Abstract
Introduction
Real-world evidence is important in regulatory and funding decisions. Manual data extraction from electronic health records (EHRs) is time-consuming and challenging to maintain. Automated extraction using natural language processing (NLP) and artificial intelligence may facilitate this process. Whereas NLP offers a faster solution than manual methods of extraction, the validity of extracted data remains in question. The current study compared manual and automated data extraction from the EHR of patients with advanced lung cancer.
Methods
Previously, we extracted EHRs from 1209 patients diagnosed with advanced lung cancer (stage IIIB or IV) between January 2015 and December 2017 at Princess Margaret Cancer Centre (Toronto, Canada) using the commercially available artificial intelligence engine, DARWEN (Pentavere, Ontario, Canada). For comparison, 100 of 333 patients that received systemic therapy were randomly selected and clinical data manually extracted by two trained abstractors using the same accepted gold standard feature definitions, including patient, disease characteristics, and treatment data. All cases were re-reviewed by an expert adjudicator. Accuracy and concordance between automated and manual methods are reported.
Results
Automated extraction required considerably less time (<1 day) than manual extraction (∼225 person-hr). The collection of demographic data (age, sex, diagnosis) was highly accurate and concordant with both methods (96%–100%). Accuracy (for either extraction approach) and concordance were lower for unstructured data elements in EHR, such as performance status, date of diagnosis, and smoking status (NLP accuracy: 88%–94%; Manual accuracy: 78%–94%; concordance: 71%–82%). Concurrent medications (86%–100%) and comorbid conditions (96%–100%), were reported with high accuracy and concordance. Treatment details were also accurately captured with both methods (84%–100%) and highly concordant (83%–99%). Detection of whether biomarker testing was performed was highly accurate and concordant (96%–98%), although detection of biomarker test results was more variable (accuracy 84%–100%, concordance 84%–99%). Features with syntactic or semantic variation requiring clinical interpretation were extracted with slightly lower accuracy by both NLP and manual review. For example, metastatic sites were more accurately identified through NLP extraction (NLP: 88%–99%; manual: 71%–100%; concordance: 70%–99%) with the exception of lung and lymph node metastases (NLP: 66%–71%; manual: 87%–92%; concordance: 58%) owing to analogous terms used in radiology reports not being included in the accepted gold standard definition.
Conclusions
Automated data abstraction from EHR is highly accurate and faster than manual abstraction. Key challenges include poorly structured EHR and the use of analogous terms beyond the accepted gold standard definition. The application of NLP can facilitate real-world evidence studies at a greater scale than could be achieved with manual data extraction
Keywords: Real-world evidence, Real-world data, Natural language processing, Health records, Artificial intelligence, Validation
Introduction
Real-world data describe patient health and experiences outside of a structured clinical trial setting. As patients receive medical care, large quantities of health care data are generated through the maintenance of health records, which has been accelerated by the widespread adoption of electronic health record (EHR) systems over the past decade. The currently accepted standard for generating real-world data from structured and unstructured EHR fields is manual data abstraction. Whereas this approach has been proven effective, there are drawbacks, such as being time-consuming, labor-intensive, and expensive, making it an arduous process that is highly susceptible to human error. These drawbacks often limit the scale and scope of real-world evidence studies.
To overcome these barriers, natural language processing (NLP) has been explored as an alternate method of data extraction from health records.1,2 NLP-based data extraction can provide results more rapidly and on a larger scale than could be achieved through manual extraction. However, uncertainty remains surrounding the validity of NLP-based extraction results, especially in the context of free-text or dictated clinical notes.3, 4, 5, 6 Recently, the commercially available artificial intelligence (AI) engine, DARWEN (Pentavere, Ontario, Canada), was evaluated against manual extraction of EHR data from a tuberculosis clinic, successfully extracting data from free-format clinical notes. The AI NLP method generated rapid results that were also accurate.2 Extracted features were grouped to evaluate their accuracy on the basis of linguistic and clinical complexity into groups of “simple,” “moderate,” and “complex” variables. To answer clinical questions, however, it is important to be able to investigate each of these features individually or grouped on the basis of the research question at hand. To this end, we compared NLP-based extraction with manual data extraction of clinical features from EHRs of patients with advanced lung cancer at a feature level.
