Abstract
To investigate the role of superoxide dismutases (SOD) in root colonization and oxidative stress, mutants of Pseudomonas putida lacking manganese-superoxide dismutase (MnSOD) (sodA), iron-superoxide dismutase (FeSOD) (sodB), or both were generated. The sodA sodB mutant did not grow on components washed from bean root surfaces or glucose in minimal medium. The sodB and sodA sodB mutants were more sensitive than wild type to oxidative stress generated within the cell by paraquat treatment. In single inoculation of SOD mutants on bean, only the sodA sodB double mutant was impaired in growth on root surfaces. In mixed inoculations with wild type, populations of the sodA mutant were equal to those of the wild type, but levels of the sodB mutant and, to a great extent, the sodA sodB mutant, were reduced. Confocal microscopy of young bean roots inoculated with green fluorescent protein-tagged cells showed that wild type and SOD single mutants colonized well predominantly at the root tip but that the sodA sodB double mutant grew poorly at the tip. Our results indicate that FeSOD in P. putida is more important than MnSOD in aerobic metabolism and oxidative stress. Inhibition of key metabolic enzymes by increased levels of superoxide anion may cause the impaired growth of SOD mutants in vitro and in planta.
Colonization of plant roots by certain fluorescent pseudomonads enhances plant growth and suppresses fungal pathogens (26, 32). However, the commercial potential of these organisms is unfulfilled in the field largely because of inefficient root colonization (32). The essential trait for root colonization is the ability to grow on nutrients from plants. Plant roots produce exudates containing sugars, organic acids, amino acids, and phenolic compounds (12, 23), and catabolism of these components would be expected. However, several metabolic enzymes within the bacterium are sensitive to superoxide anion (O2−). One of the key enzymes in the Entner-Douderoff pathway, 6-phosphogluconate dehydratase, is sensitive to O2− (13). Other O2−-sensitive enzymes are aconitase and fumarase, of the tricarboxylic acid (TCA) cycle, and dihydroxyacid dehydratase, required for branched-amino acid biosynthesis (10, 14, 33). The 4Fe-4S clusters that are in these enzymes are inactivated by O2− because of the release of Fe2+ (10, 13, 14). The Fe2+ released from 4Fe-4S clusters of the O2−-susceptible enzymes, [4Fe-4S] + O2− + 2H+ → [3Fe-4S] + Fe2+ + H2O2, may generate ·OH by the Fenton reaction (15, 18, 20). Although enzyme inactivation may cause growth inhibition, the attack of ·OH on DNA and cell membranes is lethal (18, 20, 31). Thus, the root-associated pseudomonads may require mechanisms to abate O2− to lessen metabolic effects and the possibility of cell death.
Superoxide dismutases (SODs) dismutate O2− to H2O2 and are key components of the cellular defense against O2− stress (11). SODs are distinguished by their metal cofactors, iron (Fe), manganese (Mn), or copper-zinc (Cu-Zn). Mutants of Escherichia coli lacking functional genes encoding FeSOD, sodB, and MnSOD, sodA, are unable to grow on minimal medium because O2− inactivates the enzymes necessary for synthesis of branched amino acids (4, 8). Because mutants lacking sodA are more sensitive to paraquat and to hydrogen peroxide than the sodB mutant (4), MnSOD is viewed as being more important than FeSOD in protecting E. coli cells against oxidative stress during aerobic growth. The E. coli sodA sodB mutants are highly sensitive to the internal O2− generator, paraquat, and their mutation rates are enhanced (4). The findings with Pseudomonas aeruginosa are in contrast (16). The sodB mutant of P. aeruginosa is sensitive to paraquat and hydrogen peroxide and is impaired in growth on rich and glucose minimal medium, whereas the phenotype of the sodA mutant is wild type. Consequently, Hassett et al. (16) concluded that FeSOD in P. aeruginosa is the key enzyme in aerobic metabolism and against oxidative stress.
Little is known about SODs of plant-associated bacteria. Phytopathogenic Xanthomonas oryzae pv. oryzae strains produce MnSOD as the major form detected at different stages of growth. The highest levels of SOD are detected during early logarithmic growth and decline as growth continues (5). With Xanthomonas campestris pv. campestris, the sodA gene encoding MnSOD is cloned, but no mutants lacking this SOD activity have been constructed (30).
