Abstract
This study aimed to investigate the effects of active and heat-inactivated forms of Akkermansia muciniphila, bacterium-derived outer membrane vesicles (OMVs), and cell-free supernatant on the transcription of endocannabinoid system (ECS) members, including cannabinoid receptors 1 and 2 (CB1 and CB2), fatty acid amide hydrolase (FAAH), and peroxisome proliferator-activated receptors (PPARs) genes (i.e., α, β/δ, and δ) in Caco-2 and HepG-2 cell lines. After the inoculation of A. muciniphila in brain heart infusion enriched medium, OMVs and cell-free supernatant were extracted. For the investigation of the effects of bacteria and its derivatives on the expression of ECS and PPARs genes, the aforementioned cells were treated by active and heat-inactivated bacteria, OMVs, and cell-free supernatant. Quantitative real-time polymerase chain reaction analysis revealed that both forms of the bacterium, bacterial-derived OMVs, and cell-free supernatant could affect the expression of CB1, CB2, FAAH, and PPARs genes (i.e., α, β/δ, and δ) significantly (P < 0.05). Considering the engagement of the aforementioned genes in metabolic pathways, it might be suggested that both forms of the bacterium, OMVs, and cell-free supernatant might have the potential to serve as a probiotic, paraprobiotic, and postbiotic candidate to prevent obesity, metabolic disorders, and liver diseases.
Subject terms: Extracellular signalling molecules, Microbiology, Microbial communities
Introduction
One of the most remarkable habitats for microbiota is the human gastrointestinal tract (GIT), where variable microbes colonize from the mouth down to the rectum according to environmental conditions1,2. Gut microbiota executes several structural, protective, and metabolic functions and roles, such as interfering in gut barrier functions, insuring its homeostasis, supplying nutrients, participating in signaling network, regulating epithelial tissue development, and affecting the immune system in the intestinal mucosa3. The appropriate physiology and homeostasis of the host are critically associated with the integrity of the intestinal barrier4–6. The changes in the latter might lead to gastrointestinal4,7, metabolic4,8, and even neurological disorders4,9. Accordingly, leaky gut syndrome, as one of the most important gastrointestinal disorders, can occur by possible alteration in tight junction proteins. Leaky gut syndrome might cause the translocation of intestinal microbiota and their microbial-associated molecular patterns (MAMPs) toward the liver through the gut-liver axis. It is well documented that this phenomenon can induce numerous liver disturbances10,11.
Among diverse bacterial species in the gut, Akkermansia. muciniphila is an anaerobic Gram-negative and a mucus colonizing bacterium12–15. It appropriates about 3–5% of the gut microbiota in healthy humans14,16, has a major contribution in gut barrier regulation, and involves in metabolic and homeostatic procedures4,17. Recently, it has been considered a candidate of the next generation probiotics12. In recent years, it has been specified that heat-inactivated bacteria might exert some active molecules and MAMPs that influence the host physiology, immunity, and gut barrier integrity18,19.
Several studies have shown that various living and nonliving forms of A. muciniphila in inflammatory conditions and metabolic diseases have been able to prevent the onset of disease or relieve the symptoms of the disease20–22. Additionally, to avoid inflammatory and antibiotic resistance side effects, the administration of bacterial cell-free supernatant is suggested as a substitution for alive bacteria since it contains all the beneficial and functional metabolites of the bacterial flora23. Outer membrane vesicles (OMVs) in Gram-negative bacteria have shown efficient roles not only in bacterial survival but also in interacting with the host through inter- and intra-kingdom communications without actual intercellular connection21,24.
The evidence suggests an interaction between the endocannabinoid system (ECS) and gut microbiota and subsequently the liver through the gut-liver axis in some leaky gut conditions25. The ECS consists of three main compartments, including endocannabinoids (eCBs), cannabinoid receptors (i.e., CB1R and CB2R), and related metabolic enzymes25–27. It participates in numerous biological processes in the brain and some peripheral tissues28. Cannabinoid receptors are coupled with Gi/o family of the G-proteins through which involve in signal transduction pathways29. A neuronal stimulation leads to release of endocannabinoids from the postsynaptic terminals to the extracellular space of the synaptic terminal in a retrograde manner and binding to CB1/2 receptors in the presynaptic terminals30. Activity of the Biosynthetic enzymes depends on intracellular Ca2+ concentration which determines the synthesis of endocannabinoids31. Fatty acid amide hydrolase (FAAH) is located in the postsynaptic terminal to hydrolyze these ligands. Since CB1/2 inhibit Ca2+ influx (through regulation of ion channels), Gamma amino butyrate (GABA) and glutamate release, and also nitric oxide (NO) production, it can modulate neurotransmission, inhibit adenylate cyclase and cAMP/Protein kinaseA pathway, respectively. They can also activate mitogen-activated protein kinases (MAPKs) such as ERK1/2, P38 and JNK which depending on the type of ligand and cell environmental conditions, the result of this signaling pathway will be different30,32. In GIT, CB1 modulates the motility, and reduces the secretion of fluids, gastric acids, neurotransmitters, and hormones along with increasing the gut permeability30. High fat diets induce cholecystokinin (CCK) production by enteroendocrine l-cells that leads to induce satiety signals via afferent sensory vagus nerves to the brain. This procedure is inhibited by CB1 probably through MAPK signaling and promotes over eating followed by diet induced obesity33,34. Within hepatocytes, CB1 is able to increase the expression of a lipogenic transcription factor of sterol regulatory element binding protein (SREBP). Enhancement of acetyl-CoA carboxylase 1(ACC) and fatty acid synthase (FAS) as SREBP targets lead to fatty liver, hypertriglyceridemia, and insulin resistance32,35. CB2 signaling in hepatocytes have shown a regulatory effect on lipid accumulation36, decreasing the inflammatory cytokines, steatosis, hepatic proliferation, and liver fibrosis progression32,37. Hence, The ECS functions might lead to increased permeability, decreasing inflammation, regulating food intake, and gut motility in the GIT. It also influences some metabolic disorders, such as obesity, type 2 diabetes, steatosis, fibrogenesis, alcoholic and nonalcoholic liver diseases, mainly through CB1R function25,26,28.
Peroxisome Proliferator-Activated Receptors (PPARs) are nuclear receptors with the affinity to bind to endocannabinoids. They make a role in gene regulation of fatty acid oxidation, metabolism of carbohydrates and lipids, cell proliferation, and inflammation32,38,39. Several studies have shown considerable interactions between the gut microbiota and peroxisome proliferator-activated receptors (PPARs) that participate in the regulation of gut barrier permeability, energy homeostasis, and liver metabolism27,40. Since there is a cross-reaction between ECS members and PPARs genes41 and regarding the functions of the ECS in the gut25,42 and liver43,44, the current study aimed to evaluate the effects of active and inactive forms of A. muciniphila, its derived OMVs, and cell-free supernatant on the expression of remarkable genes, such as CB1, CB2, fatty acid amide hydrolase (FAAH) as the representatives of the ECS, and PPARs (i.e., α, β/δ, and δ) genes which are all involved in the regulation of numerous metabolic, inflammatory, and developmental processes in the gut and liver.
