Abstract
Dispersed cellulose ribbons from bacterial cellulose were subjected to digestion with cloned Cel7A (cellobiohydrolase [CBH] I) and Cel6A (CBH II) from Humicola insolens either alone or in a mixture and in the presence of an excess of β-glucosidase. Both Cel7A and Cel6A were effective in partially converting the ribbons into soluble sugars, Cel7A being more active than Cel6A. In combination, these enzymes showed substantial synergy culminating with a molar ratio of approximately two-thirds Cel6A and one-third Cel7A. Ultrastructural transmission electron microscopy (TEM) observations indicated that Cel7A induced a thinning of the cellulose ribbons, whereas Cel6A cut the ribbons into shorter elements, indicating an endo type of action. These observations, together with the examination of the digestion kinetics, indicate that Cel6A can be classified as an endo-processive enzyme, whereas Cel7A is essentially a processive enzyme. Thus, the synergy resulting from the mixing of Cel6A and Cel7A can be explained by the partial endo character of Cel6A. A preparation of bacterial cellulose ribbons appears to be an appropriate substrate, superior to Valonia or bacterial cellulose microcrystals, to visualize directly by TEM the endo-processivity of an enzyme such as Cel6A.
Despite a large number of studies, the mechanism of the enzymatic digestion of crystalline cellulose stands as a major unsolved problem of persisting industrial and scientific significance. As early as 1950, it was realized that the degradation of cellulose required a complex of enzymes working together (36). This crucial observation has been confirmed by an extensive number of studies. Following these reports, a general picture has emerged indicating that at least three types of enzymes need to cooperate to digest efficiently crystalline cellulose into glucose: (i) endoglucanases (EC 3.2.1.4), which cut the cellulose chains randomly; (ii) cellobiohydrolases (CBH) (EC 3.2.1.91), which recurrently cleave cellobiose from the cellulose chain ends; and (iii) β-glucosidases (EC 3.2.2.21), which hydrolyze cellobiose and various soluble cellodextrins into glucose (reviewed in references 4, 15, 16, 22, 30, 42, 44, and 53).
The complementary activities of the different enzymes is thought to be responsible for synergistic effects, whereby the enzymatic activity of a mixture of two or several enzymes is substantially higher than the sum of the activities of the individual enzymes. Several types of synergy have been described, the easier to apprehend being the cooperation action of endo- and exo-acting enzymes on cellulose (21, 28, 35, 49, 50). In such a cooperation, the action of endocellulases is to increase the number of chain ends and, therefore, to enhance the action of exocellulases, which themselves appear to be the key enzymes for the digestion of crystalline cellulose. In this context, one of the main characteristics of CBH is that they act on cellulose chains in a “processive” manner (10, 23, 27, 37, 38, 45), as they progress along the polymer chain while releasing cellobiose in a recurrent fashion. The processivity of these enzymes appears to be related to the fine details of their three-dimensional structure (10, 38): in CBH Cel6A and Cel7A (formerly CBH II and CBH I, respectively [24]) from Trichoderma reesei and Humicola insolens, the catalytic site is buried inside a tunnel-shaped cavity whose roof consists of large flexible loops (13, 14, 38, 46, 47, 54). It is believed that once a cellulose chain has entered the active site, it becomes degraded processively as it threads through the tunnel until its release (10).
Another type of synergy, more difficult to explain, was reported as early as two decades ago (17), when the two CBH, Cel6A and Cel7A, from T. reesei were combined. This synergy has been confirmed since then with fungal as well as bacterial CBH (3, 21, 25, 33, 35, 43, 52). Several explanations have been tentatively proposed to account for the cooperation between two “exocellulases.” Some have proposed that there are two types of nonreducing ends in cellulose and that each CBH acts specifically on one of these ends, resulting in increased activity when both enzymes are present (51, 53). This explanation is, however, contradicted by observations that Cel6A and Cel7A from T. reesei act, respectively, from the nonreducing and reducing ends of the substrate (3, 5, 26, 31, 48). This finding has led to the proposal that the differences in the chain end preference and in the directionality of action of the two CBH were responsible for the “exo-exo” synergy (3, 42). To account for the synergy of Cel7A and Cel6A from T. reesei, other authors have envisaged the formation of an optimized “loose complex” of these two enzymes in solution prior to their adsorption (43).
The apparent exo-exo synergy could also be explained if, in addition to their processive CBH activity, Cel6A and/or Cel7A from T. reesei or H. insolens also occasionally had an endo character. Such an activity has been envisaged following the analysis of the pattern of interaction of exo-acting enzymes with soluble substrates ranging from oligo- to polysaccharides (1, 2, 20, 33, 39, 41, 46). So far, however, no experimental evidence of endo-processivity for Cel7A and/or Cel6A from T. reesei or H. insolens has been observed in the action of these enzymes on insoluble crystalline cellulose substrates. To assess this point, we have prepared model cellulose substrates consisting of long bacterial cellulose ribbons of high crystallinity. For the purpose of our study, these model substrates should be superior to the currently used Valonia or bacterial cellulose microcrystals. Indeed, the mode of digestion of these very long ribbons should reveal the action of given enzymes, as an endo attack should correspond to ribbon cutting, whereas processivity should be translated as ribbon thinning. In order to visualize these effects, bacterial cellulose ribbons were digested with recombinant Cel7A and Cel6A from H. insolens, either alone or in combination. The assays were quantified by the release of soluble sugars, while the topology of degradation was monitored by transmission electron microscopy (TEM).
MATERIALS AND METHODS
Purification of cellulases.
Cel7A and Cel6A were cloned and expressed in Aspergillus oryzae under standard fermentation conditions, where no cellulolytic activity was present (9). These two cellulases were purified to homogeneity as already described (40), yielding a single band in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE).
The cellobiase Novozyme 188 produced by Aspergillus niger was fractionated by size exclusion chromatography using a Sephacryl S-200 column equilibrated with 50 mM Tris-HCl buffer (pH 8.0) and 0.5 M sodium chloride. The activity of the fractions was analyzed using p-nitrophenyl-β-d-glucopyranoside (Sigma). The active fractions were pooled, concentrated using a GR81PP membrane (Dow) with a molecular mass cutoff of 10 kDa, and finally freeze-dried. The protein appeared pure on SDS-PAGE, with a single band corresponding to a molecular mass of about 100 kDa.
