Abstract
The Hi-C technique has been shown to be a promising method to detect structural variations (SVs) in human genomes. However, algorithms that can use Hi-C data for a full-range SV detection have been severely lacking. Current methods can only identify interchromosomal translocations and long-range intrachromosomal SVs (>1 Mb) at less-than-optimal resolution. Therefore, we develop EagleC, a framework that combines deep-learning and ensemble-learning strategies to predict a full range of SVs at high resolution. We show that EagleC can uniquely capture a set of fusion genes that are missed by whole-genome sequencing or nanopore. Furthermore, EagleC also effectively captures SVs in other chromatin interaction platforms, such as HiChIP, Chromatin interaction analysis with paired-end tag sequencing (ChIA-PET), and capture Hi-C. We apply EagleC in more than 100 cancer cell lines and primary tumors and identify a valuable set of high-quality SVs. Last, we demonstrate that EagleC can be applied to single-cell Hi-C and used to study the SV heterogeneity in primary tumors.
Deep-learning–based framework enables the prediction of a full range of structural variations from chromatin interactions.
INTRODUCTION
Structural variations (SVs), including deletions, inversions, duplications, and translocations, can directly contribute to tumorigenesis and other diseases through multiple mechanisms. SVs can lead to the deletion of tumor suppressor genes or duplication of proto-oncogenes (1) or promote the formation of oncogenic fusion genes (2). More recently, it has been shown that SVs can bring distal enhancers to the proximity of proto-oncogenes and cause the up-regulation of oncogenic gene expression through a mechanism termed enhancer hijacking (3, 4). The discovery of recurrent SVs has greatly advanced our knowledge about tumorigenesis and led to effective targeted therapy (5).
Despite their importance, genome-wide detection of SVs remains a challenging problem. Traditionally, karyotyping has been the major method to detect various genetic disorders in the clinic; however, it is an inherently low-throughput and low-resolution method (6). Microarray has been used to identify gains and losses of genetic materials, but it has limitations in detecting copy number neutral events such as inversions and balanced translocations (7). More recently, short-read whole-genome sequencing (WGS) has been widely used to identify a variety of genomic variations due to their high resolution, high throughput, and simplicity (8–13). However, because of the mappability issue of short reads, it is difficult to detect SVs at repetitive regions using WGS (11). The advent of long-read sequencing such as PacBio and Nanopore has partly alleviated the mappability issue (14, 15). However, these technologies have a relatively high sequencing error rate and also need deep sequencing for SV detection (>20×) (16).
Recently, we and other groups showed that Hi-C, a technique that was originally proposed to study three-dimensional (3D) genomic architectures, can also be used for systematic SV detection with as little as 1× genome coverage (11, 17–20). As SVs induce de novo chromatin interactions across the breakpoints, when Hi-C reads are mapped to the reference genome, different types of SVs are characterized by aberrant interaction blocks with different orientations. Identifying SVs is essentially the same as identifying and annotating such blocks on a Hi-C map. Compared to WGS and nanopore that require direct breakpoint spanning reads to detect SVs, such property of Hi-C substantially decreases the sequencing depths that are needed for SV detection and also gives Hi-C higher chances to detect SVs at repetitive regions, as long as the adjacent regions of breakpoints are mappable. So far, three methods have been proposed to predict SVs with Hi-C data. The Hi-C breakfinder that we codeveloped is the first algorithm of this kind, where we use an iterative approach to search for abnormal interaction blocks with significantly higher interaction frequencies compared with a background model (18). HiCtrans identifies translocation breakpoints by searching for signal changepoints on interchromosomal contact matrices of each chromosome pair (17). More recently, Wang et al. (19) proposed a new method called HiNT-TL for translocation detection, which is based on the identification of regions with both unusually high interaction frequencies and uneven distribution of interaction strengths.
However, all current methods have their limitations. HiCtrans and HiNT-TL cannot predict intrachromosomal SVs, which usually accounts for a large portion of all SVs in a cancer genome (13). Although Hi-C breakfinder can identify interchromosomal translocations, it can only detect large intrachromosomal SVs with a size >1 Mb (Table 1). The challenge for short-range SV detection is that Hi-C maps typically contain features such as topologically associating domains (TADs) and chromatin loops (18), which are usually less than 1 Mb, and such patterns make the accurate detection of SV challenging. Furthermore, all three methods still have less-than-optimal resolution. Therefore, we develop EagleC, a framework that combines deep-learning and ensemble-learning strategies to predict a full range of SVs at high resolution. We show that EagleC outperforms existing methods in both precision and recall rates. Furthermore, we demonstrate that EagleC can be used as a general framework to predict SVs in many other 3C-based platforms, such as HiChIP, ChIA-PET, capture Hi-C, and even single-cell Hi-C (scHi-C). With the pretrained models, we predicted SVs in over 100 cancer cell lines or primary tumors. Pan-cancer analysis of these datasets showed that the location and formation of SVs are closely associated with 3D chromatin architectures.
Table 1. Comparing methods for detecting SVs.
|
Interchromosomal
translocation |
Intrachromosomal
SVs (>1 Mb) |
Intrachromosomal
SVs (<1 Mb) |
Applicable to
other 3C-based techniques (ChIA-PET, HiChIP, capture Hi-C, and scHi-C) |
Support
nonhuman genome |
Gene fusions | |
| HiCtrans | √ | √ | ||||
| HiNT-TL | √ | |||||
| Hi-C breakfinder | √ | √ | ||||
| EagleC | √ | √ | √ | √ | √ | √ |
RESULTS
Overview of the EagleC framework
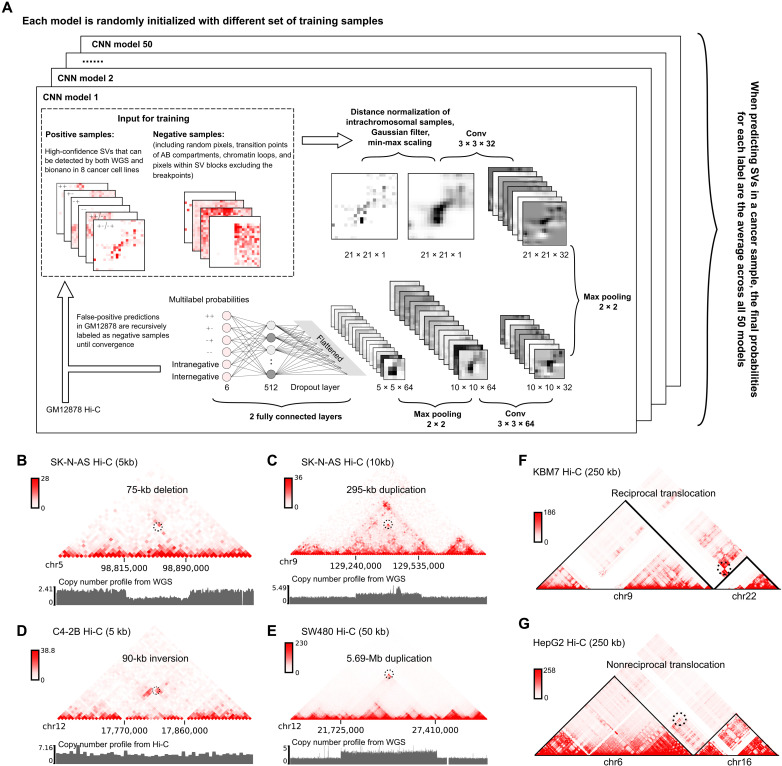
Identifying SVs from a Hi-C map is essentially a multilabel image classification problem in machine learning. There are multiple types of SVs, and each type is characterized by a unique pattern on a Hi-C contact map (figs. S1 to S4) (18). For example, we draw three consecutive fragments A, B, and C in fig. S1. A deletion of fragment B will result in the junction of the 3′-end of fragment A and the 5′-end of fragment C (left of fig. S1A). Because of the spatial proximity, there will be strong chromatin interaction signals between the 3′-end of fragment A and the 5′-end of fragment C. However, when we map the Hi-C reads of the sample to the reference genome, we will see an abnormal increase in the interactions between fragment A and fragment C (middle of fig. S1A). As a result, in the submatrix centered at the breakpoints, there will be an increase in interactions in the upper-right quadrant. Similarly, tandem duplication sequentially links the original DNA fragment and the duplicated fragment, resulting in strong interactions in the lower-left quadrant of the submatrix (fig. S1B); inverted duplication causes aberrant signals either in the upper-left or lower-right quadrants (fig. S1, C and D), depending on the direction of the inverted DNA fragment. For inversions (fig. S1E) and reciprocal translocations (fig. S2), de novo interactions are formed on the opposite sides of the breakpoints, resulting in a “butterfly shape” on the Hi-C map. In our framework, an SV with the “+−” label corresponds to the fusion of the 3′-end of a fragment to the 5′-end of another fragment, while the “++” label corresponds to the 3′-to-3′ fusion, “−+” corresponds to the 5′-to-3′ fusion, and “−−” corresponds to the 5′-to-5′ fusion.
Figure 1A describes the overall design of the EagleC framework. The positive training samples are defined as the Hi-C contact matrices surrounding a set of high-confidence SVs, which were detected by both WGS and optical mapping in eight cancer cell lines (A549, Caki2, K562, LNCaP, NCI-H460, PANC-1, SK-N-MC, and T47D) (18). We found that the original samples demonstrate severely imbalanced class distributions (Materials and Methods and table S1). To avoid the model biased toward any specific classes during the training, we proposed a data augmentation algorithm based on Poisson distributions to make sure each class has a similar number of samples (Materials and Methods). Furthermore, to make the model able to distinguish real SV signals from false-positive signals induced by normal 3D genomic features, we sampled similar numbers of intrachromosomal and interchromosomal submatrices from the Hi-C map of a normal cell line GM12878 (21) and labeled them as “intranegative” and “internegative,” respectively. These negative samples include matrices surrounding random pixels, chromatin loops, and the transition points of A/B compartments. We also included matrices from the cancer Hi-C data that are located in an SV block but not overlapping with the breakpoint as an additional negative dataset.
