Abstract
Infection is one of the major causes of mortality in patients with systemic lupus erythematosus (SLE). We previously found that CD38, an ectoenzyme that regulates the production of NAD+, is up-regulated in CD8+ T cells of SLE patients and correlates with the risk of infection. Here, we report that CD38 reduces CD8+ T cell function by negatively affecting mitochondrial fitness through the inhibition of multiple steps of mitophagy, a process that is critical for mitochondria quality control. Using a murine lupus model, we found that administration of a CD38 inhibitor in a CD8+ T cell–targeted manner reinvigorated their effector function, reversed the defects in autophagy and mitochondria, and improved viral clearance. We conclude that CD38 represents a target to mitigate infection rates in people with SLE.
CD38 suppresses mitophagy and mitochondrial fitness in lupus CD8 T cells, and inhibition of its activity enhances cytotoxicity.
INTRODUCTION
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by multiorgan damage (1). Infection represents one of the major causes of mortality in SLE patients (2), and the use of immunosuppressive drugs further augments the risk (3). Functional defects of cytotoxic T cells in SLE patients explain the dampened antiviral responses (4) and contribute to uncontrolled Epstein-Barr virus infection (5, 6). Defects in lupus T cell cytotoxicity are often recorded in SLE patients and contribute directly to disease by failing to remove autoreactive B cell clones (7). This causal relation is demonstrated by accelerated disease progression in perforin gene knockout lupus-prone mice (8). The impaired cytolytic T cell response appears to be limited to SLE patients as it is absent in healthy subjects and people with other autoimmune diseases (9). Gene expression differences between SLE and non-SLE T cells were noted in a transcriptomic analysis (10). Several key molecules linked to their inherent SLE T cell cytotoxicity defects have been reported, including reduced expression of signaling through signaling lymphocytic activation molecules family member 7 (4) and increased expression of the ectonucleotidase CD38 (11).
Mitochondria are indispensable to CD8+ T cell function and have been linked to their activation, fate, and metabolic reprogramming. Mitochondrial translation selectively regulates the expression of a subset of proteins, including those involved in the cytotoxic T lymphocyte killer response, such as cytokines, granzyme B, and perforin (12). Accumulation of depolarized mitochondria as the result of decreased mitophagy results in a phenotype of terminally exhausted T cells in tumor-infiltrating lymphocyte, and supplementation with the nicotinamide adenine dinucleotide (NAD+) precursor nicotinamide riboside reinvigorates the effector response of CD8+ T cells by improving mitochondrial fitness (13). Increased mitochondrial reactive oxygen intermediate production from electron transport chain activity at complex I and transmembrane hyperpolarization have been reported in SLE T cells (14, 15). These features are often early events in the ensuing breakdown of mitochondrial transmembrane potential (16), which signifies substantial mitochondrial damage and predispose to cell death (17). These data argue strongly that mitochondria are dysfunctional in SLE T cells. However, the mechanism responsible for the reduced mitochondrial fitness and whether this could represent a therapeutic target to improve cytotoxic T cell response against infection is largely unexplored.
Here, we present evidence on how CD38 regulates mitochondrial fitness by suppressing multiple steps of mitophagy, which directly links to CD8+ T cell dysfunction (schematic abstract; fig. S1). T cell–targeted inhibition of CD38 by 78c, or the novel CD38 inhibitor MK-0159, not only reverses mitochondrial dysfunction and reinvigorates CD8+ T cell but also substantially improves the pathology linked to viral infection.
RESULTS
CD38hiCD8+ T cells harbor functionally and morphologically defective mitochondria
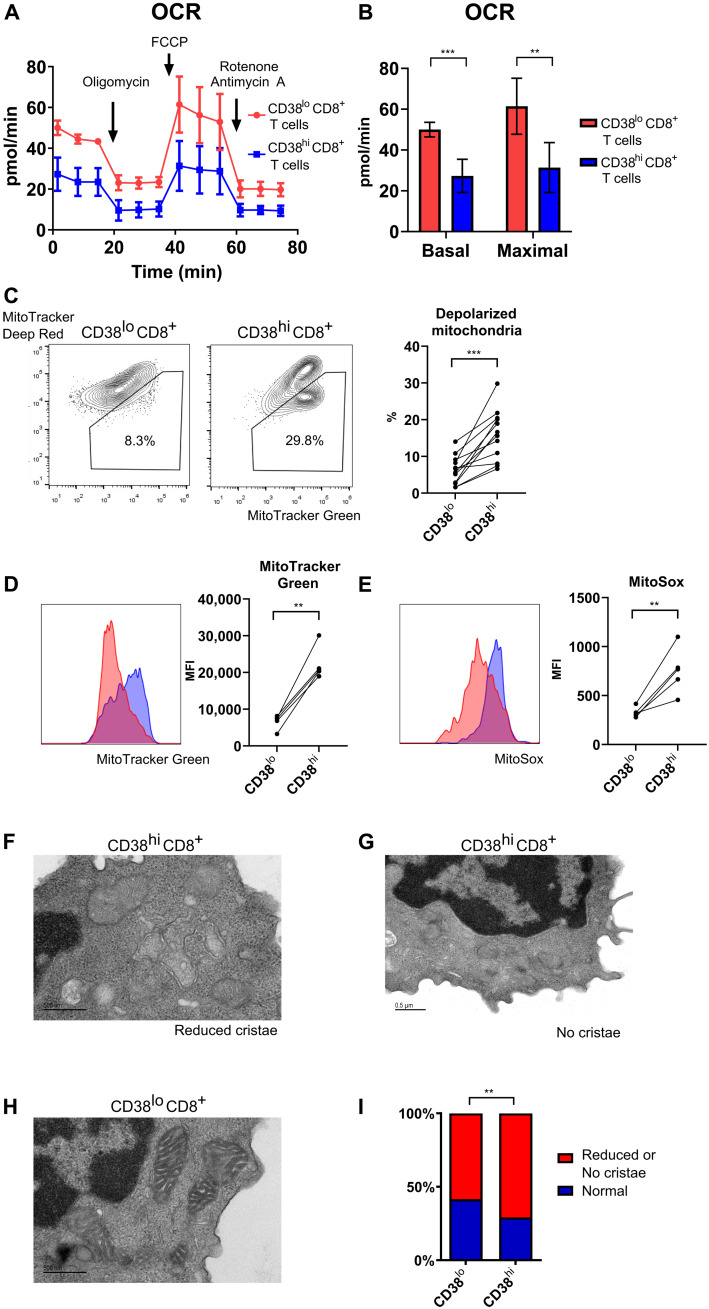
We previously demonstrated that CD38hiCD8+ T cell (gating strategy as shown in fig. S2), a population with reduced cytotoxic function and closed chromatin accessibility to relevant genes due to reduction in Sirtuin 1 (SIRT1) activity as the result of NAD+ depletion, was expanded in SLE patients and correlated strongly with the risk of infection (11). To evaluate the function of mitochondria of CD38hiCD8+ T cell, we performed Seahorse metabolic assay experiments in sorted cells. Due to restricted numbers of cells available from SLE patients because of lymphopenia and limits in total amount of peripheral blood from patient samples, metabolic assay experiments were performed only in leukocytes collected from apheresis leukoreduction collars from healthy donors. We recorded a significant reduction in both basal and maximal oxidative mitochondrial oxidative phosphorylation in the CD38hiCD8+ T cell population (Fig. 1, A and B). Similar reduction in mitochondrial respiration is also found in CD38hiCD4+ T cells (fig. S3, A and B). To further examine the mitochondria of these two CD8+ T cell populations, we used two fluorescent dyes, MitoTracker Green, indicative of total mitochondrial mass, and MitoTracker Deep Red, indicative of intact mitochondrial membrane potential. Flow cytometry analysis of CD8+ T cells from SLE patients showed a significant increase in cells with accumulation of depolarized mitochondria (Fig. 1C and fig. S2), signs of damaged mitochondria with increase in mitochondrial mass, and loss of mitochondrial transmembrane potential (MitoTracker Greenhi MitoTracker Deep Redlo). CD4+ T cells from SLE patients with low or high expression of CD38 displayed a small, comparable between the two groups, population of cells with depolarized mitochondria (fig. S3C). The difference between CD8+ and CD4+ T cells likely resulted from higher autophagy levels in SLE CD4+ T cells (18), which probably limit the accumulation of defective mitochondria. We therefore focused all the following analysis of CD38 phenotype on CD8+ T cells. We found increased total mitochondrial mass (Fig. 1D) and significant increase in mitochondrial reactive oxygen species (MitoSOX positivity) in lupus CD38hiCD8+ T cells (Fig. 1E). Electron microscopy of CD38hiCD8+ T cells from SLE patients revealed structural damage of mitochondria in this population. Specifically, a significant larger percentage of mitochondria exhibited partial or complete loss of cristae in CD38hiCD8+ compared to CD38loCD8+ T cells (Fig. 1, F to I). These data confirmed significant mitochondrial abnormalities in CD38hiCD8+ T cells, with evidence of functional defects and structural damage.
Fig. 1. CD38hiCD8+ T cells present with numerous mitochondrial defects.
(A) Oxygen consumption rate (OCR) of CD38hiCD8+ and CD38loCD8+ T cells sorted from healthy donor at baseline and after addition of oligomycin, carbonyl cyanide p-trifluoromethoxyphenylhydrazone, and rotenone/antimycin A. (B) Basal and maximal OCR of CD38hiCD8+ and CD38loCD8+ T cells sorted from healthy donor in (A). (C) Representative flow cytometry plot of MitoTracker Green and MitoTracker Deep Red staining in CD38hiCD8+ and CD38loCD8+ T cells from the peripheral blood of SLE patients. Percentage of depolarized mitochondria in CD38hiCD8+ and CD38loCD8+ T cells. (D and E) Representative flow cytometry plot of MitoTracker Green (D) and MitoSOX (E) staining and the mean fluorescence intensity (MFI) in CD38hiCD8+ and CD38loCD8+ T cells from lupus patient peripheral blood. (F to I) Representative electron microscopy of CD38hi with reduced number of cristae (F), no cristae (G), and CD38loCD8+ T cells (H) sorted from lupus patient peripheral blood. Percentage of mitochondria observed with partial or complete loss of cristae in CD38hiCD8+ and CD38loCD8+ T cells (I). Data are means ± SD; statistical analysis by two-tailed t test (B and F), paired t test (C to E), and chi-square test (I). **P < 0.01, ***P < 0.001.