Preparation of the cellulose substrate.
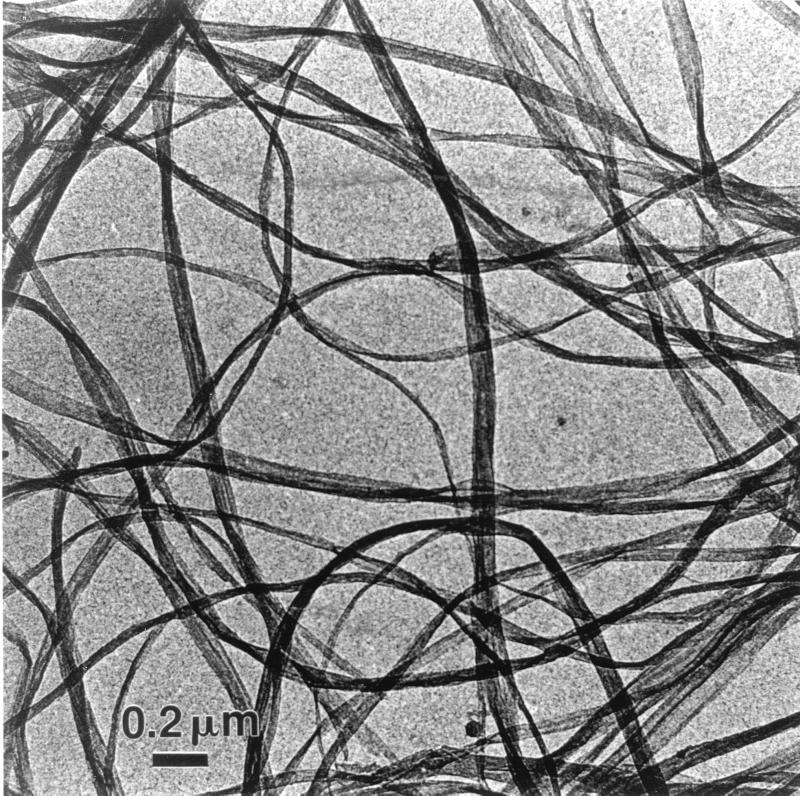
Cubes of commercial bacterial cellulose (Nata de Coco; Fujiko Co., Kobe, Japan) were used. The cubes were extensively washed with tap water in order to remove the sweet syrup. They were then cleaned for 1 week with 1% (wt/vol) aqueous sodium hydroxide. After neutralization with a few drops of concentrated HCl, the cubes were finally rinsed several times with distilled water. Some cubes were dried before and after the alkaline treatment, and it was verified by X-ray analysis that this treatment had no effect on the crystallinity of the sample. The purified cubes were then homogenized with a Waring blender. A typical preparation of bacterial cellulose ribbons is presented in Fig. 1. The final suspension, which had a concentration of 3 g/liter, was stored at 4°C after the addition of 0.01% (wt/vol) sodium azide.
FIG. 1.
Bacterial cellulose ribbons visualized by low-dose TEM.
Enzymatic hydrolysis of bacterial cellulose.
Solutions of Cel7A (72 kDa) and Cel6A (65 kDa) were prepared by dissolving freeze-dried cellulases in 50 mM sodium phosphate buffer at pH 6.5. Samples of 1 mg of bacterial cellulose were incubated without agitation at 37°C in 50 mM sodium phosphate buffer at pH 6.5. The ratio of cellulase to cellulose was varied from 0.5 to 7 nmol of enzyme/mg of cellulose, and the total reaction volume was kept at 1 ml. An appropriate amount of cellobiase was added in order to avoid the inhibition of CBH by the cellobiose released during digestion. The cellobiase amount was adjusted until thin-layer chromatography showed that the assay supernatants contained only glucose and no cellobiose.
Specimen preparation for electron microscopy.
The digested cellulose samples were centrifuged at 9,000 × g, and the supernatant was subjected to sugar analysis. The pellets were washed once with 1% aqueous sodium hydroxide to remove the adsorbed enzymes and then were extensively washed with distilled water. Drops of diluted bacterial cellulose suspensions were deposited on carbon-coated copper grids and allowed to dry. These specimens were used for imaging purpose without further treatment.
Extent of enzymatic digestion.
The extent of degradation of bacterial cellulose was deduced from the amount of soluble reducing sugars in the supernatant after centrifugation of the digestion mixture. A ferricyanide technique adapted from the method of Kidby and Davidson (29) was used: 300 mg of potassium hexacyanoferrate III and 28 g of hydrated sodium carbonate were dissolved in 1 liter of distilled water. One milliliter of 5 M aqueous sodium hydroxide was then added to alkalinize this reagent solution. One hundred microliters of the assay supernatant was added to 1 ml of reagent, and the absorbency of the solution was measured at 420 nm. A standard curve was obtained using glucose solutions of known concentrations.
TEM.
A Philips CM 200 Cryo TEM operated at an accelerating voltage of 80 kV under low-dose-mode conditions was used throughout. Images were recorded on Agfa Scientia plates.
RESULTS
When the suspensions of bacterial cellulose ribbons were tested with Cel6A and/or Cel7A, they became visually clarified, indicating substantial degradation. Quantification of the hydrolysis was achieved by measuring the amount of reducing sugar released into the supernatant. This release, as a function of the enzymatic conditions, is illustrated in the series of digestion curves presented in Fig. 2 to 5.
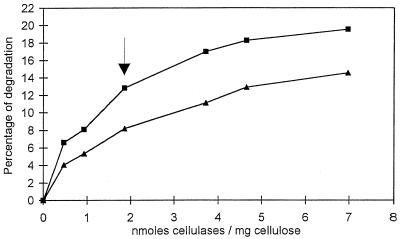
FIG. 2.
Digestion (2 h) of bacterial cellulose by Cel6A (▴) and Cel7A (■) from H. insolens in 50 mM sodium phosphate buffer at pH 6.5 and 37°C. The percentage of degradation is represented as a function of the ratio of enzyme to substrate (nanomoles of cellulase per milligram of cellulose). A quantity of 1.85 nmol of cellulase/mg of cellulose (arrow) was used for all further experiments. See the text for details.
FIG. 5.
