Abstract
A flavin reductase, which is naturally part of the ribonucleotide reductase complex of Escherichia coli, acted in cell extracts of recombinant E. coli strains under aerobic and anaerobic conditions as an “azo reductase.” The transfer of the recombinant plasmid, which resulted in the constitutive expression of high levels of activity of the flavin reductase, increased the reduction rate for different industrially relevant sulfonated azo dyes in vitro almost 100-fold. The flavin reductase gene (fre) was transferred to Sphingomonas sp. strain BN6, a bacterial strain able to degrade naphthalenesulfonates under aerobic conditions. The flavin reductase was also synthesized in significant amounts in the Sphingomonas strain. The reduction rates for the sulfonated azo compound amaranth were compared for whole cells and cell extracts from both recombinant strains, E. coli, and wild-type Sphingomonas sp. strain BN6. The whole cells showed less than 2% of the specific activities found with cell extracts. These results suggested that the cytoplasmic anaerobic “azo reductases,” which have been described repeatedly in in vitro systems, are presumably flavin reductases and that in vivo they have insignificant importance in the reduction of sulfonated azo compounds.
Various bacterial strains reduce azo dyes under anaerobic conditions. The most generally accepted hypothesis for this phenomenon is that many bacterial strains possess rather unspecific cytoplasmic enzymes, which act as “azo reductases” and under anaerobic conditions transfer electrons via soluble flavins to the azo dyes (35). We have recently suggested a different mechanism for the unspecific anaerobic reduction of azo dyes by Sphingomonas sp. strain BN6 and other bacteria. In this system, the reduction of the azo dyes is catalyzed extracellularly by the action of mediator compounds, which are either formed during the metabolism of certain substrates by the bacteria themselves or which may be added externally (e.g., compounds such as anthraquinonesulfonates). These mediators enable the transfer of redox equivalents from the cell membrane of the bacteria to azo dyes. In addition, it was demonstrated that Sphingomonas sp. strain BN6 also possesses cytoplasmic azo reductase activities, which could be assayed in vitro with cell extracts under anaerobic conditions (14, 15, 17). It was shown that the reduction rate of whole cells of Sphingomonas sp. strain BN6 for amaranth after growth with glucose was 0.3 μmol min−1 g of soluble cell protein−1 and could be increased by the addition of anthraquinone-2-sulfonate to 3 μmol min−1 g of soluble cell protein−1. In contrast, in vitro with cell extracts or solubilized membrane preparations, specific activities of 20 to 40 μmol min−1 g of soluble cell protein−1 were determined (15, 17). A possible explanation for the significantly lower reaction rates of the whole-cell preparations compared to those in the in vitro systems was that in vivo the cytoplasmic azo reductase activity does not function, because the highly polar sulfonated azo dyes are not able to penetrate through the cell membranes. In the present study, we attempted to clarify the question of whether in vivo the intracellular cytoplasmic azo reductase activity is involved at all in the metabolism of the azo compounds or whether this enzymatic activity is only active under in vitro conditions. Therefore, we increased the intracellular cytoplasmic azo reductase activity by using a genetic engineering approach and compared the resulting recombinant organism in vivo with the parent strain.
MATERIALS AND METHODS
Bacterial strains.
The isolation and characterization of Sphingomonas sp. strain BN6 have been described before (20). The strain has been deposited at the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) in Braunschweig, Germany, as DSM 6383. Escherichia coli K38 and JM109(DE3) were used for overexpression of the flavin reductase, and E. coli S17-1 (32) was used for the conjugative transfer of plasmids to strain BN6.
Culture conditions.
Sphingomonas sp. strain BN6 was routinely grown in a mineral medium according to the method of Dorn et al. (7) with glucose (15 mM) and naphthalene-2-sulfonate (0.5 mM). For Sphingomonas sp. strain BN6(pRJR34), this medium was supplemented with 10 μg of tetracycline per ml. E. coli strains were routinely cultured in nutrient broth (NB) or Luria-Bertani (LB) medium. E. coli JM109(pRJR34) was grown in LB medium plus 12.5 μg of tetracycline per ml and isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM) to induce the flavin reductase.
Plasmids and DNA manipulation techniques.
Plasmid DNA was isolated from E. coli by the method of Lee and Rasheed (18). Digestion of DNA with restriction endonucleases (Gibco BRL, Boehringer), electrophoresis, and ligation with T4 DNA ligase (Gibco BRL) were performed according to standard procedures (30). Transformation of E. coli was done by the method of Chung et al. (4).
The flavin reductase was produced by using E. coli K38(pEE1001) or E. coli JM109(pEE1001). Plasmid pEE1001 was constructed by Spyrou et al. (33) by cloning the flavin reductase gene from E. coli into the expression vector pTZ18R. In this construct, the flavin reductase gene (fre) is under the control of a phage T7 promoter.