CD38 perturbs mitochondrial turnover by inhibiting the PINK1-Parkin–dependent macroautophagy
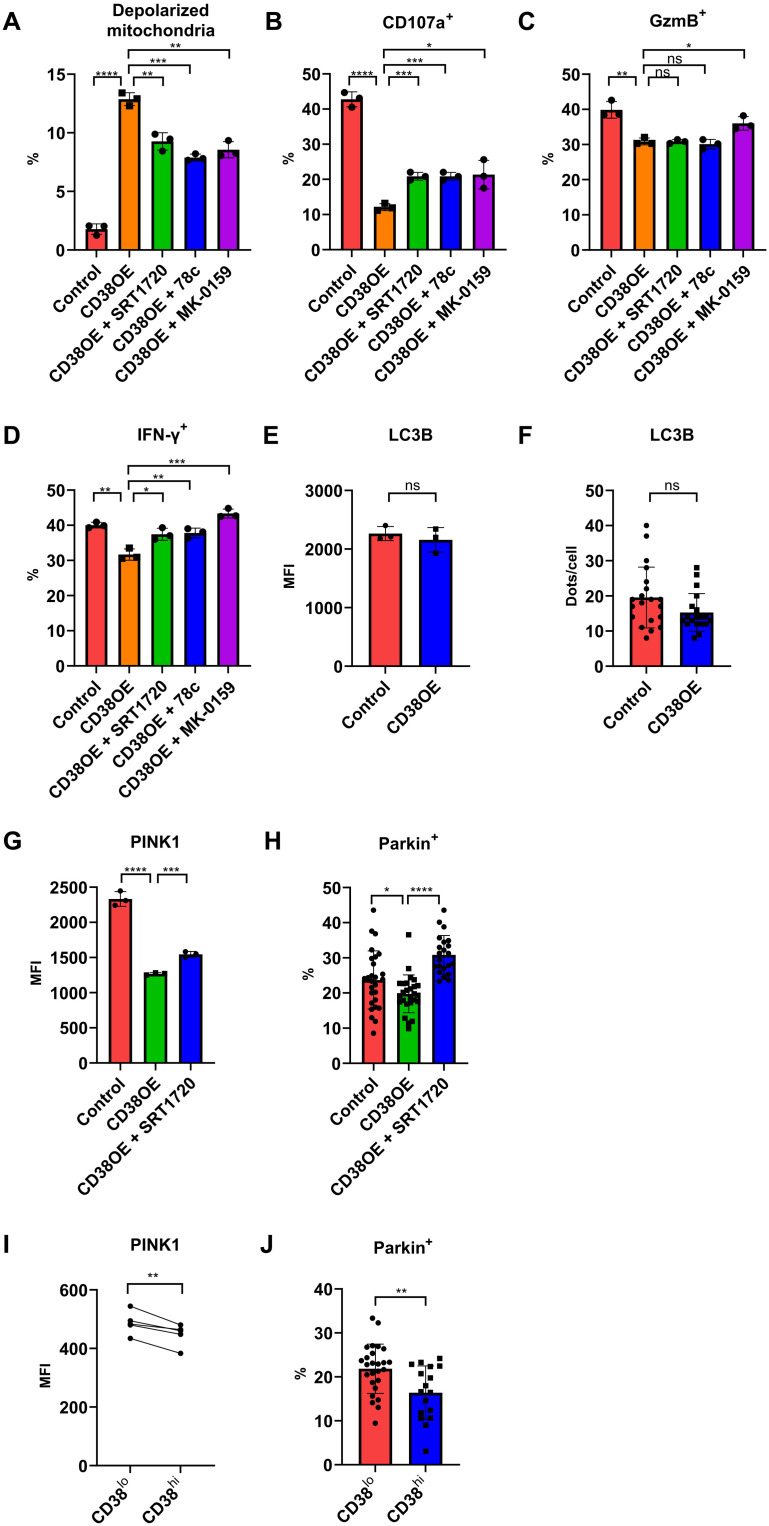
To determine whether the abovementioned mitochondrial defects were linked to CD38, we isolated CD38−CD8+ T cells from healthy donors and transfected them with either a CD38 overexpression or a control vector (fig. S4). In CD8+ T cells transfected with control vector, anti-CD3 and anti-CD28 stimulation increased the expression of CD38, which is known as a T cell activation marker linked to the T cell receptor/CD3 complex (19), but most of these T cells remained CD38lo. In contrast, CD8+ T cells transfected with CD38 overexpression vector had higher CD38 expression, which made this in vitro system suitable for the analysis of CD38hi versus CD38lo phenotype seen in SLE CD8+ T cells. After gating on the transfected cells expressing fluorescent protein, we found that overexpression of CD38 significantly increased cells with depolarized mitochondria, while addition of the SIRT1 (a NAD-dependent deacetylase) activator SRT1720 reversed this effect (Fig. 2A and fig. S5). We also tested the effects of two structurally distinct CD38 inhibitors, 78c and MK-0159, and observed a reduction in the number of cells with damaged mitochondria (Fig. 2A and fig. S5). Along with the changes in depolarized mitochondria, CD38 overexpression also led to reduction in degranulation and granzyme B and interferon-γ (IFN-γ) production by CD8+ T cells (Fig. 2, B to D). Addition of SIRT1 activator or either one of the CD38 inhibitors reversed the effect of CD38 overexpression in degranulation and IFN-γ production, while the reversal of granzyme B was seen only when MK-0159 was used (Fig. 2, B to D). These data suggest that the CD38-SIRT1 pathway directly leads to mitochondrial defects seen in SLE patients and has a major impact on the expression of functional defects in CD38hiCD8+ T cells.
Fig. 2. CD38 perturbs mitochondrial turnover by inhibiting PINK1-Parkin–dependent macroautophagy.
(A) Percentage of depolarized mitochondria in control, CD38-overexpressing CD8+ T cells, or CD38-overexpressing cells treated with SRT1720, or one of the CD38 inhibitors (78c or MK-0159). (B to D) Percentage of positive cells expressing CD107a (B), granzyme B (C), and IFN-γ (D) in control, CD38-overexpressing CD8+ T cells, or CD38-overexpressing cells treated with SRT1720, or one of the CD38 inhibitors (78c or MK-0159). (E) MFI of LC3B staining in control or CD38-overexpressed CD8+ T cells. (F) Counts of LC3B-positive dots per cell of control or CD38-overexpressed CD8+ T cells. (G) MFI of PINK1 in control, CD38-overexpressing CD8+ T cells, or CD38-overexpressing cells treated with SRT1720. (H) Percentage of Parkin positive of the TOMM20-positive area in control, CD38-overexpressing CD8+ T cells, or CD38-overexpressing cells treated with SRT1720. (I) MFI of PINK1 in CD38hiCD8+ and CD38loCD8+ T cells from lupus patient peripheral blood. (J) Percentage of Parkin positive of the TOMM20-positive area in CD38hiCD8+ and CD38loCD8+ T cells from lupus patient peripheral blood. Data are means ± SD; statistical analysis by two-tailed t test (A to H and J), paired t test (I), ns = P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Mitophagy, a macroautophagy process that targets damaged mitochondria, is the key process of maintaining mitochondrial quality by removing dysfunctional mitochondria. Since CD38 directly leads to the accumulation of depolarized mitochondria in CD8+ T cells, we examined various steps of autophagy to determine its effect on deterring mitophagic flux. We first found that CD38 overexpression did not influence autophagosome formation, as determined by the expression of LC3B using flow cytometry and immunofluorescence (Fig. 2, E and F, and fig. S6A). We then checked the next step of macroautophagy, which involves PTEN-induced kinase 1 (PINK1)–dependent ubiquitin phosphorylation to recruit Parkin to the damaged mitochondria (20), a critical process in bringing damaged organelles to the phagosome (fig. S1). A previous study has shown that high NAD+ induces mitophagy by activating SIRT1 (21), which regulates the expression of PINK1 and Parkin (22). In line with those previous findings, we tested whether CD38-SIRT1 regulates macroautophagy through PINK1 expression. Consistent with our hypothesis, CD38 overexpression significantly reduced PINK1 expression, while addition of the SIRT1 activator SRT1720 increased PINK1 (Fig. 2G). As a result, there was significantly less Parkin colocalization with mitochondria in CD38-overexpressed cells, and the presence of SRT1720 significantly increased Parkin colocalization (Fig. 2H and fig. S6B). The effect of CD38 on PINK1 and Parkin was also seen in CD38hiCD8+ T cells from SLE patients (Fig. 2, I and J). Reduction of mitophagy in SLE T cells was also reported as the result of Rab4A-mediated endosomal depletion of Drp1 (23). However, the CD38-SIRT1 pathway did not affect the expression of Rab4 (fig. S6C), suggesting that these two pathways independently contribute to the mitophagy defects seen in SLE T cells. Together, the CD38-SIRT1 pathway regulates autophagic flux by affecting the recruitment of damaged mitochondria to the autophagosome through the PINK1-Parkin pathway.
