PURPOSE
This investigation sought to evaluate the prognostic value of pretreatment of circulating tumor DNA (ctDNA) in metastatic biliary tract cancers (BTCs) treated with platinum-based first-line chemotherapy treatment.
MATERIALS AND METHODS
We performed a retrospective analysis of 67 patients who underwent ctDNA testing before platinum-based chemotherapy for first-line treatment for metastatic BTC. For analysis, we considered the detected gene with highest variant allele frequency as the dominant clone allele frequency (DCAF). Results of ctDNA analysis were correlated with patients' demographics, progression-free survival (PFS), and overall survival (OS).
RESULTS
The median age of patients was 67 (27-90) years. Fifty-four (80.6%) of 67 patients evaluated had intrahepatic cholangiocarcinoma; seven had extrahepatic cholangiocarcinoma, and six gallbladder cancers. Forty-six (68.6%) of the patients were treated with cisplatin plus gemcitabine, and 16.4% of patients received gemcitabine and other platinum (carboplatin or oxaliplatin) combinations, whereas 15% of patients were treated on a clinical trial with gemcitabine and cisplatin plus additional agents (CX4945, PEGPH20, or nab-paclitaxel). TP53, KRAS, FGFR2, ARID1A, STK11, and IDH1 were the genes with highest frequency as DCAF. The median DCAF was 3% (0%-97%). DCAF > 3% was associated with worse OS (median OS: 10.8 v 18.8 months, P = .032). Stratifying DCAF in quartiles, DCAF > 10% was significantly related to worse PFS (median PFS: 3 months, P = .014) and worse OS (median OS: 7.0 months, P = .001). Each 1% increase in ctDNA was associated with a hazard ratio of 13.1 in OS when adjusting for subtypes, metastatic sites, size of largest tumor, age, sex, and CA19-9.
CONCLUSION
DCAF at diagnosis of advanced BTC can stratify patients who have worse outcomes when treated with upfront platinum-based chemotherapy. Each increase in %ctDNA decreases survival probabilities.
INTRODUCTION
Biliary tract cancers (BTCs) include intrahepatic cholangiocarcinoma (IHC), gallbladder cancer (GBC), extrahepatic cholangiocarcinoma (EHC), and ampulla of Vater cancers.1 BTC represents 3% of gastrointestinal malignancies, with 11,980 cases expected to be diagnosed in 2021.2,3 As BTC usually present at an advanced stage, only 20% of these tumors are considered resectable.4 In patients with unresectable disease, the 5-year overall survival (OS) is about 4%.5
CONTEXT
Key Objective
In this study, we hypothesized that the dominant clone allele frequency (DCAF) on circulating tumor DNA (ctDNA) in advanced biliary tract cancer at diagnosis would be associated with overall survival (OS) and progression-free survival.
Knowledge Generated
DCAF is strongly associated with progression-free survival and OS. Each 1% increase in ctDNA was associated with a hazard ratio of 13.1 in OS when adjusting for subtypes, metastatic sites, size of largest tumor, age, sex, and CA19-9.
Relevance
DCAF at diagnosis of advanced biliary tract cancer can stratify patients who have worse outcomes when treated with upfront platinum-based chemotherapy. Each increase in %ctDNA decreases survival probability.
The survival gain with first-line chemotherapy regimens in BTC is modest since most patients develop progressive disease with a median OS of less than a year.6 This has generated interest in using next-generation tumor genomic profiling and liquid tumor biopsy on peripheral blood to look for targetable genetic alterations.7,8
Circulating tumor DNA (ctDNA) has been shown to carry tumor-specific genetic or epigenetic alterations including point mutations, copy number variations, chromosomal rearrangements, and DNA methylation. This ctDNA is released into the circulation after tumor cells undergo apoptosis or necrosis.9 Evaluation of ctDNA can identify patient-specific tumoral genetic alterations while allowing for serial monitoring of tumor genomes in a noninvasive and accurate manner.8 Therapeutically relevant alterations were seen in ctDNA in 55% of patients with BTC.8 Because of these findings, the strategy is being used in the setting of advanced disease for treatment selection.10 Furthermore, it has also been used as an early marker of response to treatment and to track mechanisms of acquired resistance.11
In colon and breast cancer, ctDNA has been used to predict response to treatment and prognosis in the adjuvant and neoadjuvant setting, respectively.12,13 One marker of interest is variant allele frequency (VAF), which is the number of mutant molecules over the total number of wild-type molecules at a specific location on the genome. Pairawan et al14 showed that VAF is a surrogate marker of tumor burden and maximum VAF (VAFmax) correlated negatively with prognosis and survival in metastatic cancer.
In this study, we hypothesized that the dominant clone allele frequency (DCAF) on ctDNA in BTC would be associated with OS and progression-free survival (PFS) and can serve as a surrogate of disease volume and severity. In addition, we looked at the relationship of DCAF to treatment response with first-line platinum-based chemotherapy and clinical demographics.
MATERIALS AND METHODS
Patients
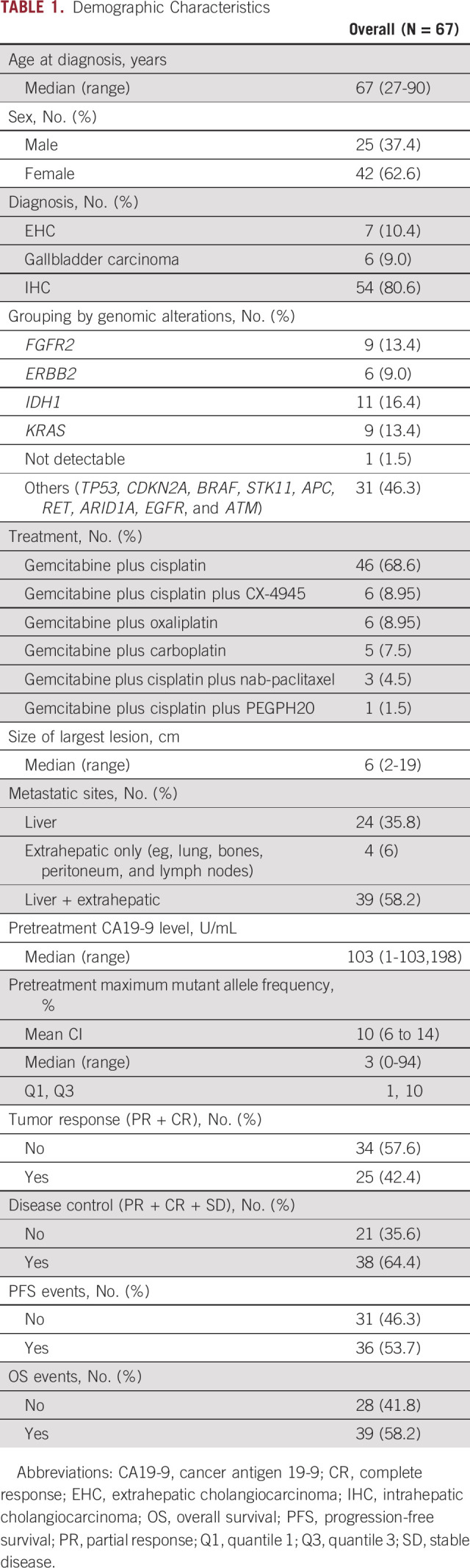
From July 2016 through June 2020, 67 patients with advanced BTC underwent ctDNA testing at diagnosis using an available assay (Guardant Health, Inc, Redwood City, CA). All the patients received care at the Mayo Clinic Cancer Center in Arizona and Florida. The analysis from this cohort was reviewed and approved by the Mayo Clinic institutional review board. Clinical and demographic information of all patients is included in Table 1.
