina HiSeq 2500 System under single-end, 100–base pair conditions.
Read files are deposited: National Centre for Biotechnology Information Gene Expression Omnibus (GEO) dataset (GSE180579). Reads were aligned to mouse genome GRCm38 and quantified against Ensembl 88 gene annotations. Tools were run using the RNAsik pipeline (https://monashbioinformaticsplatform.github.io/RNAsik-pipe/). PCR duplicates were marked with Picard (http://broadinstitute.github.io/picard), alignments were performed with STAR (70), and unstranded read counts quantified by gene using featureCounts Subread version 1.5.2 (71). BAM file manipulation was performed with SAMtools (72). The quality of single-cell data was inspected using Scater version 1.4 (73) and differential expression calculated with Monocle version 2.2.0 using the default “negbinomial.size” method (74). One outlier cell (Drp1 KO sample DG_10_S68) was removed because of low sequence diversity. Only genes with at least 10 reads in 20% of cells were included in the analysis. The DEGs of interest were thresholded with a P value threshold of 0.05 (after multiple hypothesis correction with Benjamini-Hochberg method). Expression values presented as log10 counts per million, as (fragment count +1) / total count for library among analyzed genes × 1,000,000.
Oocyte proteomic analysis
MII-stage Drp1Δ/Δ and Drp1fl/fl oocytes (300 oocytes per sample) were lysed in SDC lysis buffer [1% (w/v) sodium deoxycholate, 100 mM Hepes, pH 8.1], heated at 95°C for 5 min and then sonicated before measuring the protein concentration using the bicinchoninic acid assay method. The lysed samples were denatured and alkylated by adding tris(2-carboxyethyl) phosphine hydrochloride and 2-chloroacetamide to a final concentration of 10 and 40 mM, respectively, and the mixture was incubated at 95°C for 5 min. Sequencing grade trypsin was added at an enzyme to protein ratio of 1:100 and incubated overnight at 37°C. The reaction was stopped using 1% formic acid, and SDC was removed using 100% water-saturated ethyl acetate. The aqueous peptide–containing phase was collected and concentrated in a vacuum concentrator.
Proteomic data acquisition
Using a Dionex UltiMate 3000 RSLCnano system equipped with a Dionex UltiMate 3000 RS autosampler, an Acclaim PepMap RSLC analytical column (75 μm by 50 cm, nanoViper, C18, 2 μm, 100 Å; Thermo Fisher Scientific), and an Acclaim PepMap 100 trap column (100 μm by 2 cm, nanoViper, C18, 5 μm, 100 Å; Thermo Fisher Scientific), the tryptic peptides were separated by increasing concentrations of 80% acetonitrile/0.1% formic acid at a flow of 250 nl/min for 158 min and analyzed with a QExactive Plus mass spectrometer (Thermo Fisher Scientific). The instrument was operated in data-dependent acquisition mode to automatically switch between full-scan mass spectrometry (MS) and tandem MS acquisition. Each survey full scan [375 to 1575 mass/charge ratio (m/z)] was acquired with a resolution of 70,000 (at 200 m/z), an AGC (automatic gain control) target of 3 × 106, and a maximum injection time of 54 ms. Dynamic exclusion was set to 15 s. The 12 most intense multiply charged ions (z ≥ 2) were sequentially isolated and fragmented in the collision cell by higher-energy collisional dissociation (HCD) with a fixed injection time of 54 ms, 17,500 resolution, and an AGC target of 2 × 105.
Proteomics data analysis
The raw data files were analyzed with the MaxQuant software suite v1.6.2.10 (75) and its implemented Andromeda search engine (76) to obtain protein identifications and their respective label-free quantification (LFQ) values using standard parameters. These data were further analyzed with Perseus (77) on the ProteinGroup.txt file. First, contaminant proteins, reverse sequences, and proteins identified “only by site” were filtered out. In addition, proteins that have been only identified by a single peptide and proteins not identified/quantified consistently in same condition have been removed. The LFQ data were converted to log2 scale, samples were grouped by conditions and missing values were imputed using the “Missing not At Random” (MNAR) method, using random draws from a left-shifted Gaussian distribution of 1.8 StDev (SD) apart with a width of 0.3. Student’s t test was used to generate a list of differentially expressed proteins for each pairwise comparison. A cutoff of the adjusted P value of 0.05 (false discovery rate–adjusted) along with a log2 fold change of 1 has been applied to determine significantly regulated proteins in Drp1Δ/Δ and Drp1fl/fl comparison. The enrichment analysis was performed using gprofiler (78) for Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes, and Reactome databases against mouse reference proteome. The adjusted P value cut off of 0.05 was used to determine significantly enriched GO term or pathway. All the figures were generated in R using ggplot2 (79) and ComplexHeatmap (80) libraries.
