Abstract
Snake envenomations constitute a worldwide neglected tropical disease, with the vast majority of lethal bites inflicted by front-fanged snakes from the viperid and elapid groups. Rear-fanged snakes (colubrids) were often considered harmless and as a result, are much less studied, but several documented deaths have suggested potent venom in this group as well. The largest European snake (Malpolon monspessulanus monspessulanus), known as the “Montpellier snake”, is such a rear-fanged snake that belongs to the Lamprophiidae family. Its venom remains largely unknown but cases of envenomation with neurological symptoms have been reported. Here, we provide the first insights into the composition of its venom using mass spectrometry methods. First, liquid chromatography coupled mass spectrometry analysis of the manually collected venom samples reveals a complex profile, with the majority of masses encompassing the range 500–3000 Da, 4000–8000 Da, and 10 000–30 000 Da. Next, shotgun proteomics allowed the identification of a total of 42 different known families of proteins, including snake venom metalloproteinases, peptidase M1, and cysteine-rich secretory proteins, as the most prominent. Interestingly, three-finger toxins were not detected, suggesting that neurotoxicity may occur via other, yet to be determined, toxin types. Overall, our results provide the basis for a better understanding of the effects of a peculiar snake venom on human symptomatology, but also on the main prey consumed by this species.
Keywords: Proteomics, Opistoglyph, Toxins, Montpellier's snake
Highlights
-
•
We investigate the venom composition of the largest European venomous snake.
-
•
LC-MS analysis of the crude manually collected venom revealed a complex profile.
-
•
Shotgun proteomic analysis identified 42 different protein families.
-
•
Major components include SVMPs, consistent with the clinical features.
1. Introduction
Venomous animals produce secretions composed of highly diverse and potent pharmacological compounds (Vetter et al., 2011). Besides their ecological roles (predation and defense), venoms and toxins represent a rich source of potential therapeutics, with already several marked drugs to treat human pathologies such as pain, hypertension, and hemostatic dysfunctions (Trim et al., 2021). Among all venomous animals, snakes are arguably the most intensively studied group (Fry et al., 2008). Yet, there is a strong imbalance of research efforts across the different venomous snake families. Front-fanged elapids and viperids have trusted the vast majority of the published literature on snake venom, whereas rear-fanged snakes (e.g. Lamprophiidae, many Colubridae), despite being the most speciose groups, have remained largely over-looked (Saviola et al., 2014). Several factors may explain this discrepancy, but the fact that the front-fanged groups contain iconic species among the most lethal (e.g. cobras, mambas, vipers, and rattlesnakes) likely contributed to this narrow focus. In addition, venom collection is a much easier procedure on front-fanged compared to rear-fanged snakes, including significantly higher yields. This is directly linked to the anatomy of the venom apparatus. Indeed, the long hollow teeth of front-fanged snakes connected to the muscle-surrounded venom gland constitute a “high-pressure venom system”, which facilitates the “milking” procedure (Hayes et al., 1995). On the opposite, a low-pressure venom system occurs in rear-fanged snakes, with the secretions of the Duvernoy's gland being slowly released near the enlarged and/or grooved teeth. Milking yields were reported in the single-digit microliters, often preventing in-depth venom characterization (Hayes et al., 1995). However, the use of pilocarpine to stimulate the release of venom and the recent venomics approaches have allowed the investigation of several species of rear-fanged snake venoms (Mackessy and Saviola, 2016). Whereas the initial results suggested a reduced complexity compared to front-fanged snake venoms, other studies have reported a similar level of complexity. Clearly, there is a need for additional work on colubrid snake venoms.
The Montpellier snake (Lamprophiidae, Malpolon monspessulanus monspessulanus) is the largest venomous snake in Europe. Total body size can exceed 2 m, although it rarely reaches 1.80 m. This opistoglyph and active forager largely feeds on lizards, ranging from small species (<15 cm, Podarcis muralis), to fully grown male ocellated lizards (Timon lepidus). Yet, the diet of the Montpellier snake's is ecletic, large specimens consuming other snakes (with rare cases of cannibalism reported, see Franch and Sebastián, 2013) such as the ladder snake (Rhinechis scalaris) or the horseshoe whip snake (Hemorrhois hippocrepis), as well as birds and small mammals (rabbits and rats).
The lack of knowledge regarding the composition of its venom led to some controversial claims. Indeed, it was reported that its venom kills the prey slowly, in “twenty-four to 48 h”, which appears in stark contrast to the quick immobilization often observed in the field by some of the co-authors. Furthermore, the venom of the Montpellier snake is considered harmless to human; this standpoint is based on more than 70 documented bites (Weinstein et al., 2011). However, a recent report describes two envenomation cases of teenagers characterized by localized clinical symptoms such as pain, numbness, and swelling, but with benign consequences (Ballouard et al., in press). Moreover, at least three cases of neurotoxic envenoming have been reported. In the detailed report of such recent neurotoxic envenomation, it is indicated that the finger of the victim was deep inside the buccal cavity of the snake and likely received prolonged contact with the posterior maxillary teeth, maximizing the likelihood of envenomation (Pommier and de Haro, 2007). This rare and unfortunate situation suggests that most of the Montpellier snake's documented bites may be “dry”, likely implicating only the small teeth of the front upper or bottom jaws without envenomation. The prevalence of this phenomenon is difficult to quantify, but it may lead to an underestimation of this snake'venom neurotoxicity and potential effects/threat to humans.
In this study, we provide the first insight into the venom composition of the Montpellier snake using shotgun proteomics. The data were obtained from manually collected venom samples, and therefore reflective of a more natural venom injection as opposed to pilocarpine-induced secretions. Surprisingly, this rear-fanged snake produces highly complex venom, containing many of the major families found in other venomous snakes, except for the notable absence of the neurotoxic three-finger toxins. Overall, our results provide the basis for a better understanding of the predatory and/or defensive roles of the venom of this iconic snake and may help in the management of the clinical signs of envenoming.