Degradation of bacterial cellulose by higher concentrations of Cel7A (●, 3.5 nmol of cellulase/mg of cellulose), Cel6A (■, 3.5 nmol of cellulase/mg of cellulose), and the Cel7A-Cel6A mixture (▴, 7 nmol of cellulase/mg of cellulose).
Figure 2 shows the amount of reducing sugar released in 2 h as a function of the concentrations of Cel6A and Cel7A. Both curves have roughly the same aspect, in the sense that they increase rapidly in the range of low enzyme content (between 0 and 2 nmol of cellulase/mg of cellulose). At higher enzyme content, both curves level off to reach a plateau beyond which the increase in digestion is only marginal. Under our experimental conditions, about 20% degradation was obtained for Cel7A in the plateau region, as opposed to 15% for Cel6A at the same concentration of protein per milligram of substrate. Overall, at equivalent enzyme concentrations and for all enzyme concentrations, the curves in Fig. 2 indicate that Cel7A systematically releases more reducing sugar than does Cel6A, the ratio always being between 1.5 and 1.6.
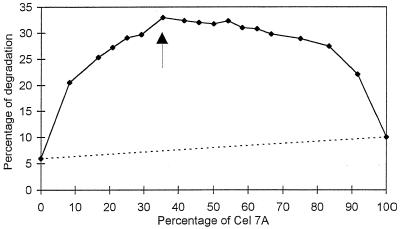
The synergy between Cel6A and Cel7A is illustrated in the curve shown in Fig. 3, which corresponds to an assay where a constant concentration of protein per milligram of substrate was used, namely, 1.85 nmol/mg of cellulose. This protein concentration was selected because it corresponds approximately to a point (arrow in Fig. 2) which is intermediate between the sharp initial increase and the plateau region observed in Fig. 2. An analysis of the curve in Fig. 3 reveals significant synergy that culminates at a ratio of 37.5% Cel7A and 62.5% Cel6A (arrow in Fig. 3). For comparison, the dotted line in Fig. 3 corresponds to the situation where no cooperative degradation would occur. Under the optimum synergistic conditions, 33% degradation of cellulose was achieved in 1 h. In this situation, the degree of synergy was greater than 4. Another aspect of the curve in Fig. 3 is that it rises rather slowly when increasing amounts of Cel7A are added to Cel6A. This behavior is quite different from that expected for classical endo-exo synergy, where the slope of synergy should be almost vertical as soon as a small quantity of endo-acting enzyme is added to a pure exocellulase (21).
FIG. 3.
Synergistic degradation of bacterial cellulose by various ratios of Cel6A and Cel7A from H. insolens at a constant total enzyme load of 1.85 nmol of cellulase/mg of cellulose. The dotted line represents the theoretical hydrolysis value expected for a noncooperative degradation. Assays were performed with 50 mM sodium phosphate buffer at pH 6.5 and 37°C for 1 h. The Cel6A-Cel7A mixture (62.5%:37.5%) indicated with an arrow was chosen for subsequent experiments. See the text for details.
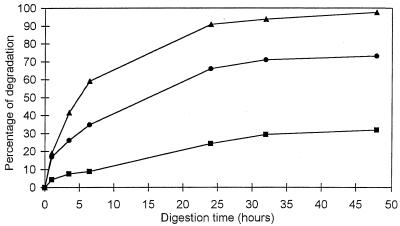
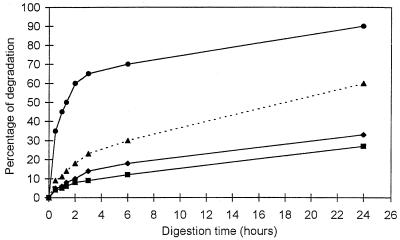
The evolution of Cel7A-Cel6A synergy as a function of time is illustrated in the series of curves in Fig. 4. The two bottom curves correspond to Cel6A alone and Cel7A alone, but with concentrations of protein per milligram of substrate different from those in Fig. 2. The dotted curve in the middle is the sum of the two bottom curves. It is the curve expected if no synergy between Cel6A and Cel7A took place. The top curve corresponds to the experimental set of data obtained when the enzymes are mixed in the optimum proportion deduced from Fig. 3 (37.5% Cel7A and 62.5% Cel6A). The top curve illustrates the extent of synergy. It reveals in particular that within 24 h, nearly all the cellulose was digested when the two enzymes were combined as opposed to 37.5 and 62.5% for Cel7A alone and Cel6A alone, respectively. A comparison between the top curve and the theoretical dotted curve indicates that the degree of synergy varied with time: at the onset of digestion, it was the highest at a value of 4; it progressively decreased to a value of 1.5 after 24 h.
FIG. 4.
Synergy between Cel7A and Cel6A in the digestion of bacterial cellulose as a function of time. Cel7A (⧫, 0.7 nmol of cellulase/mg of cellulose) and Cel6A (■, 1.15 nmol of cellulase/mg of cellulose) were incubated alone and in combination. The theoretical curve (▴) represents the nonsynergistic sum of the activities of Cel7A and Cel6A. The experimental curve (●) is the synergistic effect observed with a 37.5%:62.5% mixture of Cel7A and Cel6A at a total amount of enzyme of 1.85 nmol of cellulase/mg of cellulose. Assays were performed with 50 mM sodium phosphate buffer at pH 6.5 and 37°C.
Higher concentrations of cellulases were also used to digest bacterial cellulose. Figure 5 is an example where the concentrations of enzymes used were four times higher than those used in Fig. 4. At these high enzyme concentrations, no more synergy could be observed, as the sum of the individual activities of Cel6A and Cel7A was systematically higher than the experimental data for the mixed enzymes. This result might indicate substrate limitation and/or competition of the enzymes for the same sites. Another aspect of these high-concentration assays is that Cel7A by itself appeared quite efficient, as it digested more than 70% of the cellulose in 48 h, as opposed to 32% for Cel6A. Also, at equivalent concentrations of protein per milligram of substrate, the amount of sugar released by Cel7A was systematically more than twice as high as that released by Cel6A.
