Abstract
Mixed connective tissue disease (MCTD) is a rare systemic autoimmune disease characterized by the production of anti-U1 ribonucleoprotein antibodies and systemic symptoms similar to those of some other autoimmune diseases. HLA-DRB1 polymorphisms are important genetic risk factors for MCTD, but precise associations of DRB1 genotypes with MCTD have not been reported in Japanese people. Genotyping of HLA-DRB1 and -DQB1 was performed in Japanese MCTD patients (n = 116) and controls (n = 413). Associations of specific allele carriers and genotype frequencies with MCTD were analyzed.The following alleles were found to be associated with predisposition to MCTD: HLA-DRB1*04:01 (P = 8.66 × 10–6, Pc = 0.0003, odds ratio [OR] 7.96, 95% confidence interval [CI] 3.13‒20.24) and DRB1*09:01 (P = 0.0189, Pc = 0.5468, OR 1.73, 95% CI 1.12‒2.67). In contrast, the carrier frequency of the DRB1*13:02 allele (P = 0.0032, Pc = 0.0929, OR 0.28, 95% CI 0.11‒0.72) was lower in MCTD patients than in controls. The frequencies of heterozygosity for HLA-DRB1*04:01/*15 (P = 1.88 × 10–7, OR 81.54, 95% CI 4.74‒1402.63) and DRB1*09:01/*15 (P = 0.0061, OR 2.94, 95% CI 1.38‒6.25) were also higher in MCTD patients. Haplotype and logistic regression analyses suggested a predisposing role for HLA-DRB1*04:01, DQB1*03:03, and a protective role for DRB1*13:02. Increased frequencies of HLA-DRB1*04:01/*15 and DRB1*09:01/*15 heterozygous genotypes were found in Japanese MCTD patients.
Subject terms: Genetics, Immunology, Rheumatology
Introduction
Mixed connective tissue disease (MCTD) is a rare systemic autoimmune disease characterized by the production of anti-U1 ribonucleoprotein (RNP) antibodies and the symptoms of several other systemic autoimmune diseases, including systemic lupus erythematosus (SLE), systemic sclerosis (SSc), rheumatoid arthritis, idiopathic inflammatory myopathy with Raynaud’s phenomenon, diffuse hand edema, polyarthritis, myositis, pleuritis, pericarditis, leukopenia, esophageal dysmotility, interstitial lung disease, and pulmonary hypertension1–3. A recent study reported the prevalence and the incidence of MCTD in European populations to be 3.8 per 100,000 and 2.1 per million per year, respectively4. In 2012, about 10,000 MCTD patients were reported to reside in Japan (https://www.mhlw.go.jp/file/06-Seisakujouhou-10900000-Kenkoukyoku/0000089901.pdf). The concept of MCTD as a discrete disease was originally proposed by Sharp5. However, there has been controversy as to whether a distinct MCTD clinical entity does exist, due to the similarity to other systemic autoimmune diseases6–8. The identification of a specific serological marker (anti-U1RNP antibodies) and the unique association of MCTD with the presence of HLA-DRB1*04 support the concept of MCTD as a discrete pathological entity9,10.
The etiology of MCTD is unknown. It is affected by genetic and environmental factors, with HLA-DRB1 being an important genetic risk factor1–3. Several studies have reported that HLA-DRB1 alleles are associated with MCTD, notably in an increased frequency of DRB1*01 and DRB1*0411–17. In one recent study it was found that HLA-DRB1*04:01, DRB1*09:01, and DRB1*15:01 were associated with susceptibility to MCTD in European populations18. Conversely, HLA-DRB1*04:04, DRB1*07:01, DRB1*13:01, and DRB1*13:02 were associated with protection against MCTD17,18. HLA-DRB1*04:01 and DRB1*09:01 were also associated with susceptibility to MCTD in studies of Japanese populations19,20. However, few studies of HLA in MCTD have considered populations other than European. It was also proposed that HLA-DRB1*04 is primarily associated with the production of anti-U1RNP antibodies11,21,22. The roles of these DRB1 alleles have been analyzed in cohorts of modest size, and the effects of specific amino acid residues in the DRβ chain were not established. No assessments were made of any gene dosage effect of the predisposing alleles (i.e., homozygosity for the risk alleles conferring a higher risk than heterozygosity). The genotypes of these risk alleles were not precisely analyzed; the resolution of the genotyping methods in earlier studies was lower than is now attainable. Additionally, no genome-wide association studies have been reported for MCTD. Thus, the role of HLA-DRB1 on susceptibility to MCTD has not been well studied due to its rarity. In this study we analyzed associations of HLA-DRB1 genotypes with Japanese MCTD, using higher resolution genotyping methods.
Results
HLA-DRB1 and -DQB1 allele carrier frequency in MCTD
Genotyping of HLA-DRB1 and -DQB1 was conducted, to compare allele carrier frequencies in MCTD patients and controls (Table 1, Supplementary Table S1). (Allele carrier frequency is the frequency of individuals with a specific allele in some populations.) No deviation from the Hardy–Weinberg equilibrium was observed in the controls (HLA-DRB1: P = 0.4257; DQB1: P = 0.2162), but deviation was found in the MCTD patients (DRB1: P = 0.0130; DQB1: P = 0.2714). DRB1*04:01 was significantly associated with susceptibility to MCTD (P = 8.66 × 10–6, Pc = 0.0003, odds ratio [OR] 7.96, 95% confidence interval [CI] 3.13‒20.24). The association of HLA-DRB1*09:01 with predisposition to MCTD did not attain statistical significance after a Bonferroni correction (P = 0.0189, Pc = 0.5468, OR 1.73, 95% CI 1.12‒2.67). Neither did the association of HLA-DRB1*13:02 with protection against MCTD attain statistical significance after a Bonferroni correction (P = 0.0032, Pc = 0.0929, OR 0.28, 95% CI 0.11‒0.72). Tendencies were found for HLA-DQB1*03:03 (P = 0.0279, Pc = 0.3903, OR 1.66, 95% CI 1.08‒2.57, Supplementary Table S2) to predispose to disease, and for DQB1*06:04 (P = 0.0150, Pc = 0.2107, OR 0.33, 95% CI 0.13‒0.84, Supplementary Table S2) to protect against MCTD. Thus, higher allele carrier frequencies of HLA-DRB1*04:01 and DRB1*09:01, and a lower allele carrier frequency of DRB1*13:02, were found in MCTD patients than in controls.