For the construction of pRJR34, pEE1001 was cut with EcoRI and HindIII, the DNA fragment carrying the fre gene was isolated, and the ends were filled with the Klenow fragment. The resulting DNA fragment was ligated into the EcoRV site of plasmid pJOE890 (for a similar construct, see the study by Altenbuchner et al. [1]). From the resulting plasmid, the fre gene was removed by cleavage with EcoRI and ligated into the EcoRI cleavage site of pRK415 (16). The resulting plasmid was transformed into E. coli JM109. From eight of the recombinant clones, the flavin reductase activity was determined. The plasmid from the clone with the highest specific activity (2.4 U/mg of protein) was designated pRJR34 and used for further investigations.
Detection of the fre gene from E. coli in Sphingomonas sp. strain BN6(pRJR34) by PCR.
For the PCR, oligonucleotide primers were custom synthesized according to the sequence of the gene described by Spyrou et al. (33). From the nucleotide sequence of the fre gene, bases 16 to 38 (forward primer) and 627 to 646 (reverse primer) were used as primers. PCR mixtures (50 μl) for the amplification of DNA contained 50 pmol of each primer, 10 to 20 ng of plasmid template DNA (isolated with Pharmacia Flexiprep kits), 0.1 mM (each) deoxynucleotide triphosphate, and 1 U of Taq DNA polymerase, polymerase reaction buffer, and MgCl2 according to the manufacturer (GibcoBRL). The PCR was performed with a touchdown thermocycle program under the following conditions: initial denaturation (95°C, 3.75 min) before addition of the polymerase, 10 cycles with decreasing annealing temperature (60 to 55°C, 30 s), polymerization (72°C, 1 min), and denaturation (95°C, 45 s). Fifteen more cycles with 55°C as the annealing temperature and 72°C as the polymerization temperature were performed before the reaction was stopped, and the reaction products were analyzed by agarose gel electrophoresis. Thus, a fragment of the expected size (630 bp) was detected after amplification of the DNA from E. coli JM109(DE3)(pEE1001) and Sphingomonas sp. strain BN6(pRJR34), but not with DNA from the wild-type strain, Sphingomonas sp. strain BN6.
Anaerobic reduction of azo dyes by whole-cell preparations.
Sphingomonas sp. strain BN6 was grown aerobically in a mineral medium to the late exponential growth phase (optical density at 546 nm of about 2 to 4) and then transferred to rubber-stoppered serum bottles (100 ml). Oxygen was removed from the medium by at least 15 2-min cycles of evacuation and flushing with nitrogen gas. Glucose (10 mM) and amaranth (1.3 mM) were added anaerobically, and samples were taken at different time intervals. The decrease in the dye concentration was determined by high-performance liquid chromatography as described previously (14).
Preparation of cell extracts.
Cell suspensions in 50 mM Tris-HCl (pH 7.5) were disrupted by using a French press (Aminco, Silver Spring, Md.) at 80 MPa. Cell debris was removed by centrifugation at 100,000 × g for 30 min at 4°C. The protein concentration was determined by the method of Bradford (2), with bovine serum albumin as the standard.
Enzyme assays.
One unit of enzyme activity was defined as the amount of enzyme that converts 1 μmol of substrate per min.
(i) Flavin reductase.
The NAD(P)H:flavin oxidoreductase was measured by a modification of the method given by Fontecave et al. (9). In this aerobic assay, the flavin reductase catalyzes the reduction of riboflavin, and the reduced riboflavin is immediately reoxidized by oxygen. Cell extract was added to a solution (final volume, 1 ml) containing 10 μmol of Tris-HCl (pH 7.5), 0.25 μmol of NADPH, and 0.003 μmol of riboflavin. The decrease in A340 was measured spectrophotometrically. Reaction rates were calculated by using a molar extinction coefficient of 6.3 mM−1 cm−1.
(ii) Azo reductase.
Azo reductase activity was routinely measured by a modification of the spectrophotometric assay described previously by Haug et al. (14). The anaerobic reaction mixture contained (in 1.6 ml) 70 μmol of Tris-HCl buffer (pH 7.5), 1 μmol of NADH, 0.32 μmol of flavin adenine dinucleotide (FAD), and 0.1 μmol of amaranth. The stock solutions were made anaerobically by repeated gassing with N2. The reaction was performed in gastight cuvettes and started by the addition of cell extracts. The decrease in the concentration of amaranth was determined spectrophotometrically at 520 nm, and reaction rates were calculated by using a molar extinction coefficient of 22.6 mM−1 cm−1. In certain experiments, azo dyes different from amaranth were used. The relevant wavelengths and extinction coefficients for these dyes are summarized in Table 1.