Our previous work shows that FeSOD activity of P. putida rapidly increases upon contact with the root surface (19). No MnSOD is detected in the extracts from rhizoplane and rhizosphere P. putida cells. By Northern analysis of sodA and sodB genes in P. putida, we demonstrated that transcription of the sodA gene is induced, whereas transcription of the sodB gene is reduced, by iron deprivation (21). To understand the roles of the individual isozymes during root colonization, we constructed sodA, sodB, and sodA sodB mutants of P. putida by insertions of gentamicin and/or kanamycin resistance cassettes into the genes. To visualize colonization, the wild type strains and SOD mutants were tagged with a stable plasmid expressing a green fluorescent protein (GFP) gene (25). We report on the root colonization abilities of the P. putida SOD mutants, in single inoculation or in competition with the wild type. To understand the impaired colonization abilities of the SOD mutants, the effects of oxidative stresses and growth in different media were examined.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains used in the studies and their relevant properties are described in Table 1. All strains were stored at −80°C in 15% glycerol. When required, cultures were generated from the stock cultures by overnight growth on King's medium B agar (22) and Luria-Bertani (LB) agar (28). Bean root wash medium and minimal medium were used for measurements of the growth of the SOD mutants. Bean root wash medium was prepared using methods modified from that previously published (1). Briefly, 200 g of 14-day-old intact bean roots were washed in sterile water for 15 min. The wash was filtered through cheesecloth and centrifuged at 10,000 × g for 15 min to remove insoluble material. The root wash was lyophilized and dissolved using 0.3 g (dry weight) in 5 ml of sterilized and deionized water. The wash was stored frozen at −20°C and used as the growth medium after sterilization by membrane filtration through 0.2-μm (pore-size) filters. Minimal medium [50 mM K2HPO4, 50 mM KH2PO4, 2.5 mM (NH4)2SO4, 5 mM MgSO4] was supplemented with either 0.2% sugars or TCA intermediates as carbon source (19). When appropriate, media were amended with the appropriate antibiotics: gentamicin (7.5 μg/ml), ampicillin (50 μg/ml), tetracycline (15 μg/ml), kanamycin (25 μg/ml), nalidixic acid (50 μg/ml), and rifampin (50 μg/ml).
TABLE 1.
Strains and plasmids used in the studies
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strain | ||
| P. putida | ||
| Corvallis | Wild-type plant-growth-promoting bacteria, Nar Rir | A. J. Anderson |
| C-GFP | Strain Corvallis containing pCPP46-GFP | This study |
| SA101 | sodA, marker-exchange mutant of strain Corvallis sodA::Kmr Nar Rir Kmr | This study |
| SA-GFP | SA101 contained pCPP46-GFP | This study |
| SB101 | sodB, marker-exchanged mutant, sodB::Kmr Nar Rir Kmr | This study |
| SB-GFP | SB101 contained pCPP46-GFP | This study |
| SAB101 | sodA sodB mutant of wild type, sodA::Kmr, and sodB::Gmr, Kmr Gmr Nar Rir | This study |
| SAB-GFP | SAB101 contained in pCPP46-GFP | This study |
| E. coli DH5α | Host strain | Stratagene |
| Plasmids | ||
| pRK2073 | Helper plasmid, Mob+ Tra+ Spr | D. R. Helinski |
| pCPP46 | Broad-host-range vector, Tcr | D. Bauer |
| pCPP54 | Marker-exchange eviction plasmid, sacB-sacR, Tcr | D. Bauer |
| pBSSK+ | Cloning vector, Amr | Stratagene |
| pGEM7Zf | Cloning vector, Amr | Promega |
| pUCGM | pUC19 containing 850-bp Gmr cassette | H. B. Schweizer |
| pRL648 | pRL vector containing 0.9-kb Kmr cassette | C. P. Wolk |
| pCPP46-GFP | pCPP46 containing 800-bp gfp fragment | This study |
| pVNPTII-GFP | gfp gene with nptII promoter | S. Lindow |
| pPFSD502 | pUC19 with 7-kb SalI fragment of pPFSD501 | This study |
| pPFSD503 | pUC19 with 4.3-kb HindIII/SalI fragment of pPFSD502 | This study |
| pPFSD504 | pGEM7Zf with 2.8-kb ClaI/XbaI fragment of pPFSD503 | Fig. 1 |
| pPFSD505 | pPFSD504 with insert of 0.9-kb Kmr cassette from pRL648 within sodB gene | Fig. 1 |
| pPFSGM505 | pPFSD504 with insert of 0.8-kb Gmr cassette within sodB gene | Fig. 1 |
| pPFSAC505 | pCPP54 with 6.2-kb of ApaI/NsiI fragment containing sodB gene and Kmr cassette from pPFSD505 | Fig. 1 |
| pPFGMSAC | pCPP54 with 6.1-kb of ApaI/NsiI fragment containing sodB gene and Gmr cassette from pPFSGM505 | Fig. 1 |
| pPMSD502 | pUC19 with 4.3-kb SalI fragment of pPMSD501 | Fig. 1 |
| pPMSD503 | pUC19 with 4.3-kb SphI fragment of pPMSD501 | This study |
| pPMSD505 | pPMSD502 with insert of 0.9-kb Kmr cassette within sodA ORF | Fig. 1 |
| pPMSAC505 | pCPP54 with 5.1-kb of SalI fragment containing sodA gene and Kmr cassette from pMSD505 | Fig. 1 |
Abbreviations for phenotypes: Nar, nalidixic acid resistance; Rir, rifampin resistance; Amr, ampicillin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; Spr, spectinomycin resistance; Gmr, gentamicin resistance.