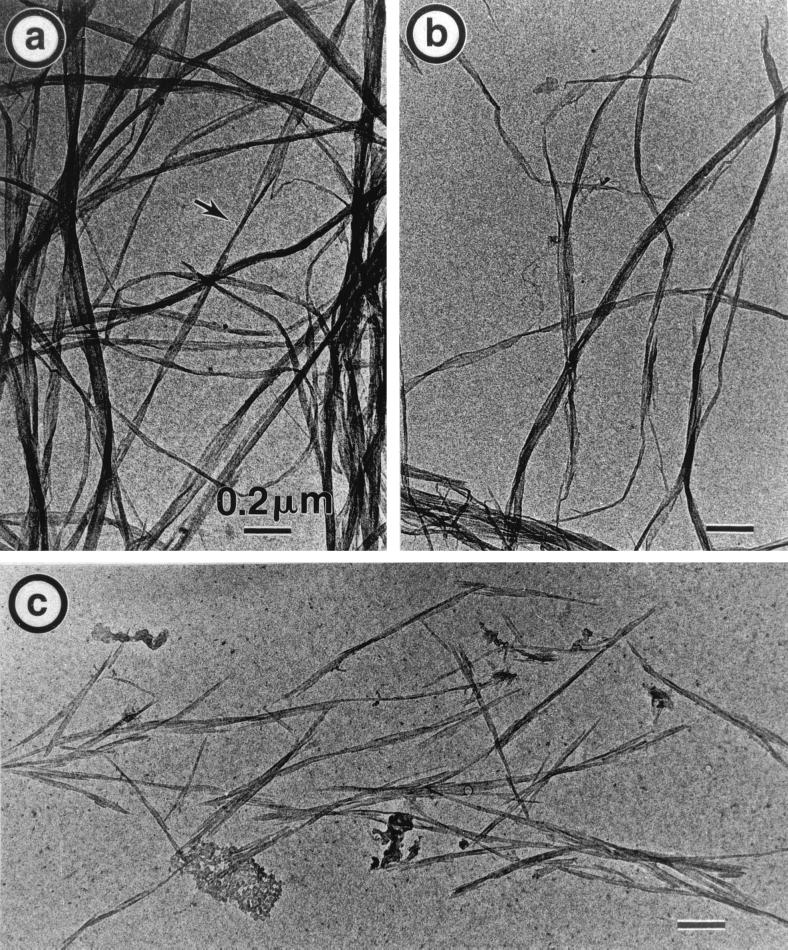
The morphological modifications imparted to bacterial cellulose by Cel6A and/or Cel7A are illustrated in the series of electron micrographs displayed in Fig. 6 and 7. Each of these micrographs should to be compared to that in Fig. 1, which corresponds to the initial sample. Figure 6 illustrates the results of 24 h of digestion with the enzyme concentration corresponding to the curves in Fig. 2 (1.85 nmol of enzyme in total). Figure 6a is the result of digestion with 0.7 nmol of Cel7A/mg of cellulose, where 30% of the sample has been solubilized. Quite interestingly, the morphology of this digested sample is very similar to that of the initial sample. Only in a few areas is it possible to detect some ribbon thinning (shown with an arrow), while no change in ribbon length is visible. In Fig. 6b, corresponding to digestion of 24% of the sample by 1.15 nmol of Cel6A/mg of cellulose, a number of ribbon ends and shorter ribbons can be seen. In addition, some ribbons appear thinner, whereas others have remained as wide as those in the initial material. In Fig. 6c, Cel6A and Cel7A have been mixed, resulting in a dramatic increase in digestion, as almost 90% of the sample has been solubilized. The morphology of the degraded sample is also drastically modified, since the ribbons are cut into micron-long rods which are far narrower than the initial ribbon elements. Thus, the sample in Fig. 6c appears to be the result of extensive ribbon thinning and cutting.
FIG. 6.
TEM of the 24-h degradation of bacterial cellulose ribbons by isolated and combined cellulases from H. insolens. (a) Incubation with 0.7 nmol of Cel7A/mg of cellulose. (b) Incubation with 1.15 nmol of Cel6A/mg of cellulose. (c) Incubation with a 37.5%:62.5% mixture of Cel7A and Cel6A (1.85 nmol of cellulase/mg of cellulose).
FIG. 7.
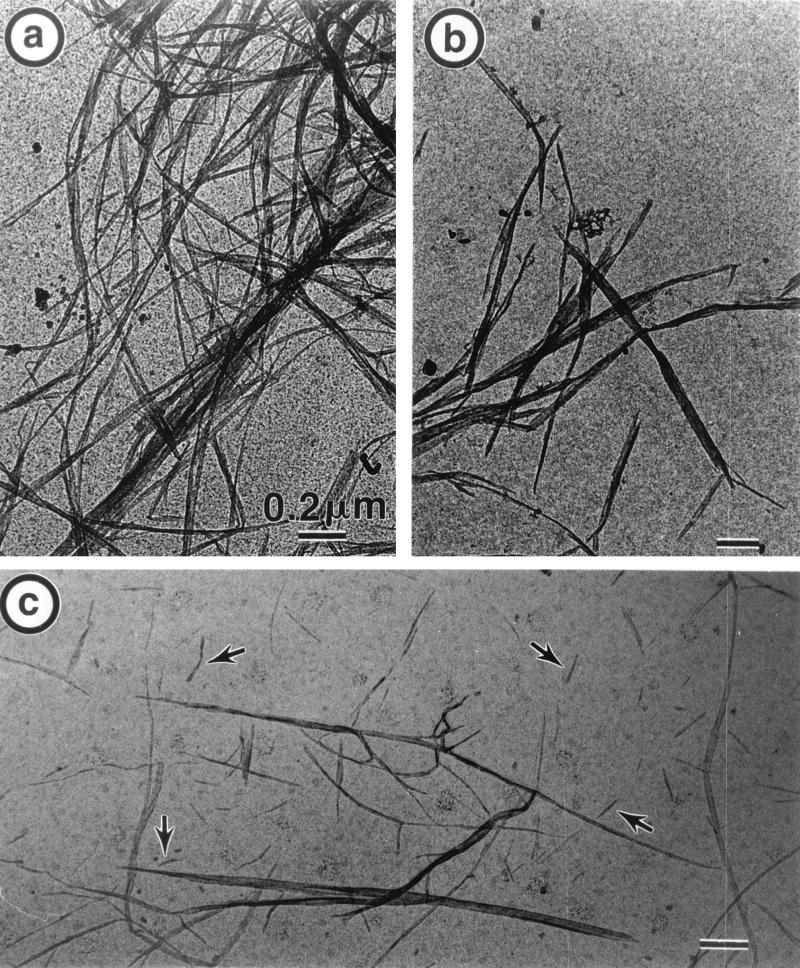
TEM of the 48-h degradation of bacterial cellulose ribbons by isolated cellulases from H. insolens. (a) Incubation with 3.5 nmol of Cel7A/mg of cellulose. (b) Incubation with 3.5 nmol of Cel6A/mg of cellulose. (c) Incubation with 3.5 nmol of Cel6A plus 3.5 nmol of Cel7A/mg of cellulose.