Table 1.
HLA-DRB1 allele carrier frequencies in MCTD patients and controls.
| MCTD (n = 116) |
Control (n = 413) |
P | OR | Pc | 95% CI | |
|---|---|---|---|---|---|---|
| DRB1*04:01 | 14 (12.1) | 7 (1.7) | 8.66 × 10–6 | 7.96 | 0.0003 | (3.13–20.24) |
| DRB1*04:04 | 2 (1.7) | 0 (0.0) | 0.0478 | 18.06 | > 1 | (0.86–378.79) |
| DRB1*08:03 | 22 (19.0) | 61 (14.8) | 0.3114 | 1.35 | > 1 | (0.79–2.31) |
| DRB1*09:01 | 43 (37.1) | 105 (25.4) | 0.0189 | 1.73 | 0.5468 | (1.12–2.67) |
| DRB1*13:02 | 5 (4.3) | 57 (13.8) | 0.0032 | 0.28 | 0.0929 | (0.11–0.72) |
| DRB1*15:01 | 27 (23.3) | 68 (16.5) | 0.1008 | 1.54 | > 1 | (0.93–2.55) |
| DRB1*15:02 | 20 (17.2) | 89 (21.5) | 0.3637 | 0.76 | > 1 | (0.44–1.30) |
| *04:01, *09:01 | 57 (49.1) | 112 (27.1) | 1.51 × 10–5 | 2.60 | (1.70–3.97) |
Allele carrier frequencies are shown in parentheses (%). Association was tested by Fisher’s exact test, using 2 × 2 contingency tables. Allele carrier frequency is the frequency of individuals with a specific allele in some populations. MCTD: mixed connective tissue disease; OR: odds ratio; CI: confidence interval; Pc: corrected P (Pc values greater than 1 are shown as “ > 1”).
HLA-DRB1*04:01 allele carrier frequencies in MCTD associated with specific clinical characteristics
Associations of HLA-DRB1*04:01 with specific clinical characteristics of MCTD were then analyzed (Table 2). It was found to be significantly associated with susceptibility to: MCTD collagen vascular disease (P = 0.0013, Pc = 0.0369, OR 9.35, 95% CI 2.81‒31.20); Raynaud’s phenomenon (P = 0.0003, Pc = 0.0073, OR 8.00, 95% CI 2.80‒22.89); hand edema (P = 3.33 × 10–5, Pc = 0.0010, OR 11.32, 95% CI 3.91‒32.80); and polyarthritis (P = 0.0008, Pc = 0.0239, OR 8.49, 95% CI 2.72‒26.45). Thus, subsets of MCTD patients with certain clinical features exhibited markedly increased frequencies of HLA-DRB1*04:01.
Table 2.
Association of HLA-DRB1*04:01 allele carrier frequencies with clinical characteristics of MCTD.
| Phenotype | Number | DRB1*04:01( +) | P | OR | Pc | 95% CI |
|---|---|---|---|---|---|---|
| Male | 12 | 1 (8.3) | 0.2063 | 5.27 | > 1 | (0.60–46.61) |
| Age at onset < 45 | 40 | 4 (10.0) | 0.0110 | 6.44 | 0.3181 | (1.80–23.06) |
| Overlap of collagen vascular disease | 36 | 5 (13.9) | 0.0013 | 9.35 | 0.0369 | (2.81–31.20) |
| Interstitial lung disease | 48 | 5 (10.4) | 0.0045 | 6.74 | 0.1263 | (2.05–22.17) |
| Pulmonary arterial hypertension | 18 | 2 (11.1) | 0.0499 | 7.25 | > 1 | (1.39–37.71) |
| Raynaud's phenomenon | 66 | 8 (12.1) | 0.0003 | 8.00 | 0.0073 | (2.80–22.89) |
| Esophageal involvement | 29 | 3 (10.3) | 0.0223 | 6.69 | 0.6481 | (1.63–27.40) |
| Hand edema | 49 | 8 (16.3) | 3.33 × 10–5 | 11.32 | 0.0010 | (3.91–32.80) |
| Polyarthritis | 47 | 6 (12.8) | 0.0008 | 8.49 | 0.0239 | (2.72–26.45) |
| Muscle weakness | 14 | 2 (14.3) | 0.0315 | 9.67 | 0.9147 | (1.81–51.51) |
| Controls | 413 | 7 (1.7) |
DRB1*04:01 allele carrier frequencies are shown in parentheses (%). Association was tested by Fisher’s exact test, using 2 × 2 contingency tables. DRB1*04:01 allele carrier frequency is the frequency of individuals with a DRB1*04:01 allele in each population. Each MCTD subset with clinical characteristics was compared with controls. MCTD: mixed connective tissue disease; OR: odds ratio; CI: confidence interval; Pc: corrected P (Pc values greater than 1 are shown as “ > 1”).
HLA-DRB1 and -DQB1 genotype frequencies in MCTD
The HLA-DRB1 genotype frequencies in the MCTD patients were compared with those in the controls (Table 3). (Genotype frequency is the frequency of individuals with a specific genotype in some populations.) Homozygosity for HLA-DRB1*09:01 was found to confer a higher risk of MCTD than heterozygosity (*09:01/not *09:01: P = 0.0666, OR 1.55, 95% CI 0.98‒2.44; *09:01/*09:01: P = 0.1523, OR 2.15, 95% CI 0.83‒5.58). This gene dosage effect was not observed for HLA-DRB1*04:01 or DRB1*13:01. Genotype frequencies of HLA-DRB1*04:01/DRB1*15:01 (P = 0.0022, OR 33.08, 95% CI 1.77‒619.03) and DRB1*04:01/DRB1*15:02 (P = 0.0001, OR 48.65, 95% CI 2.72‒870.21) were higher in MCTD patients than in controls. The frequency of HLA-DRB1*09:01/DRB1*15:01 (P = 0.0112, OR 3.75, 95% CI 1.38‒10.22) was also higher in MCTD patients, but that of DRB1*09:01/DRB1*15:02 was not. The frequencies of HLA-DRB1*04:01/*15 (P = 1.88 × 10–7, OR 81.54, 95% CI 4.74‒1402.63) and DRB1*09:01/*15 (P = 0.0061, OR 2.94, 95% CI 1.38‒6.25) were also higher in MCTD patients. Furthermore, the frequency of HLA-DRB1*04:01, *09:01/*15 (P = 3.45 × 10–7, OR 5.76, 95% CI 2.96‒11.22) in MCTD patients was higher than in the controls. Finally, the frequency of HLA-DQB1*03:03/DQB1*06:02 (P = 0.0178, OR 3.72, 95% CI 1.28‒10.85) was higher in MCTD patients than in controls (Supplementary Table S3). Thus, HLA-DRB1*04:01/DRB1*15 and DRB1*09:01/DRB1*15 heterozygous genotypes predispose to MCTD.
