Abstract
Malignant meningiomas often show invasive growth that makes complete tumor resection challenging, and they are more prone to recur after radical resection. Invasive meningioma associated transcript 1 (IMAT1) is a long noncoding RNA located on Homo sapiens chromosome 17 that was identified by our team based on absolute expression differences in invasive and non-invasive meningiomas. Our studies indicated that IMAT1 was highly expressed in invasive meningiomas compared with non-invasive meningiomas. In vitro studies showed that IMAT1 promoted meningioma cell invasion through the inactivation of the Krüppel-like factor 4 (KLF4)/hsa-miR22-3p/Snai1 pathway by acting as a sponge for hsa-miR22-3p, and IMAT1 knockdown effectively restored the tumor suppressive properties of KLF4 by preserving its tumor suppressor pathway. In vivo experiments confirmed that IMAT1 silencing could significantly inhibit the growth of subcutaneous tumors and prolong the survival period of tumor-bearing mice. Our findings demonstrated that the high expression of IMAT1 is the inherent reason for the loss of the tumor suppressive properties of KLF4 during meningioma progression. Therefore, we believe that IMAT1 may be a potential biological marker and treatment target for meningiomas.
Keywords: hsa-miR22-3p, IMAT1, Krüppel-like factor 4, meningiomas, Snai1
INTRODUCTION
Meningiomas originate from arachnoid cap cells and are the most common primary central nervous system (CNS) tumors (Perry, 2018). Based on their histopathological features, the 2021 World Health Organization (WHO) CNS tumor grading criteria classified meningiomas into three histological grades: grade I (benign), grade II (atypical), and grade III (anaplastic) tumors (Louis et al., 2021). Although the majority of meningiomas are benign and can be completely resected by surgery (Goldbrunner et al., 2016), others have a high risk of recurrence and poor prognosis, especially WHO II/III grade tumors (high-grade meningioma). Compared to benign meningiomas, the prognosis of malignant meningiomas is poor, and it seriously affects the survival and quality of life of patients (Riemenschneider et al., 2006). The recurrence rates of WHO grades I, II, and III meningiomas is 20%, 40%, and 78%, respectively (Mawrin and Perry, 2010).
Statistical analysis of 7,084 meningioma cases registered at the neurosurgical center of the Huashan Hospital affiliated with Fudan University between 2001 and 2010 revealed a high absolute number of patients with high-grade meningiomas, with a relatively high proportion of atypical meningiomas (4.59%, n = 325) compared to anaplastic meningiomas (2.34%, n = 166) (Wang et al., 2013). Meningiomas with certain molecular characteristics are prone to be aggressive, for example, NF2 mutation, loss of chromatin 22 (Suppiah et al., 2019), TERT promoter mutation, (Mirian et al., 2020), and H3K27me3 loss (Nassiri et al., 2021). Nevertheless, there is no effective molecular marker that can predict the risk grade and prognosis of meningioma invasiveness, especially in some benign tumors (Monleón et al., 2010; Prager et al., 2020). Therefore, there is an urgent need to explore novel invasion-related molecules that can assist in deciding more precise therapies for patients (Kim et al., 2021; Qiu et al., 2013; Wu et al., 2020). Long noncoding RNAs (lncRNAs) are defined as transcripts greater than 200 nucleotides with no protein-coding capacities, and they regulate tumorigenesis and progression in many tumors, providing a new prospect for studying malignant meningiomas. The focus of this study is to illuminate the biological function of a newly discovered lncRNA-IMAT1 in the progression of meningiomas.
MATERIALS AND METHODS
Cell culture
Benign (HBL52) and malignant (IOMM-Lee) meningioma cell lines were kindly provided by the Wan’s Lab (Moffitt Cancer Center, USA). 293T cell line was purchased from American Type Culture Collection (ATCC, USA). The three cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) in a humidified atmosphere of 5% CO2 at 37°C and were passaged by digestion with 0.25% trypsin (Thermo Fisher Scientific) once the cells reached a density of 70%.
Tumor tissues
We collected 28 meningioma samples (8 invasive and 20 non-invasive cases) from Huashan Neurosurgical Center between October 2016 and July 2018. There was no age limit for enrollment. All patients obtained the informed consent in accordance with the standards set by the Ethics Committee of the Huashan Hospital (KY2020-1218). The patients’ clinicopathological information and tumor classification are provided in Table 1. These cryopreserved tissues were used to detect the mRNA levels of IMAT1, hsa-miR22-3p, KLF4 (Krüppel-like factor 4), and Snai1 by quantitative reverse transcription polymerase chain reaction (RT-qPCR) and the protein levels of KLF4, Snai1, and other functional proteins by western blotting. The expression of IMAT1 and hsa-miR22-3p was also analyzed using fluorescence in situ hybridization (FISH) with the probes listed in Supplementary Table S1 according to a standard protocol (Dunagin et al., 2015).
Table 1.
Clinicopathological features of 28 meningioma patients
| Sex | Age (y) | Clinical diagnosis | Pathological diagnosis | ||
|---|---|---|---|---|---|
| F | 44 | Left frontal-parietal meningioma | WHO II | Atypical | Brain invasion |
| F | 39 | Recurrence of meningioma | WHO II | Atypical | Brain invasion |
| M | 75 | Right temporal meningioma | WHO II | Atypical | Brain invasion |
| M | 29 | Left spheno-orbital meningioma | WHO II | Atypical | Brain invasion |
| F | 58 | Left frontal meningioma | WHO II | Atypical | Brain invasion |
| F | 57 | Left spheno-orbital meningioma | WHO II | Atypical | Brain invasion |
| M | 63 | Right frontal falx meningioma | WHO II | Atypical | Brain invasion |
| F | 60 | Left frontal parasagittal meningioma | WHO II | Atypical | Brain invasion |
| F | 59 | Left anterior cranial base meningioma | WHO I | Syncytial | Brain non-invasion |
| M | 67 | Right frontal falx meningioma | WHO I | Syncytial | Brain non-invasion |
| F | 50 | Left parietal meningioma | WHO I | Syncytial | Brain non-invasion |
| M | 45 | Left frontal meningioma | WHO I | Syncytial | Brain non-invasion |
| F | 48 | Anterior cranial base meningioma | WHO I | Syncytial | Brain non-invasion |
| F | 36 | Anterior cranial base meningioma | WHO I | Syncytial | Brain non-invasion |
| F | 61 | Anterior cranial base meningioma | WHO I | Syncytial | Brain non-invasion |
| F | 65 | Sellar meningioma | WHO I | Syncytial | Brain non-invasion |
| F | 53 | Bilateral frontal-parietal meningioma | WHO I | Fibrous | Brain non-invasion |
| F | 55 | Left frontal parasagittal meningioma | WHO I | Fibrous | Brain non-invasion |
| F | 45 | Left tentorium meningioma | WHO I | Fibrous | Brain non-invasion |
| F | 69 | Left frontal falx meningioma | WHO I | Fibrous | Brain non-invasion |
| F | 62 | Left occipital parasagittal meningioma | WHO I | Fibrous | Brain non-invasion |
| F | 47 | Right frontal-parietal parasagittal meningioma | WHO I | Fibrous | Brain non-invasion |
| F | 32 | Sellar meningioma | WHO I | Fibrous | Brain non-invasion |
| F | 57 | Left tentorium meningioma | WHO I | Fibrous | Brain non-invasion |
| F | 41 | Tuberculum sellea meningioma | WHO I | Transitional | Brain non-invasion |
| M | 57 | Right frontal meningioma | WHO I | Angiomatous | Brain non-invasion |
| F | 52 | Recurrence of right frontal falx meningioma | WHO I | Secretory | Brain non-invasion |
| F | 40 | Left temporal meningioma | WHO I | Secretory | Brain non-invasion |
Clinicopathological data pertaining to 28 meningioma patients with or without invasion were obtained from the Department of Neurosurgery at the Huashan Hospital between October 2016 and July 2018. The clinical stage was determined according to the 2016 WHO classification of tumors of the CNS.
F, female; M, male.
Recombinant lentivirus packaging
The siRNA sequence complementary to IMAT1 or the coding sequence (CDS) of the human KLF4 gene was chosen, and the corresponding shRNA oligonucleotide DNA was synthesized and cloned into the pSIH1-shRNA vector (System Biosciences, USA) after annealing of the double-strands. A scrambled sequence of siRNA was used as a negative control (NC). The constructed vectors were named pSIH1-shRNA-IMAT1, pSIH1-shRNA-KLF4, and pSIH1-NC. The CDS of the KLF4 gene (1542 bp) was amplified using human complementary DNA (cDNA) as the template. The product was digested with EcoRI and BamHI (Takara Bio, China) and cloned into and cloned into pCDH vector (System Biosciences) to construct the recombinant vector pCDH-KLF4. The IMAT1 overexpression vector pCDH-IMAT1 was constructed according to the same procedure. The siRNA, siRNA-NC and primer sequences are listed in Supplementary Table S1. All recombinant vectors were sequenced, and plasmids were prepared by using an EndoFree Plasmid Kit (Qiagen, Germany). Recombinant vectors (2 μg each of pCDH-KLF4, pCDH-IMAT1, pSIH1-shRNA- KLF4, pSIH1-shRNA-IMAT1, or pSIH1-NC) and 10 μg of the pPACK Packaging Plasmid Mix (System Biosciences) were co-transfected into 293T cells using Lipofectamine 3000 (Thermo Fisher Scientific) transfection reagent. The culture medium was completely replaced with DMEM containing 1% FBS before transfection. After 48 h of transfection, the supernatants were harvested and centrifuged at 5,000 × g for 10 min at 4°C and then passed through a 0.45 µm polyvinylidene difluoride membrane (Millipore, USA). The viral titer of lentiviruses was determined by a gradient dilution method and named Lv-KLF4, Lv-IMAT1, Lv-shRNA-KLF4, and Lv-NC, respectively.
