PURPOSE
Comprehensive genomic profiling (CGP) assay is increasingly used in low-middle–income countries to detect clinically relevant genomic alterations despite its clinical benefits not being well known. Here, we describe the proportion of patients with advanced cancer in India who received targeted therapy for an actionable genetic alteration identified on CGP assays.
METHODS
This was a multicenter, retrospective cohort study in adult patients with advanced nonhematologic malignancies who underwent a CGP test. If patients received a targeted therapy for ≥ 6 months, they were considered to have obtained a clinical benefit from the medication, whereas those continuing for ≥ 12 months were considered to have attained an exceptional response. Descriptive statistics were used to describe the proportion of patients with subsequent targeted therapy.
RESULTS
During 2019-2020, 12 medical oncologists provided CGP reports for 297 patients; 221 met the inclusion criteria. Patients received a median of two lines (range: 0-5) of prior systemic therapy. On the basis of the CGP assay, 21 patients (10%) received targeted therapy. Among them, 33% was for human epidermal growth factor receptor 2 (HER2) amplification (nonbreast cancer) and 19% for HER2 or epidermal growth factor receptor exon 20 insertion mutation (lung cancer). After excluding patients with HER2 or epidermal growth factor receptor exon 20 insertions, 8% of 217 patients received targeted therapy. In the overall cohort of 221 patients, clinical benefit was seen in nine patients (4%), of whom two were exceptional responders (1%).
CONCLUSION
We observed that in a low-middle–income country setting, 10% of patients received targeted therapy on the basis of CGP assay. Only 4% of patients who underwent CGP testing obtained a clinical benefit.
INTRODUCTION
Cancer arises because of mutations in the human genome, resulting in the development of neoplastic cells. Next-generation sequencing (NGS) techniques provide us with the genetic profile of the patient's cancer, which helps in identifying clinically relevant genetic alterations that can serve as targets for potential therapies. The first molecular targeted therapy on the basis of genetic profiling was trastuzumab in patients with ERBB2-overexpressed breast cancer introduced in 1998. Since then, a large number of novel targeted therapies were discovered including bcr/abl inhibitors in chronic myeloid leukemia, BRAF inhibitors in melanoma, epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors in non–small-cell lung cancer, which yielded robust therapeutic responses. As of 2019, there are 64 Food and Drug Administration–approved molecular targeted therapies used in cancer treatment.
CONTEXT
Key Objective
We aimed to study the clinical benefit of comprehensive genomic profiling in advanced cancers in India.
Knowledge Generated
We observed that 10% of patients who underwent testing received a targeted therapy and 4% obtained a clinical benefit, defined as receiving a targeted therapy for at least 6 months.
Relevance
Our findings are unique from the perspective of clinical practice in a low- and middle-income country. It would inform shared decision making on the role of such testing in advanced cancers.
Several comprehensive genomic profiling (CGP) testing platforms such as MedGenome, Caris, and Foundation One are currently available, each assessing different types and numbers of genes. Remarkable antineoplastic activity associated with targeted therapies has led to a rise in the usage of CGP to detect clinically relevant genomic alteration. Although a large number of patients profiled have genomic alterations that could be targeted by molecular therapies, only a very small fraction of them actually receive these sequencing-directed therapies.1-8 In a recent study by Cobain, 80.5% of the profiled patients had actionable genetic variations but only 16.2% received targeted therapy on the basis of CGP assay.1 Among those patients who received targeted therapy, only 37.1% (4.82% of total patients profiled) reported clinical benefits.1 Similar results were also stated by other studies where clinical benefits were seen in only 2%-8% of the total patients profiled.1,4-6 SHIVA trial, the sole randomized study which assessed the therapeutic benefits of targeted therapy on the basis of CGP assay, reported that treatment with matched molecular therapy did not yield any significant increase in progression-free survival of patients with cancer.9 In contradiction to this, there are other nonrandomized studies that report superior clinical outcomes associated with sequencing-directed molecular therapies.3,5,8,10-13
Despite the availability and clinical use of CGP among patients with cancer in low-middle–income countries (LMICs), there is a dearth of systemic studies that assess its clinical utility. This calls for further studies that evaluate the influence of CGP assay on treatment decisions so that its therapeutic benefits could be determined. In our multicenter retrospective study, we describe the proportion of patients with advanced cancers in India who eventually received targeted molecular therapy on the basis of CGP assay. We also estimate the clinical benefit of such a testing and therapeutic strategy.