Assay of SOD activity in bacterial cells.
SOD activity was determined in extracts from bacterial cells grown in liquid cultures. At defined times during culture, cells were harvested by centrifugation, resuspended in 50 mM phosphate buffer (pH 7.8), and sonicated. The sonicates were centrifuged at 13,000 × g for 30 min to obtain the supernatant used in the assays (19). The SOD activity was measured spectrophotometrically, and the isozyme composition was determined by staining for activity the 7.5% nondenaturing gels used to separate the isozymes (19). Protein concentrations in cell extracts were estimated by using the bicinchoninic acid protein assay kit (Pierce Company, Rockford, Ill.).
DNA manipulation.
DNA manipulations for cloning and subcloning, including transformations, restriction enzyme digestions, ligations, and electrophoresis, were carried out as described by Ausubel et al. (2) and Sambrook et al. (28). Genomic DNA used for Southern analysis was isolated from P. putida and SOD-deficient mutants by the cetyltrimethylammonium bromide (CTAB)-DNA precipitation method (2). Southern hybridization analysis was performed by using the nonradioactive Genius System (Boehringer-Mannheim Biochemicals, Indianapolis, Ind.). Chemiluminescent immunodetection was performed at room temperature using Fuji X-ray film. Probes for Southern analysis were constructed by PCR of sodA and sodB genes using specific primers as described in our previous study (21).
Mutagenesis of sodA and sodB genes of P. putida.
The scheme to mutagenize sodA and sodB genes of P. putida is represented in Fig. 1. Briefly, a 0.9-kb EcoRI fragment containing a kanamycin resistance gene, derived from pRL648 (7), was inserted into an unique EcoRI site of the sodB open reading frame (ORF). A 0.9-kb SmaI-fragment containing the kanamycin cassette was blunt-end ligated into a unique PshAI site of 4.3-kb SalI fragment containing the sodA ORF. The mutated sod genes were transferred to the broad-host-range vector, pCPP54, to produce pPFSAC505 (sodB::Kmr) and pPMSAC505 (sodA::Kmr). The pCPP54 has a sacB-sacR locus that rendered kanamycin-resistant colonies unable to grow on 5% sucrose (27). The plasmids pPFSAC505 and pPMSAC505 were used in triparental mating with P. putida, using the helper plasmid pRK2073. The kanamycin-resistant colonies that survived on 5% sucrose plates were selected for further study. These colonies had expelled the plasmid containing the sacB-sacR locus, thus permitting growth on 5% sucrose, but had retained the kanamycin marker in the mutated gene because of homologous exchange. To create the sodA sodB mutant, a 0.8-kb EcoRI-fragment containing a gentamicin cassette derived from pUCGM (29), was ligated into the EcoRI site of the sodB gene ORF (pPFSGM505). The mutated sodB gene with the gentamicin cassette was transferred to pCPP54 (pPFGMSAC). The pPFGMSAC was mated with the P. putida sodA mutant (P. putida SA101) by triparental mating. Homologous-exchange double mutants were selected on LB agar plates containing kanamycin, gentamicin, and 5% sucrose for further analysis.
FIG. 1.
Strategy for homologous marker exchange mutagenesis to obtain SOD-deficient mutants of P. putida. (A) Construction of sodB and sodA sodB mutants. (B) Construction of sodA mutant. Abbreviations: E, EcoRI; H, HindIII; C, ClaI; X, XbaI; B, BamHI; S, SalI; A, ApaI; N, NsiI; Ps, PshAI; P, PstI.
Growth of the SOD-deficient mutants.
Growth of the P. putida and SOD-deficient mutants was measured by monitoring optical density at 600 nm (OD600) or by counting the CFU per milliliter on LB plates containing appropriate antibiotics. For growth experiments in liquid culture, cells from −80°C freezer stocks were grown at 26°C with shaking at 220 rpm in LB medium until achieving an OD600 of 2.5. The cultures were harvested by centrifugation at 13,000 × g for 10 min and were washed with growth medium used for the next culturing to remove traces of the LB medium. The cell pellets were resuspended in the appropriate medium and adjusted to a OD600 of 0.6. Cultures were diluted 1:100 in fresh media and incubated with shaking at 225 rpm at 26°C. For measurement of growth, ODs were recorded or serial dilutions of the culture were plated onto appropriate plates at defined times. The media used for the growth experiments were LB medium, glucose or succinate minimal medium, and bean root wash medium. Identities of wild-type and SOD mutants were checked by growth on LB plates containing appropriate antibiotics and by staining gels for SOD activity of extracts from these cells.