Table 3.
HLA-DRB1 genotype frequency in MCTD patients and controls.
| MCTD (n = 116) |
Control (n = 413) |
P | OR | 95% CI | |
|---|---|---|---|---|---|
| *04:01 | 14 (12.1) | 7 (1.7) | 8.66 × 10–6 | 7.96 | (3.13–20.24) |
| *04:01/not *04:01 | 13 (11.2) | 6 (1.5) | 1.24 × 10–5 | 8.56 | (3.18–23.07) |
| *04:01/*04:01 | 1 (0.9) | 1 (0.2) | 0.3908 | 3.58 | (0.22–57.72) |
| *09:01 | 43 (37.1) | 105 (25.4) | 0.0189 | 1.73 | (1.12–2.67) |
| *09:01/not *09:01 | 36 (31.0) | 93 (22.5) | 0.0666 | 1.55 | (0.98–2.44) |
| *09:01/*09:01 | 7 (6.0) | 12 (2.9) | 0.1523 | 2.15 | (0.83–5.58) |
| *13:02 | 5 (4.3) | 57 (13.8) | 0.0032 | 0.28 | (0.11–0.72) |
| *13:02/not *13:02 | 5 (4.3) | 52 (12.6) | 0.0103 | 0.31 | (0.12–0.80) |
| *13:02/*13:02 | 0 (0.0) | 5 (1.2) | 0.5909 | 0.32 | (0.02–5.81) |
| *04:01/*13:02 | 0 (0.0) | 1 (0.2) | 1.0000 | 1.18 | (0.05–29.17) |
| *09:01/*13:02 | 1 (0.9) | 9 (2.2) | 0.6987 | 0.39 | (0.05–3.11) |
| *04:01/*15:01 | 4 (3.4) | 0 (0.0) | 0.0022 | 33.08 | (1.77–619.03) |
| *09:01/*15:01 | 8 (6.9) | 8 (1.9) | 0.0112 | 3.75 | (1.38–10.22) |
| *04:01/*15:02 | 6 (5.2) | 0 (0.0) | 0.0001 | 48.65 | (2.72–870.21) |
| *09:01/*15:02 | 5 (4.3) | 9 (2.2) | 0.2017 | 2.02 | (0.66–6.16) |
| *04:01/*15 | 10 (8.6) | 0 (0.0) | 1.88 × 10–7 | 81.54 | (4.74–1402.63) |
| *09:01/*15 | 13 (11.2) | 17 (4.1) | 0.0061 | 2.94 | (1.38–6.25) |
| *04:01, *09:01 | 57 (49.1) | 112 (27.1) | 1.51 × 10–5 | 2.60 | (1.70–3.97) |
| *04:01, *09:01/*15:01 | 12 (10.3) | 8 (1.9) | 0.0002 | 5.84 | (2.33–14.66) |
| *04:01, *09:01/*15:02 | 11 (9.5) | 9 (2.2) | 0.0010 | 4.70 | (1.90–11.64) |
| *04:01, *09:01/*15 | 23 (19.8) | 17 (4.1) | 3.45 × 10–7 | 5.76 | (2.96–11.22) |
Genotype frequencies are shown in parentheses (%). Association was tested by Fisher’s exact test, using 2 × 2 contingency tables. Genotype frequency is the frequency of individuals with a specific genotype in some populations. Genotype frequency of the heterozygous genotype of DRB1*04:01 is shown in the row “*04:01/not *04:01” and that of the homozygous genotype of DRB1*04:01 is shown in the row “*04:01/*04:01”. MCTD: mixed connective tissue disease; OR: odds ratio; CI: confidence interval.
HLA-DRB1-DQB1 haplotypes in MCTD
HLA-DRB1-DQB1 haplotype carrier frequencies in MCTD patients were compared with controls (Table 4). (Haplotype carrier frequency is the frequency of individuals with a specific haplotype in some populations.) Higher haplotype carrier frequencies of HLA-DRB1*04:01-DQB1*03:01 (P = 2.88 × 10–5, OR 7.32, 95% CI 2.85‒18.81) and DRB1*09:01-DQB1*03:03 (P = 0.0090, OR 1.84, 95% CI 1.19‒2.86) and lower DRB1*13:02-DQB1*06:04 (P = 0.0050, OR 0.26, 95% CI 0.09‒0.73) were found in MCTD patients than in controls. However, no differences were seen for HLA-DRB1*09:01-DQB1*03:01 or DRB1*13:02-DQB1*06:09 haplotype carrier frequencies. These data suggest primary associations of HLA-DRB1*04:01, DQB1*03:03, and DRB1*13:02 with MCTD.
Table 4.
HLA-DRB1-DQB1 haplotype carrier or diplotype frequency in MCTD patients and controls.
