Abstract
The interspecific variability in the sensitivity of marine bacterial isolates to UV-B (295- to 320-nm) radiation and their ability to recover from previous UV-B stress were examined. Isolates originating from different microenvironments of the northern Adriatic Sea were transferred to aged seawater and exposed to artificial UV-B radiation for 4 h and subsequently to different radiation regimens excluding UV-B to determine the recovery from UV-B stress. Bacterial activity was assessed by thymidine and leucine incorporation measurements prior to and immediately after the exposure to UV-B and after the subsequent exposure to the different radiation regimens. Large interspecific differences among the 11 bacterial isolates were found in the sensitivity to UV-B, ranging from 21 to 92% inhibition of leucine incorporation compared to the bacterial activity measured in dark controls and from 14 to 84% for thymidine incorporation. Interspecific differences in the recovery from the UV stress were also large. An inverse relation was detectable between the ability to recover under dark conditions and the recovery under photosynthetic active radiation (400 to 700 nm). The observed large interspecific differences in the sensitivity to UV-B radiation and even more so in the subsequent recovery from UV-B stress are not related to the prevailing radiation conditions of the microhabitats from which the bacterial isolates originate. Based on our investigations on the 11 marine isolates, we conclude that there are large interspecific differences in the sensitivity to UV-B radiation and even larger differences in the mechanisms of recovery from previous UV stress. This might lead to UV-mediated shifts in the bacterioplankton community composition in marine surface waters.
UV radiation can penetrate to considerable depth in the water column. In the subtropical open Atlantic, the 10% radiation level for 340- and 380-nm wavelength is at depths of 35 and 60 m, respectively (27). The shortest-wavelength fraction of UV reaching the Earth's surface, the UV-B range (295 to 320 nm), has been found to impair organisms and consequently affect the carbon and energy flow through the aquatic food web (for a review, see the work of Karentz et al. [16]). The plankton organisms which are most severely affected by UV radiation are bacterioplankton (13). In the phytoplankton size fraction (>0.8 μm), significantly less damage was found than in the bacterioplankton fraction (<0.8 μm) (13). However, UV not only affects organisms but also photochemically alters dissolved organic matter (DOM). Photolytic cleavage of DOM by UV produces a whole suite of compounds (17, 18, 30, 31, 35, 36). Some of these cleavage products (i.e., low-molecular-weight organic acids) are taken up efficiently by the bacterioplankton, leading to enhanced bacterial production (22, 23, 29), while some other photoproducts, like free radicals, have detrimental effects on the plankton organisms (10, 28, 35).
In a recent study, it has been shown that bacterioplankton are severely inhibited by solar radiation (15). Furthermore, this inhibition is more pronounced if bacterioplankton are separated from the phytoplankton community (34), indicating some compensatory effect on bacterial metabolism induced by phytoplankton activity. Bacterioplankton communities are severely inhibited by UV, but they also efficiently recover from UV stress (15). This recovery from the previous UV stress is higher under irradiation conditions where UV-B has been excluded than in the dark (15). It has been concluded, therefore, that the photoenzymatic repair of DNA damage is more important for the bulk bacterioplankton than the dark repair. This photoenzymatic repair is activated by the longer UV-A wavelength range (360 to 400 nm) and by blue light (400 to 430 nm) (8). Due to the differential attenuation of UV in the water column (15), bacterioplankton mixed into deeper layers are exposed to an altered radiation environment, with damaging UV-B being attenuated more rapidly than UV-A and the blue light range, which are responsible for recovery. Phytoplankton frequently synthesize UV-absorbing pigments to shield themselves from UV-B; however, this has not been observed with bacterioplankton, which therefore rely exclusively on repairing damage caused by UV. It has been shown that bacterioplankton efficiently repair the UV-induced damage (15). This conclusion, however, is based on bulk measurements of metabolic activity for natural bacterioplankton communities.
Differences in the sensitivity to UV-B or in the repair efficiency of the UV-induced damage among different bacterioplankton species could result in a shift in the activity pattern and ultimately in the species distribution of bacterioplankton species. Recently, Joux et al. (14) showed that there are substantial interspecific differences in the survival and recovery of five bacterial strains subsequent to UV-B radiation. These authors monitored the time course of the formation of cyclobutane pyrimidine dimers upon exposure to UV-B radiation and determined bacterial growth after exposure to UV-B radiation during the subsequent recovery phase. In this study, we focused on the quantification of biomass synthesis (measured via leucine incorporation) and cell division (via thymidine incorporation) as influenced by UV-B stress and after recovery.
