PURPOSE
Guidelines for prostate cancer (PCA) germline testing (GT) have expanded, with impact on clinical management and hereditary cancer assessment. African American (AA) men have lower engagement in GT, with concern for widening disparities in genetically informed care. We evaluated the germline spectrum in a cohort of men with PCA enriched for AA men who underwent GT to inform tailored genetic evaluation strategies.
METHODS
Participants included AA and White men with PCA tested with a 14-gene PCA panel: ATM, BRCA1, BRCA2, CHEK2, EPCAM, HOXB13, MLH1, MSH2, MSH6, NBN, PALB2, PMS2, RAD51D, and TP53. Germline analysis was performed per standard clinical testing and variant classification protocols. Data were compared with Fisher's exact, chi-squared, or two sample t-tests, as appropriate. Multivariable analysis was conducted using Akaike's Information Criterion. The significance level was set a priori at .05.
RESULTS
The data set included 427 men all tested using the 14-gene PCA panel: AA (n = 237, 56%) and White (n = 190, 44%). Overall, the pathogenic/likely pathogenic (P/LP) variant rate was 8.2%, with AA men having lower rates of P/LP variants then White men (5.91% v 11.05%, respectively; P = .05). Borderline associations with P/LP variant status were observed by race (AA v White; odds ratio = 0.51; P = .07) and age (> 50 v ≤ 50 years; odds ratio = 0.48; P = .06). The P/LP spectrum was narrower in AA men (BRCA2, PALB2, ATM, and BRCA1) than White men (BRCA2, ATM, HOXB13, CHEK2, TP53, and NBN). A significant difference was noted in rates of variants of uncertain significance (VUSs) between AA men and White men overall (25.32% v 16.32%; P = .02) and for carrying multiple VUSs (5.1% v 0.53%, P = .008).
CONCLUSION
Germline evaluation in a cohort enriched for AA men highlights the narrower spectrum of germline contribution to PCA with significantly higher rates of multiple VUSs in DNA repair genes. These results underscore the imperative to engage AA men in GT, the need for larger panel testing in AA men, and the necessity to incorporate novel genomic technologies to clarify VUS to discern the germline contribution to PCA. Furthermore, tailored genetic counseling for AA men is important to ensure understanding of VUS and promote equitable genetics care delivery.
BACKGROUND
Germline testing (GT) for prostate cancer (PCA) has become central for PCA treatment, screening, and hereditary cancer assessment.1-4 In the metastatic setting, a major indication for GT is to inform eligibility for targeted therapies, such as poly (ADP-ribose) polymerase inhibitors, among men with metastatic, castration-resistant prostate cancer (mCRPC) who carry pathogenic/likely pathogenic variants (P/LPVs) in DNA repair genes.5,6 Two poly (ADP-ribose) polymerase inhibitors have been currently approved by the US Food and Drug Administration for men with mCRPC who have progressed on initial therapies.5,6 Olaparib was approved for men with mCRPC who progressed after prior treatment with abiraterone or enzalutamide and who carry P/LPVs in a host of DNA repair genes, some of which include BRCA1, BRCA2, ATM, CHEK2, PALB2, and RAD51B/C/D.5 Rucaparib was approved for men with mCRPC who progressed on prior androgen receptor–directed therapy and taxane-based chemotherapy and who carry P/LPVs in BRCA1 or BRCA2.6 Therefore, therapeutic decision making for men with mCRPC has become a major driver of GT.
CONTEXT
Key Objective
What is the germline spectrum contributing to prostate cancer (PCA) among African American (AA) men? This study performed in a cohort enriched for AA men sought to characterize the germline pathogenic variant spectrum, along with the spectrum of variants of uncertain significance (VUSs), among AA and White men who had undergone a 14-gene PCA genetic test.
Knowledge Generated
The results highlight the narrower spectrum of germline contribution to PCA with significantly higher rates of multiple VUSs in DNA repair genes among AA men.
