Abstract
Steroid nuclear receptor coactivator 2 (SRC2) is a member of a family of transcription coactivators. While SRC1 inhibits the differentiation of regulatory T cells (Tregs) critical for establishing immune tolerance, we show here that SRC2 stimulates Treg differentiation. SRC2 is dispensable for the development of thymic Tregs, whereas naive CD4+ T cells from mice deficient of SRC2 specific in Tregs (SRC2fl/fl/Foxp3YFP-Cre) display defective Treg differentiation. Furthermore, the aged SRC2fl/fl/Foxp3YFP-Cre mice spontaneously develop autoimmune phenotypes including enlarged spleen and lung inflammation infiltrated with IFNγ-producing CD4+ T cells. SRC2fl/fl/Foxp3YFP-Cre mice also develop severer experimental autoimmune encephalomyelitis (EAE) due to reduced Tregs. Mechanically, SRC2 recruited by NFAT1 binds to the promoter and activates the expression of Nr4a2, which then stimulates Foxp3 expression to promote Treg differentiation. Members of SRC family coactivators thus play distinct roles in Treg differentiation and are potential drug targets for controlling immune tolerance.
SRC2 controls the scale of immune responses via activating Nr4a2 gene to promote CD4+Foxp3+ iTreg differentiation.
INTRODUCTION
Regulatory T cells (Tregs) are essential to protect against autoimmune responses, maintain homeostasis, and damp immune responses after clearance of infection (1). However, Tregs are often found in tumor microenvironment to effectively prevent antitumor immunity (2). The essential physiological function of Tregs for induction and maintenance of peripheral tolerance is demonstrated by the uncontrollable autoimmunity in mice and human that lack functional Tregs due to a mutation in forkhead box P3 (Foxp3) gene (3–5). Foxp3 is a lineage-specific transcription factor that determines the generation, maintenance, and function of Tregs (6). Natural Tregs develop in the thymus mostly with T cell receptors (TCRs) recognizing self-antigens (7, 8), whereas induced Tregs (iTregs) are generated from activation of naive CD4+ T cells in the presence of transforming growth factor–β (TGFβ) (6, 9). Naive CD4+ T cells can also differentiate into inflammatory effector T cells including T helper 1 (TH1), TH2, and TH17 (10, 11), which are inhibited by Tregs. A fine balance between inflammatory T cells and Tregs is required for a functional immune system. Skewing to inflammatory T cells leads to autoimmunity, whereas development of tumor often associates with the dominance of Tregs. Thus, understanding the mechanisms that regulate the differentiation of naive T cells into inflammatory T cells and Tregs facilitates the development of previously unknown immunotherapies for controlling immune responses.
The steroid receptor coactivator (SRC) family consists of three members, SRC1 (or NCOA1), SRC2 (or NCOA2/TIF2/GRIP1), and SRC3 (or NCOA3/pCIP/ACTR/AIB1). Although SRCs do not directly bind to target DNA, they function as coactivators for steroid nuclear receptors and other transcription factors by interacting with them to stimulate gene transcription (12). Hence, SRCs are believed to orchestrate transcription programs critical for multiple cellular processes (12). However, the function of SRCs in immune system has long been ignored until recently. Our previous study illustrated that SRC1 can reciprocally regulate the differentiation of inflammatory TH17 cells and Tregs by promoting TH17, whereas it inhibits Treg differentiation (13). Similar to SRC1, our other research showed that SRC3 promotes TH17 differentiation (14), which was also demonstrated by an independent study that further indicated the selective role of SRC3 in the differentiation of pathogenic TH17 cells (15). Therefore, SRC1 and SRC3 have nonredundant function in stimulating TH17 differentiation. With regard to Tregs, a recent report using germline SRC3−/− mice and SRC3 inhibitor hinted at a possible function of SRC3 in Tregs (16). However, SRC3 was found dispensable for Treg differentiation using T cell–specific SRC3 knockout mice (15). In contrast to SRC1 and SRC3, the function of SRC2 in T cells remains unknown.
Using Treg-specific SRC2 knockout mice (SRC2fl/fl/Foxp3YFP-Cre) and T cell–specific SRC2 knockout mice (SRC2fl/fl/CD4Cre), we demonstrated the essential function of SRC2 in the maintenance of immune balance via regulating the generation of iTregs. Naive CD4+ T cells from SRC2fl/fl/Foxp3YFP-Cre and SRC2fl/fl/CD4Cre mice were defective in Treg differentiation in vitro and in vivo. Consistently, aged SRC2fl/fl/Foxp3YFP-Cre mice displayed enlarged spleens, weight loss, and damaged lung tissues that were infiltrated with lymphocytes producing inflammatory cytokines. In addition, SRC2fl/fl/Foxp3YFP-Cre mice developed more severe EAE associated with reduced Tregs and increased inflammatory CD4+ T cells. RNA sequencing (RNA-seq) analysis showed that after polarizing under Treg conditions, SRC2fl/fl/Foxp3YFP-Cre CD4+ cells had lower levels of Nr4a2, a transcription factor known to directly regulate Foxp3 expression, and forced expression of Nr4a2 rescued Treg differentiation in both SRC2fl/fl/Foxp3YFP-Cre and SRC2fl/fl/CD4Cre CD4+ cells. Mechanistically, SRC2 interacted with NFAT1, and both were recruited to the promoter region of Nr4a2. Furthermore, CRISPR-Cas9–mediated deletion of the DNA promoter region that binds SRC2 and NFAT1 reduced Nr4a2 expression and further impaired Treg differentiation. Therefore, SRC2 recruited by NFAT1 stimulates the expression of Nr4a2, which then promotes Treg differentiation via up-regulation of Foxp3. Together with our previously reported negative role of SRC1 in Treg differentiation, different members of SRC family, SRC1 and SRC2, have opposite functions in Treg differentiation.
RESULTS
SRC2 is not required for thymic Treg development but is essential for Treg differentiation from naive CD4+ T cells
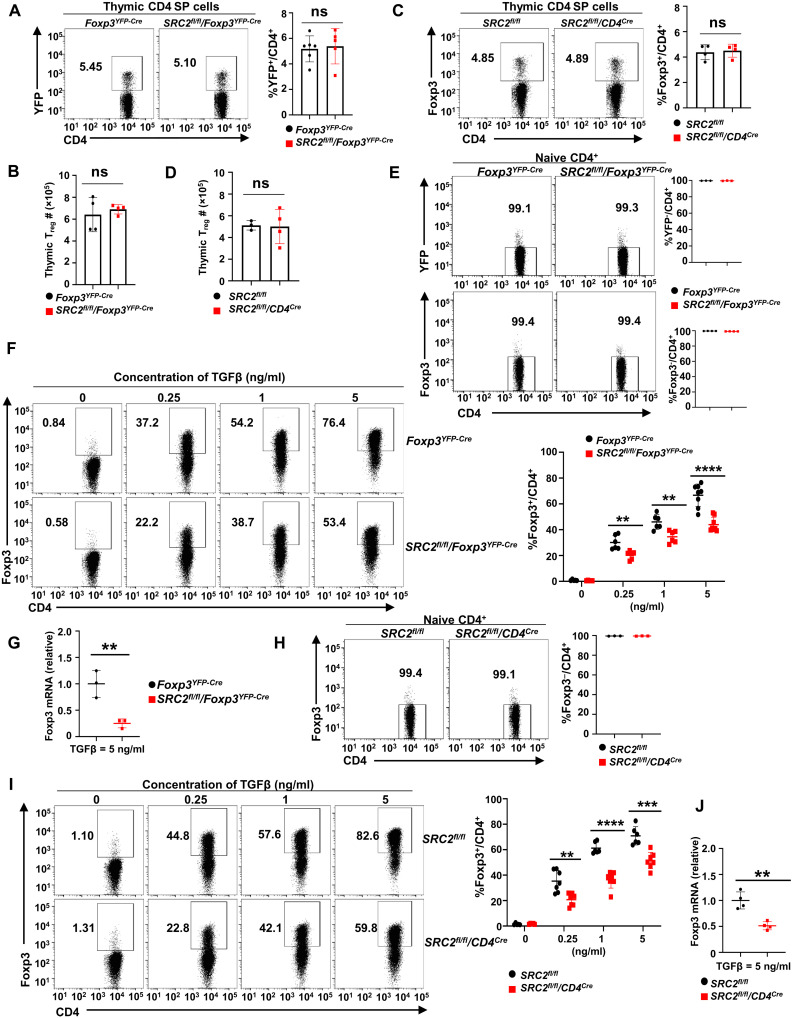
To determine the function of SRC2 in Tregs, we generated two strains of mice that deleted Ncoa2 (encoding SRC2) in Tregs (SRC2fl/fl/Foxp3YFP-Cre) or T cells (SRC2fl/fl/CD4Cre), respectively. Ncoa2 gene deletion in Tregs of SRC2fl/fl/Foxp3YFP-Cre mice (fig. S1A) and CD4+ cells of SRC2fl/fl/CD4Cre mice (fig. S1B) was confirmed by the lack of SRC2 protein assessed by immunoblot. Furthermore, greatly reduced Ncoa2 mRNA was observed in SRC2fl/fl/Foxp3YFP-Cre CD4+ T cells than in Foxp3YFP-Cre CD4+ T cells as early as 20 hours after polarization (fig. S1C), and in SRC2fl/fl/CD4Cre CD4+ T cells than in SRC2fl/fl CD4+ T cells (fig. S1D) polarized under Treg differentiation conditions.
Since Tregs develop in the thymus, thymic Treg development was first examined. Overall, thymocyte development was normal in SRC2fl/fl/Foxp3YFP-Cre and SRC2fl/fl/CD4Cre mice, as indicated by thymic cellularity (fig. S1, E and G) and percentage of thymocyte subsets: CD4−CD8− double-negative (early thymocytes), CD4+CD8+ double-positive, and CD4+/CD8+ single-positive (mature T cells) cells compared to Foxp3YFP-Cre mice (fig. S1F) and SRC2fl/fl mice (fig. S1H), respectively. There was no significant difference in the percentage and the number of thymic Tregs between Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cre mice (Fig. 1, A and B) and between SRC2fl/fl and SRC2fl/fl/CD4Cre mice (Fig. 1, C and D). Thus, SRC2 is not essential for thymocyte development, including thymic natural Treg development.
Fig. 1. SRC2 is not required for nature Treg development but essential for Treg differentiation from naive Foxp3−CD4+ T cells.