METHODS
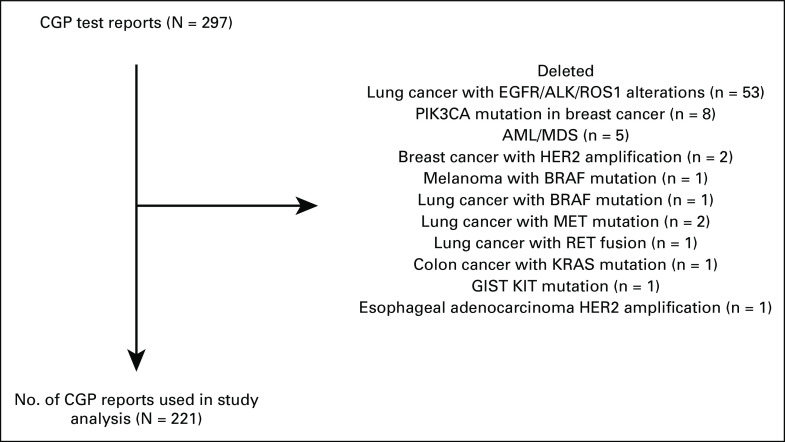
We conducted a multicenter retrospective cohort study conducted among patients with advanced cancer in India. Adult patients (age ≥ 18 years) with histologically confirmed nonhematologic advanced malignancies who underwent CGP assay were eligible for enrollment in the study. Patients with any type of tumor, stage of the disease, and undergoing any standard therapy were eligible for inclusion. No restrictions were placed on the number of lines of systemic therapy received by patients before CGP assay. Patients with genetic alterations that have well-validated standardized targeted therapy were excluded (eg; EGFR exon 18, 19, 21 mutations for lung cancer, human epidermal growth factor receptor 2 (HER2) amplification and PIK3CA mutation in breast cancer, BRAF mutation in melanoma and lung cancer, KRAS mutation in colon cancer, KIT mutation in GI stromal tumors, and HER2 amplification in esophageal adenocarcinoma; Fig 1).
FIG 1.
Study profile. AML, acute myelomonocytic leukemia; CGP, comprehensive genomic profiling; EGFR, epidermal growth factor receptor; GIST, GI stromal tumors; HER2, human epidermal growth factor receptor 2; MDS, myelodysplastic syndrome.
Sequencing-directed therapies was said to be provided only when CGP could identify an actionable mutation in a patient for which either an off-label therapy or clinical trial was available. Institutional Ethics Committee approval was obtained. Data on the eligible patients were collected from 12 participating oncologists from different parts of the country. Data were collected between August 2020 and December 2021. The oncologists provided anonymized information of consecutive CGP test reports along with patient's demographic characteristics through an online data capture form. Details on patient demographics, biopsy type, prior lines of systemic therapy, type and number of mutations, CGP platform used, genome-directed therapy received, and rationale behind the rejection of sequencing-directed therapies were extracted from data capture forms.
Proportion of profiled patients receiving sequencing-directed therapies and the reasons for declining targeted therapies were then evaluated using descriptive statistics.