Relevance
These findings underscore the imperative to engage AA men in germline testing, consideration of comprehensive panel testing in AA men, and the necessity to incorporate novel genomic technologies to clarify VUS to discern the germline contribution to PCA. Tailored genetic counseling for AA men is essential to promote equitable genetics care delivery.
Furthermore, GT can also inform PCA screening. Informed by results from large PCA screening studies focused on BRCA carriers such as the IMPACT trial,7 National Comprehensive Cancer Network (NCCN) guidelines recommend that PCA screening starts at age 40 years for men who carry P/LPVs in BRCA2, with a similar consideration for men who carry P/LPVs in BRCA1.2 A national study led by the National Cancer Institute is currently recruiting men who carry P/LPVs in a host of genes linked with PCA including BRCA1, BRCA2, and DNA Mismatch Repair (MMR) genes (MLH1, MSH2, MSH6, PMS2, and EPCAM) and HOXB13, ATM, NBN, TP53, CHEK2, PALB2, RAD51, RAD51D, BRIP1, and FANC genes to study optimal PCA screening approaches that incorporate mpMRI.8 Therefore, PCA screening strategies will be expected to further incorporate germline results in the future.
GT can also serve as a doorway to identify a hereditary cancer syndrome in a family. Men presenting for GT on the basis of personal PCA history, tumor features, or family history may be the index case to uncover hereditary cancer syndromes such as hereditary breast and ovarian cancer (linked with P/LPVs in BRCA1 and BRCA2) and Lynch syndrome (associated with P/LPVs in DNA MMR genes).1,2,9 Furthermore, multiple genes on PCA panels confer risk for additional cancers affecting males and females.4,9 Many of these cancer risks can be addressed by heightened screening or surveillance strategies as per NCCN guidelines.1,2 Current NCCN guidelines recommend that all men with metastatic, high-risk, or advanced PCA, men with PCA of Ashkenazi Jewish ancestry, men with known familial germline P/LPVs in their family, and men with strong family cancer history are offered GT, establishing the standard of care.1,2 Furthermore, targeted therapy eligibility constitutes a major indication for GT.
Given the clinical indications and hereditary cancer implications of GT for PCA, it is imperative to address engagement of diverse populations in GT. With the emergence of multigene testing, initial reports detailing the germline variant spectrum from clinical cohorts had 6% to 15% African American (AA) men.10-12 These studies uncovered important aspects of multigene testing; yet under-representation in clinical genetic studies hinders our understanding of the germline P/LPV spectrum among AA men, who have the highest PCA incidence and mortality in the United States and globally.13 To address this gap, multiple previous studies have specifically focused on characterizing the germline variant spectrum associated with PCA with attention to AA men.14-17 One study reported that among men engaged in PCA GT from a large commercial data set, AA men represented 8% of men tested and AA men had lower rates of P/LPVs in non-BRCA genes compared with White men.14 The report noted limitations such as lack of data to assess association of P/LPV rates with clinical features and family history. Furthermore, rates of variants of uncertain significance (VUSs) were not reported.14 A multi-institutional study also reported on differences in P/LPVs between AA and White men undergoing GT for PCA.15 This report included 867 patients, of whom 22% were AA. Although no differences were noted overall in P/LPV rates between AA and White men, this study also reported lower rates of P/LPVs specifically in non-BRCA genes among AA men than White men.15 Limitations of this study noted that patients had GT from various laboratories and had a modest sample size.15 Another study sequenced BRCA1/2 to define the pathogenic spectrum among AA and White men in a cohort of 1,240 patients with PCA, of whom 30% were AA.16 The study showed that rates of BRCA1/2 VUS were higher in AA men than White men. No difference in prevalence of BRCA1/2 P/LPVs were reported between race groups; however, non-BRCA genes were not evaluated.16 Another study was conducted among men undergoing GT through a commercial genetics laboratory that included a 30-gene hereditary cancer panel.17 This study included 1,351 men with a history of PCA, of whom 3% were AA/Canadian. The study reported no association of pathogenic/likely pathogenic (P/LP) with family history, ethnicity, or age at diagnosis. There was a significant difference in VUS rates, with AA/Canadian men or Asian/Pacific Islander men having higher rates of VUS.17 These studies have provided insights into the PCA germline spectrum among AA men; however, limitations of a low proportion of AA men in these cohorts and variability in genes included in panel testing in some of the studies support the need for greater investigation regarding the germline spectrum among AA men with PCA.