Fig. 1. EagleC predicts a full range of high-resolution SVs from chromatin interaction data.
(A) Workflow of the EagleC framework. (B to G) Examples showing different types of SVs predicted by EagleC. The black dashed circle indicates the SV breakpoint position in each case. The resolution of each Hi-C map is labeled within the parentheses. (B) A short-range (75 kb) heterozygous deletion predicted in the SK-N-AS cells. (C) A short-range (295 kb) duplication predicted in the SK-N-AS cells. (D) A short-range (90 kb) inversion predicted in the C4-2B cells. (E) A long-range (5.69 Mb) duplication predicted in the SW480 cells. (F) A reciprocal translocation predicted in the KBM7 cells. (G) A nonreciprocal translocation predicted in the HepG2 cells.
Because the strong diagonal Hi-C signals can confound the detection of short-range SVs, in the preprocessing steps, EagleC corrects distance effects for intrachromosomal matrices by using the distance-averaged signals (fig. S3, A and B). To alleviate potential data noise, each input matrix is then convolved with a 2D Gaussian filter followed by min-max scaling.
The inputs to the convolutional neural network (CNN) are 21 × 21 grayscale images, which go through two convolutional layers, each followed by a max-pooling layer. The probabilities of each label (++, +−, −+, −−, intranegative, and internegative) are calculated from two fully connected layers using the sigmoid activation. Before the output layer, we insert a dropout layer with a dropout probability of 0.5 to avoid overfitting.
One important component of the EagleC framework is that it performs an iterative learning procedure to gradually improve the model specificity. After each round of training, the model is used to perform a genome-wide prediction in GM12878 Hi-C. As GM12878 is a karyotypically normal cell line, all the predictions will be considered as false positives and randomly selected as additional negative samples in the next round of training. Such processes are repeated until the convergence is observed.
To further optimize the sensitivity and specificity of the framework, we perform an ensemble learning procedure. In total, 50 models are independently trained using the same iterative approach described above, with each model randomly initialized with different set of training samples. When predicting SVs in a novel sample, the final probability scores are determined as the average across all 50 models, and a pixel will be reported as an SV breakpoint if the probability of at least one positive label (++, +−, −+, and −−) is greater than a predefined cutoff (Materials and Methods).
We trained a series of EagleC models optimized for various sequencing depths using down-sampled versions of the training samples (Materials and Methods). To investigate the performance of EagleC, we predicted SVs (unless noted, all SVs reported in this study are at the 5-kb resolution) in other cancer Hi-C datasets that were not used in the training procedure (table S2). EagleC successfully predicted different types of SVs, including short-range SVs with breakpoint distance less than 1 Mb or even 100 kb (Fig. 1, B to D), large intrachromosomal SVs (Fig. 1E), reciprocal interchromosomal translocations (Fig. 1F), and nonreciprocal interchromosomal translocations (Fig. 1G).
EagleC outperforms existing methods in detecting SVs on Hi-C maps
We first visually inspected the predictions and found that nearly all blocks with abnormally high interaction frequencies were predicted as SVs, suggesting high sensitivity of the framework (Fig. 2A). We then examined closely individual loci and compared the predictions from EagleC and Hi-C breakfinder (18). In many cases, although EagleC and Hi-C breakfinder predicted the same SV blocks, the exact coordinates of the predicted breakpoints were different, and the EagleC-predicted breakpoints were more likely to be validated by WGS (Fig. 2A, regions “A,” “C,” “D,” and “E”). Further, EagleC predicted more precise breakpoints at the 5-kb resolution than Hi-C breakfinder predictions, which are usually 100-kb resolution (block with a dashed line in regions “B” and “D” in Fig. 2A).
Fig. 2. EagleC outperforms existing methods in both precision and recall.
(A) (Left) Genome-wide Hi-C map and the predicted SVs (black circles) in BT-474. (Right) Enlarged Hi-C maps of the indicated SV regions. The SV breakpoints detected by different methods are highlighted using different marks on each map. (B) Number of SVs predicted by EagleC and Hi-C breakfinder in BT-474, HCC1954, and MCF7 cells with levels of validation by orthogonal methods. For Hi-C breakfinder, we only counted SVs reported at the 10-kb resolution. (C) Precision and recall rates of SVs that can only be predicted by EagleC, SVs that can only be predicted by Hi-C breakfinder, and SVs that can be predicted by both methods. (D) The number of SVs predicted by EagleC and Hi-C breakfinder in additional 26 cancer samples with both Hi-C and WGS data available. (E and F) Recall rates and precision rates of SVs predicted by EagleC and Hi-C breakfinder in 26 cancer samples. Each dot represents an individual sample. The P values were computed using the two-sided Wilcoxon signed-rank test. (G) Size distributions of intrachromosomal SVs detected by EagleC and Hi-C breakfinder. Data were merged from 29 cancer samples. (H) Number of different range of SVs predicted by EagleC and Hi-C breakfinder with validation ratios by WGS. Short SVs, intrachromosomal SVs with breakpoint distance less than 1 Mb; long SVs, intrachromosomal SVs with breakpoint distance greater than 1 Mb; TL, interchromosomal translocations.
Next, we systematically evaluated the performance of EagleC by comparing it with all existing methods, HiCtrans (17), Hi-C breakfinder (18), and HiNT-TL (19) (Table 1). We used three breast cancer cell lines BT-474, HCC1954, and MCF7 as the benchmark datasets, as there are Hi-C, WGS, and nanopore data available in the same cell lines. Because HiCtrans and HiNT-TL can only detect interchromosomal translocations, we first focused on interchromosomal translocations alone. The reported results by each method differed greatly (fig. S5, A and B). First of all, different methods predicted a different number of translocation candidates. For example, in the MCF7 cell line, EagleC and Hi-C breakfinder predicted 154 and 116 translocations, respectively. HiCtrans reported the largest number of translocations (n = 520), while HiNT-TL detected the smallest number of translocations (n = 28). In terms of the resolutions at which the translocations were reported, EagleC predicted translocations at the highest resolution among all methods at 5 kb. The translocations reported by HiCtrans were at 10 or 20 kb; translocations reported by Hi-C breakfinder were at a mixture of 10-kb, 100-kb, and 1-Mb resolutions; and nearly all translocations reported by HiNT-TL were at the 100-kb resolution (fig. S5A). To further investigate the performance of each method, we compared the translocation predictions from each method with a reference translocation set defined by WGS and nanopore for each cell line (Materials and Methods). As shown in fig. S5B, EagleC outperforms all the other methods with both higher precision rates and higher recall rates in all three cell lines. Specifically, although HiCtrans detected three times as many interchromosomal translocations as EagleC, it recalled fewer validated SVs due to its redundant false-positive predictions within a single SV block (fig. S5C).
More in-depth analysis between EagleC and Hi-C breakfinder that includes both inter- and intrachromosomal SVs
Then, we performed more in-depth comparisons between EagleC and Hi-C breakfinder, as they are currently the only methods that can identify intrachromosomal SVs. Notably, EagleC detected 2.4-fold (244 versus 100), 2.6-fold (410 versus 157), and 4.8-fold (244 versus 51) as many SVs (including interchromosomal translocations and intrachromosomal SVs) as Hi-C breakfinder in BT-474, HCC1954, and MCF7, respectively (Fig. 2B). At the same time, EagleC achieved notably higher precision rates than Hi-C breakfinder in these cell lines. When allowing 20-kb mismatches for either side of the breakpoints, 84.8, 76.3, and 73.8% of SVs predicted by EagleC in BT-474, HCC1954, and MCF7 can be validated by either WGS or nanopore, while corresponding rates for Hi-C breakfinder are only 55.0, 55.4, and 54.9% (Fig. 2B and fig. S6, A to C). When we increased the allowed mismatch from 20 to 100 kb, the validation rates for EagleC nearly stayed the same, while the rates for Hi-C breakfinder increased by 11.0, 10.8, and 7.8% in the three cell lines, respectively, which suggests that Hi-C breakfinder failed to predict the exact breakpoint positions within the SV block for a portion of SVs (Fig. 2A and figs. S6, A to C). In BT-474, 24.2% (59 of 244) of the EagleC-predicted SVs matched 59.0% (59 of 100) of the Hi-C breakfinder predictions. Of the 185 SVs that are unique to EagleC, 83.2% (154 of 185) can be validated by either WGS or nanopore, compared with 2.4% (1 of 41) for Hi-C breakfinder unique SVs (Fig. 2C). Similarly, in HCC1954 and MCF7, 73.4 (232 of 316) and 71.0% (152 of 214) of EagleC-unique SVs can be validated, compared with 7.9 and 0.0% for SVs that are specific to Hi-C breakfinder. On average, EagleC-unique SVs have a 21.9-fold higher precision rate and a 61.0-fold higher recall rate than Hi-C breakfinder unique SVs in these three cell lines (Fig. 2C).