2. Material and methods
2.1. Venom collection
The specimens of Malpolon monspessulanus monspessulanus used in this study were collected from the Isles of Port-Cros and Porquerolles (south-east France) using protocols approved by the Direction Départementale des Territoires et de la Mer (DDTM 83 2015-01), as part of a long-term capture-recapture ecological study. These access-restricted islands support a healthy population of Montpellier snakesthanks to favorable conditions, including the presence of suitable biotopes, low human disturbance, and the absence of motorized vehicles and large predators (e.g. foxes). During the springs of 2018–2019, several adult specimens of M. monspessulanus monspessulanus were collected and manual venom extraction was occasionally attempted. Although now largely applied and validated, the pilocarpine injection method could not be employed in this study, since the venom collection had to be carried out on-site and within the few minutes of the capture-release manipulations. Interestingly, reported volumes using pilocarpine were higher but of lower protein concentration (Rosenberg et al., 1985), suggesting a possible altered composition. Therefore, we used a manual method consisting of a pipette tip covering the grooved posterior maxillary teeth and a gentle massage to the Duvernoy's gland in an attempt to stimulate a more “natural” release of venom. Following field collection, samples (4–5 μL) were diluted in distilled water then later frozen before lyophilization and stored at – 20 °C until use.
2.2. Liquid chromatography coupled mass spectrometry
Reversed-phase ultrahigh performance liquid chromatography (RP-UPLC) was operated on an Acquity H-Class ultrahigh performance liquid chromatography system (Waters, Corp., Milford, MA, United States) fitted with a UV detector (Photodiode-Array Detection) under the control of Waters MassLynx software v4.1. Separation of the crude M. monspessulanus monspessulanus venom (300 μg) was achieved using a Kinitex C18 100 Å column (2.1 × 150 mm, 3 μm) fitted with a pre-column. Elution was carried out using a gradient of 0–80% solution B (0.1% formic acid in Acetonitrile) in 80 min. Samples eluting from the UPLC were introduced into the mass spectrometer at a flow rate of 50 μL/min. Acquisitions were carried out over the range of 50 Da–1800 Da m/z every second on a Synapt-G2-S high-definition MS system (Waters, Corp., Milford, MA, United States). To obtain the molecular masses of the venom components eluting between 0 and 70 min, each peak from the total ion current (TIC) chromatogram was analyzed with Waters Mass Lynx software v4.1.
2.3. Shotgun proteomics
2.5 μL of crude M. monspessulanus monspessulanus venom at 20.5 mg/mL in 18 Mega-Ohm quality water were diluted 20 times in 100 mM NH4HCO3 pH 7.8 and reduced with 5 μL of 500 mM dithiothreitol in 18 Mega-Ohm quality water for 1 h, at 56 °C, under shaking at 300 rpm. The sample was then alkylated with 10 μL of 500 mM iodoacetamide for 1 h, at room temperature, in the dark. The venom sample was submitted to trypsin digestion by adding 5 μL of Pierce™ mass spectrometry grade trypsin (Thermo Scientific, Waltham, MA, USA) at 0.1 mg/mL in 100 mM ammonium bicarbonate buffer (4 h at 37 °C). The reaction was stopped by acidifying the media with the addition of 6 μL of trifluoroacetic acid (TFA) 10% (v/v) in 18 Mega-Ohm quality water to the reaction mixture. The sample was desalted on a ZipTip™ pipette tip filled with C18 resin, using an acetonitrile/water/TFA (49.8/50/0.2 v/v) solution as eluent. A total of 0.5 μg from the digested material was analyzed by the Acquity UPLC® M-Class (Waters, Milford, MA, USA) coupled to the Q-Exactive™ Plus Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo Scientific, Bremen, Germany). The chromatographic system was equipped with a monolithic PepSwift Capillary column 100 μm × 25 cm (Thermo Scientific, Waltham, MA, USA). Peptides were eluted using a gradient of 3–50% of solution B in 100 min and 50–80% of solution B in 30 min (A: water/0.1% formic acid; B: acetonitrile), at a flow rate of 0.6 mL/min, and data were acquired in the positive-ion mode. Concerning mass spectrometry, all the analyses were performed in data-dependent analysis (DDA) mode that automatically triggers the MS/MS experiments. The automatic gain control (AGC) target values were 3.106 for MS spectra and 2.105 for MS/MS spectra. The maximum injection times were set at 200 ms for the MS step and 1000 ms for MS/MS events. For MS/MS, a “Top 12” experiment was applied, meaning that the twelve most intense ions of each MS scan have been selected for fragmentation. Singly charged ions, ions with undetermined charge (for example, electronic noise), and ions with signal intensities below the AGC threshold set at 1e3, were excluded from the selection. For precursors ions, the selection windows were 2.0 m/z, the AGC target was 1e5 (or 50 ms as a maximum of injection time), and the resolving power of 17 500 @m/z 200. The normalized collision energy was 25. A dynamic exclusion of 10s was also applied to avoid the redundancy of MS/MS spectra of the same ions.