IOMM-Lee and HBL52 cells growing in logarithmic phase were plated in 6-well plates at a density of 1 × 105 cells/well. One day later, lentiviruses (Lv-NC, Lv-KLF4, Lv-KLF4 + Lv-IMAT1, and Lv-KLF4 + Lv-shRNA-IMAT1) were added to the cells at a multiplicity of infection of 20. The infection efficiency was evaluated by the fluorescence of green fluorescent protein 72 h after infection.
Validation of the transcription factor binding site (TFBS) of KLF4 in the promoter of hsa-miR22-3p by luciferase assay
We searched for the location of the precursor of hsa-miR22-3p in the human genome and selected a 2.5 kb DNA sequence upstream of the transcription start site as the promoter region. Then, we predicted the promoter sequence using Promoter 2.0 software (http://www.cbs.dtu.dk/services/Promoter). A TFBS for KLF4 in the hsa-miR22-3p promoter was predicted by JASPAR (https://jaspar.genereg.net/). The predicted promoter of hsa-miR22-3p (312 bp) was amplified by PCR. Next, we cloned the predicted promoter upstream of the red fluorescent protein (RFP) gene to construct the fluorescent expression vector pcDNA-pro(miR22)-RFP, which was transfected into 293T cells. 48 h after transfection, promoter activity was evaluated by RFP expression. We also cloned the promoter into the luciferase reporter vector pGL3-Enhancer (Promega Corporation, USA) upstream of the luciferase gene to construct pGL3-TFBS (wt)-miR22, which carried the wild-type TFBS of KLF4. Then, the TFBS in pGL3-TFBS (wt)-miR22 was mutated from 5’-CACCTG-3’ to 5’-TCGACC-3’ to construct pGL3-TFBS (mt)-miR22, which carried a mutated TFBS. The primers used are provided in Supplementary Table S1. Luciferase experiments were performed in 293T cells co-transfected with plasmids according to the manufacturer’s instructions and detected by the Dual Luciferase Reporter Assay System (Promega Corporation) 48 h after transfection.
Verification of the binding sites of hsa-miR22-3p in the Snai1-3’-UTR
We used TargetScan 7.1 (http://www.targetscan.org/) to predict potential hsa-miR22-3p binding sites in the 3’-UTR of human Snai1 mRNA (NM_005985.4). The 3’-UTR of Snai1 amplified by PCR was digested and cloned into the pGL3-promoter luciferase vector (Promega Corporation) to generate the vector pGL3-wt (wild-type)-Snai1. The target sites of hsa-miR22-3p in the pGL3-wt-Snai1 vector were mutated from 5’-GGCAGCT-3’ to 5’-CAGGGCT-3’ to construct the mutated reporter vector pGL3-mt-Snai1 using a site-directed mutagenesis kit (Takara Bio). The primers used are provided in Supplementary Table S1. Luciferase experiments were performed in 293T cells by co-transfection with hsa-miR22-3p mimics, inhibitor or NC (Supplementary Table S1), and the plasmids and luciferase activity in 293T cells were detected by the Dual Luciferase Reporter Assay System 48 h after transfection. The effect of IMAT1 depletion on the inhibition of luciferase activity by hsa-miR22-3p mimics was evaluated in 293T cells using the same method as that used in the validation of the binding sites of hsa-miR22-3p.
Chromatin immunoprecipitation-PCR (ChIP-PCR)
HBL52 cells infected with Lv-KLF4 for 72 h were harvested and subjected to ChIP-PCR using the EZ ChIP Kit (Millipore) according to the manufacturer’s instructions. The 52 bp PCR product carrying the predicted KLF4 TFBS was amplified and subjected to gelelectrophoresis and cloned into a simple T-vector (Takara Bio) for sequencing. The primers used for RT-PCR are provided in Supplementary Table S1. KLF4 primary antibody (4 μg; Abcam, UK) was used for purification of the KLF4 protein.
Cell viability, invasion, and wound healing assays
The experiment to detect cell viability using a CCK-8 assay kit (Dojindo, Japan) was implemented in both IOMM-Lee and HBL52 cells. Cell invasion was detected using the QCMTM 24-well Fluorimetric Cell Invasion Assay kit (Chemicon International, USA) according to the manufacturer’s instructions in IOMM-Lee cells. Migration was assessed using wound healing assay in HBL52 cells. All cells used in the above experiments were infected with the recombinant lentiviruses (Lv-NC, Lv-KLF4, Lv-IMAT1 + Lv-KLF4, or Lv-shRNA-IMAT1 + Lv-KLF4) for 72 h.
Animal xenografts
Ninety-six female BALB/c nude mice (7 weeks old) were purchased from Shanghai Laboratory Animal (SLAC, China). All protocols were approved by the Huashan Hospital Experiment Animal Ethics Committee (KY2020-1218). HBL52 cells (1 × 105) were suspended in 200 μl DMEM (10% FBS) and injected subcutaneously into the flank regions of the nude mice. Two weeks after inoculation, approximately 3 mm subcutaneous tumors were observed. All animals were randomly divided into four groups (24 mice per group): model, model + Lv-NC, model + Lv-KLF4, and model + Lv-KLF4 + Lv-shRNA-IMAT1 groups. For the intervention groups, each animal received 50 μl of recombinant lentivirus (5 × 107 Infectious Unit [IFU]) twice a week from the second week for five weeks, whereas the mice of the model group received the same volume of saline instead. Tumor volume was measured weekly from the second week and five weeks after viral injection. Then, the animals (12 randomly selected per group, 48 in total) were sacrificed, and tumors were removed. Tissue immunostaining of Ki67 was performed with Ki67 antibodies (1:500; Abcam), and the proportion of positive cells was calculated. The density of new lymphatic vessels was detected by tissue immunofluorescence stainingwith an antibody against the lymphatic specific marker CD34 (1:700; Abcam). The expression of KLF4, Snai1, α-SMA, and vimentin in the tumors was analyzed by western blotting, and the relative levels of KLF4, hsa-miR22-3p, IMAT1, and Snai1 were detected by RT-qPCR. Another 48 mice were used for survival analysis. The survival rate of the animals was monitored for 13 weeks. Once the subcutaneous tumor reached 18 mm in length, it was recorded as a death event, and the data were used to analyze the survival period of the tumor-bearing animals.
RT-qPCR
Total RNA was isolated from cells or tissues and reverse-transcribed into cDNA using M-MLV reverse transcriptase (Takara Bio). RT-qPCR was performed using the SYBR Premix Ex TaqTM kit and TP800 System (Takara Bio). The levels of IMAT1, KLF4, and Snai1 were calculated using β-actin as an internal control, and hsa-miR22-3p levels were normalized by U6 as a reference. The PCR primers used are provided in Supplementary Table S1.
Western blotting
Total protein was extracted from cells using the M-PER mammalian protein extraction reagent or from tissues using the T-PER tissue protein extraction reagent (Pierce, USA). Equal amounts of protein (15 μg) were separated by SDS-PAGE (11% gel) and transferred onto nitrocellulose membranes. The blots were incubated with primary antibodies against human KLF4 (1:400), Snai1 (1:500), α-SMA (1:200), E-cadherin (1:500), MMP2 (1:200), MMP9 (1:400), and β-actin (1:1,200) (Abcam), followed by incubation with the corresponding secondary HRP-conjugated anti-rabbit/mouse antibody (Abcam). The bands were detected by chemiluminescence and imaged with X-ray films. β-Actin was used as an endogenous reference for normalization.
Statistical analysis
All error bars in the graphs represent the mean ± SD. Statistical data were analyzed using SPSS GradPack statistical software (ver. 20.0; IBM, USA) and GraphPad Prism 7.0 (GraphPad Software, USA). Two-tailed Student’s t-test or one-way ANOVA with a post-hoc Tukey test were used for comparisons between groups. P < 0.05 was considered statistically significant.
RESULTS
Meningiomas with high levels of IMAT1 are prone to tumor invasion
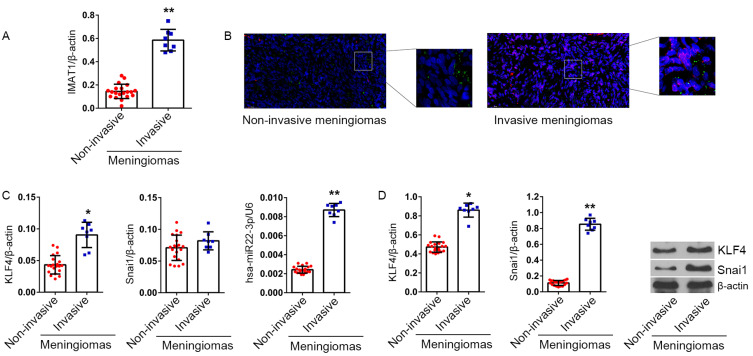
RT-qPCR data showed that the level of IMAT1 was significantly higher in invasive meningiomas than in non-invasive meningiomas (P < 0.01 vs non-invasive meningioma group, Fig. 1A). FISH data showed that the level of IMAT1 (Cy3-red) was significantly enhanced in invasive meningiomas. However, the level of hsa-miR22-3p did not appear to be significantly different between invasive and non-invasive meningiomas (Fig. 1B).