Furthermore, we evaluated the performance of EagleC and Hi-C breakfinder at various sequencing depths by down-sampling the original BT-474 and HCC1954 Hi-C data to nine different depths (ranging from 5 to 175 million contact pairs) (fig. S6D). Notably, EagleC achieved obviously higher precision and recall rates than Hi-C breakfinder at all sequencing depths. In addition, while the recall rates for Hi-C breakfinder reached a plateau at the depth with around 75 million contact pairs, the rates for EagleC kept increasing along with higher sequencing depths, which suggests that the power of Hi-C in SV detection might have been underestimated by previous studies. To evaluate the impact of tumor heterogeneity on SV prediction, we simulated a series of Hi-C datasets by mixing the BT-474/HCC1954 Hi-C with HMEC (human mammary epithelial cells, a normal breast cell line) Hi-C at various fractions while keeping the total sequencing depth at around 200 million contact pairs. Similarly, we observed that EagleC predicted much more SVs with higher accuracy than Hi-C breakfinder at all tumor heterogeneity levels (fig. S6E).
We next extended the analysis to 26 additional cancer cell lines or patient samples with both Hi-C and WGS data available (table S2). Again, we observed that compared with Hi-C breakfinder, EagleC achieved significantly higher recall rates and precision rates in all the 26 cancer samples (Fig. 2, D to F, and fig. S6F). Because of the inherent limitations of the algorithm, Hi-C breakfinder can only detect large intrachromosomal SVs greater than 1 Mb. However, as shown in Fig. 2G, 39.5% of intrachromosomal SVs predicted by EagleC are short-range SVs, with a minimum size of 35 kb. To our surprise, although SVs at this range have been thought hard to be distinguished from other Hi-C contact patterns, they were predicted with even higher accuracy than long-range SVs and translocations (Fig. 2H).
EagleC detects novel fusion genes in cancer
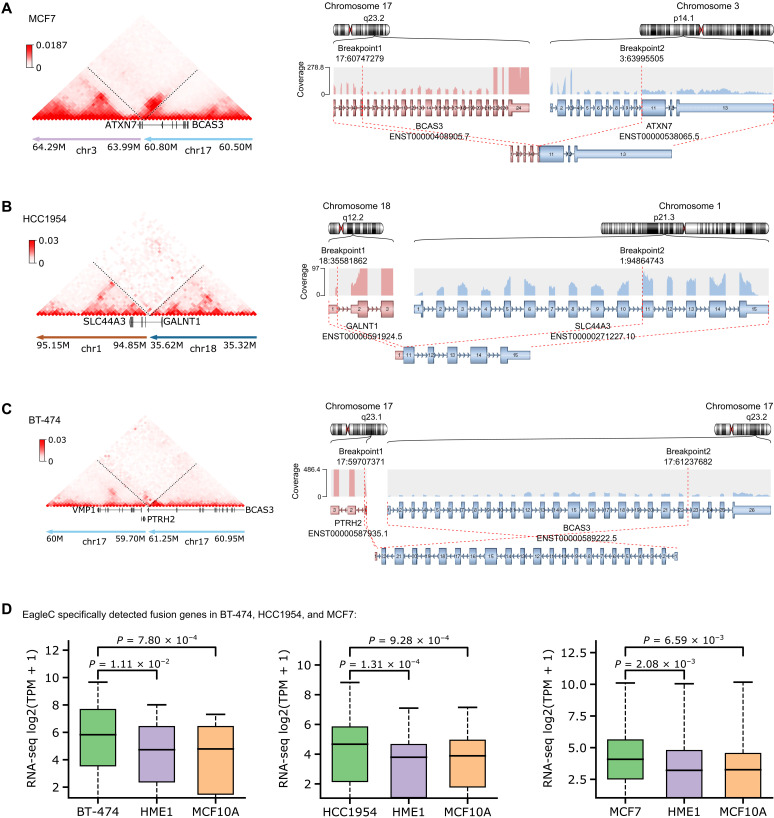
Because we noticed that a sizable portion of EagleC-predicted SVs were missed by both short-read WGS and nanopore (Fig. 2B), we investigated whether such detection can be supported by other evidence. As the RNA sequencing (RNA-seq) data are available for these three cell lines, we predicted fusion genes with the Arriba software (22). As shown in Fig. 3A, EagleC detected breakpoints inside the ATXN7 and BCAS3 genes in MCF7, while the arriba software also predicted the fusion of these two genes (Fig. 3A, right). We showed two more such examples in Fig. 3 (B and C), demonstrating that because of the high-resolution nature of EagleC, it can uniquely predict fusion genes that are missed by WGS and nanopore. We also wanted to point out that the sequencing depth of Hi-C data in these three cell lines is much lower (BT-474, 17×; HCC1954, 11×; and MCF7, 16×) than the WGS (BT-474, 44×; HCC1954, 38×; and MCF7, 38×) and nanopore (BT-474, 31×; HCC1954, 49×; and MCF7, 26×), suggesting that Hi-C can detect a unique set of SVs even with low sequencing depths. Last, we noticed that genes involved in these fusion events were significantly overexpressed in cancer cells, compared with their expression levels in nonmalignant cell lines without the fusion (Fig. 3D).
Fig. 3. Fusion genes uniquely detected by EagleC are overexpressed in cancer cells.
In this analysis, we only included fusion events that were detected by EagleC but missed by both WGS and nanopore. (A to C) Examples of novel gene fusions detected by EagleC (left) and the supporting evidence from RNA-seq data (right). The fusion gene partners, their orientations, and the retained exons in the fusion transcripts were detected and plotted using the arriba software. (D) Normalized expression signals for novel fusion genes detected by EagleC in different breast cancer cell lines versus two nonmalignant breast cell lines (HME1 and MCF10A). TPM, transcripts per kilobase million. The P values were computed using the two-sided Wilcoxon signed-rank test. In each boxplot, the center line indicates the median, the box limits represent the upper and lower quartiles, and the box whiskers indicate the 1.5× interquartile range.
EagleC can accurately predict SVs using other 3C-based techniques
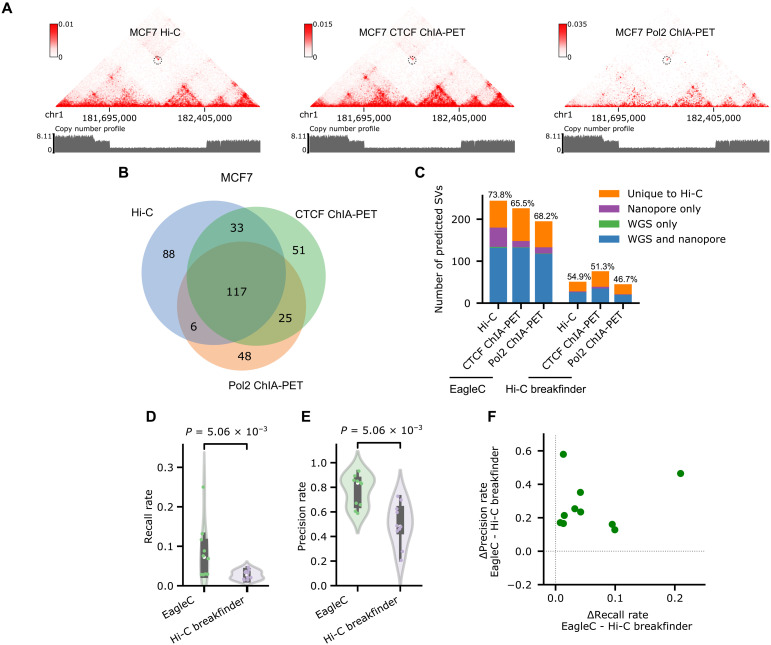
In addition to Hi-C, there are several other 3C-derived techniques. Among them, pulldown-based 3C assays, including Chromatin interaction analysis with paired-end tag sequencing (ChIA-PET) (23), HiChIP (24), Proximity Ligation-Assisted ChIP-seq (PLAC-Seq) (25), and Capture Hi-C (26), are gaining more and more interests because of their efficiency in detecting genome-wide chromatin interactions mediated by a protein or a set of genes of interest. However, the potential of these techniques in SV detection has never been explored by previous studies (Table 1). We hypothesized that the rules we learned for predicting SVs on Hi-C maps are common among all 3C-based platforms. To validate this hypothesis, we focused on the breast cancer cell line MCF7, in which there are WGS, nanopore, Hi-C, CTCF ChIA-PET, and Pol2 ChIA-PET data available (tables S2 and S3). We directly applied the EagleC models trained on Hi-C data to CTCF ChIA-PET and Pol2 ChIA-PET. Overall, EagleC predicted a similar number of SVs in Hi-C, CTCF ChIA-PET, and Pol2 ChIA-PET, and there is a large overlap between the three datasets (Fig. 4, A and B). For instance, EagleC predicted 226 SVs in CTCF ChIA-PET, 66.4% of which were predicted in Hi-C as well. Similarly, 62.8% (123 of 196) of SVs predicted in Pol2 ChIA-PET matched 50.4% (123 of 244) of predictions from Hi-C. We found that EagleC achieved comparable precision rates in both ChIA-PET datasets (CTCF ChIA-PET, 65.5%; and Pol2 ChIA-PET, 68.2%) compared to Hi-C (73.8%) (Fig. 4C). Moreover, we observed that EagleC-predicted SVs have significantly higher recall rates and precision rates than Hi-C breakfinder in all the 10 HiChIP/ChIA-PET datasets with matched WGS data (Fig. 4, D to F, and table S3).
Fig. 4. EagleC accurately predicts SVs on HiChIP and ChIA-PET contact maps.
