Abstract
Extracellular vesicles (EVs) play an essential role in the communication between cells and the tumor microenvironment. However, the effect of tumor-derived EVs on the growth and metastasis of lung adenocarcinoma (LUAD) remains to be explored. This study aimed to elucidate the role of miR-153-3p-EVs in the invasion and migration capabilities of LUAD cells and explore its mechanism through in vivo and in vitro experiments. We found that miR-153-3p was specifically and highly expressed in LUAD and its secreted EVs. Furthermore, the expression of BANCR was negatively regulated by miR-153-3p and identified as a target gene of miR-153-3p using luciferase reporter assays. Through further investigation, we found that the downregulation of BANCR activates the PI3K/AKT pathway and accelerates the process of epithelial-mesenchymal transition (EMT), which ultimately leads to the aggravation of LUAD. The orthotopic xenograft mouse model was established to illustrate the effect of miR-153-3p-EVs on LUAD. Animal studies showed that miR-153-3p-EVs accelerated tumor growth in mice. Besides, we found that miR-153-3p-EVs could damage the respiratory ability of mice and produce a mass of inflammatory cells around the lung tissue of mice. Nevertheless, antagomir-153-3p treatment could inhibit the deterioration of respiratory function and inhibit the growth of lung tumors in mice. In conclusion, our study reveals the potential molecular mechanism of miR-153-3p-EVs in the development of LUAD and provides a potential strategy for the treatment of LUAD.
Keywords: epithelial-mesenchymal transition, extracellular vesicles, lung adenocarcinoma, miRNAs, PI3/Akt
INTRODUCTION
Lung cancer is one of the most diagnosed cancers worldwide, 75% of patients were diagnosed at an advanced stage. Due to its poor prognosis, lung cancer is the most common cause of cancer death (Bade and Dela Cruz, 2020; Wadowska et al., 2020). Lung cancer is divided into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), of which NSCLC patients account for the vast majority (Herbst et al., 2018; Zhong et al., 2021). NSCLC is subdivided into adenocarcinoma, squamous cell carcinoma, and large cell carcinoma (Hu et al., 2020a; Wang et al., 2020). Compared with squamous cell carcinoma and large cell carcinoma, adenocarcinoma is the most common type of NSCLC due to its high incidence and rapid metastasis. Furthermore, lung adenocarcinoma (LUAD) has a poor chemotherapy efficacy and a low 5-year survival rate compared with lung squamous cell carcinoma (Zhang et al., 2020). Thus, it is imperative to elucidate the mechanism of NSCLC metastasis and find effective treatments to improve patients’ quality of life. The communication between the tumor and its surrounding cells is the leading cause of tumor metastasis, but the interaction mechanism remains unclear (Kim et al., 2020).
Extracellular vesicles (EVs) are vesicular bodies of lipid membranes secreted and released by various types of cells. EVs are generally classified into three categories: exosomes (about 100 nm), microvesicles (MV, about 1 μm), and apoptotic bodies (Abs, > 1 μm) (Mathieu et al., 2019). EVs are involved in regulating various biological processes and are the key to mediating cell-to-cell communication (Hu et al., 2020b). EVs carry substances such as RNA, proteins, lipids, and DNA so that they can be absorbed by other cells near the source cell or by biological fluids far away in the body and trigger various phenotypic reactions (O'Brien et al., 2020). In recent years, non-coding RNAs (ncRNAs) have gradually become a hotspot in EV-RNA research, especially EV-miRNAs, which have played a vital role in intercellular communication. MicroRNAs (miRNAs) are small, ncRNAs that can modulate genes expression by inhibiting mRNA translation or promoting mRNA degradation (Jana et al., 2020). Current studies have demonstrated the dysregulation of miRNA expression in cancer and the carcinogenic or anticancer effect of miRNA (Ali Syeda et al., 2020). MiRNA has prognostic and tumor-suppressive effects, and its therapeutic value can be directly applied in clinical practice (Guo et al., 2021; Liu and Zhang, 2017). Besides, miRNA also plays an important role in the immune system (Jia and Wei, 2020). EV-miRNAs were found to convey information between tumor cells and stromal cells. The exosomal miR-1247-3p secreted by high-metastatic hepatocellular carcinoma cells directly targets B4GALT3, which leads to the activation of the β1-integrin-NF-κB signaling pathway in fibroblasts (Fang et al., 2018). In addition, EV-miRNA as a biomarker can predict the occurrence of disease. An increase of EV-miR-301a-3p enrichment and a decrease of EV-miR-1293 enrichment have been proposed as potential biomarkers for metastatic disease of patients with clear cell renal cell carcinoma (Dias et al., 2020). However, the function of more EV-miRNAs in NSCLC is unclear.
BRAF-activated non-coding RNA (BANCR) is a type of long non-coding RNA (lncRNA) that plays a vital role in the initiation and progression of different cancers. Current study showed that BANCR was an oncogene in colorectal cancer, its silencing hampered colorectal cancer progression and enhanced Adriamycin sensitivity by regulating miR-203/CSE1L (Ma et al., 2018). Conversely, BANCR has an anti-tumor effect in human bladder cancer, which may be related to differences in tissues and microenvironment (He et al., 2016). Besides, BANCR can regulate the epithelial-mesenchymal transition (EMT) process by regulating miR-338-3p in esophageal squamous cell carcinoma (Song et al., 2020). EMT is a cellular program related to tumor initiation, invasion, metastasis, and treatment resistance (Yilmaz and Christofori, 2009). However, the regulatory mechanism of BANCR on invasion and migration of LUAD remains unclear.
In this study, we focused on the effect of miR-153-3p-EVs on the survival and invasion of LUAD and further explored the mechanism. This study offers a potential treatment to alleviate the progression of LUAD.