Materials and Methods
Study Setting
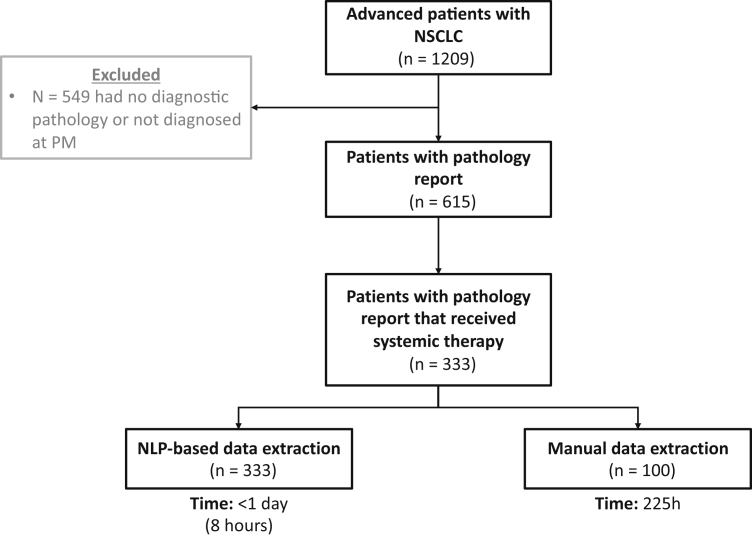
A cohort of 1209 patients diagnosed with advanced lung cancer (documented as stage IIIB or stage IV at diagnosis) was identified through an institutional cancer registry. DARWEN identified a subset of patients who were diagnosed and treated at the Princess Margaret Cancer Centre (PM) between January 2015 and December 2017, allowing for a minimum of 2 years of follow-up. The resulting study cohort consisted of 333 adult patients with advanced lung cancer who had received any systemic treatment at PM during this time (Fig. 1). DARWEN extracted data from EHRs of these patients between their dates of diagnosis until March 30, 2019. This study was conducted in alignment with the approved protocol by the University Health Network Research Ethics Board. As this is a retrospective review of patient records, individual patient consent was waived.
Figure 1.
Study population. NLP, natural language processing; PM, Princess Margaret Cancer Centre.
NLP Approach
Pentavere’s commercially available AI engine, DARWEN, was used for NLP-based data extraction.7, 8, 9, 10 This AI engine combines linguistic (lexical, syntactic, and semantic) rules-based algorithms, machine learning models, and neural networks to extract relevant data from structured and unstructured EHR fields. DARWEN’s capabilities have been previously described in detail.2 Key innovations since then include the use of transformer-based models for classification and named entity recognition, and new techniques to facilitate and accelerate model training with low volumes of training data.
Establishing the Ground Truth
All feature definitions and the ground truth were developed and modified through an iterative process whereby initial definitions were established in partnership with an expert clinical team from the PM. These definitions were then manually tested using a subset of data to identify any discrepancies between the definitions and the actual text, which were resolved with further input from the clinical team. This process allowed for multiple points of clinical input and resulted in a comprehensive final set of definitions that captured clinically relevant language for each feature (Supplementary Table 1).
Training and Fine-Tuning Algorithms
DARWEN’s algorithms were pretrained on other data sets and were then fine-tuned using a subset of patients from the present cohort of 333 patients.2 Algorithms were tuned on the basis of the feature definitions until accuracy, precision (positive predictive value), recall (sensitivity), and F1-score targets were achieved. Accuracy measured the overall effectiveness of the NLP algorithm by calculating the ratio of correctly predicted outputs as a proportion of the total. F1 score is the harmonic mean of precision and recall and was used to evaluate the performance of the algorithm . Algorithm precision and recall were balanced on a case-by-case basis, favoring precision over recall, when necessary, as dictated by research needs. Once the stability of these targets was confirmed on other subsets of unseen data from the study cohort, the algorithms were run on the entire study cohort (n = 333) and the results were independently validated as described below.
Independent Manual Validation
The independent manual validation of these algorithms included 100 randomly selected patient records from the study cohort, which were not used for algorithm training or fine-tuning. Two trained manual abstractors from PM used the final set of feature definitions to extract clinical features from these same 100 patients. A third expert adjudicator reviewed the EHRs in the case of disagreements between abstractors or discrepancies between NLP and manual methods.
Clinical Features
Data extracted (described further below and in Supplementary Table 1) included patient demographics, smoking status, date of diagnosis, Eastern Cooperative Oncology Group (ECOG) performance status closest to date of diagnosis, tumor pathologic subtype, biomarker testing, and results, comorbid conditions, number, and location of metastases, types of systemic therapy, and line of therapy (grouped by first-line or any line), and concomitant immunosuppressive medication.