Comprehensive Genomic Sequencing
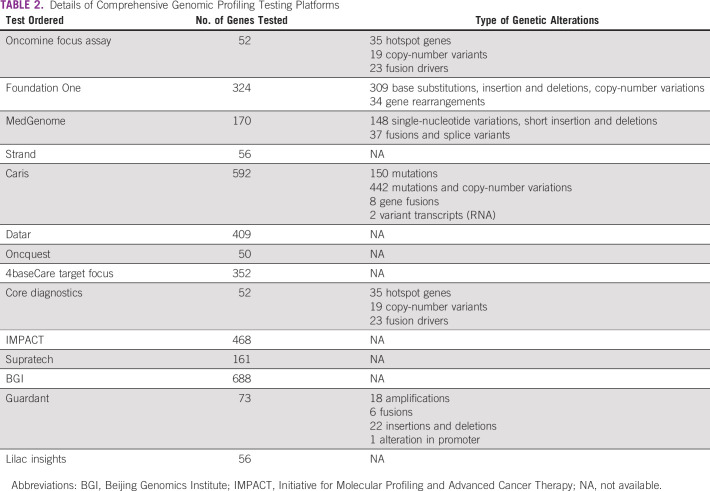
CGP assay could be performed on both tissue and liquid biopsy. Either archived tissue or fresh tissue specimens were used for tissue analysis. The decision to undertake CGP assays was made by the treating oncologist. CGP by NGS techniques could simultaneously detect different types of genetic alterations such as insertion, deletion, fusion, amplification, rearrangement, and mutations of genes. Many different platforms such as MedGenome, Foundation One, and Oncomine Focus assay were used for CGP. Each of these platforms assessed different genes and types of genetic alterations.
Report Interpretation
CGP report identifies all the actionable genetic alterations present in the patient along with possible treatment modalities. Therapeutic intervention on the basis of CGP assay was at the discretion of primary oncologists, which could be in the form of administration of an on-label or off-label drug or enrollment in a clinical trial. If the results were considered ineffective by the treating oncologist, other treatment modalities were explored.
Clinical End Points
The primary end point of the study was to assess the clinical utility of NGS by determining the proportion of patients with advanced cancer in India who eventually receive targeted therapy for an actionable genetic alteration identified on the CGP assay. Secondary objective was to determine the clinical benefit of genome-directed therapy. If patients received a targeted therapy for ≥ 6 months, they were considered to have obtained a clinical benefit from the medication. Patients on targeted therapy for ≥ 12 months were considered to have obtained an exceptional response from the medication.
Statistical Analysis
Descriptive statistics were used to describe the proportion of patients with subsequent targeted therapy.
RESULTS
We received CGP reports of 297 patients from 12 participating oncologists from different parts of the country. As per our inclusion criteria, 221 patients were included in our study (Table 1). Remaining CGP reports were excluded as they identified genetic alterations that have well-validated Food and Drug Administration–approved standard-of-care therapy. A description of the study profile is given in Figure 1. Fourteen different testing panels were used by the oncologists (Table 2).
TABLE 1.