Fig. 1. IMAT1, KLF4, hsa-miR22-3p, and Snai1 levels in non-invasive and invasive meningiomas.
(A) Relative levels of IMAT1 in invasive (n = 8) and non-invasive (n = 20) meningiomas were analyzed using RT-qPCR. β-Actin served as an internal reference. The 2-ΔCt method was used to analyze inter-group differences. (B) IMAT1 and hsa-miR22-3p levels in non-invasive and invasive meningiomas were detected using FISH, the probes for IMAT1 and hsa-miR22-3p labeled with Cy3 and DIG, with excitation wavelengths of 550 and 488 nm, and emission wavelengths of 650 and 520 nm, respectively, which are shown in red and green, respectively. (C) KLF4, Snai1, hsa-miR22-3p mRNA levels in 28 meningioma samples (20 cases of non-invasive meningiomas and 20 cases of invasive meningiomas) were detected by RT-qPCR. β-Actin and U6 served as internal references for analysis of inter-group differences using the 2-ΔCt method. (D) KLF4 (54 kDa) and Snai1 (68 kDa) proteins in 28 meningioma samples were detected by western blotting. β-Actin (43 kDa) served as an internal reference. *P < 0.05, **P < 0.01, vs non-invasive meningiomas (t-test).
The high level of IMAT1 indicates that hsa-miR22-3p and Snai1 protein no longer have a negative correlation in invasive meningiomas
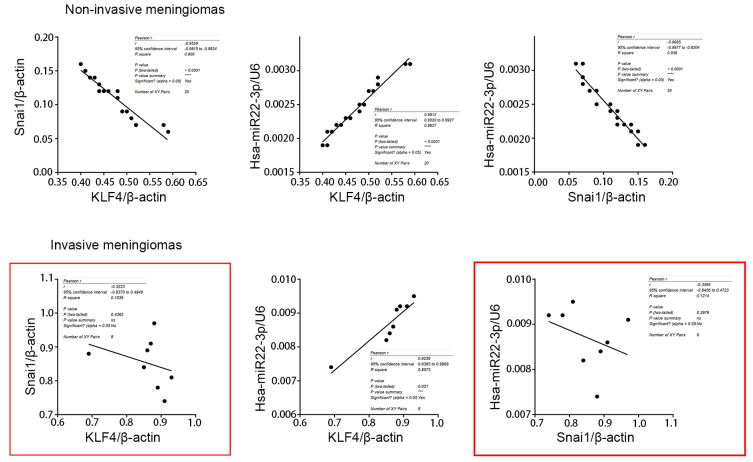
Hsa-miR22-3p, KLF4 mRNA and IMAT1 levels showed a similar change trend that was higher in invasive meningiomas than in non-invasive (P < 0.05 vs non-invasive meningioma). However, there was no significant difference in Snai1 mRNA levels between the two groups (P > 0.05) (Fig. 1C). Analysis of proteins showed that KLF4 and Snai1 proteins were significantly higher in invasive meningiomas than in non-invasive meningiomas (P < 0.05 vs non-invasive meningiomas), and the difference was more obvious for Snai1 protein (Fig. 1D). Comprehensive analysis of hsa-miR22-3p, KLF4 and Snai1 proteins revealed that in non-invasive meningiomas, hsa-miR22-3p was positively correlated with KLF4 protein and negatively correlated with Snai1 protein. However, in invasive meningiomas, hsa-miR22-3p was positively correlated with KLF4 but not correlated with Snai1 (Fig. 2).
Fig. 2. Correlation analysis in non-invasive and invasive meningiomas.
The correlation analysis of expression of Snai1, hsa-miR22-3p, and KLF4 in non-invasive and invasive meningiomas, respectively.
Hsa-miR22-3p inhibits the expression of the Snai1 protein by interacting with the 3’-UTR of Snai1 mRNA
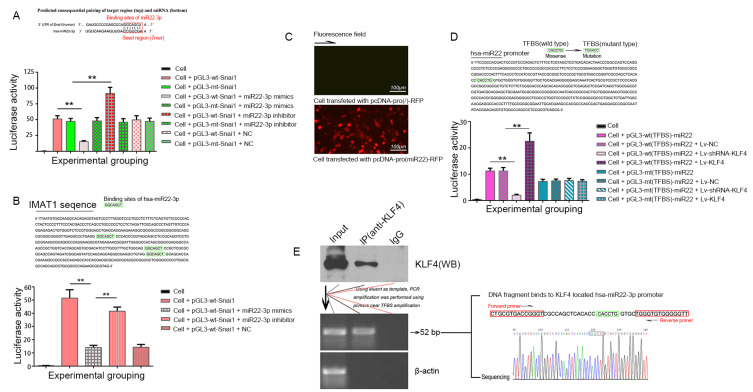
Bioinformatics analysis identified a seven-base binding site, 5’-AGCUGCC-3’, for hsa-miR22-3p in the 3’UTR of Snai1 mRNA (Fig. 3A). Both pGL3-wt-Snai1 and pGL3-mt-Snai1 expressed high levels of luciferase in 293T cells. However, an hsa-miR22-3p mimic significantly inhibited the luciferase activity in 293T cells transfected with pGL3-wt-Snai1 (52.13 ± 3.08 vs 13.42 ± 1.71; P < 0.01), while an hsa-miR22-3p inhibitor significantly increased luciferase activity in these cells (52.13 ± 3.08 vs 93.28 ± 7.12; P < 0.01). Conversely, in 293T cells transfected with pGL3-mt-Snai1, neither the hsa-miR22-3p mimic nor the inhibitor had any effect on luciferase activity (P > 0.05, vs pGL3-mt-Snai1 transfected group). Co-transfection of hsa-miR22-3p-NC had no effect on the luciferase activity in 293T cells transfected with either of the vectors (P > 0.05, vs pGL3-wt-Snai1 transfected group or pGL3-mt-Snai1 transfected group) (Fig. 3A).
Fig. 3. Luciferase assay and ChIP-PCR.
(A) Hsa-miR22-3p binds to Snai1 3’-UTR. Upper, predicted binding site of hsa-miR22-3p in 3’-UTR of Snai1. Down, the histogram shows the relative firefly luciferase activity in the different experimental groups. **P < 0.01, compared to the group transfected with the same vector but without the hsa-miR22-3p mimics or inhibitor. Data are expressed as mean ± SD of at least three independent experiments. (B) Hsa-miR22-3p binds to Snai1 3’-UTR, which is inhibited by IMAT1. Upper, predicted binding site of hsa-miR22-3p in IMAT1. Down, expression of a luciferase cassette encoding the IMAT1. **P < 0.01, compared with the group transfected with pGL3-wt-Snai1 and hsa-miR22-3p mimics. Data are expressed as mean ± SD of at least three independent experiments. (C) Fluorescence microscopic images showing the expression of RFP in 293T cells 48 h after transfection with plasmids carrying hsa-miR22-3p promoter upstream of RFP gene. (D) Verification of the TFBS of KLF4 in the hsa-miR22-3p promoter by luciferase assay. Renilla luciferase was used as reference. **P < 0.01. The tests were carried out on three biological triplicates, and data are expressed as the mean ± SD. (E) ChIP-PCR assay. Immunoprecipitation of KLF4 (left), PCR amplification of the predicted KLF4 binding sequence in the hsa-miR22-3p promoter, and sequencing of the PCR product for the predicted site (right). Cell is 293T cell.
IMAT1 interferes with the binding of hsa-miR22-3p to the Snai1 3’-UTR
Bioinformatics analysis indicated the existence of a 7-base hsa-miR22-3p binding site in the 3’ UTR of Snai1 (Fig. 3A) and three binding sites on IMAT1 (Fig. 3B). Therefore, we speculated that IMAT1 acts as a miRNA sponge by binding to hsa-miR22-3p, thereby disabling hsa-miR22-3p-mediated negative regulation of Snai1. This finding was validated by the results of the luciferase assay, which showed that hsa-miR22-3p mimics lost their inhibitory effect on luciferase activity in 293T cells transfected with pGL3-wt-Snai1 following IMAT1 overexpression (Fig. 3B).
KLF4 can bind to the miR22 promoter through the TFBS site and positively regulate its transcription
We predicted a 579 bp promoter of miR22-3p using Promoter2.0 (Fig. 3D). Based on bioinformatics analysis, we also found a TFBS of KLF4, 5’-CACCTG-3’ (Fig. 3D), in the hsa-miR22-3p promoter. Four-eight hours after transfection, RFP expression was evaluated using fluorescence microscopy (Fig. 3C). The promoter showed the ability to fully activate transcription of the downstream gene. Luciferase assays showed that overexpression or silencing of KLF4 significantly increased or decreased the luciferase activity in 293T cells transfected with pGL3-wt (TFBS)-miR22 but had no effect on the luciferase in 293T cells transfected with pGL3-mt (TFBS)-miR22 (Fig. 3D). ChIP-PCR further validated the TFBS of KLF4 in the hsa-miR22-3p promoter. The PCR primers designed for the binding site amplified the target sequence, including the TFBS (Fig. 3E), and DNA sequencing confirmed that the target sequence was the same as the reference sequence (Fig. 3E).