Materials and methods
Bacterial culture condition
Akkermansia muciniphila (ATCC BAA-835) was provided from the DSMZ institute (German Collection of Microorganisms and Cell Cultures GmbH). A. muciniphila strains were cultured in brain heart infusion (BHI) agar medium (Quelab, Canada) supplemented with 0.5% mucin-type III (Sigma-Aldrich, St. Louis, Missouri, USA), hemin (5 μg/ml), menadione (1 μg/ml)45 and 0.05% L-cysteine46 under anaerobic conditions (80% N2, 10% H2, and 10% CO2) at 37 °C for 3–7 days as described previously12.
Polymerase chain reaction (PCR) test was performed based on 16 s ribosomal ribonucleic acid (rRNA) sequence recognition to confirm the A. muciniphila, in addition to macroscopic and microscopic (Gram staining) assays (Supplementary Table 1).
Cell-free supernatant was obtained from the inoculation of A. muciniphila in 100 mL BHI broth with the above-mentioned supplementations incubated in the same condition reaching optical density (OD) of 1.5 in 600 nm, centrifuged in 8000 × g for 5 min; the pH was adjusted in 7.4 and then purified by passing through 2.22 nm filters and kept at −70 °C until usage.
OMVs extraction and confirmation
12 × 108 colony-forming units (CFU/ml) of bacteria equal to 4 McFarland were inoculated in above-mentioned supplemented BHI broth overnight, and when the OD reached 1.5 in 600 nm4, the bacterial suspension were centrifuged at 6000 × g at 4 °C. The supernatant was poured out, and the pellets were washed twice with phosphate-buffered saline (PBS) followed by centrifugation at 6000 × g at 4 °C, suspended with 9% sodium chloride solution at 6000 × g at 4 °C for an hour and then mixed with ethylenediaminetetraacetic acid-sodium deoxycholate (Sigma Aldrich, USA) buffers47. In advance, centrifugation was performed at 20,000 × g at 4 °C for an hour and ultracentrifugation at 125,000 × g twice for 2 h sequentially, and then it was kept in sucrose 3% at −70 °C48.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) procedure was performed for the evaluation of the protein content of the sample, and transmission electron microscopy (TEM) was performed as a confirmation for the presence and sizes of OMVs. The sample was prepared by negative staining and then observed by PHILIPS (Netherlands) EM 208.
Cell culture
Caco-2 (IBRC C10094) and Hep-G2 (C10096) cell lines were obtained from Iranian Biological Resource Center, Tehran, Iran, cultured in Dulbecco’s Modified Eagle’s Medium (Bioidea, Iran) supplemented by 10% heat-inactivated (56 °C, 30 min), fetal bovine serum (Bioidea, Iran), 1% nonessential amino acids (Bioidea, Iran), and 1% penicillin–streptomycin (Bioidea, Iran), which were incubated at 37 °C and 5% CO2 condition49,50. After reaching enough confluency (minimum 70%) and transferring 5 × 105 cells into every well of six well plates (Sorfa, China)12, with some modifications, compared to previous studies, four types of treatments, including active and inactivated (20 min in 56 °C) A .municiphila, OMVs, and cell-free supernatant, were administered on both Caco-2 and HepG-2 cells. The cells were separately infected by active and inactivated bacteria at the multiplicity of infection (MOI) both in 10, 50, and 100 ratios (i.e., 10, 50, and 100 bacteria per cell, respectively). Some other cells in other wells were treated by 50 and 100 μg/mL OMVs and others by a well-optimized concentration of 7% (v/v [medium/cell-free supernatant]) cell-free supernatant 12,47,49,51. Equal volumes of PBS, sucrose 3%, and supplemented BHI broth were used in separated wells as controls for comparison with their related wells. All treated cells were incubated for 24 h12,47.
RNA extraction and cDNA synthesis
The cells were collected from six wells (5 × 105 cells per well), and their total RNA was extracted using RNX-Plus Solution (2000 ng/μl; Sinacolon, Iran). In order to purify the extracted RNAs, they were treated by DNAse for 1 h at 37 °C. The presence and quality of the extracted RNAs were checked out by agarose gel electrophoresis and spectrophotometry using NanoDrop 2000 (Thermo Fisher Scientific, USA). After balancing the concentrations of RNAs, complementary deoxyribonucleic acid (cDNA) was synthesized using a cDNA synthesis kit (Parstous, Iran) according to manufacturers’ instructions. A no reverse transcriptase control cDNA was included in qRT-PCR analysis. The concentrations were checked out by spectrophotometry using a Nanodrop device47.
Quantitative real-time PCR analysis of ECS-related (i.e., CB1, CB2, and FAAH) and PPARs genes
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using SYBR Green real-time master mix (Parstous, Iran), specific primers, and newly synthesized cDNAs in LightCycler®96 SW 1.1 instrument (Roche, Germany). Glyceraldehyde 3-phosphate dehydrogenase was used as a reference gene. The program conditions consisted 1 cycle of 94 °C for 10 min followed by 40 cycles, including denaturation at 95 °C for 15 s, annealing temperatures of each set of primers for 30 s, and 72 °C as an extension time for 30 s followed by 10 min at 72 °C for the complementation of the procedure.
Statistical analysis
The data extracted from qRT-PCR were analyzed by GraphPad Prism software (version 8.4.3; GraphPad Software Inc., San Diego, CA, USA) using an independent sample t-test (between two groups) and one-way analysis of variance (with Tukey’s multiple comparison test to estimate differences between means which was used to compare means among more than two groups for each parameter). All results were considered statistically significant with a p-value less than 0.0512,47.
Results
Confirmation of A. municiphila and Its OMVs with size
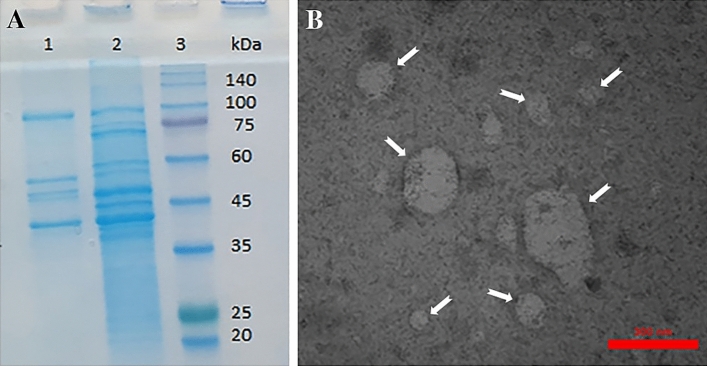
A. municiphila was confirmed by PCR methods (16 s rRNA gene PCR). The SDS-PAGE displayed the protein content of the bacteria, and TEM analysis confirmed the presence of A. muciniphila-derived OMVs in size (30–300 nm vesicles; Fig. 1).
Figure 1.