To deepen our understanding and address current gaps, we conducted a retrospective analysis of the germline spectrum comparing AA with White men with PCA, all of whom had undergone multigene panel testing using a 14-gene PCA panel from a large commercial genetic laboratory in the United States. A previous publication of the germline spectrum from this cohort reported that higher Gleason score; personal history of breast or pancreatic cancer; family history of breast, ovarian, or pancreatic cancer; and family history of Lynch syndrome–associated cancers were predictors of carrying P/LPVs among 1,812 men with PCA.18 This cohort included only 6% of AA men and included patients who underwent testing using a variety of panels. Therefore, the current study was conducted to enrich for AA men with PCA who underwent GT using the same 14-gene panel as White men from the previous study.18 The goal of the current study is to inform PCA genetic counseling and testing strategies for AA men and to promote equity in genetics care.
METHODS
IRB Approval
The study was approved by the Institutional Review Board at Thomas Jefferson University.
Cohort
The data set included genetic and clinical data on men with PCA, all of whom had undergone GT using a 14-gene PCA panel test at a clinical diagnostic laboratory (Ambry Genetics, Aliso Viejo, CA). Data on AA men were collected and curated between September 2016 and December 2020 (n = 251). Curated data on White men with PCA were extracted from the previous publication and was restricted to men who had the 14-gene PCA panel; these patients were tested between April 2012 and December 2016 (White: n = 193).18 Subsequent exclusions were men with a known BRCA1/2 PV reported in their family (AA: n = 1; White: n = 2) and patients who had prior genetic evaluation such as BRCA1/2 or DNA MMR GT, immunohistochemical screening for MMR deficiency in tumors, and other somatic testing (AA: n = 13; White: n = 1) to minimize bias in P/LPV detection rates. Therefore, the final data set included 237 AA men and 190 White men. Demographic and clinical data including age at testing, ethnicity, age of diagnosis, Gleason score, metastatic status, and personal and family history of cancer were collected through retrospective review of test requisition forms and other clinical documentation (eg, pedigrees and consult notes) provided to the laboratory.
Laboratory Methods
Analysis included the following genes on the 14-gene PCA panel: ATM, BRCA1, BRCA2, CHEK2, EPCAM, HOXB13, MLH1, MSH2, MSH6, NBN, PALB2, PMS2, RAD51D, and TP53. Sanger or next-generation sequencing analysis was performed for all coding domains and well into the flanking 5′ and 3′ ends of all introns and untranslated regions, along with gross deletion/duplication analysis of covered exons and untranslated regions, with the exception being EPCAM, for which only gross deletions encompassing the 3′ end of the gene are reported.
Variant Classification
Variants were classified using a five-tier variant classification system (pathogenic variant; likely pathogenic variant; VUS; variant, likely benign; and benign), on the basis of published recommendations from the American College of Medical Genetics and Genomics and the International Agency for Research on Cancer.19-21
Statistical Analysis
Study data were summarized with descriptive statistics including frequency counts with percentages for categorical variables and means with standard deviations for continuous variables. Distributions were compared between White and AA men using the Fisher's exact test, chi-squared test, or two-sample t-test, as appropriate. Odds ratios (ORs) for carrying P/LPVs were evaluated using multiple logistic regression considering race (AA v White), age (≤ 50 years v older), family history of PCA in first-degree/second-degree relative, and family history of other cancer in first-degree/second-degree relative as potential predictors. The Gleason score was missing for 119 participants, so it was excluded from consideration. There were too few positive cases to model all four predictors simultaneously. As such, Akaike's Information Criterion22 was used to choose among models having combinations of three predictors.