2.4. Bioinformatic analysis of proteomic data
PEAKS Studio v8.5 (Bioinformatics solutions, Waterloo, ON, Canada) a de novo assisted database software (Ma et al., 2003; Zhang et al., 2012) was chosen to analyze MS/MS data from M. monspessulanus monspessulanus venom using a snake venom protein database derived from Uniprot/SwissProt r2021_04 (The UniProt Consortium, 2021). Briefly, PEAKS studio initially produces de novo sequences from MS/MS spectra without relying on a database. The confidence of each peptide sequence obtained by this process is given by an Average Local Confidence (ALC) score. Then, these de novo sequences are corrected by comparison against the database to provide additional information about post-translational modifications (PTM's), mutations, homologous peptides, and novel peptides. Carbamidomethylation was set as fixed modification, while oxidation (M) was set as variable modifications, with maximum missed cleavages at 3 for trypsin digestion. Parent mass and fragment mass error tolerance were set at 5 ppm and 0.015 Da, respectively. False discovery rate (FDR) of 1% (Amorim et al., 2018; Degueldre et al., 2017) and unique peptide ≥2 were used for filtering out inaccurate proteins. A −10lgP >120 indicates that the detected proteins is represented by enough reliable peptides MS/MS spectra. In order to identify more relevant sequences, the Spider algorithm (Han et al., 2005) from PEAKS Studio software was used to find additional mutations or to correct the sequences. This algorithm corrects the sequences stored in the database with de novo sequences based on MS/MS spectra, which allowed us to identify PTM's and mutations. Minimum ion intensity for mutation and PTM's was set to 5%, and ALC score ≥90 for de novo sequences leading to low precursor mass error in order to identify reliable PTM's and potential mutations.
Furthermore, to visualize the matched regions by PEAKS in each protein, we developed an in-house Perl web application, that takes the comma-separated value result files from PEAKS and the supplied database in fasta format to produce an output where matched regions are highlighted in a graphical user interface. Briefly, this tool highlights in bold the matching peptides for each sequence in fasta format. Then, a sequence alignment allows for a rapid comparison of the matching regions (appearing in bold). This step was not systematically applied to all sequences but helped to evaluate whether the same peptides matched several related sequences leading to an overestimation of the total validated sequences. A final annotation step with the objective of classifying the matched sequences in the snake venom protein families was conducted using hmmcompete v0.1 (Koua and Kuhn-Nentwig, 2017). In this step, validated matches were subjected to a hmmcompete search against a custom database of Hidden Markov Models (HMMs) constructed using snake venom toxins from Uniprot/Swiss-Prot r2021_04. HMMs were built and calibrated using tools from the HMMER suite v3.3 (Eddy, 2011). Briefly, the snake venom proteins were regrouped into snake proteins families using the Uniprot/Swiss-Prot annotation associated with each sequence record. Each snake proteins families were filtered to remove sequence redundancy using CD-HIT v4.8.1 using a sequence identity threshold of 100%. The remaining sequences were aligned using the linsi strategy from MAFFT v7.487. Finally, the alignments were manually corrected in Jalview v2.11.5 before being processed using hmmbuild.
3. Results
3.1. Venom collection
The venom of the Montpellier snake is virtually unknown. The main reason for such lack of investigation remains the difficulty of venom collection. Indeed, our initial attempts to collect venom samples from the Montpellier's snake proved challenging, as expected for any rear-fanged snake. As mentioned in the method section, the use of pilocarpine to stimulate venom release was not possible, due to the quick catch-and-release procedure imposed for the ongoing population study, as well as because of the protected status of this species. Therefore, despite having enlarged and deeply grooved teeth (Fig. 1), the venom collection procedure was tedious. However, over the course of this study, several adult specimens (usually not the largest, typically about 1.20 m long) were milked. They secreted a yellowish venom that could clearly be seen slowly filling the tip of the cone tip (Fig. 1).
Fig. 1.
The Montpellier snake, Malpolon monspessulanus monspessulanus, from France in its natural environment (top panels show a male specimen, photo credit to Jean-Marie Ballouard). Bottom left panel shows the venom collection procedure (female specimen, photo credit to Sébastien Caron), facilitated by the presence of enlarged grooved teeth (bottom right panel).
The yield was relatively low per specimen: 4–6 μl.
3.2. LC-MS analysis of crude Malpolon monspessulanus venom
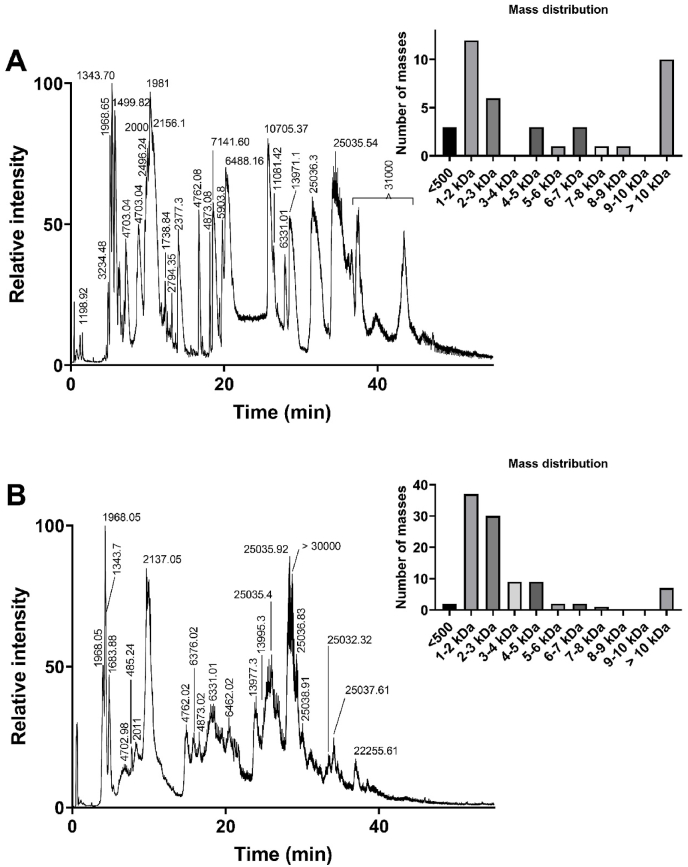
All collected venom samples were analyzed by LC-MS and showed a remarkable and unexpected complexity (Fig. 2). Most of the 40 to 100 major detected components of the venom eluted between 5 and 40 min, with three main groups based on mass ranges (Fig. 2). Indeed, the largest number of masses were detected in the range 500–3000 Da (52–69% of detected masses), followed by 10 000–30 000 Da (7–25%), and finally 4000–8000 Da (14–20%).