(A) A heterozygous deletion (chr1, 181,695,000 to 182,405,000) on chromosome 1 is correctly predicted by EagleC on Hi-C, CTCF ChIA-PET, and Pol2 ChIA-PET contact maps of MCF7 cells. The same copy number profile calculated from WGS is shown below each contact map. (B) Venn diagram of SVs predicted by EagleC on different contact maps of MCF7 cells. (C) Number of SVs predicted by Hi-C breakfinder and EagleC on different contact maps of MCF7 cells with levels of validation by orthogonal methods. The ratio of SVs that can be validated by orthogonal methods is indicated above each bar. (D and E) Performance of EagleC and Hi-C breakfinder on 10 HiChIP/ChIA-PET contact maps in cancer samples. Each dot represents an individual sample. The P values were computed using the two-sided Wilcoxon signed-rank test. (F) Differences of precision rates and recall rates between EagleC-predicted and Hi-C breakfinder–predicted SVs on 10 HiChIP/ChIA-PET datasets in cancer.
To investigate whether EagleC models are also transferable to other 3C-based platforms, we collected nine capture Hi-C datasets in mice, with each dataset containing one and only one known SV: (i) a series of duplications ranging from 420 kb to 1.74 Mb that were originally used to study the impact of duplications on TAD structures (fig. S7A) (27); (ii) a 115-kb inversion in mouse forelimb at embryonic day 11.5 (E11.5) (fig. S7B) (28); (iii) a 1.14-Mb inversion in mouse limb buds at E12.5 (fig. S7C) (29); and (iv) a series of inversions ranging from 620 kb to 1.10 Mb, which have an invariable downstream breakpoint and a variable upstream breakpoint (fig. S7D) (30). We found that EagleC was able to predict the known SVs in all these datasets. No other pixels were predicted as SVs, suggesting both high sensitivity and high specificity of EagleC in predicting SVs on capture Hi-C maps.
Detection of SVs in 105 cancer samples
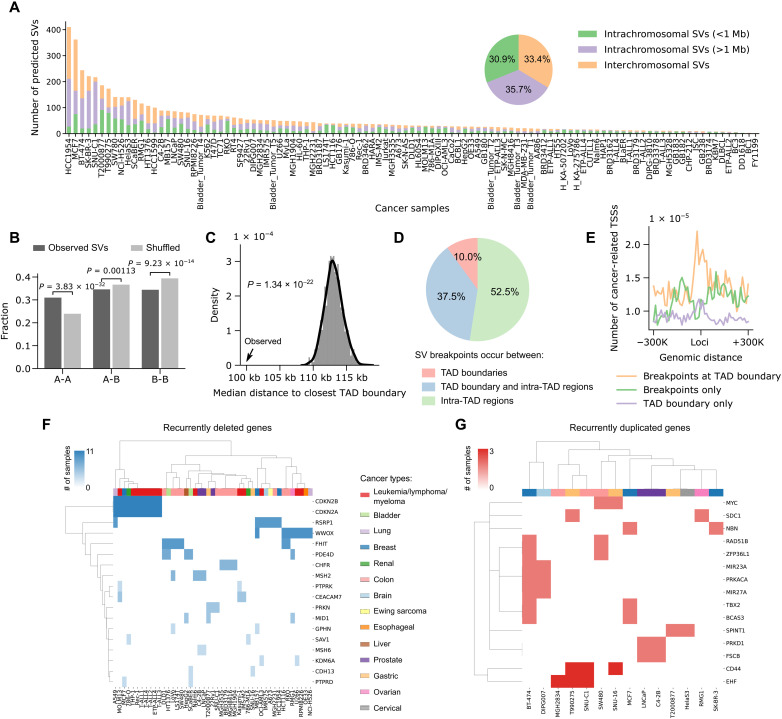
After we have validated our framework in various 3C-based platforms, we applied the trained models to 91 Hi-C datasets and 25 HiChIP/ChIA-PET datasets from 105 cancer cell lines or primary tumors (tables S2 and S3). If multiple datasets are available in the same sample, we combined their results to achieve a more comprehensive set of SV annotations. In total, we predicted 5620 SVs across all the samples, with the number in each sample ranging from 2 to 410 (Fig. 5A and table S4). The highest numbers of SVs are observed in breast cancer cell lines, consistent with previous findings that breast cancer cells frequently contain genomic instability driven chromosomal variations (31). Combining data from all samples, 30.9% of the predicted SVs are short-range SVs (<1 Mb), 35.7% are long-range SVs, and 33.4% are interchromosomal translocations.
Fig. 5. Pan-cancer analysis of SVs in 105 cancer cell lines or patient samples.
(A) Number of short-range intrachromosomal SVs, long-range intrachromosomal SVs, and interchromosomal translocations predicted in each sample. The pie chart shows the percentages of different SV categories based on the data combined from all samples. (B) SVs are significantly enriched in A-to-A and depleted in B-to-B compartments compared to randomly shuffled controls. The P values were calculated using two-sided Z test. (C) SV breakpoints occur significantly closer to TAD boundaries compared to random shuffled controls. The P values were calculated using two-sided Z test. (D) Percentages of SVs with breakpoints occurring between TAD boundaries, between a TAD boundary and intra-TAD regions, and between intra-TAD regions. (E) The TSSs of cancer-related genes are specifically enriched near SV breakpoints at TAD boundaries. (F) Unsupervised clustering of recurrently deleted genes and cancer samples based on the occurrence of genes in each sample. Different cancer types are coded by different colors. (G) Similar to (F), the clustering of recurrently duplicated genes and cancer samples.
Next, we investigated how 3D genome architectures can influence the location and formation of SVs. As genomic variations such as SVs and copy number variations (CNVs) can confound the interpretation of contact maps in cancer, we computed 3D genome features including A/B compartments and TADs for different cancer types using Hi-C data in normal cells/tissues with similar cell of origin (table S5). It has been widely known that the genome can be partitioned into two compartments, with the A compartment associated with open chromatin, and the B compartment associated with closed chromatin, and chromatin interactions within the same compartments (A-A/B-B) are stronger than interactions between different compartments (A-B) (32). We hypothesized that the preexisting chromatin interactions between distal compartments would increase the probability of SV formation between these compartments. To this end, we quantified the proportions of SVs that occurred within the same compartments (A-A/B-B) and between different compartments (A-B). As a control, we randomly shuffled the SV breakpoints in the mappable genome regions 1000 times for each cancer sample, controlling for the ratio of interchromosomal versus intrachromosomal SVs and the sizes of the intrachromosomal SVs. Compared with random controls, SVs are preferentially formed between A-A compartments rather than B-B or A-B compartments (Fig. 5B), and such patterns are largely conserved for different cancer types and different ranges of SVs (figs. S8 and S9).
At the megabase scale, it has been shown that mammalian genomes are organized into TADs (33). TAD boundaries, which are enriched for CTCF binding sites, provide an insulated environment for proper gene regulation. In comparison to an expected distribution derived from randomly shuffled SVs, we found that SV breakpoints are located significantly closer to TAD boundaries, consistent with previous findings that DNA topoisomerase II beta (TOP2B)-mediated DNA double-strand breaks are enriched at anchors of chromatin loops (Fig. 5C and figs. S8 and S9) (34). Overall, around 10% of SVs are formed between TAD boundaries, 37.5% are formed between a TAD boundary and an intra-TAD region, and 52.5% are formed between intra-TAD regions (Fig. 5D and figs. S8 and S9). Moreover, we found that transcription start sites (TSSs) of cancer-related genes are specifically enriched at breakpoint-associated TAD boundaries (Fig. 5E), suggesting that the disruption of TAD boundaries by genomic rearrangements might be an important mechanism for oncogene dysregulation and tumorigenesis.
To further explore the value of our SV annotations, we identified genes that are recurrently affected by short-range SVs in different samples. As expected, we found that the majority of deleted genes are tumor suppressor genes (Fig. 5F), such as CDKN2A/2B (35), WWOX (36), CHFR (37), and MSH2 (38) genes. On the other hand, a lot of genes within the duplicated regions are oncogenes (Fig. 5G), such as MYC, which has been reported to be associated with cell proliferation in multiple cancer types (39), and the CD44 gene, which is a common biomarker of cancer stem cells and encodes a cell-surface glycoprotein involved in tumor initiation and progression (40).
EagleC predicts known interchromosomal translocations in single cells
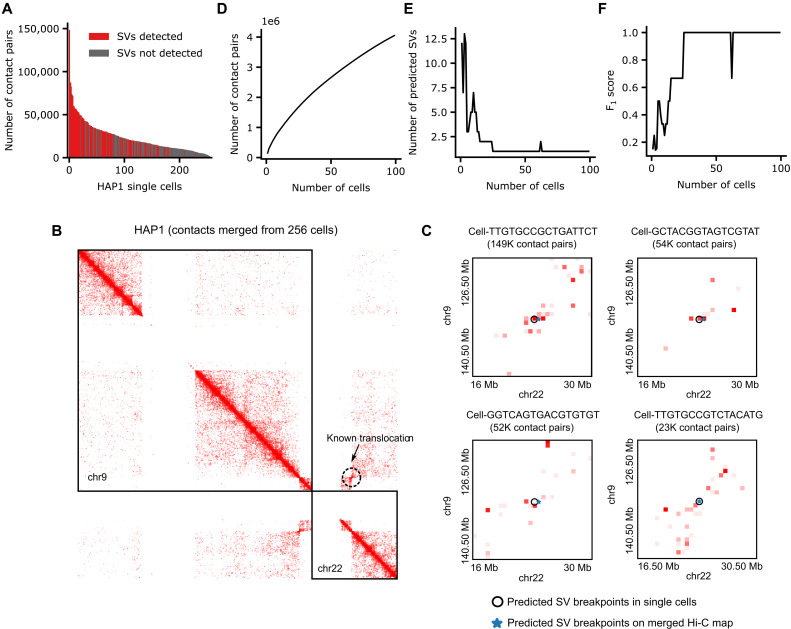
To make EagleC work for scHi-C with limited contact information per cell, we down-sampled contact maps of the same eight cancer cell lines and GM12878 cells to comparable sequencing depths, and retrained the models at the 500-kb resolution (Materials and Methods). Then, we tested EagleC on published scHi-C datasets in HAP1 and K562 (41), both of which are chronic myeloid leukemia cell lines. HAP1 cells contain a reciprocal translocation between chromosome 9 and chromosome 22 (42), while K562 cells contain a nonreciprocal translocation between chromosome 9 and chromosome 22 (43). The HAP1 dataset contains 256 single cells, with a median of 18,793 contacts per cell, while the K562 dataset contains 337 cells, with a median of merely 3974 contacts per cell (Fig. 6A and fig. S10A). Notably, we found that even with these extremely sparse contact matrices, EagleC was able to predict the known chr9-chr22 translocations in single cells (Fig. 6, B and C, and fig. S10, B and C).