A.muciniphila OMVs confirmation. (A) SDS-PAGE. (1) OMVs proteins (2) supernatant (3) molecular weight Protein Marker (B) TEM image × 18,000. Arrows indicate OMVs in different sizes (30–300 nm).
qRT-PCR analysis of the associated genes expression in Caco-2 cell line
Effects of active and inactivated A. muciniphila, derived OMVs, and cell-free supernatant on transcription level of the studied genes Involved in endocannabinoid system
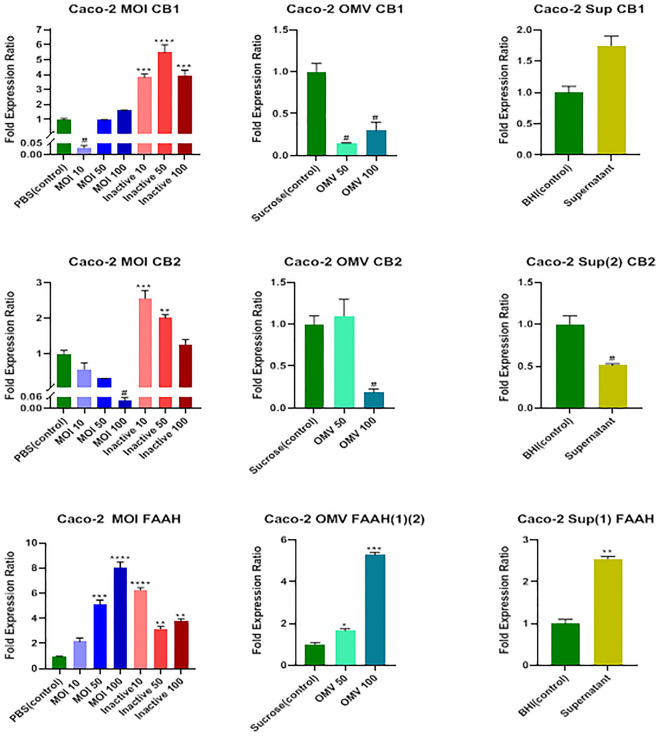
Data analysis showed a significant decrease in the messenger ribonucleic acid (mRNA) level of the CB1 receptor by MOI 10 of active A. muciniphila (P = 0.04) and both 50 and 100 μg/mL concentrations of OMVs (P = 0.01) (Fig. 2). All three MOIs of 10, 50, and 100 of the inactivated form of the bacterium could remarkably (P = 0.0008, P = 0.0001, and P = 0.0006, respectively) increase the mRNA level of the CB1 receptor; nevertheless, the cell-free supernatant did not have a significant effect on the expression of the CB1 gene (P = 0.053).
Figure 2.
Effects of active and inactive A.muciniphila (at MOIs of 10, 50 and 100), its derived OMVs (50 and 100 μg/mL), and 7% cell free supernatant on the expression of endocannabinoid system related genes (CB1, CB2 receptors, and FAAH) in Caco-2 cells. Significancy is evaluated in comparison with control. Data are shown as the mean ± SEM. (*) and (#) represent significant increase and decrease, respectively.*/# P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by one-way ANOVA and t-test statistical analysis. MOI: multiplicity of infection, OMV: outer membrane vesicle, Sup: supernatant.
A noticeable decrease occurred in the mRNA level of the CB2 receptor by MOI 100 of active A. muciniphila, 100 μg/ml concentrations of OMV, and concentration of 7% (v/v [medium/cell-free supernatant]) cell-free supernatant (P = 0.01, P = 0.03, and P = 0.04, respectively); however, the MOIs of 10 and 50 of the inactivated form of the bacterium increased the mRNA level of the CB2 receptor significantly (P = 0.0007 and P = 0.01, respectively) (Fig. 2).
As shown in Fig. 2, MOIs 50 and 100 of active A .muciniphila (P = 0.0002 and P < 0.0001, respectively), all three MOIs of 10, 50, and 100 of the inactivated form of the bacterium (P < 0.0001, P = 0.0075, and P = 0.0018, respectively), both 50 and 100 μg/ml concentrations of OMVs (P = 0.033 and P = 0.0002, respectively), and 7% (v/v [medium/cell-free supernatant]) concentration of cell-free supernatant (P = 0.0057) increased the mRNA level of the FAAH gene remarkably.
Effects of the above-mentioned treatments on transcription level of PPARs (i.e., α, β/δ, and ϒ) genes
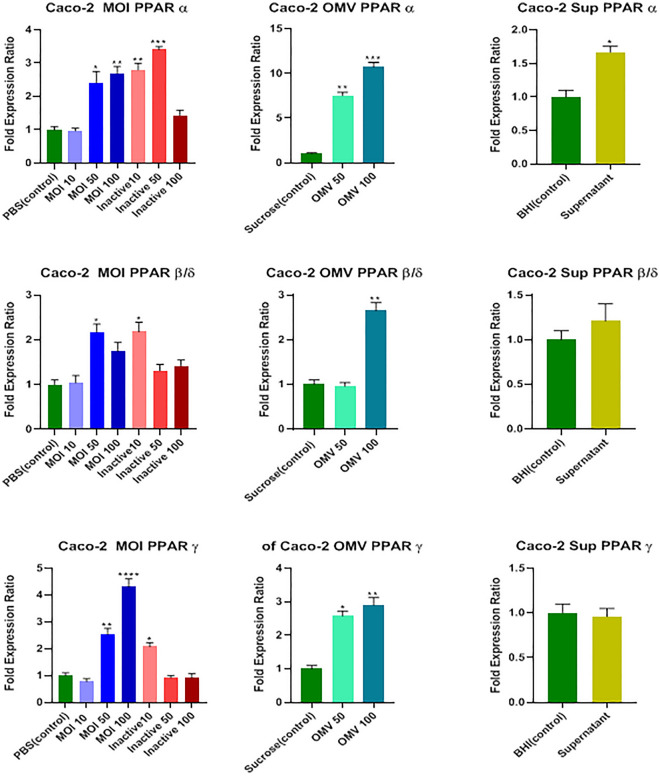
In reference to Fig. 3, MOIs 50 and 100 of active A. muciniphila (P = 0.011 and P = 0.004, respectively), MOIs of 10 and 50 of the inactivated form of the bacterium (P = 0.002 and P = 0.0004, respectively), both 50 and 100 μg/ml concentrations of OMVs (P = 0.002 and P = 0.0008, respectively), and 7% (v/v [medium/cell-free supernatant]) concentration of cell-free supernatant (P = 0.038) could significantly increase the mRNA level of the PPARα gene.
Figure 3.
Effects of active and inactive A.muciniphila (at MOIs of 10, 50, and 100), its derived OMVs (50 and 100 μg/mL) and 7% (v/v [medium/cell-free supernatant]) cell free supernatant on the expression of PPARS (α, β/δ, and ϒ) genes in Caco-2 cells. Significancy is evaluated in comparison with control. Data are shown as the mean ± SEM. (*) and (#) represent significant increase and decrease, respectively.*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by one-way ANOVA and t-test statistical analysis. MOI: multiplicity of infection, OMV: outer membrane vesicle, Sup: supernatant.
The MOI 50 of active A. muciniphila, MOI 10 of the inactivated form of the bacterium, and 100 μg/ml concentration of OMV increased the mRNA level of the PPARβ/δ gene remarkably (P = 0.015, P = 0.014, and P = 0.005, respectively); nonetheless, the cell-free supernatant could not affect PPARβ/δ transcriptome level (P = 0.43; Fig. 3).