Baseline Characteristics (N = 221)
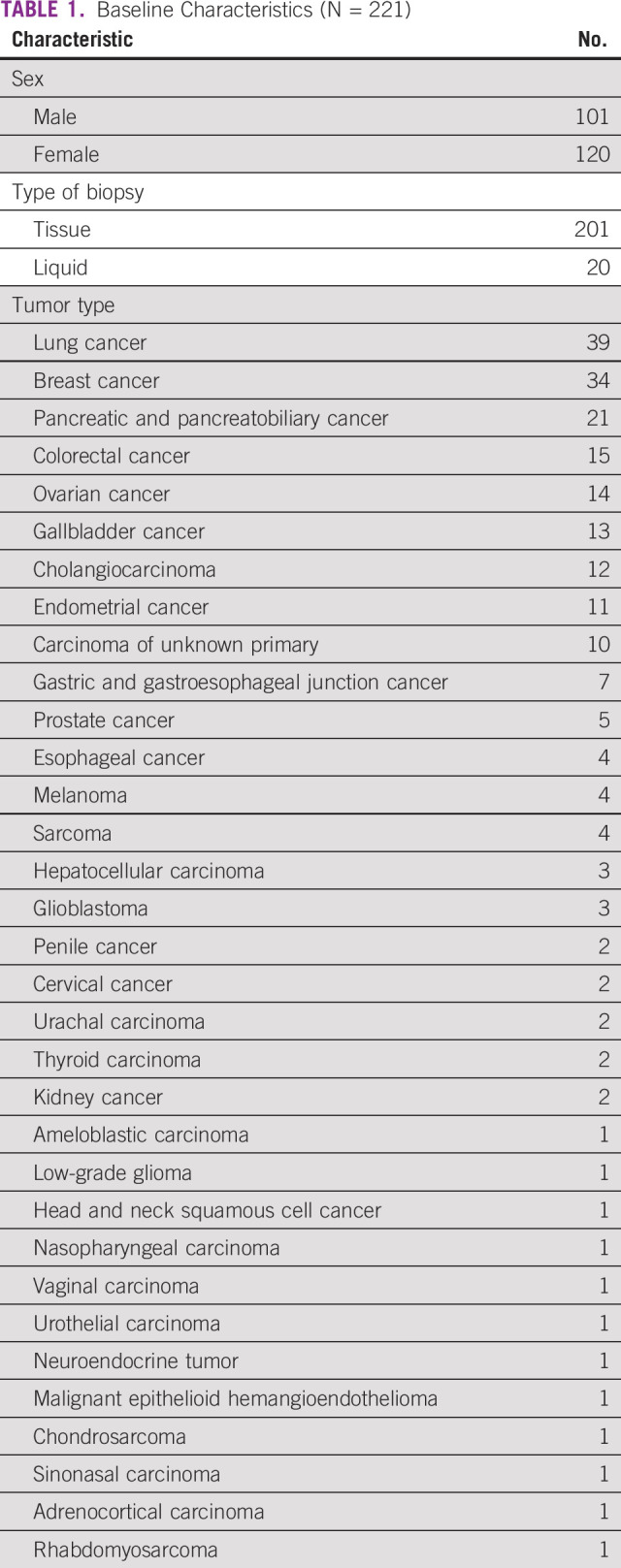
TABLE 2.
Details of Comprehensive Genomic Profiling Testing Platforms
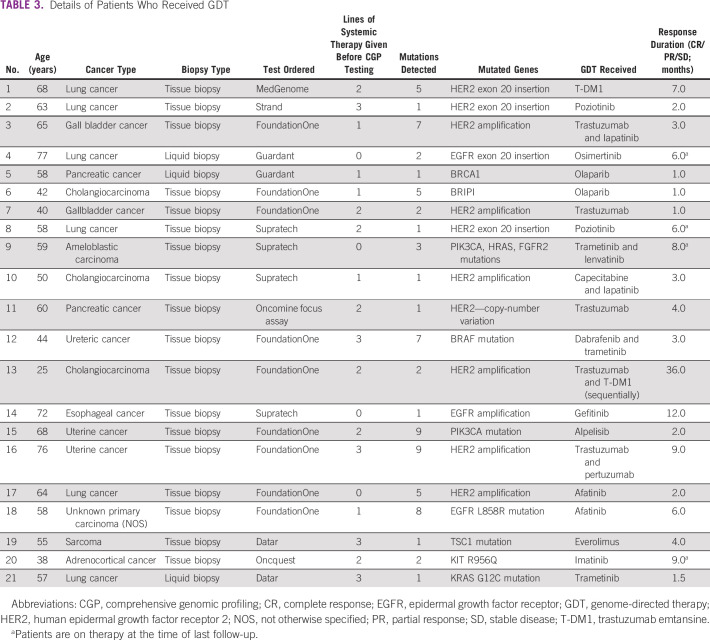
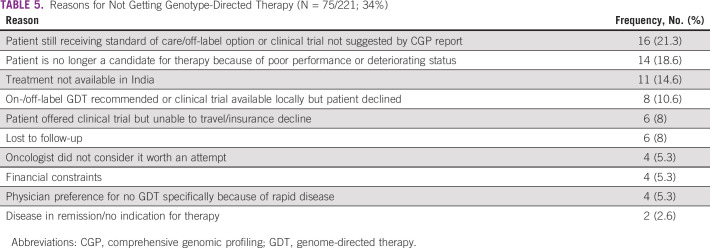
Among the study cohort, 46% were male. Ninety-one percent underwent tumor tissue biopsy and 9% had liquid biopsy. The most common cancers were lung (18%), breast (15%), pancreatobiliary (9%), and colorectal (7%). Patients received a median of two lines (range: 0-5) of prior systemic therapy (Table 1). Ninety-six patients (43%) had a targeted therapy option in the form of an approved drug or the availability of a globally recruiting clinical trial. On the basis of the CGP assay, 21 patients (10%) received targeted therapy (Table 3). Among them, 33% (n = 7) was for HER2 amplification (nonbreast cancer) and 19% for HER2 or EGFR exon 20 insertion mutation (lung cancer; n = 4; Table 4). EGFR mutation was seen in two patients. After excluding patients with HER2 or EGFR exon 20 insertion (four patients), which are emerging targets in lung cancer, 17 patients of 217 (8%) received targeted therapy. Most common reason for patients not receiving targeted therapy was because of availability of a standard-of-care therapy (21%) and declining functional status (18%; Table 5). In the overall cohort of 221 patients, clinical benefit was seen in nine patients (4%), of whom two were exceptional responders (1%). Excluding the emerging targets in lung cancer, clinical benefit was seen in six of 217 patients (3%), of whom two were exceptional responders.