All statistical analyses were conducted using SAS v9.4 (SAS Institute, Cary, NC). The significance level for hypothesis testing was set a priori at a nominal α = .05.
RESULTS
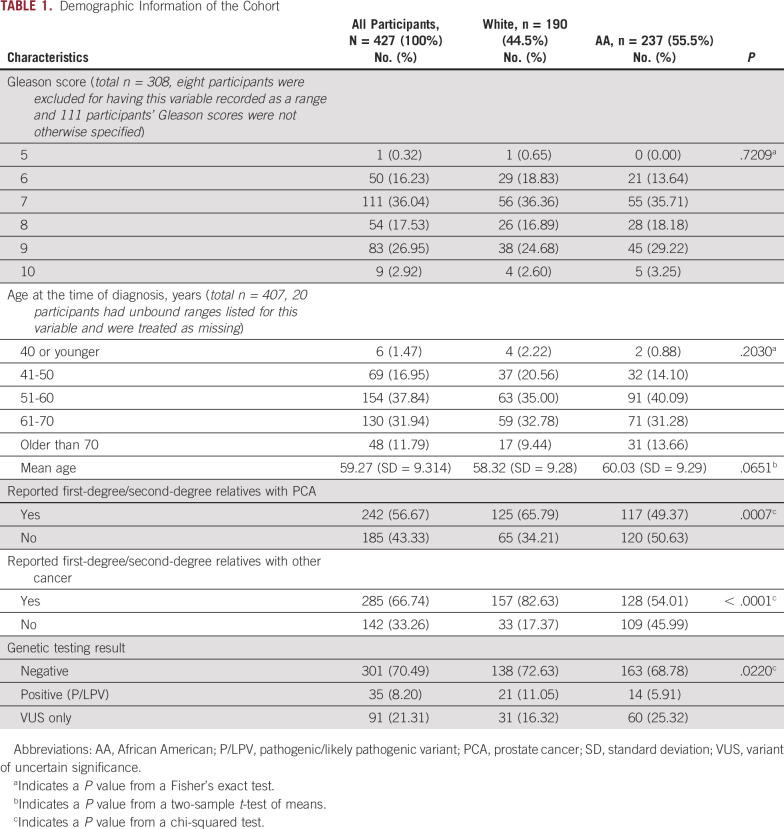
The data set included genetic and clinical data from 427 men who had undergone testing using a 14-gene PCA panel: White men, n = 190 (44%) and AA men, n = 237 (56%; Table 1). The mean age at diagnosis of the entire cohort was 59 + 9.3 years. Among men whose Gleason score was indicated (72%), 47% had Gleason ≥ 8. Among White participants, n = 36 (18.95%) had no reported value for Gleason score. Among AA participants, n = 83 (35.02%) had no reported value for Gleason score. A majority of men indicated having a first-degree/second-degree relative with PCA (56.67%) or first-degree/second-degree relative with other cancers excluding PCA (66.74%). A significant difference was observed in the reported close family history of prostate (P = .0007) and other cancers (P < .0001) between White and AA men (Table 1). No other significant differences were noted in clinical characteristics between AA and White men (Table 1).
TABLE 1.