The MOIs 50 and 100 of active A. muciniphila (P = 0.004 and P < 0.0001, respectively), MOI 10 of the inactivated form of the bacterium (P = 0.03), and both 50 and 100 μg/mL concentrations of OMVs (P = 0.012 and P = 0.007, respectively) significantly increased the mRNA level of PPARϒ gene; nevertheless, the cell-free supernatant could not affect its expression significantly (P = 0.79; Fig. 3).
qRT-PCR analysis of the associated genes expression in HepG-2 cell line
Effects of active and inactivated A. muciniphila, derived OMVs, and cell-free supernatant on transcription level of the studied genes involved in endocannabinoid system
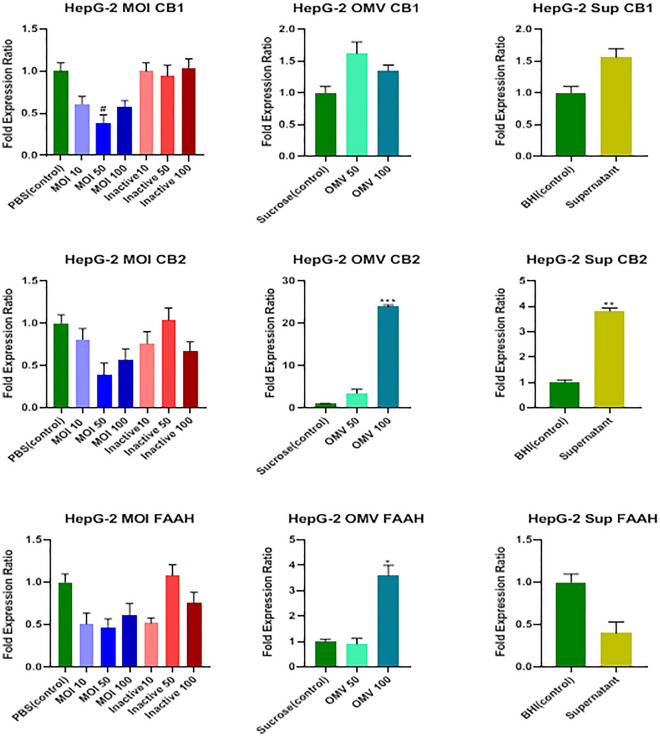
Data analysis showed that only MOI 50 of the active form of the bacterium significantly decreased the CB1 receptor transcription level (P = 0.033), and none of the inactive bacteria, OMVs, and cell-free supernatant could affect the transcription level of the CB1 receptor (P > 0.05; Fig. 4).
Figure 4.
Effects of active and inactive A.muciniphila (at MOIs of 10, 50, and 100), its derived OMVs (50 and 100 μg/mL), and 7% (v/v [medium/cell-free supernatant]) cell free supernatant on the expression of endocannabinoid system related genes (CB1, CB2 receptors, and FAAH) in HepG-2 cells. Significancy is evaluated in comparison with control. Data are shown as the mean ± SEM. (*) and (#) represent significant increase and decrease, respectively.*P < 0.05, **P < 0.01, ***P < 0.001 by one-way ANOVA and t-test statistical analysis. MOI: multiplicity of infection, OMV: outer membrane vesicle, Sup: supernatant.
According to the results shown in Fig. 4, 100 μg/mL concentrations of OMVs and 7% (v/v [medium/cell-free supernatant]) concentration of cell-free supernatant remarkably increased the mRNA level of the CB2 receptor (P = 0.0002 and P = 0.0034, respectively); nonetheless, neither active nor inactive forms of the bacterium affected its transcription level (P > 0.05). Only 100 μg/mL concentrations of OMVs increased FAAH mRNA level significantly (P = 0.013), and none of the concentrations of the active and inactive forms and cell-free supernatant affected it (P > 0.05; Fig. 4).
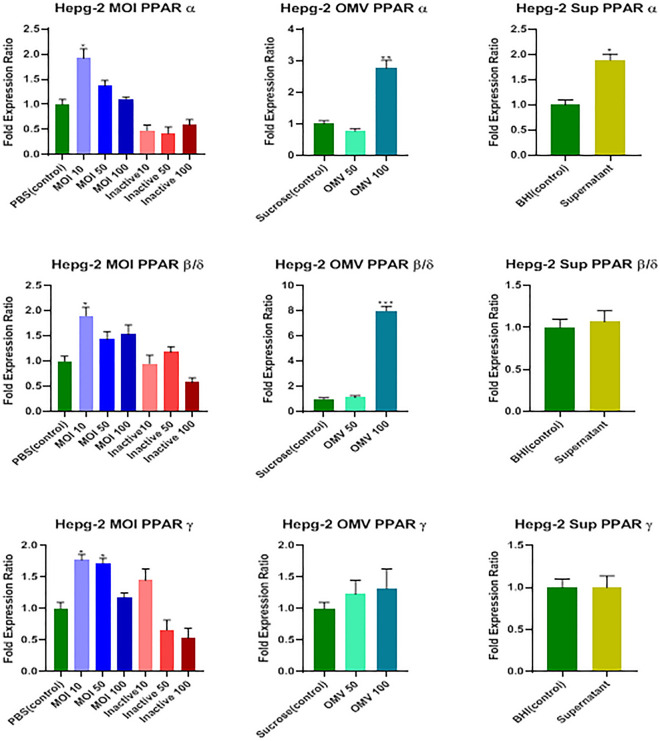
Effects of the above-mentioned treatments on transcription level of PPARs (i.e., α, β/δ, and ϒ) genes
The MOI 10 of the active form of the bacterium, 100 μg/mL concentrations of OMVs, and 7% (v/v [medium/cell-free supernatant]) concentration of cell-free supernatant noticeably increased the transcriptome level of the PPARα gene (P = 0.01, P = 0.009, and P = 0.029, respectively); however, the inactive form of the bacterium did not affect it (P > 0.05; Fig. 5).
Figure 5.
Effects of active and inactive A.muciniphila (at MOIs of 10, 50, and 100), its derived OMVs (50 and 100 μg/mL) and 7% (v/v [medium/cell-free supernatant]) cell free supernatant on the expression of PPARS (α, β/δ, and ϒ) genes in HepG-2 cells. Significancy is evaluated in comparison with control. Data are shown as the mean ± SEM. (*) and (#) represent significant increase and decrease, respectively. *P < 0.05, **P < 0.01 and***P < 0.001 by one-way ANOVA and t-test statistical analysis. MOI: multiplicity of infection, OMV: outer membrane vesicle, Sup: supernatant.
As shown in Fig. 5, MOI 10 of the active form and 100 μg/mL concentrations of OMVs could remarkably increase the mRNA level of the PPARβ/δ gene (P = 0.032 and P = 0.0005, respectively); nevertheless, neither inactive form nor the cell-free supernatant affected its transcription level (P > 0.05).
Finally, there was a significant increase in the mRNA level of the PPARϒ gene only at MOI 10 and 50 treatments of the active form of the bacterium (P = 0.034 and P = 0.047, respectively), and none of the inactive form, OMVs, and cell-free supernatant affected it (P > 0.05; Fig. 5).
Discussion
A. muciniphila is an anaerobic Gram-negative bacterium that appropriates about 3–5% of the gut bacteria in healthy humans12,14. The evidence has confirmed its role in gut barrier regulation and its involvement in metabolic and homeostatic procedures4,17. Recently, a great interest has been attracted on this subject due to its potential of introducing as the next generation probiotics12. A. muciniphila mainly colonizes in the gut, and due to some leaky gut conditions, the bacterium and related derivatives52 might pass through the gut barrier and enter some other organs, such as the liver through the gut-liver axis4,10,11. This might happen based on the interaction between the ECS, gut microbiota, and the liver25, and the involvement of PPARs genes in the regulation of gut barrier permeability27,40 and liver metabolism42–44. Therefore, this study investigated the effects of A. muciniphila and its derivatives on ECS-related and PPARs genes in Caco-2 and Hep-G2 cell lines in parallel.