Fig. 2.
LC-MS analysis of Malpolon monspessulanus monspessulanus venom. The Total Ion Current (TIC) traces obtained from two representative venom samples are shown. Panel A was from the most complex sample, whereas panel B represents the most frequently observed venom profile. Inserts show the mass distribution for each sample.
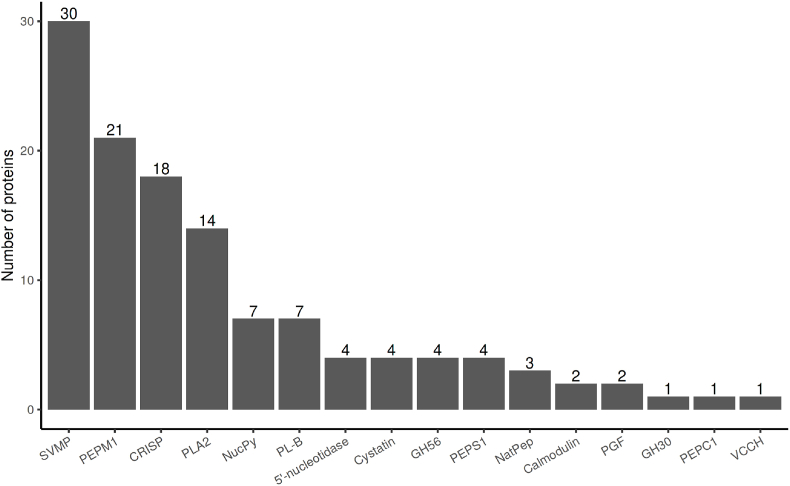
To get a more detailed insight into its composition, the venom was subjected to shotgun proteomics. A total of 262 proteins out of 126 protein groups were initially identified, from which some (36) were not annotated nor classified into a known snake protein family by the HMMs (Supplementary File). Furthermore, some protein sequences, despite having a different identifier, were redundant in the database and were removed (34 sequences). Finally, different protein sequences were matched by the same peptides in their conserved regions, also artificially inflating the number of identified proteins (see illustrated example as Suppl Fig 1). Therefore, the total numbers of identified proteins shown in Fig. 3 should be interpreted with caution, and in the absence of venom gland transcriptome from this species, the primary aim of this study is restricted to provide an overview of the families of secreted proteins in the venom. Overall, the bioinformatic analysis of the results revealed 42 different families of proteins, from which 16 were annotated as “snake venom families” (Fig. 3). The three most represented proteins families were the snake venom metalloproteinase (SVMP) family (30 proteins, Table 1), Peptidase M1 family (21 proteins), and cysteine-rich secretory protein (CRISP) family (18 proteins). Also of interest, the proteomic analysis of the venom of M. monspessulanus monspessulanus did not reveal the presence of three-finger toxins, which are commonly involved in the neurotoxicity of the venom.
Fig. 3.
16 Snake venom protein families identified in the proteome of M. monspessulanus monspessulanus. SVMP: Snake Venom Metalloproteinase, PEPM1: Peptidase M1, PLA2: Phospholipase A2, CRISP: cysteine-rich secretory protein, NucPyro: Nucleotide pyrophosphatasephosphodiesterase, PL-B: Phospholipase B-like, GH30: Glycosyl hydrolase 30, GH56: Glycosyl hydrolase 56, NatPep: Natriuretic peptide, PGF: PDGFVEGF growth factor, PEPS1: Peptidase S1, VCCH: Venom complement C3 homolog.
Table 1.
Snake Venom Metalloproteinases identified in the M. monspessulanus monspessulanus venom by PEAKS studio software.
| Protein ID | Accession number | −10lgP | Coverage (%) | #Unique | #Spec Malpolon | PTM | Avg. Mass | Description | Snake Protein Family |
|---|---|---|---|---|---|---|---|---|---|
| 659 | A0A2D4H2X0 | 162.07 | 5 | 6 | 32 | Carbamidomethylation (DHKE X@N-term) | 41 044 | Uncharacterized protein (Fragment) OS = Micrurus corallinus OX = 54 390 PE = 4 SV = 1 | P-III subfamily, P-IIIa sub-subfamily |
| 680 | A0A2D4H2Y2 | 162.07 | 6 | 6 | 32 | Carbamidomethylation (DHKE X@N-term) | 33 050 | Uncharacterized protein (Fragment) OS = Micrurus corallinus OX = 54 390 PE = 4 SV = 1 | P-III subfamily, P-IIIa sub-subfamily |
| 294 | Q10749 | 137.13 | 7 | 3 | 298 | Carbamidomethylation (DHKE X@N-term); Dethiomethyl; Tyrosine oxidation to 2-aminotyrosine | 68 176 | Snake venom metalloproteinase-disintegrin-like mocarhagin OS=Naja mossambica OX = 8644 PE = 1 SV = 3 | P-III subfamily, P-IIIa sub-subfamily |
| 382 | D5LMJ3 | 124.66 | 10 | 2 | 13 | Carbamidomethylation (DHKE X@N-term); Tyrosine oxidation to 2-aminotyrosine | 68 254 | Zinc metalloproteinase-disintegrin-like atrase-A OS=Naja atra OX = 8656 PE = 2 SV = 1 | P-III subfamily, P-IIIa sub-subfamily |
| 448 | A8QL49 | 151.56 | 8 | 5 | 52 | Carbamidomethylation; Sodium adduct; Carbamidomethylation (DHKE X@N-term); Ubiquitin; Dehydration; Pyro-glu from E | 68 988 | Zinc metalloproteinase-disintegrin-like BmMP OS=Bungarus multicinctus OX = 8616 PE = 1 SV = 1 | P-III subfamily, P-IIIa sub-subfamily |
| 564 | D3TTC2 | 121.44 | 11 | 3 | 8 | Carbamidomethylation; Carbamidomethylation (DHKE X@N-term); Tyrosine oxidation to 2-aminotyrosine | 69 181 | Zinc metalloproteinase-disintegrin-like atragin OS=Naja atra OX = 8656 PE = 1 SV = 1 | P-III subfamily, P-IIIa sub-subfamily |