To determine whether there are pronounced interspecific differences in the sensitivity and recovery from UV stress in marine bacteria, we performed experiments similar to those described in the work of Kaiser and Herndl (15) for natural bacterial assemblages. We used only artificial light sources and 11 bacterial isolates which we isolated from the northern Adriatic Sea to determine whether interspecific differences in the sensitivity to UV and in the ability to recover from UV stress might lead to alterations in the bacterial community composition.
MATERIALS AND METHODS
Experimental approach.
The interspecific variability of UV-induced inhibition of activity and the influence of different wavelength ranges on the recovery of selected marine bacterial isolates following UV-B stress were determined in laboratory experiments using artificial radiation. Heterotrophic bacterial isolates (culture conditions are described below) were harvested from the medium in their exponential growth phase. Subsequently, they were inoculated in aged, filtered (0.2-μm-pore-size polycarbonate Millipore filters, 47-mm diameter), autoclaved (121°C for 30 min), and irradiated seawater (same radiation intensity as that in the subsequent bacterial incubation, for 12 h). The seawater was irradiated in order to minimize the possible stimulatory effects of photoreactivated dissolved organic carbon in the treatments exposed to UV-B radiation compared to the dark treatments. The dissolved organic carbon content of the aged, irradiated seawater was ≈75 μM C. Each bacterial isolate was inoculated in a 1-liter quartz tube at an initial abundance of ≈106 cells ml−1 and allowed to acclimate for 10 h in the dark before starting the experiments. The quartz tubes were closed at both ends with an autoclaved Teflon-lined silicone stopper. For each isolate, the initial bacterial activity was measured (described below) and the quartz tubes were exposed to artificial UV-B radiation (wavelength range, 300 to 320 nm; 0.4 W m−2; Philips TL 100 W/01 lamps) in a temperature-controlled water bath (20°C) for 4 h to mimic the dose received by bacterioplankton in the surface layers of the ocean. A dark control treated exactly in the same way as the UV-B-exposed sample was wrapped in aluminum foil. After exposure to UV-B, the bacterial activity was measured for each isolate; subsequently, the sample was split, distributed equally into smaller quartz tubes (50 ml), and exposed to different radiation regimens. After 4 h, the bacterial activity of each isolate exposed to the different radiation regimens was assessed again. This 4-h exposure period to different radiation regimens was chosen since preliminary experiments showed that this period is long enough to discern differences in the recovery from previous UV-B stress. Longer incubation times (>1 day) would lead to an artificially high dose compared to in situ exposure conditions. The different light conditions used were UV-A plus photosynthetic active radiation (PAR; 400 to 700 nm; this condition was made by wrapping the quartz tubes in Mylar-D foil to exclude UV-B; 50% transmittance at 320 nm), PAR (condition created by wrapping the tubes in vinyl chloride foil [CI Kasei Co., Tokyo, Japan; 50% transmittance at 405 nm]), or darkness (condition created by wrapping tubes in aluminum foil under the same conditions). UV-A was provided by Philips TL 100 W/10R lamps (wavelength range, 350 to 400 nm; 0.25 W m−2). The lower intensity of UV-A than UV-B was meant to simulate the lower radiation levels in deeper layers of the upper water column. PAR was provided by white cool lamps (80 microeinsteins m−2 s−1, Philips TLD 58 W/84) corresponding to the UV-A radiation regimen found in layers below 30 m in the oligotrophic open ocean and at 5 to 10 m in oligotrophic to mesotrophic coastal waters (27).
The light sources used in this investigation closely resembled natural near-surface (<5-m depth) solar radiation only in the wavelength range from 300 to 320 nm (3). The intensity of the UV-A and the PAR range was chosen to mimic the radiation intensity at a 5- to 10-m depth in coastal marine systems (15). Therefore, we simulated the diurnal mixing event after the breakup of the diurnal thermocline (7) when the near-surface water layers are mixed with the underlying waters. Although the radiation intensity was similar to that of natural solar radiation of different depth layers of the water column, the spectral composition was different from that of solar radiation, especially in the UV-A and the PAR range, where narrow bands prevailed in our radiation setup. In the UV-A range, no significant fluence rate was provided by our lamp system in the 380- to 400-nm range. This wavelength range is partly responsible for inducing photoenzymatic repair in bacteria (8, 15). Our study, however, was not meant to determine UV-induced damage and recovery under ideal natural conditions but focused rather on the interspecific response of selected bacterial isolates from different marine environments to UV stress and the ability to recover from this stress.
All the experiments were performed in triplicate at 20 ± 1°C.
Isolation of marine bacterial strains.