Sensitivity to paraquat.
P. putida and the SOD-deficient mutants were grown and inoculated into fresh LB medium as described above. When the cells reached an OD600 of 0.2, subsamples were transferred into several sterilized flasks, treated with different concentrations of paraquat (1, 10, or 100 μM), and incubated at 26°C with shaking. ODs and CFU per milliliter were measured at defined time points. Identities of wild-type strains and SOD mutants were checked by growth on LB plates containing appropriate antibiotics and by staining gels for SOD activity in extracts from these cells.
Root colonization.
Bean seeds (Phaseolus vulgaris L. cv. Dark Red Kidney, Idaho Seed Bean Co., Twin Falls, Idaho) were surface sterilized with 10% sodium hypochlorite and washed extensively with sterile distilled water as described previously (19). Inocula were grown to an OD600 of 2.5 in LB broth, centrifuged, and resuspended in sterile 50 mM potassium phosphate buffer (pH 7.5) to an OD600 of 0.2. The sterilized seeds were soaked for 30 min in the bacterial suspensions. For coinoculation studies, the resuspended wild type and each SOD mutant were mixed using a 1:1 (vol/vol) ratio before soaking the sterilized bean seeds. Each seed was then planted into a sterilized plastic pot containing 300 ml of growth matrix. The growth matrix (pH 7.0) was prepared by mixing of 10 parts of peat moss, 7 parts of vermiculite, 3 parts of sand, and 7 parts of Perlite (vol/vol). The growth matrix was autoclaved at 121°C twice for 90 min on two successive days. Sterile water was added to runoff, and the seeds were planted. The pots were maintained under a 14-h photoperiod at 22 ± 4°C. Sterile water was provided at rate of 20 ml pot−1 each day. At defined times, three seeds or seedlings were gently removed. Seeds or excised roots were transferred into 20 ml of sterile 50 mM potassium phosphate buffer in 50-ml disposable centrifuge tubes. The tubes then were vortexed vigorously for 1 min, diluted, and plated onto LB plates containing appropriate antibiotics. CFU per g of root were scored after incubation at 26°C. Identities of wild-type strains and SOD mutants were checked by growth on LB plates containing appropriate antibiotics and by staining gels for SOD activity of extracts from these cells.
Bacterial populations were assessed at each time point using analysis of variance of a two-way factorial in a completely randomized design. Data were log transformed prior to analysis to better meet normality assumptions. We compared means of the CFU per g of root using Tukey mean comparisons. All computations were done using PROC GLM in SAS release 6.12 (SAS Institute, Cary, N.C.). Means with the same letter are not significantly different at P > 0.05.
Confocal microscopic analysis of the GFP-tagged bacteria.
An 800-bp sequence from the gfp gene, derived from pGreenTIR (25) by restriction enzyme digestion, was ligated into a broadhost vector pCPP46 to produce pCPP46-GFP. The plasmid pCPP46-GFP was transferred to the P. putida strains by triparental mating using tetracycline as a selection marker. Green fluorescence of the GFP-tagged bacteria was measured by using a fluorometer (Fluoro IV; Gilford, Oberlin, Ohio) at 489 nm for excitation and 510 nm for emission. An E. coli strain containing pVLACGreen (25) was used as a reference strain. The GFP-tagged wild-type strains and SOD-deficient mutants were inoculated into bean seeds as described above. Three days after planting, the plant roots were removed and visualized by confocal microscopy (MRC1024; Bio-Rad, Hercules, Calif.). An argon laser was used for the detection of GFP (488 nm for excitation and 507 nm for emission). The same settings of ibis (5.0), laser power (10%), gain (1,400), and black (0) for the confocal microscope were used for examination of the different roots. The fluorescent intensity of each treatment was quantified by the NIH Image Program (National Institutes of Health, Bethesda, Md.) and the Confocal Assistant Program (Bio-Rad). A histogram of 900 square pixels was measured for at least five different replicates for each treatment to determine variability within the treatment.
RESULTS
Construction of the P. putida SOD-deficient mutants.
The sodA or sodB mutants were produced by homologous exchange after gene interruption by insertion of a kanamycin resistance gene into the ORFs of each gene (Fig. 1). The sodA sodB mutant was generated by homologous marker exchange of a sodB gene, disrupted with a gentamicin resistance cassette, into the kanamycin-resistant sodA mutant. To confirm mutagenesis of SOD genes, Southern analysis was performed with digoxigenin-labeled sodA or sodB probes on SalI-digested genomic DNA of wild type or of sodA, sodB, or sodA sodB mutants. As expected because of the insertions, the restriction fragments that hybridized with the sod probes were larger from P. putida SOD mutants than those from the wild type (Fig. 2A). Three different mutants were selected for further studies: sodA mutant (SA101), sodB mutant (SB101), and sodA sodB mutant (SAB101).