The ECS consists of three main compartments, including cannabinoids, cannabinoid receptors, and metabolic enzymes, concentrated mainly in the brain and some peripheral tissues. They play various roles, such as involvement in the regulation of hunger and satiety, relaxation, protection, immunity, metabolism, decreasing inflammation, and increasing permeability in the GIT. It also influences some diseases, such as obesity, type 2 diabetes, steatosis, fibrogenesis, and alcoholic and nonalcoholic liver diseases, mainly through CB1 function25–28,53. Regarding the important role of A. muciniphila in gut barrier integrity and decreasing permeability which prevents metabolic disorders associated with obesity54,55, the present study showed that MOI 10 of A. muciniphila and both concentrations of 50 and 100 μg/mL of A. muciniphila-derived OMVs could decrease the level of CB1 mRNA in Caco-2 cells which promotes the idea of using the active A. muciniphila as a probiotic candidate and bacterial-derived OMVs for CB1 expression regulation to prevent metabolic disorders associated with obesity.
This study demonstrated that all MOIs (i.e., 10, 50, and 100) of the inactivated form of the bacterium increased the CB1 mRNA level in Caco-2 cells. Consistent with the results of the present study, Everard et al. (2013) observed that in spite of the active bacterium, heat-killed A. muciniphila could not improve the thickness of the mucus layer and counteract the fat mass development56. Russo et al. (2004) reported that CB1 deficiency might lead to some disorders, such as inflammatory bowel diseases (known as irritable bowel syndrome [IBS]), migraine, fibromyalgia, and psychological disorders57. Therefore, the inactivated A. muciniphila might be considered a paraprobiotic candidate for the treatment of IBS, especially in conditions that the treatment of live bacteria might exacerbate the inflammation condition.
On the same side, MOI 100 of active bacteria, OMV 50 μg/mL, and cell-free supernatant could decrease the CB2 mRNA level. In contrast, MOI 10 and 50 of inactivated bacteria increased the mRNA level of CB2 significantly. Since the functions of CB1 and CB2 receptors in the gut are in the same direction26,27,58,59, the latter results are in line with the present study hypothesis about different effects of active A. muciniphila, its derived OMVs, and cell-free supernatant in comparison to those of inactive A. muciniphila on ECS function as required. In similar studies on cannabinoid receptors , Rousseaux et al. in 2007 reported that among a diversity of bacteria used in the study (including: L. acidophilus NCFM, L. salivarius Ls-33, L. paracasei Lpc-37, B. lactis Bi-07 and B. lactis Bl-04, and two E. coli strains), only active and heat-inactivated Lactobacillus acidophilus NCFM , known as a probiotic, could increase the CB2 mRNA level in HT-29 epithelial cells that made it a considerable candidate for the treament of irritable bowel syndrome and abdominal pains60.
In 2016, Scarpelli et al. concluded that the inhibition of FAAH could protect anandamides from degradation leading to more activation of cannabinoid receptors61. Therefore, FAAH function has a reverse relationship with cannabinoid receptors. The current study data showed that MOI 50 and 100 of active A. muciniphila, both concentrations of OMV 50 and 100 μg/ml, and cell-free supernatant could significantly increase the level of FAAH mRNA in Caco-2 cells.
The MOI 10, 50, and 100 of inactive bacteria also increased FAAH mRNA significantly. Probably, there is a modulatory function of FAAH, since Murakami et el. (2007) discussed that bacterial lipopolysaccharide (LPS) as one of the magic components of MAMPs19 in A. muciniphila62 induces the production of anandamide which is mentioned as a ligand for the CB1 receptor. Meanwhile, the LPS stimulation could not affect FAAH; therefore, it seems natural that the rate of FAAH mRNA increased by more anandamide production to control the overproduction of these ligands63. Considerably, there is a reverse relationship in the treatment results of MOI 50 of inactive A. muciniphila between CB1 and FAAH in Caco-2 cells.
The PPARs belong to the nuclear receptor superfamily serving as transcription factors that regulate numerous transcriptional activities, such as metabolic, inflammatory, and developmental processes. The PPARs are composed of three isotypes, including PPARα, PPARβ/δ, and PPARϒ, which have different distributions in the human body; however, all three are highly expressed in the colon. The evidence has shown that there is a direct interaction between gut microbiota and PPARs in a way that gut microbiota might induce PPARs expression and activation40,64. In 2011, Goto et al. reported that PPARα stimulates fatty acid oxidation in adipocytes and ameliorates metabolic disorders65. In 2003, prior to this study, Wang et al. declared that PPARβ/δ prevents obesity through fat metabolism and energy expenditure66. La Cour Poulsen et al. in 2012 explained that numerous types of fatty acids and their derivatives could activate PPARs. Furthermore, it has been suggested that there is a relationship between the microbiota, fatty acid metabolism, and PPARs.
In 2019, Ashrafian et al. reported that A. muciniphila and its derived OMVs stimulated fatty acid oxidation and energy metabolism. These processes occurred in addition to the increased expression of PPARα and PPARϒ49. Recently, in 2020, Wang et al. reviewed that A. muciniphila produces short-chain fatty acids (SCFAs), which affect glucose and lipid homeostasis. It also increases fatty acid oxidation in the intestine and adipose tissue54,67,68. Consistent with these findings, we observed that MOIs 50 and 100 of the active form, MOIs 10 and 50 of the inactivated form, both 50 and 100 μg/mL concentrations of OMVs, and cell-free supernatant could significantly increase the rate of PPARα transcriptome. The PPARβ mRNA level was almost increased by the same treatments significantly by the effects of MOI 50 of the active and MOI 10 of inactivated forms and OMV 100 μg/mL. The result received for PPARϒ was in the same direction as previous isotypes in such a way that mRNA level was increased significantly by MOIs 50 and 100 of the active and MOI 10 of the inactivated forms and both 50 and 100 μg/mL concentrations of OMVs. All these results confirmed the positive effects of A. muciniphila and related derivatives on the transcription of PPARs genes, probably through affecting fatty acids oxidation and energy metabolism.
In gut microbiota overgrowth conditions or tight junctions’ impairments, the integrity of the gut decreases, which leads to increasing the gut permeability and translocation of bacteria from the gut lumen to the portal and/or systemic circulation. In such conditions, the bacteria might enter the liver through the gut-liver axis11. In this study, MOI 50 of active A. muciniphila decreased the mRNA level of the CB1 receptor significantly in Hep-G2 cell lines; nevertheless, inactivated A. muciniphila, OMVs, and cell-free supernatant could not affect CB1 receptor transcription. Active A. muciniphila induces beneficial effects on hepatocytes through the downregulation of the CB1 receptor since Mallat et al. in 2013 explained that the CB1 receptor is expressed in hepatocytes and hepatic myofibroblasts and involved in numerous liver diseases, such as alcohol-induced liver disease, nonalcoholic fatty liver disease, fibrogenesis, and cardiovascular alterations associated with cirrhosis43.
Studies have shown that the expression of CB2 receptors in hepatocytes is modest, mainly contributes to hepatoprotective, anti-inflammatory, antioxidant, and immunomodulatory effects69,70, and plays a prohibiting role in liver fibrosis and alcohol-induced liver damage, compared to CB1 receptor28,43,71. In this study, neither active nor inactivated A. muciniphila could affect CB2 receptor expression; however, 100 μg/mL concentration of OMVs and cell-free supernatant noticeably increased the CB2 expression in mRNA level representing the more influence of the bacterium derivatives and metabolites rather than the bacterium itself. These results might have the root in A. muciniphila-derived OMVs and related cell-free supernatant that might preserve as therapeutic agents to improve liver health conditions. Moreover, it was observed that none of the treatments had an influence on FAAH expression except 100 μg/mL concentration of OMVs. This finding might be considered satisfactory since FAAH is a hydrolyzing enzyme for the ligands of both receptors and naturally balances the ECS-dependent health of the liver.