TABLE 1.
Demographic Characteristics
Comprehensive Genomic Testing in Plasma
Circulating tumor DNA was extracted from whole blood. ctDNA fragments, both leukocyte-derived and tumor-derived, were simultaneously sequenced. The VAF was calculated as the proportion of ctDNA harboring the variant in a background of wild-type ctDNA. Analytical information, bioinformatics analysis, and Guardant360 database have been previously described.15,16
Outcomes
Assessments regarding response to therapy (complete response [CR], partial response [PR], stable disease, and disease progression) were retrospectively collected by review of patient's charts. Positive response to therapy was considered PR and CR by RECIST. Disease control rate was determined on the basis of CR, PR, and stable disease. PFS was determined during treatment with chemotherapy and after without disease progression. OS was determined by the time of diagnosis of advanced disease until death or last day of follow-up for patients on treatment and alive.
Statistical Analysis
We summarized categorical data as frequency counts and percentages, and continuous measures as means, standard deviations, medians, and ranges. Categorical variables were compared using the chi-square test or Fisher's exact test. Continuous variables were compared using the one-way analysis of variance test or Kruskal-Wallis test. Multivariate logistic regressions were performed to assess the association of ctDNA with response rate and disease control rate with adjustment for disease subtype, age, sex, CA19-9, lesion size, and metastatic site. The distributions of time-to-event outcomes were estimated using the Kaplan-Meier methods and compared between low and high ctDNA dichotomized by the median DCAF (ie, low < 3% v high ≥ 3% ctDNA) using the log-rank test. Hazard ratios (HRs) and 95% CIs were estimated using a multivariate Cox model adjusting for disease subtype, age, sex, CA19-9, lesion size, and metastatic site. Sensitivity analysis was performed to explore either 3 quantiles (≤ 33% percentile, 34%-66% percentile, and > 66% percentile) or quartiles as the cutoffs in DCAF.
Ethics
The study was reviewed and approved by the Mayo Clinic Institutional Review Board. The informed consent was waived after IRB evaluation. This study was conducted in accordance with the Declaration of Helsinki.
RESULTS
Patient Demographics
A total of 67 patients were included in the analysis. 80.6% (54) had IHC, 10.4% (seven) of patients had EHC, and 9% (six) had GBC. All patients included had ctDNA collected before the first-line chemotherapy regimen for advanced disease. The median age of all patients was 67 (27-90) years, and the majority were female (62.6%). All patients were treated with platinum-based chemotherapy regimens as first-line treatment. Most patients (68.6%) were treated with cisplatin plus gemcitabine, and 11 (16.4%) patients received gemcitabine plus other platinum (carboplatin or oxaliplatin) combinations, whereas 10 (15%) patients were treated on a clinical trial with gemcitabine and cisplatin plus additional agents (CX4945, PEGPH20, or nab-paclitaxel). The median size of largest lesion was 6 cm (2-19 cm), and more than half (58.2%) had multiple metastatic sites including liver and extrahepatic sites. Lungs, bones, lymph nodes, and peritoneum were the sites with most extrahepatic metastasis identified. Other clinical information is given in Table 1.
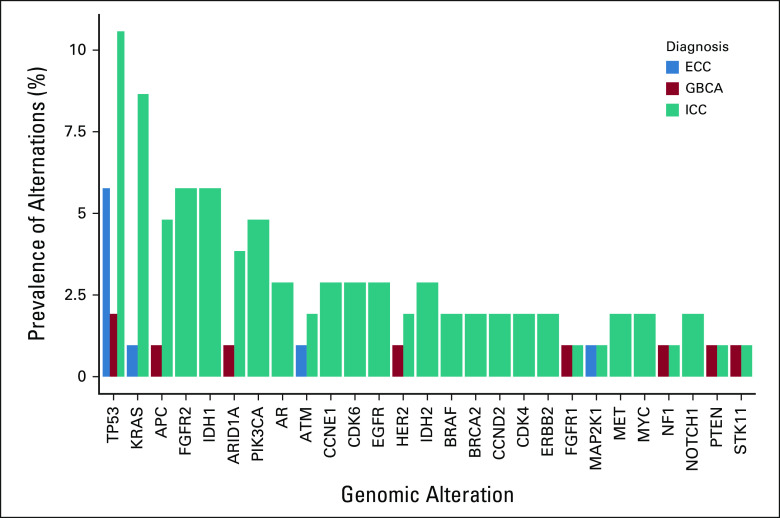
Several potential targetable genes were detected with ctDNA including FGFR2, HER2, IDH, MET, EGFR, BRAF, and KRAS. A higher prevalence of TP53 was observed among the three subtypes. Homologous recombinant repair genes were identified in IHC and EHC, including ATM and BRCA2. Prevalence of all genomic alterations according to primary tumor is presented in Figure 1.
FIG 1.
Prevalence of genomic alterations according to primary tumor. ECC, extrahepatic cholangiocarcinoma; GBCA, gallbladder cancer; ICC, intrahepatic cholangiocarcinoma.