Chemicals.
All chemicals were obtained from Merck, Fluka, Sigma, or Aldrich, except for Mordant yellow 3, which was kindly provided by Bayer AG.
RESULTS
The flavin reductase from E. coli as azo reductase.
It was previously suggested that, under anaerobic conditions, unspecific cytoplasmic azo reductases act via the intermediate formation of free reduced flavins (35). This hypothesis has been substantiated by the observation that photochemically generated reduced FAD (FADH2) reduced the sulfonated azo compound amaranth (10). In this system, the only function of the “cytoplasmatic azoreductases” would be the supply of reduced flavins for the subsequent purely chemical reduction of the azo compounds by the reduced flavins. It was therefore reasoned that any flavin reductase [NAD(P)H:flavin oxidoreductase] could also act as an azo reductase. Recently, a flavin reductase gene (fre) was cloned. The gene product, which is naturally part of the ribonucleotide reductase complex of E. coli, was produced in large amounts with the expression plasmid pEE1001. In this construct, the flavin reductase gene is transcribed from the phage T7 promoter (26, 33). This system was applied to assay the flavin reductase for its azo reductase activity. E. coli JM109(DE3)(pEE1001), E. coli K38(pEE1001), and, as a negative control, E. coli JM109 were grown in LB medium, cell extracts were prepared, and the flavin reductase and the azo reductase activities were determined under anaerobic conditions. Thus, it was confirmed that the E. coli strains with the recombinant plasmid really produced the flavin reductase with very high specific activities, and it was demonstrated that the flavin reductase could act under anaerobic conditions as azo reductase (Table 2). Surprisingly, the flavin reductase activity was constitutively expressed in E. coli JM109(DE3)(pEE1001), and the activity could not be increased by the addition of IPTG (up to 2 mM). The proposed function of FAD as a mediator compound transferring redox equivalents from the flavin reductase to the azo dye was shown in a control experiment, in which the cell extract was incubated with NADPH and amaranth with or without added FAD. Thus, it was found that, in the absence of externally added FAD, less than 5% of the azo reductase activity could be detected.
Comparison of the cytoplasmic azo reductase activities in E. coli K38(pEE1001) and Sphingomonas sp. strain BN6 with different industrially relevant azo dyes.
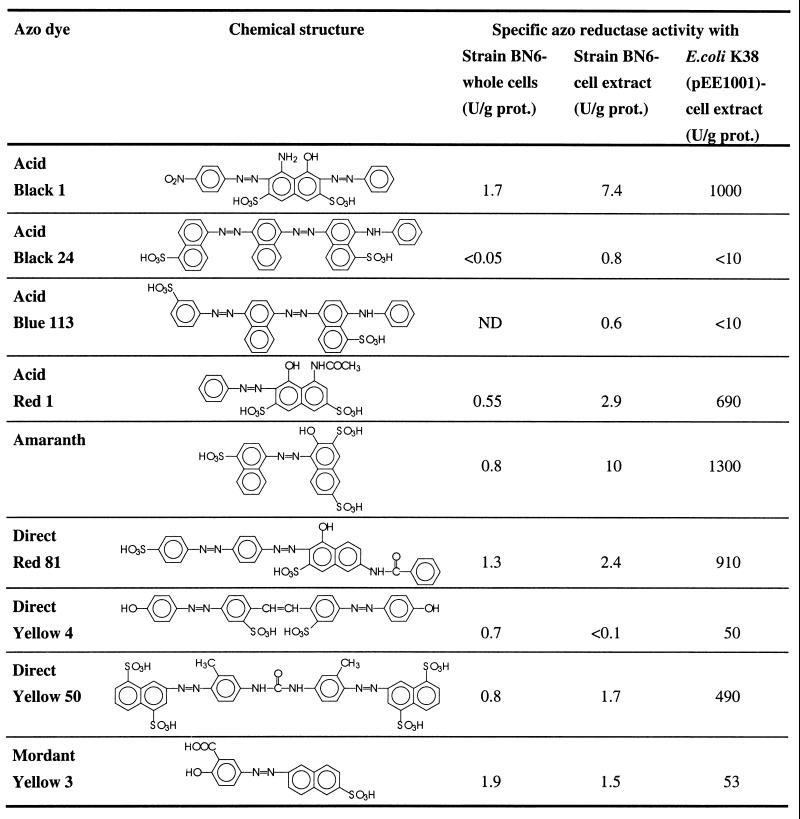
It has been previously reported that azo reductase activity was present in cell extracts of Sphingomonas sp. strain BN6 (14, 15, 17). Therefore, the azo reductase activities of cell extracts from E. coli K38(pEE1001) and Sphingomonas sp. strain BN6 were compared and tested for their ability to reduce different industrially relevant azo dyes (Table 3). Thus, it was found that the azo reductase activity in the cell extract of E. coli K38(pEE1001) for many azo dyes was more than 100-fold higher than the activities found in cell extracts of strain BN6. The cell extracts from both strains had high relative activities for those dyes which carried in the ortho position to the azo group a hydroxyl group and in general had somehow lower activities for dyes with a hydroxy group in the para position to the azo bond. The hydrogen of the hydroxy group in the ortho position can form an intramolecular hydrogen bridge with the nitrogen atom of the azo group. This reduces the electron density at the azo bond and thus facilitates a reductive cleavage. Furthermore, it was observed that an increasing number of sulfonic acid substituents also resulted in an increased reduction rate. This may be caused by the electron withdrawal effect of the sulfonic acid substituents, which also reduces the electron density at the azo bond.