IMAT1 knockdown and overexpression can enhance and weaken the effect of KLF4 on inhibiting the proliferation, invasion and migration of meningioma cells, respectively
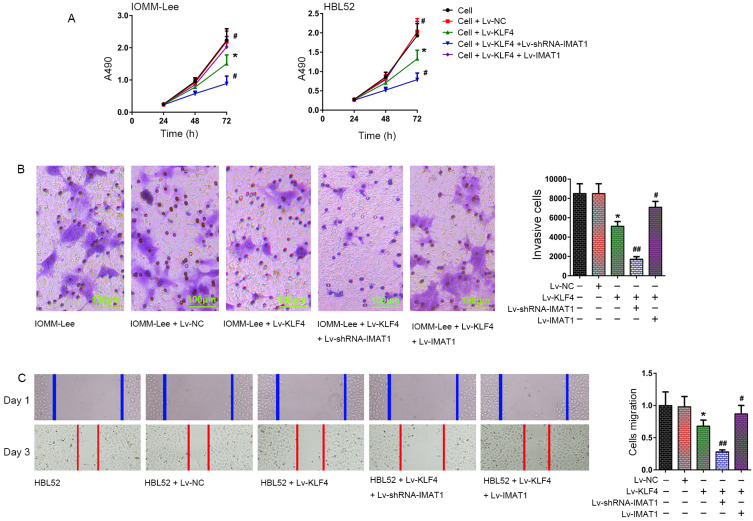
Cell proliferation assay data showed that overexpression of KLF4 suppressed the proliferation of IOMM-Lee and HBL52 cells (P < 0.05 vs cell group or Lv-NC group, 72 h). Compared to the Lv-KLF4 group, IMAT1 knockdown further enhanced the inhibitory effect of KLF4 on the proliferation of the two meningioma cell lines (P < 0.05 vs Lv-KLF4 group, 72 h). However, overexpression of IMAT1 completely reversed the inhibitory effect of KLF4 on the proliferation of meningioma cells (P < 0.05 vs Lv-KLF4 group, 72 h) (Fig. 4A). Transwell assays showed that the invasive ability of IOMM-Lee cells was significantly weakened in the cells of the Lv-KLF4 group compared to the control group (P < 0.05 vs IOMM-Lee group or IOMM-Lee + Lv-NC group). IMAT1 silencing further enhanced the KLF4-mediated decrease in the invasive ability of IOMM-Lee cells compared to the cells of the Lv-KLF4 group (P < 0.05 vs IOMM-Lee + Lv-KLF4 group), whereas IMAT1 overexpression effectively inhibited KLF4-mediated reduction in the invasive ability of IOMM-Lee cells with KLF4 overexpression (P < 0.01 vs IOMM-Lee + Lv-KLF4 group). There was no significant difference in the invasive abilities of the IOMM-Lee and IOMM-Lee + Lv-NC groups (P > 0.05; Fig. 4B). Analysis of cell migration showed that the migration ability of HBL52 cells was significantly suppressed in the Lv-KLF4 group (P < 0.05 vs HBL52 group or HBL52 + Lv-NC group). Compared to the Lv-KLF4 group, IMAT1 silencing further enhanced the KLF4-mediated suppression of migration (P < 0.01 vs HBL52 + Lv-KLF4 group). However, IMAT1 overexpression effectively inhibited the KLF4-mediated suppression of migration (P < 0.01 vs the HBL52 + Lv-KLF4 group; Fig. 4C).
Fig. 4. Effect of overexpression and knockdown of IMAT1 on effects of KLF4 to cellular processes in meningioma cells.
(A) Proliferation of IOMM-Lee and HBL52 cells was determined 72 h after infection with the indicated viruses using CCK-8 assay. (B) Invasion of IOMM-Lee cells was determined using the transwell assay 72 h after infection with the indicated viruses. (C) Migration of HBL52 cells was determined by scratch wound healing assay 72 h after infection with the indicated viruses. Images of cells were used to observe the number of cells growing across the scratched lines. The number of cells growing across the scratched line was defined as the number of migrated cells, and used to compare the changes in the migration ability in each group. *P < 0.05, vs cell group; #P < 0.05, ##P < 0.01, vs cell + Lv-KLF4 group (t-test). The tests were carried out in biological triplicates, and data are expressed as the mean ± SD.
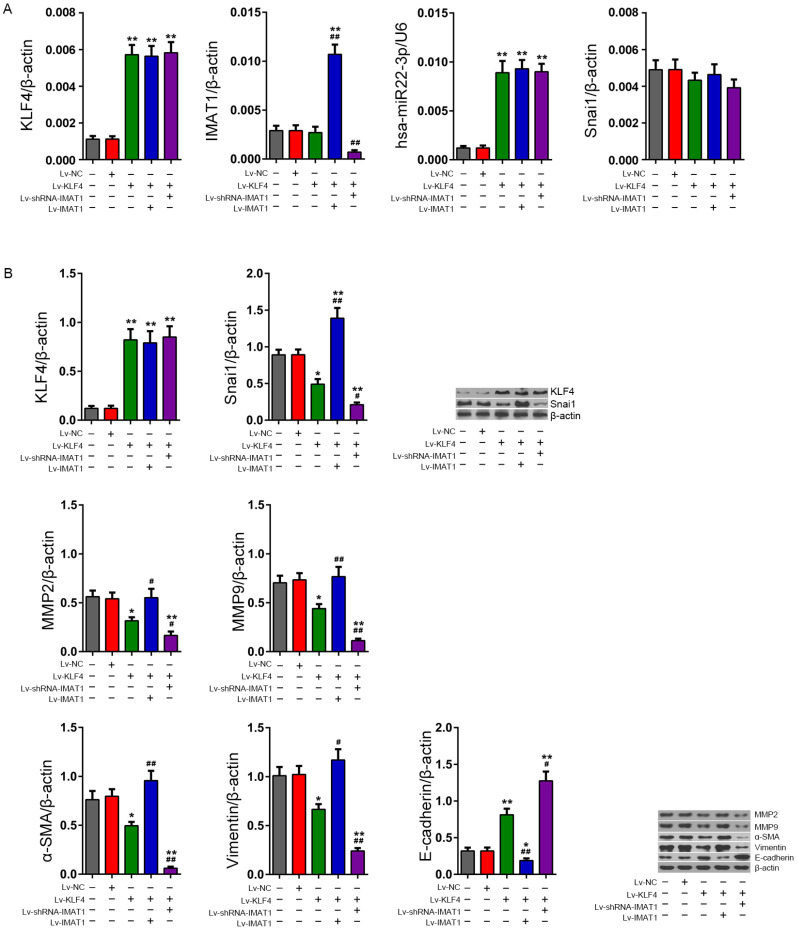
IMAT1 silencing enhances the effect of KLF4 on the inhibition of Snai1 and its downstream invasion-related proteins in meningioma cells
RT-qPCR data showed that the KLF4 mRNA content in HBL52 cells infected with Lv-KLF4 increased significantly (P < 0.01 vs the HBL52 group), and IMAT1 overexpression and silencing had no effect on KLF4 mRNA in HBL52 cells infected with Lv-KLF4 (P > 0.05, vs the HBL52 + Lv-KLF4 group). The change trend of hsa-miR22-3p content in each treatment group was consistent with KLF4 mRNA. RT-qPCR data also showed that Lv-KLF4 infection had no significant effect on the IMAT1 content in HBL52 cells (P > 0.05, vs the HBL52 group), while IMAT1 overexpression and silencing could significantly up-regulate or down-regulate the content of IMAT1 in HBL52 cells infected with HBL52 + Lv-KLF4 (P < 0.01, vs the HBL52 + Lv-KLF4 group). In addition, there was no significant difference in the content of Snai1 mRNA in each treatment group (Fig. 5A). Western blotting showed that while there was no significant difference in the expression of KLF4 protein between the HBL52 + Lv-NC and HBL52 groups (P > 0.05 vs the HBL52 group), KLF4 was significantly increased in the HBL52 + Lv-KLF4 group (P < 0.01 vs the HBL52 group), and there was no significant difference between the HBL52 + Lv-KLF4 + Lv-shRNA-IMAT1 and HBL52 + Lv-KLF4 + Lv-IMAT1 and HBL52 + Lv-KLF4 groups (P > 0.05). Analysis of invasion-related proteins showed that the expression of Snai1, α-SMA and vimentin decreased in the HBL52 + Lv-KLF4 group (P < 0.05 vs HBL52 group) and decreased even further in the HBL52 + Lv-KLF4+Lv-shRNA-IMAT1 group (P < 0.05 vs HBL52 + Lv-KLF4 group), whereas it was significantly increased in the HBL52 + Lv-IMAT1 + Lv-KLF4 group (P < 0.05 vs HBL52 + Lv-KLF4 group). Moreover, western blotting results also showed that the changes in the expression of MMP2 and MMP9 in each group were consistent with those of Snai1, while E-cadherin showed an expression pattern that was opposite that of α-SMA and vimentin (Fig. 5B).
Fig. 5. Levels of invasion-related functional proteins in HBL52 cells following manipulation of KLF4 and IMAT1 expression.
(A) RT-qPCR assay. Relative levels of KLF4, IMAT1, hsa-miR22-3p, and Snai1 were calculated using the 2-ΔCt method. (B) Western blotting. Quantitative analyses of the KLF4, Snai1, and invasion-related functional proteins (MMP2/9, α-SMA, vimentin, and E-cadherin) in HBL52 cells in various groups after 72 h of infection. *P < 0.05, **P < 0.01, vs HBL52 group; #P < 0.05, ##P < 0.01, vs HBL52 + Lv-KLF4 group (t-test). The tests were carried out in biological triplicates, and data are expressed as the mean ± SD.