| MCTD (n = 116) |
Control (n = 413) |
P | OR | 95% CI | |
|---|---|---|---|---|---|
| DRB1-DQB1 haplotype | |||||
| *04:01-*03:01 | 13 (11.2) | 7 (1.7) | 2.88 × 10–5 | 7.32 | (2.85–18.81) |
| *09:01-*03:01 | 0 (0.0) | 4 (1.0) | 0.5809 | 0.39 | (0.02–7.31) |
| *09:01-*03:03 | 43 (37.1) | 100 (24.2) | 0.0090 | 1.84 | (1.19–2.86) |
| *13:02-*06:04 | 4 (3.4) | 50 (12.1) | 0.0050 | 0.26 | (0.09–0.73) |
| *13:02-*06:09 | 1 (0.9) | 6 (1.5) | 1.0000 | 0.59 | (0.07–4.95) |
| *15:01-*03:01 | 1 (0.9) | 3 (0.7) | 1.0000 | 1.19 | (0.12–11.53) |
| *15:01-*06:02 | 25 (21.6) | 65 (15.7) | 0.1616 | 1.47 | (0.88–2.46) |
| *15:02-*06:01 | 19 (16.4) | 89 (21.5) | 0.2429 | 0.71 | (0.41–1.23) |
| DRB1-DQB1 diplotype | |||||
| *04:01-*03:01/*15:01-*06:02 | 3 (2.6) | 0 (0.0) | 0.0103 | 25.50 | (1.31–497.34) |
| *09:01-*03:03/*15:01-*06:02 | 7 (6.0) | 7 (1.7) | 0.0178 | 3.72 | (1.28–10.85) |
| *04:01-*03:01/*15:02-*06:01 | 6 (5.2) | 0 (0.0) | 0.0001 | 48.65 | (2.72–870.21) |
| *09:01-*03:03/*15:02-*06:01 | 4 (3.4) | 9 (2.2) | 0.4953 | 1.60 | (0.48–5.30) |
| *04:01-*03:01, *09:01-*03:03/*15:01-*06:02 | 10 (8.6) | 7 (1.7) | 0.0009 | 5.47 | (2.03–14.71) |
| *04:01-*03:01, *09:01-*03:03/*15:02-*06:01 | 10 (8.6) | 9 (2.2) | 0.0027 | 4.23 | (1.68–10.69) |
| *04:01-*03:01/*15:01-*06:02, *15:02-*06:01 | 9 (7.8) | 0 (0.0) | 9.13 × 10–7 | 73.08 | (4.22–1265.70) |
| *09:01-*03:03/*15:01-*06:02, *15:02-*06:01 | 11 (9.5) | 16 (3.9) | 0.0281 | 2.60 | (1.17–5.77) |
| *04:01-*03:01, *09:01-*03:03/*15:01-*06:02, *15:02-*06:01 | 20 (17.2) | 16 (3.9) | 4.97 × 10–6 | 5.17 | (2.58–10.35) |
Haplotype carrier or diplotype frequencies are shown in parentheses (%). Association was tested by Fisher’s exact test, using 2 × 2 contingency tables. Haplotype carrier frequency is the frequency of individuals with a specific haplotype in some populations. Diplotype frequency is the frequency of individuals with a specific diplotype in some populations. MCTD: mixed connective tissue disease; OR: odds ratio; CI: confidence interval.
HLA-DRB1-DQB1 diplotype frequencies were also compared between MCTD patients and controls (Table 4). (Diplotype frequency is the frequency of individuals with a specific diplotype in some populations.) HLA-DRB1*04:01-DQB1*03:01/DRB1*15:01-DQB1*06:02 (P = 0.0103, OR 25.50, 95% CI 1.31‒497.34) and DRB1*04:01-DQB1*03:01/DRB1*15:02-DQB1*06:01 (P = 0.0001, OR 48.65, 95% CI 2.72‒870.21) conferred susceptibility to MCTD. The diplotype frequency of HLA-DRB1*09:01-DQB1*03:03/DRB1*15:01-DQB1*06:02 was higher in MCTD patients (P = 0.0178, OR 3.72, 95% CI 1.28‒10.85), but DRB1*09:01-DQB1*03:03/DRB1*15:02-DQB1*06:01 was not. Diplotype frequencies of HLA-DRB1*04:01-DQB1*03:01/DRB1*15:01-DQB1*06:02, DRB1*15:02-DQB1*06:01 (P = 9.13 × 10–7, OR 73.08, 95% CI 4.22‒1265.70), DRB1*09:01-DQB1*03:03/DRB1*15:01-DQB1*06:02, DRB1*15:02-DQB1*06:01 (P = 0.0281, OR 2.60, 95% CI 1.17‒5.77) were also higher in MCTD patients than in controls. Additionally, the frequency of HLA-DRB1*04:01–DQB1*03:01, DRB1*09:01–DQB1*03:03/DRB1*15:01–DQB1*06:02, DRB1*15:02–DQB1*06:01 was higher in MCTD patients (P = 4.97 × 10–6, OR 5.17, 95% CI 2.58‒10.35). Thus, some specific heterozygous diplotypes predispose to MCTD.
Associations of amino acid residues in the HLA-DRβ chain with MCTD
We next investigated whether the presence of any specific amino acid residues in the HLA-DRβ chain were associated with the occurrence of MCTD. Histidine at position 32 (32H, P = 1.83 × 10–6, OR 0.30, Pc = 6.22 × 10–5, 95% CI 0.18‒0.51) and serine at position 13 (13S, P = 1.48 × 10–5, OR 0.33, Pc = 0.0005, 95% CI 0.19‒0.56) were found to be associated with protection against MCTD (Fig. 1). Conversely, lysine at position 71 (71 K, P = 4.03 × 10–5, OR = 6.16, Pc = 0.0014, 95% CI 2.59‒14.63) was associated with a predisposition to MCTD. Associations of amino acid residues in the HLA-DQβ chain were also analyzed (Supplementary Figure S1) but none was detected. Thus, certain amino acid residues in the HLA-DRβ chain but not in the HLA-DQβ chain were associated with susceptibility to or protection against MCTD.
Figure 1.
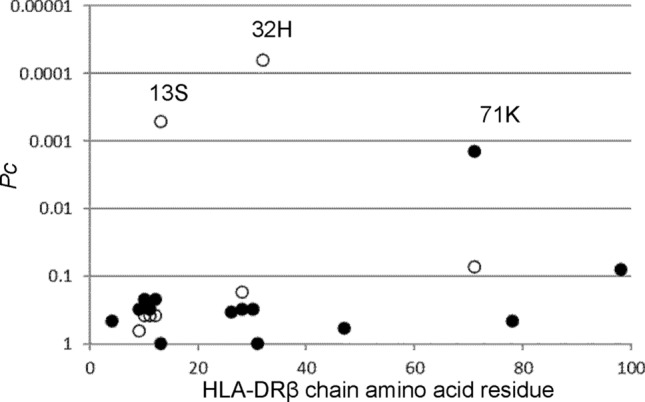
Associations of amino acid residues in the DRβ chain with MCTD. Significance of the associations was established by Fisher’s exact test, using 2 × 2 contingency tables. Corrected P (Pc) values were generated by multiplying P values by the number of analyzed amino acid residues. Predisposing associations are indicated by filled circles and protective associations by open circles. MCTD: mixed connective tissue disease.