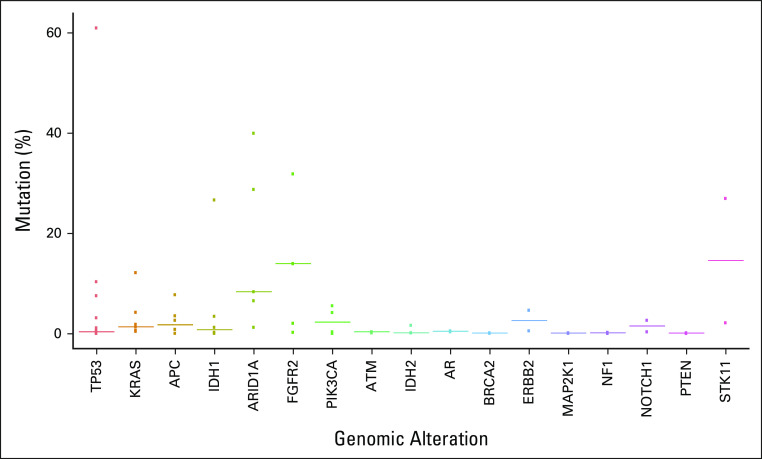
TP53, KRAS, FGFR2, ARID1A, STK11, and IDH1 were the genes with highest VAF as dominant clone (Fig 2). Most ERBB2 (HER2) genomic alterations detected were amplifications, identified in four patients. Other genes with detected amplifications included KRAS, EGFR, BRAF, MET, CCNE1, CCND1, CCND2, MYC, FGFR1, FGFR2, CDK4, CDK6, PIK3CA, and AR. For analysis, we considered the detected genomic alteration with the highest VAF as the DCAF.
FIG 2.
Variant allele frequency of detected genes.
DCAF and Prognostic Factors
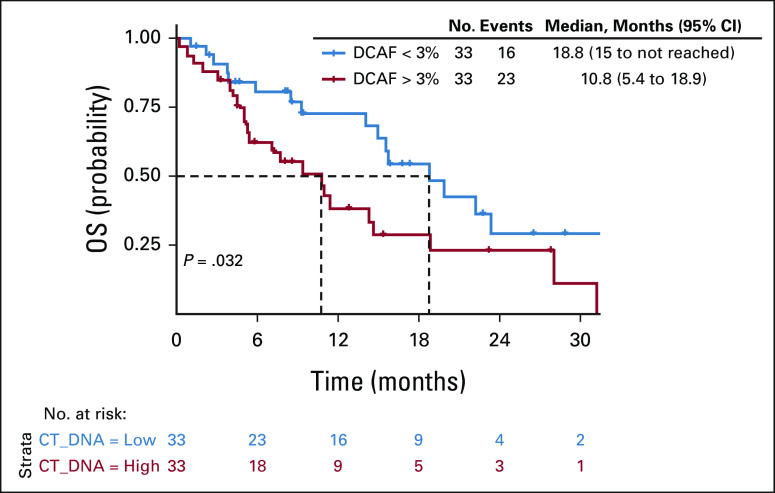
One patient with no tumor genomic alteration detected was excluded from this analysis. The median DCAF was 3% (0%-97%). DCAF > 3% was associated with inferior PFS (median PFS: 4.7 v 7.7 months, P = .087; Data Supplement) and significantly worse OS (median OS: 10.8 v 18.8 months, P = .032. Fig 3).
FIG 3.
Kaplan-Meier curve for OS by DCAF > 3%. DCAF, dominant clone allele frequency; OS, overall survival.
We further analyzed DCAF using either three quantiles or quartiles as the cutoffs. DCAF distributed across three quantiles (Q1: ctDNA ≤ 1%, Q2: ctDNA 1%-7%, and Q3: ctDNA ≥ 7%) was significantly associated with PFS (P = .022), with a shorter median PFS of 3.2 months for patients with ctDNA ≥ 7%, compared with 10.5 months for patients with ctDNA ≤ 1% and 10.7 months for patients with ctDNA 1%-7% (Data Supplement). DCAF distributed across three quantiles was not statistically associated with OS differences (P = .065; Data Supplement). DCAF divided by quartiles (Q1: ctDNA ≤ 0.6%, ctDNA Q2: 0.6%-3%, ctDNA Q3: 3%-10%, and ctDNA Q4: ≥ 10%) was significantly associated with both PFS (P = .014; Fig 4) and OS (P = .001; Fig 5).
FIG 4.
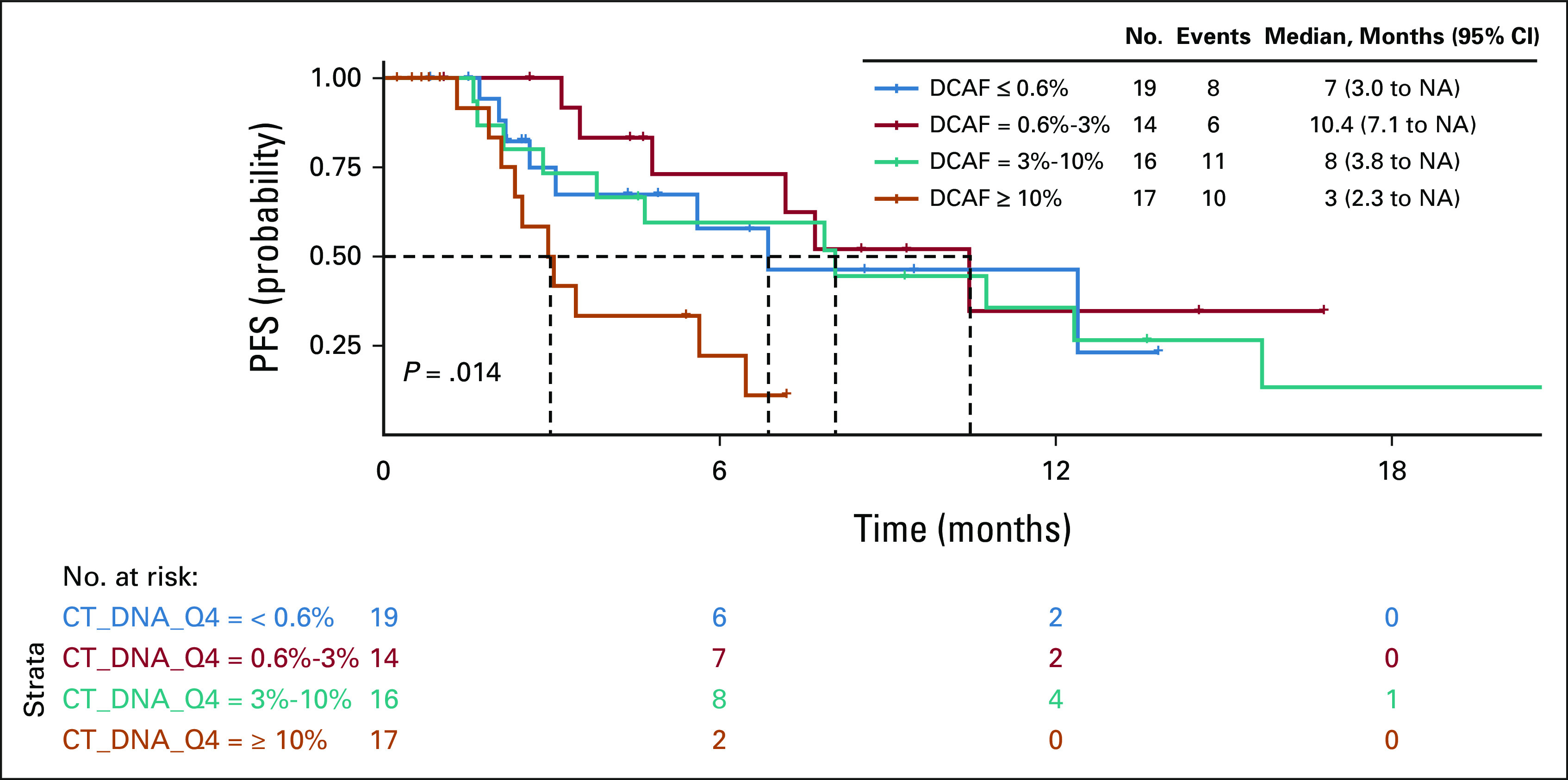
Kaplan-Meier curve for PFS by ctDNA. ctDNA, circulating tumor DNA; DCAF, dominant clone allele frequency; PFS, progression-free survival.