MATERIALS AND METHODS
Experimental animals
BALB/c mice were purchased from Shanghai Animal Laboratory Center and housed in Beijing Rehabilitation Hospital. All animal experiments complied with ethical regulations and were approved by the Medical Ethics Committee of the Beijing Rehabilitation Hospital (approval No. 2020-bjkfyy-120).We turned on the lights at 7:00 am and turned them off at 7:00 pm in the incubator. In the first experiment, SW1271 cells (5 × 106 each) were orthotopically injected transpleurally into 6 mice. After 7 days of injection, the mice were equally divided into two groups (n = 3 in each group), and the same dose of miR-NC-EVs or miR-153-3p-EVs was injected via the tail vein once every 7 days for 35 days. Seven days after the last injection treatment, all mice were euthanized, and tumors were collected. Tumor lengths and widths were measured using calipers. Tumor volumes were calculated using V (mm3) = (L × W2)/2 (L = tumor length, W = tumor width). In the second experiment, SW1271 cells (5 × 106 each) were orthotopically injected transpleurally into 6 mice. After 7 days of injection, mice were equally divided into two groups (n = 3 each group), and the same dose of antagomir-153-3p or antagomir-NC was injected into the tail vein once every 7 days for 35 days. This study was performed in strict accordance with the requirements in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Cell culture
Human LUAD cell lines (NCI-H1993, SW1271, NCI-H522) and human lung squamous cell carcinoma (LUSC) cell lines (NCI-H226, YTMLC-90, SK-MES-1, NCI-H1915) were purchased from ATCC (USA). All cells were cultured in RPMI-1640 medium with 10% fetal bovine serum (FBS). The cell lines were cultured in an incubator at 37°C and 5% CO2.
RNA sequencing analysis
Gene expression data of lung cancer were downloaded from the Gene Expression Omnibus (GEO) dataset (GSE135918, GSE111803, GSE151963, and GSE74190). We used RStudio software for data analysis. Raw data were normalized and analyzed by limma package. Significant differential expressed transcripts were screened by fold change ≥ 2 or ≤ −2 and P value ≤ 0.05.
The online database gene
Based on The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) projects, the RNA sequencing expression data related to our project was analyzed using the GEPIA web tool (http://gepia.cancer-pku.cn) (Tang et al., 2017). Based on PAR-CLIP, HITS-CLIP, iCLIP, CLASH data sets, miRNA-mRNA and miRNA-lncRNA interaction networks related to our project were analyzed using StarBase 3.0 (http://starbase.sysu.edu.cn) (Li et al., 2014) and Diana (https://diana.e-ce.uth.gr/lncbasev3) (Karagkouni et al., 2020).
Lentiviral vector construction, production and transfection
Human BANCR full-length cDNA was amplified by PCR from the mRNA of LUAD cells. Then, the shBANCR sequences were designed with shLuc as the negative control (NC). The objective products were cloned into pLVX-shRNA2. The constructed vectors and the lentivirus packaging vectors were cotransfected into SW1271 for 48 h, respectively. Lentiviruses were produced, harvested, and purified with ultracentrifugation. SW1271 cells seeded in 24-well plates were transfected with lentivirus, using 8 μg/ml polybrene (Sigma, USA). Stable expression cells were screened in a medium containing 800 μg/ml G418 (Sigma). LV-BANCR, LV-hsa-miR-153-3p or their corresponding NC was transduced into LUAD cells. Hsa-miR-153-3p mimics, inhibitors or their parental NCs were transfected into the LUAD cells using Lipofectamine 2000 reagent (Invitrogen, USA).
Quantitative real-time polymerase chain reaction
The cells were plated in 24-well plates and treated according to the experimental design. To extract RNA, cells were washed with phosphate-buffered saline (PBS). Then the total RNA was isolated using Trizol reagent (Life Technologies, USA) according to the manufacturer’s instructions. And the reverse transcription was performed using SuperScript II (Invitrogen). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed with SYBR Prime Script RT-PCR Kits (Takara, Japan) based on the manufacturer’s instructions. For analysis, the expression level of mRNA was normalized against GAPDH mRNA measured per sample. Specific primers were shown in Supplementary Table S1.
Western blot
The protein samples were extracted and separated by 10% SDS-PAGE gel, and then transferred to a PVDF membrane (Millipore, USA). The membrane was then blocked with 5% skimmed milk and incubated overnight, using the following main detection antibodies at 4°C: anti-CD63 (1:1,000; Abcam, UK), anti-TSG101 (1:1,000; Abcam), anti-Alix (1:1,000; Abcam), anti-Hsp90 (1:1,000; Abcam), anti-GRP94 (1:1,000; Abcam), anti-Cytochrome (1:1,000; Abcam), anti-p-pi3k (1:1,000; Abcam), anti-pi3k (1:1,000; Abcam), anti-p-akt (1:1,000; Abcam), anti-akt (1:1,000; Abcam), anti-E-cadherin (1:1,000; Abcam), anti-vimentin (1:1,000; Abcam), or anti-b-actin (1:5,000; Proteintech, USA). We washed 3 times with TBS-T and the membranes were cultured with the secondary antibody at 24°C for 1 h. The western blots were pictured using an ECL Reagent (Pierce, USA) and the density was verified using ImageJ software (NIH, USA).
CCK-8 assay
According to the experimental procedure instruction, we used the CCK-8 kit to evaluate cell proliferation activity. Cells were seeded into 96-well plates at a density of 1,000 cells/well. To investigate the effect of miR-153-3p on LUAD, the cells were transfected with miR-153-3p and NC. To investigate the effect of BANCR on LUAD, the cells were transfected with inh-miR-153-3p, BANCR-KD, inh-miR-153-3p+BANCR-KD, and NC. To investigate the effect of BANCR/PI3K/AKT axis on LUAD, the cells were transfected with inh-miR-153-3p, BANCR-KD, inh-miR-153-3p+LY294002, BANCR-KD+LY294002, and NC. The cell proliferation activity was measured at 0, 1, 2, 3, and 4 days after transfection. Before detection, the medium was removed, and the cells were washed with PBS to remove dead cells. Then the CCK8 dilution was added to 96-well plates (100 μl per well) and incubated at 37°C for about 1.5 h. After incubation, we took out the plate, and the absorption value of 450 nm was measured.
Transwell assay
Transwell assay was used to investigate the migration and invasion of LUAD cells. We plated the cells in the upper part of the Matrigel-coated invasion chamber or the uncoated migration chamber (BD Biosciences, USA). Then added 500 μl of DMEM medium containing 10% FBS to the lower chamber and added serum-free medium to the upper chamber. After 48 h of incubation, we wiped off non-invasive cells with cotton swabs. We used 95% ethanol to fix the invading or migrating cells and stained the cells with 0.1% crystal violet. Finally, counted the migration and invasion cells in the lower chamber under an inverted microscope.
Wound-healing assay
Cells were seeded in 6-well plates and cultured to the subfusion state. After being starved in a serum-free medium for 24 h, the monolayer cells were scraped linearly to introduce an artificial wound that was captured at 0 h and 24 h.