TABLE 3.
Details of Patients Who Received GDT
TABLE 4.
Common Targets That Were Used
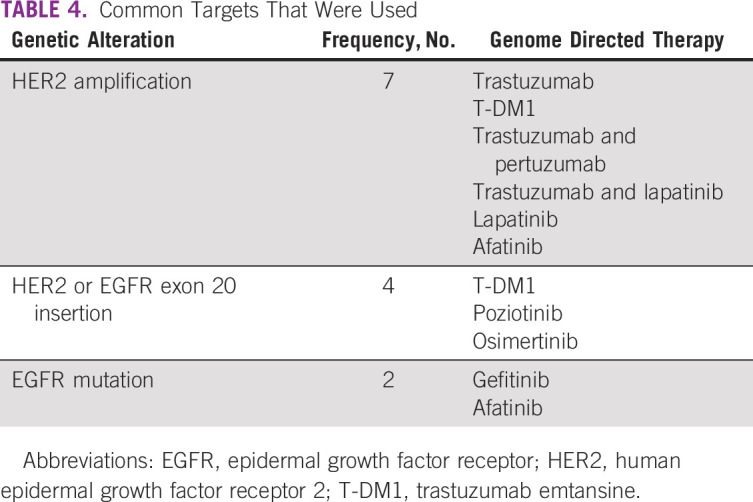
TABLE 5.
Reasons for Not Getting Genotype-Directed Therapy (N = 75/221; 34%)
DISCUSSION
Personalized medicine on the basis of alteration in patients' genome is an area of promising development in cancer therapy.14 Replacing nonselective conventional standard therapies with targeted genomic drugs tailored against a specific mutation on tumor cells is appealing to both doctors as well as the general public. Improved clinical benefits associated with certain specific targeted therapies and the dropping cost of CGP have resulted in a greater demand for the assays. Although there are some studies evaluating the clinical utility of CGP in regular practice, the results are largely contradictory.1,8,9,15 To the best of our knowledge, there are no similar studies conducted in LMICs. The clinical benefits of CGP must be adequately studied and properly established before it can be integrated into regular clinical practice. This is especially important in a developing country such as India with a resource-constrained health sector where CGP still remains exceedingly expensive for large sections of populations.