FIG 5.
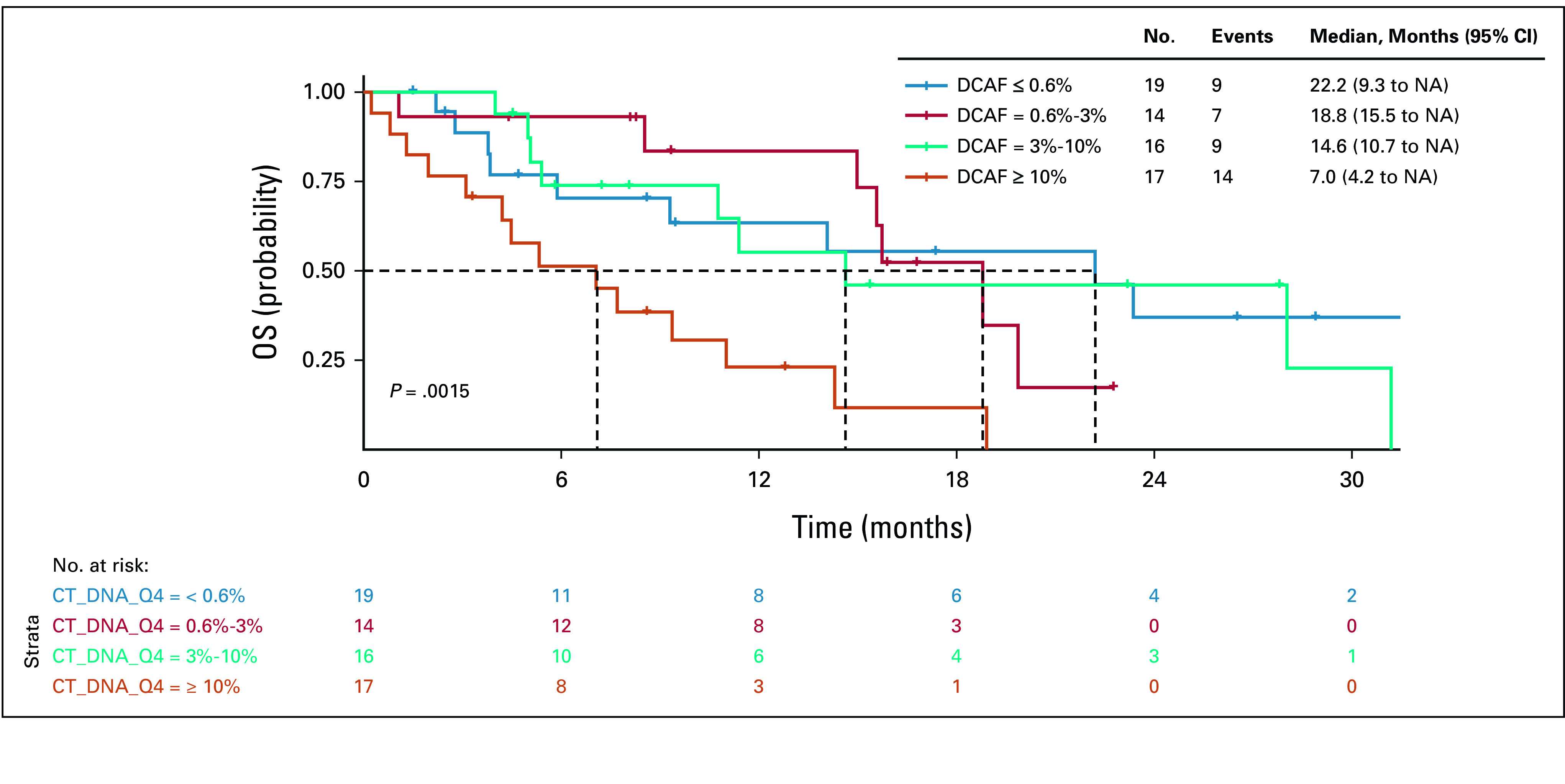
Kaplan-Meier curve for OS by ctDNA. ctDNA, circulating tumor DNA; DCAF, dominant clone allele frequency; OS, overall survival.