Whole-body plethysmography
To detect the effect of miR-153-3p-EVs on mice, awake and unrestrained female BALB/c mice were recorded under three different treatments: sham, treated with EVs, and treated with miR-153-3p-EVs using the whole-body plethysmography (WBP) system (Data Sciences International, USA). To detect the effect of antagomir-153-3p on mice, awake, unrestrained female BALB/c mice were recorded under two different treatments: treated with antagomir-153-3p or antagomir-153-3p using a WBP system (Data Sciences International). The mice were weighed and placed in one of the different plethysmography chambers, and each group of data was recorded at the same time. The WBP recording and analysis were adapted from the previous description. In short, all mice were adapted for 10 min and then recorded for 45 min. Ponemah 5 (Data Sciences International) was used to analyze the data collection of the ventilation parameters respiration rate (RR), tidal volume (VT) and minute ventilation (VE) of each group of mice within 2 min. VT and VE are normalized to mouse body weight, expressed as ml/g and ml/g/min–1, respectively. Data are mean ± SEM.
EVs purification
We purified LUAD-derived EVs using ultracentrifugation. EVs were cultured in Ultra medium (12-725 F; Lonza, USA) containing 10% EVs-free FBS (50A-1; System Biosciences, USA) for 72 h prior to isolation. According to published protocols (Théry et al., 2006), the supernatants were centrifuged at 300 × g for 10 min, 2,000 × g for 10 min and 10,000 × g for 30 min at 4°C to eliminate cell debris. The supernatant was precipitated by centrifugation at 110,000 × g for 70 min. The EVs precipitate was washed in 3 ml PBS, then recentrifuged at 110,000 × g for 70 min and resuspended in PBS.
EVs transfection
We used an Exo-Fect™ Exosome Transfection kit (System Biosciences) to load miRNAs into EVs according to the manufacturer’s instructions. Briefly, EVs were mixed with Exo-Fect solution and miRNA mimics at 37°C in a shaker for 10 min. ExoQuick-TC was then added and incubated on ice for 30 min to stop the reaction. Then centrifuge the mixture at 13,000 rpm for 4 min to obtain EVs with transfected miRNA.
EVs internalization
NCI-H1993, SW1271, and NCI-H522 were inoculated in confocal dishes overnight, respectively. For EVs tracking, EVs were marked using PKH26 according to the manufacturer’s protocols. Next, we added the marked EVs. The cells were incubated at 37°C with 5% CO2 for 4 h, washed with PBS for 3 times. The nuclei were labeled with Hoechst staining. Finally, we observed the internalization of EVs into recipient cells using confocal microscopy.
Transmission electron microscopy (TEM)
We used differential centrifugation to obtain EVs secreted by LUAD. The separated EVs were diluted with PBS and then fixed in 2% paraformaldehyde. Then, the samples were transferred to a formvar copper mesh. Blotted, and contrast stained with uranyl acetate (Electron Microscopy Sciences, USA) at room temperature, then blotted on filter paper and air-dried before analysis. The FEI Tecnai 110 kV transmission electron microscope was used to examine the formulation at 80 kV.
Nanoparticle tracking analysis
We used differential centrifugation to obtain EVs secreted by LUAD. EVs were purified by ultracentrifugation and quantified using the NanoSight NS300 (NanoSight, UK). The sample was diluted to the optimal concentration in PBS. Each sample was recorded 3 times for 30 s, the temperature was manually monitored, and the camera level was set to 10. Data were represented as mean ± SD of three replicates. All measurements were performed at room temperature.
In vivo bioluminescence imaging
We used IVIS Spectrum (Perkin Elmer, USA) to monitor tumor growth and metastasis development at 7, 21, and 35 days through in vivo bioluminescence imaging (BLI). Mice were intraperitoneally injected with D-luciferin (Solarbio, China). Sodium pentobarbital was used to anesthetize mice. The fluorescence emitted by tumors was detected according to the manufacturer’s instructions and analyzed using Living Image software (ver. 4.3; Caliper Lifesciences, USA).
Immunohistochemical studies
Tissues needed were removed from mice at the time of sacrifice, fixed the tissue in paraffin, and sectioned after fixation. Immunohistochemical (IHC) analysis was performed using heat-induced antigen retrieval in sodium citrate buffer (pH 6.0). IHC staining was performed with primary anti-E-cadherin and primary anti-vimentin antibodies. After incubation with the secondary antibody, it was inoculated with the SABC kit (ZSGB-Bio, China) and diaminobenzidine (DAB; Sigma) to examine the expression of these markers in cells. The sections were stained with hematoxylin.
Fluorescence in situ hybridization
We cultivated SW1271 cells on the slides, washed them with PBS, and fixed them in 4% paraformaldehyde. After permeabilization, the hybridization was performed overnight with the lncRNA BANCR fluorescence in situ hybridization (FISH) probes (5'-ACAGGACTCCATGGCAAACG-3'). The cytoskeleton was stained with a fluorescent staining reagent for vinculin and cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) at room temperature. Specimens were analyzed on a Nikon inverted fluorescence microscope (Nikon, Japan).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7 software (GraphPad Software, USA). Statistical differences between the two groups were analyzed using the Student’s t-test. The comparison between multiple groups was made using the one-way ANOVA test followed by post-hoc test (least significant difference). Independent experiments were repeated at least three times for each experiment and error bars are mean ± SD. P < 0.05 was considered statistically significant.
RESULTS
Enrichment of miR-153-3p in tissues, serum and peripheral EVs in LUAD patients
To investigate abnormal miRNAs in tissues, serum and peripheral EVs in LUAD patients, we analyzed the miRNA expression profile by the public database. We first screened 60 miRNAs with the most significant differences in lung cancer tissues (GSE135918), exosomes extracted from peripheral blood (GSE111803) and plasma sample (GSE151963) of patients with LUAD. Interestingly, we found that miR-153-3p was significantly upregulated in all three tissues. (Fig. 1A). To further verify our findings, Venn analysis was used to find miRNAs that differed significantly in all three tissues. The results showed that the expression levels of miR-153-3p, miR-369-3p, and miR-4732-5p in tissues, serum and peripheral EVs of LUAD patients were significantly different from that in healthy controls (Fig. 1B). Together, these results implied that miR-153-3p is highly expressed in LUAD and plays an important role in the development of LUAD.
Fig. 1. Enrichment of miR-153-3p in tissues, serum and peripheral EVs in LUAD patients.
(A) Heat map of the significant differentially expressed miRNAs in lung cancer tissue specimens compared with non-tumor lung tissue specimens (GSE135918). Heat map of the significant differentially expressed miRNAs extracted from peripheral blood exosome in LUAD patients compared with healthy controls (GSE111803). Heat map of the significant differentially expressed miRNAs in plasma sample between LUAD patients and healthy people (GSE151963). Red represents high expression, and blue represents low expression in all heat maps. (B) Venn diagrams of differential miRNAs comparison (GSE135918, GSE111803, and GSE151963).