All samples from which bacterial strains were subsequently isolated were collected in the northern Adriatic Sea about 1 km off Rovinj (Croatia). Marine snow (from a 10- to 20-m depth), the top 0 to 2 mm of the sandy sediment (≈25-m depth), and ambient water (from a 10- to 20-m depth) were collected by scuba divers with disposable syringes (12-ml capacity). Upon return to the lab (Center for Marine Research, Ruder Boskovic Institute, Rovinj, Croatia), the samples (100 μl of seawater, one marine snow particle, or 100 μl of the sediment slurry) were spread on agar plates (1.5% [wt/vol] agar) containing ZoBell 2216E medium (5 g of peptone, 1 g of yeast extract, in 1 liter of 0.45-μm-pore-size-filtered seawater collected from the same site and autoclaved at 121°C for 30 min). After incubation in the dark at 20°C for 2 to 7 days, isolates were picked from the plates according to differences in color and shape of the colony and transferred into liquid ZoBell medium for molecular characterization (described below). The strains were named according to their origin (W for ambient water, S for sediment, and MS for marine snow). The closest relatives and the phylogenetic positions of the bacterial isolates used in this study are given in Table 1.
TABLE 1.
Bacterial isolates used in the experiments, their closest relatives,their phylogenetic affiliations (group), and the percentages of similarity to the closest relativea
| Strain | Closest relative | Group | % Similarity |
|---|---|---|---|
| MS1 | Pseudoalteromonas sp. strain S9 | γ | 93.9 |
| MS2 | Chromohalobacter marismortui | γ | 95.6 |
| MS3 | Pseudoalteromonas carrageenovora | γ | 99 |
| W5-2 | Vibrio fluvialis | γ | 100 |
| W5-3 | Vibrio mytili | γ | 98 |
| W5-4 | Vibrio sp. strain NAP-4 | γ | 98 |
| S3 | Vibrio shiloi | γ | 98 |
| S1 | Uncultured marine eubacterium Vibrio sp. strain HstpLK41 | γ | 98 |
| S8 | Shewanella gelidimarina | γ | 98 |
| W5-5 | Uncultured Micrococcus clone MT2 | gram+ | 98 |
| MS20 | Planococcus citreus | gram+ | 96 |
MS, strains isolated from marine snow; S, strains isolated from the superficial layer of the sediment; W, strains isolated form the water column. All strains were isolated from the northern Adriatic Sea. The phylogenetic affiliation is based on the comparison of 409 to 439 nucleotides. gram+, gram positive.
Molecular characterization of the strains.
Cells were collected from stationary-phase cultures in liquid ZoBell medium by centrifugation (3,200 × g, 15 min), resuspended in 2 ml of lysis buffer (400 mM NaCl, 750 mM sucrose, 20 mM EDTA, 50 mM Tris-HCl, pH 9.0), and incubated with lysozyme (final concentration, 1 mg ml−1; Merck, Darmstadt, Germany) at 37°C for 30 min. Sodium dodecyl sulfate (final concentration, 1% [wt/vol]; Sigma, St. Louis, Mo.) and proteinase K (final concentration, 100 mg ml−1; Boehringer Mannheim Biochemicals B.V., Almere, The Netherlands) were added, and the samples were incubated at 55°C for 120 min. Subsequently, the lysate was extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and an equal volume of chloroform-isoamyl alcohol (24:1). The nucleic acids were precipitated by addition of sodium acetate (pH 5.2; final concentration, 0.2 M; Sigma) and 2.5 volumes of 100% ethanol (Fluka, Buchs, Switzerland), followed by overnight storage at −20°C and centrifugation (20,000 × g, 15 min). The pellet was washed twice with 70% ethanol, air dried, and dissolved in 50 μl of autoclaved 0.2-μm-pore-size filtered double-distilled water.
DNA encoding 16S rRNA was amplified from the extract by PCR with Taq polymerase (Pharmacia Biotech, Uppsala, Sweden) using bacterial ribosomal DNA primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) in 50-μl PCR mixtures containing both primers at 0.2 mM, 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, and 200 mM deoxynucleoside triphosphate (Pharmacia Biotech). The PCR conditions were as follows: initial denaturation step of 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. Cycling was completed by a final extension at 72°C for 7 min. The PCR products were purified from an agarose gel using the Qiaex gel extraction kit (Qiagen, Hilden, Germany), and nucleotide sequences were determined by automated sequencing with a BigDye Terminator Cycle Sequencing kit (Perkin-Elmer) with primer 27F in an ABI 310 sequencer (Perkin-Elmer).
The nucleotide sequences obtained were compared to known sequences of the GenBank database by using the gapped BLAST search algorithm (2, 4) and aligned with the closest relatives in terms of nucleotide sequence similarity and RNA secondary structure using the ARB software package (24).
Preparation of the isolates for the experiments.