(A and C) Representative flow cytometric analysis (left panels) and the percentage (right panels) of Treg (YFP+ or Foxp3+) cells among CD4+ thymocytes from indicated mice (n ≥ 4 per genotype). (B and D) Absolute number of Treg in the thymus from indicated mice (n ≥ 3 per genotype). (E) Representative flow cytometric analysis (left panels) and percentage (right panels) of naive YFP− (top) and Foxp3− (bottom panels) among CD4+ cells isolated from spleens of indicated mice (n = 3 per genotype). (F) Representative flow cytometric analysis (left panels) and percentage (right panel) of Foxp3+ Tregs differentiated from naive CD4+ T cells shown in (E) in the presence of different concentrations of TGFβ for 48 hours (n ≥ 4 per treatment cohort). (G) Quantitative polymerase chain reaction (qPCR) analysis of Foxp3 mRNA in indicated CD4+ cells 48 hours after Treg differentiation in the presence of TGFβ (5 ng/ml; n = 3 per genotype). (H) Representative flow cytometric analysis (left panels) and percentage (right panel) of naive Foxp3− CD4+ cells isolated from spleens of indicated mice (n = 3 per genotype). (I) Representative flow cytometric analysis (left panels) and percentage (right panel) of Foxp3+ Tregs differentiated from naive Foxp3− CD4+ cells shown in (H) when treated with varying concentrations of TGFβ for 48 hours (n ≥ 5 per genotype). (J) qPCR analysis of Foxp3 mRNA in indicated CD4+ cells 48 hours after Treg differentiation in the presence of TGFβ (5 ng/ml; n = 4 per genotype). Boxed area: Cell population of interest. Data are from three experiments (B, D, G, J; A, C, E, F, H, and I, right panels; presented as means ± SD) or are from one representative of three independent experiments (A, C, E, F, H, and I, left panels). **P < 0.01; ***P < 0.001; ****P < 0.0005; ns, not significant (two-tailed Student’s t test).
iTregs are differentiated from peripheral naive CD4+ T cells in the presence of TGFβ. Next, we examined the function of SRC2 on iTreg differentiation. Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cre mice allow us to use yellow fluorescent protein (YFP) as a reporter for Foxp3 expression (fig. S1I). We confirmed that purified naive CD4+YFP− T cells (Fig. 1E, top panels) from spleens of Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cre mice were Foxp3− (Fig. 1E, bottom panels). Furthermore, in the presence of TGFβ, these naive CD4+YFP− T cells differentiated into Foxp3+YFP+ Tregs (Fig. 1F and fig. S1J). However, the ability of naive CD4+YFP− T cells from SRC2fl/fl/Foxp3YFP-Cre mice to generate iTregs was greatly impaired compared to the CD4+ T cells from Foxp3YFP-Cre mice at all the TGFβ concentrations that we tested (Fig. 1F). Consistently, Foxp3 mRNA was decreased in SRC2fl/fl/Foxp3YFP-Cre CD4+ cells than in Foxp3YFP-Cre CD4+ cells after Treg differentiation (Fig. 1G). The observed impaired Treg differentiation was not due to changes in cell proliferation and survival, which were comparable between Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cr cells gated on either Foxp3+ (fig. S1, K and L, top panels) or Foxp3− cells (fig. S1, K and L, bottom panels), as monitored by the proliferation marker Ki-67 (fig. S1K) and live/dead dye (fig. S1L) at 20 and 48 hours after initiation of Treg differentiation. Similarly, compared to control SRC2fl/fl CD4+ T cells, the capacity of naive SRC2fl/fl/CD4Cre Foxp3−CD4+ T cells (Fig. 1H) to generate iTregs in vitro was greatly decreased under varying TGFβ concentrations (Fig. 1I) and showed correspondingly lower Foxp3 mRNA levels in SRC2fl/fl/CD4Cre CD4+ cells after Treg differentiation (Fig. 1J). Thus, we show that SRC2, although dispensable for thymic Treg development, is essential for iTreg generation from naive CD4+ T cells in vitro.
SRC2 is required for generating iTregs in vivo
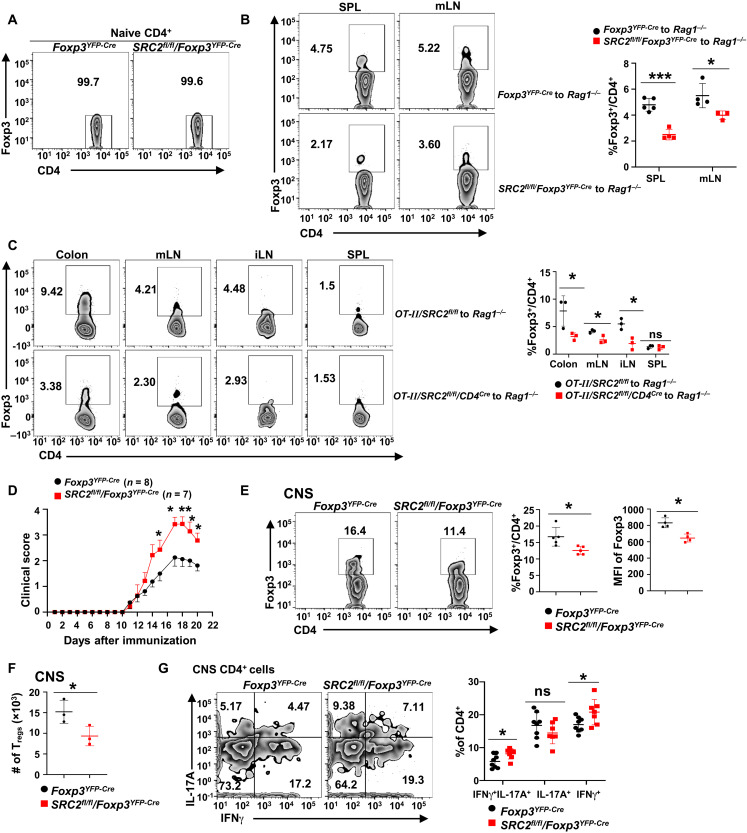
To determine the function of SRC2 in vivo in Treg generation, sorted naive Foxp3YFP-Cre or naive SRC2fl/fl/Foxp3YFP-Cre YFP−CD4+ T cells that lack Foxp3+ Tregs (Fig. 2A) were adoptively transferred to Rag1−/− mice (17, 18). Significant Tregs were detected in spleens and mesenteric lymph nodes (mLNs) 3 weeks after adoptive transfer of naive Foxp3YFP-Cre CD4+ cells (Fig. 2B, top two panels). In contrast, naive SRC2fl/fl/Foxp3YFP-Cre CD4+ T cells generated less Tregs in vivo (Fig. 2B, bottom). Next, an oral tolerance model was used to determine SRC2 function in the generation of Tregs in vivo (9). In this model, sorted naive CD4+Foxp3− T cells from OT-II/SRC2fl/fl or OT-II/SRC2fl/fl/CD4Cre mice (fig. S2A) were adoptively transferred into Rag1−/− mice, and Tregs were induced mostly in gut-associated lymphoid tissues by feeding ovalbumin peptide (OVA) in drinking water (Fig. 2C). Consistently, significantly less Tregs were generated from OT-II/SRC2fl/fl/CD4Cre CD4+ T cells than from OT-II/SRC2fl/fl CD4+ T cells in colon, mLN, and inguinal lymph nodes (iLNs), but no difference was observed in spleens. Collectively, these results demonstrate an essential role for SRC2 in promoting Treg differentiation in vivo.
Fig. 2. SRC2 is required for generating Tregs in vivo.
(A) Representative flow cytometric analysis of Foxp3− cells in sorted naive CD4+ cells from indicated mice before adoptive transfer into Rag1−/− mice. (B) Representative flow cytometric analysis (left panels) and percentage (right panel) of Foxp3+CD4+ Tregs in spleen and mLN of Rag1−/− mice 3 weeks after adoptive transfer of 0.4 × 106 naive CD4+ cells (n ≥ 3 per genotype). (C) Representative flow cytometric analysis (left panels) and percentage (right panel) of Foxp3+ CD4+ Tregs in colon, mLN, iLN, and spleen of Rag1−/− mice transferred with 3 × 106 naive OT-II/SRC2fl/fl or OT-II/SRC2fl/fl/CD4Cre CD4+ cells and subsequently treated with OVA (20 mg/ml) for 5 days (n = 3 per genotype). (D) Mean clinical EAE scores of indicated mice at different days after EAE induction with MOG35–55. (E) Representative flow cytometric analysis (left panels) and the percentage and Foxp3 MFI (right panels) of Foxp3+CD4+ Tregs recovered from the CNS of EAE-induced mice (n = 5 per genotype). (F) Number of Foxp3+CD4+ Tregs recovered from the CNS of EAE-induced mice (n = 3 per genotype). (G) Representative flow cytometric analysis (left panels) and the percentage (right panel) of interferon-γ–positive (IFNγ+) and IL-17A+ cells among CD4+ T cells recovered from the CNS of EAE-induced mice (n ≥ 7 per genotype). Boxed area: Cell population of interest. Data are from three experiments (D, presented as means ± SEM; B, C, E, and G, right panels, presented as means ± SD) or are from one representative of three independent experiments (A; B, C, E, and G, left panels). *P < 0.05, **P < 0.01, and ***P < 0.001 (two-tailed Student’s t test).
To determine whether SRC2-regulated generation of Treg plays a role in controlling autoimmune responses, we compared the development of EAE between Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cre mice (Fig. 2D). Compared to Foxp3YFP-Cre mice, SRC2fl/fl/Foxp3YFP-Cre mice developed much severer EAE and had significantly less Tregs with lower levels of Foxp3 (Fig. 2, E and F), but more inflammatory CD4+IFNγ+IL-17A+ and CD4+IFNγ+ cells but not interleukin-17A–positive (IL-17A+) cells in the central nervous system (CNS) (Fig. 2G and fig. S2B for gating strategy). Our results thus support an essential function for SRC2 in Treg differentiation in vivo, and SRC2-regulated generation of Tregs controls the scale of immune responses in vivo.