MiR‐153‐3p is highly expressed in LUAD
To further confirm the potential role of miR-153-3p in lung cancer. We performed qRT-PCR to detect the expression of miR-153-3p in LUAD cell lines and LUSC cell lines. The results showed that the expression level of miR-153-3p in LUAD cell line was significantly higher than that in LUSC cell line. Furthermore, the expression of miR-153-3p in SW1271 was significantly higher than NCI-H522 and NCI-H1993 in LUAD cell lines (Fig. 2A). To investigate the EVs derived from LUAD cells, EVs separated and purified from LUAD cell lines (NCI-H522, SW1271, NCI-H1993) were observed by TEM. The results showed that EVs derived from NCI-H522, SW1271, and NCI-H1993 supernatants were round EVs surrounded by a lipid bilayer membrane (Fig. 2B). Besides, nanoparticle tracking analysis revealed that most vesicles in NCI-H522/EVs were ranged from 80 to 140 nm in diameter with a peak at 93 nm, the majority of vesicles in SW1271/EVs ranged from 70 to 130 nm in diameter with a peak at 108 nm, and NCI-H1993/EVs ranged from 78 to 145 nm in diameter with a peak at 90 nm (Fig. 2C). The western blot analysis of EVs showed that four exosome markers (CD63, TSG101, Alix, Hsp90) were significantly enriched, while two non-exosome markers (GRP94, Cytochrome) were significantly lacking (Fig. 2D). Similar to the results of miR-153-3p in LUAD and LUSC cell lines, the qRT‐PCR analysis showed that the expression of miR-153-3p in LUSC-derived EVs was generally lower than that in LUAD-derived EVs. In particular, the expression of SW1271 was the highest in all LUAD-derived EVs (Fig. 2E). Furthermore, we detected the expression of miR-153-3p in the supernatant of EVs and EVs-removed supernatant using qRT-PCR (Supplementary Fig. S1A) and found that the expression of miR-153-3p in EVs was significantly higher than that in the supernatant. GSE74190 also confirmed that miR-153-3p was uniquely enriched in LUAD, compared with normal tissue, LUSC and SCLC (Fig. 2F, Supplementary Fig. S1B). The above evidence indicated that miR-153-3p is highly and specifically expressed in LUAD and LUAD-derived EVs.
Fig. 2. MiR-153-3p was highly expressed in LUAD.
(A) qRT-PCR analysis of miR-153-3p expression in LUSC cell lines (YTMLC-90, NCI-H266, and SK-MES-1) and LUAD cell lines (NCI-H1993, SW1271 and NCI-H522). (B) TEM of EVs derived from LUAD cell lines: EVs/ NCI-H1993, EVs/SW1271, and EVs/NCI-H522. (C) Nanoparticle tracking analysis of EVs/NCI-H1993, EVs/SW1271, and EVs/NCI-H522. (D) Western blot analysis of exosome markers (CD63, TSG101, Alix, and Hsp90) and non-exosome markers (GRP94 and Cytochrome) in LUAD cells lysate and separated EVs. (E) qRT-PCR analysis of miR-153-3p expression in EVs derived from LUSC cell lines (YTMLC-90, NCI-H266, and SK-MES-1) and LUAD cell lines (NCI-H1993, SW1271, and NCI-H522). (F) Heat map of the significant differentially expressed miRNAs extracted from selected lung carcinomas cell populations (small cell lung carcinoma, adenocarcinoma, and squamous cell carcinoma) compared with adjacent normal tissues (GSE74190). The data in the figures are represented the mean ± SD. **P < 0.01.
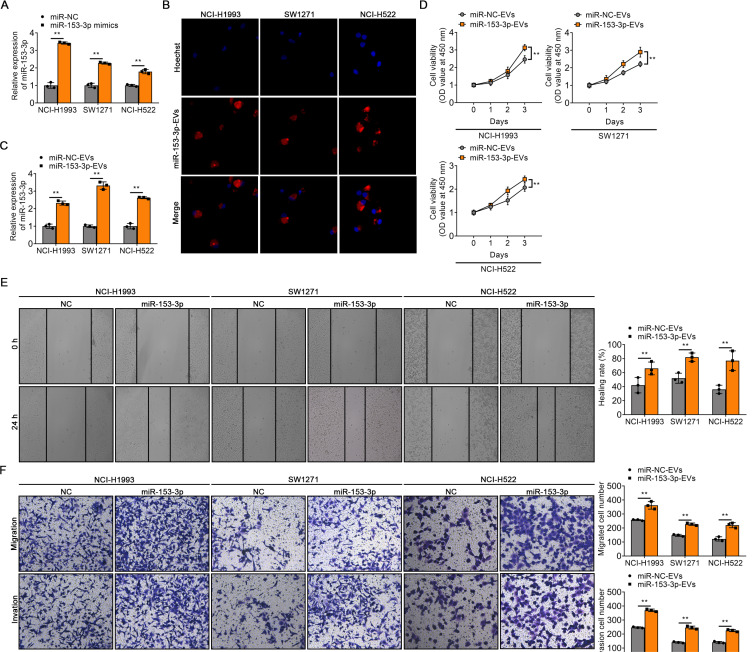
EV-transferred miR-153-3p enhances the malignant phenotype of LUAD cells
To address the potential effect of EV-transferred miR-153-3p on LUAD, miR-153-3p mimics were used to transfect EVs of LUAD cell lines. The qRT-PCR analysis showed that the expression of miR-153-3p was upregulated in EVs treated with miR-153-3p mimics (Fig. 3A). We next used miR-153-3p-EVs to cultivate three LUAD cell lines (NCI-H1993, SW1271, NCI-H522) and then detected the expression level of miR-153-3p in these cell lines. We found that most miR-153-EVs were internalized into the recipient target cell (Fig. 3B). Meanwhile, miR-153-3p-EVs treatment significantly amplified the expression of miR-153-3p in cells. Furthermore, miR-153-3p accumulated in recipient cells and had the strongest effect on SW1271 cells (Fig. 3C). CCK-8 assay was applied to detect the proliferation ability of cells, which showed that cells cultured with miR-153-3p-EVs showed a significant increase in cell proliferation (Fig. 3D). Wound-healing transwell assay revealed significant promotion of migration and invasion of cells cultured with miR-153-3p-EVs (Figs. 3E and 3F). These results indicated that EVs can deliver miR-153-3p to LUAD cells and enhance their malignant phenotype.