CD38 regulates lysosomal acidification through epigenetic control of V-ATPase expression
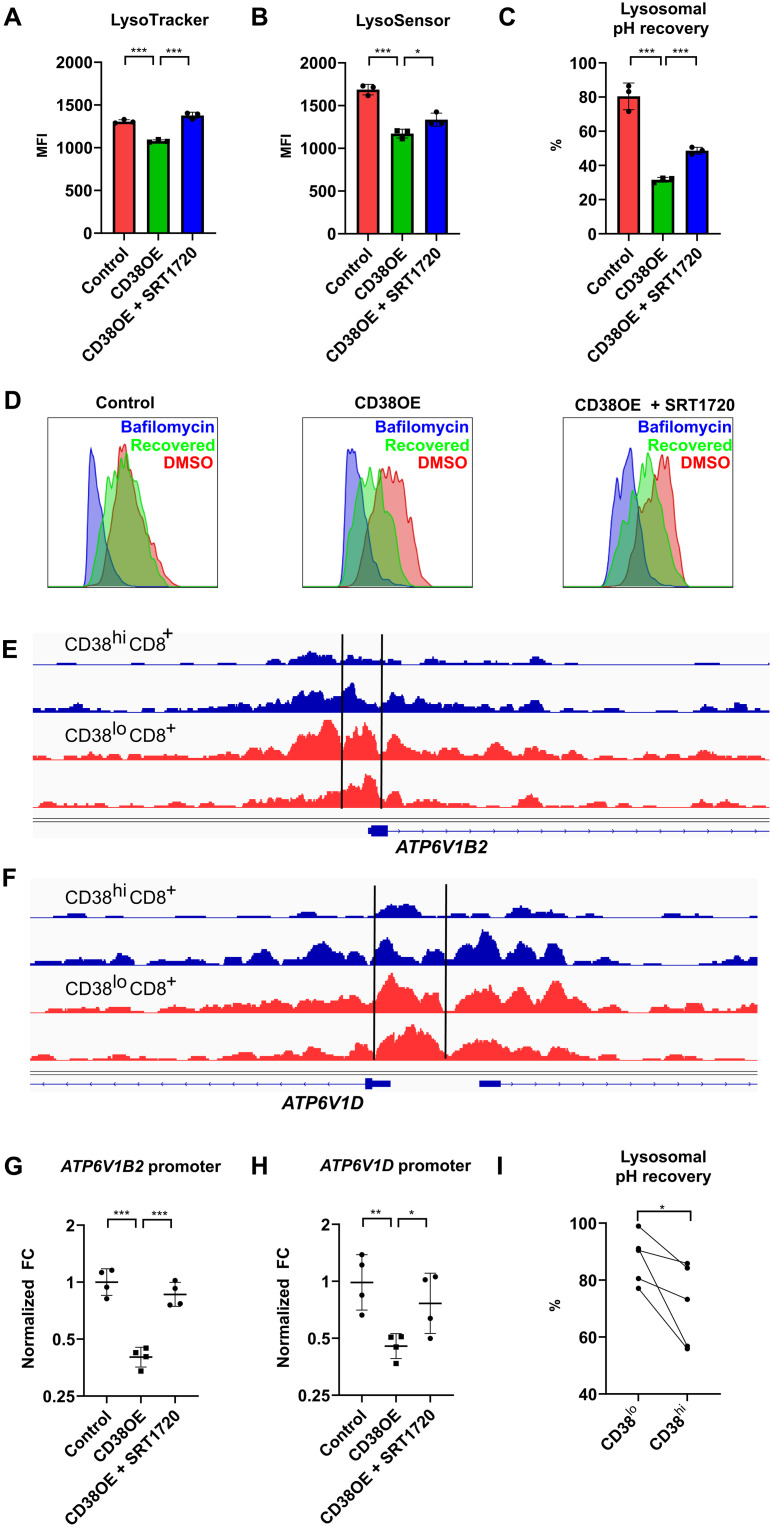
The final step of autophagic flux involves fusion of autophagosome with the lysosome, which leads to degradation of the damaged organelle. This process depends on lysosomal acidification regulated by the vacuolar-type adenosine triphosphatase (V-ATPase), and depletion of V-ATPase subunits perturbs the autophagic flux (24). We first noticed that CD38 overexpression significantly reduced the mean fluorescence intensity (MFI) of LysoTracker, a fluorescent dye that stained intracellular acidic organelles, mostly lysosomes (Fig. 3A). We further confirmed that CD38-overexpressing CD8+ T cells displayed less acidic lysosomes by using the pH-sensitive dye LysoSensor Green DND-189, while addition of the SIRT1 activator SRT1720 reversed lysosome acidification (Fig. 3B). To examine whether the difference in lysosomal pH was due to changes in V-ATPase expression, the proton pump expressed on lysosomal surface that is responsible for lysosomal acidification, a lysosomal pH recovery test was performed, which examined the percentage of cells stained with the LysoTracker after the removal of the reversible V-ATPase inhibitor bafilomycin. Compared to control cells, which almost fully recovered 1 hour after bafilomycin removal, CD38-overexpressing cells had significantly slower lysosomal pH recovery (Fig. 3, C and D), while the defects seen in CD38 overexpression can be partially reversed by adding a SIRT1 activator. These data suggested either a functional defect or numerical reduction in the expression of V-ATPase related to CD38. We had previously demonstrated that CD38 suppressed the expression of numerous genes in SLE CD8+ T cell by reducing the deacetylation of the histone-lysine N-methyltransferase EZH2 by SIRT1 and causing more chromatin methylation (fig. S1) (11). After reanalysis of our published ATAC-seq (assay for transposase accessible chromatin with high-throughput sequencing) data, we found that there were significant reductions in chromatin opening in the promoter regions of two V-ATPase subunit genes ATP6V1D and ATP6V1B2 in CD38hiCD8+ T cells from SLE patients (Fig. 3, E and F). By performing ATAC–quantitative polymerase chain reaction (qPCR) using primers that identify these ATAC-seq peaks, we confirmed that CD38 reduced chromatin opening of the promoter regions of these two V-ATPase subunit genes, and addition of a SIRT1 activator increased the chromatin accessibility (Fig. 3, G and H). These data suggest that CD38 reduces V-ATPase expression epigenetically and negatively affects lysosomal acidification in CD38hiCD8+ T cells from SLE patients (Fig. 3I), which is critical for the turnover of mitochondria.
Fig. 3. CD38 regulates lysosomal acidification through epigenetic control of V-ATPase expression.
(A and B) MFI of LysoTracker Deep Red (A) and LysoSensor Green (B) staining in control, CD38-overexpressing CD8+ T cells with and without SRT1720. (C) Percentage of lysosomal pH recovery after removing the reversible V-ATPase inhibitor bafilomycin. (D) Representative flow cytometry plot of LysoTracker Deep Red staining in lysosomal pH recovery test of control, CD38-overexpressing CD8+ T cells with and without SRT1720 in DMSO control condition (red), complete V-ATPase inhibition condition with bafilomycin (blue), and removal of bafilomycin for 1 hour to allow recovery of lysosomal pH (green). (E and F) Sequencing tracks of ATAC-seq data over the promoter region of ATP6V1B2 (E) and ATP6V1D (F) of two replicates of CD38hiCD8+ T cells (blue) and CD38loCD8+ T cells (red) from lupus patient peripheral blood. (G and H) Result of ATAC-qPCR showing normalized fold change (FC) of the two peaks located at the promoter region of ATP6V1B2 (G) and ATP6V1D (H) in control, CD38-overexpressing CD8+ T cells with and without SRT1720. (I) Percentage of lysosomal pH recovery after removing the reversible V-ATPase inhibitor bafilomycin in CD38hiCD8+ and CD38loCD8+ T cells from lupus patient peripheral blood. Data are means ± SD; statistical analysis by two-tailed t test (A to C and G and H), paired t test (I), *P < 0.05, **P < 0.01, ***P < 0.001.