FIG. 2.
(A) Southern hybridization of SalI-digested total genomic DNA from P. putida wild type and SOD-deficient mutants with the digoxigenin-labeled 280-bp sodB PCR probe or the digoxigenin-labeled 280-bp sodA PCR probe. (B) SOD activities after native polyacrylamide gel electrophoresis of extracts from cells of different growth phases of wild-type P. putida and the sodA, sodB, and sodA sodB mutants.
Analysis of SOD activity levels and isozyme composition (Table 2 and Fig. 2B) confirmed mutations in the functional genes. In extracts from cells grown to stationary phase in LB medium, only FeSOD activity was detected in the wild type and in the sodA mutant. No activity was detected in the sodB mutant or the sodA sodB mutant. When cells were grown in KB medium, which permits expression of MnSOD in stationary phase (19), an MnSOD band was observed in extracts of wild type and of the sodB mutant but not in the sodA mutant or the sodA sodB mutant (Fig. 2B). The sodA mutant cells grown on LB rich medium to stationary phase produced ca. 85% of wild-type SOD activity (Table 2). In contrast, SOD activity of the sodB mutant was below detection in LB medium-grown cells or at ca. 20% the level of wild type in KB medium-grown cells when MnSOD was expressed (Table 2). The SOD specific activities of the wild type and sodA mutant grown in either KB or LB medium were slightly less than that of the wild type (Table 2). No SOD activity was detected in extracts from the sodA sodB mutant cells grown in KB or LB medium (Table 2).
TABLE 2.
Specific activities of SOD in extracts from stationary-phase cells of P. putida wild type and sodA, sodB, and sodA sodB mutants
| P. putida isolates | Medium | Sp act of SOD (U/mg of protein)a |
|---|---|---|
| Wild type (Corvallis) | LB | 66 ± 20 |
| KB | 25 ± 15 | |
| SA101 (sodA) | LB | 47 ± 20 |
| KB | 17 ± 10 | |
| SB101 (sodB) | LB | <0.5 |
| KB | 6 ± 2 | |
| SAB101 (sodA sodB) | LB, KB | <0.5 |
The data were obtained from three separate experiments, and the standard deviations are given.
Altered growth of the P. putida SOD-deficient mutants.
Differential growth of SOD-deficient mutants was observed in a medium containing components washed from bean roots. The medium supported the growth of wild type without a lag period, but with the SOD mutant cell numbers initially declined (Fig. 3A). Growth of sodA mutant cells was initiated after 10 to 20 h, and the sodB mutant started growth about 100 h after inoculation. The sodA sodB mutant did not grow on this medium (Fig. 3A). These findings were observed in three separate studies, although the results from only one study are provided in Fig. 3A. In these studies and in the following studies the identity of the cells recovered from the growth media or the plant roots was confirmed by plating onto media containing the appropriate antibiotics, and cell extracts were prepared from the cells in representative studies to confirm the lack of SOD activity.
FIG. 3.
Growth of P. putida and SOD-deficient mutants in different media. Growth of the bacteria was determined by measuring the OD600. Media: A, bean root wash medium; B, LB medium; C, minimal medium with succinate; D, minimal medium with glucose. The means of three experiments are presented, and the vertical bars represent the standard errors.
To explore the effect of aerobic metabolism further, we measured the growth of the SOD-deficient mutants in rich media (LB and KB) and minimal medium with different carbon sources. In comparison with the wild type, the sodA mutant grew equally well, but the growth rate of the sodB mutant was slower in LB medium. The growth of the sodA sodB mutant in LB medium was delayed, with a long lag phase (Fig. 3B). Eventually all cultures reached the same OD600. These studies were repeated three times, and in all studies the sodA sodB mutants eventually grew after a long lag phase. The colony size of the sodB mutant was 654 ± 34 μm compared to the sodA mutant (3,960 ± 46 μm) and the wild type (4,752 ± 99 μm) after 3 days of growth on KB plates. The size of the sodA sodB mutant (<500 μm) was smaller than that of the sodB mutant.
Growth of the SOD-deficient mutants in minimal media varied with the carbon source (Fig. 3C and D). Growth of the sodA mutant was similar to that of the wild type in minimal medium containing glucose or succinate. Growth of the sodB mutant was delayed in minimal medium containing succinate (Fig. 3C) and even more with glucose as a sole carbon source (Fig. 3D). The sodA sodB mutant grew in minimal medium containing succinate after a lag phase, 24 h (Fig. 3C), but did not grow in minimal medium containing glucose (Fig. 3D). The lag phase before growth commenced on succinate was similar for all of three studies performed, but the data from only one of these studies are shown.