Bacterial isolates were grown in liquid ZoBell medium (1-liter Erlenmeyer flasks) on a laboratory shaker at 20°C. Growth was monitored by measuring the optical density at 650 nm with a spectrophotometer every 2 h. In the late exponential phase, cells were harvested by centrifugation (3,200 × g for 15 min), and the pellet was resuspended in 0.2-μm-pore-size-filtered, autoclaved seawater and centrifuged again. This procedure was repeated three times to remove all traces of nutrients originating from the culture medium. Thereafter, the cells were suspended in 5 ml of 0.2-μm-pore-size-filtered, autoclaved seawater, the bacterial abundance was determined as described below, and thereafter the bacterial abundance in the quartz tube was adjusted with autoclaved, irradiated seawater to ≈106 cells ml−1.
Determination of bacterial abundance and activity.
The bacterial abundance was estimated by direct counting of the bacterial cells collected on polycarbonate filters (0.2-μm pore size; Millipore) using epifluorescence microscopy on acridine orange-stained samples (12). Bacterial activity was estimated by incorporation of [3H]thymidine and [3H]leucine (each at 20 nM final concentration) in triplicate subsamples (5 ml) and two formaldehyde-killed blanks (2% final concentration) using the methods described elsewhere (9, 19, 21). All the incubations were performed in the dark and were terminated with formaldehyde (2% final concentration) after 30 min. Subsequently, the samples were filtered onto 0.45-μm-pore-size cellulose nitrate filters (Millipore HA; 25-mm filter diameter) and rinsed with 5 ml of ice-cold 5% trichloroacetic acid and 5 ml of distilled water. The filters were transferred to scintillation vials and dissolved in 1 ml of ethylacetate (Riedel-de Haën), and 8 ml of scintillation cocktail (Insta-Gel; Canberra-Packard) was added. The radioactivity was measured in a liquid scintillation counter (Packard Tri-Carb 2000) using the external standard ratio technique.
RESULTS AND DISCUSSION
Interspecific differences of UV-B-mediated inhibition of the metabolic activity.
Generally, our results agree with recent findings that there is considerable interspecific variability in the sensitivity to UV stress among different marine bacterial isolates (14). These authors measured the formation of cyclobutane pyrimidine dimers and therefore quantified the DNA damage caused by UV radiation and monitored the survival of the bacterial strains by growing them in rich medium. In contrast to their study, we measured the effect of UV stress via the incorporation of thymidine and leucine. Thymidine is incorporated into bacterial DNA and leucine is incorporated into bacterial protein; thus, we measured cell division and biomass production (9, 19, 33). These two measurements of bacterial activity are usually closely correlated in natural bacterioplankton assemblages (20), although phasing of bacterial growth has been reported to occur occasionally in marine environments, leading to large deviations between these two measurements (6). As the synthesis of new DNA and biomass can be uncoupled by UV-induced damage (26), measuring both thymidine and leucine incorporation is necessary to monitor the metabolic activities of bacteria.
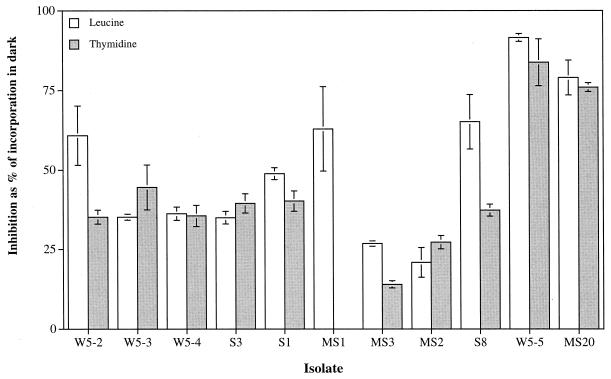
Exposure of the bacterial isolates to artificial UV-B radiation for 4 h resulted in decreased bacterial thymidine and leucine incorporation rates in the UV-B-exposed treatments compared to the rates for the controls held in the dark (Fig. 1). Large interspecific differences in the inhibition of bacterial activity after exposure to UV-B were detected, ranging from 20.9 to 91.5% inhibition of leucine incorporation rates and from 14.1 to 83.7% inhibition of thymidine incorporation rates compared to the activity measured in the dark treatment (Fig. 1). The five different isolates belonging to the genus Vibrio (W5-2, W5-3, W5-4, S3, and S1) exhibited a similar inhibition in activity subsequent to exposure to UV-B radiation (mean ± standard deviation [SD], [39 ± 3.8]% for thymidine and [43.2 ± 11.5]% for leucine). The highest sensitivity to UV-B radiation (>80% inhibition of leucine and thymidine incorporation compared to the dark treatment) was detected for the two gram-positive relatives of Micrococcus sp. (W5-5) and Planococcus citreus (MS20) (Fig. 1). No relation was found between UV sensitivity of the isolates and the environments from which they were isolated, supporting earlier findings that bacterioplankton from environments exposed to high natural UV radiation are not specifically adapted to UV radiation (11, 25). The patterns of inhibition of thymidine incorporation closely corresponded to that observed for leucine incorporation, and percentages of inhibition derived from these two incorporation measurements deviated from each other, on average, by only 10.7%.