A. muciniphila produces SCFAs, such as propionate, butyrate, and acetate54 which can activate PPAR α expression through which prevents lipid accumulation in the liver72. The PPARα is mostly found in hepatocytes, plays a critical role in fatty acid uptake and fatty acid oxidation, decreases the production of very-low-density lipoprotein, and increases high-density lipoprotein. It also downregulates the hepatic inflammatory processes32. In the current study, MOI 10 of the active A. muciniphila, 100 μg/mL concentration of OMVs, and the-cell free supernatant considerably increased the PPARα expression in mRNA level. This result suggests that less amount of the active bacterium might promote PPARα expression better than higher amounts which might cause no inflammation in liver tissues. Similar results have been obtained for PPARβ/δ as it was observed that MOI 10 of the active A. muciniphila and 100 μg/mL concentration of OMVs significantly increased the mRNA level of PPARβ/δ.
Previous studies revealed that the potential role of PPARβ/δ in hepatocytes is apparent; they assumed that this gene is highly expressed in these cells and regulates glucose utilization and fatty acid metabolism. It also participates in the alleviation of inflammation and fibrosis32,72,73. Transcription analysis showed that A. muciniphila’s MOIs of 10 and 50 could enhance the PPARϒ transcriptome level, which is according to the results of studies of several researchers who concluded that gut microbiota is associated with metabolism, including PPARs expression related pathways72,74. Wagnerberger et al. in 2013 reported that intake of Lactobacillus casei upregulated hepatic PPARϒ leading to inhibition of Toll-like receptor 4 and suppression of steatosis75. Additionally, in previous studies in 2011, Nan et al. declared that the overexpression of PPARϒ can influence some liver diseases, such as reducing effect on steatosis, inflammation, and fibrosis in steatohepatitis murine model76. In a recent study, Keshavarz et al. reported that MOI 10 of heat-killed bacteria increased the expression of PPARϒ remarkably18. The effect of PPARϒ on fatty acid oxidation and glucose homeostasis as an insulin sensitizer was also lately reviewed by Wu et al.72.
In normal conditions, the bacterial derivatives might pass through the gut barrier either directly or via dynamin-dependent endocytosis and then translocate toward the liver52. In some leaky gut conditions, the bacteria might translocate from the gut to the liver through the gut-liver axis10,11. Some associations might be observed in the gene transcription of the cells in both organs. For example, as shown in Figs. 2 and 4, active A. muciniphila could decrease the level of CB1 transcription in both cell lines. Furthermore, OMVs treatments in both cell lines increased the FAAH mRNA level. The transcription of PPARα, β/δ, and ϒ was increased by active A. muciniphila in both cell lines (Figs. 3 and 5). The increased level of PPARα by the treatment of inactive bacteria and derived OMVs in Caco-2 cells was in line with HepG-2 cells (Figs. 3 and 5). A similar result was observed by the effect of OMVs treatments on PPARβ/δ transcription in two cell lines (Figs. 3 and 5). One contradiction was the effects of OMVs and cell-free supernatant treatments on the CB2 transcription level, which were contradictory in two cell lines. They might affect CB2 gene transcription with different mechanisms in the aforementioned cell lines, which requires more investigation.
The limitations of the current study were accomplishing these experiments in expression level and performing the same procedure under in vivo conditions, especially to investigate the correlation of gut and liver gene expressions affected by the aforementioned treatments.
In conclusion, we considered the positive effects of A. muciniphila and its derivatives, such as OMVs and bacterial metabolites, on controlling the activity of ECS compartments which might influence obesity, metabolic disorders, and liver diseases depending on their type. According to the present study results, A. muciniphila and its derivatives might be considered probiotic, paraprobiotic, and postbiotic candidates to protect organs against metabolic syndromes and liver diseases.
It is worth mentioning that there is an interaction between eCBs and PPARs genes which introduces some possible pathways of PPARs activation by eCBs either directly or indirectly41. Considering the essential roles of PPARs as nuclear receptors in the regulation of energy homeostasis, metabolism, cell differentiation, and inflammation77–80, the evidence suggests that numerous ECB functions, such as analgesic, neuroprotective, neuronal function modulation, anti-inflammatory, metabolic, anti-tumor, gastrointestinal, and cardiovascular effects of some cannabinoids are mediated by PPARs41. This association attracts the attention to scrutinize all possible pathways that might influence this study’s findings.
Supplementary Information
Acknowledgements
We would like to thank all of the patients who participated in the study.
Author contributions
F.GH.: clinical sample and data acquisition and performed the experiments; F.S., and Z.H.: analyzed data, interpreted data; A.F. and S.D.S.: designed and supervised clinical study, interpreted data, read and approved manuscript. All authors reviewed the manuscript.
Data availability
All data that support all the experimental findings in this article is available in the Supplementary Data File provided.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: In the original version of this Article, Fateh Abolfazl was incorrectly listed as a corresponding author. The correct corresponding author for this Article is Seyed Davar Siadat. Correspondence and request for materials should only be addressed to d.siadat@gmail.com.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/19/2022
A Correction to this paper has been published: 10.1038/s41598-022-16591-8
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-13840-8.
References
- 1.Ferrario C, et al. How to feed the mammalian gut microbiota: Bacterial and metabolic modulation by dietary fibers. Front. Microbiol. 2017;8:1749. doi: 10.3389/fmicb.2017.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turroni F, Ribbera A, Foroni E, van Sinderen D, Ventura M. Human gut microbiota and bifidobacteria: From composition to functionality. Antonie Van Leeuwenhoek. 2008;94:35–50. doi: 10.1007/s10482-008-9232-4. [DOI] [PubMed] [Google Scholar]
- 3.Monda V, et al. Exercise modifies the gut microbiota with positive health effects. Oxid. Med. Cell. Longev. 2017;2017:1–8. doi: 10.1155/2017/3831972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chelakkot C, et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018;50:e450–e450. doi: 10.1038/emm.2017.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wells JM, et al. Homeostasis of the gut barrier and potential biomarkers. Am. J. Physiol. Gastrointestinal Liver Physiol. 2017;312:G171–G193. doi: 10.1152/ajpgi.00048.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson LW, Artis D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 7.Piche T, et al. Impaired intestinal barrier integrity in the colon of patients with irritable bowel syndrome: Involvement of soluble mediators. Gut. 2009;58:196–201. doi: 10.1136/gut.2007.140806. [DOI] [PubMed] [Google Scholar]
- 8.Karl JP, et al. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am. J. Physiol. Gastrointestinal Liver Physiol. 2017;312:559–571. doi: 10.1152/ajpgi.00066.2017. [DOI] [PubMed] [Google Scholar]
- 9.de Magistris L, et al. Alterations of the intestinal barrier in patients with autism spectrum disorders and in their first-degree relatives. J. Pediatr. Gastroenterol. Nutr. 2010;51:418–424. doi: 10.1097/MPG.0b013e3181dcc4a5. [DOI] [PubMed] [Google Scholar]
- 10.Vajro P, Paolella G, Fasano A. Microbiota and gut-liver axis: A mini-review on their influences on obesity and obesity related liver disease. J. Pediatr. Gastroenterol. Nutr. 2013;56:461. doi: 10.1097/MPG.0b013e318284abb5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ilan Y. Leaky gut and the liver: A role for bacterial translocation in nonalcoholic steatohepatitis. World J. Gastroenterol: WJG. 2012;18:2609. doi: 10.3748/wjg.v18.i21.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashrafian F, Behrouzi A. Comparative study of effect of Akkermansia muciniphila and its extracellular vesicles on toll-like receptors and tight junction. Gastroenterol. Hepatol. Bed Bench. 2019;12:163. [PMC free article] [PubMed] [Google Scholar]
- 13.Derrien M, et al. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Microbiol. 2011;2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evolut. Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 15.Zhang T, Li Q, Cheng L, Buch H, Zhang F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019;12:1109–1125. doi: 10.1111/1751-7915.13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belzer C, De Vos WM. Microbes inside—from diversity to function: The case of Akkermansia. ISME J. 2012;6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 2007;73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keshavarz Azizi Raftar S, et al. The anti-fibrotic effects of heat-killed Akkermansia muciniphila MucT on liver fibrosis markers and activation of hepatic stellate cells. Probiot. Antimicrob. Proteins. 2021;13:776–787. doi: 10.1007/s12602-020-09733-9. [DOI] [PubMed] [Google Scholar]
- 19.Negi S, Das DK, Pahari S, Nadeem S, Agrewala JN. Potential role of gut microbiota in induction and regulation of innate immune memory. Front. Immunol. 2019;10:2441. doi: 10.3389/fimmu.2019.02441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fábrega MJ, et al. Activation of immune and defense responses in the intestinal mucosa by outer membrane vesicles of commensal and probiotic Escherichia coli strains. Front. Microbiol. 2016;7:705. doi: 10.3389/fmicb.2016.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmadi Badi S, et al. Microbiota-derived extracellular vesicles as new systemic regulators. Front. Microbiol. 2017;8:1610. doi: 10.3389/fmicb.2017.01610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang C-S, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS ONE. 2013;8:e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moradi, M., Tajik, H., Mardani, K. & Ezati, P. In Veterinary Research Forum. 193 (Faculty of Veterinary Medicine, Urmia University, Urmia, Iran).