Statistical analysis
Statistical analyses were performed in GraphPad Prism 8. P values were calculated with Student’s t test or chi-square test as indicated in respective figure or table legends. Not significant, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001. All data are presented as means ± SD of biological replicates, and error bars represent SD of biological repeats unless otherwise stated in the figure legends.
Acknowledgments
We thank Monash Histology Platform, Monash Bioinformatics Platform, Monash Proteomics and Metabolomics Platform, and Monash Micro Imaging for enabling many of the experiments described herein. We thank T. Kono, Tokyo University of Agriculture for help with oocyte RNA-seq. Figures were created using BioRender.com.
Funding: This work was supported by National Health and Medical Research Council (NHMRC) Australia 1165627 (D.A. and J.C.), NHMRC Peter Doherty–Australian Biomedical Fellowship APP1112766 (D.A.), NHMRC Research Fellowship 1117975 (R.L.R.), and NIH GM144103 (H.S.).
Author contributions: Conceptualization: D.A. and J.C. Methodology/Investigation: D.A., I.-w.L., J.L., U.A.-Z., Q.-H.Z., W.S.Y., L.H., Y.W., J.R.M., and R.L.R. Resources: H.S. provided Drp1 floxed mice. Funding acquisition: D.A. and J.C. Writing—original draft: D.A. and J.C. Writing—review and editing: D.A., J.C., J.R.M., and R.L.R.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All sequencing data have been deposited in the GEO under accession number GSE180579. All other data are available in the main text or the Supplementary Materials. We obtained code MiNa from https://github.com/StuartLab/MiNA.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Tables S1, S4 and S5
Other Supplementary Material for this manuscript includes the following:
Tables S2 and S3
REFERENCES AND NOTES
- 1.Hamanaka R. B., Chandel N. S., Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 35, 505–513 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra P., Chan D. C., Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15, 634–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T. W., Iijima M., Sesaki H., The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186, 805–816 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraus F., Ryan M. T., The constriction and scission machineries involved in mitochondrial fission. J. Cell Sci. 130, 2953–2960 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S. O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y. I., Taguchi N., Morinaga H., Maeda M., Takayanagi R., Yokota S., Mihara K., Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11, 958–966 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Uo T., Dworzak J., Kinoshita C., Inman D. M., Kinoshita Y., Horner P. J., Morrison R. S., Drp1 levels constitutively regulate mitochondrial dynamics and cell survival in cortical neurons. Exp. Neurol. 218, 274–285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh K., Nakamura K., Iijima M., Sesaki H., Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 23, 64–71 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rambold A. S., Pearce E. L., Mitochondrial dynamics at the interface of immune cell metabolism and function. Trends Immunol. 39, 6–18 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Buck M. D., O’Sullivan D., Klein Geltink R. I., Curtis J. D., Chang C. H., Sanin D. E., Qiu J., Kretz O., Braas D., van der Windt G. J. W., Chen Q., Huang S. C. C., O’Neill C. M., Edelson B. T., Pearce E. J., Sesaki H., Huber T. B., Rambold A. S., Pearce E. L., Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khacho M., Clark A., Svoboda D. S., Azzi J., MacLaurin J. G., Meghaizel C., Sesaki H., Lagace D. C., Germain M., Harper M. E., Park D. S., Slack R. S., Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell 19, 232–247 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Wai T., Langer T., Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. 27, 105–117 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Chandel N. S., Mitochondria as signaling organelles. BMC Biol. 12, 34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adhikari D., Liu K., Molecular mechanisms underlying the activation of mammalian primordial follicles. Endocr. Rev. 30, 438–464 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Jansen R. P., Germline passage of mitochondria: Quantitative considerations and possible embryological sequelae. Hum. Reprod. 15 ( Suppl. 2), 112–128 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Piko L., Taylor K. D., Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev. Biol. 123, 364–374 (1987). [DOI] [PubMed] [Google Scholar]