Demographic Information of the Cohort
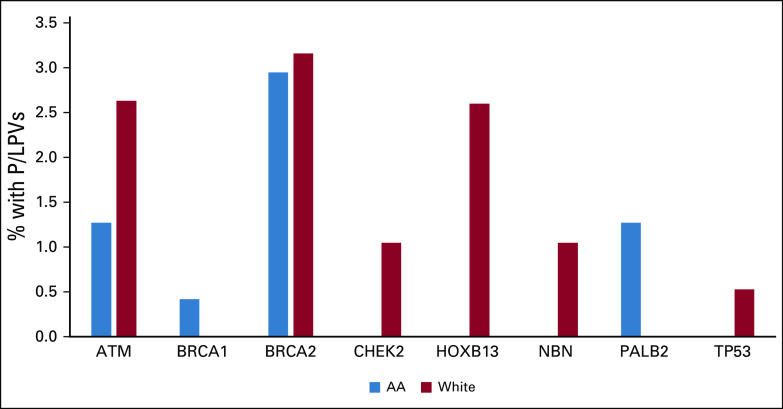
A significant difference was noted in the distribution of genetic test results betweeen AA and White males (P = .02) (Table 1). In the entire cohort, 8.2% tested positive for P/LPV, with a trend toward lower rates of P/LP variants among AA compared to White males (5.91% v 11.05%, P = .05), respectively. The P/LPV spectrum in AA men included BRCA2 (n = 7), PALB2 (n = 3), ATM (n = 3), and BRCA1 (n = 1). Among White men, a wider spectrum of mutations was observed: BRCA2 (n = 6), ATM (n = 5), HOXB13 (n = 5), CHEK2 (n = 2), TP53 (n = 1), and NBN (n = 2) (Fig 1). Multiple logistic regression showed borderline associations of the following variables for carrying P/LPVs: race (AA v White; OR = 0.51; 95% CI, 0.24 to 1.06; P = .07) and age (> 50 v ≤ 50; OR = 0.48; 95% CI, 0.22 to 1.05; P = .06), adjusted for family history of PCA in first-degree/second-degree relative.
FIG 1.
Spectrum of germline P/LPVs among AA and White men. AA, African American.
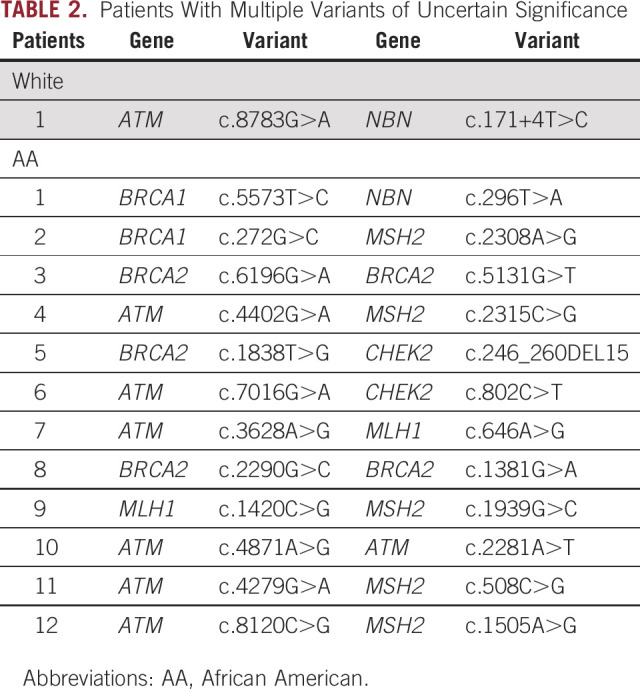
VUS only was reported in 21.31% of the overall cohort, with a significant difference noted between AA and White men (25.32% v 16.32%, respectively; P = .02). Of interest, 12 of 237 AA men had multiple VUSs reported compared with one of 190 White men (5.1% v 0.53%; P = .008). Table 2 shows the genes and variant information for patients with multiple VUSs observed. Among AA men, multiple VUSs were all in DNA repair genes including BRCA2, BRCA1, ATM, MLH1, MSH2, and CHEK2.
TABLE 2.
Patients With Multiple Variants of Uncertain Significance
DISCUSSION
There is growing recognition of the need to engage diverse populations in GT to address disparities and develop strategies for equity in cancer genetics care delivery. Multiple national organizations have emphasized the need to address cancer disparities, which now integrally involve genetic evaluation, including ASCO, American Association for Cancer Research, American Cancer Society, and National Cancer Institute.23,24 However, AA men have had significantly lower engagement in clinical GT for PCA and in clinical genetic studies, which limits the ability to provide tailored GT and management.