NGS is often prescribed for patients when we seek a treatment option in the form of a targeted therapy. Most often, it is done when patients have exhausted all available standard-of-care therapeutic options. However, in some situations, even upfront testing is routinely done—for instance, for patients with adenocarcinoma of the lung. It is also recommended in the context of clinical trial recruitment for studies of novel investigative strategies such as targeted therapy. In this study, we aimed to assess the benefits of CGP in routine clinical practice in India. Patients with solid organ malignancies who underwent CGP were included in our study. Patients found to have genetic alterations, which otherwise would have been detected with a targeted genetic testing, were not included in the study population—for example, patients with HER2 amplification that would have been detected with a HER2 fluorescence in situ hybridization test, or patients with EGFR sensitizing mutation in exon 19 that would have been detected with a targeted EGFR mutation testing.
Of the 221 who were enrolled in our study, actionable mutations were found only in 96 (43%). No mutations or nonactionable mutations were detected in the remaining patients. This is much less compared with studies conducted elsewhere where the proportion of clinically relevant genomic alteration was more than 80%.1,16,17 However, the proportion of patients who eventually received sequencing-directed therapies on the basis of an actionable genetic alteration identified by CGP assay was 10%. This is comparable to similar studies from other parts of the world.1,16-18 Clinical benefit, as defined as being on the drug for ≥ 6 months, was noted in only 4% of patients. Although the scope of precision therapies continues to be immense, the patient needs to be made aware of the clinical reality that only a small fraction of patients who undergo CGP receive sequencing-directed therapies and a still smaller fraction actually gain benefit from it.1 Awareness of these data would help in patient counseling so that the expectations match the real-world scenario. Effective communication between the patient and their oncologist would help in the curbing of exaggerated expectations associated with precision therapy.
Clinically relevant genomic alteration reported by CGP platforms can either be a mutation with well-validated targeted treatment or a clinically unproven hypothetical target against which a potential therapy may be successful.19 Many of the CGP platforms do not prioritize the various targetable mutation discovered and it is often up to the oncologist to select the clinically efficient evidence-based targeted therapy.19 The use of clinical value-based ranking of actionable mutation similar to European Society for Medical Oncology Scale for Clinical Actionability of molecular Targets would enable the oncologist in selecting actionable mutations with proven clinical benefits.19 Some highly recommended targets in the European Society for Medical Oncology Scale for Clinical Actionability of molecular Targets include HER2 in breast cancer and ROS1 rearrangements.19
CGP in clinical practice should be used with utmost care. We are of the view that the nonjudicious and indiscriminate use of CGP would be of limited clinical value. Beyond the standard indications, NGS must be restricted to patient populations in specific clinical situations. It may be prescribed in patients with limited therapeutic options where the sequencing-directed therapies may help in enrolling in a clinical trial or receiving an off-label therapy.20 The clinical utility of genomic profiling is not uniform across all cancers. It is especially helpful in non–small-cell lung cancer, which often presents with several genomic alterations that can be targeted in the initial therapy.20 Rare cancers without well-defined treatment guidelines or standard drug regimens are another accepted indication for NGS.20 However, it should be avoided in terminally ill patients with aggressive cancer, and those with poor functional status.1,20 It should also not be ordered in cancers against which standard conventional therapies with proven clinical value are already available or in those cases where genomic sequencing is unlikely to detect a new targetable genomic mutation or have any substantial influence on therapeutic management.1,20
The financial aspect of NGS should also be taken into perspective. Although it is considered as a cheaper alternative to single-gene testing, it continues to remain unaffordable to a large number of patients with cancer in India.21 Apart from the cost of sequencing, the economic burden resulting from the targeted therapy that ensues must also be taken into account while determining the cost-effectiveness of NGS. The economic impact of precision therapy has added importance in a country like India with limited resources, poor insurance coverage, and high out-of-pocket expenditure. There can also be physical toxicities especially when combination therapies with no previous data are used.