Logistic regression analysis of HLA alleles for MCTD
Because DRB1 and -DQB1 alleles are in strong linkage disequilibrium, a conditional logistic regression analysis between them in MCTD was performed to determine which locus was responsible for the observed associations (Supplementary Table S4). The association of HLA-DRB1*04:01 remained significant (Padjusted = 4.49 × 10–6, ORadjusted 12.46, 4.24‒36.62), when conditioned on DQB1*03:01. A protective association of HLA-DQB1*03:01 was detected (Padjusted = 0.0106, ORadjusted 0.44, 0.24‒0.83) when conditioned on DRB1*04:01, suggesting that the primary predisposing association for MTCD is with DRB1*04:01. The association of HLA-DRB1*09:01 was no longer apparent (Padjusted = 0.8031, ORadjusted 1.15, 0.39‒3.32) when conditioned on DQB1*03:03, but the association of DQB1*03:03 was still present when conditioned on DRB1*09:01, (Padjusted = 0.5116, ORadjusted 1.43, 0.49‒4.11), suggesting a primary role for DQB1*03:03. We also found that the protective association of HLA-DRB1*13:02 remained when conditioned on DQB1*06:04 (Padjusted = 0.1522, ORadjusted 0.16, 95% CI 0.01‒1.99), although it was no longer statistically significant. Finally, the protective association of HLA-DQB1*06:04 was no longer present when conditioned on DRB1*13:02 (Padjusted = 0.5962, ORadjusted 2.01, 0.15‒26.73), suggesting that the latter was the primary protective association. These suggested primary associated alleles in the logistic regression analysis were the same as those identified by haplotype analysis. Thus, certain HLA-DRB1 or DQB1 alleles seem to be primarily responsible for susceptibility to or protection against MCTD.
HLA-DRB1*04:01allele carrier frequencies in SLE or SSc with anti-U1RNP antibodies
HLA-DRB1*04:01 allele carrier frequencies in SLE or SSc patients with anti-U1RNP antibodies were analyzed, to determine whether DRB1*04:01 was primarily associated with the production of anti-U1RNP antibodies or with susceptibility to MCTD (Supplementary Table S5). It is evident that HLA-DRB1*04:01 is associated with the production of anti-U1RNP antibodies in SLE. Thus, HLA-DRB1*04:01 may be associated with the production of anti-U1RNP antibodies in general.
Discussion
In the present study we confirmed the associations of HLA-DRB1*04:01 and DRB1*09:01 with Japanese MCTD. DRB1*13:02 was also confirmed to be protective against MCTD. The genotype frequencies of HLA-DRB1*04:01/DRB1*15 and DRB1*09:01/DRB1*15 were increased in MCTD patients. The predisposing role of HLA-DRB1*04:01, DQB1*03:03, and the protective role of DRB1*13:02, were suggested.
Previous studies have reported that HLA-DRB1*04:01, DRB1*09:01, and DRB1*15:01 are associated with susceptibility to MCTD18, and that DRB1*04:04, DRB1*07:01, DRB1*13:01, and DRB1*13:02 are associated with protection17,18 in European populations. HLA-DRB1*04:01 and DRB1*09:01 seem to be the common predisposing alleles in both European and Japanese populations, and DRB1*04:05 was a protective allele, at least in Japanese19,20. However, HLA-DRB1*15:01 was not found to be predisposing, and neither DRB1*04:04 nor DRB1*04:05 were protective in this study. These data suggest that it is HLA-DRB1*13:02 that is protective against MCTD in both European and Japanese populations, and that it is also the protective allele for multiple other systemic autoimmune diseases23.
The susceptibility alleles for MCTD were distinct from those for SLE (HLA-DRB1*15:01)24, SSc (DRB1*10:01)25, or idiopathic inflammatory myopathy (DRB1*08:03)26 in the Japanese. The susceptibility alleles for MCTD were similar to those for rheumatoid arthritis (HLA-DRB1*01:01, DRB1*04:01, DRB1*04:05, DRB1*04:10, DRB1*09:01, DRB1*10:01)27, except that DRB1*04:05 is the most important risk allele for Japanese rheumatoid arthritis, which was not shared with MCTD. Thus, different DRB1 alleles were associated with susceptibility to or protection against MCTD, when compared with other collagen vascular diseases including SLE, SSc, idiopathic inflammatory myopathy, and rheumatoid arthritis. This supports the notion that MCTD is a distinct disease entity. A gene dosage effect was detected in the associations between HLA-DRB1 risk alleles and rheumatoid arthritis27.
In the present study on MCTD, no gene dosage effect was found for HLA-DRB1*04:01 but it was found for DRB1*09:01, suggesting differential roles for DRB1*04:01 and DRB1*09:01 in the pathogenesis of MCTD.