Dysfunctional CD8+ T cells in BXD2 lupus–prone mice accentuate viral hepatitis after infection with LCMV
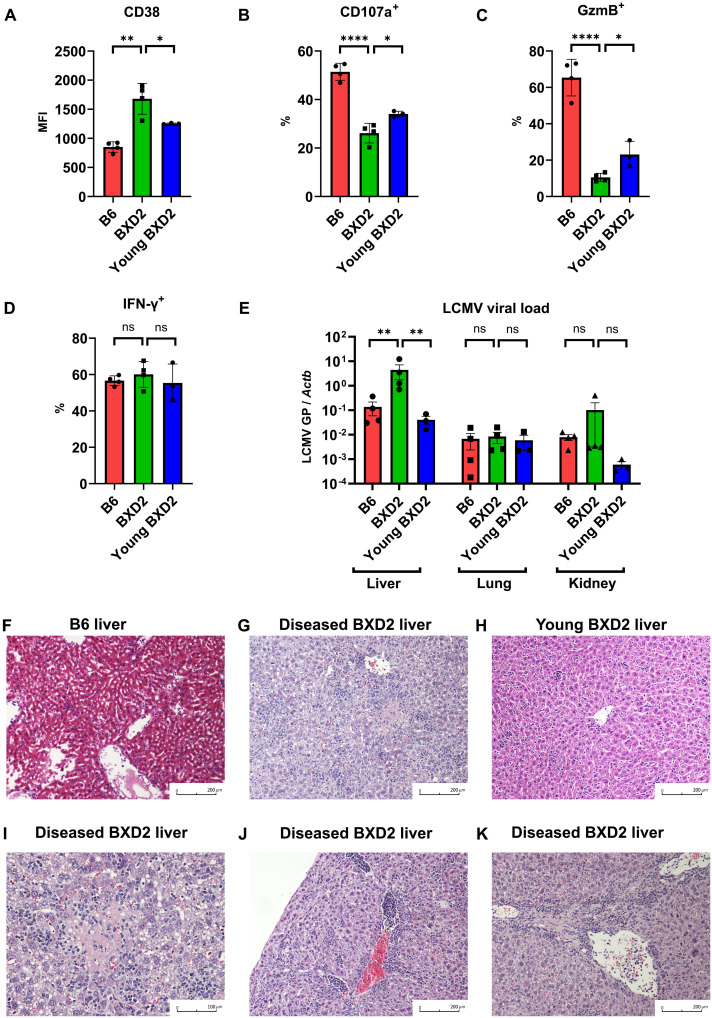
After confirming the role of CD38 in limiting the function of CD8+ T cells and mitochondrial fitness in vitro, we sought to address whether CD38 compromises their function in vivo and whether it affects the clearance of viruses. We used BXD2 lupus–prone mice, which develop spontaneously numerous autoimmune disease manifestations, including high titers of autoantibodies, splenomegaly, spontaneous arthritis, and glomerulonephritis (25). BXD2 mice have the H-2Db haplotype, similar to one of its progenitors, C57BL/6J, a feature that makes the construction of H-2Db gp33 tetramers suitable for the identification of lymphocytic choriomeningitis virus (LCMV)–specific CD8+ T cells. We infected diseased, 6- to 7-month-old BXD2 and nondiseased, young, 2-month-old BXD2, along with age-matched 6-month-old C57BL/6J with the LCMV Armstrong virus, and sacrificed them 8 days later. Despite having higher numbers of total splenocytes due to increased spleen size for both young and old BXD2 mice (fig. S7, A and B), there were no difference in the total number of LCMV-specific gp33+ CD8+ T cells (fig. S7C). Detailed analysis on these gp33+ CD8+ T cells showed that LCMV-specific CD8+ T cells from the diseased BXD2 mice had the highest expression of CD38 as compared to those from age-matched control C57BL/6J and young BXD2 mice (Fig. 4A and fig. S7D). Although we did not observe an obvious separation of the CD8+ T cell population in BXD2 lupus mice based on CD38 expression, as what we had seen in human samples (figs. S2 and S7D), CD38 expression was significantly higher in BXD2 as compared with control C57BL/6J mice, and it increased as disease progressed (Fig. 4A and fig. S7D). Consistent with what we found in human SLE CD8+ T cells, the increase in CD38 expression in gp33+ CD8+ T cells from BXD2 mice coincided with a marked reduction in degranulating capacity and granzyme B production, but this reduction in CD8+ T cell function was significantly less in nondiseased young BXD2 mice (Fig. 4, B and C, and fig. S7, E and F). IFN-γ production, however, was comparable among all mice (Fig. 4D). The reduction in CD8+ T cell function correlated significantly with an increase in the LCMV viral load in the liver but not in other organs including kidneys and lungs (Fig. 4E). This increase in LCMV viral load in the liver of diseased BXD2 mice corresponded to the presence of numerous inflammatory foci, which are barely seen in either age-matched B6 mice or young, nondiseased BXD2 mice (Fig. 4, F to H). Detailed analysis of the pathological features in infected BXD2 mice revealed hepatocyte necrosis (Fig. 4I) and portal and periportal inflammation (Fig. 4, J and K). However, there were no obvious differences in inflammation or tissue damage in the kidneys and lungs (fig. S7, G to J). These extensive features of hepatic inflammation and hepatocyte necrosis were consistent with changes in effector function of antigen-specific CD8+ T cells, and pathology in diseased BXD2 mice was consistent with the lack of viral clearance in the liver. Meanwhile, these features of extensive liver inflammation were not seen in age-matched noninfected BXD2 mice (fig. S7K), suggesting that the nature of this inflammation was viral hepatitis rather than autoimmune hepatitis associated with lupus disease activity (26). These findings of restricted LCMV Armstrong–induced tissue inflammation in the liver but not in other organs were consistent with those reported in an immunosuppressed murine LCMV infection model (27). Increased susceptibility of developing overt tissue damage in the liver can also be attributed to the fact that higher mitochondrial oxidative stress is observed in the liver of lupus-prone mice (28). Diseased BXD2 mice had higher baseline mitochondrial reactive oxygen species as compared with B6 mice, and this difference was exacerbated after LCMV infection (fig. S7L), which likely aggravated the degree of tissue damage. Together, we found up-regulation of CD38 and reduction in CD8+ T cell function in diseased BXD2 lupus–prone mice, but not in age-matched control B6 or young nondiseased BXD2 mice. The drastic reduction in degranulation and granzyme production correlated with up-regulation of CD38, similar to what was seen in SLE patients, and the change in T cell function was consistent with the presentation of viral hepatitis.
Fig. 4. Dysfunctional CD8+ T cells in BXD2 lupus mice cause viral hepatitis in LCMV infection.
(A) MFI of CD38 in gp33+ CD8+ T cells of B6, diseased BXD2, or young nondiseased BXD2 mice 8 days after LCMV Armstrong infection. (B to D) Percentage of positive cells expressing CD107a (B), granzyme B (C), and IFN-γ (D) in gp33+ CD8+ T cells of B6, diseased BXD2, or young nondiseased BXD2 mice 8 days after LCMV Armstrong infection. (E) LCMV viral load 8 days after LCMV Armstrong infection using quantitative PCR of glycoprotein gene normalized by tissue ACTB in liver, kidney, and lung of B6, diseased BXD2, or young nondiseased BXD2 mice. (F to H) Representative liver histology of B6 (F), diseased BXD2 (G), or young nondiseased BXD2 (H) mice 8 days after LCMV Armstrong infection. (I to K) Representative liver histology of diseased BXD2 mice 8 days after LCMV Armstrong infection showing focal lesion of necrosis (I) and portal and periportal inflammation (J and K). Data are means ± SD (A to D), mean ± SE (E); statistical analysis by two-tailed t test. ns = P > 0.05, *P < 0.05, **P < 0.01, ****P < 0.0001.
CD38 inhibition reverses mitochondrial defect and restores CD8+ T cell function in BXD2 lupus–prone mice against LCMV infection
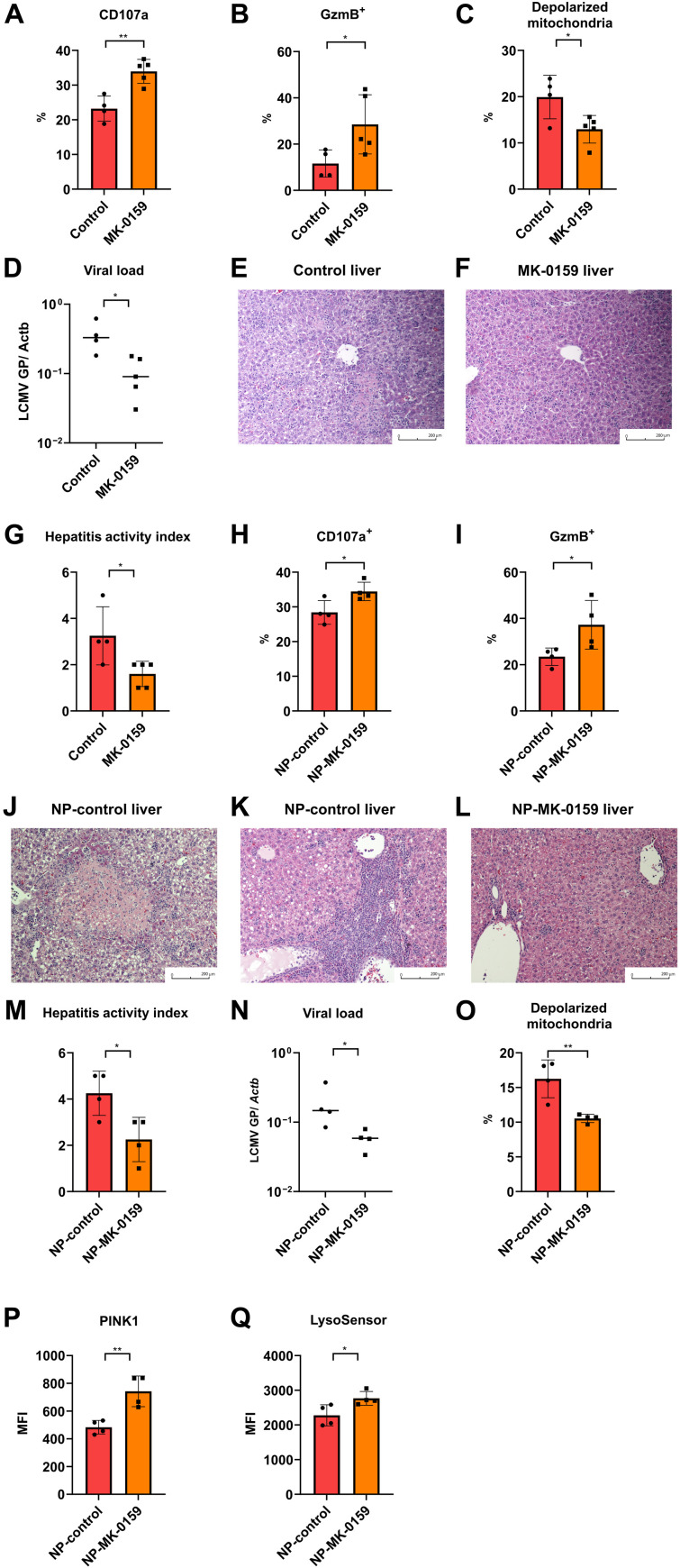
To test whether CD38 inhibition represents an option to improve infection rates and outcome in SLE, we treated diseased BXD2 mice with the CD38 inhibitor MK-0159 or distilled water via oral gavage. Systemic CD38 inhibition did not have any effect on spleen size, total numbers of splenocytes, splenic plasma cells (fig. S8, A to D), or the expansion of gp33+ CD8+ T cells (fig. S8, E and F). Eight days after LCMV infection, we found that the effector function of antigen-specific CD8+ T cells was significantly reversed (Fig. 5, A and B, and fig. S8, G and H), depolarized mitochondria were reduced (Fig. 5C), and the liver LCMV viral load was reduced (Fig. 5D). The reinvigoration of antigen-specific gp33+ CD8+ T cells along with the reduction in liver LCMV viral load was consistent with the marked improvement of pathological features of viral hepatitis in the MK-0159–treated group. Liver specimens from infected and MK-0159–treated mice showed significantly reduced inflammation and lacked hepatocyte necrosis (Fig. 5, E to G). This experiment demonstrated that systemic inhibition of CD38 can reinvigorate CD8+ T cell cytotoxicity and limit virally induced organ inflammation.
Fig. 5. CD38 inhibition reverses mitochondrial defect and restores CD8+ T cell function in BXD2 lupus mice against LCMV infection.