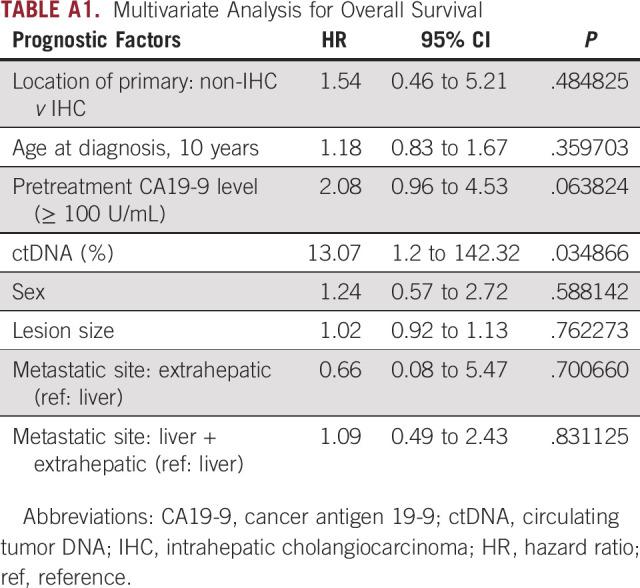
Each 1% increase in ctDNA is associated with a hazard ratio of 13.1 in OS when adjusting for primary tumor, size of the largest lesion, metastatic sites, sex, age, and CA19-9 (Appendix Table A1). No significant differences in response or disease control rate to chemotherapy were observed in patients with low or high ctDNA (Data Supplement). No statistical significance was found between DCAF and the presence of potential actionable targets including FGFR2, IDH1/2, ERBB2, and KRAS (Data Supplement). No association between ctDNA and survival in the context of FGFR2 alterations or IDH mutation was identified (Data Supplement).
The interaction between CA19-9 and DCAF was not statistically significant (OS: Pinteraction = 0.12; PFS: Pinteraction = .06). Although cholangiocarcinoma patients with high DCAF and high CA19-9 (DFCA ≥ 3%, CA19-9 ≥ 100) had worst OS, no statistical significance was found for PFS (P = .19) or OS (P = .13; Data Supplement).
DISCUSSION
In this study, we assessed whether the highest VAF detected by ctDNA, namely, DCAF, could be a prognostic factor in patients with advanced BTC at diagnosis. On the basis of the findings, patients with DCAF > 3% at diagnosis had worse OS when treated with upfront platinum-based chemotherapy. Furthermore, DCAF> 10% was significantly related to worse PFS and OS. However, no differences in response rate were observed among patients with high or low DCAF. Moreover, ctDNA proved to be an independent factor related to OS in multivariate analysis. Collectively, these data suggest a prognostic and not predictive role for DCAF in patients with advanced BTC undergoing platinum-based therapy.
The landscape of ctDNA genomic alterations of BTCs has already been previously described.8,17 Similar to our findings, these studies included more patients with IHC and the genes with the highest detection with ctDNA included principally KRAS, TP53, FGFR2, IDH1, and ARID1A.8,17 In our cohort, we observed different patterns of prevalence, with ATM and MAP2K1 detected in EHC and ERBB2 and NF1 and PTEN in GBC.
VAF is related to outcomes, which is more prominent in metastatic disease and is associated with tumor volume.14,16,18 In metastatic pancreatic cancer, detectable ctDNA and high VAF were associated with worse OS.18-20 Prognostic significance was observed in other solid tumors including colorectal cancer, breast cancer, and prostate cancer.21-23 Little is known about VAF and prognosis in BTCs. Lower values of VAF were associated with prolonged PFS in a cohort of 24 patients with cholangiocarcinoma.18 Considering the highest VAF value, we showed that the DCAF> 3% is related to numerically inferior PFS (but not statistically significant) and worse OS in patients with BTCs treated with standard upfront platinum-based chemotherapy for advanced disease. Interestingly, the DCAF was determined by multiple different genes among the cases, including TP53, KRAS, FGFR2, ARID1A, STK11, and IDH1, suggesting as previously stated by other colleagues that the highest VAF would be a surrogate of disease burden not related specifically to the gene detected. In agreement with this, evaluating the presence of specific genes of interest in the overall analysis of ctDNA, we did not find any association with DCAF and possible targetable genes including FGFR2, ERBB2, IDH1, and KRAS.
Prognostic factors related to PFS and OS in advanced BTCs were evaluated from the ABC-02 trial and an international data set.24 In this analysis, the authors evaluated prognostic factors in a combined sample size of more than 1,000 patients.24 Although the results suggest multiple factors in multivariate analysis including hemoglobin, sex, and neutrophils, receiver operator curve analysis suggested that the model generated had a limited prognostic value.24 Even the primary tumor site was not significant, in contrast to the findings of other groups.25 After multiple efforts evaluating scores and factors to prognostication of advanced BTCs,26-28 the ability to predict prognosis needs improvement. In our analysis, the OS impact of ctDNA was observed after stratifying with other possible prognostic factors including size of largest lesion, locally advanced/metastatic designation, primary tumor location, metastatic sites, sex, cancer antigen 19-9, and age. Evaluating ctDNA as a continuous variable, higher values are related to inferior survival probabilities. On the basis of the findings, ctDNA and DCAF could be a reliable assay to collect as a prognostic instrument in prospective trials.
Considering the investigative nature of ctDNA in BTCs, larger, multicentered prospective studies would be necessary to address the application of ctDNA in various disease assessment junctures, considering early diagnosis, minimal residual disease assessment, monitoring in advanced stages during systemic treatment, and assessment of mutations that are associated with resistance during treatment with targeted therapies. Future application of ctDNA and DCAF in metastatic disease would be a tool for genomic profiling in prospective trials, can be a surrogate of disease volume, and will assist in stratification of patients with advanced BTC in randomized studies, beyond currently known factors such as locally advanced vs. metastatic disease and CA19-9 levels. DCAF could also be used for selecting patients eligible for more intensive regimens of chemotherapy on the basis of higher levels of ctDNA.
Some limitations of this study include the number of patients, limited institution aspect, inherent limitations associated with a targeted gene panel, and the retrospective nature of data collection. Furthermore, most of the patients included had tumor arising from the intrahepatic duct. This limitation is shared with studies in BTCs, and efforts for future studies should be made to include patients with extrahepatic and gallbladder carcinomas in initiatives for genomic profiling and ctDNA. Despite the limited number of patients, strong association of ctDNA and OS was observed and provides the impetus for broader evaluation. This study only evaluated patients treated with upfront chemotherapy. Although this is the standard of care in the first-line setting, at this time, multiple trials are evaluating targeted treatments including FGFR2 inhibitors in the first-line therapy for advanced disease and prospective studies may need to account for the emerging therapeutic landscape in BTCs. In this study, ctDNA collection was evaluated before first-line platinum-based chemotherapy and was not powered to evaluate DCAF for further lines of systemic treatment. On the other hand, as stated previously, the presence of targetable genes had no association with DCAF results and impact of ctDNA on OS.