DARWEN extracted patient demographics, including date of birth and sex, from unstructured clinical notes. Date of birth and date of diagnosis, extracted from pathology reports, were used to calculate age at diagnosis. All mentions of smoking status were extracted from unstructured clinical notes and contextualized to the date of diagnosis. ECOG was extracted longitudinally, with corresponding timestamps, from the date of diagnosis to the end of the study period. For the analyses included in this study, the ECOG status documented the closest to each patient’s date of diagnosis was used. All tumor histologies identified as lung-related within pathology reports or unstructured clinical notes were extracted and grouped into adenocarcinoma, large cell, non–small cell, small cell, or squamous carcinomas (based on the American Joint Committee on Cancer, eighth edition). All documented biomarker tests and results were extracted from pathology reports or clinical notes, specifically for ALK, BRAF, EGFR, KRAS, programmed death-ligand 1 (PD-L1), and ROS1. PD-L1 results were extracted on the basis of explicit mention of positive or negative findings (e.g., “patient is PD-L1–positive”) and the tumor proportion score (TPS) (PD-L1 <1%, 1%–49%, and ≥50%); both features were not always present in the EHRs simultaneously. Any diagnosis or positive history of the comorbid conditions of interest were extracted from clinical notes (see Supplementary Table 2). Any mention of metastases from the predefined anatomical locations was extracted from radiology reports at any time point after the date of diagnosis. Systemic therapies were extracted from clinical notes at any time point after the date of diagnosis and were grouped into chemotherapy, immunotherapy, or targeted therapy. First-line therapy was identified as the first treatment(s) a patient received after diagnosis. For analyses, the line of therapy was categorized as first-line or any line. Immunosuppressive medications specified as being of interest by the study investigators were extracted from the “current medications” section of the clinical notes (Supplementary Table 3).
Statistical Analysis
The results of NLP and manual data extraction were compared for accuracy against the expert adjudicator’s final response. The concordance rate was calculated as the percentage of agreement between the two extraction methods. When applicable, sensitivity and specificity were calculated for both methods.
Results
Before extracting data through either method, clinical gold standard definitions were developed and used to train both manual abstractors and NLP algorithms. Once trained, NLP-based data extraction for this study cohort (n = 333) took less than 1 day, whereas manual data extraction (n = 100) took approximately 225 hours.
Of the 100 patients included in the validation, 54% were men, 66% had adenocarcinoma, 70% had an ECOG of 0 to 1 at diagnosis, and 35% were nonsmokers. All 100 patients received systemic therapy, with 59 on chemotherapy, 36 on targeted therapy, and six on immunotherapy in the first-line setting (Table 1).
Table 3.
Metastatic Sites of Disease
| Site of Metastasis | Positive Cases | Accuracy (%) |
Concordance (%) | |
|---|---|---|---|---|
| NLP | Manual | |||
| Abdominal | 15 | 88.0 | 86.0 | 74.0 |
| Adrenal | 18 | 96.0 | 77.0 | 73.0 |
| Brain | 29 | 99.0 | 71.0 | 70.0 |
| Bone | 52 | 95.0 | 81.0 | 76.0 |
| Liver | 23 | 96.0 | 95.0 | 91.0 |
| Lung | 55 | 71.0 | 87.0 | 58.0 |
| Lymph | 57 | 66.0 | 92.0 | 58.0 |
| Pericardium | 3 | 99.0 | 100 | 99.0 |
| Renal | 3 | 99.0 | 99.0 | 98.0 |
| Spleen | 4 | 99.0 | 97.0 | 96.0 |
NLP, natural language processing.
Demographics and Disease Characteristics
In general, patient demographics were reported with high accuracy and concordance across extraction methods as expected, given these elements were captured with low linguistic complexity (Table 1). Age at diagnosis and sex were extracted from unstructured sources with high accuracy by NLP (100% for both features) and manual extraction methods (99% and 100%, respectively). NLP and manually extracted data were 99% concordant for age and 100% concordant for sex (Fig. 2A-F).
Figure 2.
Concordance between NLP data extraction and manual data extraction results. (A) Demographics and disease characteristics, (B) Metastatic sites, (C) Biomarker testing performed, (D) Biomarker status, (E) Systemic therapy type (first line), and (F) Systemic therapy type (any line). The dashed lines indicate % concordance between NLP and manual data extraction results. NLP, natural language processing; PD-L1, programmed death-ligand 1.