(A and B) Percentage of positive cells expressing CD107a (A) and granzyme B (B) in gp33+ CD8+ T cells 8 days after LCMV Armstrong infection from diseased BXD2 treated with control water or MK-0159. (C) Percentage of depolarized mitochondria in gp33+ CD8+ T cells 8 days after LCMV Armstrong infection from diseased BXD2 treated with control water or MK-0159. (D) LCMV viral load 8 days after LCMV Armstrong infection using qPCR of glycoprotein gene normalized by tissue ACTB in diseased BXD2 treated with control water or MK-0159. (E to G) Representative liver histology 8 days after LCMV Armstrong infection from diseased BXD2 treated with control water (E) or MK-0159 (F) and their respective hepatitis activity index (G). (H and I) Percentage of positive cells expressing CD107a (H) and granzyme B (I) in gp33+ CD8+ T cells 8 days after LCMV Armstrong infection from diseased BXD2 treated with control NP or NP-MK-0159. (J to M) Representative liver histology 8 days after LCMV Armstrong infection from diseased BXD2 treated with NP-control with the presentation of necrosis (J) or portal and periportal inflammation (K) or treated with NP-MK-0159 (L) and their respective hepatitis activity index (M). (N) LCMV viral load 8 days after LCMV Armstrong infection using qPCR of glycoprotein gene normalized by tissue ACTB in diseased BXD2 treated with NP-control or NP-MK-0159. (O) Percentage of depolarized mitochondria in gp33+ CD8+ T cells 8 days after LCMV Armstrong infection from diseased BXD2 treated with NP-control or NP-MK-0159. (P and Q) MFI of PINK1 (P) and LysoSensor Green (Q) in gp33+ CD8+ T cells 8 days after LCMV Armstrong infection from diseased BXD2 treated with NP-control or NP-MK-0159. Data are means ± SD; statistical analysis by two-tailed t test. *P < 0.05, **P < 0.01.
Because CD38 is expressed by multiple cells, its systemic inhibition would alter the function of other cells and cause side effects. To deliver a CD38 inhibitor to CD8+ T cells in a targeted manner, we encapsulated MK-0159 in biodegradable polymeric nanoparticles (NPs) tagged with the CD8a antibody, clone YTS 105, which is a nondepleting antibody and does not occupy the major histocompatibility complex (MHC) class I/CD8αβ complex to affect CD8+ T cell function (29). NP delivery of MK-0159 significantly increased degranulation and granzyme B production in the antigen-specific CD8+ T cells (Fig. 5, H and I, and fig. S8, I to J). The improved cytotoxic function of antigen-specific CD8+ T cells was reflected in improved liver histology features (Fig. 5, J to M) and reduced viral load (Fig. 5N). Compared to the control-treated group, mice treated with CD8a antibody–tagged and MK-0159–loaded NPs had a significant reduction in inflammation. Meanwhile, targeted drug delivery to CD8+ T cell significantly improved mitochondrial quality by reducing depolarized mitochondria (Fig. 5O and fig. S8K), increased expression of PINK1(Fig. 5P), and improved lysosomal acidifications (Fig. 5Q). Together, these data support the role of systemic and cell-targeted inhibition of CD38 in reversing mitochondrial, autophagic, and lysosomal defects in CD8+ T cells of lupus-prone mice, and reinvigorated their effector functions to combat infection.
DISCUSSION
Here, we have demonstrated that the NAD+-modulating ectoenzyme CD38 regulates mitochondrial fitness in SLE CD8+ T cells and negatively affects their function and ability to combat infection. By reducing cellular NAD+ levels, CD38 suppresses the activity of sirtuins and limits the recruitment of damaged mitochondria to the phagophore, which subsequently is encapsulated into the autophagosome through the PINK1-Parkin pathway. CD38 also diminishes lysosomal acidification by reducing the expression of V-ATPase epigenetically. These events result in the accumulation of damaged depolarized mitochondria in CD38hiCD8+ T cells and reduce their ability to clear viruses effectively. Using lupus-prone mice infected with LCMV, we confirmed the role of CD38 in compromising the cytotoxic function of CD8+ T cells. At the translational level, we demonstrated the therapeutic potential of a novel small-molecule CD38 inhibitor administered systemically or, more importantly, in a targeted manner to CD8+ T cells, in restoring their cytotoxic function and suppressing viral hepatitis.
Numerous mitochondrial defects, including transmembrane hyperpolarization, have been reported in SLE T cells (14). However, the impact of these mitochondrial abnormalities on SLE T cell function and the consequences on infection risk are largely unexplored. Mitochondria are involved in various aspects of CD8+ T cell function, including the regulation of translation of key effector function proteins (12) and the control of the fate of CD8+ T cell differentiation (30). Accumulation of depolarized mitochondria has been described as one of the causes of functional defect of tumor-infiltrating T cells, leading to impaired ability to kill tumor cells (13). We found a similar CD8+ T cell dysfunction caused by mitochondrial defects in SLE CD8+ T cells. The altered mitochondrial dynamics of tumor-infiltrating T cells can be reversed by administering the NAD+ precursor nicotinamide riboside, which acts synergistically with anti–PD-1 treatment in controlling tumor growth (13). CD38 has also been reported to cause age-related NAD+ decline and cellular senescence, which further leads to mitochondrial dysfunction (31). Since CD38 inhibition prevents degradation of NAD+, the effect of MK-0159 on SLE CD8+ T cells is consistent with that of NAD+ supplementation on tumor-infiltrating T cells. Moreover, we have dissected mechanistically the role of CD38 on autophagy and found that CD38 suppresses mitophagy by inhibiting the recruitment of damaged mitochondria to the phagophore through the SIRT1-PINK1-Parkin pathway. This pathway has no effect on Rab4a-mediated endosomal depletion of Drp1 (23) and presents a separate pathway in reducing mitophagy. Reduction of mitophagy contributes to the accumulation of oxidative stress generating mitochondria in these T cells (Fig. 1E), which are sensitive to mammalian target of rapamycin (mTOR) blockade treatment (32, 33). The CD38-SIRT1 pathway also suppresses lysosomal acidification via epigenetic control of CD38 expression, previously unknown in CD8+ T cells.
One issue that remains to be determined is the cause of up-regulation of CD38 in lupus T cells. The inflammatory milieu in SLE patients is most likely the cause because the expression of CD38 increases gradually in lupus-prone mice as disease progresses. Among numerous cytokines that are increased in SLE, type I IFN has been linked to multiple immune defects. Recent work by Buang et al. (34) has shown that SLE patients with increased type I IFN–stimulated genes have aberrant mitochondria accumulated in CD8+ T cells, and exposure of CD8+ T cells to IFN-α increases NAD+ consumption through the up-regulation of CD38. The mechanistic link between type 1 IFN and aberrant mitochondrial function complements our findings and helps explain the paradox of SLE patients experiencing a higher risk of infection despite high levels of type 1 IFN. Type 1 IFN, a component of the innate immunity response, combats viral infections by inducing numerous effector mechanisms that dampen virus replication and promote viral clearance (35). However, persistent exposure to type 1 IFN can have a negative impact on viral clearance. In a model of chronic LCMV infection, type 1 IFN signaling regulates immunosuppressive signals from dendritic cells, and blocking its signaling pathway improves viral clearance by promoting IFN-γ–expressing CD4+ T cells (36). Persistent up-regulation of type 1 IFN is prominent in SLE (37), and the higher susceptibility of these patients to infection suggests that chronic exposure to type 1 IFN has a negative effect on T cell function. The up-regulation of CD38 in lupus T cells could serve as an important mechanistic link as type 1 IFN up-regulates CD38, which directly leads to T cell dysfunction by perturbing mitochondrial fitness. Considering the potential bidirectional effects of type 1 IFN, both antiviral and immunosuppressive, it is challenging to apply this treatment to reverse this mitochondrial immune cell pathology to combat infection. A randomized control trial of sifalimumab, an anti–IFN-α monoclonal antibody, showed better SLE responder index response at the expense of increased Herpes zoster infection (38). Tumor necrosis factor–α (TNF-α), another cytokine long known to contribute to lupus pathogenesis (39), causes increased expression of CD38 (40). This TNF-α–regulated CD38 expression requires nuclear factor κB (NF-κB) activation (41) and is abrogated by corticosteroids (41). Since the use of corticosteroids increases significantly the risk of infections in SLE patients (39), their use to suppress CD38 to combat infection is not justified. Therefore, our proposed treatment of target CD38, which is directly responsible to the mitochondrial defects, could be an appealing approach for curbing infection risk in SLE patients. One limitation of administering CD38 inhibitors to SLE patients, like any novel drug that is first in class, is the lack of knowledge of its potential long-term side effects. However, initial reports on the use of the anti-CD38 monoclonal antibody daratumumab showed promising effect in depleting CD38-expressing plasma cells, restoring cytotoxic function of T cells, without obvious adverse events up to 1 year after treatment (40).
Together, our findings demonstrate the mechanistic role of CD38 in perturbing mitochondrial fitness, which further leads to limited ability of CD8+ T cells to fight viral infections. By depleting NAD+, CD38 suppresses the activity of SIRT1, which leads to a reduction in PINK1-Parkin–regulated macroautophagy and V-ATPase–dependent lysosomal acidification. Since blockade of type I IFN limits both inflammation and antiviral immunity in patients with SLE, our studies launch the use of CD8+ T cell–targeted delivery of a small drug inhibitor of CD38 to fend off viral infections.
MATERIALS AND METHODS
Study design
We performed mitochondrial analysis of peripheral blood CD8+ T cells from patients with SLE to determine mitochondrial abnormalities. Functional study of Seahorse metabolic flux CD38hiCD8+ versus CD38loCD8+ T cells was performed in sorted cells isolated from healthy donors due to limitation in available number of cells. Mitochondrial structural defect was confirmed by electron microscopy in CD38hiCD8+ T cells sorted from lupus patients. To determine the effect of CD38 on autophagic flux and lysosomal acidification, CD38−CD8+ T cells were isolated from healthy donor and transfected with control or CD38 overexpression vector. Either CD38 inhibitors (78c or MK-0159) or SIRT1 activator (SRT1720) was added to test whether the effect of CD38 overexpression on autophagic flux and lysosomal acidifications could be reversed to determine the pathway in regulation. After confirming the role of CD38 seen in CD38hiCD8+ T cells from lupus patients using the in vitro culture system, the role of CD38 in vivo was tested in murine lupus mice BXD2 with LCMV Armstrong infection. We used pharmacological blockade of CD38, either given systemically or targeted specifically to CD8+ T cells through NP delivery, and concluded that CD38 inhibition could reverse mitochondrial defects, reinvigorate CD8+ T cell function, and enhance viral clearance.