In conclusion, ctDNA is a powerful prognostic tool in advanced BTCs. DCAF at diagnosis of advanced disease for patients who would receive platinum-based systemic therapy identifies patients with a worse prognosis. ctDNA should be evaluated in prospective trials as a stratification factor for advanced disease and as a surrogate for tumor burden.
APPENDIX
TABLE A1.
Multivariate Analysis for Overall Survival
Jun Yin
Employment: Mayo Clinic
Mohamad Bassam Sonbol
Research Funding: Lilly (Inst), Taiho Oncology (Inst)
Daniel H. Ahn
Stock and Other Ownership Interests: Natera
Consulting or Advisory Role: Eisai, Exelixis, Genentech/Roche, Advanced Accelerator Applications, Novartis, Daiichi Sankyo/Astra Zeneca
Research Funding: Bayer, AstraZeneca
Jason S. Starr
Consulting or Advisory Role: Natera, Ipsen, Pfizer, Taiho Oncology, TerSera, Advanced Accelerator Applications
Research Funding: Incyte (Inst), Merus (Inst), Rafael Pharmaceuticals (Inst), Molecular Templates (Inst), MacroGenics (Inst), Vedanta Biosciences (Inst), Leap Therapeutics
Hani Babiker
Consulting or Advisory Role: Endocyte, Celgene, Idera, Myovant Sciences, Novocure
Speakers' Bureau: Guardant Health
Tanios S. Bekaii-Saab
Consulting or Advisory Role: Amgen (Inst), Ipsen (Inst), Lilly (Inst), Bayer (Inst), Roche/Genentech (Inst), AbbVie, Incyte (Inst), Immuneering, Seattle Genetics (Inst), Pfizer (Inst), Boehringer Ingelheim, Janssen, Eisai, Eisai, Daiichi Sankyo/UCB Japan, AstraZeneca, Exact Sciences, Natera, Treos Bio, Celularity, SOBI, BeiGene, Foundation Medicine, Arcus Biosciences (Inst), Stemline Therapeutics, Kanaph Therapeutics, Deciphera, Illumina, Foundation Medicine
Patents, Royalties, Other Intellectual Property: Patent WO/2018/183488, Patent WO/2019/055687
Other Relationship: Exelixis, Merck (Inst), AstraZeneca, Lilly, Pancreatic Cancer Action Network, FibroGen, Suzhou Kintor Pharmaceuticals, 1-Globe, Imugene, Xilis, Replimune, Sun Biopharma, UpToDate
Open Payments Link: https://openpaymentsdata.cms.gov/physician/636276
Gregory J. Gores
Honoraria: Sagimet Biosciences
Rory Smoot
Consulting or Advisory Role: AstraZeneca
Bolni Nagalo
Patents, Royalties, Other Intellectual Property: Patent “Chimeric Vesiculoviruses and Methods of Use” Mitesh J. Borad, and Bolni M. Nagalo
James Bogenberger
Stock and Other Ownership Interests: Xpecting Diagnostics Inc
Research Funding: Agios, RedHill Biopharma, Tolero Pharmaceuticals, Lexicon
Aaron Mansfield
Honoraria: Roche
Consulting or Advisory Role: Genentech (Inst), Bristol Myers Squibb (Inst), AbbVie (Inst), AstraZeneca (Inst)
Research Funding: Novartis (Inst)
Travel, Accommodations, Expenses: AbbVie, Roche
Kabir Mody
Stock and Other Ownership Interests: CytoDyn, ONCOtherapeutics
Consulting or Advisory Role: Celgene, Genentech/Roche, Merrimack, Eisai, AstraZeneca, Vicus Therapeutics, Ipsen, Boston Scientific, BTG, BTG, Exelixis, Exelixis, Incyte (Inst), QED Therapeutics
Research Funding: FibroGen (Inst), Senhwa Biosciences (Inst), ARIAD (Inst), TRACON Pharma (Inst), MedImmune (Inst), Agios (Inst), ArQule (Inst), Taiho Pharmaceutical (Inst), Gritstone Bio (Inst), Incyte (Inst), Merck (Inst), Vyriad (Inst), Turnstone Bio (Inst), AstraZeneca (Inst), Basilea (Inst)
Mitesh J. Borad
Stock and Other Ownership Interests: Gilead Sciences, AVEO, Intercept Pharmaceuticals, Spectrum Pharmaceuticals
Consulting or Advisory Role: G1 Therapeutics, Fujifilm (Inst), Agios (Inst), Insys Therapeutics (Inst), Novartis (Inst), ArQule (Inst), Celgene (Inst), Inspyr Therapeutics, Halozyme (Inst), Pieris Pharmaceuticals (Inst), Taiho Pharmaceutical (Inst), Immunovative Therapies, Exelixis, Lynx Group, Genentech, Western Oncolytics, Klus Pharma, De Novo Pharmaceuticals, Merck, Imvax
Research Funding: Boston Biomedical (Inst), miRNA Therapeutics (Inst), Senhwa Biosciences (Inst), MedImmune (Inst), BioLineRx (Inst), Agios (Inst), Halozyme (Inst), Celgene (Inst), Threshold Pharmaceuticals (Inst), Toray Industries (Inst), Dicerna (Inst), SillaJen (Inst), Eisai (Inst), Taiho Pharmaceutical (Inst), EMD Serono (Inst), Isis Pharmaceuticals (Inst), Incyte (Inst), Sun Biopharma (Inst), ARIAD (Inst), ImClone Systems (Inst), QED Therapeutics (Inst), Incyte (Inst), Puma Biotechnology (Inst), Adaptimmune (Inst), Merck Serono (Inst), RedHill Biopharma (Inst), Basilea (Inst), AstraZeneca (Inst)
Travel, Accommodations, Expenses: ArQule, Celgene, AstraZeneca
No other potential conflicts of interest were reported.
DISCLAIMER
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
SUPPORT
The Mayo Clinic Hepatobiliary SPORE (P50CA 210964) funded the statistical analysis for this project (to M.J.B.). Supported by the National Institute of Health (NIH) through a DP2 Award CA195764 (to M.J.B.), National Cancer Institute (NCI) K12 award CA090628 (to M.J.B.) and K01 award CA234324 (to B.N.), SPORE Project Award 5P50CA210964-03 (to G.J.G. and M.J.B.), SPORE Supplement Award 3P50CA210964-02S1 (to O.B.), Mayo Clinic Center for Individualized Medicine (CIM) Precision Cancer Therapeutics Program, and Mayo Clinic Cancer Center.