Figure 7 shows the morphology of the sample degraded for 48 h and described in Fig. 5. In this case, which corresponds to a massive dose of enzymes, the morphological effects are even more pronounced. Figure 7a is the result of the action of 3.5 nmol of Cel7A/mg of cellulose. Here, only a few ribbon ends are observed, despite the fact that 70% of the sample has been hydrolyzed. On the other hand, all the ribbons are markedly narrower than those in the initial sample. When 3.5 nmol of Cel6A/mg of cellulose was used (Fig. 7b), all the ribbons were cut into micron-long elements substantially wider than the ribbons in Fig. 7a and only moderately narrower than those in the initial sample. The micrograph shown in Fig. 7c corresponds to samples where 1 mg of cellulose was digested with 3.5 nmol of Cel6A and 3.5 nmol of Cel7A. After 48 h, approximately 98% of the sample has been solubilized. The insoluble particles in Fig. 7c are heterogeneous, some of them consisting of very fine needles; elements such as those in Fig. 7b or 6c are also observed.
DISCUSSION
The goal of this study was to define a crystalline cellulose substrate tailor-made to reveal the difference in the action of the two CBH, Cel6A and Cel7A, from H. insolens as well as the synergy existing when these two enzymes are mixed. Suspensions of bacterial cellulose ribbons appear to be well suited for this purpose. Indeed, these suspensions consist of virtually endless thin ribbons of high crystallinity (32) and high molecular weight (34). As there is only a limited number of chain ends along the ribbons, their cutting into short fragments should indicate an endo mode of action of a given enzyme. On the other hand, cellulases having a pure processive mode should leave endless ribbons, but ribbons that are thinner than the initial ones. A combination of the two types of enzymes should therefore yield short thin ribbons indicative of substantial synergy. In view of this hypothesis, the results presented in this study clearly indicate that the processive character dominates the activity of Cel7A from H. insolens on crystalline cellulose. This finding is best illustrated in Fig. 7a, which shows that after 2 days of degradation and with a high enzyme-to-substrate ratio, all the bacterial cellulose ribbons have been thinned, whereas only a very few cuts can be observed. On the other hand, the pattern of degradation of bacterial cellulose ribbons by Cel6A indicates that this enzyme is able to cut substantially the substrate without much thinning, following what appears to be an endo type of degradation. The dual activities of Cel6A—processivity and endo attack—are nevertheless present and clearly illustrated in Fig. 7b, where a high enzyme concentration and an extended digestion time led to short ribbons somewhat thinner than the initial ones.
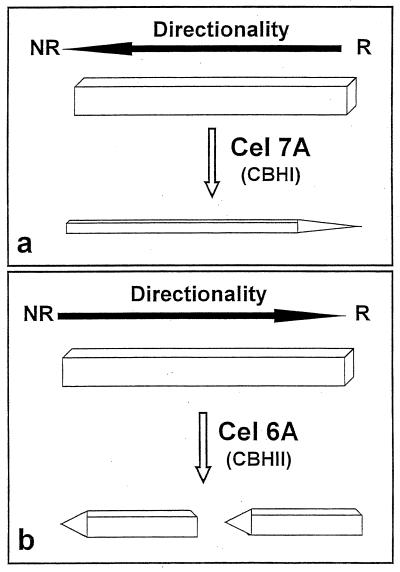
Altogether, the present observations of the digestion of bacterial cellulose ribbons and those of the degradation of Valonia cellulose microcrystals (7, 8, 26, 31) suggest the schematic representation shown in Fig. 8. In this scheme, a crystal or a microfibril of cellulose treated with Cel7A from H. insolens (Fig. 7a, schematized in Fig. 8a) or from T. reesei is essentially thinned down by the high processivity of this enzyme. As the erosion of the crystal occurs from the reducing end (26, 48), this end becomes pointed. When a cellulose crystal is digested by Cel6A, it becomes cut in several areas by the endo mode of attack of the enzyme (Fig. 6b and 7b, schematized in Fig. 8b). Each subcrystal is then eroded by the processive action of the enzyme, with the result of pointed tips oriented toward the nonreducing ends of the crystals (31). In this action, the degraded crystals retain a width roughly the same as that of the initial crystal.
FIG. 8.
Schematic representation of the topology of action of Cel7A (a) and Cel6A (b) toward an isolated crystal of cellulose. NR and R, nonreducing and reducing ends of the crystals, respectively.
Although the term processivity has only recently been introduced to describe the action of cellulases, the observation of the marked processive character of Cel7A, resulting in thinning of the substrate, is not new. It was already observed clearly in the digestion of Valonia cellulose microfibrils by Cel7A from T. reesei (7) and H. insolens (6). The action of Cel6A on crystalline cellulose substrates has been also described as processive (10, 38), the only obvious difference between the two enzymes being the directionality of their action (10, 38, 42). The present study shows, for the first time we believe, the endo character of the action of Cel6A on a solid, crystalline cellulose substrate. Our results indicate that this enzyme is better described as an endo-processive CBH. It should be noted that the ability of CBH to perform endo cuts does not prevent these enzymes from acting on chain ends (when available), like true exocellulases, as well (23).