| 761 | A7X437 | 104.78 | 13 | 6 | 18 | Carbamidomethylation (DHKE X@N-term); Dihydroxy; Hydroxylation; 2-amino-3-oxo-butanoic_acid; Proline oxidation to pyroglutamic acid | 11 043 | SVMP-PP-Psa1 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 762 | A7X447 | 104.78 | 13 | 6 | 18 | Carbamidomethylation (DHKE X@N-term); Dihydroxy; Hydroxylation; 2-amino-3-oxo-butanoic_acid; Proline oxidation to pyroglutamic acid | 10 971 | SVMP-PP-Psa3 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 763 | A7X457 | 104.78 | 13 | 6 | 18 | Carbamidomethylation (DHKE X@N-term); Dihydroxy; Hydroxylation; 2-amino-3-oxo-butanoic_acid; Proline oxidation to pyroglutamic acid | 11 049 | SVMP-PP-Psa5 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 764 | A7X443 | 104.78 | 13 | 6 | 18 | Carbamidomethylation (DHKE X@N-term); Dihydroxy; Hydroxylation; 2-amino-3-oxo-butanoic_acid; Proline oxidation to pyroglutamic acid | 10 985 | SVMP-PP-Psa2 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 765 | A7X4A6 | 104.78 | 13 | 6 | 18 | Carbamidomethylation (DHKE X@N-term); Dihydroxy; Hydroxylation; 2-amino-3-oxo-butanoic_acid; Proline oxidation to pyroglutamic acid | 11 118 | SVMP-PP-Psa17 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 766 | A7X497 | 104.78 | 13 | 6 | 18 | Carbamidomethylation (DHKE X@N-term); Dihydroxy; Hydroxylation; 2-amino-3-oxo-butanoic_acid; Proline oxidation to pyroglutamic acid | 11 087 | SVMP-PP-Psa15 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 928 | A7X493 | 104.78 | 13 | 6 | 18 | Carbamidomethylation (DHKE X@N-term); Dihydroxy; Hydroxylation; 2-amino-3-oxo-butanoic_acid; Proline oxidation to pyroglutamic acid | 11 003 | SVMP-PP-Psa14 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 929 | A7X452 | 104.78 | 13 | 6 | 18 | Carbamidomethylation (DHKE X@N-term); Dihydroxy; Hydroxylation; 2-amino-3-oxo-butanoic_acid; Proline oxidation to pyroglutamic acid | 11 046 | SVMP-PP-Psa4 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 932 | A7X465 | 104.78 | 10 | 6 | 18 | Carbamidomethylation (DHKE X@N-term); Dihydroxy; Hydroxylation; 2-amino-3-oxo-butanoic_acid; Proline oxidation to pyroglutamic acid | 14 748 | SVMP-PP-Psa7 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 933 | A7X461 | 104.78 | 10 | 6 | 18 | Carbamidomethylation (DHKE X@N-term); Dihydroxy; Hydroxylation; 2-amino-3-oxo-butanoic_acid; Proline oxidation to pyroglutamic acid | 14 786 | SVMP-PP-Psa6 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 1048 | A7X4C4 | 103.74 | 13 | 4 | 24 | Carbamidomethylation; Deamidation (NQ); Carbamidomethylation (DHKE X@N-term); Trifluoroleucine | 10 713 | SVMP-PP-Psa21 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 1101 | A7X4B5 | 103.74 | 13 | 4 | 24 | Carbamidomethylation; Deamidation (NQ); Carbamidomethylation (DHKE X@N-term); Trifluoroleucine | 10 687 | SVMP-PP-Psa19 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 1102 | A7X4E9 | 103.74 | 13 | 4 | 24 | Carbamidomethylation; Deamidation (NQ); Carbamidomethylation (DHKE X@N-term); Trifluoroleucine | 10 625 | SVMP-PP-Psa26 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 1103 | A7X4C0 | 103.74 | 13 | 4 | 24 | Carbamidomethylation; Deamidation (NQ); Carbamidomethylation (DHKE X@N-term); Trifluoroleucine | 10 717 | SVMP-PP-Psa20 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 1104 | A7X4C9 | 103.74 | 13 | 4 | 24 | Carbamidomethylation; Deamidation (NQ); Carbamidomethylation (DHKE X@N-term); Trifluoroleucine | 10 615 | SVMP-PP-Psa22 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 1105 | A7X4D4 | 103.74 | 13 | 4 | 24 | Carbamidomethylation; Deamidation (NQ); Carbamidomethylation (DHKE X@N-term); Trifluoroleucine | 10 653 | SVMP-PP-Psa23 OS=Psammophis mossambicus OX = 234 064 PE = 2 SV = 1 | P-III subfamily |
| 280 | A0A098M208 | 120.1 | 10 | 5 | 30 | Carbamidomethylation; Amidation; Carbamidomethylation (DHKE X@N-term); Pyro-glu from E; Mutation | 67 736 | Metalloproteinase (Type III) 9a OS=Hypsiglena sp. JMG-2014 OX = 1550645 PE = 3 SV = 1 | P-III subfamily |
| 388 | A0A346CIB8 | 110.29 | 10 | 2 | 15 | Carbamidomethylation; Amidation; O-Diisopropylphosphorylation | 72 160 | Metalloproteinase (Fragment) OS=Spilotes sulphureus OX = 1899469 PE = 2 SV = 1 | P-III subfamily |
| 427 | A0A346CM41 | 202.65 | 12 | 2 | 36 | Carbamidomethylation; Oxidation (M); Amidation; Sodium adduct; Carbamidomethylation (DHKE X@N-term); 3 more | 68 148 | Metalloproteinase 37 (Fragment) OS = Ahaetulla prasina OX = 499 056 PE = 2 SV = 1 | P-III subfamily |
| 429 | A0A346CM36 | 202.65 | 12 | 2 | 36 | Carbamidomethylation; Oxidation (M); Amidation; Sodium adduct; Carbamidomethylation (DHKE X@N-term); 3 more | 68 120 | Metalloproteinase 31 (Fragment) OS = Ahaetulla prasina OX = 499 056 PE = 2 SV = 1 | P-III subfamily |
| 462 | A0A098M137 | 102.68 | 7 | 2 | 20 | Carbamidomethylation; Carbamidomethylation (DHKE X@N-term); 2-amino-3-oxo-butanoic_acid; Pyro-glu from E; Mutation | 68 654 | Metalloproteinase (Type III) 6c OS=Hypsiglena sp. JMG-2014 OX = 1550645 PE = 3 SV = 1 | P-III subfamily |