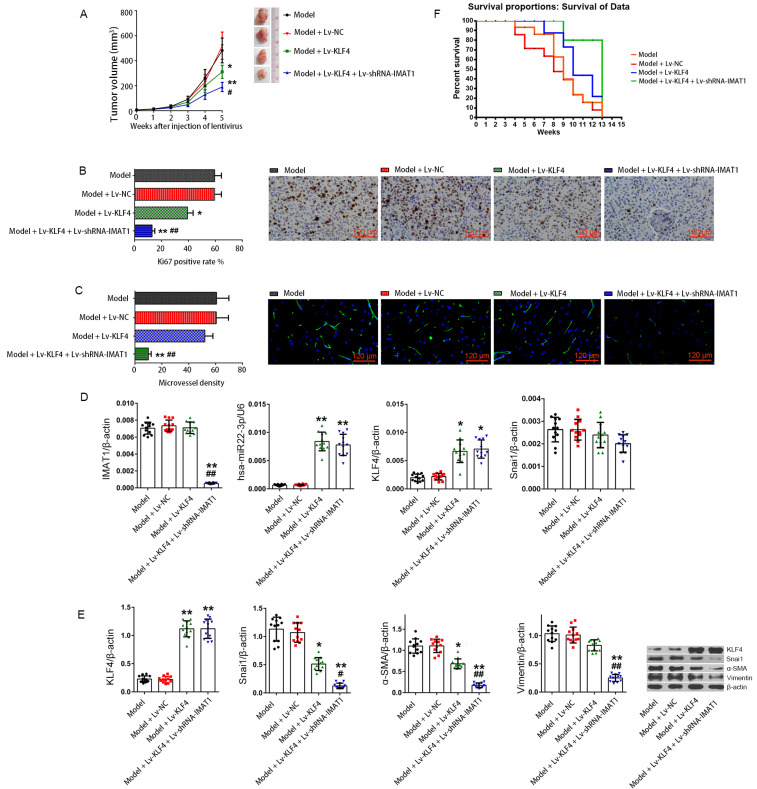
Knockdown of IMAT1 enhances the effect of KLF4 on inhibiting the proliferation of subcutaneous tumors and prolonging the survival of tumor-bearing nude mice
Five weeks after administration of lentiviruses, the tumor volumes in the model (HBL52), model + Lv-NC, model + Lv-KLF4, and model + Lv-KLF4 + Lv-shRNA-IMAT1 groups were 451.41 ± 65.62, 488.72 ± 61.95, 303.83 ± 65.78, and 179.77 ± 42.55 mm3, respectively. Compared to the model group, the tumor inhibition rates in the model + Lv-NC, model + Lv-KLF4, and model + Lv-KLF4 + Lv-shRNA-IMAT1 groups were 0%, 32.69%, and 60.18%, respectively. These results suggest that KLF4 overexpression significantly suppressed the subcutaneous tumor growth of HBL52 cells (P < 0.05 vs model or model + Lv-NC groups), and IMAT1 silencing significantly enhanced the tumor suppressor activity of KLF4 (P < 0.05 vs model + Lv-KLF4 group; Fig. 6A). RT-qPCR data showed that the IMAT1 level was the lowest in the Lv-KLF4 + Lv-shRNA-IMAT1 group (P < 0.01 vs the model or model + Lv-NC or model + Lv-KLF4 groups), and there was no significant difference in the other three groups (P > 0.05; Fig. 6D). KLF4 mRNA was significantly lower in the model and NC groups than in the other two groups (P < 0.01 vs model + Lv-KLF4 or model + Lv-KLF4 + Lv-shRNA-IMAT1 group), and there was no significant difference between the model and NC groups or between the model + Lv-KLF4 and model + Lv-KLF4 + Lv-shRNA-IMAT1 groups (P > 0.05). RT-qPCR also showed that the level of hsa-miR22-3p in tumor-bearing tissues was completely consistent with KLF4 mRNA, while the content of Snai1 mRNA was not significantly different in all groups (P > 0.05; Fig. 6D). Western blotting data showed that KLF4 protein was significantly enhanced in tumors from the model + Lv-KLF4 and model + Lv-KLF4 + Lv-shRNA-IMAT1 groups compared to the model and model + Lv-NC groups (P < 0.01 vs model and model + Lv-NC groups), and there were no significant differences between the model and model + Lv-NC groups or between the model + Lv-KLF4 and model + Lv-KLF4 + Lv-shRNA-IMAT1 groups (P > 0.05). The expression of invasion-related proteins, including Snai1, α-SMA, and vimentin, showed the same trend in the tumors from each group, with the lowest level in the model + Lv-KLF4 + Lv-shRNA-IMAT1 group (P < 0.01 vs the other three groups) and the highest in the model and model + LV-NC groups (P < 0.01 vs the other two groups) and at the mid-level in the model + Lv-KLF4 group. There was no significant difference in the level between the model and NC groups (P > 0.05; Fig. 6E). Immunohistochemical staining showed that the changes in Ki67 positivity rates in the tumors from each group were consistent with the changes in Snai1 protein expression (Fig. 6B). Immunofluorescence staining of CD34 in the subcutaneous tumors showed that the density of new lymphatic vessels in the tumors was consistent with the change in the expression of Snai1 protein in tumors from each group (Fig. 6C). Survival data of tumor-bearing mice showed that animals in the model + Lv-KLF4 + Lv-shRNA-IMAT1 group had the highest survival rate, while those in the model and model + Lv-NC groups had the lowest survival rate, and there was no significant difference in survival rate between the model and model + NC groups; the survival rate of the model + Lv-KLF4 group was between that of the model and model + Lv-KLF4 + Lv-shRNA-IMAT1 groups (Fig. 6F). Taken together, the in vivo data indicated that invasion-related proteins induced by Snai1 were disadvantageous for survival of tumor-bearing mice, whereas silencing of IMAT1 significantly improved the survival of the tumor-bearing mice by indirectly inhibiting Snai1 protein in the HBL52 cells.
Fig. 6. Subcutaneous tumors in nude mice.
(A) Growth curves of tumors in vivo. The formula for calculating the tumor volume was as follows: V = 0.5 × a × b × b, where a and b represent the long and short diameters of the tumor. The number of animals in one group was 12 (n = 12). *P < 0.05, **P < 0.01, vs model group; #P < 0.05, vs model + Lv-KLF4 group. Data are expressed as the means ± SD. (B) Immunohistochemical staining of Ki67 protein in tumors cells from each group. For statistics, 5 visual fields of each sample were randomly selected for counting and calculating the average value, the Ki67 positive rate is expressed as the percentage of positive cells in all. *P < 0.05,**P < 0.01, vs model group; ##P < 0.01, vs model + Lv-KLF4 group (t-test). Data are expressed as the means ± SD. (C) Immunofluorescence staining of CD34 in tumors from each group. Five visual fields of each sample were randomly selected for counting, microvessel density was expressed as the number of microvessels in a unit field of view. **P < 0.01, vs model group; ##P < 0.01, vs model + Lv-KLF4 group (t-test). Data are expressed as the means ± SD. (D) RT-qPCR was used to detect the relative levels of IMAT1, hsa-miR22-3p, KLF4, and Snai1 in tumors from each group. β-actin and U6 were used as reference genes, respectively. *P < 0.05, **P < 0.01, vs model group; ##P < 0.01, vs model + Lv-KLF4 group (t-test). Data are expressed as the mean ± SD. (E) Changes in the of relevant protein expressions (KLF4, Snai1, α-SMA, and vimentin) in tumors from each group were analyzed by western blotting. The x-coordinate represents the experimental groups, and the y-coordinate represents the relative protein level. β-actin was used as the reference protein. The results are shown as the mean ± SD. *P < 0.05, **P < 0.01, vs model group; #P < 0.05, ##P < 0.01, vs model + Lv-KLF4 group (t-test). Data are expressed as the means ± SD. (F) Survival analysis of the tumor-bearing mice. Survival events of mice were recorded once a week until the 13th week following recombinant virus injection.
DISCUSSION
Malignant meningiomas tend to grow in an invasive pattern. Attaining invasiveness may be the main cause of malignant transformation, metastasis, and postoperative recurrence of meningiomas (Banan et al., 2021; Jalali et al., 2015). In fact, for most cancers, inhibition of tumor cell invasion is considered to be an effective and fundamental strategy to combat malignant progression (Chuang et al., 2021; Zhang et al., 2020a). Inhibition of the regulatory factors related to meningioma cell invasion may effectively inhibit the proliferation and survival of meningioma cells (Tummalapalli et al., 2007). To date, the mechanism of meningioma invasion has not been elucidated and needs further investigation.
In our previous study, we found that the nuclear transcription factor KLF4 is closely associated with the progression of meningiomas. As a tumor suppressor, KLF4 affects the progression of meningiomas through the regulation of the tumor suppressor gene, and p53 and KLF4 overexpression effectively inhibits the invasion and migration of meningioma cells (Tang et al., 2017). In recent years, several studies have demonstrated that Snai1 (Wang et al., 2021a), an epithelial mesenchymal transition (EMT) regulator, plays an important role in tumor cell invasion and metastasis (Choi et al., 2015; Greening et al., 2015). Snai1 functions as a transcription factor by binding to the E-box element (5’-CANNTG-3’) in the promoter region of many tumor-associated genes (Carter et al., 2021; Nieto, 2002; Wang et al., 2021a). Snail promotes or inhibits the transcription of E-cadherin, IL-8, and CD147 to regulate EMT in various epithelial tumor cells (Yin et al., 2021; Zada et al., 2021).