FIG. 1.
Interspecific variation in the percentage of UV-B-mediated inhibition of thymidine and leucine incorporation of selected bacterial isolates originating from different environments of the northern Adriatic Sea (Table 1) compared to corresponding dark incubations. Samples were exposed to artificial UV-B radiation for 4 h. Symbols indicate the means ± SDs of triplicate measurements.
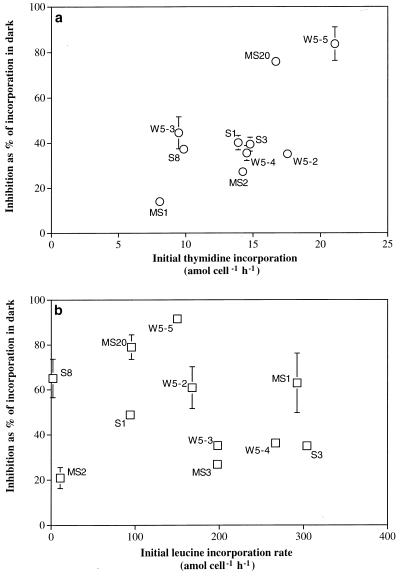
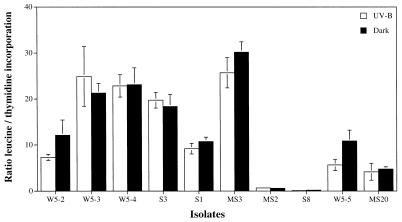
The rate of thymidine incorporation of the different isolates measured before the exposure to UV-B correlated with the percentage of inhibition detectable after exposure to UV-B for 4 h (Fig. 2a), indicating that those cells dividing faster are also most inhibited in their cell division. No such relation, however, was detectable for leucine incorporation (Fig. 2b). Again, no tendency was discernible for strains isolated from a particular environment. Although both thymidine and leucine incorporation rates declined upon UV-B exposure (Fig. 1), there were some differences detectable in the uptake rates of thymidine and leucine, as shown in Fig. 2. This becomes particularly obvious if the molar ratio between leucine and thymidine incorporation is calculated for the individual isolates. The molar ratio of leucine versus thymidine incorporation of the tested bacterial isolates was generally within the range reported elsewhere for bulk bacterioplankton communities (5, 32). Some very low ratios, however, were obtained for a close relative of Chromohalobacter marismortui (MS2) and for Shewanella gelidimarina (S8) isolated from marine snow and from the surface sediment layer, respectively (Fig. 3). Despite large interspecific differences in the leucine-to-thymidine incorporation ratios ranging from 0.2 to 30.2 in the dark incubations (Fig. 3), only 4 out of 10 isolates exhibited slightly, although significantly, lower ratios (Wilcoxon sum rank test, P < 0.05) after UV-B exposure compared to incorporation in the dark (Fig. 3), indicating that both DNA and protein synthesis were impaired to the same extent. No distinct response pattern for the different taxa was detectable, although the interspecific variability in the leucine/thymidine ratios within the genus Vibrio was lower than that for the other isolates belonging to different genera.
FIG. 2.
Relation between the percentage of inhibition of thymidine (a) and leucine (b) incorporation in bacterial isolates due to UV-B exposure for 4 h (compared to the corresponding dark incubation) and the initial incorporation rates prior to exposure to UV-B. Error bars indicate the SDs of triplicate measurements; where they are missing, the SD is smaller than the symbol.
FIG. 3.
Molar ratio of leucine to thymidine incorporation of bacterial isolates after exposure to UV-B for 4 h or after being held in the dark. Error bars indicate the SDs of triplicate measurements.
Interspecific differences in the recovery of bacterial isolates from UV stress under different radiation conditions.
Following 4 h of exposure to UV-B radiation, the samples were split and exposed to different radiation regimens (UV-A, PAR plus UV-A, and dark) for another 5 h, and subsequently, the thymidine and leucine incorporation was measured again. In previous work, it has been found that bacterioplankton activity is reduced when they are directly exposed to UV radiation (1, 11, 25). However, this UV-induced reduction of the bacterial activity might be compensated for in coastal water by UV-mediated photolysis of the DOM pool, leading to the formation of labile DOM. Fractions of the photolyzed DOM are readily taken up by bacterioplankton after relief from the UV stress (15, 27). To reduce this interference of photolysis of DOM, all the water used in these experiments was previously irradiated (see Materials and Methods); thus, the recovery of the bacterial isolates observed in these experiments is not the result of different DOM quality.