Transfer of the flavin reductase gene to Sphingomonas sp. strain BN6.
The plasmid pEE1001, which carries the fre gene, is a pBR322 derivative and has only a narrow host range. It has been previously reported that IncP1 plasmids can be conjugatively transferred to Sphingomonas sp. strain BN6 (28). Therefore, plasmid pRJR34 was constructed, which contains the fre gene cloned into the broad-host-range plasmid pRK415 (16). After the preparation of cell extracts from JM109(pRJR34), flavin reductase activities of 1.7 U/mg of protein were found. Thus, the flavin reductase was expressed in this system too. Finally, E. coli S17-1 (32) was transformed with plasmid pRJR34, and the resulting transformant was used to conjugatively transfer pRJR34 to Sphingomonas sp. strain BN6. After selection on a mineral medium with glucose (10 mM) plus tetracycline (5 μg/ml), a transfer frequency of plasmid (pRJR34) from E. coli S17-1 to strain BN6 of almost 100% was found.
Expression of the flavin reductase and the azo reductase activity in Sphingomonas sp. strain BN6.
Sphingomonas sp. strain BN6(pRJR34) was grown in liquid culture with NB and tetracycline (10 μg/ml), and cell extracts were prepared. The flavin reductase activity in the recombinant strain was 0.7 U/mg of protein, compared to a flavin reductase activity in wild-type strain BN6 of 0.008 U/mg of protein. The presence of the fre gene from E. coli in Sphingomonas sp. strain BN6(pRJR34) was also shown by PCR (see Materials and Methods). These results demonstrated that the recombinant organism indeed expressed the flavin reductase gene originating from E. coli. Therefore, whether these cell extracts also showed an increased azo reductase activity under anaerobic conditions was also tested. Thus, it could be demonstrated that the azo reductase activity in the recombinant strain was almost 30-fold higher than that in the wild-type strain (Table 2).
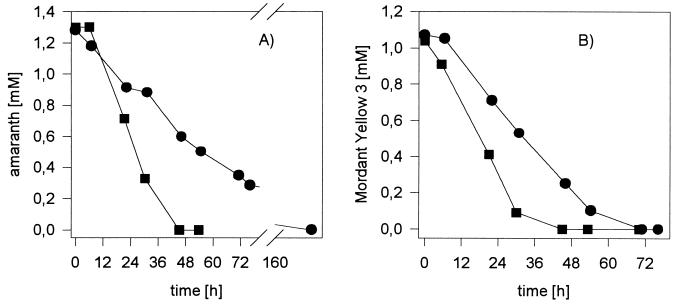
In contrast to the cell-free system, whole cells of Sphingomonas sp. BN6(pRJR34) showed only an approximately threefold increase in the reduction rate for amaranth and Mordant yellow 3 compared to the wild type BN6 strain (Fig. 1). Furthermore, the specific activities of whole cells (0.002 U mg of protein−1) were significantly lower for strain BN6(pRJR34) than the activities determined with cell extracts from the same strain (0.14 U mg of protein−1). This suggested that either the cell membranes limited the uptake of the highly polar sulfonated azo compounds or the lack of some cofactors (e.g., free flavins) limited the reduction of the azo compounds by whole cells. The addition of flavin mononucleotide (FMN) or FAD to cell suspensions of the recombinant strain BN6(pRJR34) did not significantly increase the azo reductase activity. In contrast, an almost 100-fold increase in the reduction rate was observed after the addition of anthraquinone-2-sulfonate (0.5 mM).
FIG. 1.