K.M. and M.J.B. contributed equally to this work.
DATA SHARING STATEMENT
The data sets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
AUTHOR CONTRIBUTIONS
Conception and design: Pedro Luiz Serrano Uson Junior, Umair Majeed, Jun Yin, Gehan Botrus, Mohamad Bassam Sonbol, Jeremy C. Jones, Ashton W.R. Boyle, Tanios S. Bekaii-Saab, Rory Smoot, Bolni Nagalo, Joachim Petit, Kabir Mody, Mitesh J. Borad
Administrative support: Gehan Botrus, Mansi Arora
Provision of study materials or patients: Gehan Botrus, Tanios S. Bekaii-Saab
Collection and assembly of data: Pedro Luiz Serrano Uson Junior, Umair Majeed, Gehan Botrus, Daniel H. Ahn, Jeremy C. Jones, Samantha R. Inabinett, Natasha Wylie, Ashton W.R. Boyle, Tanios S. Bekaii-Saab, Bolni Nagalo, Joachim Petit, Oumar Barro, Kabir Mody, Mitesh J. Borad
Data analysis and interpretation: Pedro Luiz Serrano Uson Junior, Umair Majeed, Jun Yin, Gehan Botrus, Mohamad Bassam Sonbol, Daniel H. Ahn, Jason S. Starr, Jeremy C. Jones, Hani Babiker, Samantha R. Inabinett, Tanios S. Bekaii-Saab, Gregory J. Gores, Michael Barrett, Bolni Nagalo, Nathalie Meurice, Natalie Elliott, Joachim Petit, Yumei Zhou, Mansi Arora, Chelsae Dumbauld, Alexander Baker, James Bogenberger, Kenneth Buetow, Aaron Mansfield, Kabir Mody, Mitesh J. Borad
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jun Yin
Employment: Mayo Clinic
Mohamad Bassam Sonbol
Research Funding: Lilly (Inst), Taiho Oncology (Inst)
Daniel H. Ahn
Stock and Other Ownership Interests: Natera
Consulting or Advisory Role: Eisai, Exelixis, Genentech/Roche, Advanced Accelerator Applications, Novartis, Daiichi Sankyo/Astra Zeneca
Research Funding: Bayer, AstraZeneca
Jason S. Starr
Consulting or Advisory Role: Natera, Ipsen, Pfizer, Taiho Oncology, TerSera, Advanced Accelerator Applications
Research Funding: Incyte (Inst), Merus (Inst), Rafael Pharmaceuticals (Inst), Molecular Templates (Inst), MacroGenics (Inst), Vedanta Biosciences (Inst), Leap Therapeutics
Hani Babiker
Consulting or Advisory Role: Endocyte, Celgene, Idera, Myovant Sciences, Novocure
Speakers' Bureau: Guardant Health
Tanios S. Bekaii-Saab
Consulting or Advisory Role: Amgen (Inst), Ipsen (Inst), Lilly (Inst), Bayer (Inst), Roche/Genentech (Inst), AbbVie, Incyte (Inst), Immuneering, Seattle Genetics (Inst), Pfizer (Inst), Boehringer Ingelheim, Janssen, Eisai, Eisai, Daiichi Sankyo/UCB Japan, AstraZeneca, Exact Sciences, Natera, Treos Bio, Celularity, SOBI, BeiGene, Foundation Medicine, Arcus Biosciences (Inst), Stemline Therapeutics, Kanaph Therapeutics, Deciphera, Illumina, Foundation Medicine
Patents, Royalties, Other Intellectual Property: Patent WO/2018/183488, Patent WO/2019/055687
Other Relationship: Exelixis, Merck (Inst), AstraZeneca, Lilly, Pancreatic Cancer Action Network, FibroGen, Suzhou Kintor Pharmaceuticals, 1-Globe, Imugene, Xilis, Replimune, Sun Biopharma, UpToDate
Open Payments Link: https://openpaymentsdata.cms.gov/physician/636276
Gregory J. Gores
Honoraria: Sagimet Biosciences
Rory Smoot
Consulting or Advisory Role: AstraZeneca
Bolni Nagalo
Patents, Royalties, Other Intellectual Property: Patent “Chimeric Vesiculoviruses and Methods of Use” Mitesh J. Borad, and Bolni M. Nagalo
James Bogenberger
Stock and Other Ownership Interests: Xpecting Diagnostics Inc
Research Funding: Agios, RedHill Biopharma, Tolero Pharmaceuticals, Lexicon
Aaron Mansfield
Honoraria: Roche
Consulting or Advisory Role: Genentech (Inst), Bristol Myers Squibb (Inst), AbbVie (Inst), AstraZeneca (Inst)
Research Funding: Novartis (Inst)
Travel, Accommodations, Expenses: AbbVie, Roche
Kabir Mody
Stock and Other Ownership Interests: CytoDyn, ONCOtherapeutics
Consulting or Advisory Role: Celgene, Genentech/Roche, Merrimack, Eisai, AstraZeneca, Vicus Therapeutics, Ipsen, Boston Scientific, BTG, BTG, Exelixis, Exelixis, Incyte (Inst), QED Therapeutics
Research Funding: FibroGen (Inst), Senhwa Biosciences (Inst), ARIAD (Inst), TRACON Pharma (Inst), MedImmune (Inst), Agios (Inst), ArQule (Inst), Taiho Pharmaceutical (Inst), Gritstone Bio (Inst), Incyte (Inst), Merck (Inst), Vyriad (Inst), Turnstone Bio (Inst), AstraZeneca (Inst), Basilea (Inst)
Mitesh J. Borad
Stock and Other Ownership Interests: Gilead Sciences, AVEO, Intercept Pharmaceuticals, Spectrum Pharmaceuticals
Consulting or Advisory Role: G1 Therapeutics, Fujifilm (Inst), Agios (Inst), Insys Therapeutics (Inst), Novartis (Inst), ArQule (Inst), Celgene (Inst), Inspyr Therapeutics, Halozyme (Inst), Pieris Pharmaceuticals (Inst), Taiho Pharmaceutical (Inst), Immunovative Therapies, Exelixis, Lynx Group, Genentech, Western Oncolytics, Klus Pharma, De Novo Pharmaceuticals, Merck, Imvax
Research Funding: Boston Biomedical (Inst), miRNA Therapeutics (Inst), Senhwa Biosciences (Inst), MedImmune (Inst), BioLineRx (Inst), Agios (Inst), Halozyme (Inst), Celgene (Inst), Threshold Pharmaceuticals (Inst), Toray Industries (Inst), Dicerna (Inst), SillaJen (Inst), Eisai (Inst), Taiho Pharmaceutical (Inst), EMD Serono (Inst), Isis Pharmaceuticals (Inst), Incyte (Inst), Sun Biopharma (Inst), ARIAD (Inst), ImClone Systems (Inst), QED Therapeutics (Inst), Incyte (Inst), Puma Biotechnology (Inst), Adaptimmune (Inst), Merck Serono (Inst), RedHill Biopharma (Inst), Basilea (Inst), AstraZeneca (Inst)
Travel, Accommodations, Expenses: ArQule, Celgene, AstraZeneca
No other potential conflicts of interest were reported.