Fig. 6. EagleC predicts interchromosomal translocations in HAP1 scHi-C.
(A) Number of Hi-C contact pairs per cell. Red bars and gray bars represent cells with and without SV detected, respectively. (B) There is a known reciprocal translocation between chr9 and chr22 in the HAP1 cell line. (C) EagleC accurately predicts breakpoint coordinates of the known translocation in single cells. The cell ID and corresponding number of contacts are indicated in the title of each contact map. (D to F) The number of contact pairs (D), the number of predicted SVs (E), and the F1 scores of the predicted SVs (F) in a series of pooled single-cell maps.
To systematically investigate the lower limit of contact number for accurately predicting SVs in single cells, we ranked all the 256 HAP1 cells by their sequencing depths and generated a series of contact matrices (contact pairs ranging from 148,635 to 4.05 million) by pooling up to 99 deepest single cells (Fig. 6D). As expected, the number of predicted SVs decreases along with the increasing number of cells (Fig. 6E). By using SVs predicted from the merged Hi-C map as the gold standard SV set, we found that the F1 scores increased with the cell number and reached a plateau of 1 when the cell number reached 25 (1.68 million contact pairs) (Fig. 6F). For K562 cells, we performed a similar analysis by pooling up to 300 deepest K562 single cells, but this time, we counted interchromosomal translocations and intrachromosomal SVs separately (fig. S10, D to H). Again, by using SVs predicted from the merged Hi-C map of all 337 K562 single cells as the gold standard, we observed that the F1 scores for both intrachromosomal SVs and interchromosomal translocations increased with the cell number. However, predicting intrachromosomal SVs needed a higher number of usable reads from more cells to achieve a reasonable performance (fig. S10F).
In conclusion, EagleC can identify both interchromosomal translocations and intrachromosomal SVs in scHi-C data. However, because of insufficient usable reads per cell, pooling contacts from multiple cells can help achieve the most accurate predictions at current stages.
DISCUSSION
Although several methods have been developed to detect SVs using Hi-C data, the power of Hi-C in detecting short-range SVs with breakpoint distance less than 1 Mb has not been achieved, mainly due to the challenge of distinguishing SV signals from other chromatin interaction signals within this range. Here, by taking the advantage of CNNs in image recognition and ensemble learning in avoiding the overfitting problem, we developed EagleC to fill this important gap. For individual models, we applied an iterative training approach to gradually improve their specificity by incorporating negative samples from a normal cell line. We showed that EagleC not only predicted unique short-range SVs but also greatly improved the overall prediction power over existing methods. We demonstrated the feasibility of using Hi-C to detect fusion genes, some of which were missed by both WGS and nanopore. Although our current framework cannot achieve the base-pair resolution, we observed that Hi-C has unique ability in detecting fusion points within introns compared with RNA-seq (Fig. 3, A and B). Moreover, EagleC can serve as a general model to predict SVs using other 3C-based contact maps including ChIA-PET, HiChIP/PLAC-Seq, capture Hi-C, and even scHi-C. With unique properties of different platforms in enriching different set of chromatin interactions, we envision that the application of EagleC in these platforms will boost SV-related discoveries, such as enhancer hijacking (4). Furthermore, by applying EagleC to 116 Hi-C/HiChIP/ChIA-PET datasets, we predicted SVs in 105 cancer samples and found the distributions of SVs on the genome are closely associated with 3D chromatin architectures.
We note that existing methods such as Hi-C breakfinder (18) and HiNT-TL (19) can only be applied to human samples (Table 1), as they rely on the identification of interaction blocks that deviate from the expected interaction frequencies, which were only precalculated for human genomes. In comparison, the contact patterns learned by EagleC are genome agnostic and can be used to predict SVs or judge the accuracy of genome assemblies in any species (fig. S7) (44). Because the data we collected in this study had various sequencing depths and quality, we limited our analyses at the 5-kb resolution and predicted SVs with a minimum size of 35 kb. However, our framework should be able to predict SVs at higher resolutions (1 kb) when sequencing depths are sufficient.
Recent progress in single-cell sequencing techniques has enabled the studies of molecular changes and evolutionary trajectories during cancer development. In this course, multiple algorithms have been developed for identifying single-nucleotide variants (SNVs) and CNVs in single cells (45–47). However, predicting SVs at the single-cell level is still relatively unexplored. Here, by applying EagleC to scHi-C datasets in cancer cell lines (41), we demonstrated that the lower limit of contact number for EagleC to accurately predict SVs is between 1 and 2 million usable reads (Fig. 6F and fig. S10F). It has been shown that several biotin-free scHi-C protocols, such as Dip-C (48), can achieve such level of sequencing depths per cell. On the other hand, single-nucleus methyl-3C sequencing, another method without biotin pulldown of ligation junctions, can simultaneously measure chromatin contacts and DNA methylation levels in the same cells (49). Combining these technologies and EagleC in primary samples will enable the study of SV heterogeneity and potentially identify SVs that are critical for cancer studies.
MATERIALS AND METHODS
Hi-C data processing
For Hi-C/HiChIP/ChIA-PET datasets, if the data had been mapped to hg38 and processed into contact matrices at multiple resolutions, we directly downloaded and used the processed contact matrices in our study; if only raw sequencing data were available, we processed the data using the runHiC Python package (https://pypi.org/project/runHiC/), which is based on the 4DN Hi-C data processing pipeline; otherwise, if the data were originally mapped to hg19 and raw sequencing data were not available, then we converted the coordinates to hg38 using pairLiftOver (https://pypi.org/project/pairLiftOver/; see description below). For Hi-C datasets, we used the CNV-normalized matrices calculated by our recently developed toolkit NeoLoopFinder (4) as input for EagleC. For HiChIP and ChIA-PET datasets, we used the iterative correction and eigenvector decomposition (ICE)-normalized matrices as input for EagleC (50).
For other Hi-C–based SV detection methods, we installed and ran Hi-C breakfinder following https://github.com/dixonlab/hic_breakfinder. We installed and ran HiNT-TL (v2.2.8) following the official guidelines at https://github.com/parklab/HiNT. We downloaded, installed, and ran HiCtrans (hictrans.v3.R) following https://github.com/ay-lab/HiCtrans.
pairLiftOver
To facilitate the processing of Hi-C/HiChIP data that were mapped to a different reference genome (hg19) and did not have raw sequencing data available, we developed a command line tool called pairLiftOver to convert the 2D genomic coordinates of chromatin contacts between assemblies. pairLiftOver is based on the UCSC chain files (https://genome.ucsc.edu/goldenPath/help/chain.html), which describes pairwise alignment between two assemblies. The input to pairLiftOver can be two kinds of pairs files: (i) the pairs format defined by 4DN DCIC (https://github.com/4dn-dcic) and (ii) allValidPairs defined by HiC-Pro (https://nservant.github.io/HiC-Pro/RESULTS.html). Both formats define contact pairs in plain text, with each row representing 2D coordinates of a single pair. pairLiftOver iterates each row of a pairs file and converts the coordinates of both sides using the pyliftover package (https://github.com/konstantint/pyliftover). A pair is retained only if both sides can be uniquely mapped to the target genome. For each row, only columns pertaining to genomic coordinates (columns 2 to 5 for 4DN pairs; columns 2 and 3 and columns 5 and 6 for allValidPairs) are converted and all other columns remain unchanged. The input pairs file can be plain text file, gzip/bgzip compressed file (.gz), or lz4 compressed file (.lz4). By default, pairLiftOver will output a sorted pairs file in the standard 4DN pairs format (https://github.com/4dn-dcic/pairix), containing seven columns: “readID,” “chr1,” “pos1,” “chr2,” “pos2,” “strand1,” and “strand2.” However, users can also choose to output a matrix file in “.mcool” (50) or “.hic” (51) format by setting the parameter “--output-format.”
scHi-C data processing
The FASTQ files of the scHi-C data were downloaded from GSE84920. The cellular demultiplexing was performed by following the pipeline described in the original paper (41), and cells with a low number of reads were filtered out. Then, the demultiplexed reads were aligned to hg19 using BWA-MEM (52) with the parameter “-SP5M.” The BAM files were then parsed into the 4DN pairs format, and the polymerase chain reaction duplications were removed by using pairtools (https://github.com/open2c/pairtools). Only contact pairs with UU/UR/RU flags in the pairs file and with both sides mapped to different restriction fragments were kept for further analysis. Last, we generated contact matrices at the 500-kb resolution by using the cooler Python package (50).