Aged SRC2fl/fl/Foxp3YFP-Cre mice develop inflammation-associated lung tissue damages
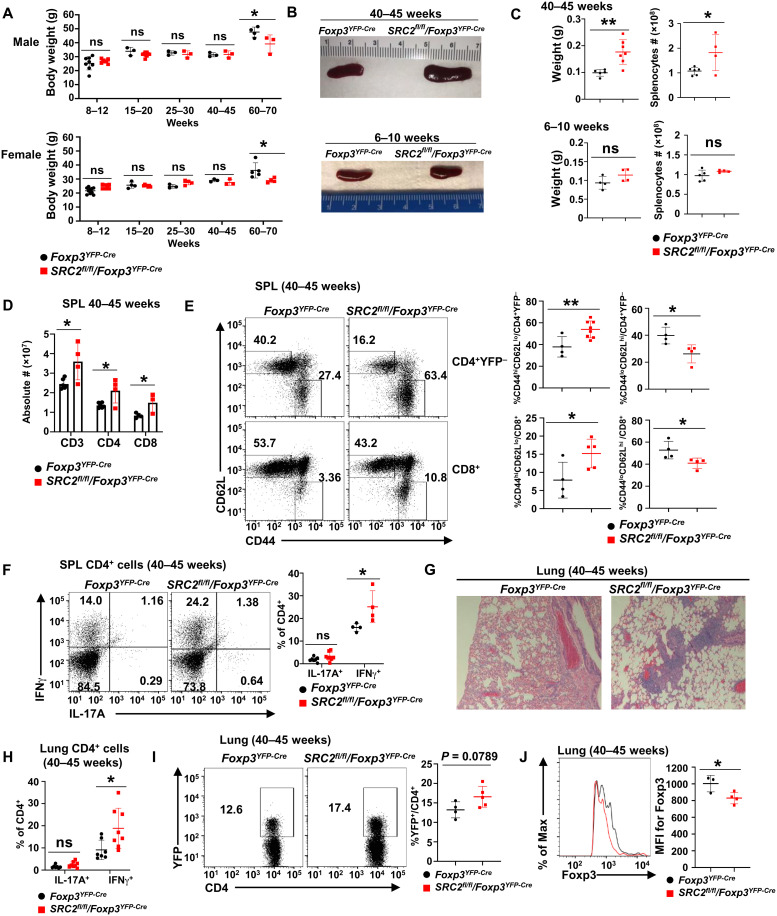
We noticed that aged SRC2fl/fl/Foxp3YFP-Cre mice were smaller and suffered from hair loss compared to their age-matched SRC2fl/fl counterparts (fig. S3A). Sixty- to 70-week-old SRC2fl/fl/Foxp3YFP-Cre mice suffered modest weight loss than Foxp3YFP-Cre mice, regardless of sex, compared to younger mice (Fig. 3A). In addition, 40- to 45-week-old SRC2fl/fl/Foxp3YFP-Cre mice had enlarged spleen (splenomegaly) (Fig. 3B), which was further confirmed by increased spleen weight and cellularity (Fig. 3C) when compared to age-matched Foxp3YFP-Cre mice (Fig. 3, B and C), whereas no differences in spleen size, weight, and cellularity were observed between the cohorts at 6- to 10-week-old mice (Fig. 3, B and C). Increased numbers of CD3+ T cells, including both CD4+ and CD8+ T cell subsets, contributed to the increased cellularity of the spleens from older SRC2fl/fl/Foxp3YFP-Cre mice (Fig. 3D). Since splenomegaly is a sign of inflammation, it indicates that the inflammatory disease resulted from defective Tregs because of specific deletion of SRC2 in aged SRC2fl/fl/Foxp3YFP-Cre mice. The percentage of CD44hiCD62lo memory-like cells were significantly increased, while CD44loCD62hi naive CD4+ and CD8+ T cells were significantly decreased in spleens of 40- to 45-week-old SRC2fl/fl/Foxp3YFP-Cre mice (Fig. 3E) than in Foxp3YFP-Cre mice; no differences in these cell subtypes were observed in spleens at 6 to 10 weeks between the cohorts (fig. S3B). At 40 to 45 weeks, SRC2fl/fl/Foxp3YFP-Cre mice had increased percentage of IFNγ+CD4+ cells but not IL-17A+CD4+ splenocytes (Fig. 3F). Furthermore, compared to Foxp3YFP-Cre mice, SRC2fl/fl/Foxp3YFP-Cre mice had severe lung damage (Fig. 3G) associated with increased inflammatory IFNγ+CD4+ cells (Fig. 3H) at 40 to 45 weeks. Increased Tregs (Fig. 3I) were found in the damaged lung of SRC2fl/fl/Foxp3YFP-Cre mice. However, these SRC2fl/fl/Foxp3YFP-Cre Tregs had significantly reduced mean florescence intensity (MFI) for Foxp3 than Foxp3YFP-Cre Tregs (Fig. 3J). Increased Tregs were also found in spleens and lymph nodes of older (40 to 45 weeks) SRC2fl/fl/Foxp3YFP-Cre mice with obvious lung inflammation but not in younger mice (6 to 25 weeks) (fig. S3C), suggesting that increased Tregs in older SRC2fl/fl/Foxp3YFP-Cre mice are likely a compensatory mechanism trying to inhibit the observed inflammation. This is consistent with what is observed in other mice with defective Tregs (19). Consistent with reduced MFI for Foxp3 found in Tregs from lungs of older SRC2fl/fl/Foxp3YFP-Cre mice, MFI for Foxp3 in Tregs in the spleens and lymph nodes showed the same trend of decrease with increase in age of SRC2fl/fl/Foxp3YFP-Cre mice compared to Foxp3YFP-Cre mice (fig. S3D). Analysis of CD62Lhi cells, a marker for naive Tregs, indicated a significant decrease in CD62Lhi naive Tregs in the spleen, lymph nodes, and lungs of older SRC2fl/fl/Foxp3YFP-Cre mice (fig. S3, E and F), supporting the notion that increased Tregs in older SRC2fl/fl/Foxp3YFP-Cre mice likely result from Treg proliferation (20). These results demonstrate the critical function of SRC2 in Treg-dependent maintenance of immune tolerance in vivo.
Fig. 3. Aged SRC2fl/fl/Foxp3YFP-Cre mice develop inflammation-associated lung tissue damages.
(A) Body weight measurement of indicated male (top panel) and female (bottom panel) mice at different ages (n ≥ 3 per genotype per group). (B) Representative image of spleens from older (top panel) and younger (bottom panel) mice of indicated genotypes. (C) Weight (left panels) and cellularity (right panels) of the spleens from older (top panels) and younger (bottom panels) indicated mice (n ≥ 4 per genotype per group). (D) Absolute number of CD3+, CD4+, and CD8+ T cells in the spleens from indicated older mice (n ≥ 4 per genotype per group). (E) Representative flow cytometric analysis (left panels) and percentage (right panels) of CD62L and CD44 cells among splenic CD4+YFP− or CD8+ T cells from indicated aged mice (n ≥ 4 per genotype per group). (F) Representative flow cytometric analysis (left panels) and percentage (right panel) of IL-17A+ and IFNγ+ cells among CD4+ cells from spleens of indicated aged mice (n ≥ 4 per genotype). (G) Section of hematoxylin and eosin (H&E)–stained lung from indicated aged mice. (H) Percentage of IL-17A+ and IFNγ+ cells among CD4+ cells recovered from lung of indicated aged mice (n ≥ 7 per genotype). (I) Representative flow cytometric analysis (left panels) and percentage (right panel) of Tregs (YFP+) among CD4+ cells recovered from lung of indicated aged mice (n ≥ 4 per genotype). (J) Representative flow cytometric analysis (left panel) and the MFI (right panel) for Foxp3 in Foxp3+CD4+ cells recovered from lung of indicated aged mice (n ≥ 3 per genotype). Boxed area: Cell population of interest. Data are from three experiments (A, C, D, and H; E, F, I, and J, right panels; presented as means ± SD) or are from one representative of three independent experiments (B and G; E, F, I, and J, left panels). *P < 0.05 and **P < 0.01 (two-tailed Student’s t test).
SRC2 is dispensable for the suppressive function of Tregs in younger mice
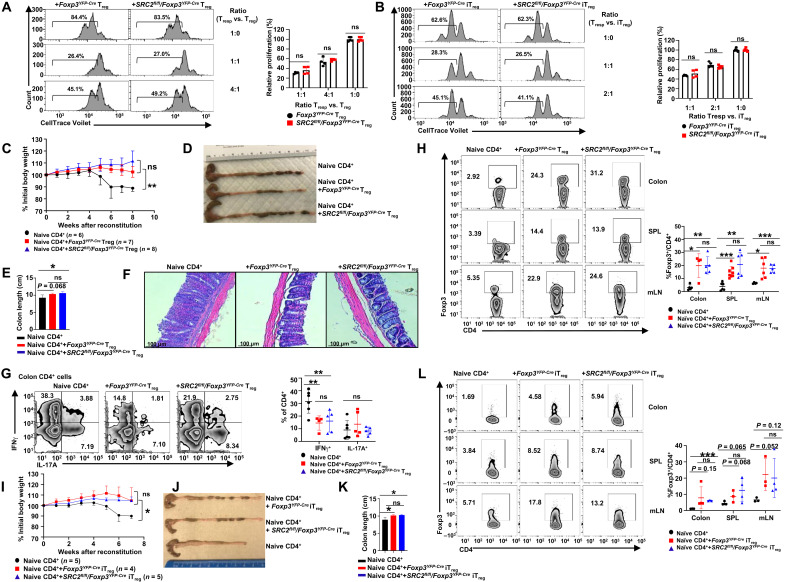
We first examined the expression of several surface markers, CD73, CD39, CD25, and CTLA-4, which are indicators for the suppressive function of Tregs (21–23). There were no significant differences in the expression for all these markers between Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cre Tregs (CD4+YFP+) from spleens and mLNs in 6- to 8-week-old mice (fig. S4A). To determine the suppressive function of Tregs, we assessed the ability of Tregs to inhibit CD4+ T cell proliferation in vitro. SRC2 deficiency did not alter the suppressive function of CD4+YFP+ Tregs sorted from the spleen of 6- to 8-week-old SRC2fl/fl/Foxp3YFP-Cre mice or in vitro derived SRC2fl/fl/Foxp3YFP-Cre iTregs (Fig. 4, A and B), suggesting that SRC2 is not essential for the suppressive function of Tregs. Last, the in vivo function of Tregs was tested in the prevention of colitis. In the absence of Tregs, adoptive transfer of naive CD4+ T cells (CD45RBhiCD25−CD4+) into Rag1−/− mice induced severe colitis, as indicated by weight loss (Fig. 4C), shortened colon (Fig. 4, D and E), damaged intestinal tissues (Fig. 4F), and greatly increased proinflammatory IFNγ+CD4+ T cells in the colon (Fig. 4G) and mLN (fig. S4B). In contrast, cotransfer of CD4+YFP+ Foxp3YFP-Cre Tregs or SRC2fl/fl/Foxp3YFP-Cre Tregs with naive CD4+ T cells rescued these severe colitis phenotypes in Rag1−/− mice (Fig. 4, C to F). In these rescued mice, we also observed a significant reduction in proinflammatory IFNγ+CD4+ T cells in the colon and mLN (Fig. 4G and fig. S4B). Furthermore, higher levels of adoptively transferred Tregs from either Foxp3YFP-Cre or SRC2fl/fl/Foxp3YFP-Cre mice were found in the gut-associated tissues and spleens of recipients (Fig. 4H), thus contributing to the prevention of colitis.
Fig. 4. SRC2 is dispensable for the suppressive function of Tregs in younger mice.
(A and B) Representative flow cytometric analysis (left panels) and the relative proliferation (right panel) of responder T (Tresp) cells cultured with splenic YFP+CD4+ Tregs isolated from indicated 6- to 8-week-old mice (A) or YFP+CD4+ iTregs differentiated in vitro (B) (n = 4 per genotype). (C) Body weight of Rag1−/− recipients over time after adoptive transfer of WT CD45RBhiCD25−CD4+ naive T cells alone or in combination with splenic YFP+CD4+ Tregs from 6- to 8-week-old mice. (D to F) Representative image of colons (D), colon length (E) (n = 5 per genotype), and H&E-stained colon section (F) from Rag1−/− recipients 8 weeks after adoptive transfer. (G) Representative flow cytometric analysis (left panels) and percentage (right panel) of CD4+IL-17A+ and CD4+IFNγ+ cells recovered from colons of Rag1−/− recipients (n ≥ 4 per group). (H) Representative flow cytometric analysis (left panels) and percentage (right panel) of Foxp3+CD4+ Tregs recovered from colon, spleen, and mLN of Rag1−/− recipients (n ≥ 4 per group). (I) Body weight of Rag1−/− recipients over time after adoptive transfer of WT CD45RBhiCD25−CD4+ naive T cells alone or in combination with in vitro differentiated YFP+CD4+ iTregs. (J and K) Representative image of colons (J) and colon length (K) (n ≥ 4 per genotype) from Rag1−/− recipients 7 weeks after adoptive transfer. (L) Representative flow cytometric analysis (left panels) and percentage (right panel) of Foxp3+CD4+ Tregs recovered from colon, spleen, and mLN of Rag1−/− recipients (n ≥ 4 per group). Boxed area: Cell population of interest. Data are from three experiments (C and I, presented as means ± SEM; E, I, K, A, B, G, H, and L, right panels, presented as means ± SD) or are from one representative of three independent experiments (D, F, and J; A, B, G, H, and L, left panels). *P < 0.05, **P < 0.01, and ***P < 0.001 (two-tailed Student’s t test).
Similar to natural Tregs sorted from younger mice, in vitro derived iTregs also prevented colitis; Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cre iTregs showed comparable inhibitory efficacy in preventing weight loss (Fig. 4I) and shortening of the colon (Fig. 4, J and K). Adoptively transferred iTregs derived from either Foxp3YFP-Cre or SRC2fl/fl/Foxp3YFP-Cre naive CD4+ T cells were maintained at comparable levels in the colon, spleen, and mLN of the recipients (Fig. 4L). These results suggest that SRC2, although required for Treg differentiation, is not essential for suppressive Treg function.