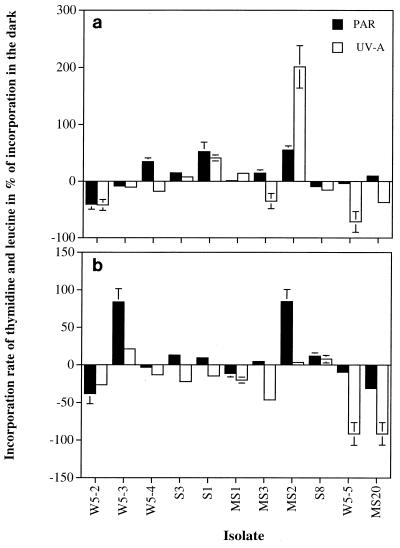
Recovery of bacterial isolates was stimulated by UV-A and PAR in 6 out of 11 isolates for thymidine incorporation (Fig. 4a) and in 3 out of the 11 isolates for leucine incorporation (Fig. 4b). Considerable differences in the magnitude of the recovery were generally observable between thymidine and leucine incorporation for the different isolates, leading to an uncoupling of the biomass and DNA synthesis rates (Fig. 4). In contrast to previous findings for marine communities (15), the UV-A treatment resulted in an enhancement of thymidine and leucine incorporation in only four and three of the isolates tested, respectively. This might be due to the spectral composition of our light source, where no significant fluence rate was provided in the 380- to 400-nm range (see Materials and Methods). Despite that, a relative of C. marismortui, MS2, exhibited twofold-higher thymidine incorporation rates after UV-A exposure than after the dark treatment. Significantly enhanced thymidine incorporation was also observed for three other strains (Fig. 4). The two gram-positive relatives of Micrococcus sp. (W5-5) and P. citreus (MS20), however, recovered more efficiently from the previous UV stress under dark conditions as revealed by thymidine as well as leucine incorporation (except for a slightly higher thymidine incorporation in P. citreus under PAR [Fig. 4a]).
FIG. 4.
Percentage of recovery of thymidine (a) and leucine (b) incorporation following incubation under PAR and UV-A radiation for 4 h compared to the respective incorporation rate measured after incubation in the dark for 4 h following UV-B exposure. The response measured in the PAR treatment was subtracted from that measured in the PAR plus UV-A treatment to distinguish between the photoreactivation induced by UV-A and that induced by PAR.
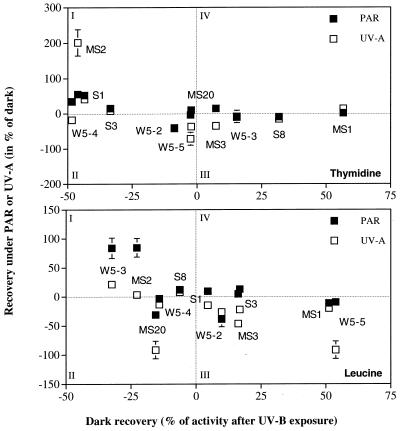
An apparent relationship was detectable between the ability to recover under dark conditions and the extent of recovery in the presence of PAR (Fig. 5). To illustrate this, we plotted the recovery under PAR or UV-A as a percentage of the dark treatment, against the recovery in the dark as a percentage after UV-B exposure (Fig. 5). We divided the plot into four areas according to the metabolic response: I, isolates recovering under the light treatment (UV-A or PAR) but continuing to lose metabolic activity in the dark; II, strains that did not exhibit recovery under either light or dark conditions; III, strains that showed the ability to recover in the dark but were inhibited by the light treatments (PAR or UV-A); and IV, strains able to recover under both dark and light conditions. Most of the isolates fell into categories I and III, for both leucine and thymidine incorporation, indicating that bacterial isolates significantly recovering under PAR or UV-A poorly recovered under dark conditions and vice versa. The most efficient recovery in the dark was observed for the isolate MS1 (a relative of Pseudoalteromonas sp.) while a close relative of C. marismortui, MS2, showed a very high efficiency in recovering from UV stress under UV-A as well as PAR but did not recover in the dark. There was no strain falling in category II for both leucine and thymidine incorporation. This indicates that all the tested strains showed the ability to enhance their metabolic rates after UV damage under either light or dark conditions. The most UV-sensitive isolate tested in this study was the gram-positive relative of P. citreus MS20 (Fig. 5), exhibiting only a low recovery in thymidine incorporation under PAR conditions. Despite the limited number of bacterial isolates tested and the artificial radiation used in this study, we conclude that bacteria quite rapidly resume metabolic activity subsequent to UV stress.
FIG. 5.