Precision medicine has the potential to revolutionize oncology in the coming years. Genomic profiling was instrumental in improving our understanding of many of the advanced cancers. It has provided us with a wide variety of therapeutic options for cancers that were considered largely untreatable. We must now focus on rational implementation of precision medicine in routine clinical practice. We encourage the integration of CGP into routine clinical practice, especially when it is affordable. However, we must be able to estimate the real-world worth of the assay. To the best of our knowledge, this is the first study from an LMIC context that contextualizes the utility of the assay in a clinical practice setting. Setting up of molecular tumor boards is another implementation measure that can enhance the science of precision medicine from the laboratory to the clinic. It would help oncologists choose therapeutic options with the meaningful clinical benefit. Greater access to clinical trials would enhance our ability to translate the results of genomic profiling to clinical practice.
Our study has some limitations. First, it is a retrospective cohort study and may be subject to recall bias. However, being a multicenter study, our results can be considered more generalizable and valid. Decision to perform CGP was made at the discretion of the primary oncologist and not by institutional tumor boards. Therefore, the decision-making process may be subject to individual physician bias. Comparing the outcomes from our study with the results from molecular tumor board–based decision making will be an important step in testing the impact of such collaborative endeavors. There was also heterogeneity pertaining to the timing of CGP testing. The number of lines of systemic therapy received before CGP varied between 0 to 5. Small sample size and diversity in tumor types are other key limitations.
In conclusion, in our study, we aimed to analyze the clinical value of CGP testing via NGS in routine clinical practice in India. Of the 221 patients sequenced, only 21 patients (10%) received sequencing-directed therapies. Only 4% gained some clinical benefit from the CGP testing. Evidence synthesized in this study could help in the development of clinical interventions aimed at improving the practice of precision medicine in India.
Nitesh Rohatgi
Honoraria: Lilly
Consulting or Advisory Role: Pfizer
Bhawna Sirohi
Honoraria: Roche Pharma AG, Eisai
Consulting or Advisory Role: Sanofi (Inst), Merck Serono (Inst)
Research Funding: Dr. Reddy's (Inst), Roche (Inst), Novartis (Inst)
Patents, Royalties, Other Intellectual Property: Elsevier Book on Modern GI Oncology, Harper Collins—book on cancer
No other potential conflicts of interest were reported.
DATA SHARING STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
AUTHOR CONTRIBUTIONS
Conception and design: Aju Mathew, Serena Joseph
Financial support: Aju Mathew, Nirmal Raut
Administrative support: Aju Mathew, Nirmal Raut
Provision of study materials or patients: Nitesh Rohatgi, Reetu Jain, Vivek Agarwala, Deepak Kumar Shukla, Anurag Mehta, Vineet Talwar, Nirmal Raut
Collection and assembly of data: Aju Mathew, Serena Joseph, Nitesh Rohatgi, Bhawna Sirohi, Reetu Jain, Vivek Agarwala, Deepak Kumar Shukla, Anurag Mehta, Raja Pramanik, Vineet Talwar, Vinayak Maka, Nirmal Raut
Data analysis and interpretation: Aju Mathew, Serena Joseph, Jeffrey Boby, Steve Benny, Janeesh Veedu, Senthil Rajappa, Nitesh Rohatgi, Bhawna Sirohi, Vivek Agarwala, Vinayak Maka, Nirmal Raut
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/go/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Nitesh Rohatgi
Honoraria: Lilly
Consulting or Advisory Role: Pfizer
Bhawna Sirohi
Honoraria: Roche Pharma AG, Eisai
Consulting or Advisory Role: Sanofi (Inst), Merck Serono (Inst)
Research Funding: Dr. Reddy's (Inst), Roche (Inst), Novartis (Inst)
Patents, Royalties, Other Intellectual Property: Elsevier Book on Modern GI Oncology, Harper Collins—book on cancer
No other potential conflicts of interest were reported.