WGS and nanopore data processing
For WGS, the paired-end reads were first mapped to hg38 by BWA-MEM (v0.7.17), and duplicate reads were removed by Picard (v2.6.0) (https://github.com/broadinstitute/picard). Then, we used two methods to detect SVs from the same BAM files: (i) We ran Delly (v0.8.7) with parameters “-t ALL -q 20 -s 15” (53), and (ii) we ran smoove (v0.2.6) (https://github.com/brentp/smoove) with default parameters, which is an optimized pipeline based on lumpy (54). When we evaluated the precision rate for SV predictions from Hi-C (Figs. 2, D, F, and H, and 4, E and F; and fig. S6F), the union of SVs detected by delly and smoove was used as the reference SV set; when we evaluated the recall rate (Figs. 2E and 4, D and F; and fig. S6F), the intersect of delly and smoove was used as the reference SV set. We also inferred copy number profiles from WGS using Control-FREEC (v11.6) (55). Multiple ploidy values (“ploidy = 1,2,3,4”) were specified in the configuration file to enable the program to automatically select the one that explains the most observed copy number alterations.
For nanopore, we applied three methods for SV detection: sniffles (56), Picky (57), and svim (58). To run sniffles (v1.0.12) and svim (1.4.2), the reads were aligned to hg38 using minimap2 (v2.20) (59) with parameters “-ax map-ont -L,” and after we have obtained the alignments in BAM format, we ran both methods with default parameters. For Picky, we used LAST (v1256) to align reads. To speed up the calculation, we followed the official pipeline to split the raw FASTQ files into multiple chunks, with each chunk containing 800,000 reads (https://github.com/TheJacksonLaboratory/Picky/wiki/Cluster-Support). We then ran Picky on these chunk files separately and combined results from all chunks. To evaluate the precision rates in Figs. 2 (B and C) and 4C and fig. S6 (A to E), the union of WGS-detected SVs and nanopore-detected SVs was used as the reference SV set, where WGS SVs were defined as the union of SVs from delly and smoove, and nanopore SVs were defined as the union of SVs from svim, sniffles, and Picky. To evaluate the recall rates in Fig. 2C and fig. S6 (D and E), the intersect of WGS-detected SVs and nanopore-detected SVs was used as the reference SV set.
RNA-seq data processing
The RNA-seq data for BT-474, HCC1954, MCF7, HME1, and MCF10A cell lines were downloaded from GSE152908. The raw FASTQ files were first processed using fastp (v0.20.1) with parameters “--detect_adapter_for_pe --trim_poly_x --correction.” The trimmed reads were then processed using the ENCODE long-read RNA-seq pipeline (https://github.com/ENCODE-DCC/long-rna-seq-pipeline) with default parameters to calculate both the genome-wide plus and minus strand signal tracks and gene quantifications.
The gene fusions were detected using Arriba (v2.2.1) (22) with suggested parameters using chimeric alignments outputted by STAR (v2.7.10a) as input (https://arriba.readthedocs.io/en/latest/workflow/).
Down-sample Hi-C contact maps to a specified sequencing depth
Our down-sampling procedure assumes that the number of contacts between two genomic regions follows a binomial distribution. Suppose there are totally Ntotal contact pairs in the original matrix M, and we want to generate a down-sampled matrix M′ with around Nsample contact pairs, i.e., with a down-sample rate of α = Nsample/Ntotal, Nsample < Ntotal. To this end, for each nonzero pixel in M, we designate the corresponding contact frequency in M′ a random integer number generated from a binomial distribution with parameters Mij and α, where Mij is the contact count of the 100% Hi-C matrix between bin i and bin j. The same algorithm was also used when we mixed BT-474/HCC1954 Hi-C with HMEC Hi-C at different fractions.
Collection of training samples and data augmentation
In our previous work (18), we have compiled comprehensive SV lists for eight cancer lines (A549, Caki2, K562, LNCaP, NCI-H460, PANC-1, SK-N-MC, and T47D) from multiple experimental platforms. To create a high-quality positive training set for EagleC, we manually curated a set of high-confidence SVs that can be detected by both WGS and optical mapping, and have Hi-C signals surrounding the breakpoints. In total, we obtained 243 such SVs in eight cell lines. We noticed that this original SV set demonstrated severely imbalanced distributions in two aspects: (i) the numbers of SVs with different orientations (++, 37; +−, 96; −+, 61; −−, 29; ++/−−, 15; +−/−+, 5) and (ii) the numbers of SVs at different ranges (short-range SVs, 67; and long-range SVs and translocations, 176). To avoid any biases introduced by such imbalance during the training, and to boost the number of samples, we proposed a data augmentation algorithm as follows: Given a submatrix Mij with a size of 21 × 21, where each entry represents the raw contact frequency between bin i and bin j, we can generate a matrix of the same size, where the value of each entry follows a Poisson distribution with λ = Mij. By using this algorithm as the core, we increased the positive training set to ~3000 samples for each individual model of the EagleC framework and made sure that these samples had balanced distributions in both different SV orientations and different genomic ranges.
For negative training samples, the chromatin loops in GM12878 were downloaded from a previous study (21) with the coordinates converted from hg19 to hg38 using LiftOver (60), and A/B compartments were identified using cooltools (v0.3.2, https://github.com/open2c/cooltools) at the 10-kb resolution.
Implementation of the EagleC framework
EagleC is an ensemble-learning framework that makes predictions based on 50 different models and uses CNN as the individual model. Each CNN model takes 21 × 21 grayscale images as input. Sequentially, the CNN architecture includes the following components: (i) convolution with 32 filters of a kernel size 3 × 3 and stride size 1, followed by the ReLU activation and a 2 × 2 max pooling; (ii) convolution with 64 filters of a kernel size 3 × 3 and stride size 1, followed by the ReLU activation and a 2 × 2 max pooling; and (iii) two fully connected layers with 512 hidden units. The first fully connected layer is followed by the ReLU activation and a dropout layer with a dropout probability of 0.5 to avoid overfitting, and the second fully connected layer acts as the final sigmoid output layer, which computes probability scores for each of the six labels: ++, +−, −+, −−, intranegative, and internegative. Each individual model is randomly initialized with a different set of training samples and trained using an iterative approach (Fig. 1A). During each round of training, the model is optimized against the accuracy using the Adam algorithm. We built the whole framework in Python and the neural network part was implemented using the TensorFlow Keras API (v2.3.0).
Computationally, it is impractical to perform predictions for the submatrix surrounding every pixel of a genome-wide contact map at high resolutions. To speed up the calculation, we perform several prefiltering procedures based on our prior knowledge that SVs usually induce abnormal signals with both high intensity and high density: (i) We filter out pixels where there are fewer than five nonzero values within their 21 × 21 window, and (ii) for intrachromosomal maps, we only consider pixels within the 3 × 3 window of significant interactions. Here, the significant interactions are identified using a model that accounts for the distance-dependent decay of interaction frequencies. Specifically, the expected interaction frequency at given genomic distance k is calculated as follows
where is a CNV-normalized or ICE-normalized intrachromosomal contact matrix with a size of n × n. Note that all pixels with genomic distance greater than 100 bins at a given resolution will have the same expected background. Then, the P value for each observed interaction frequency Mij is calculated on the basis of the Poisson process with expected value , where Wi is the bias vector extracted either from the “weight” (50) (in case of ICE normalization) or “sweight” (4) (in case of CNV normalization) column of the “.cool” file. To reduce potential false negatives, we apply a loose P value cutoff of 0.05 to include as many pixels as possible. In addition to the filtering procedures, different intrachromosomal and interchromosomal matrices can be automatically processed in parallel. All that users need to do is to submit the same command for a certain number of times. According to our test on a computational cluster [CPU information: Intel(R) Xeon(R) Gold 6230 CPU @ 2.10GHz], with 16 parallelized jobs, the program can be generally finished within 3 hours for a dataset with ~200 million contact pairs, which is comparable to or even faster than existing methods (table S6).
The final probability scores for each pixel are calculated by averaging the values across all 50 models. A pixel is identified as a candidate SV breakpoint if the probability of at least one positive label (++, +−, −+, and −−) is greater than a predefined cutoff (we set different cutoffs for different resolutions; see details below). We perform DBSCAN to identify any local clusters of highly scored pixels, and within each cluster, the pixel with the highest probability score will be reported in the final SV list.
To optimize the prediction performance for various sequencing depths, we down-sampled contact matrices of training samples to a series of sequencing depths and independently trained EagleC models for each depth. Specifically, we trained models for six levels of sequencing depths, including 300-800 million (M), 200-300M, 100-200M, 50-100M, 10-50M, and 5-10M contact pairs. In prediction, the most appropriate models were selected according to the number of contacts in the target contact map; for example, SVs in BT-474 and HCC1954 were predicted using the “100-200M” models because the Hi-C maps in these two cell lines contain 192M and 188M contact pairs, respectively.
Combining SV predictions from multiple resolutions
To optimize the performance of our SV prediction pipeline in both specificity and sensitivity, we propose a strategy that combines predictions from multiple resolutions including 5, 10, and 50 kb. Basically, high-resolution contact matrices (5 or 10 kb) usually achieve higher accuracy and have unique advantages in predicting short-range SVs, while low-resolution contact matrices at 50 kb can complement the predictions when sequencing depths are not sufficient to cover the real SV breakpoints at high resolutions. In our pipeline, we first predict SVs at 5-, 10-, and 50-kb resolutions separately. The default probability cutoffs are empirically set to 0.8, 0.8, and 0.99999 for 5, 10, and 50 kb, respectively. However, according to our test against the benchmark datasets used in this study, EagleC is pretty robust to different cutoff values, but generally tuning down the cutoffs detects more SVs with slightly lower accuracy, while tuning up the cutoffs detects fewer SVs with slightly higher accuracy (fig. S11). For 10- and 50-kb predictions, we further search for the most probable breakpoint coordinates within a local region on 5-kb contact maps so that all the reported SVs will have the 5-kb resolution. After we have obtained SV predictions from individual resolutions, we merge the SV coordinates from different resolutions together and only report nonredundant SVs for each sample.