Type 1 diabetes is an organ-specific autoimmune disease that affects pancreatic β cells; the frequencies of HLA-DRB1*03/DRB1*04 and DRB1*04/DRB1*08 heterozygous genotypes are increased in this disease28. HLA-DRB1*04/DRB1*08 heterozygous genotype frequencies are also increased in autoimmune hepatitis, another organ-specific autoimmune disease29. Analogously, the frequencies of HLA-DRB1*04:01/DRB1*15 and DRB1*09:01/DRB1*15 heterozygous genotypes are increased in Japanese MCTD, as found in the present study. Heterozygous genotypes of HLA-DRB1*04:01/DRB1*15 and DRB1*09:01/DRB1*15 might increase the probability of presentation of self-antigens and increase the risk of MCTD. Thus, the manner in which DRB1 is associated with MCTD seems similar to type 1 diabetes and autoimmune hepatitis. The frequencies of heterozygous genotypes HLA-DRB1*03/DRB1*04 and DRB1*04/DRB1*08 were reported to be increased in type 1 diabetes, and it was proposed that HLA-DRB1, -DQA1, and -DQB1 loci independently contributed to susceptibility to this disease28. These heterozygous risk genotypes were explained by trans-complementing DQα-β heterodimer molecules encoded by the HLA-DQA1 allele of one haplotype and the DQB1 allele of the other haplotype28. The frequencies of HLA-DRB1*04:01/*15 and DRB1*09:01/*15 heterozygous genotypes were also increased in Japanese MCTD, as shown here. HLA-DRB1*04:01 would be responsible for predisposition to MCTD in the DRB1*04:01-DQB1*03:01 haplotype and DQB1*03:03 in the DRB1*09:01-DQB1*03:03 haplotype, based on the data obtained from haplotype and logistic regression analyses in the present study. These data analogously suggest pathogenic roles of trans-complementing DQα-β heterodimer molecules in MCTD. HLA-DRB1-DQB1 diplotype analyses showed that DRB1*09:01-DQB1*03:03/DRB1*15:01-DQB1*06:02 was associated with susceptibility to MCTD. The HLA-DQA1 allele in the DRB1*09:01-DQB1*03:03 haplotype is likely to be DQA1*01:01, given haplotype distributions in Japanese30. However, the DQA1 allele in DRB1*15:01-DQB1*06:02 is DQA1*01:02. The risk diplotype HLA-DRB1*09:01-DQB1*03:03/DRB1*15:01-DQB1*06:02 would encode DQA1*01:01-DQB1*06:02 and DQA1*01:02-DQB1*03:03 molecules in trans. Other culprit genes in linkage disequilibrium with HLA-DRB1-DQB1 loci could also cause MCTD. Thus, several explanations can be proposed for the mechanisms of MCTD pathogenesis, based on results of analyses of associations with HLA-DRB1 and -DQB1.
The present study found associations of amino acid residues 13S, 32H, and 71 K in the HLA-DRβ chain with MCTD. The 13S and 32H residues are encoded by HLA-DRB1*13:02, and 71 K by DRB1*04:01. The strongest protective association of 32H amino acid residue would reflect the protective effects of HLA-DRB1*13:02.
There has been some controversy as to whether DRB1*04 is primarily associated with the production of anti-U1RNP antibodies11,21,22. In the present study, HLA-DRB1*04:01 was associated with the production of anti-U1RNP antibodies in SLE, suggesting that DRB1*04:01 in MCTD was primarily associated with the production of anti-U1RNP antibodies, not for the pathogenesis.
In the present study, the predisposing association of HLA-DRB1*04:01 and DRB1*09:01 and the protective association of DRB1*13:02 with Japanese MCTD have been documented. To the best of our knowledge, this is the first report on associations of HLA-DRB1*04:01 and DRB1*09:01/DRB1*15 heterozygous genotypes with MCTD. HLA-DRB1*04:01-DQB1*03:01 and DRB1*09:01-DQB1*03:03 haplotypes were associated with MCTD. The association of HLA-DRB1*04:01-DQB1*03:01, DRB1*09:01-DQB1*03:03/DRB1*15:01-DQB1*06:02, DRB1*15:02-DQB1*06:01 diplotypes was also detected. It is suggested that specific combinations of HLA-DRB1 and -DQB1 alleles or haplotypes play important roles in the pathogenesis of MCTD.
The present study has some limitations. No replication analysis was conducted. The sample size was modest, although it is the largest in an Asian population. The distribution pattern of HLA alleles would differ in other ethnic populations. Other loci in the HLA region might contribute to the pathogenesis of MCTD. Thus, larger-scale studies of the HLA region in multiethnic populations should be performed to confirm the findings of the present study, despite the obstacle of the rarity of MCTD.
Materials and methods
Patients and controls
MCTD patients (n = 116; mean age ± SD, 59.1 ± 15.9 years, 12 male [11.5%]) were recruited at: Tokyo Metropolitan Tama Medical Center, Sagamihara National Hospital, Kyushu Medical Center, Teikyo University, Nagoya Medical Center, Yokohama City University, Yokohama Minami Kyosai Hospital, Nagasaki Medical Center, and Fukushima Medical University. SLE and SSc patients were also recruited at: Tokyo Metropolitan Tama Medical Center, Sagamihara National Hospital, Kyushu Medical Center, Teikyo University, Nagoya Medical Center, Yokohama City University, Yokohama Minami Kyosai Hospital, and Nagasaki Medical Center. The controls (n = 413; mean age ± SD, 41.4 ± 12.6 years, 62 male [14.0%]) were recruited at Sagamihara Hospital, Kanazawa University, Teikyo University, or by the Pharma SNP Consortium (Tokyo, Japan)31,32. The controls and patients were native Japanese. All patients fulfilled the MCTD criteria of Kasukawa et al.33 or the criteria for MCTD established by the Research Committee of the Ministry of Health, Labour, and Welfare of Japan34,35.
This study was reviewed and approved by the Research Ethics Committees of: Tokyo Metropolitan Tama Medical Center, Sagamihara National Hospital, Kyushu Medical Center, Teikyo University, Nagoya Medical Center, Yokohama City University, Yokohama Minami Kyosai Hospital, Nagasaki Medical Center, Fukushima Medical University, and Tokyo National Hospital. Written informed consent was obtained from all participants. This study was conducted in accordance with the principles expressed in the Declaration of Helsinki.
Genotyping
HLA genotyping of HLA-DRB1 and -DQB1 loci was conducted by polymerase chain reaction, using reverse sequence-specific oligonucleotide probes (WAKFlow HLA typing kits, Wakunaga, Akitakata, Japan) with the Bio-Plex system (Bio-Rad, Hercules, CA). Genotyping results for some of the SLE and SSc patients and Japanese controls were previously published24,25,29.
Statistical analysis
Deviation from the Hardy–Weinberg equilibrium was analyzed by Genepop (http://genepop.curtin.edu.au/)36. Associations of allele carrier frequencies, genotype frequencies, haplotype carrier frequencies, diplotype frequencies, and amino acid residue carrier frequencies were tested by Fisher’s exact test, using 2 × 2 contingency tables. A multiple logistic regression analysis under the additive model was conducted to determine whether each allele primarily contributes to predisposition to or protection against MCTD. The Bonferroni method was used to correct for multiple comparisons, and corrected P (Pc) values were generated by multiplying P values by the number of analyzed alleles or amino acid residues.