Fig. 3. EV-transferred miR-153-3p enhances the malignant phenotype of LUAD cells.
(A) The expression of miR-153-3p in EVs transfected with miR-NC or miR-153-3p mimics, respectively. (B) Confocal microscopy images of miR-153-3p-EVs uptaking by NCI-H1993, SW1271, and NCI-H522, respectively. MiR-153-3p-EVs were marked using PKH26 and the nuclei were marked using Hoechst. (C) qRT-PCR analysis of miR-153-3p accumulation in LUAD cell lines treated with miR-NC-EVs or miR-153-3p-EVs, respectively. (D) The viability of LUAD cell lines (NCI-H1993, SW1271, and NCI-H522) was assessed at 1, 2, and 3 days after treatment with miR-NC-EVs or miR-153-3p-EVs. (E) Quantification of the wound-healing assay in NCI-H1993, SW1271, and NCI-H522 cells treated with miR-NC-EVs or miR-153-3p-EVs. (F) Quantification of migration and invasion assays in NCI-H1993, SW1271, and NCI-H522 cells treated with miR-NC-EVs or miR-153-3p-EVs. The data in the figures are represented the mean ± SD. **P < 0.01.
BANCR is an antioncogene of LUAD regulated by miR-153-3p
To explore the mechanism of miR-153-3p on LUAD, we first used the miRNA-lncRNA interactions database Diana and StarBase to identify the potential downstream targets of miR-153-3p. Venn analysis indicated three potential downstream targets (AC005261.1, BANCR, and ZBTB20-AS5) of miR-153-3p in both databases (Fig. 4A). Among them, BANCR overlaps in the two databases. Interestingly, BANCR has been shown to be a tumor suppressor in lung cancer (Yu et al., 2017). Therefore, we hypothesized that miR-153-3p could affect the progression of LUAD by regulating BANCR. To verify our hypothesize, we next performed luciferase reporter assays to identify downstream targets of miR-153-3p. We found that miR-153-3p mimics reduced luciferase activity of the wild-type BANCR reporter gene, but not the BANCR MUT (Figs. 4B and 4C). The qRT-PCR analysis showed that the expression of BANCR in SW1271 was significantly down-regulated compared with that in NCI-H1993 and NCI-H522 (Fig. 4D). Kaplan–Meier analysis using GEPIA indicated that BANCR was associated with poor clinical outcomes in LUAD, while LUSC was not (Fig. 4E). We found that illustrated that transfection of miR-153-3p inhibitors enhanced BANCR expression in LUAD cell lines, especially in SW1271 using qRT-PCR (Fig. 4F). The lack of BANCR has been revealed to activate the PI3K pathways to promote the aggressiveness of papillary thyroid carcinoma (PTC) (Zhang et al., 2018). Therefore, to explore whether the role of BANCR in LUAD is similar to that in PTC, the western blot analysis was used to verify PI3K pathways activation in LUAD. Knockdown of BANCR increased PI3K and AKT phosphorylation, whereas inh-miR-153-3p inhibited this process. Interestingly, BANCR-KD+inh-miR-153-3p treatment reactivated the phosphorylation of PI3K and AKT, suggesting that miR-153-3p activated the PI3K/AKT signaling pathway via inhibiting BANCR (Fig. 4G). CCK-8 assay indicated that knockdown of BANCR significantly promoted tumor growth, while treatment with inh-miR-153-3p suppressed tumor growth. Moreover, the BANCR-KD+inh-miR-153-3p group neutralized both effects (Fig. 4H). To further explore whether BANCR regulates LUAD progression through the PI3K/AKT pathway, we used PI3K/AKT inhibitor LY294002 to detect the involvement of PI3K and AKT in cell viability and invasion upon treatment with inh-miR-153-3p and BANCR-KD. CCK-8 assay indicated that inh-miR-153-3p significantly alleviated the tumor growth, and LY294002 and inh-miR-153-3p treatment reinforced this effect. Furthermore, BANCR knockout significantly accelerated the tumor growth, while LY294002 inhibited this growth (Fig. 4I). Transwell assay also confirmed that miR-153-3p alleviated the migration and invasion of tumor cells via the BANCR/PI3K/AKT axis (Supplementary Fig. S2A). We performed the western blot to identify the EMT process in LUAD. We found that the expression of E-cadherin was downregulated in the BANCR-KD group but upregulated in the inh-miR-153-3p group. Vimentin was upregulated in the BANCR-KD group and reversed in the inh-miR-153-3p group. In addition, the expression of E-cadherin and vimentin showed no significant change in the inh-miR-153-3p+BANCR group compared with the NC group (Fig. 4J). Furthermore, StarBase demonstrated that miR-153-3p was negatively correlated with BANCR in LUAD (Fig. 4K). However, the other two target genes had no significant correlation with miR-153-3p (Supplementary Figs. S2B and S2C). Taken together, these findings revealed that the level of BANCR affects the PI3K pathway and the process of EMT, which is regulated by miR-153-3p.
Fig. 4. BANCR is an antioncogene of LUAD regulated by miR-153-3p.
(A) Venn diagrams of potential downstream targets of miR-153-3p in two databases (Diana and StarBase). (B) The predicted miR-153-3p binding sites in the BANCR transcript. (C) Relative luciferase activity of reporter containing the 3’ UTR of BANCR upon transfection with miR-152-3p mimics or mimics NC in LUAD cells. (D) qRT-PCR analysis of BANCR expression in NCI-H1993, SW1271, and NCI-H522 cells. (E) Overall survival of LUAD and LUSC patients in different expression of BANCR from GEPIA database. (F) qRT-PCR analysis of BANCR expression in NCI-H1993, SW1271, and NCI-H522 cells after transfection with miR-152-3p inhibitor. (G) Western blot analysis of p-pi3k, pi3k, p-akt, akt in the treatment with inh-miR-153-3p, BANCR-KD, or inh-miR-153-3p+BANCR-KD. (H) The cell viability was assessed at 1, 2, and 3 days after treatment with inh-miR-153-3p, BANCR-KD, and inh-miR-153-3p+BANCR-KD. (I) The cell viability was assessed at 1, 2, and 3 days after treatment with inh-miR-153-3p, BANCR-KD, inh-miR-153-3p+LY294002, or BANCR-KD+ LY294002. (J) Western blot analysis of E-cadherin and vimentin in the treatment with inh-miR-153-3p, BANCR-KD, or inh-miR-153-3p+BANCR-KD. (K) The relationship between BANCR and miR-153-3p was detected by StarBase. The data in the figures are represented the mean ± SD. *P < 0.05; **P < 0.01.