Sensitivity of the SOD mutants to oxidative stress.
To test the role of SOD activities in protection against O2− stress, the SOD mutants and the wild type were treated with paraquat to generate O2− within the cell. When treated with 100 μM paraquat, both the wild type and the sodA mutant continued to grow, whereas growth of the sodB mutant stopped and the sodA sodB mutant was killed (Fig. 4). The data shown are for three studies that showed the same sensitivity to paraquat treatment. Further studies showed that growth was inhibited by 10 μM paraquat for the sodB mutant and by >1 μM paraquat for the double mutant (data not shown). Thus, the sensitivity to paraquat was in the following order: wild type < sodA mutant < sodB mutant < sodA sodB mutant.
FIG. 4.
Paraquat sensitivity of P. putida and SOD-deficient mutants. After the bacteria were treated with 100 μM paraquat, bacterial cell numbers were determined by plating serial dilutions on LB agar plates. The means of three experiments are presented, and the vertical bars represent the standard errors.
Root colonization of the SOD mutants.
The colonization data shown in Fig. 5A are the means of three different studies. On the bean roots, the wild type, the sodA mutant, and the sodB mutant reached maximum cell densities, ca. 109 CFU/g of root, 3 to 5 days after planting (Fig. 5A).
FIG. 5.
Colonization of P. putida SOD-deficient mutants on bean roots inoculated singly (A) or coinoculated with wild type (B). At defined times, cell numbers were determined by plating serial dilutions of root washes on LB agar plates containing appropriate antibiotics. The means of three separate experiments are presented, and the standard deviations are represented by the vertical bars. Different letters indicate significant differences for the means among treatments at each time point at P > 0.05.
Populations of these bacteria then gradually decreased to ca. 107 CFU/g of root. The populations of the sodA sodB mutant were statistically lower, by about 10-fold, than that of the wild type and the single SOD mutants at each time point sampled (Fig. 5A).
During colonization of P. putida in native soil, P. putida must compete with other soil microorganisms for nutrients. To examine the effect of bacterial competition on colonization potential, mixed inocula of the wild type and each SOD-deficient mutant were used. The growth of the wild type in coinoculation with SOD mutants was not affected (Fig. 5B). Both the sodA mutant and the wild type grew equally well on the bean roots in coinoculation (Fig. 5B). However, the growth of the sodB mutant was decreased by coinoculation with the wild type at each of the time points. The growth of the sodA sodB mutant in coinoculation with the wild type was severely impaired, with cells being recovered at levels >100-fold less than the population level of the wild type (Fig. 5B). The letters a, b, and c in Fig. 5B indicate that the means at each of the time points were statistically different at P > 0.05.
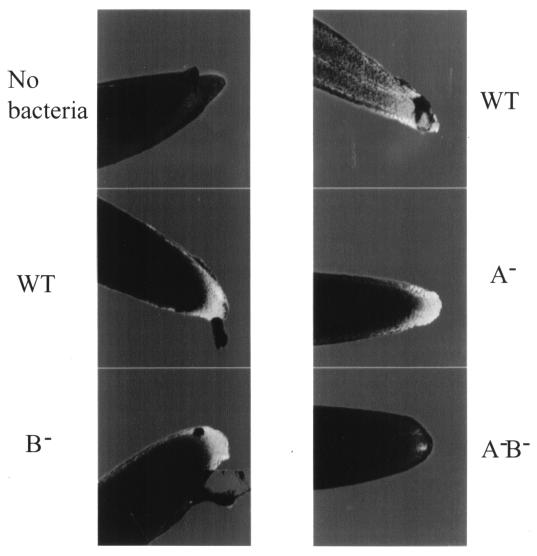
To visualize colonization, we tagged the bacteria with a stable plasmid expressing GFP, pCPP46-GFP. The relative fluorescences of the GFP-tagged wild type and SOD mutants were the same in cells grown to stationary phase in LB medium (data not shown). Bean roots were examined by confocal microscopy at 3 days after planting (Fig. 6). The most intense green fluorescence for each bacterium was detected at the root tips, with weaker green fluorescence from the rest of the root surfaces. Fluorescence was measured over a defined area for the root tip and the tissue above the tip for a minimum of five different roots for each treatment. The fluorescence intensities at the root tip colonized with the wild type (221 ± 33) was similar to that colonized with the sodA mutant (218 ± 33) and the sodB mutant (195 ± 23). Fluorescence with the sodA sodB mutant (83 ± 21) was weaker, being comparable to controls lacking any inoculum (35 ± 21). The regions above the root tip showed less fluorescence, in agreement with the root tip being the dominant site for colonization. When colonized by the wild type, fluorescence was 39 ± 31 compared to 7 ± 4 when the roots were colonized with the sodA sodB mutant; the background was 3.4 ± 3.0.