Mice
Mice were housed in the pathogen-free Animal Resources Facility at Beth Israel Deaconess Medical Center. Animal handling and experimental protocols were approved by the Institutional Animal Care and Use Committee. C57BL/6 (B6) mice and BXD2 mice were purchased from the Jackson Laboratory.
CD38 inhibitors
CD38 inhibitors, 78c and MK-0159, were both synthesized and provided by Mitobridge Inc. (patent WO 2021/087087 A1) (41, 42). Purities of 99.1 and 99.8% for 78c and MK-0159, respectively, were confirmed by high-performance liquid chromatography (HPLC).
LCMV infection and MK-0159 treatment
C57BL/6 (B6) mice and BXD2 mice of the indicated age were housed in the Biosafety Level 2 (BSL2) Animal facility at Beth Israel Deaconess Medical Center and infected with LCMV Armstrong (courtesy of N. Joshi and J. Craft, Yale University) at the dose of 2 × 105 plaque-forming units intraperitoneally. For mice treated with the CD38 inhibitor, MK-0159 provided by Mitobridge was given to diseased BXD2 mice at the dose of 30 mg/kg via oral gavage twice a day for 9 days (days −1 to 7 of LCMV infection). For NP delivery, MK-0159 was encapsulated in poly-lactic acid-co-glycolic acid–based NP and intravenously injected on days 0, 3, and 6 of LCMV infection (a total of three doses) at the dose of 0.6 mg/kg for MK-0159 encapsulated in NP at the dose of 14.6 mg/kg for each injection. The concentration of encapsulated MK-0159 was confirmed by HPLC.
Preparation of NPs
The poly(d,l-lactic-co-glycolic) acid (PLGA)–based copolymers were purchased from PolySciTech Akina Inc. Methoxy poly(ethylene glycol)-b-PLGA copolymer [mPEG-PLGA; molecular weight, 5000:30,000 Da, 50:50 lactic acid:glycolic acid (LA:GA) (w/w)] and PLGA-b-poly(ethylene glycol)-maleimide [PLGA-PEG-MAL; molecular weight, ~30,000 of PLGA and 5000 Da of PEG, 50:50 LA:GA (w/w)] functioned as the core polymers of NP. NPs were prepared by our previous nanoprecipitation protocol with minor modifications (43). PEG-PLGA and MAL-PEG-PLGA were dissolved in acetone. MK-0159 (Mitobridge Inc., USA) was added to the polymer mixture. As a control, we prepared only polymer mixture without MK-0159. Mixtures prepared as organic phase were added dropwise to 0.015% aqueous solution of polyvinyl alcohol (Sigma-Aldrich) under vigorous stirring to formulate water-soluble NP-control or NP–MK-0159. Then, NP suspensions were stirred for 2 hours to evaporate acetone and harden NPs. We spin down NP solution by ultracentrifuge (L-80 Optima, Beckman) at 21,000 rpm for 30 min. Resulting pallets of NP were resuspended with Dulbecco’s phosphate-buffered saline (DPBS; Mediatech Inc., Manassas, VA). Next, we conjugated anti-CD8a (MA1-81180, Invitrogen) on the surface of NPs via thiol-MAL chemistry. Tris(2-carboxyethyl) phosphine hydrochloride (TCEP; 0.5 M; Sigma-Aldrich) was used to chemically reduce disulfide bonds of antibody. Anti-CD8a (100 μg) was mixed with 100 μl of TCEP and incubated for 15 min at room temperature. The reduced anti-CD8a was mixed with the NP suspension overnight at 4°C, and we purified NP solution again to remove nonreacted anti-CD8. Final samples, NP-control and NP-MK-0159, were stored at 4°C before use.
Flow cytometry
For flow cytometry, CD8+ T cells from human peripheral blood were stained with the following antibodies or reagents, listed with clones, vendor, and catalog numbers: anti-human CD4 (clone RPA-T8, 300521), anti-human CD8a (clone SK1, BioLegend, 344718), anti-human CD8a (clone HIT8a, BioLegend, 300908), anti-human CD38 (clone HIT2, BioLegend, 303516 and 303510), allophycocyanin (APC) mouse immunoglobulin G1 (IgG1), κ isotype control (BD, 555751), anti–green fluorescent protein (GFP) antibody (Rockland, 600-102-215), anti–IFN-γ (clone 4S.B3, BioLegend, 502509), anti–granzyme B (clone GB11, BD, 560212), anti-CD107a (clone H4A, BioLegend, 328608), PINK1 antibody (Rockland, 600-401-GU5), anti-Rab4 antibody (EPR3043, Abcam, ab109009), goat anti-rabbit IgG secondary antibody, and APC (Invitrogen, A-10931). Mouse CD8+ T cell analysis was stained with the following antibodies: anti-mouse CD8a (clone 53-6.7, BioLegend, 344718), anti-mouse CD8b (clone Ly3, BioLegend, 126629), anti-mouse CD38 (clone 90, BioLegend, 102718), anti–interferon regulatory factor 4 (IRF4) (clone IRF4.3E4, BioLegend, 646408), anti-mouse CD138 (clone 281-2, BioLegend, 142508), anti–IFN-γ (clone XMG1.2, BioLegend, 505808), anti–granzyme B (clone GB11, BD, 560212), and anti-CD107a (clone 1D4B, BioLegend, 121608). H-2Db LCMV gp33 (C9M) tetramer-KAVYNFATM was provided by the National Institutes of Health Tetramer Core Facility. Other reagents used for staining include MitoTracker Green (Thermo Fisher Scientific, M7514), MitoTracker Deep Red (Thermo Fisher Scientific, M22426), MitoSOX Red (Thermo Fisher Scientific, M36008), LysoSensor Green DND-189 (Thermo Fisher Scientific, L7535), and LysoTracker Deep Red (Thermo Fisher Scientific, L12492). The data were acquired by BD FACSAria and Beckman Coulter CytoFLEX at Flow Cytometry Core Facility, Beth Israel Deaconess Medical Center, and analyzed by FlowJo 10.0.7.
Immunofluorescence and confocal microscopy
For confocal microscopy imaging, transfected cells and lupus patient peripheral blood mononuclear cells were stained with the indicated antibodies, including LC3B (Invitrogen, PA5-30598), TOMM20 (EPR15581-39, Abcam, ab205487), and Parkin (clone PRK8, Santa Cruz Biotechnology, sc-32282), and sorted by BD FACSAria. Cells were resuspended in PBS and allowed to be adhered to cover slide coated with Corning Cell-Tak Cell and Tissue Adhesive (Fisher Scientific, CB-40240). Cover slides were then mounted with ProLong Gold Antifade Mountant with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific, P36931). Confocal microscopy was done with a Zeiss LSM 880 upright confocal microscope at Confocal Imaging and Immunohistochemistry Core Facility, Beth Israel Deaconess Medical Center.
In vitro T cell culture and transfection
CD8+ T cells were isolated from apheresis leukoreduction collars using RosetteSep Human CD8+ T Cell Enrichment Cocktail (STEMCELL Technologies, 15063). CD38+CD8+ T cells were removed by using EasySep Human Biotin Positive Selection Kit II (STEMCELL Technologies, 17663) and Biotin anti-CD38 monoclonal antibody (clone HIT2, BioLegend, 303518). The remaining CD38−CD8+ T cells were cultured overnight in six-well plates coated with plate-bound anti-CD3 (1 μg/ml; clone OKT3, BioLegend, 317326) and anti-CD28 (2 μg/ml; clone CD28.2, BioLegend, 302934). After overnight stimulation, activated CD8+ T cells were transfected with control vector or CD38 overexpression vector with C-GFPSpark tag (Sino Biological, HG10818-ACG) by using the Human T Cell Nucleofector Kit (Lonza, VPA-1002). For MitoTracker and immunofluorescence staining, control OFPSpark Vector (Sino Biological, CV025) and CD38 overexpression vector with OFPSpark (Sino Biological, HG10818-ACR) were used because of the consideration of fluorescence colors. Transfected cells were rested for 6 hours and then cultured overnight with control dimethyl sulfoxide (DMSO) (0.01%), 0.5 μM SRT1720, 20 μM 78c, or 20 μM MK-0159 as indicated in 48-well plates coated with plate-bound anti-CD3 and anti-CD28.
Transmission electron microscopy
CD38hiCD8+ and CD38loCD8+ T cells were freshly isolated from human SLE patients’ peripheral blood using FACSAria for transmission electron microscopy experiments. The sorted cells were fixed in 2.5% glutaraldehyde, 2% paraformaldehyde, in 0.1 M cacodylate buffer (pH 7.4) (modified Karnovsky’s fixative) at room temperature for at least 1 hour at room temperature, followed by 4°C overnight. Cells were washed in 0.1 M cacodylate buffer (pH 7.4) the following day and then postfixed in 1% osmium tetroxide in 0.1 M cacodylate buffer (pH 7.4) for 1 hour at 4°C. After washing the fixed cells with distilled water, cells were then incubated overnight in 2% aqueous uranyl acetate at 4°C. The samples were then dehydrated in ethanol and propylene oxide and embedded in SeaKem LE Agarose. Agarose blocks of each sample were sectioned using a Leica Ultracut E ultramicrotome. Formvar- and carbon-coated slot grids were used to collect ultracut sections, and these sections were contrast-stained with 2% uranyl acetate and lead citrate. Transmission electron microscopy pictures were taking using a JEOL 1400 TEM (JEOL Inc.) equipped with an Orius SC1000 digital charge-coupled device camera (Gatan Inc.).