Reduction of amaranth (A) or Mordant yellow 3 (B) under anaerobic conditions by whole cells of Sphingomonas sp. strain BN6 (●) or BN6(pRJR34) (■). The cells were grown aerobically in a mineral medium with glucose (10 mM) and naphthalene-2-sulfonate (0.5 mM) [plus 10 μg of tetracycline per ml for BN6(pRJR34)]. When the cells reached the late-exponential growth phase, 25 ml of the cultures was transferred into serum bottles (100-ml volume) with a nitrogen atmosphere, and glucose (10 mM) and Mordant yellow 3 (1 mM) or amaranth (1.3 mM) were added. At the indicated intervals, aliquots were taken, cells were removed by centrifugation, and the concentrations of the azo dyes were determined by high-performance liquid chromatography.
Aerobic function of the flavin reductase as azo reductase.
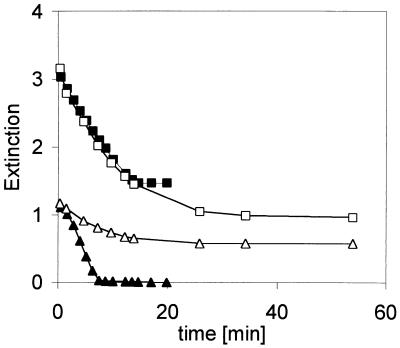
All of the experiments described above were performed under anaerobic conditions, because it is well known that reduced flavins react rapidly with molecular oxygen. Since the recombinant strains expressed the flavin reductase with high specific activities, we assumed that there could be an azo reductase activity of the flavin reductase as well in the presence of molecular oxygen. Therefore, the azo reductase assays with cell extracts were compared under anaerobic and aerobic conditions. The reactions were analyzed spectrophotometrically at λ = 340 nm to measure the oxidation of NADH and at λ = 520 nm to assay the reduction of amaranth (Fig. 2). Thus, it was found that, also under aerobic conditions, some decrease in the concentration of amaranth was observed, although the reaction rates were significant lower than those of the anaerobic control. In contrast, almost identical oxidation rates for NADH were observed under aerobic and anaerobic conditions (Fig. 2). This observation may explain the results of some sporadic reports about the presence of unspecific aerobic azo reductases in bacteria such as E. coli (11, 12).
FIG. 2.
Reduction of amaranth by cell extracts from E. coli K38(pEE1001) under aerobic (□, ▵) and anaerobic conditions (■, ▴). The reaction mixtures contained (per milliliter) 50 μmol of NaK phosphate buffer (pH 7.7), 0.4 μmol of NADH, 0.1 μmol of FAD, and 0.05 μmol of amaranth. The reactions were started by the addition of 20 μl of a cell extract from E. coli K38(pEE1001) (88 μg of protein), and the decrease in absorbance was simultaneously determined spectrophotometrically at λ = 340 nm (■, □) and λ = 520 nm (▴, ▵). For the reaction under anaerobic conditions, all solutions were made anaerobically by repeated gassing with N2, and the reaction was performed in gastight cuvettes.
DISCUSSION
There are numerous reports which describe the reduction of azo compounds by bacteria under anaerobic conditions. The main interest in this field has focused on bacteria from the intestine which are involved in the metabolism of azo dyes ingested as food additives, and there are some studies which refer to bacteria isolated from soil or sewage treatment systems (5, 6, 8, 27, 29, 31). The earlier studies were mainly performed with facultatively anaerobic bacteria (e.g., Proteus vulgaris, Streptococcus faecalis, or Bacillus sp.). It had been repeatedly suggested that, in these strains, cytosolic flavin-dependent reductases were responsible for this unspecific reaction and that the low permeability of the cell membranes for the highly polar sulfonated azo compounds would cause the significantly lower reduction rates observed with whole cells compared to those in cell-free systems (19, 27, 36). Because none of these cytosolic azo reductases has ever been purified, until now it has been speculative whether bacterial flavin reductases were responsible for the azo reductase activity observed with bacterial cell extracts. The present study proved that flavin reductases are indeed able to act as azo reductases and that cell extracts containing the cloned flavin reductase decolorized a broad range of industrially relevant azo dyes.
Our results suggested that the reduction of sulfonated azo dyes by reduced flavins formed by cytosolic flavin-dependent azo reductases is mainly observed in vitro and in vivo is of insignificant importance. This became evident with the recombinant strain BN6(pRJR34), which in vitro showed significantly increased azo reductase activities compared to those of the wild type, although in vivo only a very small increase in azo reductase activity was found. Theoretically, this could also have been caused by a lack of the required cofactors (NADH and/or free flavins) in the resting cells. This seems to be improbable, because the anaerobically incubated cells should possess a rather high concentration of NADH. There is only limited information available about the free flavin content of bacterial cells. However, it has been observed that recombinant FMN-dependent luciferases are fully functional in different bacterial backgrounds without the concurrent transfer of recombinant FMN reductases (34). This has also been shown recently for strain BN6 (J. Klein, personal communication). This suggests that reduced free flavins are present in various bacteria and also in strain BN6 in quantities which allow the support of enzymatic reactions which require reduced flavins.