The strength of our study was the curated clinical and genetic data enriched for AA men with PCA to have a comparable sample size with the White male cohort along with uniform panel testing between groups. Our results show that AA men had half the P/LP rate and 1.5x the VUS rate compared with White men. Our findings confirm results from previous studies that among the genes that are commonly tested on PCA panels, AA men have a narrower spectrum of P/LPVs than White men. For example, P/LPVs in CHEK2, HOXB13, NBN, and TP53 were found exclusively in White individuals and accounted for nearly half (47.6%) of positive results in this group. Importantly, the P/LPVs identified in AA men were in key DNA repair genes including BRCA2, PALB2, ATM, and BRCA1, which have major implications for therapy in the metastatic setting and hereditary cancer implications for families.1-7 These genetic findings highlight important avenues for precision therapy, enhanced screening, and cancer risk assessment that AA men and families may miss from lack of engagement in genetic evaluation. Also consistent with previous studies is that rates of VUS were higher among AA men compared with White men in our study. Our study contributes data that AA men can have multiple VUSs reported in their genetic results, which are substantially higher than their White male counterparts. Importantly, the VUSs among AA men were in key DNA repair genes including BRCA2, BRCA1, ATM, MLH1, MSH2, and CHEK2, where resolution of pathogenicity would be critical toward affecting management recommendations and delineating hereditary cancer syndromes in these patients and their families.1-7 These findings highlight the need for expanded research to clarify VUS and identify novel genes and variants contributing to PCA risk and inform precision therapy among AA men.
Current approaches to GT have involved DNA testing technologies to identify P/LPVs. For AA men who have higher rates of VUS and multiple VUSs in an individual patient, next-generation genomic technologies, such as RNA testing, may need to be given greater consideration to determine the pathogenicity of variants.25 This testing involves evaluation of a blood specimen and therefore requires infrastructure support to coordinate blood collection, which may be challenging in the COVID-19 pandemic and postpandemic era since much of genetic evaluation has shifted to virtual visits with remote saliva sample collection for GT.26,27 Mobile phlebotomy and other types of care processes will likely need to be considered more diligently in diverse populations when addressing implementation of RNA testing.27 In addition, expanded panel testing may be needed to systematically test a larger set of genes among AA men to identify and characterize variants in genes of high or moderate penetrance given that multiple studies have now shown that AA men have a narrow spectrum of germline variants from standard multigene panels.14-17 Genetics research findings from genomewide association studies and large population studies also provide insights into the PCA germline spectrum regarding common variants and need further study to determine the clinical role in AA men.28,29
Our study also supports the need to adapt genetic counseling strategies for AA men considering PCA GT. Multiple studies including our study have now reported that AA men have higher rates of VUS, and our study showed that some patients have multiple VUS findings.14-17 Pretest counseling therefore needs to emphasize the higher chance of VUS results among AA men undergoing genetic counseling, with post-test counseling reinforcing how VUSs do not determine management but can be reclassified over time.30 Previous work from our research team reported that a subset of men who have VUS in the context of PCA GT may not fully understand VUS and need additional education to limit propagation of misinformation for patients and their families.31 Creating educational aids that incorporate literacy, numeracy, and cultural barriers is important to consider when adapting counseling models.32,33
There are some limitations of this study to consider. Although we had a sizable comparative cohort of AA men regarding germline and clinical results, the data set relied on extracting clinical information from data entered on test requisition forms, which led to variability in missing data. The smaller sample sizes of P/LPVs limited the ability to conduct logistic regression modeling and identify predictors of positive genetic results. Despite this limitation, our multivariable modeling showed interesting, although not statistically significant, estimates of AA men having lower odds of a P/LPV identified from a standard PCA multigene panel relative to White men, again highlighting the need for expanded panel testing for AA men, greater engagement of AA men in clinical genetic and research efforts, and incorporating novel technologies to gain deeper insights into the germline spectrum. A strength of our study was uniformity of the PCA panel tested in the cohort, percentage of curated clinical information that was available, and relatively comparable sample sizes between AA and White men evaluated.