Application of EagleC to scHi-C
We made the following modifications when we applied EagleC to scHi-C: (i) All the training and predicting steps were based on contact matrices at the 500-kb resolution; (ii) raw contact signals were used instead of ICE/CNV-normalized signals; (iii) the models were trained for eight levels of sequencing depths with lower number of contacts, including 10-20M, 5-10M, 3-5M, 1-3M, 750K-1M, 500-750K, 250-500K, and 100-250K contact pairs; and (iv) in prediction, the probability cutoff was set to 0.95.
Annotations of duplications and deletions
We defined duplications and deletions by using both the orientation information of SV breakpoints and copy number profiles. Specifically, duplications were defined as intrachromosomal SVs with −+, ++, or −− orientations, and the genomic interval between breakpoints had a copy number ratio larger than 1.5, while deletions were defined as intrachromosomal SVs with the +− orientation, and the genomic interval had a copy number ratio smaller than 0.3. Copy number profiles calculated from WGS were used if WGS was available; otherwise, we used copy number profiles inferred from Hi-C in this calculation (4). In addition, in Fig. 5 (F and G), we only considered short-range SVs with a breakpoint distance of less than 1 Mb.
Identification of compartments and TADs
We identified A/B compartments and TADs on Hi-C contact maps of several normal cell lines or tissues (table S5) to investigate the associations between 3D genomic architectures and SV formation. The A/B compartments were identified using cooltools (v0.3.2). Briefly, the eigenvalue decomposition was performed on the 100-kb intrachromosomal contact maps, and the first eigenvector (PC1) was used to capture the “plaid” contact pattern. The original PC1 was oriented according to gene densities (Ensembl 93) so that positive values correspond to active genomic regions (A compartment) and negative values correspond to inactive regions (B compartment). For TADs, we ran HiTAD at the 25-kb resolution and defined TADs as the bottom-level domains returned by HiTAD (61).
Acknowledgments
Funding: F.Y. is supported by NIH grants 5R01HG011207, 5R35GM124820, 5R01HG009906, and 1U24HG012070.
Author contributions: F.Y. conceived and supervised the project. X.W. implemented the EagleC framework and performed the data analysis. Y.L. helped process the scHi-C data.
Competing interests: F.Y. and X.W. are listed as inventors of a provisional patent based on this work. F.Y. is a scientific cofounder for Sariant Therapeutics Inc. The remaining author declares no competing interests.
Data and materials availability: A list of high-confidence SVs we compiled for training EagleC models is provided in table S1. Detailed information of cancer Hi-C datasets collected in this study is summarized in table S2. Detailed information of HiChIP/ChIA-PET datasets is summarized in table S3. Breakpoint coordinates predicted in each sample are provided in table S4. Hi-C datasets in normal cell lines or tissues are summarized in table S5. The list of cancer-related genes was obtained from the Bushman Lab (http://bushmanlab.org/assets/doc/allOnco_May2018.tsv). The EagleC software and detailed documentation are available at Zenodo (https://doi.org/10.5281/zenodo.6482060) and GitHub (https://github.com/XiaoTaoWang/EagleC).
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Table S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5
REFERENCES AND NOTES
- 1.Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J. S., Dobson J., Urashima M., McHenry K. T., Pinchback R. M., Ligon A. H., Cho Y. J., Haery L., Greulich H., Reich M., Winckler W., Lawrence M. S., Weir B. A., Tanaka K. E., Chiang D. Y., Bass A. J., Loo A., Hoffman C., Prensner J., Liefeld T., Gao Q., Yecies D., Signoretti S., Maher E., Kaye F. J., Sasaki H., Tepper J. E., Fletcher J. A., Tabernero J., Baselga J., Tsao M. S., Demichelis F., Rubin M. A., Janne P. A., Daly M. J., Nucera C., Levine R. L., Ebert B. L., Gabriel S., Rustgi A. K., Antonescu C. R., Ladanyi M., Letai A., Garraway L. A., Loda M., Beer D. G., True L. D., Okamoto A., Pomeroy S. L., Singer S., Golub T. R., Lander E. S., Getz G., Sellers W. R., Meyerson M., The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitelman F., Johansson B., Mertens F., The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 7, 233–245 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Weischenfeldt J., Dubash T., Drainas A. P., Mardin B. R., Chen Y., Stutz A. M., Waszak S. M., Bosco G., Halvorsen A. R., Raeder B., Efthymiopoulos T., Erkek S., Siegl C., Brenner H., Brustugun O. T., Dieter S. M., Northcott P. A., Petersen I., Pfister S. M., Schneider M., Solberg S. K., Thunissen E., Weichert W., Zichner T., Thomas R., Peifer M., Helland A., Ball C. R., Jechlinger M., Sotillo R., Glimm H., Korbel J. O., Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 49, 65–74 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X., Xu J., Zhang B., Hou Y., Song F., Lyu H., Yue F., Genome-wide detection of enhancer-hijacking events from chromatin interaction data in rearranged genomes. Nat. Methods 18, 661–668 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasty P., Montagna C., Chromosomal rearrangements in cancer: Detection and potential causal mechanisms. Mol. Cell. Oncol. 1, e29904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan T. S., Cancer cytogenetics: Methodology revisited. Ann. Lab. Med. 34, 413–425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zack T. I., Schumacher S. E., Carter S. L., Cherniack A. D., Saksena G., Tabak B., Lawrence M. S., Zhsng C. Z., Wala J., Mermel C. H., Sougnez C., Gabriel S. B., Hernandez B., Shen H., Laird P. W., Getz G., Meyerson M., Beroukhim R., Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell P. J., Stephens P. J., Pleasance E. D., O’Meara S., Li H., Santarius T., Stebbings L. A., Leroy C., Edkins S., Hardy C., Teague J. W., Menzies A., Goodhead I., Turner D. J., Clee C. M., Quail M. A., Cox A., Brown C., Durbin R., Hurles M. E., Edwards P. A., Bignell G. R., Stratton M. R., Futreal P. A., Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 40, 722–729 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mardis E. R., Wilson R. K., Cancer genome sequencing: A review. Hum. Mol. Genet. 18, R163–R168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L. B., Martin S., Wedge D. C., Van Loo P., Ju Y. S., Smid M., Brinkman A. B., Morganella S., Aure M. R., Lingjaerde O. C., Langerod A., Ringner M., Ahn S. M., Boyault S., Brock J. E., Broeks A., Butler A., Desmedt C., Dirix L., Dronov S., Fatima A., Foekens J. A., Gerstung M., Hooijer G. K., Jang S. J., Jones D. R., Kim H. Y., King T. A., Krishnamurthy S., Lee H. J., Lee J. Y., Li Y., McLaren S., Menzies A., Mustonen V., O’Meara S., Pauporte I., Pivot X., Purdie C. A., Raine K., Ramakrishnan K., Rodriguez-Gonzalez F. G., Romieu G., Sieuwerts A. M., Simpson P. T., Shepherd R., Stebbings L., Stefansson O. A., Teague J., Tommasi S., Treilleux I., Van den Eynden G. G., Vermeulen P., Vincent-Salomon A., Yates L., Caldas C., ‘t van Veer L., Tutt A., Knappskog S., Tan B. K., Jonkers J., Borg A., Ueno N. T., Sotiriou C., Viari A., Futreal P. A., Campbell P. J., Span P. N., Van Laere S., Lakhani S. R., Eyfjord J. E., Thompson A. M., Birney E., Stunnenberg H. G., van de Vijver M. J., Martens J. W., Borresen-Dale A. L., Richardson A. L., Kong G., Thomas G., Stratton M. R., Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harewood L., Kishore K., Eldridge M. D., Wingett S., Pearson D., Schoenfelder S., Collins V. P., Fraser P., Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biol. 18, 125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghandi M., Huang F. W., Jane-Valbuena J., Kryukov G. V., Lo C. C., McDonald E. R. III, Barretina J., Gelfand E. T., Bielski C. M., Li H., Hu K., Andreev-Drakhlin A. Y., Kim J., Hess J. M., Haas B. J., Aguet F., Weir B. A., Rothberg M. V., Paolella B. R., Lawrence M. S., Akbani R., Lu Y., Tiv H. L., Gokhale P. C., de Weck A., Mansour A. A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J. M., Porter D. A., Jones M. D., Golji J., Caponigro G., Taylor J. E., Dunning C. M., Creech A. L., Warren A. C., McFarland J. M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y. E., Cherniack A. D., Tsherniak A., Vazquez F., Jaffe J. D., Lane A. A., Weinstock D. M., Johannessen C. M., Morrissey M. P., Stegmeier F., Schlegel R., Hahn W. C., Getz G., Mills G. B., Boehm J. S., Golub T. R., Garraway L. A., Sellers W. R., Next-generation characterization of the cancer cell line encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akdemir K. C., Le V. T., Chandran S., Li Y., Verhaak R. G., Beroukhim R., Campbell P. J., Chin L., Dixon J. R., Futreal P. A.; P. S. V. W. Group, Consortium P., Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat. Genet. 52, 294–305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Logsdon G. A., Vollger M. R., Eichler E. E., Long-read human genome sequencing and its applications. Nat. Rev. Genet. 21, 597–614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Zhao Y., Bollas A., Wang Y., Au K. F., Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou A., Lin T., Xing J., Evaluating nanopore sequencing data processing pipelines for structural variation identification. Genome Biol. 20, 237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty A., Ay F., Identification of copy number variations and translocations in cancer cells from Hi-C data. Bioinformatics 34, 338–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon J. R., Xu J., Dileep V., Zhan Y., Song F., Le V. T., Yardimci G. G., Chakraborty A., Bann D. V., Wang Y., Clark R., Zhang L., Yang H., Liu T., Iyyanki S., An L., Pool C., Sasaki T., Rivera-Mulia J. C., Ozadam H., Lajoie B. R., Kaul R., Buckley M., Lee K., Diegel M., Pezic D., Ernst C., Hadjur S., Odom D. T., Stamatoyannopoulos J. A., Broach J. R., Hardison R. C., Ay F., Noble W. S., Dekker J., Gilbert D. M., Yue F., Integrative detection and analysis of structural variation in cancer genomes. Nat. Genet. 50, 1388–1398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S., Lee S., Chu C., Jain D., Kerpedjiev P., Nelson G. M., Walsh J. M., Alver B. H., Park P. J., HiNT: A computational method for detecting copy number variations and translocations from Hi-C data. Genome Biol. 21, 73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim K., Kim M., Kim Y., Lee D., Jung I., Hi-C as a molecular rangefinder to examine genomic rearrangements. Semin. Cell Dev. Biol. , (2022). [DOI] [PubMed] [Google Scholar]