FIG. 6.
Confocal images of GFP-tagged strains of P. putida on bean root tips 3 days after planting. A minimum of five roots for each inoculum was examined under the same setting of ibis, black, and power as described in the text.
DISCUSSION
The performance of P. putida mutants lacking either or both FeSOD and MnSOD on plant roots confirmed the previous biochemical findings with P. putida (19) that FeSOD is the dominant isozyme expressed during aerobic growth as well as during root association. The reduction in SOD activity in sodB or sodA sodB mutants may cause considerable O2−-mediated stress that impaired growth of these mutants in vitro and in planta. Confocal microscopy of GFP-labeled bacteria revealed that P. putida colonized mainly on the root tip area in 3-day-old bean plants. The root tips may release more nutrients than other root parts because of sloughing of root cap cells, a lack of the secondary cell walls to limit secretion, and the active metabolic state of the meristem (9, 17). We have two explanations for the reduced populations of SOD-deficient mutants on the plant roots especially at the root tip. First, if the active metabolism of the root tip generates a high level of reactive oxygen species, growth of the mutants may be impaired by their enhanced sensitivity to reactive oxygen species, as demonstrated here by exposure to paraquat. Our previous work has shown that bean root surface enzymes produce O2− during normal metabolism with increased levels upon colonization with soil bacteria (34). The second possibility is that accumulated O2− within the mutants inhibited enzymes required for metabolism of the root surface components. We find that glucose (89 μg/ml) is the major sugar present in the components washed from the bean roots, although myoinositol (23 μg/ml) and xylose and fructose (13 to 19 μg/ml) are also present (D. Bishop, personal communication). However, we predict that the sodA sodB mutant would be unable to utilize these sugars because of inactivation of the 6-phosphogluconate dehydratase involved in the Entner-Douderoff pathway. Indeed, we found that the sodA sodB mutant cannot grow on minimal medium containing glucose, sucrose, or fructose (data not shown). Another instance where accumulated O2− in the mutants appears deleterious is in the rapid loss of CFU observed upon transfer of cells into the root wash medium. This observation supports the concept developed by Broomfield et al. (3) and Dodd et al. (6) that transfer of bacteria from one environment to another causes an imbalance in metabolism, instantaneously producing O2− and free radicals.
However, the P. putida sodA sodB mutant grew on succinate and other TCA cycle acids (fumarate, citrate, and malate) (data not shown) after a long lag phase. This result suggests to us that any inhibition of the TCA enzymes or the enzymes for branched amino acids by excessive O2− is overcome. Thus, we speculate that the continuous supply of organic acids on the bean root permitted the growth of the sodA sodB or sodB mutant, albeit at a reduced rate. Four times more organic acids than sugars are reported in tomato root exudates (24). This composition correlates with the finding that colonization of tomato roots by mutants of P. fluorescens defective in the utilization of organic acids was impaired but that colonization at wild-type levels occurred with mutants defective in the utilization of sugars (24).
Our findings demonstrate that P. putida requires at least wild-type levels of SOD activity to survive and perform with competence in new environments. Our findings also suggest to us that further understanding of the basic metabolic pathways supporting growth of the pseudomonads in the rhizosphere and at the root surface is required.
ACKNOWLEDGMENTS
This research was supported by the Utah Agricultural Experiment Station, Utah State University, Logan.
We thank S. E. Lindow and G. A. Beattie (University of California, Berkeley) for the gift of the GFP constructs. We also thank Joseph Shope for his guidance in the confocal microscopic analysis and Susan Durham for assistance in the statistical analysis.
Footnotes
Journal paper number 7163 of the Utah Agricultural Experiment Station.