Lysosomal pH recovery test
Lysosomal pH recovery test was carried out as described in the published paper (44), with the only modification to collect fluorescent signals on flow cytometer. Control or CD38-overexpressed cells with or without SRT1720 treatment were cultured in medium with either DMSO or 100 nM bafilomycin A. For the recovery group, cells were treated with 100 nM bafilomycin A for 1 hour before washing three times with culture medium and rested for another 1 hour to allow recovery. All three groups of cells (DMSO, bafilomycin, and recovery groups) were then stained with LysoTracker Deep Red for 1 hour, and the recovery percentage was calculated on the basis of changes in MFI (MFIrecovered − MFIBafilomycin/MFIDMSO − MFIBafilomycin).
Analysis for ATAC-seq
To assay chromatin accessibility, we reanalyzed our previously published ATAC-seq dataset on CD38hiCD8+ and CD38loCD8+ T cells from lupus patients (GSE141197) (11). Low-quality and mitochondrial DNA sequences were first filtered, and the remaining high-quality sequences were mapped to human genome (GRCh37/hg19) using Bowtie2 (45). Low mapping quality, mismatched sequences, and duplicates were further excluded. Peak calling was done by MACS2 (version 2.1.1) (46), and the aligned reads were normalized by CPM (counts per million). To compare differential chromatin accessibility between CD38hiCD8+ and CD38loCD8+ T cells, edgeR was used to calculate statistically significant differences in binding regions (47). Sequencing tracks of ATAC-seq data were visualized by Integrative Genomics Viewer (Broad Institute).
ATAC-qPCR
CD8+ T cells (50,000) transfected with either control vector or CD38 overexpression vector were sorted using BD FACSAria, and the samples were prepared by following the Omni-ATAC protocol (48). Primers for quantifying the ATAC-seq peak region of interest were designed on the basis of the published protocol (49) with the use of the same cycling conditions: 98°C for 2 min, followed by 40 cycles of 98°C for 10 s, 60°C for 20 s, and 72°C for 30 s. The SYBR Green dye signals were detected using Roche LightCycler 480. Primers were listed in table S1, and the reads were normalized by the universal normalization primers of KIF26B.
LCMV quantification
To quantify LCMV viral load in the tissue with a higher sensitivity, we performed LCMV quantification PCR based on the two published protocols (50, 51). Before harvesting organs from each mouse, surgical equipment was cleaned with 70% alcohol to avoid contamination. The harvested organs were immediately preserved in RNAlater solutions (Thermo Fisher Scientific, AM7024) and stored on ice. Tissue RNA was extracted using the RNeasy Plus Mini Kit (Qiagen, 74136), and complementary DNA (cDNA) was generated by RNA to cDNA EcoDry Premix (Takara, 639549). qPCR was performed with the primers indicated in table S2.
Histology
Mouse lung, kidney, and liver samples were immediately fixed in 10% formalin after sacrificing the mice. The fixed tissues were embedded with paraffin by the histology core at Beth Israel Deaconess Medical Center, and the tissue sections were stained with hematoxylin and eosin. The severity of hepatitis was quantified on the basis of the modified hepatitis activity index (52).
Seahorse extracellular flux assay
For seahorse extracellular flux assay, CD38hi and CD38lo population from either CD8+ or CD4+ T cells was sorted from healthy donor apheresis leukoreduction collars and cultured overnight in 96-well plates coated with plate-bound anti-CD3 (1 μg/ml; clone OKT3, BioLegend, 317326) and anti-CD28 (2 μg/ml; clone CD28.2, BioLegend, 302934). The cultured cells were washed and resuspended in Seahorse XF RPMI 1640 medium (Agilent, 103336-100), and Seahorse analysis experiments were performed per the manufacturer’s protocol.
Human samples
Peripheral blood from lupus patients was collected at Rheumatology Outpatient Clinic, Beth Israel Deaconess Medical Center after informed consent, and the patient sample collection protocol was approved by Institutional Review Board, Beth Israel Deaconess Medical Center. Leukocytes from healthy donors were collected from apheresis leukoreduction collars from Blood Donor Center, Boston Children Hospital.
Hepatocyte isolation
Mouse hepatocyte isolation protocol was adapted from a published method (28). In short, the mice were sedated and euthanized after isoflurane inhalation, followed by dissection of abdomen. Liver perfusion was performed with 25 ml of buffer A [Hanks’ balanced salt solution (HBSS) with 0.5 EGTA], followed by buffer B [HBSS with 1 mM CaCl2, 15 mM Hepes (Gibco, 15630080), and 100 U of collagenase D (Roche, 11088866001)]. The isolated suspension was filtered through a 70-μm filter and washed with culture medium.
Statistical analysis
Statistical analysis was performed by GraphPad Prism version 9. Bar graphs in all figures indicate the mean, and the error bars represent the SD. To compare data between two groups, statistical analysis was performed by two-tailed t test. All analyses were conducted at the significance level of 0.05. Statistical results were labeled in each figure as ns = P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Acknowledgments
We thank S. Krishfield and C. Ioannidis for collecting patient samples, and K. Smith from Electron Microscopy (EM) Core, Beth Israel Deaconess Medical Center for EM sample preparation and imaging.
Funding: This research was supported by NIH grant R01AI148161.
Author contributions: Conceptualization: P.-M.C., E.K., S.A., and G.C.T.; methodology: P.-M.C., E.K., A.S., H.L., J.R., S.J., R.A., S.A., and J.D.B.; investigation: P.-M.C. and E.K.; writing: P.-M.C. and G.C.T.; review and editing: M.G.T. and G.C.T.; funding acquisition: G.C.T.; and supervision: G.C.T.
Competing interests: Mitobridge Inc. filed a patent application for the CD38 inhibitor MK-0159 related to this work (WO 2021/087087 A1, filed on 29 October 2020, published on 06 May 2021), but none of the authors were listed as inventors of this patent. The authors declare no other competing interests.
Data and materials availability: MK-0159 can be provided by Mitobridge Inc. pending scientific review and a completed material transfer agreement. Requests for MK-0159 should be directly submitted to Mitobridge Inc. Phone: 617-401-9100. Email: info@mitobridge.com. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 and S2
REFERENCES AND NOTES
- 1.Tsokos G. C., Systemic lupus erythematosus. N. Engl. J. Med. 365, 2110–2121 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Bultink I. E. M., de Vries F., van Vollenhoven R. F., Lalmohamed A., Mortality, causes of death and influence of medication use in patients with systemic lupus erythematosus vs matched controls. Rheumatology (Oxford) 60, 207–216 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feldman C. H., Hiraki L. T., Winkelmayer W. C., Marty F. M., Franklin J. M., Kim S. C., Costenbader K. H., Serious infections among adult Medicaid beneficiaries with systemic lupus erythematosus and lupus nephritis. Arthritis Rheumatol. 67, 1577–1585 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comte D., Karampetsou M. P., Yoshida N., Kis-Toth K., Kyttaris V. C., Tsokos G. C., Signaling lymphocytic activation molecule family member 7 engagement restores defective effector CD8+ T cell function in systemic lupus erythematosus. Arthritis Rheumatol. 69, 1035–1044 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang I., Quan T., Nolasco H., Park S. H., Hong M. S., Crouch J., Pamer E. G., Howe J. G., Craft J., Defective control of latent Epstein-Barr virus infection in systemic lupus erythematosus. J. Immunol. 172, 1287–1294 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Larsen M., Sauce D., Deback C., Arnaud L., Mathian A., Miyara M., Boutolleau D., Parizot C., Dorgham K., Papagno L., Appay V., Amoura Z., Gorochov G., Exhausted cytotoxic control of Epstein-Barr virus in human lupus. PLOS Pathog. 7, e1002328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P. M., Tsokos G. C., The role of CD8+ T-cell systemic lupus erythematosus pathogenesis: An update. Curr. Opin. Rheumatol. 33, 586–591 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng S. L., Moslehi J., Robert M. E., Craft J., Perforin protects against autoimmunity in lupus-prone mice. J. Immunol. 160, 652–660 (1998). [PubMed] [Google Scholar]
- 9.Stohl W., Impaired polyclonal T cell cytolytic activity. A possible risk factor for systemic lupus erythematosus. Arthritis Rheum. 38, 506–516 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Bradley S. J., Suarez-Fueyo A., Moss D. R., Kyttaris V. C., Tsokos G. C., t cell transcriptomes describe patient subtypes in systemic lupus erythematosus. PLOS ONE 10, e0141171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katsuyama E., Suarez-Fueyo A., Bradley S. J., Mizui M., Marin A. V., Mulki L., Krishfield S., Malavasi F., Yoon J., Sui S. J. H., Kyttaris V. C., Tsokos G. C., The CD38/NAD/SIRTUIN1/EZH2 axis mitigates cytotoxic CD8 T cell function and identifies patients with SLE prone to infections. Cell Rep. 30, 112–123.e114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lisci M., Barton P. R., Randzavola L. O., Ma C. Y., Marchingo J. M., Cantrell D. A., Paupe V., Prudent J., Stinchcombe J. C., Griffiths G. M., Mitochondrial translation is required for sustained killing by cytotoxic T cells. Science 374, eabe9977 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Yu Y. R., Imrichova H., Wang H., Chao T., Xiao Z., Gao M., Rincon-Restrepo M., Franco F., Genolet R., Cheng W. C., Jandus C., Coukos G., Jiang Y. F., Locasale J. W., Zippelius A., Liu P. S., Tang L., Bock C., Vannini N., Ho P. C., Disturbed mitochondrial dynamics in CD8+ TILs reinforce T cell exhaustion. Nat. Immunol. 21, 1540–1551 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Gergely P. Jr., Grossman C., Niland B., Puskas F., Neupane H., Allam F., Banki K., Phillips P. E., Perl A., Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum. 46, 175–190 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doherty E., Oaks Z., Perl A., Increased mitochondrial electron transport chain activity at complex I is regulated by N-acetylcysteine in lymphocytes of patients with systemic lupus erythematosus. Antioxid. Redox Signal. 21, 56–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banki K., Hutter E., Gonchoroff N. J., Perl A., Elevation of mitochondrial transmembrane potential and reactive oxygen intermediate levels are early events and occur independently from activation of caspases in Fas signaling. J. Immunol. 162, 1466–1479 (1999). [PMC free article] [PubMed] [Google Scholar]