- 16.Kono T., Obata Y., Yoshimzu T., Nakahara T., Carroll J., Epigenetic modifications during oocyte growth correlates with extended parthenogenetic development in the mouse. Nat. Genet. 13, 91–94 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Stewart K. R., Veselovska L., Kelsey G., Establishment and functions of DNA methylation in the germline. Epigenomics 8, 1399–1413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumollard R., Campbell K., Halet G., Carroll J., Swann K., Regulation of cytosolic and mitochondrial ATP levels in mouse eggs and zygotes. Dev. Biol. 316, 431–440 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Dumollard R., Ward Z., Carroll J., Duchen M. R., Regulation of redox metabolism in the mouse oocyte and embryo. Development 134, 455–465 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Dalton C. M., Carroll J., Biased inheritance of mitochondria during asymmetric cell division in the mouse oocyte. J. Cell Sci. 126, 2955–2964 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Zubaidi U., Liu J., Cinar O., Robker R. L., Adhikari D., Carroll J., The spatio-temporal dynamics of mitochondrial membrane potential during oocyte maturation. Mol. Hum. Reprod. 25, 695–705 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Blerkom J., Runner M. N., Mitochondrial reorganization during resumption of arrested meiosis in the mouse oocyte. Am. J. Anat. 171, 335–355 (1984). [DOI] [PubMed] [Google Scholar]
- 23.Dumollard R., Marangos P., Fitzharris G., Swann K., Duchen M., Carroll J., Sperm-triggered [Ca2+] oscillations and Ca2+ homeostasis in the mouse egg have an absolute requirement for mitochondrial ATP production. Development 131, 3057–3067 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Wakai T., Mehregan A., Fissore R. A., Ca2+ signaling and homeostasis in mammalian oocytes and eggs. Cold Spring Harb. Perspect. Biol. 11, a035162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Udagawa O., Ishihara T., Maeda M., Matsunaga Y., Tsukamoto S., Kawano N., Miyado K., Shitara H., Yokota S., Nomura M., Mihara K., Mizushima N., Ishihara N., Mitochondrial fission factor Drp1 maintains oocyte quality via dynamic rearrangement of multiple organelles. Curr. Biol. 24, 2451–2458 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Lan Z. J., Xu X., Cooney A. J., Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol. Reprod. 71, 1469–1474 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Manczak M., Sesaki H., Kageyama Y., Reddy P. H., Dynamin-related protein 1 heterozygote knockout mice do not have synaptic and mitochondrial deficiencies. Biochim. Biophys. Acta 1822, 862–874 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Garcia V., Fineberg E., Wilson R., Murray A., Mazzeo C. I., Tudor C., Sienerth A., White J. K., Tuck E., Ryder E. J., Gleeson D., Siragher E., Wardle-Jones H., Staudt N., Wali N., Collins J., Geyer S., Busch-Nentwich E. M., Galli A., Smith J. C., Robertson E., Adams D. J., Weninger W. J., Mohun T., Hemberger M., Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature 555, 463–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D., Liu J., Yang X., Zhou C., Guo J., Wu C., Qin Y., Guo L., He J., Yu S., Liu H., Wang X., Wu F., Kuang J., Hutchins A. P., Chen J., Pei D., Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell 21, 819–833.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Cervera A. M., Bayley J. P., Devilee P., McCreath K. J., Inhibition of succinate dehydrogenase dysregulates histone modification in mammalian cells. Mol. Cancer 8, 89 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacFarlane A. J., Perry C. A., Girnary H. H., Gao D., Allen R. H., Stabler S. P., Shane B., Stover P. J., Mthfd1 is an essential gene in mice and alters biomarkers of impaired one-carbon metabolism. J. Biol. Chem. 284, 1533–1539 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beaudin A. E., Perry C. A., Stabler S. P., Allen R. H., Stover P. J., Maternal Mthfd1 disruption impairs fetal growth but does not cause neural tube defects in mice. Am. J. Clin. Nutr. 95, 882–891 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Etchegaray J. P., Mostoslavsky R., Interplay between metabolism and epigenetics: A nuclear adaptation to environmental changes. Mol. Cell 62, 695–711 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu C., Thompson C. B., Metabolic regulation of epigenetics. Cell Metab. 16, 9–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma U., Rando O. J., Metabolic inputs into the epigenome. Cell Metab. 