Supplementary Information
Author contributions
H.F. and S.T. conceived and designed the study. S.O., T.H., and H.F. performed the experiments. S.O. and H.F. analyzed the data. H.F., K.S., A.H., A.K., T.M., N.F., E.S., S.O., H.K., M.K., S.N., K.M., and S.T. contributed reagents, materials, and analysis tools. S.O., H.F., and S.T. contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Funding
The work was supported by Grants-in-Aid for Scientific Research (B, C) (26293123, 22591090, 15K09543, 18K08402) and for Young Scientists (B) (24791018) from the Japan Society for the Promotion of Science, Health and Labour Science Research Grants from the Ministry of Health, Labour, and Welfare of Japan, Grants-in-Aid of the Practical Research Project for Allergic Diseases and Immunology (Research on Allergic Diseases and Immunology) from Japan Agency for Medical Research and Development, Grants-in-Aid for Clinical Research from National Hospital Organization, Research Grants from Daiwa Securities Health Foundation, Research Grants from Japan Research Foundation for Clinical Pharmacology, Research Grants from The Nakatomi Foundation, Research Grants from Takeda Science Foundation, Research Grants from Mitsui Sumitomo Insurance Welfare Foundation, Bristol-Myers K.K. RA Clinical Investigation Grant from Bristol-Myers Squibb Co., and research grants from the following pharmaceutical companies: Abbott Japan Co., Ltd., Astellas Pharma Inc., Chugai Pharmaceutical Co., Ltd., Eisai Co., Ltd., Mitsubishi Tanabe Pharma Corporation, Merck Sharp and Dohme Inc., Pfizer Japan Inc., Takeda Pharmaceutical Company Limited, and Teijin Pharma Limited. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
The data that support the findings of this study are not publicly available due to privacy and ethical restrictions. The data are available from the corresponding author upon reasonable request.
Competing interests
HF has the following conflicts, and the following funders are supported wholly or in part by the indicated pharmaceutical companies. The Japan Research Foundation for Clinical Pharmacology is run by Daiichi Sankyo, the Takeda Science Foundation is supported by an endowment from the Takeda Pharmaceutical Company, and the Nakatomi Foundation was established by Hisamitsu Pharmaceutical Co., Inc. The Daiwa Securities Health Foundation was established by Daiwa Securities Group Inc. and Mitsui Sumitomo Insurance Welfare Foundation was established by Mitsui Sumitomo Insurance Co., Ltd. HF was supported by research grants from Bristol-Myers Squibb Co. HF received honoraria from Ajinomoto Co., Inc., Daiichi Sankyo Co., Ltd., Dainippon Sumitomo Pharma Co., Ltd., Pfizer Japan Inc., Takeda Pharmaceutical Company, Luminex Japan Corporation Ltd., and Ayumi Pharmaceutical Corporation. ST was supported by research grants from nine pharmaceutical companies: Abbott Japan Co., Ltd., Astellas Pharma Inc., Chugai Pharmaceutical Co., Ltd., Eisai Co., Ltd., Mitsubishi Tanabe Pharma Corporation, Merck Sharp and Dohme Inc., Pfizer Japan Inc., Takeda Pharmaceutical Company Limited, and Teijin Pharma Limited. ST received honoraria from Asahi Kasei Pharma Corporation, Astellas Pharma Inc., AbbVie GK., Chugai Pharmaceutical Co., Ltd., Ono Pharmaceutical Co., Ltd., Mitsubishi Tanabe Pharma Corporation, and Pfizer Japan Inc. The other authors declare no financial or commercial conflicts of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-14116-x.
References
- 1.Kasukawa R. Mixed connective tissue disease. Intern. Med. 1999;38:386–393. doi: 10.2169/internalmedicine.38.386. [DOI] [PubMed] [Google Scholar]
- 2.Gunnarsson R, Hetlevik SO, Lilleby V, Molberg Ø. Mixed connective tissue disease. Best. Pract. Res. Clin. Rheumatol. 2016;30:95–111. doi: 10.1016/j.berh.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Alves MR, Isenberg DA. "Mixed connective tissue disease": A condition in search of an identity. Clin. Exp. Med. 2020;20:159–166. doi: 10.1007/s10238-020-00606-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunnarsson R, Molberg O, Gilboe IM, Gran JT. The prevalence and incidence of mixed connective tissue disease: A national multicentre survey of Norwegian patients. Ann. Rheum. Dis. 2011;70:1047–1051. doi: 10.1136/ard.2010.143792. [DOI] [PubMed] [Google Scholar]
- 5.Sharp GC, Irvin WS, Tan EM, Gould RG, Holman HR. Mixed connective tissue disease–an apparently distinct rheumatic disease syndrome associated with a specific antibody to an extractable nuclear antigen (ENA) Am. J. Med. 1972;52:148–159. doi: 10.1016/0002-9343(72)90064-2. [DOI] [PubMed] [Google Scholar]
- 6.Reichlin M. Problems in differentiating SLE and mixed connective-tissue disease. N. Engl. J. Med. 1976;295:1194–1195. doi: 10.1056/NEJM197611182952112. [DOI] [PubMed] [Google Scholar]
- 7.Bennett RM, O'Connell DJ. Mixed connective tisssue disease: a clinicopathologic study of 20 cases. Semin. Arthritis Rheum. 1980;10:25–51. doi: 10.1016/0049-0172(80)90013-X. [DOI] [PubMed] [Google Scholar]
- 8.LeRoy EC, Maricq HR, Kahaleh MB. Undifferentiated connective tissue syndromes. Arthritis Rheum. 1980;23:341–343. doi: 10.1002/art.1780230312. [DOI] [PubMed] [Google Scholar]
- 9.Aringer M, Steiner G, Smolen JS. Does mixed connective tissue disease exist? Yes. Rheum. Dis. Clin. North Am. 2005;31:411–420. doi: 10.1016/j.rdc.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Swanton J, Isenberg D. Mixed connective tissue disease: still crazy after all these years. Rheum. Dis. Clin. North Am. 2005;31:421–436. doi: 10.1016/j.rdc.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Genth E, Zarnowski H, Mierau R, Wohltmann D, Hartl PW. HLA-DR4 and Gm(1,3;5,21) are associated with U1-nRNP antibody positive connective tissue disease. Ann. Rheum. Dis. 1987;46:189–196. doi: 10.1136/ard.46.3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black CM, et al. HLA and immunoglobulin allotypes in mixed connective tissue disease. Arthritis Rheum. 1988;31:131–134. doi: 10.1002/art.1780310119. [DOI] [PubMed] [Google Scholar]