REFERENCES
- 1.Albert F G, Benett L W, Anderson A J. Peroxidase associated with the root surface of Phaseolus vulgaris. Can J Bot. 1986;64:573–578. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989. [Google Scholar]
- 3.Bloomfield S F, Stewart G S A B, Dodd C E R, Booth I R, Power E G M. The viable but non-culturable phenomenon explained? Microbiology. 1998;144:1–3. doi: 10.1099/00221287-144-1-1. [DOI] [PubMed] [Google Scholar]
- 4.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chamnongpol S, Mongkolsuk S, Vattanaviboon P, Fuangthong M. Unusual growth phase and oxygen tension regulation of oxidative stress protection enzymes, catalase and superoxide dismutase, in the phytopathogen Xanthomonas oryzae pv. oryzae. Appl Environ Microbiol. 1995;61:393–396. doi: 10.1128/aem.61.1.393-396.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dodd C E R, Sharman R L, Bloomfield S F, Booth I R, Stewart G S A B. Inimical process: bacterial self-destruction and sub-lethal injury. Trends Food Sci Technol. 1997;8:238–241. [Google Scholar]
- 7.Elhai J, Wolk C P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 8.Farr S B, D'Ari R, Touati D. Oxygen-dependent mutagenesis in Escherichia coli lacking superoxide dismutase. Proc Natl Acad Sci USA. 1986;83:8268–8272. doi: 10.1073/pnas.83.21.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldman L J. Regulation of root development. Annu Rev Plant Physiol. 1984;35:223–242. doi: 10.1146/annurev.pp.35.060184.001255. [DOI] [PubMed] [Google Scholar]
- 10.Flint D H, Emptage M H, Finnegan M G, Fu W, Johnson M K. The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. J Biol Chem. 1993;268:14732–14742. [PubMed] [Google Scholar]
- 11.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 12.Gamliel A, Katan J. Influence of seed and root exudates on fluorescent pseudomonads and fungi in solarized soil. Phytopathology. 1992;82:320–327. [Google Scholar]
- 13.Gardner P R, Fridovich I. Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J Biol Chem. 1991;266:1478–1483. [PubMed] [Google Scholar]
- 14.Gardner P R, Fridovich I. Superoxide sensitivity of the Escherichia coli aconitase. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 15.Halliwell B, Gutterridge J M C. Oxygen free radicals and iron in relation to biology and medicine some problems and concepts. Arch Biochem Biophys. 1986;246:501–514. doi: 10.1016/0003-9861(86)90305-x. [DOI] [PubMed] [Google Scholar]
- 16.Hassett D J, Schweizer H P, Ohman D E. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawes M C. Sloughed root cap cells: a regulator of microbial populations in the rhizosphere? Plant Soil. 1990;129:19–27. [Google Scholar]
- 18.Imlay J A, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 19.Katsuwon J, Anderson A J. Catalase and superoxide dismutase of root-colonizing saprophytic fluorescent pseudomonads. Appl Environ Microbiol. 1990;56:3576–3582. doi: 10.1128/aem.56.11.3576-3582.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keyer K, Imlay J A. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y C, Miller C D, Anderson A J. Transcriptional regulation by iron of genes encoding iron (Fe) and manganese (Mn) superoxide dismutases from Pseudomonas putida. Gene. 1999;239:129–135. doi: 10.1016/s0378-1119(99)00369-8. [DOI] [PubMed] [Google Scholar]
- 22.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 23.Klein D A, Frederick B A, Biondini M, Trlica M J. Rhizosphere microorganisms effects on soluble amino acids, sugars and organic acids in the root zone of Agropyron cristatum, A. smithii and Bouteloua gracilis. Plant Soil. 1988;110:19–25. [Google Scholar]
- 24.Lugtenberg B, van der Bij A, Bloemberg G, Woeng T C A, Dekkers L, Kravchenko L, Mulders I, Phoelich C, Simons M, Spaink H, Tikhonovich I, de Weger L, Wijffelman C. Molecular basis of rhizosphere colonization by Pseudomonas bacteria. In: Stacey G, Mullin B, Gresshoff P M, editors. Biology of plant-microbe interactions. St. Paul, Minn: International Society for Molecular Plant-Microbe Interactions; 1996. pp. 433–440. [Google Scholar]
- 25.Miller W G, Lindow S E. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene. 1997;191:149–153. doi: 10.1016/s0378-1119(97)00051-6. [DOI] [PubMed] [Google Scholar]
- 26.O'Sullivan D J, O'Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid J L, Collmer A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schweizer H P. Small broad-host-range gentamycin resistant gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 30.Smith S G, Wilson T J G, Dow J M, Daniels M J. A gene for superoxide dismutase from Xanthomonas campestris pv. campestris and its expression during bacterial-plant interactions. Mol Plant-Microbe Interact. 1996;9:584–593. doi: 10.1094/mpmi-9-0584. [DOI] [PubMed] [Google Scholar]
- 31.Stadtman E R. Metal ion-catalyzed oxidation of protein: biochemical mechanism and biological consequence. Free Rad Biol Med. 1990;9:315–325. doi: 10.1016/0891-5849(90)90006-5. [DOI] [PubMed] [Google Scholar]
- 32.Weller D M. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu Rev Phytopathol. 1988;26:379–407. [Google Scholar]
- 33.Woods S A, Schwartzbach S D, Guest J R. Two biochemically distinct classes of fumarase in Escherichia coli. Biochem Biophys Acta. 1988;954:14–16. doi: 10.1016/0167-4838(88)90050-7. [DOI] [PubMed] [Google Scholar]
- 34.Zdor R E, Anderson A J. Influence of root colonizing bacteria in the defense responses of bean. Plant Soil. 1992;140:99–107. [Google Scholar]