- 17.Susin S. A., Zamzami N., Castedo M., Daugas E., Wang H. G., Geley S., Fassy F., Reed J. C., Kroemer G., The central executioner of apoptosis: Multiple connections between protease activation and mitochondria in Fas/APO-1/CD95- and ceramide-induced apoptosis. J. Exp. Med. 186, 25–37 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alessandri C., Barbati C., Vacirca D., Piscopo P., Confaloni A., Sanchez M., Maselli A., Colasanti T., Conti F., Truglia S., Perl A., Valesini G., Malorni W., Ortona E., Pierdominici M., T lymphocytes from patients with systemic lupus erythematosus are resistant to induction of autophagy. FASEB J. 26, 4722–4732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morra M., Zubiaur M., Terhorst C., Sancho J., Malavasi F., CD38 is functionally dependent on the TCR/CD3 complex in human T cells. FASEB J. 12, 581–592 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., Endo T., Fon E. A., Trempe J. F., Saeki Y., Tanaka K., Matsuda N., Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Jang S. Y., Kang H. T., Hwang E. S., Nicotinamide-induced mitophagy: Event mediated by high NAD+/NADH ratio and SIRT1 protein activation. J. Biol. Chem. 287, 19304–19314 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao Z. Q., Zhang X., Zhen Y., He X. Y., Zhao S., Li X. F., Yang B., Gao F., Guo F. Y., Fu L., Liu X. Z., Duan C. Z., A novel small-molecule activator of Sirtuin-1 induces autophagic cell death/mitophagy as a potential therapeutic strategy in glioblastoma. Cell Death Dis. 9, 767 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caza T. N., Fernandez D. R., Talaber G., Oaks Z., Haas M., Madaio M. P., Lai Z. W., Miklossy G., Singh R. R., Chudakov D. M., Malorni W., Middleton F., Banki K., Perl A., HRES-1/Rab4-mediated depletion of Drp1 impairs mitochondrial homeostasis and represents a target for treatment in SLE. Ann. Rheum. Dis. 73, 1888–1897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauvezin C., Nagy P., Juhász G., Neufeld T. P., Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 6, 7007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mountz J. D., Yang P., Wu Q., Zhou J., Tousson A., Fitzgerald A., Allen J., Wang X., Cartner S., Grizzle W. E., Yi N., Lu L., Williams R. W., Hsu H. C., Genetic segregation of spontaneous erosive arthritis and generalized autoimmune disease in the BXD2 recombinant inbred strain of mice. Scand. J. Immunol. 61, 128–138 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Liu Y., Yu J., Oaks Z., Marchena-Mendez I., Francis L., Bonilla E., Aleksiejuk P., Patel J., Banki K., Landas S. K., Perl A., Liver injury correlates with biomarkers of autoimmunity and disease activity and represents an organ system involvement in patients with systemic lupus erythematosus. Clin. Immunol. 160, 319–327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Araki K., Gangappa S., Dillehay D. L., Rouse B. T., Larsen C. P., Ahmed R., Pathogenic virus-specific T cells cause disease during treatment with the calcineurin inhibitor FK506: Implications for transplantation. J. Exp. Med. 207, 2355–2367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oaks Z., Winans T., Caza T., Fernandez D., Liu Y., Landas S. K., Banki K., Perl A., Mitochondrial dysfunction in the liver and antiphospholipid antibody production precede disease onset and respond to rapamycin in pupus-prone mice. Arthritis Rheumatol. 68, 2728–2739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shore D. A., Issafras H., Landais E., Teyton L., Wilson I. A., The crystal structure of CD8 in complex with YTS156.7.7 Fab and interaction with other CD8 antibodies define the binding mode of CD8 alphabeta to MHC class I. J. Mol. Biol. 384, 1190–1202 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Windt G. J., Everts B., Chang C. H., Curtis J. D., Freitas T. C., Amiel E., Pearce E. J., Pearce E. L., Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camacho-Pereira J., Tarragó M. G., Chini C. C. S., Nin V., Escande C., Warner G. M., Puranik A. S., Schoon R. A., Reid J. M., Galina A., Chini E. N., CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 23, 1127–1139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai Z. W., Hanczko R., Bonilla E., Caza T. N., Clair B., Bartos A., Miklossy G., Jimah J., Doherty E., Tily H., Francis L., Garcia R., Dawood M., Yu J., Ramos I., Coman I., Faraone S. V., Phillips P. E., Perl A., N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: A randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 64, 2937–2946 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai Z. W., Kelly R., Winans T., Marchena I., Shadakshari A., Yu J., Dawood M., Garcia R., Tily H., Francis L., Faraone S. V., Phillips P. E., Perl A., Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: A single-arm, open-label, phase 1/2 trial. Lancet 391, 1186–1196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buang N., Tapeng L., Gray V., Sardini A., Whilding C., Lightstone L., Cairns T. D., Pickering M. C., Behmoaras J., Ling G. S., Botto M., Type I interferons affect the metabolic fitness of CD8+ T cells from patients with systemic lupus erythematosus. Nat. Commun. 12, 1980 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwasaki A., Pillai P. S., Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14, 315–328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson E. B., Yamada D. H., Elsaesser H., Herskovitz J., Deng J., Cheng G., Aronow B. J., Karp C. L., Brooks D. G., Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340, 202–207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaussabel D., Quinn C., Shen J., Patel P., Glaser C., Baldwin N., Stichweh D., Blankenship D., Li L., Munagala I., Bennett L., Allantaz F., Mejias A., Ardura M., Kaizer E., Monnet L., Allman W., Randall H., Johnson D., Lanier A., Punaro M., Wittkowski K. M., White P., Fay J., Klintmalm G., Ramilo O., Palucka A. K., Banchereau J., Pascual V., A modular analysis framework for blood genomics studies: Application to systemic lupus erythematosus. Immunity 29, 150–164 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khamashta M., Merrill J. T., Werth V. P., Furie R., Kalunian K., Illei G. G., Drappa J., Wang L., Greth W.; CD1067 study investigators , Sifalimumab, an anti-interferon-α monoclonal antibody, in moderate to severe systemic lupus erythematosus: A randomised, double-blind, placebo-controlled study. Ann. Rheum. Dis. 75, 1909–1916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrinton L. J., Liu L., Goldfien R., Michaels M. A., Tran T. N., Risk of serious infection for patients with systemic lupus erythematosus starting glucocorticoids with or without antimalarials. J. Rheumatol. 43, 1503–1509 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Ostendorf L., Burns M., Durek P., Heinz G. A., Heinrich F., Garantziotis P., Enghard P., Richter U., Biesen R., Schneider U., Knebel F., Burmester G., Radbruch A., Mei H. E., Mashreghi M. F., Hiepe F., Alexander T., Targeting CD38 with daratumumab in refractory systemic lupus erythematosus. N. Engl. J. Med. 383, 1149–1155 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Haffner C. D., Becherer J. D., Boros E. E., Cadilla R., Carpenter T., Cowan D., Deaton D. N., Guo Y., Harrington W., Henke B. R., Jeune M. R., Kaldor I., Milliken N., Petrov K. G., Preugschat F., Schulte C., Shearer B. G., Shearer T., Smalley T. L. Jr., Stewart E. L., Stuart J. D., Ulrich J. C., Discovery, synthesis, and biological evaluation of thiazoloquin(az)olin(on)es as potent CD38 inhibitors. J. Med. Chem. 58, 3548–3571 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Tarragó M. G., Chini C. C. S., Kanamori K. S., Warner G. M., Caride A., de Oliveira G. C., Rud M., Samani A., Hein K. Z., Huang R., Jurk D., Cho D. S., Boslett J. J., Miller J. D., Zweier J. L., Passos J. F., Doles J. D., Becherer D. J., Chini E. N., A potent and specific CD38 inhibitor ameliorates age-related metabolic dysfunction by reversing tissue NAD+ decline. Cell Metab. 27, 1081–1095.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bahmani B., Uehara M., Ordikhani F., Li X., Jiang L., Banouni N., Ichimura T., Kasinath V., Eskandari S. K., Annabi N., Bromberg J. S., Shultz L. D., Greiner D. L., Abdi R., Ectopic high endothelial venules in pancreatic ductal adenocarcinoma: A unique site for targeted delivery. EBioMedicine 38, 79–88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Latifkar A., Ling L., Hingorani A., Johansen E., Clement A., Zhang X., Hartman J., Fischbach C., Lin H., Cerione R. A., Antonyak M. A., Loss of Sirtuin 1 alters the secretome of breast cancer cells by impairing lysosomal integrity. Dev. Cell 49, 393–408.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., Orloff L. A., Risca V. I., Kundaje A., Khavari P. A., Montine T. J., Greenleaf W. J., Chang H. Y., An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yost K. E., Carter A. C., Xu J., Litzenburger U., Chang H. Y., ATAC primer tool for targeted analysis of accessible chromatin. Nat. Methods 15, 304–305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCausland M. M., Crotty S., Quantitative PCR technique for detecting lymphocytic choriomeningitis virus in vivo. J. Virol. Methods 147, 167–176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welsh R. M., Seedhom M. O., Lymphocytic choriomeningitis virus (LCMV): Propagation, quantitation, and storage. Curr. Protoc. Microbiol. Chapter 15, Unit 15A.1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishak K., Baptista A., Bianchi L., Callea F., De Groote J., Gudat F., Denk H., Desmet V., Korb G., MacSween R. N., Phillips M. J., Portmann B. G., Poulsen H., Scheuer P. J., Schmid M., Thaler H., Histological grading and staging of chronic hepatitis. J. Hepatol. 22, 696–699 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 and S2