| 465 | A0A098M214 | 102.68 | 7 | 2 | 20 | Carbamidomethylation; Carbamidomethylation (DHKE X@N-term); 2-amino-3-oxo-butanoic_acid; Pyro-glu from E; Mutation | 68 684 | Metalloproteinase (Type III) 6a OS=Hypsiglena sp. JMG-2014 OX = 1550645 PE = 3 SV = 1 | P-III subfamily |
| 466 | A0A098M216 | 102.68 | 7 | 2 | 20 | Carbamidomethylation; Carbamidomethylation (DHKE X@N-term); 2-amino-3-oxo-butanoic_acid; Pyro-glu from E; Mutation | 68 670 | Metalloproteinase (Type III) 6b OS=Hypsiglena sp. JMG-2014 OX = 1550645 PE = 3 SV = 1 | P-III subfamily |
| 468 | A0A182C6B6 | 120.7 | 10 | 4 | 43 | Carbamidomethylation; Carbamidomethylation (DHKE X@N-term); Acetylation (N-term); Acetylation (K); Replacement of proton with ammonium ion; Mutation | 69 622 | Metalloproteinase OS=Phalotris mertensi OX = 1260334 PE = 3 SV = 1 | P-III subfamily |
Among the most represented venom proteins are the SVMPs. The matching SVMP sequences are listed in Table 1, and interestingly, out of the 30 sequences, 16 are from the related species Psammophis mossambicus (Lamprophiidae familly). This species, known as olive grass snake, occurs in the northeast of South Africa and shows similar behavior and diet as the Montpellier's snake.
4. Discussion
The Montpellier snakes live around the Mediterranean region including, for the European species (Malpolon monspessulanus monspessulanus), most of Spain and southern France, while it remains strangely absent from Italy (Carranza et al., 2006). This impressive can adopt a defensive behavior that can be intimidating (capable of standing up like a cobra). Montpellier snake venom has not been investigated with recent omic methods, but previous studies have shown that it is lethal to mice at 200 μg/mg. Phospholipase, phosphodiesterase, and caseinase activity were demonstrated in early studies (Rosenberg et al., 1985). This venom also induced hemorrhages in the lungs of mice with leakage of blood cells and plasma into the pulmonary alveoli (Rosenberg et al., 1992). Interestingly, a 24 000 Da protein, as determined by SDS-PAGE, called CM-b causes profuse bleeding from the nostrils of mice. It is still not known whether this effect is due to a direct action of the protein on the blood vessels or via a mediator.
Clearly, the venom apparatus found in the Montpellier snake is not adequate for delivering venom to a human-sized mammal. Indeed, like in many opistoglyph snakes, the teeth used to deliver venom are posteriorly in the upper jaw (typically below the eye) and they are smaller compared to the large fangs of front-fanged snake. However, a case of envenomation with cranial nerve disturbances has been recorded in the south of France, where the victim had his finger inserted deep into the mouth of the reptile for prolonged time (Pommier and de Haro, 2007). Interestingly, and unlike the reptile's typical prey, the patient mostly had neurological symptoms such as visual disturbances rather than hemorrhagic symptoms.
In this study, the venom of M. monspessulanus monspessulanus was investigated for the first time using proteomics. This study revealed a viper-like enzymatic composition with a majority of compounds being SVMPs. Among rear-franged snakes, based on the venom compounds we can distinguish the viperid venoms and the elapid venoms (Mackessy and Saviola, 2016; Modahl and Mackessy, 2019). Like M. monspessulanus monspessulanus, several rear-franged snakes have been found to also have a viperid venom including Ahaetulla prasina (Modahl et al., 2018), Borikenophis portoricensis (Modahl et al., 2018; Weldon and Mackessy, 2012) (Modahl et al., 2018; Weldon and Mackessy, 2012), and Hydrodynastes gigas (Hill and Mackessy, 2000).
Initially, some SVMPs found in the venom of M. monspessulanus monspessulanus were classified by PEAKS into two different subclasses: P-III which is constituted of a protease domain, a disintegrin, and a cysteine-rich domain, and P-II which lost the cysteine-rich domain. Intriguingly, P-II SVMPs were not reported for any rear-fanged snakes so far. Upon careful inspection of the peptides and the matching region for each SVMP sequence, thanks to our in-house tool, it appeared that the cysteine-rich domain was not covered by any peptide, only the disintegrin and protease domains. Our results highlight the limitations of shotgun proteomics in absence of transcriptomic sequences from the same species. Interestingly, SVMPs from the P-III subfamily, are known to causes hemorrhages (Table 1) and their presence is consistent with some of the effects described in mice.