Since aged SRC2fl/fl/Foxp3YFP-Cre mice showed inflammation, an indication of defective Treg function, we thus determined the inhibitory function of Tregs from older mice. Tregs from 26-week-old SRC2fl/fl/Foxp3YFP-Cre mice started to show slightly decreased inhibitory activity at 4:1 Tresp (responder T cell)/Treg ratio than those from age-matched Foxp3YFP-Cre mice (fig. S4C), suggesting that the impaired inhibitory function of Tregs from older SRC2fl/fl/Foxp3YFP-Cre mice likely also contributes to the observed lung tissue inflammation.
SRC2 stimulates the expression of Nr4a2 critical for Treg differentiation
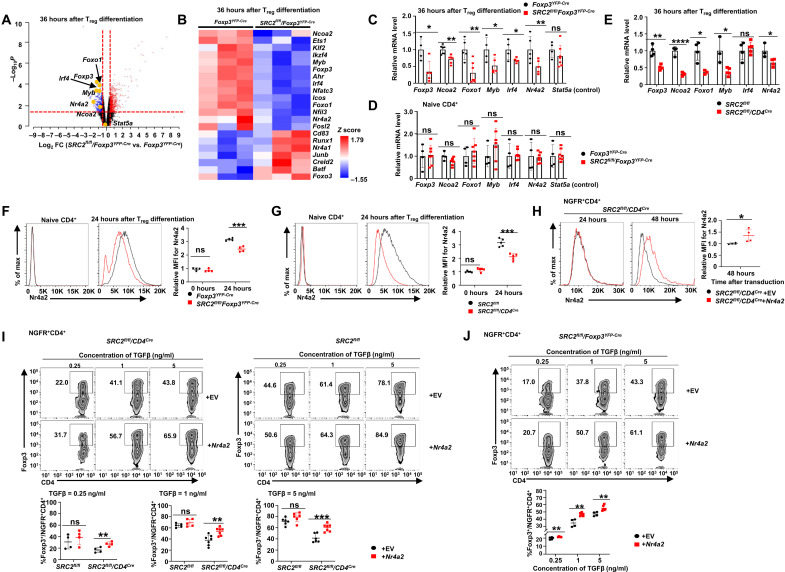
We next determined mechanisms for SRC2-regulated Treg differentiation. We first excluded the function of SRC2 in the regulation of Foxp3 stability, as the degradation rate of Foxp3 in Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cre Tregs was equivalent (fig. S5A). SRC2 is a transcriptional coactivator that is believed to regulate cellular function by controlling gene expression. Thus, we next performed RNA-seq analysis to detect the transcriptome of the following four groups of cells (fig. S5B, left panel): (i) Foxp3YFP-Cre naive CD4+ T cells, (ii) SRC2fl/fl/Foxp3YFP-Cre naive CD4+ T cells, (iii) Foxp3YFP-Cre CD4+ cells polarized in TGFβ for 36 hours, and (iv) SRC2fl/fl/Foxp3YFP-Cre CD4+ cells polarized in TGFβ for 36 hours. The expression of Foxp3 was significantly lower in polarized SRC2fl/fl/Foxp3YFP-Cre CD4+ than the wild-type (WT) control (fig. S5B, middle panel). Principal components analysis of transcriptomes clustered three repeats within each group together, whereas the naive CD4+ T cells and polarized CD4+ cells showed the biggest differences in gene expression patterns (fig. S5B, right panel), indicating the excellent quality and reproducibility of RNA-seq results. Comparing transcriptomes between differentiated Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cre cells, we identified many differentially expressed genes known to regulate Treg differentiation (Fig. 5, A and B, and fig. S5C). Not surprisingly, Ncoa2 (encoding SRC2) was among the most down-regulated genes due to gene deletion. Foxp3 was also down-regulated in SRC2fl/fl/Foxp3YFP-Cre cells, confirming impaired Treg differentiation.
Fig. 5. SRC2 stimulates the expression of Nr4a2 critical for Treg differentiation.
(A) Volcano plot comparison of gene expression between Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cre CD4+ cells (n = 3 per genotype). Differentially up-regulated genes (red) and down-regulated genes (blue) with a cutoff at P < 0.05 and fold change (FC) > 1.4 are shown. (B) Heatmap of Ncoa2 and other genes known to regulate Treg differentiation. (C to E) qPCR analysis of Foxp3, Ncoa2, Foxo1, Myb, Irf4, Nr4a2, and Stat5a (control) mRNA in CD4+ cells 36 hours after Treg polarization (C and E) and naive CD4+ cells (D) derived from indicated mice (n ≥ 4 per genotype per group). (F and G) Representative flow cytometric analysis of protein levels (left panels) and relative MFI (right panel) for Nr4a2 in indicated genotypes of naive CD4+ cells versus CD4+ cells 24 hours after Treg polarization [n = 4 per genotype for (F) and n = 5 per genotype for (G)]. (H) Representative flow cytometric analysis of protein levels (left panels) and relative MFI (right panel) for Nr4a2 in SRC2fl/fl/CD4Cre CD4+ T cells transduced with retrovirus expressing ± Nr4a2 and differentiated under Treg polarization conditions for 24 or 48 hours (n = 4 per genotype). (I and J) Representative flow cytometric analysis (top panels) and the percentage (bottom panels) of Foxp3+CD4+ Tregs among indicated genotypes of NGFR+CD4+ cells transduced with retrovirus expressing ± Nr4a2 and polarized for 48 hours under Treg conditions (n ≥ 4 per genotype per group). EV, empty vector; NGFR, marker of transduction; boxed region, cell population of interest. Data are from three experiments (A to H, right panels; I and J, bottom panels; presented as means ± SD) or are from one representative of three independent experiments (F to H, left panels; I and J, top panels). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0005 (two-tailed Student’s t test).
Since SRC2 is a transcriptional coactivator, we paid particular attention to the down-regulated transcription factor genes. Four transcription factors—Myb, Irf4, Foxo1, and Nr4a2—were down-regulated in the absence of SRC2 and are known to positively regulate Treg differentiation (9, 24–26). Individual quantitative polymerase chain reaction (qPCR) confirmed the down-regulation of these four transcription factors together with Ncoa2 and Foxp3 in SRC2fl/fl/Foxp3YFP-Cre CD4+ Tregs, whereas Stat5a served as a control that did not show any significant changes in qPCR analysis (Fig. 5C). These four transcription factors did not show obvious differential mRNA expression between Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cre in naive CD4+ T cells (Fig. 5D), suggesting that the observed changes were induced upon Treg differentiation. Ncoa2 also did not show differences between Foxp3YFP-Cre and SRC2fl/fl/Foxp3YFP-Cre naive CD4+ T cells, as its gene deletion is only induced when Foxp3 starts to express during Treg differentiation. Our results suggest that SRC2 is required for the up-regulation of these four transcription factors in Tregs at the mRNA level. Since SRC2fl/fl/CD4Cre CD4+ T cells also showed impaired Treg differentiation, we compared the expression of above transcription factors between differentiated SRC2fl/fl and SRC2fl/fl/CD4Cre CD4+ cells (Fig. 5E). Except Irf4, the other three transcription factors were also down-regulated together with Foxp3 in SRC2fl/fl/CD4Cre CD4+ cells after polarization under Treg conditions. Therefore, SRC2 stimulates the expression of some transcription factors critical for Treg differentiation.
We next evaluated the effects of forced expression of above down-regulated transcription factors on Treg differentiation from CD4+ T cells that are deficient in SRC2. For this purpose, retrovirus expressing individual transcription factor was transduced into SRC2fl/fl/CD4Cre CD4+ T cells that then differentiated into Tregs in the presence of TGFβ. Forced expression of Foxo1, Irf4, or Myb together with green fluorescent protein (GFP) did not rescue Treg differentiation in SRC2fl/fl/CD4Cre CD4+ T cells compared to the cells transduced with virus expressing only GFP [empty vector (EV); fig. S5D]. Examining transcription factor protein expression, Foxo1 (fig. S5E) and interferon regulatory factor 4 (IRF4; fig. S5F) protein levels were not obviously down-regulated upon Treg differentiation in the absence of SRC2 (Myb protein could not be reliably detected).
Consistent with prior reports that Nr4a2 is up-regulated upon T cell activation (27, 28), polarized Foxp3YFP-Cre (Fig. 5F, middle panel) and SRC2fl/fl (Fig. 5G, middle panel) CD4+ cells in the presence of TGFβ expressed higher levels of Nr4a2 at 24 hours than their corresponding respective naive CD4+ counterparts without stimulation (Fig. 5, F and G, left panel). However, after 24-hour polarization under Treg conditions, Nr4a2 in both SRC2fl/fl/Foxp3YFP-Cre (Fig. 5F, middle panel) and SRC2fl/fl/CD4Cre (Fig. 5G, middle panel) CD4+ cells failed to up-regulate to the levels detected in their corresponding WT counterparts Foxp3YFP-Cre or SRC2fl/fl CD4+ cells, respectively. This result suggests that SRC2 can promote Nr4a2 expression. Retrovirus expressing Nr4a2 together with nerve growth factor receptor (NGFR) was used to transduce SRC2fl/fl CD4+ cells (Fig. 5H); this greatly increased Nr4a2 levels at 48 hours, but not 24 hours, after transduction when compared to control cells transduced with EV expressing only NGFR (Fig. 5H). Correspondingly, transduction with Nr4a2 significantly elevated Foxp3 expression in CD4+ cells at 48 hours, but not 24 hours, after transduction and differentiation compared to EV control cells (fig. S5G). Moreover, forced expression of Nr4a2 significantly stimulated Treg differentiation in SRC2fl/fl/CD4Cre (Fig. 5I, left panels), but not in SRC2fl/fl (Fig. 5I, right panels), CD4+ T cells at all concentrations of TGFβ that we tested, suggesting that exogenous Nr4a2 is able to overcome SRC2 deficiency and rescue the Treg differentiation defect. In addition, forced expression of Nr4a2 was also able to stimulate the differentiation of Tregs from SRC2fl/fl/Foxp3YFP-Cre CD4+ cells (Fig. 5J). However, Nr4a2, together with IRF4 (fig. S5H) or Foxo1 (fig. S5I), did not further stimulate Treg differentiation compared to Nr4a2 alone in SRC2fl/fl/CD4Cre CD4+ T cells. Our results thus support a model that SRC2 promotes Treg differentiation via up-regulation of Nr4a2, which is known to stimulate Foxp3 gene expression (26).