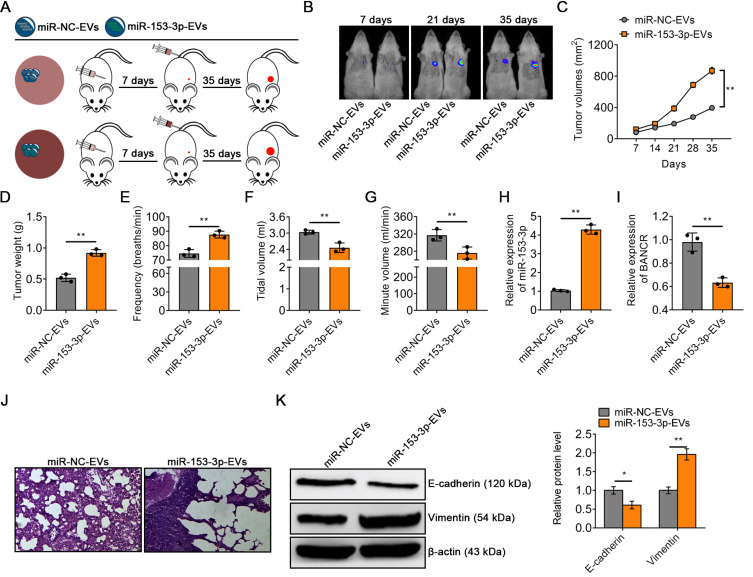
EVs modified with miR-153-3p mimics contribute to the progression of lung cancer
To explore the role of miR-153-3p-EVs mediating intercellular communications in the tumor microenvironment, we established an orthotopic xenograft mouse model by transpleural injecting SW1271 cells into BALB/c nude mice. Seven days after the injection of tumor cells, the mice were divided into two groups and injected with equal amounts of miR-NC-EVs or miR-153-3p-EVs at 7-day intervals (Fig. 5A). In vivo imaging of tumors showed that glowing tumor cells were detected in 6 mice 7 days after injection of tumor cells. In addition, more tumor proliferation and metastasis occurred in mice after injection of miR-153-3p-EVs compared with the control group, suggesting that the injection of miR-153-3p-EVs significantly accelerated the progression of LUAD (Fig. 5B). Besides, compared with the miR-NC-EVs group, miR-153-3p-EVs markedly accelerated tumor growth (Figs. 5C and 5D). We evaluated the effects of miR-NC-EVs and miR-153-3p-EVs on ventilatory function in mice by using the Whole Body Plethysmograph. The results showed that compared with the miR-NC-EVs, miR-153-3p-EVs accelerated the RR of mice (Fig. 5E). On the contrary, miR-153-3p-EVs decreased VE and VT in mice (Figs. 5F and 5G). The expression level of miR-153-3p was also significantly increased, while BANCR was significantly reduced after miR-153-3p-EVs treatment (Figs. 5H and 5I). H&E staining showed that miR-153-3p-EVs enhanced the infiltration of lung tumors in mice and produced excessive inflammatory cells (Fig. 5J). Besides, the western blot indicated that miR-153-3p-EVs markedly suppressed the expression of E-cadherin and enhanced the expression of vimentin, suggesting a promoting effect of EMT in LUAD (Fig. 5K). These results suggested that the damaged respiratory function of the mice can cause plenty of inflammatory cells and promote EMT around the lung tissues to further worsen LUAD.
Fig. 5. EVs modified with miR-153-3p mimics contributed to the progression of lung cancer.
(A) Experimental design of mouse model treated with miR-NC-EVs or miR-153-3p-EVs. (B) Representative BLI images of tumor in 6 mice at 7 days, 21 days, and 35 days treated with miR-NC-EVs or miR-153-3p-EVs. (C) Tumor volume was calculated every 7 days after injection. (D) Tumor weight was calculated after tumor excision. Ventilatory function ((E) respiratory rate, (F) tidal volume and (G) minute volume) of mice in miR-NC-EVs group or miR-153-3p-EVs group. (H) qRT-PCR analysis of miR-153-3p expression in the tumor treated with miR-NC-EVs or miR-153-3p-EVs in vivo. (I) qRT-PCR analysis of BANCR expression in the tumor treated with miR-NC-EVs or miR-153-3p-EVs in vivo. (J) H&E staining of lung tissue morphology of mice in the miR-NC-EVs group or miR-153-3p-EVs group. (K) Western blot analysis of E-cadherin and vimentin in tumor in vivo treated with miR-NC-EVs or miR-153-3p-EVs. The data in the figures are represented the mean ± SD. *P < 0.05; **P < 0.01.
Targeting miR-153-3p effectively mitigates the progression of LUAD and restored lung ventilation function
To further explore the effect of miR-153-3p-EVs on the progression of LUAD in vivo, we established mouse orthotopic xenograft by transpleural injecting SW1271 cells. One week after injection, two groups of mice were treated with antagomir-NC or antagomir-153-3p administration (Fig. 6A). We found that after the antagomir-153-3p administration, the tumor volume and weight of the mice were significantly reduced compared to the control group (Figs. 6B and 6C). By detecting the ventilation function of mice, we found that the respiratory frequency of mice treated with antagomir-153-3p was significantly reduced (Fig. 6D). Meanwhile, the tidal volume was significantly increased in mice after antagomir-153-3p administration compared with the antagomir-NC group (Fig. 6E). Besides, compared with the antagomir-NC group, the minute ventilation was higher in mice after antagomir-153-3p administration (Fig. 6F). The qRT-PCR analysis showed that antagomir-153-3p administration significantly reduced the expression of miR-153-3p and prevented the loss of BANCR in tumors in vivo (Figs. 6G and 6H). The FISH analysis also revealed that the level of BANCR was upregulated after antagomir-153-3p administration in mice (Fig. 6I). H&E staining showed that the lung tissue morphology was almost invisible, with severe tumor infiltration and many inflammatory cells in antagomir-NC group. Nevertheless, lung tissue morphology was acceptable, with a low degree of tumor invasion and few inflammatory cells in the antagomir-153-3p group (Fig. 6J). Ki67 IHC results revealed that the IHC score of Ki67 was significantly reduced by antagomir-153-3p treatment (Fig. 6K). The western blot indicated that antagomir-153-3p treatment markedly enhanced the expression of E-cadherin and suppressed the expression of vimentin (Fig. 6L). In summary, these results indicated that treatment targeting miR-153-3p effectively alleviates the progression of LUAD in mice and restores its lung ventilation function.