Given the domination of the venom compounds by the SVMPs, our results suggest that the venom from this species can induce hemorrhage, myonecrosis, and edema, presumably in its main preys. In humans, the paucity of robust information implies that the relationship between the amount of venom injected, circulating in the bloodstream, and clinical effects cannot be evaluated.. Nonetheless, prolonged bites can induce symptoms and this species should not be handled by inexperienced people (Ballouard et al., in press). The SVMPs may have adapted to be prey-specific and their activity optimized for targeting reptiles' physiology rather than mammals. However, this first insight into the Montpellier snake's venom also points towards possible components that can cause harm to humans. Indeed, the presence of Phospholipases A2 could manifest in different ways including neurotoxicity, myotoxicity, cardiotoxicity, and anticoagulant activity. Although these are usually not commonly reported in rear-fanged snakes, 12 sequences of this family were found in the venom of the Montpelier snake and can serve a pre-digestive or prey capture role. In contrast, the presence of the CRISP as the second most abundant protein family is not a surprise as it was reported to be abundant in the venom of other rear-franged species (Hill and Mackessy, 2000; Peichoto et al., 2007). Nevertheless, their ecological role remains unclear although likely important due to their abundance in the snake venom (Modahl and Mackessy, 2019).
The absence of three-finger toxins in the proteomic analysis of the venom potentially complicates the comprehension of the neurological symptoms induced by M. monspessulanus monspessulanus. Three-finger toxins exhibit a postsynaptic neurotoxic activity as antagonists of nicotinic acetylcholine receptors (Bourne et al., 2005), muscarinic acetylcholine receptors (Chung et al., 2002), adrenergic receptors (Rajagopalan et al., 2007), and GABA receptors (Rosso et al., 2015). Recently, a molecular phylogenetic study has reported the presence of a three-finger toxin from M. monspessulanus that was very similar to the related Psammophis subtaeniatus, but also to the very neurotoxic elapid species Naja kaouthia (Xie et al., 2022). Therefore, an ongoing study on the venom gland transcriptomic analysis of M. monspessulanus monspessulanus is needed to clarify and identify the toxin genes produced by this species and to provide greater insight into the venom composition and mode of action of this venom.
Credit author statement
Dominique Koua: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft preparation. Anicet Ebou: Methodology, Software, Formal analysis, Visualization, Writing – review & editing. Zeinab Habbouche: Investigation, Ressources, Data curation. Jean-Marie Ballouard: Investigation, Ressources. Sébastien Caron: Investigation, Ressources. Xavier Bonnet: Investigation, Ressources. Sebastien Dutertre: Conceptualization, Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Ethical statement
The authors declare that no animal experiments were carried out in the work reported in this paper. The specimens of Malpolon monspessulanus monspessulanus from which the venom was obtained in this study were collected from the Isles of Port-Cros and Porquerolles (south-east France) using protocols approved by the Direction Départementale des Territoires et de la Mer (DDTM 83 2015-01), as part of a recurrent and long-term capture-recapture ecological study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank the Parc National de Port-Cros for the partnership and providing accessibility to the study sites, all students for helping us to collect the snakes and the Plateforme de Protéomique Fonctionnelle (FPP) of Montpellier.
Handling Editor: Ray Norton
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.toxcx.2022.100130.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
References
- Amorim F.G., Costa T.R., Baiwir D., De Pauw E., Quinton L., Sampaio S.V. Proteopeptidomic, functional and immunoreactivity characterization of Bothrops moojeni snake venom: influence of snake gender on venom composition. Toxins. 2018;10:177. doi: 10.3390/toxins10050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballouard J.M., Schmitt C, Bonnet X, Renet J, Caron S, Reynoard J, de Haro L, Deso G., (in press). Envenomation by montpellier snake, Malpolon monspessulanus following prolonged bites. Wilderness and Environmental Medecine. https://doi: 10.1016/j.wem.2022.02.011. [DOI] [PubMed]
- Bourne Y., Talley T.T., Hansen S.B., Taylor P., Marchot P. Crystal structure of a Cbtx–AChBP complex reveals essential interactions between snake α-neurotoxins and nicotinic receptors. EMBO J. 2005;24:1512–1522. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranza S., Arnold E.N., Pleguezuelos J.M. Phylogeny, biogeography, and evolution of two Mediterranean snakes, Malpolon monspessulanus and Hemorrhois hippocrepis (Squamata, Colubridae), using mtDNA sequences. Mol. Phylogenet. Evol. 2006;40:532–546. doi: 10.1016/j.ympev.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Chung C., Wu B.-N., Yang C.-C., Chang L.-S. Venom: Purification, Characterization and Gene Organization. Vol. 383. 2002. Muscarinic toxin-like proteins from Taiwan banded Krait (Bungarus multicinctus) pp. 1397–1406. [DOI] [PubMed] [Google Scholar]
- Degueldre M., Verdenaud M., Legarda G., Minambres R., Zuniga S., Leblanc M., Gilles N., Ducancel F., De Pauw E., Quinton L. Diversity in sequences, post-translational modifications and expected pharmacological activities of toxins from four Conus species revealed by the combination of cutting-edge proteomics, transcriptomics and bioinformatics. Toxicon. 2017;130:116–125. doi: 10.1016/j.toxicon.2017.02.014. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7 doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franch M., Sebastián O.S. A case of cannibalism by an extra large female of Malpolon monspessulanus (Montpellier snake) in the Iberian Peninsula. Herpetol. Notes. 2013;6:379–380. [Google Scholar]