SRC2 recruited by NFAT1 binds to the promoter and activates gene expression of Nr4a2
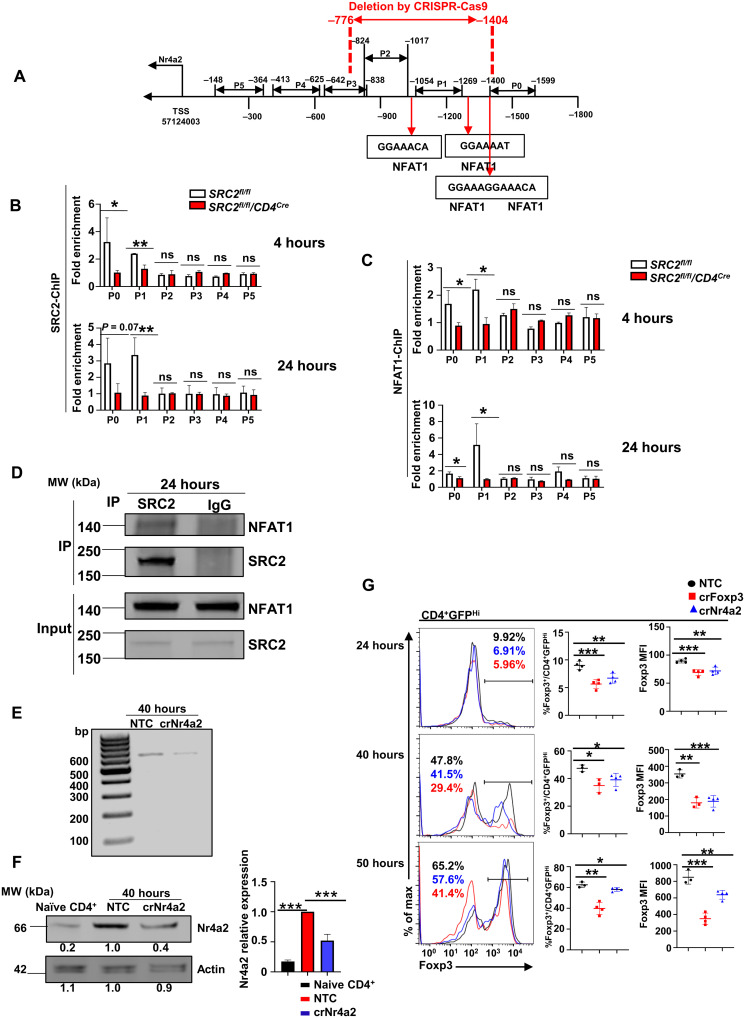
We next determined how SRC2 regulates Nr4a2 expression. Since Nr4a2 mRNA is decreased in the absence of SRC2, we hypothesized that SRC2 is critical for the transcriptional up-regulation of Nr4a2 expression. SRC2 ChIP-seq (chromatin immunoprecipitation and DNA sequencing) analysis detected stronger DNA binding signals at the Nr4a2 promoter region in SRC2fl/fl CD4+ cells than in SRC2fl/fl/CD4Cre CD4+ cells after 36-hour polarization under Treg conditions (fig. S6A). Several pairs of primers were then designed to cover the 1.6-kb promoter region (P0 to P5) upstream of Nr4a2 gene transcription starting site (Fig. 6A). ChIP assays with these primers detected signals indicating interaction of SRC2 at the distal promoter elements (P0 and P1) but not at the proximal P3 to P5 regions at 4 hours (Fig. 6B, top panel) and increasingly at 24 hours (Fig. 6B, bottom panel) after Treg differentiation. As a coactivator, SRC2 does not directly bind but is recruited to DNA by transcription factors. We thus searched for potential transcription factor–binding sites in the promoter region by PROMO, a virtual laboratory for the identification of putative transcription factor binding sites (TFBS) in DNA sequences from a species or groups of species of interest, and identified a few transcription factor–binding sites including signal transducer and activator of transcription 4 (Stat4), NFAT, and Stat6 (fig. S6B). Several conserved NFAT1-binding sites surrounding the P1 region (Fig. 6A) drew our attention, as NFAT1 is a known regulator of Treg differentiation (29, 30) and has been reported to stimulate Nr4a2 expression in CD8+ cells (31). Thus, ChIP assays were performed using the same P0 to P5 primers (Fig. 6A and fig. S6A) to determine whether NFAT1 interacts with Nr4a2 promoter. NFAT1-binding signals were detected at the distal P0 and P1 regions at 4 hours (Fig. 6C, top panel) and increasingly at P1 at 24 hours (Fig. 6C, bottom panel) after Treg differentiation. This was similar to the SRC2-binding patterns, suggesting that NFAT1 recruits SRC2 to the Nr4a2 promoter. Furthermore, NFAT1 was also detected in anti-SRC2 antibody immunoprecipitated complexes from SRC2fl/fl CD4+ cells 24 hours after Treg differentiation (Fig. 6D and fig. S6C for full blot image).
Fig. 6. SRC2 recruited by NFAT1 binds to the promoter and active gene expression of Nr4a2.
(A) Schematic representation of the six regions on Nr4a2 promoter covered by P0 to P5 primers, the locations of identified NFAT1-binding sites, and the region deleted using CRISPR-Cas9. (B and C) ChIP-qPCR analysis of SRC2 binding (B) or NFAT1 binding (C) to the Nr4a2 promoter (P0 to P5 regions) in CD4+ cells from indicated genotypes under Treg polarization at the indicated time points (n ≥ 3 per genotype per group). (D) Immunoblot analysis of NFAT1 among anti-SRC2 antibody immunoprecipitated (IP) SRC2 complexes from SRC2fl/fl CD4+ cells 24 hours after Treg polarization. Bottom blots, whole-cell lysate input control. (E) PCR analysis of the abundance of NFAT1/SRC2 binding region on Nr4a2 promoter in CD4+ cells transduced with virus expressing nontarget control (NTC) or the region containing NFAT1/SRC2-binding guiding RNAs shown in (A) (crNr4a2) and polarized under Treg condition for 40 hours. (F) Immunoblot analysis of Nr4a2 in naive CD4+ cells or CD4+ T cells transduced with virus expressing NTC and crNr4a2 shown in (E) and polarized under Treg conditions for 40 hours. The number in the bottom of the blots is the relative mean intensity of each band, and the right panel is the summary of the relative mean intensity. (G) Representative flow cytometric analysis of Foxp3 (left panels), percentages of Foxp3+ cells (middle panels), and MFI for Foxp3 (right panels) among GFPhiCD4+ cells transduced with virus expressing NTC, crFoxp3, and crNr4a2 guiding RNAs and polarized under Treg condition at the indicated time points (n ≥ 3 per treatment per group). Data are from three experiments (B, C, and F, right panels; G, middle and right panels; presented as means ± SD) or are from one representative of three independent experiments (D to G, left panels). *P < 0.05, **P < 0.01, and ***P < 0.001 (two-tailed Student’s t test). MW, molecular weight; bp, base pairs.
To further determine whether the NFAT1/SRC2 binding is functionally important for Treg differentiation via Nr4a2 regulation, the region containing the potential NFAT1/SRC2-binding sites was deleted using CRISPR-Cas9 with two guiding RNAs (crNr4a2) in CD4+ T cells from mice expressing Cas9 (Fig. 6A and fig. S6D). At the same time, we used a nontargeting construct (NTC) as a negative control and a Foxp3 gene deletion construct (crFoxp3) as a positive control. Deletion of the region containing NFAT1/SRC2-binding sites was confirmed in crNr4a2-transduced cells by PCR analysis (Fig. 6E). Nr4a2 levels were greatly higher in differentiated cells (NTC control) than in naive CD4+ T cells (Fig. 6F and fig. S6E for full blot image), confirming our previous observation that Nr4a2 is up-regulated during Treg differentiation (Fig. 5, F and G). Furthermore, Nr4a2 levels were reduced significantly following deletion of the region containing NFAT1/SRC2-binding sites compared to the NTC control (Fig. 6F and fig. S6E for full blot image), supporting the notion that NFAT1/SRC2 recruitment to the Nr4a2 promoter stimulates gene expression. Compared to NTC control, Foxp3 expression was down-regulated using the crFoxp3 gene deletion construct at 24, 40, and 50 hours after transduction and Treg differentiation (Fig. 6G), confirming successful deletion of Foxp3 gene with CRISPR-Cas9. Deletion of the NFAT1/SRC2-binding region on Nr4a2 promoter by crNr4a2 led to impaired Treg differentiation in terms of both percentage of Foxp3+ Tregs (Fig. 6G, middle panels) and MFI of Foxp3 (Fig. 6G, right panels), as compared to that of NTC control. As an additional control, we also deleted an adjacent DNA fragment (fig. S6, F and G), which did not affect Nr4a2 expression (fig. S6H) and also did not affect Foxp3 differentiation (fig. S6I). Therefore, Nr4a2 promoter region that binds SRC2/NFAT1 is a critical regulatory element that controls Nr4a2 expression and Treg differentiation. Together, NFAT1 recruits SRC2 to Nr4a2 locus to stimulate Nr4a2 expression, which then activates Foxp3 expression, resulting in the promotion of Treg differentiation.
DISCUSSION
SRC2 has long been known to have an anti-inflammatory function in the innate immune system (32). Nuclear factor κB (NF-κB) and activating protein 1 (AP1) are the transcription factors responsible for the activation of the majority of the inflammatory genes in response to the stimulus by inflammatory signals and cytokines (33). By acting as a cofactor for glucocorticoid receptor (GR) that is tethered to the promoter regions by protein-protein interaction with NF-κB or AP1, SRC2 is able to inhibit the expression of such inflammatory genes (34, 35). This partially explains the immune inhibitory effects of GR that is the target of broadly used immunosuppressive drugs in clinics. In addition, GR can also constrain inflammation by sequestrating SRC2 from IRFs that use SRC2 as coactivator (36). IRFs are the critical transcription factors stimulating the expression of the inflammatory genes including chemokines and cytokines in response to type I interferons (IFNs) (37). When SRC2 is sequestrated by GR, IRFs, in the absence of its coactivator, fail to stimulate their target genes, resulting in the suppression of inflammation (38, 39). Furthermore, SRC2 also inhibits the expression of the genes encoding inflammatory cytokines, tumor necrosis factor–α (TNFα), IL-6, and IL-1, when working with estrogen receptors (40–42). Our novel findings suggest that SRC2 suppresses immune responses by promoting the generation of iTregs, which is an important regulatory component of adaptive immunity. Tregs repress the function of effector T cells by direct contact via inhibitory surface molecules PD-1 and CTLA-4 and by production of anti-inflammatory cytokines including TGFβ and IL-10 (43). These anti-inflammatory cytokines also impair innate immune responses. Therefore, SRC2 functions in both innate and adaptive immune cells to balance the overall immune responses by promoting the inhibitory arm of the immunity.
Three members of SRC family coactivators are all approximately 160 kDa in size, share overall similar structure, and are very conservative in amino acid sequence (44, 45). They often act as coactivators for the same transcription factors. However, we previously showed that SRC1 inhibited Treg differentiation. In contrast, we show here that SRC2 stimulates iTreg differentiation, indicating that different members of SRC family could have totally opposite functions in Tregs. Upon phosphorylation by TCR signaling molecule protein kinase C–θ (PKC-θ), SRC1 dissociates Foxp3 from Foxp3-RORγt complexes, resulting in accelerated degradation of Foxp3 (13). Thus, a posttranslational phosphorylation event of SRC1 is the critical mechanism for SRC1-inhibited Treg differentiation. Here, we show that by acting as a coactivator for NFAT1, SRC2/NFAT1-mediated transcription activation of Nr4a2 stimulates the Foxp3 gene expression, resulting in promoting Treg differentiation. Therefore, SRC2-controlled transcription program is critical for Treg differentiation. Together, our results demonstrated that the function of SRCs is not solely dependent on their associated transcription factors; the highly conserved members of SRC family can control diverse function via regulating gene expression in a context-dependent manner: (i) SRCs recruit different cofactors including epigenetic modification enzymes, allowing formation of distinct transcriptional complexes. (ii) SRCs are subjected to different posttranslational modifications including phosphorylation, ubiquitination, sumoylation, acetylation, and methylation, which regulates SRC-interacting proteins and/or stability (46). (iii) SRCs sense different environmental cues that instruct SRCs to make corresponding changes. Our results thus demonstrate how different members of SRCs, although highly conserved, can control the same function such as Treg differentiation via distinct mechanisms, resulting in totally different outcomes.