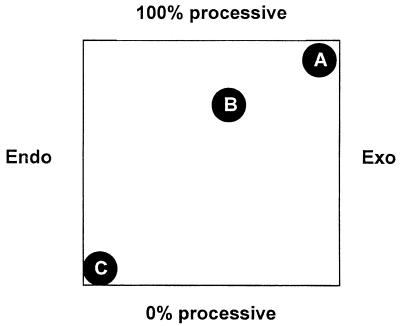
One could tentatively position cellulolytic enzymes on a bidimensional map where one axis would vary as a function of processivity and the other would vary as a preference for internal glycosidic bonds or for chain ends (Fig. 9). On this qualitative map, a typical endo-acting enzyme, such as, for instance, Cel45A (formerly called EGV [24]) from H. insolens, would be located closer to the 100% endo and 0% processive markers. On the other hand, Cel7A would be located close to the 100% processive and 100% exo markers. Cel6A would be in between, more likely closer to Cel7A than to Cel45A. A more precise location of cellulases on this map awaits methods able to measure their degree of processivity, i.e., the average number of subsequent cuts performed on the same substrate chain following the initial cut. It is also possible that the degree of processivity or even the preference for internal bonds versus chain ends varies as a function of the substrate used (23).
FIG. 9.
Bidimensional map for cellulases. Endo, preference for intrachain bonds only; Exo, preference for chain ends only; 0% processive, after the initial attack, a second attack on the same chain is not more probable than an attack on another chain; 100% processive, an infinite number of cuts on the same chain may occur after the initial attack. (A) Cel7A. (B) Cel6A. (C) Cel45A.
The functional classification of Cel7A as highly processive and Cel6A as endo-processive explains quite well the apparent exo-exo synergy that results when these two enzymes act together on crystalline substrates. Indeed, under the optimum conditions resulting from the curves shown in Fig. 3 and 4, the combination of the processive action of Cel7A and the endo-processive behavior of Cel6A should result in thin, short, needle-like elements. This is exactly what is seen in Fig. 6c when Cel6A and Cel7A were combined for the digestion of bacterial cellulose ribbons.
The structures of Cel6A from H. insolens and Cel6A from T. reesei have been experimentally determined (38, 46). The structure of H. insolens Cel7A has not yet been solved, but the strong sequence similarity (63%) of this enzyme to Cel6A from T. reesei, whose structure has been solved (13), predicts an identical folding geometry. All these CBH have tunnel-shaped active sites. The roof of the tunnels is made of pairs of large loops (one pair for Cel6A and two pairs for Cel7A) significantly longer than those in their endoglucanase counterparts. With endoglucanases such as Cel45A from H. insolens, a large loop adjacent to the catalytic site shows substantial flexibility upon substrate binding (11, 12). Significantly, similar substrate-induced movements, albeit not as large as that shown for Cel45A, have been observed recently in the loops closing the active site of Cel6A from H. insolens (47). The ability of CBH to occasionally “open” to perform their initial attack, first envisioned (10) and then proposed (23, 46), has recently found an elegant demonstration (54): the structures of T. reesei Cel6A in complex with oligosaccharides have shown that one of the loops has substantial mobility and that the resulting tunnel could be either more tightly closed or almost fully open. The open conformation is likely to be responsible for the endo character of Cel6A and for the observed synergy of Cel6A with Cel7A (54).
Despite differences in their topologies, the active sites of CBH (tunnel) and of endoglucanases (cleft) can accommodate only one cellulose chain. An endo attack, whether from an endoglucanase or from an endo-processive CBH such as Cel6A, is unlikely to occur on an extended cellulose chain bound to its neighbors by hydrogen bonds at the surface of a perfect crystal. Instead, the endo cuts probably take place at areas containing bent, flexible, and hydrated disordered chains. In selecting model substrates susceptible to showing visible morphological changes during digestion, one is confronted with the diversity of cellulose samples available. In the past, our laboratory as well as other laboratories have advocated the use of Valonia microcrystals to visualize the exo mode of action of CBH (6–8, 18, 19, 26). These studies have led to the images of pointed-tip digested microcrystals, with the points toward the reducing ends for Cel7A digestion and the nonreducing ends for Cel6A digestion. Bacterial microcrystalline cellulose (BMCC) has also been developed as a substrate cleaner than Avicel or filter paper and more reactive than Valonia microcrystals (21). However, both BMCC and Valonia microcrystals are prepared through a harsh acid treatment, which hydrolyzes the structural defects distributed along the parent microfibrils. These substrates are therefore somewhat resistant to an endo mode of hydrolysis as opposed to an exo mode, since a large number of chain ends have been created by the acid treatment. Thus, neither Valonia microcrystals nor BMCC is appropriate for the visualization of the result of endocellulase attack in processive enzymes. We believe that the use of the bacterial cellulose ribbons described here or any microfibrillated cellulose is best suited to demonstrate the endo-processivity of cellulases. Such a substrate could also be used with other cellulolytic systems.
A final aspect of the digestion of bacterial cellulose ribbons by the combination of Cel6A and Cel7A is the aspect of synergistic degradation as a function of the enzyme composition (Fig. 3). In a classical synergy between pure endo- and exocellulases acting on a high-molecular-weight substrate, the greatest synergistic effect is observed at an extremely low endocellulase content (21). With enzymes differing in their directionality of action, differing in their ability to make endo-processive cuts, and yet able to also perform exo cuts, one expects a shift of the maximal synergistic effect to more balanced mixtures, as shown here, where the increase in degradation culminates at approximately a 2:1 molar ratio of Cel6A and Cel7A. It remains to be seen whether this synergistic feature can be also observed in other enzymatic systems.
ACKNOWLEDGMENT
This work was funded by the Eurocell contract from the European Commission (Biotechnology Programme contract BIO4-CT97-2303).