25, 544–558 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Barlow D. P., Bartolomei M. S., Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6, a018382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue A., Jiang L., Lu F., Zhang Y., Genomic imprinting of Xist by maternal H3K27me3. Genes Dev. 31, 1927–1932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X., Wang C., Liu W., Li J., Li C., Kou X., Chen J., Zhao Y., Gao H., Wang H., Zhang Y., Gao Y., Gao S., Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Zheng H., Huang B., Zhang B., Xiang Y., du Z., Xu Q., Li Y., Wang Q., Ma J., Peng X., Xu F., Xie W., Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 63, 1066–1079 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Ferguson-Smith A. C., Sasaki H., Cattanach B. M., Surani M. A., Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362, 751–755 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Inoue A., Chen Z., Yin Q., Zhang Y., Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes Dev. 32, 1525–1536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H., Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Xu Q., Xiang Y., Wang Q., Wang L., Brind’Amour J., Bogutz A. B., Zhang Y., Zhang B., Yu G., Xia W., du Z., Huang C., Ma J., Zheng H., Li Y., Liu C., Walker C. L., Jonasch E., Lefebvre L., Wu M., Lorincz M. C., Li W., Li L., Xie W., SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat. Genet. 51, 844–856 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Lu X., Gao Z., Qin D., Li L., A maternal functional module in the mammalian Oocyte-To-Embryo transition. Trends Mol. Med. 23, 1014–1023 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Yu X. J., Yi Z., Gao Z., Qin D., Zhai Y., Chen X., Ou-Yang Y., Wang Z. B., Zheng P., Zhu M. S., Wang H., Sun Q. Y., Dean J., Li L., The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat. Commun. 5, 4887 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alazami A. M., Awad S. M., Coskun S., al-Hassan S., Hijazi H., Abdulwahab F. M., Poizat C., Alkuraya F. S., TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 16, 240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin J., Xu H., Chen B., Wang W., Wang L., Sun X., Sang Q., Expanding the genetic and phenotypic spectrum of female infertility caused by TLE6 mutations. J. Assist. Reprod. Genet. 37, 437–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demond H., Anvar Z., Jahromi B. N., Sparago A., Verma A., Davari M., Calzari L., Russo S., Jahromi M. A., Monk D., Andrews S., Riccio A., Kelsey G., A KHDC3L mutation resulting in recurrent hydatidiform mole causes genome-wide DNA methylation loss in oocytes and persistent imprinting defects post-fertilisation. Genome Med. 11, 84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Docherty L. E., Rezwan F. I., Poole R. L., Turner C. L. S., Kivuva E., Maher E. R., Smithson S. F., Hamilton-Shield J. P., Patalan M., Gizewska M., Peregud-Pogorzelski J., Beygo J., Buiting K., Horsthemke B., Soellner L., Begemann M., Eggermann T., Baple E., Mansour S., Temple I. K., Mackay D. J. G., Mutations in NLRP5 are associated with reproductive wastage and multilocus imprinting disorders in humans. Nat. Commun. 6, 8086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahadevan S., Sathappan V., Utama B., Lorenzo I., Kaskar K., van den Veyver I. B., Maternally expressed NLRP2 links the subcortical maternal complex (SCMC) to fertility, embryogenesis and epigenetic reprogramming. Sci. Rep. 7, 44667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanin D. E., Matsushita M., Geltink R. I. K., Grzes K. M., van Teijlingen Bakker N., Corrado M., Kabat A. M., Buck M. D., Qiu J., Lawless S. J., Cameron A. M., Villa M., Baixauli F., Patterson A. E., Hässler F., Curtis J. D., O’Neill C. M., O’Sullivan D., Wu D., Mittler G., Huang S. C.-C., Pearce E. L., Pearce E. J., Mitochondrial membrane potential regulates nuclear gene expression in macrophages exposed to prostaglandin E2. Immunity 49, 1021–1033.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martínez-Reyes I., Diebold L. P., Kong H., Schieber M., Huang H., Hensley C. T., Mehta M. M., Wang T., Santos J. H., Woychik R., Dufour E., Spelbrink J. N., Weinberg S. E., Zhao Y., DeBerardinis R. J., Chandel N. S., TCA cycle and mitochondrial membrane potential are necessary for diverse biological functions. Mol. Cell 61, 199–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ducker G. S., Rabinowitz J. D., One-carbon metabolism in health and disease. Cell Metab. 