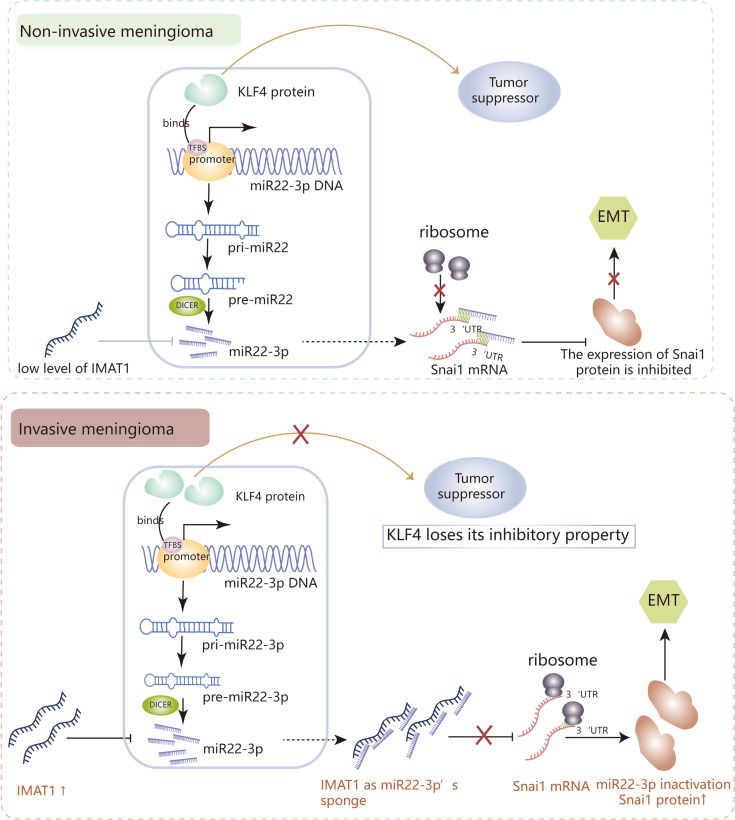
The ideal treatment for malignant meningiomas must be based on the inhibition of tumor cell invasion (Albini and Colacci, 1993). In this study, we found that IMAT1 binds to hsa-miR22-3p in the form of a miRNA sponge, thus significantly weakening its negative regulation of Snai1 protein expression. The interaction between lncRNAs and miRNAs has been shown to influence the progression of tumor invasion and metastasis through post-transcriptional regulation of gene expression (Wang et al., 2021b; Zhang et al., 2021). According to existing reports, lncRNAs can regulate miRNA functions by acting as endogenous miRNA sponges to suppress miRNA function, thereby disabling the post-transcriptional regulation of its target genes (Zhu et al., 2020). Abnormal lncRNA expression has been found in various cancers and these studies indicate that the influence of lncRNA on miRNA function is widespread in the regulation of tumor progression. LncRNAs, as competing endogenous RNAs (ceRNA), interact with miRNAs to regulate target genes, and play important roles in the onset and development of cancers (Chen et al., 2016; Lin et al., 2021). Our research data show that IMAT1 levels were significantly higher in invasive meningiomas than in non-invasive meningiomas. Compared with invasive meningiomas, the negative correlation between hsa-miR22-3p and its target protein Snai1 was better maintained in non-invasive meningiomas with lower levels of IMAT1. For many years, meningiomas have been considered to have the pathological characteristics of benign tumors (Robert et al., 2022), but the malignant progression of meningiomas should not be ignored. Although the data used for IMAT1 screening came from 8 invasive meningiomas, the uniformity of the detection indicators in the sample makes us no longer doubt the sample size. After all, invasive meningiomas are rare in clinical patients. Even so, we believe that the collection of enrolled sample specimens should be continuously implemented in the future. Molecular and functional experiments showed that IMAT1 knockout and overexpression significantly inhibited or enhanced proliferation and invasion in human meningioma cells expressing KLF4, respectively. These results suggest that the effect of IMAT1 on promoting meningioma invasion is related to the maintenance of the relationship between hsa-miR22-3p and its target protein Snai1. IMAT1 can significantly inhibit the antitumor effect of KLF4. Therefore, whether there is crosstalk between IMAT1 and the KLF4/hsa-miR22-3p/Snai1 pathway is an interesting question. RNA immunoprecipitation experiments confirmed that IMAT1 and Snai1 did not interact directly with each other (data not shown). A series of bioinformatics analyses showed a seven-base hsa-miR22-3p binding site of the seed region in the 3'-UTR of Snai1, while in IMAT1, there were several binding sites of hsa-miR22-3p seed regions. We speculate that IMAT1 competitively binds to hsa-miR22-3p as a miRNA sponge, thereby inhibiting hsa-miR22-3p-mediated negative regulation of the Snai1 protein. Hsa-miR22-3p is located on human chromosome 17, and there are a few reports concerning its role in tumor invasion and metastasis. In this study, we first identified high levels of hsa-miR22-3p in invasive meningiomas than in non-invasive meningiomas and demonstrated that the high expression of Snai1 is due to the inactivation of hsa-miR22-3p function (not because of its reduced levels). Correlation analysis showed that in both types of meningiomas, the expression of KLF4 protein was positively correlated with hsa-miR22-3p levels, and luciferase and ChIP-PCR experiments also showed that KLF4 can positively regulate transcription by binding to the hsa-miR22-3p promoter. Overall, we believe that IMAT1 is independent of the meningioma inhibitory pathway KLF4/hsa-miR22-3p/Snai1, but it can determine whether the tumor inhibitory pathway works normally (Fig. 7). To verify the universality of the conclusions, we used both benign and malignant meningioma cell lines for our in vitro studies.
Fig. 7. Schematic model of the IMAT1 mechanism in meningiomas.
In non-invasive meningiomas, the expression of IMAT1 is low, and KLF4 functions as tumor suppressor. IMAT1 competitively binds to hsa-miR22-3p as a miRNA sponge, thereby inhibiting hsa-miR22-3p-mediated negative regulation of Snai1 protein, and then the KLF4/hsa-miR22-3p/Snai1 pathway inactivates, promoting the invasion of meningiomas.
The results in tumor-bearing mice confirmed that KLF4 overexpression can significantly inhibit the proliferation of subcutaneous tumors and increase thesurvivaltimeoftumor-bearinganimals, while IMAT1 knockdown can significantly enhance the above effects of KLF4, which was confirmed again by microvessel density count and Ki67-positive assay data. The results of Snai1 and its downstream or associated functional factors in cells and subcutaneous tumors showed that KLF4 could inhibit the expression of Snai1 and further inhibit the expression of the invasion genes α-SMA (Shukal et al., 2020), vimentin (Tang et al., 2020), MMP2/9 (Singh et al., 2021), and promote the expression of invasion inhibitor gene E-cadherin (Liu et al., 2022), while IMAT1 silencing and overexpression further enhanced or reversed the antitumor effect of KLF4, respectively. As an invasion regulator, the linkage changes between Sani1 and its downstream functional factors are in full compliance with relevant research reports.
Our study is the first to propose a potential clinical solution for the malignant transformation of meningiomas based on targeting KLF4. In meningioma patients with low or no IMAT1, KLF4 overexpression may effectively inhibit malignant transformation of meningiomas through the KLF4/hsa-miR22-3p/Snai1 pathway. Correspondingly, targeted knockdown of IMAT1 is an effective means to obtain KLF4/hsa-miR22-3p/Snai1 inhibitory activity in meningioma patients with high expression of IMAT1. In the past, lncRNAs were thought to be limited by their long single-stranded structure, making them unable to easily enter serum and prone to degradation; hence, they were not considered suitable as tumor diagnostic markers. However, with a better understanding of their mechanisms and advances in detection methods, researchers have found that some lncRNAs can be used as molecular markers for tumor diagnosis. PCA3 was the first lncRNA to be identified as a clinical diagnostic marker for prostate cancer through experimental data (Hessels et al., 2003). Through clinical comparative experiments, Hessels et al. (2003) found that the detection rate of PCA3 in 108 prostate cancer patients with serum PSA values > 3 ng/ml was approximately 67% (RT-PCR) (Lee et al., 2011). This study demonstrated that RT-qPCR was a promising tool for the quantitative detection of PCA3 and that lncRNA has great potential as a clinical diagnostic marker to reduce the number of unnecessary biopsies. In addition, MALAT-1 (Tano et al., 2010), SUMOIP3 (Mei et al., 2013), and PCAT-1 (Prensner et al., 2011) also have potential as tumor diagnostic markers and prognostic factors. However, research on lncRNAs is still in the initial stages, and the functional lncRNAs identified thus far represent only the tip of the iceberg compared to the numbers predicted by bioinformatics.