We showed that Nr4a2 (Nurr1) is a critical target gene of SRC2 in the regulation of Treg differentiation. Consistent with this result, both SRC2 and Nr4a2 are selectively required for Treg differentiation from naive T cells but are not required for natural Treg development in the thymus (27, 28). A previous study showed that Nr4a2 is essential for the inhibitory function of Tregs (27). However, the inhibitory function of both natural Tregs from younger mice and iTregs was not impaired when SRC2 was deleted in Tregs in our current study. This is likely due to lower but not complete absence of Nr4a2 expression in SRC2fl/fl/Foxp3Cre-YFP T cells, and these relatively lower levels of Nr4a2 are sufficient for the inhibitory function but not for the differentiation of iTregs. Furthermore, Tregs from older, but not younger, SRC2fl/fl/Foxp3Cre-YFP mice start to display impaired inhibitory function, suggesting that the SRC2 function in Tregs is age dependent and the impaired inhibitory function of Tregs likely contributes to the observed lung tissue inflammation in old SRC2fl/fl/Foxp3Cre-YFP mice. This impaired inhibitory function of Tregs may result from the accumulation of the defects with growing age, which is worthy of further investigation. Nr4a2 is an orphan nuclear receptor that lacks a classical ligand-binding pocket and thus functions as a ligand-independent transcription factor (28). Nr4a2 activity is therefore believed to be largely regulated by its expression. We found that Nr4a2 is up-regulated by SRC2, which likely results in increased Nr4a2 activity required for Treg differentiation. We showed that SRC2 physically interacts with NFAT1, and both bind to the same promoter region of Nr4a2 gene during Treg differentiation. Ca2+/calcineurin/NFAT signals activated by TCR stimulation are reported to be required for up-regulating Nr4a2 gene expression in T cells (31, 47). Thus, our results support a model that SRC2 associated with NFAT1 is recruited to the Nr4a2 promoter to stimulate the expression of Nr4a2, which, in turn, promotes Treg differentiation by activating Foxp3 gene expression. SRC2 can be recruited by multiple transcription factors; thus, it is worth investigating whether SRC2 can also be recruited by other transcription factors to coordinate the overall transcription program essential for Treg differentiation.
Increased iTregs in tumor microenvironment are responsible for failed antitumor immune responses (48). Checkpoint inhibitors that disrupt the function of Tregs via blocking inhibitory PD-1 and CTLA-4 have shown efficacy in the treatment of cancers. Intensive research has been focused on preventing the function and/or reducing the number of Tregs for boosting immune responses against tumors. Our results show that SRC2 stimulates the generation of iTregs, and thus, inhibition of SRC2, similar to SRC2 knockout mice, is expected to reduce the number of Tregs. Therefore, SRC2 is a potential target for boosting antitumor immunity. All members of SRC family are considered oncogenes, as they play roles in tumorigenesis in different types of cancers (49, 50). Small-molecule SRC inhibitors have already been developed for the treatment of cancers (51, 52). It would be interesting to test whether SRC2 inhibitors can boost antitumor immunity by preventing the generation of Tregs. Together with our results, it raises the possibility that SRC2 inhibitors can be used to treat cancers by targeting both cancer cells and immune system.
MATERIALS AND METHODS
Mice
Transgenic CD4Cre (TgCd4cre, 022071), Rag1−/− (Rag1tm1Mom, 002216), Cas9 (Rosa26LSL-Cas9, 028551), and C57BL (B6, 000664) mice were purchased from the Jackson Laboratory. SRC2fl/fl mice were obtained from Jianming Xu Lab (Molecular and Cell Biology, Baylor College of Medicine, TX). OT-II mice were obtained from Jianhua Yu Lab (Department of Hematology and Hematopoietic Cell Transplantation, City of Hope, CA), and Foxp3YFP-Cre mice were obtained from Mark Boldin Lab (Molecular and Cellular Biology, Beckman Research Institute, City of Hope, CA). All mice were bred at the C57BL/6j background and housed under specific pathogen–free conditions in the Animal Resource Center at the Beckman Research Institute of City of Hope under protocols approved by the Institutional Animal Care and Use Committee (IACUC#07023). Mice were 10 to 12 weeks of age for EAE studies and 6 to 10 weeks of age for other experiments, unless indicated otherwise, with littermates age- and sex-matched across experimental groups.
Antibodies and cytokines
Monoclonal antibodies against mouse CD3 (145-2C11), CD28 (37.51), IL-4 (11B11), and IFNγ (XMG1.2), as well as phycoerythrin (PE)–conjugated anti-CD8 (dilution ratio, 1:100; 53-6.7), allophycocyanin (APC)–conjugated anti-Foxp3 (dilution ratio, 1:100; FJK-16s), PE-indotricarbocyanine (Cy7)–conjugated anti–IL-17A (dilution ratio, 1:100; eBio17B7), APC-conjugated anti-IFNγ (dilution ratio, 1:100; XMG1.2), and LIVE/DEAD Fixable Near-IR Dead Cell Stain (dilution ratio, 1:1000; L34976) were from Invitrogen. PE-conjugated anti-CD25 (dilution ratio, 1:100; PC61), Brilliant Violet (BV) 605–conjugated anti-CD4 (dilution ratio, 1:100; RM4-5), BV 421–conjugated anti-CD3 (dilution ratio, 1:100; 145-2C11), APC-conjugated anti-CD45 (dilution ratio, 1:100; I3/2.3), PE-Cy7–conjugated anti-CD45RB (dilution ratio, 1:100; C363-16A), PE-conjugated anti–Ki-67 (dilution ratio, 1:100; 16A8), PE-Cy7–conjugated anti-CD62L (dilution ratio, 1:100; MEL-14), APC-Cy7–conjugated anti-CD44 (dilution ratio, 1:100; IM7), Alexa Fluor 488–conjugated anti-IRF4 (dilution ratio, 1:100; IRF4.3E4), PE-Cy7–conjugated anti-NGFR (dilution ratio, 1:100; ME20.4), PE-conjugated anti-CD73 (dilution ratio, 1:100; TY/11.8), PE-Cy7–conjugated anti-CD39 (dilution ratio, 1:100; Duha59), APC-Cy7–conjugated anti-CD45 (dilution ratio, 1:100; 30-F11), PE-Cy7–conjugated anti–CTLA-4 (dilution ratio, 1:100; UC10-4B9), and recombinant murine IL-2 were from BioLegend. APC-conjugated anti-CD25 (dilution ratio, 1:100; PC61) was from BD. Fluorescein isothiocyanate (FITC)–conjugated anti-Nr4a2 (dilution ratio, 1:150; orb464231) was from Biorbyt. FITC-conjugated anti-Foxo1 (dilution ratio, 1:100; 83N7F8) was from Novus. PE-conjugated anti–IL-10 (dilution ratio, 1:100; JES5-16E3) was from eBioscience. Rabbit anti-hamster antibody (55398) was from MP Biomedicals. Antibodies against SRC2 (dilution ratio, 1:2000; A300-346A, Bethyl), rabbit IgG (P120-101, Bethyl), NFAT1 (dilution ratio, 1:1000; 5861S, Cell Signaling Technology), Nr4a2 (dilution ratio, 1:150; sc-376984, Santa Cruz Biotechnology), and β-actin (dilution ratio, 1:1000; SC-8422, Santa Cruz Biotechnology) were used for immunoblot analysis. Recombinant mouse TGFβ was from Miltenyi Biotec.
Plasmids
The retroviral vector murine stem cell virus (MSCV)–internal ribosomal entry site (IRES)–GFP was a gift from W. S. Pear (University of Pennsylvania). Complementary DNA (cDNA) encoding Foxo1 was cloned into MSCV-IRES-GFP vector. IRF4-MIEG-GFP was a gift from Mark H. Kaplan Lab (Indiana University School of Medicine), and MSCV-HA-Nr4a2-IRES-NGFR was a gift from Joyce Chen Lab (La Jolla Institute for Immunology, La Jolla, CA, USA). MSCV-IRES-NGFR (plasmid #27489), MSCV-PIG-Myb (plasmid #66988), and retro–guide RNA (gRNA)–eGFP (plasmid #116926) were purchased from Addgene.
Flow cytometry
For surface staining, cells isolated from mice or in vitro culture were directly stained with antibodies and/or fixable live/dead dye with 2% fetal bovine serum (FBS) and 1 mM EDTA at 4°C for 15 min. For transcription factor staining, cells prestained with surface markers were fixed and permeabilized in TF Fix/Perm buffer (BD Biosciences) at 4°C for 20 min, washed once with TF Perm/Wash buffer, and stained with target markers in the TF Perm/Wash buffer at 4°C for 15 min. For intracellular cytokine analysis, cells were stimulated with phorbol 12-myristate 13-acetate (50 ng/ml; Sigma-Aldrich) and ionomycin (750 ng/ml; Sigma-Aldrich) at 37°C for 3 hours in the presence of GolgiStop (BD Biosciences) before staining. After stimulation, cells were stained with surface markers and then fixed and permeabilized with Cytofix/Cytoperm buffer (BD Biosciences) for 20 min followed by staining cytokines in the Perm/Wash buffer (BD Biosciences) after washing. The expression of surface and intracellular markers was analyzed with a BD LSRFortessa flow cytometer.
Isolation of naive CD4+ T cells and in vitro Treg differentiation
Naive CD4+ T cells were isolated from mouse spleens by negative selection using the Naive CD4+ T Cell Isolation Kit (Miltenyi Biotec). Suspensions of 5 × 105 cells per milliliter of RPMI 1640 medium (Corning Inc.) containing 2 mM l-glutamine, 50 μM β-mercaptoethanol, penicillin (100 U/ml), streptomycin (100 mg/ml), and 10% FBS (Corning Inc.) were cultured in 24-well plates or 48-well plates precoated with rabbit anti-hamster (0.1 mg/ml). The medium was supplemented with hamster anti-CD3 (0.25 μg/ml), hamster anti-CD28 (1 μg/ml), TGFβ (0.25, 1, or 5 ng/ml), anti–IL-4 (2.5 μg/ml), and anti-IFNγ (2.5 μg/ml) for Treg differentiation for up to 48 hours.
In vivo induction of iTregs by adoptively transferring naive CD4+ cells
Splenic cells were collected from Foxp3YFP-Cre or SRC2fl/fl/Foxp3YFP-Cre mice (6 to 8 weeks). Naive CD4+ T cells were first enriched by negative selection using the Naive CD4+ T Cell Isolation Kit, and then CD4+YFP− cells were sorted via FACSAria Fusion (BD) to enable a high purity of ≥99.0%. A total of 4 × 105 naive CD4+YFP− cells were intraperitoneally injected into sex-matched Rag1−/− mice. Three weeks after adoptive transfer, cells from spleen and mLN of Rag1−/− recipient mice were collected and analyzed.
In vivo induction of iTregs by oral tolerance
Splenic cells were collected from OT-II/SRC2fl/fl or OT-II/SRC2fl/fl/CD4Cre mice (6 to 8 weeks), naive CD4+ T cells were first enriched by negative selection using the Naive CD4+ T Cell Isolation Kit, and CD4+CD25− cells were then sorted via FACSAria Fusion to enable a high purity of ≥99.0%. A total of 3 × 106 cells were intraperitoneally injected to sex-matched Rag1−/− mice. After 24 hours, recipient mice were provided with grade VI OVA (20 mg/ml; Sigma-Aldrich) ad libitum in drinking water for 5 days. Drinking water containing OVA was changed every 2 days. Cells were collected from colon, spleen, iLN, and mLN at day 6 for analysis.