- 24.Fateh A, et al. New insight into the application of outer membrane vesicles of Gram-negative bacteria. Vaccine Res. 2016;3:1–4. doi: 10.18869/acadpub.vacres.3.7.1. [DOI] [Google Scholar]
- 25.Cani PD, et al. Endocannabinoids—at the crossroads between the gut microbiota and host metabolism. Nat. Rev. Endocrinol. 2016;12:133. doi: 10.1038/nrendo.2015.211. [DOI] [PubMed] [Google Scholar]
- 26.Maccarrone M, et al. Endocannabinoid signaling at the periphery: 50 years after THC. Trends Pharmacol. Sci. 2015;36:277–296. doi: 10.1016/j.tips.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiPatrizio NV. Endocannabinoids in the gut. Cannabis Cann. Res. 2016;1:67–77. doi: 10.1089/can.2016.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basu P, Aloysius M, Shah N, Brown R., Jr The endocannabinoid system in liver disease, a potential therapeutic target. Aliment. Pharmacol. Ther. 2014;39:790–801. doi: 10.1111/apt.12673. [DOI] [PubMed] [Google Scholar]
- 29.Howlett, A. Cannabinoid receptor signaling. Cannabinoids, 53–79 (2005). [DOI] [PubMed]
- 30.Zou S, Kumar U. Cannabinoid receptors and the endocannabinoid system: Signaling and function in the central nervous system. Int. J. Mol. Sci. 2018;19:833. doi: 10.3390/ijms19030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy KL. Interaction between cannabinoid system and toll-like receptors controls inflammation. Mediat. Inflamm. 2016;2016:1–18. doi: 10.1155/2016/5831315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bazwinsky-Wutschke I, Zipprich A, Dehghani F. Endocannabinoid system in hepatic glucose metabolism, fatty liver disease, and cirrhosis. Int. J. Mol. Sci. 2019;20:2516. doi: 10.3390/ijms20102516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Argueta DA, Perez PA, Makriyannis A, DiPatrizio NV. Cannabinoid CB1 receptors inhibit gut-brain satiation signaling in diet-induced obesity. Front. Physiol. 2019;10:704. doi: 10.3389/fphys.2019.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hotamisligil GS, Davis RJ. Cell signaling and stress responses. Cold Spring Harb. Perspect. Biol. 2016;8:a006072. doi: 10.1101/cshperspect.a006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chanda D, et al. Activation of cannabinoid receptor type 1 (Cb1r) disrupts hepatic insulin receptor signaling via cyclic AMP-response element-binding protein H (Crebh)-mediated induction of Lipin1 gene. J. Biol. Chem. 2012;287:38041–38049. doi: 10.1074/jbc.M112.377978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Gottardi A, Spahr L, Ravier-Dall'Antonia F, Hadengue A. Cannabinoid receptor 1 and 2 agonists increase lipid accumulation in hepatocytes. Liver Int. 2010;30:1482–1489. doi: 10.1111/j.1478-3231.2010.02298.x. [DOI] [PubMed] [Google Scholar]
- 37.Reichenbach V, et al. Prevention of fibrosis progression in CCl4-treated rats: Role of the hepatic endocannabinoid and apelin systems. J. Pharmacol. Exp. Ther. 2012;340:629–637. doi: 10.1124/jpet.111.188078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 39.Wahli W, Michalik L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol. Metab. 2012;23:351–363. doi: 10.1016/j.tem.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Hasan AU, Rahman A, Kobori H. Interactions between host PPARs and gut microbiota in health and disease. Int. J. Mol. Sci. 2019;20:387. doi: 10.3390/ijms20020387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Sullivan SE. An update on PPAR activation by cannabinoids. Br. J. Pharmacol. 2016;173:1899–1910. doi: 10.1111/bph.13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veilleux A, Di Marzo V, Silvestri C. The expanded endocannabinoid system/endocannabinoidome as a potential target for treating diabetes mellitus. Curr. Diab.Rep. 2019;19:1–12. doi: 10.1007/s11892-019-1248-9. [DOI] [PubMed] [Google Scholar]
- 43.Mallat A, Teixeira-Clerc F, Lotersztajn S. Cannabinoid signaling and liver therapeutics. J. Hepatol. 2013;59:891–896. doi: 10.1016/j.jhep.2013.03.032. [DOI] [PubMed] [Google Scholar]
- 44.Mallat A, Teixeira-Clerc F, Deveaux V, Manin S, Lotersztajn S. The endocannabinoid system as a key mediator during liver diseases: New insights and therapeutic openings. Br. J. Pharmacol. 2011;163:1432–1440. doi: 10.1111/j.1476-5381.2011.01397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elhenawy W, Debelyy MO, Feldman MF. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. MBio. 2014 doi: 10.1128/mBio.00909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shanson D, Singh J. Effect of adding cysteine to brain-heart infusion broth on the isolation of Bacteroides fragilis from experimental blood cultures. J. Clin. Pathol. 1981;34:221–223. doi: 10.1136/jcp.34.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Badi SA, Khatami S, Irani S, Siadat SD. Induction effects of bacteroides fragilis derived outer membrane vesicles on toll like receptor 2, toll like receptor 4 genes expression and cytokines concentration in human intestinal epithelial cells. Cell J. (Yakhteh) 2019;21:57. doi: 10.22074/cellj.2019.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Claassen I, et al. Production, characterization and control of a Neisseria meningitidis hexavalent class 1 outer membrane protein containing vesicle vaccine. Vaccine. 1996;14:1001–1008. doi: 10.1016/0264-410X(96)00020-5. [DOI] [PubMed] [Google Scholar]
- 49.Ashrafian F, et al. Akkermansia muciniphila-derived extracellular vesicles as a mucosal delivery vector for amelioration of obesity in mice. Front. Microbiol. 2019;10:2155. doi: 10.3389/fmicb.2019.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yaghoubfar R, et al. Effect of Akkermansia muciniphila, Faecalibacterium prausnitzii, and their extracellular vesicles on the serotonin system in intestinal epithelial cells. Probiot. Antimicrob. Prot. 2021;13:1–11. doi: 10.1007/s12602-020-09640-z. [DOI] [PubMed] [Google Scholar]