A further indication that the flavin-dependent azo-reductases are almost completely laboratory artifacts was shown by the effects of externally added flavins on the rates of azo reduction under anaerobic conditions. Thus, it was found that the addition of FAD to a resting cell suspension of strain BN6 resulted in no significant increase in the rate of azo dye reduction. In contrast, the addition of the same concentration of anthraquinone-2,6-disulfonate resulted in an almost 10-fold increase in the dye reduction rate (17). Similar observations were also made with Sphingomonas sp. strain BN6(pRJR34) and E. coli JM109(DE3)(pEE1001) (unpublished results). This suggested that the bacterial membranes not only are efficient barriers for the uptake of sulfonated azo dyes, but also are hardly permeable for flavin-containing cofactors and therefore restrict the transfer of reduction equivalents by flavins from the cytosol of intact cells to sulfonated azo dyes in the culture medium. Thus, in living cells with intact cell membranes, other enzyme systems and/or other redox mediators are presumably responsible for the reduction of azo dyes. In bacteria which possess electron transport systems in their membranes, such as the purely aerobic or facultatively anaerobic bacteria studied in the present report, the transfer of the redox equivalents from the respiratory chain of the cell membranes to appropriate redox mediators could take place directly. If intracellular reductases should be involved in this process, it may be assumed that mediators different from flavin cofactors must be involved. A prerequisite for these mediators would be a higher ability to pass the bacterial membranes than that of flavins.
A different concept for the reduction of sulfonated azo compounds which also does not require a membrane transport of the dyes has been suggested for bacterial strains from the strictly anaerobic microflora of the intestine. Rafii and coworkers isolated different bacteria from the human intestine (e.g., Eubacterium sp., Clostridium sp., Butyrivibrio sp., or Bacteroides sp.), which decolorized sulfonated azo dyes during growth on solid or liquid complex media. It was shown that at least part of the azo reductase activities were extracellular, because the culture supernatants were able to decolorize the dyes under anaerobic conditions (24, 25). This extracellular azo reductase activity was due to the activity of specific proteins, because after nondenaturing polyacrylamide gel electrophoresis and activity staining under anaerobic conditions, usually only one distinct protein band in the gels was able to decolorize the dyes (21). In most isolates, flavin compounds were required for azo reductase activity, but in some clostridia, the azo reductase activity was independent of externally added flavins (24). Recently, a DNA fragment from Clostridium perfringens was cloned in phage λgt11, which increased the azo reductase activity of lytic and lysogenic cultures of E. coli infected with the recombinant phage (23). The azo reductase from C. perfringens was detected by immunoelectron microscopy throughout the cytoplasm and in the vicinity of the cells (22). This suggested that the protein is not a typical extracellular enzyme, but that it is presumably released only from lysed cells. The azo reductase activity from C. perfringens is clearly different from the flavin reductase investigated in the present study, because the enzyme activity was described as being independent of that from added flavins, and furthermore, the enzyme was rapidly and irreversibly inactivated by oxygen (24). It is still unclear in this system how the supposed extracellular azoreductases should gain the NADH necessary for the reduction of the azo dyes in their extracellular environment, and there may be some effects from the complex growth media of the bacteria. Furthermore, there are some other publications about the reduction of azo compounds by whole-cell preparations of strictly anaerobic bacteria which suggested the involvement of redox mediators (3, 4). Therefore, it can't be excluded at present that extracellular mediator compounds also participate in the reduction of azo compounds by strictly anaerobic bacteria.
The third possibility for the extracellular reduction of azo dyes caused by microorganisms is the action of reduced inorganic compounds (Fe2+, H2S), which are formed as end products of certain strictly anaerobic bacteria. Also, in these cases, mediator compounds may have an important function (13, 37).
It may be concluded that, depending on the environmental conditions (or the reaction conditions of a biotechnological process), different reactions catalyzed by bacteria can accomplish the reduction of azo compounds under anaerobic conditions, but that intracellular reactions have only insignificant importance in these reactions.
TABLE 1.
Color index registration numbers, absorption maxima, purity, and calculated molar extinction coefficients of the sulfonated azo dyes useda
| Azo dye | Color index no. | Dye purity (%) | Absorption maximum (nm) | Extinction coefficient (mM−1 cm−1) |
|---|---|---|---|---|
| Acid black 1 (naphthol blue black) | 20470 | 85 | 618 | 53.2 |
| Acid black 24 | 26370 | 50 | 560 | 20.1 |
| Acid black 52 (Palatines fast black) | 15711 | 25 | 565 | 63.2 |
| Acid blue 113 | 26360 | 70 | 557 | 20.1 |
| Acid red 1 | 18050 | 60 | 530 | 21.1 |
| Direct red 81 | 28160 | 50 | 509 | 55.2 |
| Direct yellow 4 (brillant yellow) | 24890 | 70 | 404 | 74.2 |
| Direct yellow 50 | 29025 | 60 | 398 | 38.3 |
| Amaranth (acid red 27) | 16185 | 90 | 520 | 22.6 |
The dyes were purchased from Aldrich or Sigma and not further purified. The absorption maxima were determined in NaK phosphate buffer (pH 7.4, 54 mM). The molar extinction coefficients were calculated at the dye purities indicated.