Induction and assessment of EAE
EAE was induced and assessed according to the manufacturer’s instructions (Hooke Laboratories, Lawrence, MA). Briefly, Foxp3YFP-Cre or SRC2fl/fl/Foxp3YFP-Cre mice were immunized with 200 mg of MOG35–55 (Hooke Laboratories) in complete Freund’s adjuvant by subcutaneous injection at two dorsal sites of mice, followed by two intraperitoneal injections of 80 ng of pertussis toxin at days 0 and 1. The severity of EAE was monitored and evaluated on a scale from 0 to 5 according to Hooke Laboratories’ guideline. Briefly, 0 represents no disease. 1 represents paralyzed tail. 2 represents hindlimb weakness. 3 represents hindlimb paralysis. 4 represents hindlimb and forelimb paralysis, and 5 represents moribund and death. When a mouse was euthanized because of severe paralysis, a score of 5 was entered for that mouse for the rest of the experiment.
In vivo Treg suppression assay
Colitis was induced in sex-matched Rag1−/− mice by intraperitoneally injecting 4 × 105 CD45RBhiCD25−CD4+ naive T cells sorted from the spleen of C57BL mice (8 to 10 weeks). For natural Treg suppression assay, 2 × 105 CD4+YFP+ Tregs sorted from the spleen of 6- to 8-week-old Foxp3YFP-Cre or SRC2fl/fl/Foxp3YFP-Cre mice were mixed with 4 × 105 CD45RBhiCD25−CD4+ naive T cells from C57BL mice and injected into sex-matched Rag1−/− mice. For iTreg suppression assay, iTregs were first induced in vitro from naive CD4+ cells from Foxp3YFP-Cre or SRC2fl/fl/Foxp3YFP-Cre mice for 48 hours, and CD4+YFP+ iTregs were sorted out. In total, 2 × 105 CD4+YFP+ iTregs were mixed with 4 × 105 CD45RBhiCD25−CD4+ naive T cells from C57BL mice and injected to sex-matched Rag1−/− mice as above. Mice were weighed immediately following T cell transfer and weekly thereafter. Seven to 8 weeks after cell transfer, colon, spleen, and mLN were removed from Rag1−/− recipient mice for analysis.
In vitro Treg suppression assay
Sorted CD4+CD25− T cells were labeled with CellTrace Violet (C34557, Invitrogen) and served as Tresp cells. Tresp cells (6 × 105 cells/ml) were cocultured with CD4+YFP+ Tregs sorted from the spleens of Foxp3YFP-Cre or SRC2fl/fl/Foxp3YFP-Cre mice in 48-well plates [precoated with rabbit anti-hamster (0.1 mg/ml)] in culture medium supplemented with hamster anti-CD3 (0.25 μg/ml), hamster anti-CD28 (1 μg/ml), and IL-2 (20 ng/ml) for 3 days. For iTreg, naive CD4+ cells were purified and cultured under the Treg differentiation condition for 48 hours, and CD4+YFP+ iTregs were sorted and cocultured with Tresp cells for 3 days. The ratios of Tresp cells to Tregs were 1:0, 1:1, and 4:1 for Tregs sorted from mice and 1:0, 1:1, and 2:1 for iTregs sorted from in vitro differentiation. Proliferation of Tresp cells was assessed by flow cytometry.
Histology study
Tissues were cleaned and fixed with 4% paraformaldehyde, embedded in paraffin, and then sectioned and stained with hematoxylin and eosin.
RNA-seq and analysis
Naive CD4+ T cells isolated from Foxp3YFP-Cre or SRC2fl/fl/Foxp3YFP-Cre mice were differentiated into Tregs in 24-well plates in the presence of TGFβ (5 ng/ml), anti–IL-4, and anti-IFNγ for 36 hours. Naive CD4+ T cells and CD4+ cells after 36 hours of Treg differentiation were collected and subjected to RNA extraction with the RNeasy Mini Kit (QIAGEN). Each group has three replicates from different mice. Quality control, library preparation, and sequencing were performed at Novogene. The analysis was performed through Partek Flow. Briefly, the sequence reads were aligned to the mouse whole genome (GRCm38) with validation of quality through prealignment and postalignment quality assurance (QA)/quality control (QC). Aligned reads were further subjected to quantification using the Partek E/M algorithm and normalization to counts per million (CPM) with 0.001 added to each. The identification of differentially expressed features was performed through the Partek GSA algorithm that applies multiple statistical models to each gene. Genes with total counts over 10 were considered to be statistically expressed in the cells.
Reverse transcription quantitative real-time PCR
Total RNA of cells was extracted according to the manufacturer’s guide using the RNeasy Mini Kit (QIAGEN). The first-strand cDNA synthesis was performed by reverse transcription using a Tetro cDNA synthesis kit (Bioline). Subsequent qPCR was performed using PowerUp SYBR Green Master Mix (Applied Biosystems) in the QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific). The primers used for qPCR are listed in table S1. The amplification efficiency of all primers has been tested, and the optimized conditions were used in all qPCRs. Gene expression was calculated with the ∆∆Ct method normalized to the control gene encoding β-actin, and all measurements were performed in triplicate.
Retroviral transduction
Vectors were firstly transfected to Platinum-E (Plat-E; Cell Biolabs) retroviral packaging cells by using BioT transfection reagent (Bioland Scientific) followed by a changing fresh medium at 24 hours. The virus-containing medium collected at 48 and 72 hours was filtered with a 0.45-μm polyvinylidene difluoride (PVDF) syringe filter (Millipore), followed by either direct transduction to T cells or storing at −80°C for later use. Naive CD4+ cells were activated by hamster anti-CD3 (0.25 μg/ml) and hamster anti-CD28 antibodies (1 μg/ml) in precoated plates for 20 hours before transduction. Transduction to activated CD4+ T cells was performed by spin infection with viral supernatants (2500g, 30°C for 2 hours) in the presence of polybrene (10 μg/ml; Sigma-Aldrich). Afterward, the plates were kept in the incubator at 37°C for 3 hours. The viral supernatant was replaced by a fresh culture medium with polarizing cytokines and antibodies for Treg differentiation.
Chromatin immunoprecipitation and DNA sequencing
ChIP was performed with the ChIP-IT High Sensitivity Kit (Active Motif, 53040). Briefly, a total of 2 × 107 CD4+ cells from SRC2fl/fl or SRC2fl/fl/CD4Cre after Treg differentiation were fixed and sheared as described in the ChIP-IT High Sensitivity manual. ChIP reactions were then performed on 30 μg of the prepared chromatin using specific antibodies (anti-SRC2 from Bethyl or anti-NFAT1 from Cell Signaling Technology) overnight, followed by precipitation with protein G agarose beads. DNA was recovered for sequencing or quantitative reverse transcription PCR to quantify specific DNA fragments that were precipitated. For sequencing, ChIP-enriched samples were sequenced on NovaSeq PE100 at TGen. The analysis was performed through Partek Flow. Briefly, the sequence reads were aligned to the mm10 mouse genome with validation of quality through prealignment and postalignment QA/QC. The enrichment of SRC2 binding sites across the genome was analyzed using MACS2. The primers used for RT-qPCR are listed in table S1.
Western blotting and immunoprecipitation
For Western blotting, cells were lysed in radioimmunoprecipitation assay buffer containing 20 mM tris-HCl (pH 7.4), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and leupeptin (1 μg/ml) on ice for 45 min and spun down at 15,000 rpm for 10 min at 4°C to collect the extract. The 2× Laemmli sample buffer (Bio-Rad) containing β-mercaptoethanol was mixed with cell extract and heated at 95°C for 5 min. Protein was separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membrane (Millipore). Target proteins were sequentially immunoblotted with relevant primary antibodies and fluorescent secondary antibodies (LI-COR Biosciences) followed by measuring fluorescent intensity with LI-COR Odyssey blot imager (LI-COR Biosciences).
For immunoprecipitation, 1 × 107 cells were lysed in 300 μl of Pierce IP Lysis Buffer (Invitrogen) containing 1% Triton X-100, 20 mM tris-Cl (pH 7.4), 150 mM NaCl, and 5 mM EDTA supplemented with protease inhibitor cocktail (Sigma-Aldrich) on ice for 45 min and spin down at 15,000 rpm for 10 min at 4°C to collect the extract. Five percent of the cell lysate was saved for pre-IP samples. Cell lysates were incubated overnight with the relevant antibodies, and proteins were immunoprecipitated for an additional 4 hours at 4°C with protein A/G Sepharose beads (Millipore). Beads were washed twice by phosphate-buffered saline and by lysis buffer for the last wash. Beads were then suspended in 2× Laemmli sample buffer containing β-mercaptoethanol and heated at 95°C for 5 min. The supernatant containing precipitated proteins was subjected to SDS-PAGE and analyzed by immunoblot.
Deletion of SRC2/NFAT1-binding region on Nr4a2 promoter by CRISPR-Cas9
The CRISPR-Cas9 system was used for deleting the SRC2/NFAT1-binding region on mouse Nr4a2 promoter. Pairs of primers containing sequences of nontargeting control single guide RNAs (sgRNAs), sgRNAs targeting mouse Foxp3, and sgRNAs targeting the upstream or downstream of SRC2/NFAT1-binding region on Nr4a2 promoter were designed and cloned into retro-gRNA-eGFP vector. Plasmids containing sgRNAs targeting the upstream or downstream of SRC2/NFAT1-binding region on Nr4a2 promoter were used together to generate retrovirus (crNr4a2) to delete the target region in CD4+ cells. Similarly, retroviruses were also produced with the plasmids containing nontargeting control sgRNAs (NTC) or sgRNAs targeting mouse Foxp3 (crFoxp3) to serve as a negative and a positive control to monitor the knocking down of Foxp3 upon infection. To confirm the deletion of SRC2/NFAT1-binding region on Nr4a2 promoter, the genomic DNA from infected CD4+ cells was extracted and the SRC2/NFAT1-binding region abundance was assessed by PCR. As a control, we also deleted an adjacent DNA fragment (crNeg) on Nr4a2 promoter using the same approach. The sgRNA sequences for NTC, crNr4a2, crFoxp3, and crNeg and the primers used to evaluate the abundance of targeted deletion fragment on Nr4a2 promoter from genomic DNA are listed in table S1.
Statistics and reproducibility
The results were analyzed for statistical significance with unpaired Student’s t test or one-way analysis of variance (ANOVA) where appropriate. All data are presented as means ± SD. P values are calculated using GraphPad Prism and presented where the statistical significance (P < 0.05) was found.
Acknowledgments
We thank W. S. Pear, M. H. Kaplan, and J. Chen for sharing the plasmids of MSCV-IRES-GFP, IRF4-MIEG-GFP, and MCSV-HA-Nr4a2-IRES-NGFR, respectively. We also thank J. Xu, J. Yu, and M. Boldin for sharing the mice strain of SRC2fl/fl, OT-II, and Foxp3YFP-Cre, respectively. Besides, we appreciate the help from the following City of Hope core facilities: Animal Resource Center, Integrative Genomics Core, Pathology Solid Tumor Core, and Bioinformatics Core. We thank C. S. Jayasena and Z. He for critically reviewing and editing the manuscript. The high-throughput sequence data can be accessed at GEO via record #GSE201431.