Relation between the recovery of thymidine and leucine incorporation under PAR and UV-A irradiation (as percentage of the incorporation rate after incubation for 4 h in the dark subsequent to UV-B exposure) and the dark recovery (calculated as percentage of activity measured immediately after UV-B exposure and at the end of the dark incubation for 4 h). The response measured in the PAR treatment was subtracted from that measured in the PAR plus UV-A treatment to distinguish between the photoreactivation induced by UV-A and that induced by PAR. Dotted lines denote the boundaries among the four categories of recovery from UV stress.
Conclusion.
In summary, we have shown that large interspecific differences in the sensitivity to UV-B and in the recovery from UV stress exist in bacterial isolates originating from a coastal marine environment. No direct correlation was found between the magnitude of the inhibition and the recovery from UV-B stress for the bacterial isolates examined which would be a prerequisite for the development of more UV-resistant bacterial communities in the surface layers of the water column. Thus, the results reported here confirm previous findings for bulk bacterioplankton communities (11, 25) that surface bacterioplankton are as sensitive to UV radiation as are deep-water bacterioplankton. Also, the large interspecific variability in the recovery from previous UV stress reported recently (14) has been confirmed in this study. We found, however, a previously unnoticed inverse relation between the recovery ability in the dark and that under PAR conditions. The observed large interspecific variability among the marine bacterial isolates found in this study with respect to their sensitivity to UV and their different strategies to recover from previous UV stress might indicate that UV potentially influences the species composition of marine bacterioplankton in surface waters.
ACKNOWLEDGMENTS
The hospitality of the staff of the Center for Marine Research at the Ruder Boskovic Institute at Rovinj (Croatia) is gratefully acknowledged. The comments of three anonymous reviewers substantially improved the manuscript.
Funding support was provided by grants from the Austrian Science Foundation (Fonds zur Förderung der wissenschaftlichen Forschung, FWF project no. 10023 to G.J.H.) and by the Environment and Climate Program of the European Union (Microbial Community Response to UV-B Stress in European Waters, project no. EV5V-CT94-0512). J.M.A. was supported by a TMR grant (MAS3-CT96-5004) of the European Union and by a predoctoral grant from the Basque Government.
Footnotes
Publication no. 3471 of the Netherlands Institute for Sea Research (NIOZ).
REFERENCES
- 1.Aas P, Lyons M M, Pledger R, Mitchell D L, Jeffrey W H. Inhibition of bacterial activities by solar radiation in nearshore waters and the Gulf of Mexico. Aquat Microb Ecol. 1996;11:229–238. [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behr H D. Net total and UV-B radiation at the sea surface. J Atmos Chem. 1992;15:299–314. [Google Scholar]
- 4.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger B, Hoch B, Kavka G, Herndl G J. Bacterial metabolism in the River Danube: parameters influencing bacterial production. Freshw Biol. 1995;34:601–616. [Google Scholar]
- 6.Chin-Leo G, Kirchman D L. Unbalanced growth in natural assemblages of marine bacterioplankton. Mar Ecol Prog Ser. 1990;63:1–8. [Google Scholar]
- 7.Doney S C, Najjar R G, Stewart S. Photochemistry, mixing and diurnal cycles in the upper ocean. J Mar Res. 1995;53:341–369. [Google Scholar]
- 8.Friedberg E C. DNA repair. New York, N.Y: W. H. Freeman and Company; 1985. p. 614. [Google Scholar]
- 9.Fuhrman J A, Azam F. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar Biol. 1982;66:109–120. [Google Scholar]
- 10.Helz G R, Zepp R G, Crosby D G. Aquatic and surface photochemistry. Boca Raton, Fla: Lewis Publishers; 1994. p. 552. [Google Scholar]
- 11.Herndl G J, Müller-Niklas G, Frick J. Major role of ultraviolet-B in controlling bacterioplankton growth in the surface layer of the ocean. Nature. 1993;361:717–719. [Google Scholar]
- 12.Hobbie J E, Daley R J, Jasper S. Use of Nuclepore filters for counting bacteria by epifluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffrey W H, Pledger R J, Aas P, Hager S, Coffin R B, van Haven R, Mitchell D L. Diel and depth profiles of DNA photodamage in bacterioplankton exposed to ambient solar ultraviolet radiation. Mar Ecol Prog Ser. 1996;137:283–291. [Google Scholar]