Many disease characteristics were reported with high accuracy by NLP extraction as they were described using a limited number of terms. The histologic subtype was 98% accurate across methods and was highly concordant between NLP and manual extraction (96%). NLP was more accurate than manual extraction for date of diagnosis (94% versus 83%) and ECOG performance status (93% versus 78%) with concordance between methods of 77% and 71%, respectively.
The dynamic nature of some features adds further complexity. For example, smoking status can change over time with strict definitions of ex-smokers and nonsmokers. However, detail in clinical notes may not accurately categorize patient smoking status. Given these challenges, manual extraction was slightly more accurate than NLP for smoking status (94% versus 88%) with a concordance of 82% between methods (Table 1).
Comorbidities
A total of 16 comorbidities were investigated, 11 of which were found, extracted and validated from the study cohorts’ EHRs (Supplementary Table 2). Comorbidities are reported in EHRs in a less content-rich and more straightforward manner. Synonymous terms for each comorbidity are incorporated into feature definitions and are, therefore, captured highly accurately by NLP. Comorbidities were reported with 96% to 100% accuracy for both extraction methods with concordance ranging from 93% to 100%. NLP extraction of more frequent comorbidities was more sensitive than manual extraction (50%–100% versus 20%–100%, respectively) (Supplementary Table 2). Specificity was more similar between methods (97%–100% versus 99%–100%, respectively). In the case of less frequent comorbidities (i.e., an occurrence of ≤1), specificity was 100% and sensitivity ranged from 0% to 100% for both NLP and manual extraction.
Treatment Received
Cancer Treatment
Detailed cancer treatment information in clinical notes was extracted. These treatments were then expressed as Boolean variables capturing whether chemotherapy, immunotherapy, or targeted therapy were received as first-line therapy or were ever received throughout the course of the patient’s treatment. By expressing therapeutic information in such a manner, the variability, and complexity surrounding documentation of cancer treatments in clinical notes were mitigated.
Type of first-line treatment (chemotherapy, immunotherapy, and targeted therapy) was extracted by both methods with high accuracy (NLP: 95%–99%; manual: 96%–100%) and concordance (92%–99%; Table 1). Sensitivity and specificity were high for both methods of extraction, in which sensitivity ranged from 95% to 100% (NLP: 97%–100%; manual: 95%–100%) and specificity ranged from 93% to 100% (NLP: 93%–100%; manual: 100%). Treatments received at any line were also evaluated (Table 1). Both NLP and manual methods performed well when extracting chemotherapy (n = 69) or immunotherapy (n = 12) received at any point (94%–98% accuracy across methods; 88%–96% concordance; 92%–94% sensitivity; 94%–99% specificity). However, for patients receiving targeted therapy ever (n = 40), manual extraction either missed or incorrectly reported 16 cases, resulting in lower accuracy (84% versus 99%, respectively) and specificity (73% and 100%, respectively) than NLP-based extraction, with 83% concordance.
Immunosuppressive Treatment
The study cohort was screened for 11 different immunosuppressive medications, seven of which were received by patients (Supplementary Table 3). Although all patients received systemic therapy (n = 100), only 66 patients received concurrent immunosuppressive treatment. Dexamethasone (n = 54) and prednisone (n = 7) were the most frequently administered immunosuppressive treatments, with the remaining five medications each only prescribed to one patient. When screening for the use of dexamethasone, outputs between extraction methods were 76% concordant. Manual extraction of dexamethasone was more accurate (96% versus 80%) than NLP extraction, but similar specificity was observed across methods (100% and 97.8%, respectively). Manual extraction was also more sensitive (92.6% versus 64.8%) than NLP extraction of dexamethasone. This is likely because of the inferred use of dexamethasone as part of chemotherapy treatment protocols, despite a lack of explicit mention of inclusion within the medical records. As NLP did not have this clinical insight as part of the feature definition, this contributed to missed cases. Prednisone data were reported with 90% concordance and were more accurately detected by NLP than by manual extraction (100% versus 90%). NLP extraction of prednisone data was also more sensitive (100% versus 57.1%), and more specific (100% versus 92.3%) than manual extraction. Although cyclosporine, eculizumab, hydrocortisone, hydroxychloroquine, and methotrexate were taken by one patient each, it is worth noting that manual extraction of hydrocortisone data resulted in 14 false-positive results (NLP specificity 100% versus manual 85.9%).