- 51.Chatterjee D, Chaudhuri K. Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett. 2011;585:1357–1362. doi: 10.1016/j.febslet.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 52.Jones EJ, et al. The uptake, trafficking, and biodistribution of Bacteroides thetaiotaomicron generated outer membrane vesicles. Front. Microbiol. 2020;11:57. doi: 10.3389/fmicb.2020.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McPartland JM, Guy GW, Di Marzo V. Care and feeding of the endocannabinoid system: A systematic review of potential clinical interventions that upregulate the endocannabinoid system. PLoS ONE. 2014;9:e89566. doi: 10.1371/journal.pone.0089566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y, et al. Function of Akkermansia muciniphila in obesity: Interactions with lipid metabolism, immune response and gut systems. Front. Microbiol. 2020;11:219. doi: 10.3389/fmicb.2020.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pascale A, Marchesi N, Govoni S, Coppola A, Gazzaruso C. The role of gut microbiota in obesity, diabetes mellitus, and effect of metformin: new insights into old diseases. Curr. Opin. Pharmacol. 2019;49:1–5. doi: 10.1016/j.coph.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 56.Everard A, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russo EB. CANNABIS and CANNABINOIDS-Clinical Endocannabinoid Deficiency (CECD): Can this concept explain therapeutic benefits of cannabis in migraine, fibromyalgia, irritable bowel syndrome and other. Neuroendocrinol. Lett. 2004;25:31–39. [PubMed] [Google Scholar]
- 58.Gyires K, Zádori SZ. Role of cannabinoids in gastrointestinal mucosal defense and inflammation. Curr. Neuropharmacol. 2016;14:935–951. doi: 10.2174/1570159X14666160303110150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jansma J, Brinkman F, van Hemert S, El Aidy S. Targeting the endocannabinoid system with microbial interventions to improve gut integrity. Prog. Neuro-Psychopharmacol. Biol. Psychiat. 2020;106:110169. doi: 10.1016/j.pnpbp.2020.110169. [DOI] [PubMed] [Google Scholar]
- 60.Rousseaux C, et al. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat. Med. 2007;13:35–37. doi: 10.1038/nm1521. [DOI] [PubMed] [Google Scholar]
- 61.Scarpelli R, Sasso O, Piomelli D. A double whammy–targeting both fatty acid amide hydrolase (FAAH) and cyclooxygenase (COX) to treat pain and inflammation. ChemMedChem. 2016;11:1242. doi: 10.1002/cmdc.201500395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reunanen J, et al. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl. Environ. Microbiol. 2015;81:3655–3662. doi: 10.1128/AEM.04050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murakami T, et al. Antibacterial cathelicidin peptide CAP11 suppresses the anandamide production from lipopolysaccharide-stimulated mononuclear phagocytes. FEBS Lett. 2007;581:140–144. doi: 10.1016/j.febslet.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 64.Decara J, et al. Peroxisome proliferator-activated receptors: Experimental targeting for the treatment of inflammatory bowel diseases. Front. Pharmacol. 2020;11:730. doi: 10.3389/fphar.2020.00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goto T, et al. Activation of peroxisome proliferator-activated receptor-alpha stimulates both differentiation and fatty acid oxidation in adipocytes [S] J. Lipid Res. 2011;52:873–884. doi: 10.1194/jlr.M011320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y-X, et al. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity. Cell. 2003;113:159–170. doi: 10.1016/S0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 67.Ottman N, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS ONE. 2017;12:e0173004. doi: 10.1371/journal.pone.0173004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lukovac S, et al. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio. 2014;5:e01438–e11414. doi: 10.1128/mBio.01438-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hashiesh HM, et al. A focused review on CB2 receptor-selective pharmacological properties and therapeutic potential of β-caryophyllene, a dietary cannabinoid. Biomed. Pharmacother. 2021;140:111639. doi: 10.1016/j.biopha.2021.111639. [DOI] [PubMed] [Google Scholar]
- 70.Bie B, Wu J, Foss JF, Naguib M. An overview of the cannabinoid type 2 (CB2) receptor system and its therapeutic potential. Curr. Opin. Anaesthesiol. 2018;31:407. doi: 10.1097/ACO.0000000000000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trebicka J, et al. Role of cannabinoid receptors in alcoholic hepatic injury: Steatosis and fibrogenesis are increased in CB2 receptor-deficient mice and decreased in CB1 receptor knockouts. Liver Int. 2011;31:860–870. doi: 10.1111/j.1478-3231.2011.02496.x. [DOI] [PubMed] [Google Scholar]
- 72.Wu L, et al. Crosstalk between PPARs and gut microbiota in NAFLD. Biomed. Pharmacother. 2021;136:111255. doi: 10.1016/j.biopha.2021.111255. [DOI] [PubMed] [Google Scholar]
- 73.Liu S, et al. Role of peroxisome proliferator-activated receptor δ/β in hepatic metabolic regulation. J. Biol. Chem. 2011;286:1237–1247. doi: 10.1074/jbc.M110.138115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding J, et al. Divergent selection-induced obesity alters the composition and functional pathways of chicken gut microbiota. Genet. Sel. Evol. 2016;48:1–9. doi: 10.1186/s12711-016-0270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wagnerberger S, et al. Lactobacillus casei Shirota protects from fructose-induced liver steatosis: A mouse model. J. Nutr. Biochem. 2013;24:531–538. doi: 10.1016/j.jnutbio.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 76.Nan Y-M, et al. Adenovirus-mediated peroxisome proliferator activated receptor gamma overexpression prevents nutritional fibrotic steatohepatitis in mice. Scand. J. Gastroenterol. 2011;46:358–369. doi: 10.3109/00365521.2010.525717. [DOI] [PubMed] [Google Scholar]
- 77.Friedland SN, et al. The cardiovascular effects of peroxisome proliferator-activated receptor agonists. Am. J. Med. 2012;125:126–133. doi: 10.1016/j.amjmed.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 78.Menendez-Gutierrez MP, Roszer T, Ricote M. Biology and therapeutic applications of peroxisome proliferator-activated receptors. Curr. Topics Med. Chem. 2012;12:548–584. doi: 10.2174/156802612799436669. [DOI] [PubMed] [Google Scholar]
- 79.Neher MD, Weckbach S, Huber-Lang MS, Stahel PF. New insights into the role of peroxisome proliferator-activated receptors in regulating the inflammatory response after tissue injury. PPAR Res. 2012;2012:1–13. doi: 10.1155/2012/728461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.la Cour Poulsen, L., Siersbæk, M. & Mandrup, S. In Seminars in cell & developmental biology. 631–639 (Elsevier). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support all the experimental findings in this article is available in the Supplementary Data File provided.