REFERENCES
- 1.Hilal T, Nakazawa M, Hodskins J, et al. : Comprehensive genomic profiling in routine clinical practice leads to a low rate of benefit from genotype-directed therapy. BMC Cancer 17:602, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zehir A, Benayed R, Shah RH, et al. : Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23:703-713, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuxen IV, Rohrberg KS, Oestrup O, et al. : Copenhagen Prospective Personalized Oncology (CoPPO)-Clinical utility of using molecular profiling to select patients to phase I trials. Clin Cancer Res 25:1239-1247, 2019 [DOI] [PubMed] [Google Scholar]
- 4.Johnson DB, Dahlman KH, Knol J, et al. : Enabling a genetically informed approach to cancer medicine: A retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next-generation sequencing panel. Oncologist 19:616-622, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsimberidou AM, Iskander NG, Hong DS, et al. : Personalized medicine in a phase I clinical trials program: The MD Anderson Cancer Center initiative. Clin Cancer Res 18:6373-6383, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltran H, Eng K, Mosquera JM, et al. : Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol 1:466-474, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirshfield KM, Tolkunov D, Zhong H, et al. : Clinical actionability of comprehensive genomic profiling for management of Rare or refractory cancers. Oncologist 21:1315-1325, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheler JJ, Janku F, Naing A, et al. : Cancer therapy directed by comprehensive genomic profiling: A single center study. Cancer Res 76:3690-3701, 2016 [DOI] [PubMed] [Google Scholar]
- 9.Le Tourneau C, Delord JP, Gonçalves A, et al. : Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 16:1324-1334, 2015 [DOI] [PubMed] [Google Scholar]
- 10.Zhou C, Wu YL, Chen G, et al. : Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol 12:735-742, 2011 [DOI] [PubMed] [Google Scholar]
- 11.Radovich M, Kiel PJ, Nance SM, et al. : Clinical benefit of a precision medicine based approach for guiding treatment of refractory cancers. Oncotarget 7:56491-56500, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tredan O, Corset V, Wang Q, et al. : Routine molecular screening of advanced refractory cancer patients: An analysis of the first 2490 patients of the ProfiLER study. J Clin Oncol 35, 2017. (18_suppl; LBA100) [Google Scholar]
- 13.Tsimberidou AM, Hong DS, Ye Y, et al. : Initiative for Molecular Profiling and Advanced Cancer Therapy (IMPACT): An MD Anderson precision medicine study. JCO Precis Oncol 10.1200/PO.17.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyman DM, Taylor BS, Baselga J: Implementing genome-driven oncology. Cell 168:584-599, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Von Hoff DD, Stephenson JJ Jr, Rosen P, et al. : Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol 28:4877-4883, 2010 [DOI] [PubMed] [Google Scholar]
- 16.Cobain EF, Wu YM, Vats P, et al. : Assessment of clinical benefit of integrative genomic profiling in advanced solid tumors. JAMA Oncol 7:525-533, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh AP, Shum E, Rajdev L, et al. : Impact and diagnostic gaps of comprehensive genomic profiling in real-world clinical practice. Cancers (Basel) 12:1156, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nesline MK, DePietro P, Dy GK, et al. : Oncologist uptake of comprehensive genomic profile guided targeted therapy. Oncotarget 10:4616-4629, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mateo J, Chakravarty D, Dienstmann R, et al. : A framework to rank genomic alterations as targets for cancer precision medicine: The ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann Oncol 29:1895-1902, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colomer R, Mondejar R, Romero-Laorden N, et al. : When should we order a next generation sequencing test in a patient with cancer? EClinicalMedicine 25:100487, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruneri G, De Braud F, Sapino A, et al. : Next-generation sequencing in clinical practice: Is it a cost-saving alternative to a single-gene testing approach? Pharmacoecon Open 5:285-298, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.