In conclusion, our study reinforces that clinical GT among AA men reveals a narrow spectrum of P/LPVs in key DNA repair genes, with important implications for precision therapy and hereditary cancer assessment. Furthermore, AA men had significantly higher rates of multiple VUSs, indicating critical need for greater inclusion of diverse populations in genetic studies to discern the pathogenic spectrum contributing to PCA aggressiveness and risk. These findings also support dedicated efforts to incorporate expanded panel testing and novel genomic technologies to clarify the PCA germline spectrum in AA men, tailor genetic counseling for AA men and their families, and enhance initiatives for greater engagement of AA men in genetic studies to reduce disparities and promote equity in genetics care delivery.
Veda N. Giri
Stock and Other Ownership Interests: Novopyxis (I)
Honoraria: Invitae, Ambry Genetics
Speakers' Bureau: Janssen
Mary Pritzlaff
Employment: Ambry Genetics/Konica Minolta
Patents, Royalties, Other Intellectual Property: CancerGene Connect
Carolyn Horton
Employment: Ambry Genetics
No other potential conflicts of interest were reported.
SUPPORT
Supported by a grant to the Sidney Kimmel Cancer Center from the National Cancer Institute: 3P30CA056036; Department of Defense Award (W81XWH-21-1-0142; V.N.G. and S.W.K.).
AUTHOR CONTRIBUTIONS
Conception and design: Veda N. Giri, Carrie Horton
Administrative support: Carrie Horton
Provision of study materials or patients: Carrie Horton
Collection and assembly of data: Veda N. Giri, Mary Pritzlaff, Carrie Horton
Data analysis and interpretation: Veda N. Giri, Rebecca Hartman, Scott W. Keith
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Veda N. Giri
Stock and Other Ownership Interests: Novopyxis (I)
Honoraria: Invitae, Ambry Genetics
Speakers' Bureau: Janssen
Mary Pritzlaff
Employment: Ambry Genetics/Konica Minolta
Patents, Royalties, Other Intellectual Property: CancerGene Connect
Carolyn Horton
Employment: Ambry Genetics
No other potential conflicts of interest were reported.
REFERENCES
- 1.National Comprehensive Cancer Network Clinical Guidelines in Oncology (NCCN Guidelines®): Prostate Cancer (Version 3.2022). NCCN.org [Google Scholar]
- 2.National Comprehensive Cancer Network Clinical Guidelines in Oncology (NCCN Guidelines®): Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic (Version 2.2022). NCCN.org [Google Scholar]
- 3.Genetics of Prostate Cancer (PDQ®)–Health Professional Version. National Cancer Institute. www.cancer.gov [PubMed] [Google Scholar]
- 4.Giri VN, Knudsen KE, Kelly WK, et al. : Implementation of germline testing for prostate cancer: Philadelphia prostate cancer consensus conference 2019. J Clin Oncol 38:2798-2811, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bono J, Mateo J, Fizazi K, et al. : Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 382:2091-2102, 2020 [DOI] [PubMed] [Google Scholar]
- 6.Abida W, Patnaik A, Campbell D, et al. : Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol 32:3763-3772, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Page EC, Bancroft EK, Brook MN, et al. : Interim results from the IMPACT study: Evidence for prostate-specific antigen screening in BRCA2 mutation carriers. Eur Urol 76:831-842, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Men at High Genetic Risk for Prostate Cancer. ClinicalTrials.gov identifier: NCT03805919 [Google Scholar]
- 9.PDQ® Cancer Information Summaries: Genetics. National Cancer Institute. https://www.cancer.gov/publications/pdq/information-summaries/genetics [Google Scholar]
- 10.Giri VN, Obeid E, Gross L, et al. : Inherited mutations in males undergoing multigene panel testing for prostate cancer—Emerging implications for personalized prostate cancer genetic evaluation. JCO Precis Oncol 10.1200/PO.16.00039 [epub ahead of print on May 4, 2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giri VN, Hegarty SE, Hyatt C, et al. : Germline genetic testing for inherited prostate cancer in practice: Implications for genetic testing, precision therapy, and cascade testing. Prostate 79:333-339, 2019 [DOI] [PubMed] [Google Scholar]
- 12.Nicolosi P, Ledet E, Yang S, et al. : Prevalence of germline variants in prostate cancer and implications for current genetic testing guidelines. JAMA Oncol 5:523-528, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Facts and Figures 2022. American Cancer Society. www.cancer.org [Google Scholar]