REFERENCES
- 1.Amano Y, Shiroishi M, Nisizawa K, Hoshino E, Kanda T. Fine substrate specificities of four exo-type cellulases produced by Aspergillus niger, Trichoderma reesei and Irpex lacteus on (1→3), (1→4)-β-d glucans and xyloglucan. J Biochem. 1996;120:1123–1129. doi: 10.1093/oxfordjournals.jbchem.a021531. [DOI] [PubMed] [Google Scholar]
- 2.Armand S, Drouillard S, Schülein M, Henrissat B, Driguez H. A bifunctionalized fluorogenic tetrasaccharide as a substrate to study cellulases. J Biol Chem. 1997;272:2709–2713. doi: 10.1074/jbc.272.5.2709. [DOI] [PubMed] [Google Scholar]
- 3.Barr B K, Hsieh Y-L, Ganem B, Wilson D B. Identification of two functionally different classes of exocellulases. Biochemistry. 1996;35:586–592. doi: 10.1021/bi9520388. [DOI] [PubMed] [Google Scholar]
- 4.Béguin P. Molecular biology of cellulose degradation. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- 5.Biely P, Vrsanska M, Claeyssens M. Mode of action of Trichoderma reesei β-1,4 glucanases on cellooligosaccharides. In: Suominen P, Reinikainen T, editors. Proceedings of the Second TRICEL Symposium on Trichoderma reesei Cellulases and other Hydrolases. Espoo, Finland: Foundation for Biotechnical and Industrial Fermentation Research; 1993. pp. 99–108. [Google Scholar]
- 6.Boisset C, Armand S, Drouillard S, Chanzy H, Driguez H, Henrissat B. Structure-function relationships in cellulases: the enzymatic degradation of insoluble cellulose. In: Claeyssens M, Nerinckx W, Piens K, editors. Carbohydrases from Trichoderma reesei and other microorganisms. Structure, biochemistry, genetics and applications. Cambridge, United Kingdom: The Royal Society of Chemistry; 1998. [Google Scholar]
- 7.Chanzy H, Henrissat B. Electron microscopy study of the enzymic hydrolysis of Valonia cellulose. Carbohydr Polym. 1983;3:161–173. [Google Scholar]
- 8.Chanzy H, Henrissat B. Unidirectional degradation of Valonia cellulose microcrystals subjected to cellulase action. FEBS Lett. 1985;184:285–288. [Google Scholar]
- 9.Christensen T, Woeldike H, Boel E, Mortensen S B, Hjortshoej K, Thim L, Hansen M T. High level expression of recombinant genes in Aspergillus oryzae. Bio/Technology. 1988;6:1419–1422. [Google Scholar]
- 10.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 11.Davies G J, Dodson G G, Hubbard R E, Tolley S P, Dauter Z, Wilson K S, Hjort C, Mikkelsen J M, Rasmussen G, Schülein M. Structure and function of endoglucanase V. Nature. 1993;365:362–364. doi: 10.1038/365362a0. [DOI] [PubMed] [Google Scholar]
- 12.Davies G J, Tolley S P, Henrissat B, Hjort C, Schülein M. Structures of oligosaccharide-bound forms of endoglucanase V from Humicola insolens at 1.9 Å resolution. Biochemistry. 1995;34:16210–16220. doi: 10.1021/bi00049a037. [DOI] [PubMed] [Google Scholar]
- 13.Divne C, Ståhlberg J, Reinikainen T, Ruohonen L, Pettersson G, Knowles J K C, Teeri T T, Jones T A. The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science. 1994;265:524–528. doi: 10.1126/science.8036495. [DOI] [PubMed] [Google Scholar]
- 14.Divne C, Ståhlberg J, Teeri T T, Jones T A. High resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei. J Mol Biol. 1998;275:309–325. doi: 10.1006/jmbi.1997.1437. [DOI] [PubMed] [Google Scholar]
- 15.Eriksson K-E L, Blanchette R A, Ander P. Biodegradation of cellulose. In: Eriksson K-E L, Blanchette R A, Ander P, editors. Microbial and enzymatic degradation of wood and wood components. Berlin, Germany: Springer-Verlag; 1990. pp. 89–180. [Google Scholar]
- 16.Eveleigh D E. Cellulase: a perspective. Philos Trans R Soc London Ser A. 1987;321:435–447. [Google Scholar]
- 17.Fägerstam L G, Pettersson L G. The 1.4-β-glucan cellobiohydrolases of Trichoderma reesei QM 9414. FEBS Lett. 1980;119:97–100. [Google Scholar]
- 18.Hayashi N, Sugiyama J, Okano T, Ishihara M. Selective degradation of the cellulose Iα component in Cladophora cellulose with Trichoderma viride cellulase. Carbohydr Res. 1998;305:109–116. [Google Scholar]
- 19.Hayashi N, Sugiyama J, Okano T, Ishihara M. The enzymatic susceptibility of cellulose microfibrils of the algal-bacterial type and the cotton-ramie type. Carbohydr Res. 1998;305:261–269. [Google Scholar]
- 20.Henriksson K, Teleman A, Suortti T, Reinikainen T, Jaskari J, Teleman O, Poutanen K. Hydrolysis of barley (1→3), (1→4)-β-d glucan by a cellobiohydrolase II preparation from Trichoderma reesei. Carbohydr Polym. 1995;26:109–119. [Google Scholar]
- 21.Henrissat B, Driguez H, Viet C, Schülein M. Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Bio/Technology. 1985;3:722–726. [Google Scholar]
- 22.Henrissat B. Cellulases and their interaction with cellulose. Cellulose. 1994;1:169–196. [Google Scholar]
- 23.Henrissat B. Enzymatic cellulose degradation. Cellulose Commun. 1998;5:84–90. [Google Scholar]
- 24.Henrissat B, Teeri T T, Warren R A J. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett. 1998;425:352–354. doi: 10.1016/s0014-5793(98)00265-8. [DOI] [PubMed] [Google Scholar]
- 25.Hoshino E, Shiroishi M, Amano Y, Nomura M, Kanda T. Synergistic actions of exo-type cellulases in the hydrolysis of cellulose with different crystallinities. J Ferment Bioeng. 1997;84:300–306. [Google Scholar]
- 26.Imai T, Boisset C, Samejima M, Igarashi K, Sugiyama J. Unidirectional processive action of cellobiohydrolase Cel7A on Valonia cellulose microcrystals. FEBS Lett. 1998;432:113–116. doi: 10.1016/s0014-5793(98)00845-x. [DOI] [PubMed] [Google Scholar]
- 27.Irwin D, Shin D-H, Zhang S, Barr B K, Sakon J, Karplus P A, Wilson D B. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J Bacteriol. 1998;180:1709–1714. doi: 10.1128/jb.180.7.1709-1714.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irwin D C, Spezio M, Walker L P, Wilson D B. Activity studies of eight purified cellulases: specificity, synergism and binding domain effects. Biotechnol Bioeng. 1993;42:1002–1013. doi: 10.1002/bit.260420811. [DOI] [PubMed] [Google Scholar]