Fig. 6. Targeting miR-153-3p effectively mitigated the progression of LUAD and restored lung ventilation function.
(A) Experimental design of mouse treated with antagomir-NC or antagomir-153-3p administration. (B) Tumor volume was calculated every 7 days after injection. (C) Tumor weight was calculated after tumor excision. Ventilatory function (the respiration rate, D; tidal volume, E; and minute volume, F) of mice treated with antagomir-NC or antagomir-153-3p administration. (G) qRT-PCR analysis of miR-153-3p expression in the tumor treated with antagomir-NC or antagomir-153-3p administration in vivo. (H) qRT-PCR analysis of BANCR expression in the tumor treated with antagomir-NC or antagomir-153-3p administration in vivo. (I) FISH analysis of BANCR of mice treated with antagomir-NC or antagomir-153-3p administration. (J) The lung tissue morphology of mice treated with antagomir-NC or antagomir-153-3p administration was detected by H&E staining. (K) Representative IHC stain images of ki67 of mice treated with antagomir-NC or antagomir-153-3p administration. (L) Western blot analysis of E-cadherin and vimentin in tumor in vivo treated with antagomir-NC or antagomir-153-3p administration. The data in the figures are represented the mean ± SD. **P < 0.01.
DISCUSSION
In recent years, tumor-derived EVs have drawn much attention as a mediator in intracellular communication. Tumor-derived EVs serve as a messenger in tumor progression and metastasis, which can regulate the proliferation, migration and invasion of tumor cells (Becker et al., 2016). The previous view suggested that ITGBL1-enriched EVs stimulate the activation of the TNFAIP3-mediated NF-κB signaling pathway in fibroblasts to prepare a favorable microenvironment for colorectal cancer metastasis or metastatic growth (Ji et al., 2020). Besides, mtDNA-enriched EVs generated by ALDH2-deficient hepatocytes were proved to affect hepatocellular carcinoma progression (Seo et al., 2019). Interestingly, we also found that EVs-mediated miRNAs play an important role in tumor development. In this study, we revealed the roles of LUAD-derived EVs in regulating cancer progression. Compared with the NCI-H1993 and NCI-H522 cell lines, the SW1271 cell line showed higher invasion ability. Notably, the expression of miR-153-3p-EVs secreted by SW1271 cells was higher than NCI-H1993 and NCI-H522 cell lines. Therefore, consistent with previous views, the evidence suggested that EVs can affect the development of cancer by delivering different cargos (Xu et al., 2018).
MiR-153-3p has been reported to be involved in the progression of a variety of cancers. In thyroid cancer, miR-153-3p suppresses cells by regulating E3F3 (Deng et al., 2021). In addition, lncRNA FGD5-AS1 promotes tumor growth by regulating MCL1 via sponging miR-153-3p in oral cancer (Ge et al., 2020). Similarly, we also found that abnormal expression of miR-153-3p was associated with the development of LUAD. However, different from miR-153-3p in thyroid cancer or oral cancer, miR-153-3p was highly expressed in LUAD. Overexpression of miR-153-3p promoted the development of LUAD and further impaired ventilation in mice, while antagomir-153-3p administration restored ventilation in mice. Specifically, the high expression of miR-153-3p in LUAD inhibited the expression of BANCR. Further investigation demonstrated that BANCR was one of the downstream targets of miR-153-3p.
As an lncRNA, BANCR presents two sides in the progression of cancer. As an oncogene, BANCR promotes esophageal squamous cell carcinoma growth by activating the Wnt/β-catenin signaling pathway (Chen et al., 2019). Analogously, BANCR is also highly expressed in gastric cancer (Li et al., 2015). On the contrary, BANCR is a tumor suppressor involved in tumor proliferation, apoptosis, and migration in bladder cancer. Our study found that BANCR was negatively associated with poor clinical outcomes in LUAD. Compared with the NC group, BANCR knockdown significantly enhanced the cell viability of LUAD, which is consistent with previous research (Yu et al., 2017). Furthermore, BANCR knockdown enhanced the activation of the PI3K/AKT pathway and promoted the EMT process in cells. Compared with the NC group, the IHC score of Ki67 in the BANCR knockdown group was significantly increased. Therefore, BANCR is a tumor suppressor gene in LUAD.
In conclusion, EVs-delivered miR-153-3p potentiates the survival and invasion of LUAD by suppressing BANCR. The inhibited BANCR activates the PI3K/AKT pathway in cells, thereby promoting the EMT process of cells and ultimately enhancing the migration and invasion capabilities of LUAD. Together, drugs targeting the intervention of miR-153-3p have provided a new thought for the treatment of LUAD.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by the Science and Technology Development Special Project of Beijing Rehabilitation Hospital Affiliated to Capital Medical University (2019-002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We greatly appreciate the support from the Beijing Rehabilitation Hospital.