Meningiomas are common CNS tumors in adults, and their diagnosis is based on histopathology. Although some somatic mutations have been reported, such as those in the NF2, TRAF7, AKT1, KLF4 (Galani et al., 2017), SMARCE1 (Sievers et al., 2021), and TERT promoters (Maier et al., 2021), these molecules have limitations as diagnostic criteria. Therefore, finding new molecules to diagnose and screen early-stage meningiomas is urgently needed. We have found a new lncRNA, IMAT1, whose expression is high in aggressive meningiomas and promotes the progression of meningiomas. IMAT1 competitively binds to hsa-miR22-3p, resulting in the downstream Snai1 not being inhibited in the presence of KLF4. Therefore, IMAT1 is a potential diagnostic, therapeutic and prognostic molecule. However, the specific test program needs to be studied further. To date, there are several reports on the regulation of lncRNAs in the malignant transformation of meningiomas (Ding et al., 2020; Li et al., 2019; Xing et al., 2018; Zhang et al., 2020b; Zheng et al., 2020), but no lncRNAs have been used as clinical diagnostic markers. Our preliminary research shows that IMAT1 is highly expressed in invasive meningiomas and is closely related to the metastasis and development of meningiomas. In addition, we found that IMAT1 degradation residues with specific structures and the small peptides encoded by IMAT1 have detectable levels in the cerebrospinal fluid and serum of meningioma patients (data not shown), meaning that lncRNA-related products as body fluid markers have a theoretical basis. Therefore, IMAT1 has potential as a marker of malignant meningiomas. Of course, the update of the two small molecule detection methods and the study of the correlation with a large number of patients’ clinical progress data are still needed.
Although our study identified the involvement of IMAT1 in invasive and non-invasive meningiomas, details about the precise mechanisms need to be investigated further. The significance of our study is that it provides mechanistic insights into how some lncRNAs may destroy the inherent regulatory mechanisms between miRNAs and their target genes to functionally inhibit some tumor suppressor regulatory pathways in a specific patient population. We believe that the abnormal expression of KLF4 and IMAT1 are two independent factors in the progression of meningiomas. The experimental data show that the change in IMAT1 does not affect the KLF4 protein but can block the transmission of KLF4’s antitumor effect from hsa-miR22-3p/Snai1, and the positive correlation between KLF4 and hsa-miR22-3p was maintained in all stages of tumor progression. The reasons for the increased expression of KLF4 and IMAT1 in the invasion of meningiomas may need to be further studied from the direction of epigenetics, which undoubtedly requires a larger number of meningioma clinical samples and patient information.
In summary, our study provides a new theoretical explanation for the functional inactivation of some tumor suppressor genes (such as KLF4) under specific conditions, that is, the activity of functional proteins not only depends on their expression, but also restricted by the downstream action pathway. In addition, we should not ignore the attempt to define IMAT1 as a diagnostic and clinical screening marker of meningiomas.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81372707, 81772267).
Footnotes
AUTHOR CONTRIBUTIONS
Y.D., Y.G. (Yu Ge), and D.W. performed the experiments and wrote the paper, and Y.G. (Yu Ge) corrected it. Y.D. acquired the data (acquired and managed patient cohort data, provided facilities, etc.). Y.G. (Yu Ge) draw the graphical abstract. D.W. analyzed and interpreted the data. S.S., L.H., J.D., S.L., H.C., Q.X., and Q.L. partially participated in the experiments. Y.G. (Ye Gong) and T.Z. designed and supervised the project. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Albini A., Colacci A. Inhibition of malignant tumor cell invasion: an approach to anti-progression. Basic Life Sci. 1993;61:335–350. doi: 10.1007/978-1-4615-2984-2_32. [DOI] [PubMed] [Google Scholar]
- Banan R., Abbetmeier-Basse M., Hong B., Dumitru C.A., Sahm F., Nakamura M., Krauss J.K., Hartmann C. The prognostic significance of clinicopathological features in meningiomas: microscopic brain invasion can predict patient outcome in otherwise benign meningiomas. Neuropathol. Appl. Neurobiol. 2021;47:724–735. doi: 10.1111/nan.12700. [DOI] [PubMed] [Google Scholar]
- Carter L.E., Cook D.P., McCloskey C.W., Grondin M.A., Landry D.A., Dang T., Collins O., Gamwell L.F., Dempster H.A., Vanderhyden B.C. Transcriptional heterogeneity of stemness phenotypes in the ovarian epithelium. Commun. Biol. 2021;4:527. doi: 10.1038/s42003-021-02045-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wang W., Cao L., Li Z., Wang X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol. Cells. 2016;39:330–336. doi: 10.14348/molcells.2016.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.J., Kim N., Chang H., Lee H.S., Park S.M., Park J.H., Shin C.M., Kim J.M., Kim J.S., Lee D.H., et al. Helicobacter pylori-induced epithelial-mesenchymal transition, a potential role of gastric cancer initiation and an emergence of stem cells. Carcinogenesis. 2015;36:553–563. doi: 10.1093/carcin/bgv022. [DOI] [PubMed] [Google Scholar]
- Chuang Y.C., Hsieh M.C., Lin C.C., Lo Y.S., Ho H.Y., Hsieh M.J., Lin J.T. Pinosylvin inhibits migration and invasion of nasopharyngeal carcinoma cancer cells via regulation of epithelial-mesenchymal transition and inhibition of MMP-2. Oncol. Rep. 2021;46:143. doi: 10.3892/or.2021.8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C., Yi X., Xu J., Huang Z., Bu X., Wang D., Ge H., Zhang G., Gu J., Kang D., et al. Long non-coding RNA MEG3 modifies cell-cycle, migration, invasion, and proliferation through AKAP12 by sponging miR-29c in meningioma cells. Front. Oncol. 2020;10:537763. doi: 10.3389/fonc.2020.537763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunagin M., Cabili M.N., Rinn J., Raj A. Visualization of lncRNA by single-molecule fluorescence in situ hybridization. Methods Mol. Biol. 2015;1262:3–19. doi: 10.1007/978-1-4939-2253-6_1. [DOI] [PubMed] [Google Scholar]
- Galani V., Lampri E., Varouktsi A., Alexiou G., Mitselou A., Kyritsis A.P. Genetic and epigenetic alterations in meningiomas. Clin. Neurol. Neurosurg. 2017;158:119–125. doi: 10.1016/j.clineuro.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Goldbrunner R., Minniti G., Preusser M., Jenkinson M.D., Sallabanda K., Houdart E., von Deimling A., Stavrinou P., Lefranc F., Lund-Johansen M., et al. EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol. 2016;17:e383–e391. doi: 10.1016/S1470-2045(16)30321-7. [DOI] [PubMed] [Google Scholar]
- Greening D.W., Gopal S.K., Mathias R.A., Liu L., Sheng J., Zhu H.J., Simpson R.J. Emerging roles of exosomes during epithelial-mesenchymal transition and cancer progression. Semin. Cell Dev. Biol. 2015;40:60–71. doi: 10.1016/j.semcdb.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Hessels D., Klein Gunnewiek J.M., van Oort I., Karthaus H.F., van Leenders G.J., van Balken B., Kiemeney L.A., Witjes J.A., Schalken J.A. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 2003;44:8–15. doi: 10.1016/S0302-2838(03)00201-X. discussion 15-16. [DOI] [PubMed] [Google Scholar]
- Jalali S., Singh S., Agnihotri S., Wataya T., Salehi F., Alkins R., Burrell K., Navab R., Croul S., Aldape K., et al. A role for matrix remodelling proteins in invasive and malignant meningiomas. Neuropathol. Appl. Neurobiol. 2015;41:e16–e28. doi: 10.1111/nan.12166. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Lim K.H., Yang S., Joo J.Y. Long non-coding RNAs in brain tumors: roles and potential as therapeutic targets. J. Hematol. Oncol. 2021;14:77. doi: 10.1186/s13045-021-01088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.L., Dobi A., Srivastava S. Prostate cancer: diagnostic performance of the PCA3 urine test. Nat. Rev. Urol. 2011;8:123–124. doi: 10.1038/nrurol.2011.10. [DOI] [PubMed] [Google Scholar]
- Li T., Ren J., Ma J., Wu J., Zhang R., Yuan H., Han X. LINC00702/miR-4652-3p/ZEB1 axis promotes the progression of malignant meningioma through activating Wnt/β-catenin pathway. Biomed. Pharmacother. 2019;113:108718. doi: 10.1016/j.biopha.2019.108718. [DOI] [PubMed] [Google Scholar]
- Lin J., Li X., Zhang F., Zhu L., Chen Y. Transcriptome wide analysis of long non-coding RNA-associated ceRNA regulatory circuits in psoriasis. J. Cell. Mol. Med. 2021;25:6925–6935. doi: 10.1111/jcmm.16703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Xu P., Ke S., Dong H., Zhan M., Hu Q., Li J. Histone methyltransferase SETDB1 inhibits TGF-β-induced epithelial-mesenchymal transition in pulmonary fibrosis by regulating SNAI1 expression and the ferroptosis signaling pathway. Arch. Biochem. Biophys. 2022;715:109087. doi: 10.1016/j.abb.2021.109087. [DOI] [PubMed] [Google Scholar]