- 13.Ruuska P, et al. Differences in HLA antigens between patients with mixed connective tissue disease and systemic lupus erythematosus. Ann. Rheum. Dis. 1992;51:52–55. doi: 10.1136/ard.51.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gendi NS, et al. HLA type as a predictor of mixed connective tissue disease differentiation. Ten-year clinical and immunogenetic followup of 46 patients. Arthritis Rheum. 1995;38:259–266. doi: 10.1002/art.1780380216. [DOI] [PubMed] [Google Scholar]
- 15.Weckmann AL, et al. Immunogenetics of mixed connective tissue disease in a Mexican Mestizo population. Clin. Exp. Rheumatol. 1999;17:91–94. [PubMed] [Google Scholar]
- 16.Hassan AB, et al. MICA4/HLA-DRB1*04/TNF1 haplotype is associated with mixed connective tissue disease in Swedish patients. Hum. Immunol. 2003;64:290–296. doi: 10.1016/S0198-8859(02)00776-0. [DOI] [PubMed] [Google Scholar]
- 17.Paradowska-Gorycka A, et al. Association of HLA-DRB1 alleles with susceptibility to mixed connective tissue disease in Polish patients. Hla. 2015;87:13–18. doi: 10.1111/tan.12698. [DOI] [PubMed] [Google Scholar]
- 18.Flåm ST, Gunnarsson R, Garen T, Lie BA, Molberg Ø. The HLA profiles of mixed connective tissue disease differ distinctly from the profiles of clinically related connective tissue diseases. Rheumatology (Oxford) 2015;54:528–535. doi: 10.1093/rheumatology/keu310. [DOI] [PubMed] [Google Scholar]
- 19.Dong RP, et al. Difference in HLA-linked genetic background between mixed connective tissue disease and systemic lupus erythematosus. Tissue Antigens. 1993;41:20–25. doi: 10.1111/j.1399-0039.1993.tb01972.x. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida K, et al. Mixed connective tissue disease is distinct from systemic lupus erythematosus: Study of major histocompatibility complex class I polypeptide-related sequence A and HLA gene polymorphisms. Tissue Antigens. 2013;81:44–45. doi: 10.1111/tan.12027. [DOI] [PubMed] [Google Scholar]
- 21.Smolen JS, et al. HLA-DR antigens in systemic lupus erythematosus: Association with specificity of autoantibody responses to nuclear antigens. Ann. Rheum. Dis. 1987;46:457–462. doi: 10.1136/ard.46.6.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman RW, Sharp GC, Deutscher SL. Analysis of anti-U1 RNA antibodies in patients with connective tissue disease. Association with HLA and clinical manifestations of disease. Arthritis Rheum. 1995;38:1837–1844. doi: 10.1002/art.1780381218. [DOI] [PubMed] [Google Scholar]
- 23.Furukawa H, et al. The role of common protective alleles HLA-DRB1*13 among systemic autoimmune diseases. Genes Immun. 2017;18:1–7. doi: 10.1038/gene.2016.40. [DOI] [PubMed] [Google Scholar]
- 24.Furukawa H, et al. Human leukocyte antigens and systemic lupus erythematosus: A protective role for the HLA-DR6 alleles DRB1*13:02 and *14:03. PLoS ONE. 2014;9:e87792. doi: 10.1371/journal.pone.0087792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furukawa H, et al. Human Leukocyte Antigen and Systemic Sclerosis in Japanese: The Sign of the Four Independent Protective Alleles, DRB1*13:02, DRB1*14:06, DQB1*03:01, and DPB1*02:01. PLoS ONE. 2016;11:e0154255. doi: 10.1371/journal.pone.0154255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furukawa H, et al. Human leukocyte antigen in Japanese patients with idiopathic inflammatory myopathy. Mod. Rheumatol. 2020;30:696–702. doi: 10.1080/14397595.2019.1637593. [DOI] [PubMed] [Google Scholar]
- 27.Oka S, et al. Protective effect of the HLA-DRB1*13:02 allele in Japanese rheumatoid arthritis patients. PLoS ONE. 2014;9:e99453. doi: 10.1371/journal.pone.0099453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erlich H, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: Analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka S, et al. HLA-DRB1 and DQB1 alleles in Japanese type 1 autoimmune hepatitis: The predisposing role of the DR4/DR8 heterozygous genotype. PLoS ONE. 2017;12:e0187325. doi: 10.1371/journal.pone.0187325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakajima F, Nakamura J, Yokota T. Analysis of HLA haplotypes in Japanese, using high resolution allele typing. MHC. 2001;8:1–32. doi: 10.12667/mhc.8.1. [DOI] [Google Scholar]
- 31.Kamitsuji S, et al. Japan PGx Data Science Consortium Database: SNPs and HLA genotype data from 2994 Japanese healthy individuals for pharmacogenomics studies. J. Hum. Genet. 2015;60:319–326. doi: 10.1038/jhg.2015.23. [DOI] [PubMed] [Google Scholar]
- 32.Kamatani N, et al. Establishment of B-cell lines derived from 996 Japanese individuals. Tissue Culture Res. Commun. 2004;23:71–80. [Google Scholar]
- 33.Kasukawa RT, Miyawaki T, Yoshida S, Tanimoto H, Nobunaga K. In: Mixed connective tissue disease and anti-nuclear antibodies: proceedings of the International Symposium on Mixed Connective Tissue Disease and Anti-nuclear Antibodies. Sharp GC, Kasukawa R, editors. Berlin: Elsevier Science Publishers; 1987. pp. 41–47. [Google Scholar]
- 34.Yoshida SFS. The Cutting-edge of Medicine: Progress in the diagnosis and treatment of mixed connective tissue disease. J. Japan. Soc. Internal Med. 2012;101:1413–1419. doi: 10.2169/naika.101.1413. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka Y, et al. 2019 Diagnostic criteria for mixed connective tissue disease (MCTD): From the Japan research committee of the ministry of health, labor, and welfare for systemic autoimmune diseases. Mod Rheumatol. 2021;31:29–33. doi: 10.1080/14397595.2019.1709944. [DOI] [PubMed] [Google Scholar]
- 36.Rousset F. genepop'007: A complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are not publicly available due to privacy and ethical restrictions. The data are available from the corresponding author upon reasonable request.