- Fry B.G., Scheib H., Van Der Weerd L., Young B., McNaughtan J., Ramjan S.F.R., Vidal N., Poelmann R.E., Norman J.A. Evolution of an arsenal: structural and functional diversification of the venom system in the advanced snakes (Caenophidia) Mol. Cell. Proteomics. 2008;7:215–246. doi: 10.1074/mcp.M700094-MCP200. [DOI] [PubMed] [Google Scholar]
- Han Y., Ma B., Zhang K. Spider: software for protein identification from sequence tags with de novo sequencing error. J. Bioinf. Comput. Biol. 2005:697–716. doi: 10.1142/S0219720005001247. 03. [DOI] [PubMed] [Google Scholar]
- Hayes W.K., Lavín-Murcio P., Kardong K.V. Northern Pacific rattlesnakes (Crotalus viridis oreganus) meter venom when feeding on prey of different sizes. Copeia. 1995;1995:337–343. doi: 10.2307/1446896. [DOI] [Google Scholar]
- Hill R.E., Mackessy S.P. Characterization of venom (Duvernoy's secretion) from twelve species of colubrid snakes and partial sequence of four venom proteins. Toxicon. 2000;38:1663–1687. doi: 10.1016/S0041-0101(00)00091-X. [DOI] [PubMed] [Google Scholar]
- Koua D., Kuhn-Nentwig L. Spider neurotoxins, short linear cationic peptides and venom protein classification improved by an automated competition between exhaustive profile HMM classifiers. Toxins. 2017;9:245. doi: 10.3390/toxins9080245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., Lajoie G. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- Mackessy S.P., Saviola A.J. Understanding biological roles of venoms among the Caenophidia: the importance of rear-fanged snakes. Integr. Comp. Biol. 2016;56:1004–1021. doi: 10.1093/icb/icw110. [DOI] [PubMed] [Google Scholar]
- Modahl C.M., Mackessy S.P. Venoms of rear-fanged snakes: new proteins and novel activities. Front. Ecol. Evol. 2019;7:279. doi: 10.3389/fevo.2019.00279. [DOI] [Google Scholar]
- Modahl C.M., Frietze S., Mackessy S.P. Transcriptome-facilitated proteomic characterization of rear-fanged snake venoms reveal abundant metalloproteinases with enhanced activity. J. Proteonomics. 2018;187:223–234. doi: 10.1016/j.jprot.2018.08.004. [DOI] [PubMed] [Google Scholar]
- Peichoto M.E., Teibler P., Mackessy S.P., Leiva L., Acosta O., Gonçalves L.R.C., Tanaka-Azevedo A.M., Santoro M.L. Purification and characterization of patagonfibrase, a metalloproteinase showing α-fibrinogenolytic and hemorrhagic activities, from Philodryas patagoniensis snake venom. Biochim. Biophys. Acta Gen. Subj. 2007;1770:810–819. doi: 10.1016/j.bbagen.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Pommier P., de Haro L. Envenomation by Montpellier snake (Malpolon monspessulanus) with cranial nerve disturbances. Toxicon. 2007;50:868–869. doi: 10.1016/j.toxicon.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Rajagopalan N., Pung Y.F., Zhu Y.Z., Wong P.T.H., Kumar P.P., Kini R.M. β-Cardiotoxin: a new three-finger toxin from Ophiophagus hannah (king cobra) venom with beta-blocker activity. Faseb. J. 2007;21:3685–3695. doi: 10.1096/fj.07-8658com. [DOI] [PubMed] [Google Scholar]
- Rosenberg H.I., Bdolah A., Kochva E. Lethal factors and enzymes in the secretion from Duvernoy's gland of three colubrid snakes. J. Exp. Zool. 1985;233:5–14. doi: 10.1002/jez.1402330103. [DOI] [PubMed] [Google Scholar]
- Rosenberg H.I., Kinamon S., Kochva E., Bdolah A. The secretion of Duvernoy's gland of Malpolon monspessulanus induces haemorrhage in the lungs of mice. Toxicon. 1992;30:920–924. doi: 10.1016/0041-0101(92)90391-h. [DOI] [PubMed] [Google Scholar]
- Rosso J.-P., Schwarz J.R., Diaz-Bustamante M., Céard B., Gutiérrez J.M., Kneussel M., Pongs O., Bosmans F., Bougis P.E. MmTX1 and MmTX2 from coral snake venom potently modulate GABA(A) receptor activity. Biophys. J. 2015;108:434a. doi: 10.1016/j.bpj.2014.11.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saviola A.J., Peichoto M.E., Mackessy S.P. Rear-fanged snake venoms: an untapped source of novel compounds and potential drug leads. Toxin Rev. 2014;33:185–201. doi: 10.3109/15569543.2014.942040. [DOI] [Google Scholar]
- Trim C.M., Byrne L.J., Trim S.A. In: Progress in Medicinal Chemistry. Witty D.R., Cox B., editors. Elsevier; 2021. Chapter One - utilisation of compounds from venoms in drug discovery; pp. 1–66. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium The. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter I., Davis J.L., Rash L.D., Anangi R., Mobli M., Alewood P.F., Lewis R.J., King G.F. Venomics: a new paradigm for natural products-based drug discovery. Amino Acids. 2011;40:15–28. doi: 10.1007/s00726-010-0516-4. [DOI] [PubMed] [Google Scholar]
- Weinstein S.A., Warrell D.A., White J., Keyler D.E., editors. Venomous” Bites from Non-venomous Snakes. Elsevier; London: 2011. Front-matter. i–iii. [DOI] [Google Scholar]
- Weldon C.L., Mackessy S.P. Alsophinase, a new P-III metalloproteinase with α-fibrinogenolytic and hemorrhagic activity from the venom of the rear-fanged Puerto Rican Racer Alsophis portoricensis (Serpentes: dipsadidae) Biochimie. 2012;94:1189–1198. doi: 10.1016/j.biochi.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Xie B., Dashevsky D., Rokyta D., Ghezellou P., Fathinia B., Shi Q., Richardson M.K., Fry B.G. Dynamic genetic differentiation drives the widespread structural and functional convergent evolution of snake venom proteinaceous toxins. BMC Biol. 2022;20:4. doi: 10.1186/s12915-021-01208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G.A., Ma B. Peaks DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.010587. M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.