TABLE 2.
Specific activities of the flavin reductase and the azo reductase in cell extracts
| Enzyme | Sp act (U/mg of protein) of cell extract
|
||||
|---|---|---|---|---|---|
| E. coli JM109 | E. coli JM109(DE3)(pEE1001) | E. coli K38(pEE1001) | Sphingomonas sp. strain BN6 | Sphingomonas sp. strain BN6(pRJR34) | |
| Flavin reductase | 0.09 | 2.0 | 4.0 | 0.008 | 0.7 |
| Azo reductase | 0.007 | 0.3 | 1.3 | 0.004 | 0.14 |
TABLE 3.
Specific reductive activities of different industrially relevant azo dyesa
ND, not done; prot., protein.
REFERENCES
- 1.Altenbuchner J, Viell P, Pelletier I. Positive selection vectors based on palindromic DNA sequences. Methods Enzymol. 1992;216:457–466. doi: 10.1016/0076-6879(92)16042-i. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Brown J P. Reduction of polymeric azo and nitro dyes by intestinal bacteria. Appl Environ Microbiol. 1981;41:1283–1286. doi: 10.1128/aem.41.5.1283-1286.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung K-T, Fulk G E, Egan M. Reduction of azo dyes by intestinal anaerobes. Appl Environ Microbiol. 1978;35:558–562. doi: 10.1128/aem.35.3.558-562.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung K-T, Stevens S E, Jr, Cerniglia C E. The reduction of azo dyes by the intestinal microflora. Crit Rev Microbiol. 1992;18:175–190. doi: 10.3109/10408419209114557. [DOI] [PubMed] [Google Scholar]
- 7.Dorn E, Hellwig M, Reineke W, Knackmuss H-J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99:61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- 8.Dubin P, Wright K L. Reduction of azo food dyes in cultures of Proteus vulgaris. Xenobiotica. 1975;5:563–571. doi: 10.3109/00498257509056126. [DOI] [PubMed] [Google Scholar]
- 9.Fontecave M, Eliasson R, Reichard P. NAD(P)H:flavin oxidoreductase of E. coli: a ferric iron reductase participating in the generation of the free radical of ribonucleotide reductase. J Biol Chem. 1987;262:12325–12331. [PubMed] [Google Scholar]
- 10.Fujita M, Peisach J. The stimulation of microsomal azoreduction by flavins. Biochim Biophys Acta. 1982;719:178–189. doi: 10.1016/0304-4165(82)90087-3. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh D K, Ghosh S, Sadhukhan P, Mandal A, Chaudhuri J. Purification of two azoreductases from Escherichia coli K12. Indian J Exp Biol. 1993;31:951–954. [PubMed] [Google Scholar]
- 12.Ghosh D K, Mandal A, Chaudhuri J. Purification and partial characterization of two azoreductases from Shigella dysenteriae type 1. FEMS Microbiol Lett. 1992;98:229–234. doi: 10.1016/0378-1097(92)90161-g. [DOI] [PubMed] [Google Scholar]
- 13.Glaus M A, Heijman C G, Schwarzenbach R P, Zeyer J. Reduction of nitroaromatic compounds mediated by Streptomyces sp. exudates. Appl Environ Microbiol. 1992;58:1945–1951. doi: 10.1128/aem.58.6.1945-1951.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haug W, Schmidt A, Nörtemann B, Hempel D C, Stolz A, Knackmuss H-J. Mineralization of the sulfonated azo dye Mordant Yellow 3 by a 6-aminonaphthalene-2-sulfonate-degrading bacterial consortium. Appl Environ Microbiol. 1991;57:3144–3149. doi: 10.1128/aem.57.11.3144-3149.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keck A, Klein J, Kudlich M, Stolz A, Knackmuss H-J, Mattes R. Reduction of azo dyes by mediators originating in the naphthalenesulfonic acid degradation pathway of Sphingomonas sp. strain BN6. Appl Environ Microbiol. 1997;63:3684–3690. doi: 10.1128/aem.63.9.3684-3690.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keen K T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 17.Kudlich M, Keck A, Klein J, Stolz A. Localization of the enzyme system involved in the anaerobic reduction of azo dyes by Sphingomonas sp. strain BN6 and effect of artificial redox mediators on the rate of azo dye reduction. Appl Environ Microbiol. 1997;63:3691–3694. doi: 10.1128/aem.63.9.3691-3694.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S, Rasheed S. A simple procedure for maximum yield of high-quality plasmid DNA. BioTechniques. 1990;9:676–679. [PubMed] [Google Scholar]