Footnotes
AUTHOR CONTRIBUTIONS
H.C. designed the study, wrote the manuscript, and corrected the manuscript. P.Z. conceived the experiments, provided reagents, performed experiments, and analyzed the data. H.Y. provided expertise, validated of experimental design, and supervised the progress of the study. J.X. managed project and secured funding.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ali Syeda Z., Langden S., Munkhzul C., Lee M., Song S.J. Regulatory mechanism of microRNA expression in cancer. Int. J. Mol. Sci. 2020;21:1723. doi: 10.3390/ijms21051723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bade B.C., Dela Cruz C.S. Lung cancer 2020: epidemiology, etiology, and prevention. Clin. Chest Med. 2020;41:1–24. doi: 10.1016/j.ccm.2019.10.001. [DOI] [PubMed] [Google Scholar]
- Becker A., Thakur B.K., Weiss J.M., Kim H.S., Peinado H., Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zheng Y., Wu B., Chen X., Sun F., Ge P., Wang P. BANCR regulates the cell invasion and migration in esophageal squamous cell carcinoma through Wnt/beta-catenin signaling pathway. Onco Targets Ther. 2019;12:9319–9327. doi: 10.2147/OTT.S227220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Guo B., Fan Y. MiR-153-3p suppresses cell proliferation, invasion and glycolysis of thyroid cancer through inhibiting E3F3 expression. Onco Targets Ther. 2021;14:519–529. doi: 10.2147/OTT.S267887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias F., Teixeira A.L., Nogueira I., Morais M., Maia J., Bodo C., Ferreira M., Silva A., Vilhena M., Lobo J., et al. Extracellular vesicles enriched in hsa-miR-301a-3p and hsa-miR-1293 dynamics in clear cell renal cell carcinoma patients: potential biomarkers of metastatic disease. Cancers (Basel) 2020;12:1450. doi: 10.3390/cancers12061450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang T., Lv H., Lv G., Li T., Wang C., Han Q., Yu L., Su B., Guo L., Huang S., et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018;9:191. doi: 10.1038/s41467-017-02583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C., Dong J., Chu Y., Cao S., Zhang J., Wei J. LncRNA FGD5-AS1 promotes tumor growth by regulating MCL1 via sponging miR-153-3p in oral cancer. Aging (Albany N.Y.) 2020;12:14355–14364. doi: 10.18632/aging.103476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Yang Y., Zhao W., Yan Z., Yang X., Yan Y., Hao R., Hu J., Jiao F. MiR-16-5p plays an inhibitory role in human non-small cell lung cancer through Fermitin family member 2. Biocell. 2021;45:627–638. doi: 10.32604/biocell.2021.013496. [DOI] [Google Scholar]
- He A., Liu Y., Chen Z., Li J., Chen M., Liu L., Liao X., Lv Z., Zhan Y., Zhuang C., et al. Over-expression of long noncoding RNA BANCR inhibits malignant phenotypes of human bladder cancer. J. Exp. Clin. Cancer Res. 2016;35:125. doi: 10.1186/s13046-016-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- Hu K., Li K., Lv J., Feng J., Chen J., Wu H., Cheng F., Jiang W., Wang J., Pei H., et al. Suppression of the SLC7A11/glutathione axis causes synthetic lethality in KRAS-mutant lung adenocarcinoma. J. Clin. Invest. 2020a;130:1752–1766. doi: 10.1172/JCI124049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Liu C., Bi Z.Y., Zhou Q., Zhang H., Li L.L., Zhang J., Zhu W., Song Y.Y., Zhang F., et al. Comprehensive landscape of extracellular vesicle-derived RNAs in cancer initiation, progression, metastasis and cancer immunology. Mol. Cancer. 2020b;19:102. doi: 10.1186/s12943-020-01199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S., Krishna M., Singhal J., Horne D., Awasthi S., Salgia R., Singhal S.S. Therapeutic targeting of miRNA-216b in cancer. Cancer Lett. 2020;484:16–28. doi: 10.1016/j.canlet.2020.04.020. [DOI] [PubMed] [Google Scholar]
- Ji Q., Zhou L., Sui H., Yang L., Wu X., Song Q., Jia R., Li R., Sun J., Wang Z., et al. Primary tumors release ITGBL1-rich extracellular vesicles to promote distal metastatic tumor growth through fibroblast-niche formation. Nat. Commun. 2020;11:1211. doi: 10.1038/s41467-020-14869-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Wei Y. Modulators of microRNA function in the immune system. Int. J. Mol. Sci. 2020;21:2357. doi: 10.3390/ijms21072357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagkouni D., Paraskevopoulou M.D., Tastsoglou S., Skoufos G., Karavangeli A., Pierros V., Zacharopoulou E., Hatzigeorgiou A.G. DIANA-LncBase v3: indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2020;48(D1):D101–D110. doi: 10.1093/nar/gkz1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Kim H.K., Lee K., Hong Y., Cho J.H., Choi J.W., Lee J.I., Suh Y.L., Ku B.M., Eum H.H., et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 2020;11:2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue) doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhang L., Zhang Y., Zhou F. Increased expression of LncRNA BANCR is associated with clinical progression and poor prognosis in gastric cancer. Biomed. Pharmacother. 2015;72:109–112. doi: 10.1016/j.biopha.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Liu M., Zhang X. An integrated analysis of mRNA-miRNA transcriptome data revealed hub regulatory networks in three genitourinary cancers. Biocell. 2017;41:19–26. doi: 10.32604/biocell.2017.41.019. [DOI] [Google Scholar]
- Ma S., Yang D., Liu Y., Wang Y., Lin T., Li Y., Yang S., Zhang W., Zhang R. LncRNA BANCR promotes tumorigenesis and enhances adriamycin resistance in colorectal cancer. Aging (Albany N.Y.) 2018;10:2062–2078. doi: 10.18632/aging.101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M., Martin-Jaular L., Lavieu G., Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- O'Brien K., Breyne K., Ughetto S., Laurent L.C., Breakefield X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020;21:585–606. doi: 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo W., Gao Y., He Y., Sun J., Xu H., Feng D., Park S.H., Cho Y.E., Guillot A., Ren T., et al. ALDH2 deficiency promotes alcohol-associated liver cancer by activating oncogenic pathways via oxidized DNA-enriched extracellular vesicles. J. Hepatol. 2019;71:1000–1011. doi: 10.1016/j.jhep.2019.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Wang K., Yang X., Dai W., Fan Z. Long noncoding RNA BANCR mediates esophageal squamous cell carcinoma progression by regulating the IGF1R/Raf/MEK/ERK pathway via miR3383p. Int. J. Mol. Med. 2020;46:1377–1388. doi: 10.3892/ijmm.2020.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;Chapter 3 doi: 10.1002/0471143030.cb0322s30. Unit 3.22. [DOI] [PubMed] [Google Scholar]
- Wadowska K., Bil-Lula I., Trembecki Ł., Śliwińska-Mossoń M. Genetic markers in lung cancer diagnosis: a review. Int. J. Mol. Sci. 2020;21:4569. doi: 10.3390/ijms21134569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhao X., Wang Y., Ren F., Sun D., Yan Y., Kong X., Bu J., Liu M., Xu S. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Rai A., Chen M., Suwakulsiri W., Greening D.W., Simpson R.J. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018;15:617–638. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- Yilmaz M., Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- Yu X., Zheng H., Chan M.T., Wu W. BANCR: a cancer-related long non-coding RNA. Am. J. Cancer Res. 2017;7:1779–1787. [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Du Y., Zhang X., Li M., Li X. Downregulation of BANCR promotes aggressiveness in papillary thyroid cancer via the MAPK and PI3K pathways. J. Cancer. 2018;9:1318–1328. doi: 10.7150/jca.20150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen J., Cheng T., Yang H., Li H., Pan C. Identification of the key genes and characterizations of Tumor Immune Microenvironment in Lung Adenocarcinoma (LUAD) and Lung Squamous Cell Carcinoma (LUSC) J. Cancer. 2020;11:4965–4979. doi: 10.7150/jca.42531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W., Chen A., Tang X., Liu Y. Circular RNA circFOXM1 triggers the tumorigenesis of non-small cell lung cancer through miR-132-3p/TMEM14A axis. Biocell. 2021;45:901–910. doi: 10.32604/biocell.2021.08672. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.