- 14.Sartor O, Yang S, Ledet E, et al. : Inherited DNA-repair gene mutations in African American men with prostate cancer. Oncotarget 11:440-442, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ledet EM, Burgess EF, Sokolova AO, et al. : Comparison of germline mutations in African American and Caucasian men with metastatic prostate cancer. Prostate 81:433-439, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrovics G, Price DK, Lou H, et al. : Increased frequency of germline BRCA2 mutations associates with prostate cancer metastasis in a racially diverse patient population. Prostate Cancer Prostatic Dis 22:406-410, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon DH, Borno HT, Cheng HH, et al. : Ethnic disparities among men with prostate cancer undergoing germline testing. Urol Oncol 38:80.e1-80.e7, 2020 [DOI] [PubMed] [Google Scholar]
- 18.Pritzlaff M, Tian Y, Reineke P, et al. : Diagnosing hereditary cancer predisposition in men with prostate cancer. Genet Med 22:1517-1523, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S, Aziz N, Bale S, et al. : Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405-424, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plon SE, Eccles DM, Easton D, et al. : Sequence variant classification and reporting: Recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 29:282-291, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pesaran T, Karam R, Huether R, et al. : Beyond DNA: An integrated and functional approach for classifying germline variants in breast cancer genes. Int J Breast Cancer 2016:2469523, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akaike H: A new look at the statistical model identification. IEEE Trans Automatic Control 19:716-723, 1974 [Google Scholar]
- 23.Patel MI, Lopez AM, Blackstock W, et al. : Cancer disparities and health equity: A policy statement from the American Society of Clinical Oncology. J Clin Oncol 38:3439-3448, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polite BN, Adams-Campbell LL, Brawley OW, et al. : Charting the future of cancer health disparities research: A position statement from the American Association for Cancer Research, the American Cancer Society, the American Society of Clinical Oncology, and the National Cancer Institute. CA Cancer J Clin 67:353-361, 2017 [DOI] [PubMed] [Google Scholar]
- 25.Karam R, Conner B, LaDuca H, et al. : Assessment of diagnostic outcomes of RNA genetic testing for hereditary cancer. JAMA Netw Open 2:e1913900, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dratch L, Paul RA, Baldwin A, et al. : Transitioning to telegenetics in the COVID-19 era: Patient satisfaction with remote genetic counseling in adult neurology. J Genet Couns 30:974-983, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlmann WR, McKeon AJ, Wang C: Genetic counseling, virtual visits, and equity in the era of COVID-19 and beyond. J Genet Couns 30:1038-1045, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conti DV, Wang K, Sheng X, et al. : Two novel susceptibility loci for prostate cancer in men of African ancestry. J Natl Cancer Inst 109:djx084, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darst BF, Wan P, Sheng X, et al. : A germline variant at 8q24 contributes to familial clustering of prostate cancer in men of African ancestry. Eur Urol 78:316-320, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mersch J, Brown N, Pirzadeh-Miller S, et al. : Prevalence of variant reclassification following hereditary cancer genetic testing. JAMA 320:1266-1274, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giri VN, Obeid E, Hegarty SE, et al. : Understanding of multigene test results among males undergoing germline testing for inherited prostate cancer: Implications for genetic counseling. Prostate 78:879-888, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hann KEJ, Freeman M, Fraser L, et al. : Awareness, knowledge, perceptions, and attitudes towards genetic testing for cancer risk among ethnic minority groups: A systematic review. BMC Public Health 17:503, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giri VN, Shimada A, Leader AE: Predictors of population awareness of cancer genetic tests: Implications for enhancing equity in engaging in cancer prevention and precision medicine. JCO Precis Oncol 10.1200/PO.21.00231 [epub ahead of print on November 3, 2021] [DOI] [PMC free article] [PubMed] [Google Scholar]