REFERENCES
- 1.de Groen PC, Gores GJ, LaRusso NF, et al. : Biliary tract cancers. N Engl J Med 341:1368-1378, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Charbel H, Al-Kawas FH: Cholangiocarcinoma: Epidemiology, risk factors, pathogenesis, and diagnosis. Curr Gastroenterol Rep 13:182-187, 2011 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Fuchs HE, et al. : Cancer statistics, 2021. CA Cancer J Clin 71:7-33, 2021 [DOI] [PubMed] [Google Scholar]
- 4.Shroff RT, Kennedy EB, Bachini M, et al. : Adjuvant therapy for resected biliary tract cancer: ASCO clinical practice guideline. J Clin Oncol 37:1015-1027, 2019 [DOI] [PubMed] [Google Scholar]
- 5.Farley DR, Weaver AL, Nagorney DM: “Natural history” of unresected cholangiocarcinoma: Patient outcome after noncurative intervention. Mayo Clin Proc 70:425-429, 1995 [DOI] [PubMed] [Google Scholar]
- 6.Valle J, Wasan H, Palmer DH, et al. : Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 362:1273-1281, 2010 [DOI] [PubMed] [Google Scholar]
- 7.Valle JW, Lamarca A, Goyal L, et al. : New horizons for precision medicine in biliary tract cancers. Cancer Discov 7:943-962, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mody K, Kasi PM, Yang J, et al. : Circulating tumor DNA profiling of advanced biliary tract cancers. JCO Precis Oncol 3:1-9, 2019 [DOI] [PubMed] [Google Scholar]
- 9.Lu L, Bi J, Bao L: Genetic profiling of cancer with circulating tumor DNA analysis. J Genet Genomics 45:79-85, 2018 [DOI] [PubMed] [Google Scholar]
- 10.Hovelson DH, Liu CJ, Wang Y, et al. : Rapid, ultra low coverage copy number profiling of cell-free DNA as a precision oncology screening strategy. Oncotarget 8:89848-89866, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goyal L, Saha SK, Liu LY, et al. : Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion–positive cholangiocarcinoma. Cancer Discov 7:252-263, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reece M, Saluja H, Hollington P, et al. : The use of circulating tumor DNA to monitor and predict response to treatment in colorectal cancer. Front Genet 10:1118, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Lai H, Liu J, et al. : Circulating tumor DNA predicts the response and prognosis in patients with early breast cancer receiving neoadjuvant chemotherapy. JCO Precis Oncol 4:244-257, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pairawan S, Hess KR, Janku F, et al. : Cell-free circulating tumor DNA variant allele frequency associates with survival in metastatic cancer. Clin Cancer Res 26:1924-1931, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanman RB, Mortimer SA, Zill OA, et al. : Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS one 10:e0140712, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botrus G, Kosirorek H, Sonbol MB, et al. : Circulating tumor DNA‐based testing and actionable findings in patients with advanced and metastatic pancreatic adenocarcinoma. Oncologist 26:569-578, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamura R, Kurzrock R, Mallory RJ, et al. : Comprehensive genomic landscape and precision therapeutic approach in biliary tract cancers. Int J Cancer 148:702-712, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strijker M, Soer EC, de Pastena M, et al. : Circulating tumor DNA quantity is related to tumor volume and both predict survival in metastatic pancreatic ductal adenocarcinoma. Int J Cancer 146:1445-1456, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernard V, Kim DU, San Lucas FA, et al. : Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterology 156:108-e4, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel H, Okamura R, Fanta P, et al. : Clinical correlates of blood-derived circulating tumor DNA in pancreatic cancer. J Hematol Oncol 12:130, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dasari A, Morris VK, Allegra CJ, et al. : ctDNA applications and integration in colorectal cancer: An NCI Colon and Rectal–Anal Task Forces whitepaper. Nat Rev Clin Oncol 17:757-770, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cullinane C, Fleming C, O'Leary DP, et al. : Association of circulating tumor DNA with disease-free survival in breast cancer: A systematic review and meta-analysis. JAMA Netw Open 3:e2026921, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodall J, Assaf ZJ, Shi Z, et al. : Circulating tumor DNA (ctDNA) dynamics associate with treatment response and radiological progression-free survival (rPFS): Analyses from a randomized phase II trial in metastatic castration-resistant prostate cancer (mCRPC). J Clin Oncol 38, 2020. (suppl; abstr 5508) [Google Scholar]
- 24.Bridgewater J, Lopes A, Wasan H, et al. : Prognostic factors for progression-free and overall survival in advanced biliary tract cancer. Ann Oncol 27:134-140, 2016 [DOI] [PubMed] [Google Scholar]
- 25.Peixoto RD, Renouf D, Lim H: A population based analysis of prognostic factors in advanced biliary tract cancer. J Gastrointest Oncol 5:428-432, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park HS, Park JS, Chun YJ, et al. : Prognostic factors and scoring model for survival in metastatic biliary tract cancer. Cancer Res Treat 49:1127-1139, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salati M, Filippi R, Vivaldi C, et al. : The prognostic nutritional index predicts survival and response to first‐line chemotherapy in advanced biliary cancer. Liver Int 40:704-711, 2020 [DOI] [PubMed] [Google Scholar]
- 28.Kim BJ, Hyung J, Yoo C, et al. : Prognostic factors in patients with advanced biliary tract cancer treated with first-line gemcitabine plus cisplatin: Retrospective analysis of 740 patients. Cancer Chemother Pharmacol 80:209-215, 2017 [DOI] [PubMed] [Google Scholar]