- 14.Joux F, Jeffrey W H, Lebaron P, Mitchell D L. Marine bacterial isolates display diverse responses to UV-B radiation. Appl Environ Microbiol. 1999;65:3820–3827. doi: 10.1128/aem.65.9.3820-3827.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser E, Herndl G J. Rapid recovery of marine bacterioplankton activity after inhibition by UV radiation in coastal waters. Appl Environ Microbiol. 1997;63:4026–4031. doi: 10.1128/aem.63.10.4026-4031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karentz D, Bothwell M L, Coffin R B, Hanson A, Herndl G J, Kilham S S, Lesser M P, Lindell M, Moeller R E, Morris D P, Neale P J, Sanders R W, Weiler C S, Wetzel R G. Impact of UVB radiation on pelagic freshwater ecosystems: report of the working group on bacteria and phytoplankton. Arch Hydrobiol. 1994;43:31–69. [Google Scholar]
- 17.Kieber D J, Jiao J, Kiene R P, Bates T S. Impact of dimethylsulfide photochemistry on methyl sulfur cycling in the equatorial Pacific Ocean. J Geophys Res. 1996;101(C2):3715–3722. [Google Scholar]
- 18.Kieber R J, Zhou X, Mopper K. Formation of carbonyl compounds from UV-induced photodegradation of humic substances in natural waters: fate of riverine carbon in the sea. Limnol Oceanogr. 1990;35:1503–1515. [Google Scholar]
- 19.Kirchman D, K'Ness E, Hodson R. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol. 1985;49:599–607. doi: 10.1128/aem.49.3.599-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchman D L. Incorporation of thymidine and leucine in the subarctic Pacific: application to estimating bacterial production. Mar Ecol Prog Ser. 1992;82:301–309. [Google Scholar]
- 21.Kirchman D L, Newell S Y, Hodson R E. Incorporation versus biosynthesis of leucine: implications for measuring rates of protein synthesis and biomass production by bacteria in marine systems. Mar Ecol Prog Ser. 1986;32:47–59. [Google Scholar]
- 22.Lindell M J, Granéli H W, Tranvik L J. Effects of sunlight on bacterial growth in lakes of different humic content. Aquat Microb Ecol. 1996;11:135–141. [Google Scholar]
- 23.Lindell M J, Granéli W, Tranvik L J. Enhanced bacterial growth in response to photochemical transformation of dissolved organic matter. Limnol Oceanogr. 1995;40:195–199. [Google Scholar]
- 24.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 25.Müller-Niklas G, Heissenberger A, Puskaric S, Herndl G J. Ultraviolet-B radiation and bacterial metabolism in coastal waters. Aquat Microb Ecol. 1995;9:111–116. [Google Scholar]
- 26.Neidhardt F C, Ingraham J L, Schaechter M. Physiology of the bacterial cell: a molecular approach. Sunderland, Mass: Sinauer Associates, Inc.; 1990. p. 506. [Google Scholar]
- 27.Obernosterer I, Reitner B, Herndl G J. Contrasting effects of solar radiation on dissolved organic matter and its bioavailability to marine bacterioplankton. Limnol Oceanogr. 1999;44:1645–1654. [Google Scholar]
- 28.Palenik B, Price N M, Morel F M M. Potential effects of UV-B on the chemical environment of marine organisms: a review. Environ Pollut. 1991;70:117–130. doi: 10.1016/0269-7491(91)90084-a. [DOI] [PubMed] [Google Scholar]
- 29.Reitner B, Herzig A, Herndl G J. Role of ultraviolet-B radiation on photochemical and microbial oxygen consumption in a humic-rich shallow lake. Limnol Oceanogr. 1997;42:950–960. [Google Scholar]
- 30.Scully N M, Lean D R S, McQueen D J, Cooper W J. Photochemical formation of hydrogen peroxide in lakes: effects of dissolved organic carbon and ultraviolet radiation. Can J Fish Aquat Sci. 1995;52:2675–2681. [Google Scholar]
- 31.Scully N M, McQueen D J, Lean D R S, Cooper W J. Hydrogen peroxide formation: the interaction of ultraviolet radiation and dissolved organic carbon in lake waters along a 43-75°N gradient. Limnol Oceanogr. 1996;41:540–548. [Google Scholar]
- 32.Servais P. Bacterial production measured by 3H-thymidine and 3H leucine incorporation in various aquatic ecosystems. Arch Hydrobiol Beih Ergeb Limnol. 1992;37:73–81. [Google Scholar]
- 33.Simon M, Azam F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- 34.Sommaruga R, Obernosterer I, Herndl G J, Psenner R. Inhibitory effect of solar radiation on thymidine and leucine incorporation by freshwater and marine bacterioplankton. Appl Environ Microbiol. 1997;63:4178–4184. doi: 10.1128/aem.63.11.4178-4184.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zafiriou O C, Joussot-Dubien J, Zepp R G, Zika R G. Photochemistry of natural waters. Environ Sci Technol. 1984;18:358–371. [Google Scholar]
- 36.Zepp R G, Callaghan T V, Erickson D J. Effects of increased solar ultraviolet radiation on biogeochemical cycles. Ambio. 1995;24:181–187. [Google Scholar]