- 29.Kidby D K, Davidson D J. A convenient ferricyanide estimation of reducing sugars in the nanomole range. Anal Biochem. 1973;55:321–325. doi: 10.1016/0003-2697(73)90323-0. [DOI] [PubMed] [Google Scholar]
- 30.Klyosov A A. Trends in biochemistry and enzymology of cellulose degradation. Biochemistry. 1990;29:10577–10585. doi: 10.1021/bi00499a001. [DOI] [PubMed] [Google Scholar]
- 31.Koyama M, Helbert W, Imai T, Sugiyama J, Henrissat B. Parallel-up structure evidences the molecular directionality during biosynthesis of bacterial cellulose. Proc Natl Acad Sci USA. 1997;94:9091–9095. doi: 10.1073/pnas.94.17.9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulhshreshtha A K, Dweltz N E. Paracrystalline lattice disorder in cellulose. I. Reappraisal of the application of the two-phase hypothesis to the analysis of powder X-ray diffractograms of native and hydrolyzed cellulosic materials. J Polym Sci Polym Phys Ed. 1973;11:487–497. [Google Scholar]
- 33.Kyriacou A, MacKenzie C R, Neufeld R J. Detection and characterization of the specific and nonspecific endoglucanases of Trichoderma reesei: evidence demonstrating endoglucanase activity by cellobiohydrolase II. Enzyme Microb Technol. 1987;9:25–32. [Google Scholar]
- 34.Marx-Figini M. The control of molecular weight and molecular-weight distribution in the biogenesis of cellulose. In: Brown R M Jr, editor. Cellulose and other natural polymer systems. Biogenesis, structure and degradation. New York, N.Y: Plenum Press; 1982. pp. 243–271. [Google Scholar]
- 35.Nidetzky B, Steiner W, Hayn M, Claeyssens M. Cellulose hydrolysis by the cellulases from Trichoderma reesei: a new model for synergistic interaction. Biochem J. 1994;298:705–710. doi: 10.1042/bj2980705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reese E T, Siu R G H, Levinson H S. The biological degradation of soluble cellulose derivatives and its relationship to the mechanism of cellulose hydrolysis. J Bacteriol. 1950;59:485–497. doi: 10.1128/jb.59.4.485-497.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reverbel-Leroy C, Pages S, Belaich A, Belaich J-P, Tardif C. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J Bacteriol. 1997;179:46–52. doi: 10.1128/jb.179.1.46-52.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouvinen J, Bergfors T, Teeri T, Knowles J K C, Jones T A. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990;249:380–386. doi: 10.1126/science.2377893. [DOI] [PubMed] [Google Scholar]
- 39.Schmid G, Wandrey C. Evidence for the lack of exo-cellobiohydrolase activity in the cellulase system of Trichoderma reesei QM 9414. J Biotechnol. 1990;14:393–410. [Google Scholar]
- 40.Schülein M. Enzymatic properties of cellulases from Humicola insolens. J Biotechnol. 1997;57:71–81. doi: 10.1016/s0168-1656(97)00090-4. [DOI] [PubMed] [Google Scholar]
- 41.Ståhlberg J, Johansson G, Pettersson G. Trichoderma reesei has no true exo-cellulase: all intact and truncated cellulases produce new reducing end groups on cellulose. Biochim Biophys Acta. 1993;1157:107–113. doi: 10.1016/0304-4165(93)90085-m. [DOI] [PubMed] [Google Scholar]
- 42.Teeri T T. Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol. 1997;15:160–167. [Google Scholar]
- 43.Tomme P, Heriban V, Claeyssens M. Adsorption of two cellobiohydrolases from Trichoderma reesei to Avicel: evidence for “exo-exo” synergism and possible “loose complex” formation. Biotechnol Lett. 1990;12:525–530. [Google Scholar]
- 44.Tomme P, Warren R A J, Gilkes N R. Cellulose hydrolysis by bacteria and fungi. Adv Microb Physiol. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- 45.Tomme P, Kwan E, Gilkes N R, Kilburn D G, Warren R A J. Characterization of CenC, an enzyme from Cellulomonas fimi with both endo- and exoglucanase activities. J Bacteriol. 1996;178:4216–4223. doi: 10.1128/jb.178.14.4216-4223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varrot A, Hastrup S, Schülein M, Davies G J. Crystal structure of the catalytic core domain of the family 6 cellobiohydrolase II, Cel6A, from Humicola insolens, at 1.92 Å resolution. Biochem J. 1999;337:297–304. [PMC free article] [PubMed] [Google Scholar]
- 47.Varrot A, Schülein M, Davies G J. Structural changes of the active site tunnel of Humicola insolens cellobiohydrolase, Cel6A, upon oligosaccharide binding. Biochemistry. 1999;38:8884–8891. doi: 10.1021/bi9903998. [DOI] [PubMed] [Google Scholar]
- 48.Vrsanska M, Biely P. The cellobiohydrolase I from Trichoderma reesei QM 9414: action on cellooligosaccharides. Carbohydr Res. 1992;227:19–27. [Google Scholar]
- 49.Wood T M, McCrae S I. The purification and properties of the C1 component of Trichoderma koningii cellulase. Biochem J. 1972;128:1183–1192. doi: 10.1042/bj1281183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood T M, McCrae S I. Synergism between enzymes involved in the solubilization of native cellulose. Adv Chem Ser. 1979;181:181–209. [Google Scholar]
- 51.Wood T M. Properties of cellulolytic enzyme systems. Biochem Soc Trans. 1985;13:407–410. doi: 10.1042/bst0130407. [DOI] [PubMed] [Google Scholar]
- 52.Wood T M, McCrae S I. The cellulase of Penicillium pinophilum. Synergism between enzyme components in solubilizing cellulose with special reference to the involvement of two immunologically distinct cellobiohydrolases. Biochem J. 1986;234:93–99. doi: 10.1042/bj2340093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wood T M. Fungal cellulases. In: Haigler C H, Weimer P J, editors. Biosynthesis and biodegradation of cellulose. New York, N.Y: Marcel Dekker, Inc.; 1991. pp. 491–533. [Google Scholar]
- 54.Zou J Y, Kleywegt G J, Ståhlberg J, Driguez H, Nerinckx W, Claeyssens M, Koivula A, Teeri T T, Jones A T. Crystallographic evidence for substrate ring distortion and protein conformational changes during catalysis in cellobiohydrolase Cel6A from Trichoderma reesei. Structure. 1999;7:1035–1045. doi: 10.1016/s0969-2126(99)80171-3. [DOI] [PubMed] [Google Scholar]