- 21.Rao S. S., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., Robinson J. T., Sanborn A. L., Machol I., Omer A. D., Lander E. S., Aiden E. L., A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhrig S., Ellermann J., Walther T., Burkhardt P., Frohlich M., Hutter B., Toprak U. H., Neumann O., Stenzinger A., Scholl C., Frohling S., Brors B., Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 31, 448–460 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fullwood M. J., Liu M. H., Pan Y. F., Liu J., Xu H., Mohamed Y. B., Orlov Y. L., Velkov S., Ho A., Mei P. H., Chew E. G., Huang P. Y., Welboren W. J., Han Y., Ooi H. S., Ariyaratne P. N., Vega V. B., Luo Y., Tan P. Y., Choy P. Y., Wansa K. D., Zhao B., Lim K. S., Leow S. C., Yow J. S., Joseph R., Li H., Desai K. V., Thomsen J. S., Lee Y. K., Karuturi R. K., Herve T., Bourque G., Stunnenberg H. G., Ruan X., Cacheux-Rataboul V., Sung W. K., Liu E. T., Wei C. L., Cheung E., Ruan Y., An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462, 58–64 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mumbach M. R., Rubin A. J., Flynn R. A., Dai C., Khavari P. A., Greenleaf W. J., Chang H. Y., HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang R., Yu M., Li G., Chee S., Liu T., Schmitt A. D., Ren B., Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 26, 1345–1348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mifsud B., Tavares-Cadete F., Young A. N., Sugar R., Schoenfelder S., Ferreira L., Wingett S. W., Andrews S., Grey W., Ewels P. A., Herman B., Happe S., Higgs A., LeProust E., Follows G. A., Fraser P., Luscombe N. M., Osborne C. S., Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Franke M., Ibrahim D. M., Andrey G., Schwarzer W., Heinrich V., Schopflin R., Kraft K., Kempfer R., Jerkovic I., Chan W. L., Spielmann M., Timmermann B., Wittler L., Kurth I., Cambiaso P., Zuffardi O., Houge G., Lambie L., Brancati F., Pombo A., Vingron M., Spitz F., Mundlos S., Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Kragesteen B. K., Spielmann M., Paliou C., Heinrich V., Schopflin R., Esposito A., Annunziatella C., Bianco S., Chiariello A. M., Jerkovic I., Harabula I., Guckelberger P., Pechstein M., Wittler L., Chan W. L., Franke M., Lupianez D. G., Kraft K., Timmermann B., Vingron M., Visel A., Nicodemi M., Mundlos S., Andrey G., Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat. Genet. 50, 1463–1473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Despang A., Schopflin R., Franke M., Ali S., Jerkovic I., Paliou C., Chan W. L., Timmermann B., Wittler L., Vingron M., Mundlos S., Ibrahim D. M., Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat. Genet. 51, 1263–1271 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Kraft K., Magg A., Heinrich V., Riemenschneider C., Schopflin R., Markowski J., Ibrahim D. M., Acuna-Hidalgo R., Despang A., Andrey G., Wittler L., Timmermann B., Vingron M., Mundlos S., Serial genomic inversions induce tissue-specific architectural stripes, gene misexpression and congenital malformations. Nat. Cell Biol. 21, 305–310 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Duijf P. H. G., Nanayakkara D., Nones K., Srihari S., Kalimutho M., Khanna K. K., Mechanisms of genomic instability in breast cancer. Trends Mol. Med. 25, 595–611 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O., Sandstrom R., Bernstein B., Bender M. A., Groudine M., Gnirke A., Stamatoyannopoulos J., Mirny L. A., Lander E. S., Dekker J., Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., Ren B., Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canela A., Maman Y., Jung S., Wong N., Callen E., Day A., Kieffer-Kwon K. R., Pekowska A., Zhang H., Rao S. S. P., Huang S. C., McKinnon P. J., Aplan P. D., Pommier Y., Aiden E. L., Casellas R., Nussenzweig A., Genome organization drives chromosome fragility. Cell 170, 507–521.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clurman B. E., Groudine M., The CDKN2A tumor-suppressor locus—A tale of two proteins. N. Engl. J. Med. 338, 910–912 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Khawaled S., Nigita G., Distefano R., Oster S., Suh S. S., Smith Y., Khalaileh A., Peng Y., Croce C. M., Geiger T., Seewaldt V. L., Aqeilan R. I., Pleiotropic tumor suppressor functions of WWOX antagonize metastasis. Signal Transduct. Target. Ther. 5, 43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kashima L., Toyota M., Mita H., Suzuki H., Idogawa M., Ogi K., Sasaki Y., Tokino T., CHFR, a potential tumor suppressor, downregulates interleukin-8 through the inhibition of NF-kappaB. Oncogene 28, 2643–2653 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Zink D., Mayr C., Janz C., Wiesmuller L., Association of p53 and MSH2 with recombinative repair complexes during S phase. Oncogene 21, 4788–4800 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Chen H., Liu H., Qing G., Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 3, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C., Zhao S., Karnad A., Freeman J. W., The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 11, 64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramani V., Deng X., Qiu R., Gunderson K. L., Steemers F. J., Disteche C. M., Noble W. S., Duan Z., Shendure J., Massively multiplex single-cell Hi-C. Nat. Methods 14, 263–266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Essletzbichler P., Konopka T., Santoro F., Chen D., Gapp B. V., Kralovics R., Brummelkamp T. R., Nijman S. M., Burckstummer T., Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 24, 2059–2065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shibata Y., Malhotra A., Dutta A., Detection of DNA fusion junctions for BCR-ABL translocations by Anchored ChromPET. Genome Med. 2, 70 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H., Luan Y., Liu T., Lee H. J., Fang L., Wang Y., Wang X., Zhang B., Jin Q., Ang K. C., Xing X., Wang J., Xu J., Song F., Sriranga I., Khunsriraksakul C., Salameh T., Li D., Choudhary M. N. K., Topczewski J., Wang K., Gerhard G. S., Hardison R. C., Wang T., Cheng K. C., Yue F., A map of cis-regulatory elements and 3D genome structures in zebrafish. Nature 588, 337–343 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poirion O., Zhu X., Ching T., Garmire L. X., Using single nucleotide variations in single-cell RNA-seq to identify subpopulations and genotype-phenotype linkage. Nat. Commun. 9, 4892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mallory X. F., Edrisi M., Navin N., Nakhleh L., Methods for copy number aberration detection from single-cell DNA-sequencing data. Genome Biol. 21, 208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao R., Bai S., Henderson Y. C., Lin Y., Schalck A., Yan Y., Kumar T., Hu M., Sei E., Davis A., Wang F., Shaitelman S. F., Wang J. R., Chen K., Moulder S., Lai S. Y., Navin N. E., Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat. Biotechnol. 39, 599–608 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan L., Xing D., Chang C. H., Li H., Xie X. S., Three-dimensional genome structures of single diploid human cells. Science 361, 924–928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee D. S., Luo C., Zhou J., Chandran S., Rivkin A., Bartlett A., Nery J. R., Fitzpatrick C., O’Connor C., Dixon J. R., Ecker J. R., Simultaneous profiling of 3D genome structure and DNA methylation in single human cells. Nat. Methods 16, 999–1006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdennur N., Mirny L. A., Cooler: Scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311–316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Durand N. C., Shamim M. S., Machol I., Rao S. S., Huntley M. H., Lander E. S., Aiden E. L., Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.H. Li, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 (2013).
- 53.Rausch T., Zichner T., Schlattl A., Stutz A. M., Benes V., Korbel J. O., DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Layer R. M., Chiang C., Quinlan A. R., Hall I. M., LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 15, R84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boeva V., Popova T., Bleakley K., Chiche P., Cappo J., Schleiermacher G., Janoueix-Lerosey I., Delattre O., Barillot E., Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sedlazeck F. J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., Schatz M. C., Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gong L., Wong C. H., Cheng W. C., Tjong H., Menghi F., Ngan C. Y., Liu E. T., Wei C. L., Picky comprehensively detects high-resolution structural variants in nanopore long reads. Nat. Methods 15, 455–460 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heller D., Vingron M., SVIM: Structural variant identification using mapped long reads. Bioinformatics 35, 2907–2915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H., Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuhn R. M., Haussler D., Kent W. J., The UCSC genome browser and associated tools. Brief. Bioinform. 14, 144–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X. T., Cui W., Peng C., HiTAD: Detecting the structural and functional hierarchies of topologically associating domains from chromatin interactions. Nucleic Acids Res. 45, e163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Table S6
Tables S1 to S5