- 19.Mechsner K, Wuhrmann K. Cell permeability as a rate limiting factor in the microbial reduction of sulfonated azo dyes. Eur J Appl Microbiol Biotechnol. 1982;15:123–126. [Google Scholar]
- 20.Nörtemann B, Baumgarten J, Rast H G, Knackmuss H-J. Bacterial communities degrading amino- and hydroxynaphthalene-2-sulfonates. Appl Environ Microbiol. 1986;52:1195–1202. doi: 10.1128/aem.52.5.1195-1202.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rafii F, Cerniglia C E. An anaerobic gel assay for the detection of azoreductase from anaerobic bacteria. J Microbiol Methods. 1990;12:138–149. [Google Scholar]
- 22.Rafii F, Cerniglia C E. Localization of the azoreductase of Clostridium perfringens by immuno-electron microscopy. Curr Microbiol. 1993;27:143–145. doi: 10.1007/BF01576011. [DOI] [PubMed] [Google Scholar]
- 23.Rafii F, Coleman T. Cloning and expression in Escherichia coli of an azoreductase gene from Clostridium perfringens and comparison with azoreductase genes from other bacteria. J Basic Microbiol. 1999;39:29–35. [PubMed] [Google Scholar]
- 24.Rafii F, Franklin W, Cerniglia C E. Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl Environ Microbiol. 1990;56:2146–2151. doi: 10.1128/aem.56.7.2146-2151.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rafii F, Moore J D, Ruseler-van Embden J G H, Cerniglia C E. Bacterial reduction of azo dyes used in foods, drugs and cosmetics. Microecol Ther. 1995;25:147–156. [Google Scholar]
- 26.Reichard P. From RNA to DNA, why so many ribonucleotide reductases? Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- 27.Roxon J J, Ryan A J, Wright S E. Enzymatic reduction of tartrazine by Proteus vulgaris from rats. Food Cosmet Toxicol. 1967;5:645–656. doi: 10.1016/s0015-6264(67)83216-4. [DOI] [PubMed] [Google Scholar]
- 28.Russ R, Stolz A, Knackmuss H-J. Comparison of the efficiency to degrade naphthalene-2-sulfonate between a mixed culture and an in-vitro constructed hybrid strain. In: Schmid R D, editor; Schmid R D, editor. Biochemical engineering 3. Stuttgart, Germany: Kurz & Co.; 1995. pp. 114–116. [Google Scholar]
- 29.Ryan A J, Roxon J J, Sivayavirojana A. Bacterial azo reduction: a metabolic reaction in mammals. Nature. 1968;219:854–855. doi: 10.1038/219854a0. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Scheline R R, Nygaard R T, Longberg B. Enzymatic reduction of the azo dye, Acid Yellow, by extracts of Streptococcus faecalis, isolated from rat intestine. Food Cosmet Toxicol. 1970;8:55–58. doi: 10.1016/s0015-6264(70)80223-1. [DOI] [PubMed] [Google Scholar]
- 32.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in-vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. BioTechnology. 1983;1:784–791. [Google Scholar]
- 33.Spyrou G, Haggård-Ljungquist E, Krook M, Jörnvall H, Nilsson E, Reichard P. Characterization of the flavin reductase gene (fre) of Escherichia coli and construction of a plasmid for overproduction of the enzyme. J Bacteriol. 1991;173:3673–3679. doi: 10.1128/jb.173.12.3673-3679.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart G S, Williams P. lux genes and the applications of bacterial luminescense. J Gen Microbiol. 1992;138:1289–1300. doi: 10.1099/00221287-138-7-1289. [DOI] [PubMed] [Google Scholar]
- 35.Walker R. The metabolism of azo compounds: a review of the literature. Food Cosmet Toxicol. 1970;8:659–676. doi: 10.1016/s0015-6264(70)80455-2. [DOI] [PubMed] [Google Scholar]
- 36.Wuhrmann K, Mechsner K, Kappeler T. Investigations on rate-determining factors in the microbial reduction of azo dyes. J Appl Microbiol Biotechnol. 1980;9:325–338. [Google Scholar]
- 37.Yoo E S, Libra J, Wiesmann U. Untersuchungen zum anaeroben Entfärbungsmechanismus von Azofarbstoffen. DECHEMA Jahrestagung. 1999;1999:399–400. [Google Scholar]