Funding: This work was supported by grants from NIH R01-AI109644, R21-AI163256, institutional pilot funding, Jackie and Bruce Barrow Cancer Research Scholars’ Program, and Caltech-CoH Biomedical Initiative. Research reported in this publication included work performed in the animal, genomic, and flow cytometry cores supported under NIH grant P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author contributions: Conceptualization: Z.S., W.Z., and Y.G. Methodology: W.Z., X.C., X.Z., H.W., and I.N. Investigation: W.Z., X.C., X.Z., and H.W. Visualization: W.Z., X.C., X.Z., and H.W. Supervision: Z.S. Writing—original draft: Z.S. and W.Z. Writing—review and editing: Z.S., W.Z., M.F., Y.G., and I.N.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The reagents including DNA construct and mice will be provided by Z.S. pending scientific review and a completed material transfer agreement. Requests for such reagents should be submitted to mta@coh.org. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Table S1
REFERENCES AND NOTES
- 1.Plitas G., Rudensky A. Y., Regulatory T cells: Differentiation and function. Cancer Immunol. Res. 4, 721–725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka A., Sakaguchi S., Regulatory T cells in cancer immunotherapy. Cell Res. 27, 109–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett C. L., Christie J., Ramsdell F., Brunkow M. E., Ferguson P. J., Whitesell L., Kelly T. E., Saulsbury F. T., Chance P. F., Ochs H. D., The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27, 20–31 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Brunkow M. E., Jeffery E. W., Hjerrild K. A., Paeper B., Clark L. B., Yasayko S. A., Wilkinson J. E., Galas D., Ziegler S. F., Ramsdell F., Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27, 68–73 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Wildin R. S., Ramsdell F., Peake J., Faravelli F., Casanova J. L., Buist N., Levy-Lahad E., Mazzella M., Goulet O., Perroni L., Bricarelli F. D., Byrne G., McEuen M., Proll S., Appleby M., Brunkow M. E., X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 27, 18–20 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Curotto de Lafaille M. A., Lafaille J. J., Natural and adaptive foxp3+ regulatory T cells: More of the same or a division of labor? Immunity 30, 626–635 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Lio C. W., Hsieh C. S., A two-step process for thymic regulatory T cell development. Immunity 28, 100–111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burchill M. A., Yang J., Vang K. B., Moon J. J., Chu H. H., Lio C. W., Vegoe A. L., Hsieh C. S., Jenkins M. K., Farrar M. A., Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity 28, 112–121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma J., Ding Y., Fang X., Wang R., Sun Z., Protein kinase C-θ inhibits inducible regulatory T cell differentiation via an AKT-Foxo1/3a-dependent pathway. J. Immunol. 188, 5337–5347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bettelli E., Carrier Y., Gao W., Korn T., Strom T. B., Oukka M., Weiner H. L., Kuchroo V. K., Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Zhou L., Lopes J. E., Chong M. M., Ivanov I. II, Min R., Victora G. D., Shen Y., Du J., Rubtsov Y. P., Rudensky A. Y., Ziegler S. F., Littman D. R., TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature 453, 236–240 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh C. A., Qin L., Tien J. C., Young L. S., Xu J., The function of steroid receptor coactivator-1 in normal tissues and cancer. Int. J. Biol. Sci. 8, 470–485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen S., Wang F., Zhang J., He Z., Ma J., Gwack Y., Xu J., Sun Z., SRC1 promotes Th17 differentiation by overriding Foxp3 suppression to stimulate RORγt activity in a PKC-θ-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 115, E458–E467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Z., Zhang J., Du Q., Xu J., Gwack Y., Sun Z., SRC3 is a cofactor for RORγt in Th17 differentiation but not thymocyte development. J. Immunol. 202, 760–769 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka K., Martinez G. J., Yan X., Long W., Ichiyama K., Chi X., Kim B. S., Reynolds J. M., Chung Y., Tanaka S., Liao L., Nakanishi Y., Yoshimura A., Zheng P., Wang X., Tian Q., Xu J., O’Malley B. W., Dong C., Regulation of pathogenic T helper 17 cell differentiation by steroid receptor coactivator-3. Cell Rep. 23, 2318–2329 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Nikolai B. C., Jain P., Cardenas D. L., York B., Feng Q., McKenna N. J., Dasgupta S., Lonard D. M., O’Malley B. W., Steroid receptor coactivator 3 (SRC-3/AIB1) is enriched and functional in mouse and human Tregs. Sci. Rep. 11, 3441 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W., Xu Z., Zheng Y., Wang J., Qian W., Olsen N., Brand D., Lin J., Zheng S. G., A protocol to develop T helper and Treg cells in vivo. Cell. Mol. Immunol. 14, 1013–1016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R., Campbell S., Amir M., Mosure S. A., Bassette M. A., Eliason A., Sundrud M. S., Kamenecka T. M., Solt L. A., Genetic and pharmacological inhibition of the nuclear receptor RORalpha regulates TH17 driven inflammatory disorders. Nat. Commun. 12, 1–18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu L. F., Boldin M. P., Chaudhry A., Lin L. L., Taganov K. D., Hanada T., Yoshimura A., Baltimore D., Rudensky A. Y., Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 142, 914–929 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiault N., Darrigues J., Adoue V., Gros M., Binet B., Perals C., Leobon B., Fazilleau N., Joffre O. P., Robey E. A., van Meerwijk J. P., Romagnoli P., Peripheral regulatory T lymphocytes recirculating to the thymus suppress the development of their precursors. Nat. Immunol. 16, 628–634 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Shevach E. M., Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity 30, 636–645 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Schmidt A., Oberle N., Krammer P. H., Molecular mechanisms of treg-mediated T cell suppression. Front. Immunol. 3, 51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konopacki C., Pritykin Y., Rubtsov Y., Leslie C. S., Rudensky A. Y., Transcription factor Foxp1 regulates Foxp3 chromatin binding and coordinates regulatory T cell function. Nat. Immunol. 20, 232–242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koizumi S. I., Ishikawa H., Transcriptional regulation of differentiation and functions of effector T regulatory cells. Cell 8, 939 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvisi G., Brummelman J., Puccio S., Mazza E. M., Tomada E. P., Losurdo A., Zanon V., Peano C., Colombo F. S., Scarpa A., Alloisio M., Vasanthakumar A., Roychoudhuri R., Kallikourdis M., Pagani M., Lopci E., Novellis P., Blume J., Kallies A., Veronesi G., Lugli E., IRF4 instructs effector Treg differentiation and immune suppression in human cancer. J. Clin. Invest. 130, 3137–3150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekiya T., Kashiwagi I., Inoue N., Morita R., Hori S., Waldmann H., Rudensky A. Y., Ichinose H., Metzger D., Chambon P., Yoshimura A., The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2, 1–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekiya T., Kondo T., Shichita T., Morita R., Ichinose H., Yoshimura A., Suppression of Th2 and Tfh immune reactions by Nr4a receptors in mature T reg cells. J. Exp. Med. 212, 1623–1640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odagiu L., May J., Boulet S., Baldwin T. A., Labrecque N., Role of the orphan nuclear receptor NR4A family in T-cell biology. Front. Endocrinol. 11, 624122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tone Y., Furuuchi K., Kojima Y., Tykocinski M. L., Greene M. I., Tone M., Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9, 194–202 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Li X., Liang Y., LeBlanc M., Benner C., Zheng Y., Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell 158, 734–748 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jennings E., Elliot T. A. E., Thawait N., Kanabar S., Yam-Puc J. C., Ono M., Toellner K. M., Wraith D. C., Anderson G., Bending D., Nr4a1 and Nr4a3 reporter mice are differentially sensitive to T cell receptor signal strength and duration. Cell Rep. 33, 108328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rollins D. A., Coppo M., Rogatsky I., Minireview: Nuclear receptor coregulators of the p160 family: Insights into inflammation and metabolism. Mol. Endocrinol. 29, 502–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newton K., Dixit V. M., Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 4, a006049 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogatsky I., Luecke H. F., Leitman D. C., Yamamoto K. R., Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc. Natl. Acad. Sci. U.S.A. 99, 16701–16706 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogatsky I., Zarember K. A., Yamamoto K. R., Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. EMBO J. 20, 6071–6083 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reily M. M., Pantoja C., Hu X., Chinenov Y., Rogatsky I., The GRIP1:IRF3 interaction as a target for glucocorticoid receptor-mediated immunosuppression. EMBO J. 25, 108–117 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honda K., Takaoka A., Taniguchi T., Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Flammer J. R., Dobrovolna J., Kennedy M. A., Chinenov Y., Glass C. K., Ivashkiv L. B., Rogatsky I., The type I interferon signaling pathway is a target for glucocorticoid inhibition. Mol. Cell. Biol. 30, 4564–4574 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chinenov Y., Gupte R., Dobrovolna J., Flammer J. R., Liu B., Michelassi F. E., Rogatsky I., Role of transcriptional coregulator GRIP1 in the anti-inflammatory actions of glucocorticoids. Proc. Natl. Acad. Sci. U.S.A. 109, 11776–11781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.An J., Ribeiro R. C., Webb P., Gustafsson J. A., Kushner P. J., Baxter J. D., Leitman D. C., Estradiol repression of tumor necrosis factor-alpha transcription requires estrogen receptor activation function-2 and is enhanced by coactivators. Proc. Natl. Acad. Sci. U.S.A. 96, 15161–15166 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cvoro A., Tzagarakis-Foster C., Tatomer D., Paruthiyil S., Fox M. S., Leitman D. C., Distinct roles of unliganded and liganded estrogen receptors in transcriptional repression. Mol. Cell 21, 555–564 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Nwachukwu J. C., Srinivasan S., Bruno N. E., Parent A. A., Hughes T. S., Pollock J. A., Gjyshi O., Cavett V., Nowak J., Garcia-Ordonez R. D., Houtman R., Griffin P. R., Kojetin D. J., Katzenellenbogen J. A., Conkright M. D., Nettles K. W., Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network. eLife 3, e02057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vignali D. A., Collison L. W., Workman C. J., How regulatory T cells work. Nat. Rev. Immunol. 8, 523–532 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Malley B. W., SRC-2 coactivator: A role in human metabolic evolution and disease. Mol. Med. 26, 45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson A. B., O’Malley B. W., Steroid receptor coactivators 1, 2, and 3: Critical regulators of nuclear receptor activity and steroid receptor modulator (SRM)-based cancer therapy. Mol. Cell. Endocrinol. 348, 430–439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S., Shang Y., Regulation of SRC family coactivators by post-translational modifications. Cell. Signal. 19, 1101–1112 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Tokuoka H., Hatanaka T., Metzger D., Ichinose H., Nurr1 expression is regulated by voltage-dependent calcium channels and calcineurin in cultured hippocampal neurons. Neurosci. Lett. 559, 50–55 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Togashi Y., Shitara K., Nishikawa H., Regulatory T cells in cancer immunosuppression—Implications for anticancer therapy. Nat. Rev. Clin. Oncol. 16, 356–371 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Rohira A. D., Yan F., Wang L., Wang J., Zhou S., Lu A., Yu Y., Xu J., Lonard D. M., O’Malley B. W., Targeting SRC coactivators blocks the tumor-initiating capacity of cancer stem-like cells. Cancer Res. 77, 4293–4304 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suresh S., Durakoglugil D., Zhou X., Zhu B., Comerford S. A., Xing C., Xie X. J., York B., O’Donnell K. A., SRC-2-mediated coactivation of anti-tumorigenic target genes suppresses MYC-induced liver cancer. PLOS Genet. 13, e1006650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song X., Chen J., Zhao M., Zhang C., Yu Y., Lonard D. M., Chow D. C., Palzkill T., Xu J., O’Malley B. W., Wang J., Development of potent small-molecule inhibitors to drug the undruggable steroid receptor coactivator-3. Proc. Natl. Acad. Sci. U.S.A. 113, 4970–4975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y., Lonard D. M., Yu Y., Chow D. C., Palzkill T. G., O’Malley B. W., Small molecule inhibition of the steroid receptor coactivators, SRC-3 and SRC-1. Mol. Endocrinol. 25, 2041–2053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Table S1