Biomarkers
EHRs were screened for gene and protein alterations often observed in lung cancer patients. These biomarker reports are content-rich, and the report structure can vary both between test types and over time. However, whether a biomarker was tested is documented relatively clearly and consistently in the clinical records. NLP detected whether biomarker testing was performed with 98% to 99% accuracy for ALK (n = 71), BRAF (n = 19), EGFR (n = 72), KRAS (n = 19), PD-L1 (n = 29), and ROS-1 (n = 4; Table 2). Concordance between the methods ranged from 96% to 98% across biomarkers. NLP extraction across all biomarkers for whether testing was performed resulted in high sensitivity (94.7%–98.6%, except for ROS-1 with 50%) and high specificity (96.6%–100%). Manual extraction reported biomarker testing with 97% to 100% accuracy and was highly sensitive (89.5%–100%) and specific (96.5%–100%).
Table 1.
Demographics and Disease Characteristics
| Characteristic | Number of Cases | Accuracy (%) |
Concordance (%) | |
|---|---|---|---|---|
| NLP | Manual | |||
| Age at diagnosis | 100 | 100 | 99.0 | 99.0 |
| Sex | 100 | 100 | 100 | |
| Male | 54 | |||
| Female | 46 | |||
| Date of diagnosis (±30 d) | 100 | 94.0 | 83.0 | 77.0 |
| ECOG PS at diagnosis | 93.0 | 78.0 | 71.0 | |
| 0 | 16 | |||
| 1 | 54 | |||
| 2 | 14 | |||
| 3 | 13 | |||
| 4 | 1 | |||
| Unknown | 2 | |||
| Smoking status | 88.0 | 94.0 | 82.0 | |
| Nonsmoker | 35 | |||
| Former smoker | 34 | |||
| Smoker | 31 | |||
| Histologic subtype | 98.0 | 98.0 | 96.0 | |
| Adenocarcinoma | 66 | |||
| Large cell | 4 | |||
| Non–small cell | 3 | |||
| Small cell | 21 | |||
| Squamous | 6 | |||
| First line treatmenta | ||||
| Chemotherapy | 59 | 95.0 | 96.0 | 92.0 |
| Immunotherapy | 6 | 99.0 | 100 | 99.0 |
| Targeted Therapy | 36 | 99.0 | 99.0 | 98.0 |
| Treatment (any line) | ||||
| Chemotherapy | 69 | 94.0 | 94.0 | 88.0 |
| Immunotherapy | 12 | 98.0 | 98.0 | 96.0 |
| Targeted therapy | 40 | 99.0 | 84.0 | 83.0 |
ECOG, Eastern Cooperative Oncology Group; NLP, natural language processing; PS, performance status.
One patient received combination therapy as first line treatment.
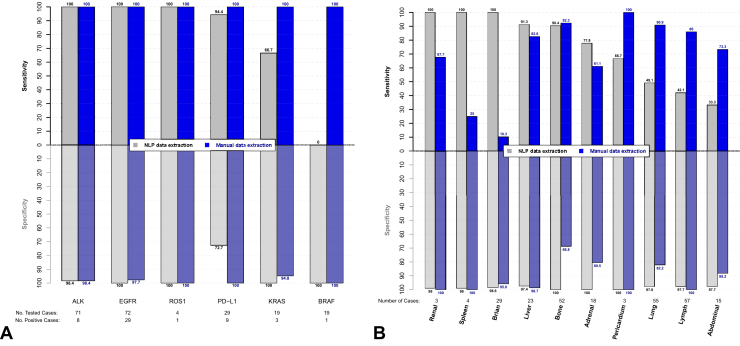
Compared with whether a test has been performed, the biomarker results may be recorded in multiple locations within a pathology report, adding a source of variability to the extraction of these data. NLP extraction of biomarker test results was highly accurate for ALK, BRAF, EGFR, KRAS, and ROS-1 (95%–100%) and was slightly less accurate for PD-L1 status (86%); accordingly, concordance between methods varied across biomarkers (86-100%). Biomarker status for ALK (n = 8), EGFR (n = 29), and ROS-1 (n = 1) was reported with high sensitivity (NLP and manual: 100%) and specificity (NLP: 98%–100%; manual: 98%–100%) across both extraction methods (Fig. 3A). Similarly, PD-L1 results were reported with high sensitivity (n = 20; NLP: 94%; manual: 100%) but with varying specificity (NLP: 73%; manual: 100%). Both BRAF (n = 1) and KRAS (n = 3) status were reported with low sensitivity (BRAF: 0%; KRAS: 67%) and high specificity (BRAF and KRAS: 100%) by NLP extraction. Manual extraction of these same features was highly sensitive (BRAF and KRAS: 100%) and specific (BRAF: 100%; KRAS: 95%). With few patients testing positive for BRAF, KRAS, or ROS-1, there is expected variability in the sensitivity of these extracted data.