- Louis D.N., Perry A., Wesseling P., Brat D.J., Cree I.A., Figarella-Branger D., Hawkins C., Ng H.K., Pfister S.M., Reifenberger G., et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A.D., Stenman A., Svahn F., Mirian C., Bartek J., Jr., Juhler M., Jr., Zedenius J., Jr., Broholm H., Jr., Mathiesen T., Jr. TERT promoter mutations in primary and secondary WHO grade III meningioma. Brain Pathol. 2021;31:61–69. doi: 10.1111/bpa.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawrin C., Perry A. Pathological classification and molecular genetics of meningiomas. J. Neurooncol. 2010;99:379–391. doi: 10.1007/s11060-010-0342-2. [DOI] [PubMed] [Google Scholar]
- Mei D., Song H., Wang K., Lou Y., Sun W., Liu Z., Ding X., Guo J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med. Oncol. 2013;30:709. doi: 10.1007/s12032-013-0709-2. [DOI] [PubMed] [Google Scholar]
- Mirian C., Duun-Henriksen A.K., Juratli T., Sahm F., Spiegl-Kreinecker S., Peyre M., Biczok A., Tonn J.C., Goutagny S., Bertero L., et al. Poor prognosis associated with TERT gene alterations in meningioma is independent of the WHO classification: an individual patient data meta-analysis. J. Neurol. Neurosurg. Psychiatry. 2020;91:378–387. doi: 10.1136/jnnp-2019-322257. [DOI] [PubMed] [Google Scholar]
- Monleón D., Morales J.M., Gonzalez-Segura A., Gonzalez-Darder J.M., Gil-Benso R., Cerdá-Nicolás M., López-Ginés C. Metabolic aggressiveness in benign meningiomas with chromosomal instabilities. Cancer Res. 2010;70:8426–8434. doi: 10.1158/0008-5472.CAN-10-1498. [DOI] [PubMed] [Google Scholar]
- Nassiri F., Wang J.Z., Singh O., Karimi S., Dalcourt T., Ijad N., Pirouzmand N., Ng H.K., Saladino A., Pollo B., et al. Loss of H3K27me3 in meningiomas. Neuro Oncol. 2021;23:1282–1291. doi: 10.1093/neuonc/noab036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M.A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Perry A. Elsevier; Philadelphia, USA: 2018. Meningiomas; pp. 259–298. [DOI] [Google Scholar]
- Prager B.C., Vasudevan H.N., Dixit D., Bernatchez J.A., Wu Q., Wallace L.C., Bhargava S., Lee D., King B.H., Morton A.R., et al. Perry A., Brat D.J., editors. Practical Surgical Neuropathology: A Diagnostic Approach (Second Edition), editor The meningioma enhancer landscape delineates novel subgroups and drives druggable dependencies. Cancer Discov. 2020;10:1722–1741. doi: 10.1158/2159-8290.CD-20-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J.R., Iyer M.K., Balbin O.A., Dhanasekaran S.M., Cao Q., Brenner J.C., Laxman B., Asangani I.A., Grasso C.S., Kominsky H.D., et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M.T., Hu J.W., Yin R., Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34:613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- Riemenschneider M.J., Perry A., Reifenberger G. Histological classification and molecular genetics of meningiomas. Lancet Neurol. 2006;5:1045–1054. doi: 10.1016/S1474-4422(06)70625-1. [DOI] [PubMed] [Google Scholar]
- Robert S.M., Vetsa S., Nadar A., Vasandani S., Youngblood M.W., Gorelick E., Jin L., Marianayanam N., Erson-Omay E.Z., Günel M., et al. The integrated multiomic diagnosis of sporadic meningiomas: a review of its clinical implications. J. Neurooncol. 2022;156:205–214. doi: 10.1007/s11060-021-03874-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukal D., Bhadresha K., Shastri B., Mehta D., Vasavada A., Johar K., Sr. Dichloroacetate prevents TGFβ-induced epithelial-mesenchymal transition of retinal pigment epithelial cells. Exp. Eye Res. 2020;197:108072. doi: 10.1016/j.exer.2020.108072. [DOI] [PubMed] [Google Scholar]
- Sievers P., Sill M., Blume C., Tauziede-Espariat A., Schrimpf D., Stichel D., Reuss D.E., Dogan H., Hartmann C., Mawrin C., et al. Clear cell meningiomas are defined by a highly distinct DNA methylation profile and mutations in SMARCE1. Acta Neuropathol. 2021;141:281–290. doi: 10.1007/s00401-020-02247-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D., Deshmukh R.K., Das A. SNAI1-mediated transcriptional regulation of epithelial-to-mesenchymal transition genes in breast cancer stem cells. Cell. Signal. 2021;87:110151. doi: 10.1016/j.cellsig.2021.110151. [DOI] [PubMed] [Google Scholar]
- Suppiah S., Nassiri F., Bi W.L., Dunn I.F., Hanemann C.O., Horbinski C.M., Hashizume R., James C.D., Mawrin C., Noushmehr H., et al. Molecular and translational advances in meningiomas. Neuro Oncol. 2019;21(Suppl 1):i4–i17. doi: 10.1093/neuonc/noy178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Zhu H., Wang X., Hua L., Li J., Xie Q., Chen X., Zhang T., Gong Y. KLF4 is a tumor suppressor in anaplastic meningioma stem-like cells and human meningiomas. J. Mol. Cell Biol. 2017;9:315–324. doi: 10.1093/jmcb/mjx023. [DOI] [PubMed] [Google Scholar]
- Tang L., Chen Y., Chen H., Jiang P., Yan L., Mo D., Tang X., Yan F. DCST1-AS1 promotes TGF-β-induced epithelial-mesenchymal transition and enhances chemoresistance in triple-negative breast cancer cells via ANXA1. Front. Oncol. 2020;10:280. doi: 10.3389/fonc.2020.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano K., Mizuno R., Okada T., Rakwal R., Shibato J., Masuo Y., Ijiri K., Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Tummalapalli P., Gond C.S., Dinh D.H., Gujrati M., Rao J.S. RNA interference-mediated targeting of urokinase plasminogen activator receptor and matrix metalloproteinase-9 gene expression in the IOMM-Lee malignant meningioma cell line inhibits tumor growth, tumor cell invasion and angiogenesis. Int. J. Oncol. 2007;31:5–17. doi: 10.3892/ijo.31.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.J., Xie Q., Gong Y., Mao Y., Wang Y., Cheng H.X., Zhong P., Che X.M., Jiang C.C., Huang F.P., et al. Histopathological classification and location of consecutively operated meningiomas at a single institution in China from 2001 to 2010. Chin. Med. J. (Engl.) 2013;126:488–493. [PubMed] [Google Scholar]
- Wang H., Chirshev E., Hojo N., Suzuki T., Bertucci A., Pierce M., Perry C., Wang R., Zink J., Glackin C.A., et al. The epithelial-mesenchymal transcription factor SNAI1 represses transcription of the tumor suppressor miRNA let-7 in cancer. Cancers (Basel) 2021a;13:1469. doi: 10.3390/cancers13061469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang L., Wang Q. Constitutive activation of the NEAT1/miR-22-3p/Ltb4r1 signaling pathway in mice with myocardial injury following acute myocardial infarction. Aging (Albany N.Y.) 2021b;13:15307–15319. doi: 10.18632/aging.203089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Mo Y., Peng M., Tang T., Zhong Y., Deng X., Xiong F., Guo C., Wu X., Li Y., et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer. 2020;19:22. doi: 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H., Wang S., Li Q., Ma Y., Sun P. Long noncoding RNA LINC00460 targets miR-539/MMP-9 to promote meningioma progression and metastasis. Biomed. Pharmacother. 2018;105:677–682. doi: 10.1016/j.biopha.2018.06.005. [DOI] [PubMed] [Google Scholar]
- Yin D., Hu Z.Q., Luo C.B., Wang X.Y., Xin H.Y., Sun R.Q., Wang P.C., Li J., Fan J., Zhou Z.J., et al. LINC01133 promotes hepatocellular carcinoma progression by sponging miR-199a-5p and activating annexin A2. Clin. Transl. Med. 2021;11:e409. doi: 10.1002/ctm2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zada S., Hwang J.S., Ahmed M., Lai T.H., Pham T.M., Elashkar O., Kim D.R. Cross talk between autophagy and oncogenic signaling pathways and implications for cancer therapy. Biochim. Biophys. Acta Rev. Cancer. 2021;1876:188565. doi: 10.1016/j.bbcan.2021.188565. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Song L.R., Huo X.L., Wang L., Zhang G.B., Hao S.Y., Jia H.W., Kong C.L., Jia W., Wu Z., et al. MicroRNA-221/222 inhibits the radiation-induced invasiveness and promotes the radiosensitivity of malignant meningioma cells. Front. Oncol. 2020a;10:1441. doi: 10.3389/fonc.2020.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Han P., Guo Q., Hao Y., Qi Y., Xin M., Zhang Y., Cui B., Wang P. Oncogenic landscape of somatic mutations perturbing pan-cancer lncRNA-ceRNA regulation. Front. Cell Dev. Biol. 2021;9:658346. doi: 10.3389/fcell.2021.658346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yu R., Li Q., Li Y., Xuan T., Cao S., Zheng J. SNHG1/miR-556-5p/TCF12 feedback loop enhances the tumorigenesis of meningioma through Wnt signaling pathway. J. Cell. Biochem. 2020b;121:1880–1889. doi: 10.1002/jcb.29423. [DOI] [PubMed] [Google Scholar]
- Zheng J., Pang C.H., Du W., Wang L., Sun L.G., Xing Z.Y. An allele of rs619586 polymorphism in MALAT1 alters the invasiveness of meningioma via modulating the expression of collagen type V alpha (COL5A1) J. Cell. Mol. Med. 2020;24:10223–10232. doi: 10.1111/jcmm.15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Wang J.Z., Chen D., He Y.T., Meng N., Chen M., Lu R.X., Chen X.H., Zhang X.L., Yan G.R. An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat. Commun. 2020;11:1685. doi: 10.1038/s41467-020-15403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.