25, 27–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ansó E., Weinberg S. E., Diebold L. P., Thompson B. J., Malinge S., Schumacker P. T., Liu X., Zhang Y., Shao Z., Steadman M., Marsh K. M., Xu J., Crispino J. D., Chandel N. S., The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat. Cell Biol. 19, 614–625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lozoya O. A., Xu F., Grenet D., Wang T., Grimm S. A., Godfrey V., Waidyanatha S., Woychik R. P., Santos J. H., Single nucleotide resolution analysis reveals pervasive, long-lasting DNA methylation changes by developmental exposure to a mitochondrial toxicant. Cell Rep. 32, 108131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kopinski P. K., Janssen K. A., Schaefer P. M., Trefely S., Perry C. E., Potluri P., Tintos-Hernandez J. A., Singh L. N., Karch K. R., Campbell S. L., Doan M. T., Jiang H., Nissim I., Nakamaru-Ogiso E., Wellen K. E., Snyder N. W., Garcia B. A., Wallace D. C., Regulation of nuclear epigenome by mitochondrial DNA heteroplasmy. Proc. Natl. Acad. Sci. U.S.A. 116, 16028–16035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L., Zhu S., Shu W., Guo Y., Guan Y., Zeng J., Wang H., Han L., Zhang J., Liu X., Li C., Hou X., Gao M., Ge J., Ren C., Zhang H., Schedl T., Guo X., Chen M., Wang Q., Characterization of metabolic patterns in mouse oocytes during meiotic maturation. Mol. Cell 80, 525–540.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guantes R., Rastrojo A., Neves R., Lima A., Aguado B., Iborra F. J., Global variability in gene expression and alternative splicing is modulated by mitochondrial content. Genome Res. 25, 633–644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shahbazi M. N., Mechanisms of human embryo development: From cell fate to tissue shape and back. Development 147, dev190629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu L. L., Russell D. L., Wong S. L., Chen M., Tsai T., John J. C. S., Norman R. J., Febbraio M. A., Carroll J., Robker R. L., Mitochondrial dysfunction in oocytes of obese mothers: Transmission to offspring and reversal by pharmacological endoplasmic reticulum stress inhibitors. Development 142, 681–691 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Igosheva N., Abramov A. Y., Poston L., Eckert J. J., Fleming T. P., Duchen M. R., McConnell J., Maternal diet-induced obesity alters mitochondrial activity and redox status in mouse oocytes and zygotes. PLOS ONE 5, e10074 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferey J. L. A., Boudoures A. L., Reid M., Drury A., Scheaffer S., Modi Z., Kovacs A., Pietka T., DeBosch B., Thompson M. D., Diwan A., Moley K. H., A maternal high-fat, high-sucrose diet induces transgenerational cardiac mitochondrial dysfunction independently of maternal mitochondrial inheritance. Am. J. Physiol. Heart Circ. Physiol. 316, H1202–H1210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saben J. L., Boudoures A. L., Asghar Z., Thompson A., Drury A., Zhang W., Chi M., Cusumano A., Scheaffer S., Moley K. H., Maternal metabolic syndrome programs mitochondrial dysfunction via germline changes across three generations. Cell Rep. 16, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Al-Zubaidi U., Adhikari D., Cinar O., Zhang Q.-H., Yuen W. S., Murphy M. P., Rombauts L., Robker R. L., Carroll J., Mitochondria-targeted therapeutics, MitoQ and BGP-15, reverse aging-associated meiotic spindle defects in mouse and human oocytes. Hum. Reprod. 36, 771–784 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pasquariello R., Ermisch A. F., Silva E., McCormick S., Logsdon D., Barfield J. P., Schoolcraft W. B., Krisher R. L., Alterations in oocyte mitochondrial number and function are related to spindle defects and occur with maternal aging in mice and humans. Biol. Reprod. 100, 971–981 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Ge Z.-J., Schatten H., Zhang C.-L., Sun Q.-Y., Oocyte ageing and epigenetics. Reproduction 149, R103–R114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McGrath J., Solter D., Mann J., Pronuclear transplantation in the mouse embryo. Cold Spring Harbor Protoc. 2017, pdb.prot094417 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Hodges C. A., Hunt P. A., Simultaneous analysis of chromosomes and chromosome-associated proteins in mammalian oocytes and embryos. Chromosoma 111, 165–169 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Reddy P., Liu L., Adhikari D., Jagarlamudi K., Rajareddy S., Shen Y., du C., Tang W., Hämäläinen T., Peng S. L., Lan Z. J., Cooney A. J., Huhtaniemi I., Liu K., Oocyte-specific deletion of Pten causes premature activation of the primordial follicle pool. Science 319, 611–613 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCarthy D. J., Campbell K. R., Lun A. T., Wills Q. F., Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N. J., Livak K. J., Mikkelsen T. S., Rinn J. L., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M., Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., Cox J., The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J., g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.H. Wickham, ggplot2: Elegant Graphics for Data Analysis (Use R, Springer, 2009).
- 80.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Tables S1, S4 and S5
Tables S2 and S3