Figure 3.
Sensitivity and specificity of (A) biomarker status results and (B) metastasis site results. PD-L1, programmed death-ligand 1.
Metastases
Metastatic sites were detected with varying concordance (70%–99%) between NLP and manual data extraction methods (Table 3). NLP extraction was more accurate than manual extraction for the detection of adrenal (96% versus 77%), brain (99% versus 71%), and bone (95% versus 81%) metastases. NLP-based extraction less accurately detected metastases in the lymph node (66% versus 92%) and lung (71% versus 87%) compared with manual extraction. For all other metastatic sites, NLP and manual data extraction were comparably accurate: abdominal (88% versus 86%), liver (96% versus 95%), pericardium (99% versus 100%), renal (99% versus 99%), and spleen (99% versus 97%). Whereas NLP-extracted data were reported with high specificity (97%–100%), sensitivity varied widely (33%–100%; Fig. 3B). Similarly, manually extracted data was more specific (69%–100%) than sensitive (10%–100%). Metastases are sometimes reported vaguely in radiology reports, with findings frequently being reported as being suspicious (and all the various ways of saying this) but not confirmed. As such, it can be difficult to identify from a passage of text alone whether a mass is explicitly considered to be metastatic. Clinical interpretation by manual abstractors can increase the accuracy of some extracted features but can also present an opportunity for incorrect interpretation of the text. In this study, clinical judgment exercised by manual abstractors when reviewing metastases resulted in low sensitivity. Whereas sensitivity of metastases extracted by NLP also varied widely, NLP was able to more consistently capture reported metastases based solely on the established definitions.
Table 2.
Biomarker Testing and Results
| Biomarker | Biomarker Testing Performed |
Biomarker Results Captureda |
||||||
|---|---|---|---|---|---|---|---|---|
| Number tested | Accuracy (%) |
Concordance (%) | Positive Cases | Accuracy (%) |
Concordance (%) | |||
| NLP | Manual | NLP | Manual | |||||
| ALK | 71 | 99.0 | 97.0 | 96.0 | 8 | 98.6 | 98.6 | 97.2 |
| BRAF | 19 | 99.0 | 98.0 | 97.0 | 1 | 94.7 | 100 | 94.7 |
| EGFR | 72 | 98.0 | 98.0 | 96.0 | 29 | 100 | 98.6 | 98.6 |
| KRAS | 19 | 99.0 | 98.0 | 97.0 | 3 | 94.7 | 94.7 | 89.5 |
| PD-L1 | 29 | 98.0 | 100 | 98.0 | 20 | 86.2 | 100 | 86.2 |
| ROS1 | 4 | 98.0 | 100 | 98.0 | 1 | 100 | 100 | 100 |
NLP, natural language processing; PD-L1, programmed death-ligand 1.
Out of the corresponding number of patients tested.
Discussion
This study illustrates the validity of a commercially available NLP tool to extract feature-level data from the EHRs of patients with advanced lung cancer. Many previous studies either grouped features on the basis of clinical and linguistic complexity,2, or extracted a single feature from clinical documentation.6,11, 12, 13 This study implemented DARWEN to extract clinical features through an automated NLP-based method. These features were validated against a manually extracted data set compiled by two extractors and reviewed by an expert adjudicator with extensive clinical knowledge. The results of NLP-based data extraction were largely comparable to those of the expert manual extraction team, with a few exceptions in which NLP outperformed manual review, or, conversely, was challenged by features requiring clinical interpretation. The sensitivity, specificity, accuracy, and concordance of both extraction methods were evaluated for all extracted features, however, from a clinical perspective, accuracy, and concordance are more important. Regardless of methodology, extracting this data from EHRs is critical for real-world evidence studies and is also necessary for identifying patient subgroups for respective analyses; NLP-based extraction achieves this more rapidly and at a larger scale than could be accomplished with manual review alone.
Despite a single set of feature definitions used across both methods of data extraction, there is an opportunity for interpretation from the set definitions by manual reviewers, leading to variability in extracted results. In some cases, this benefits manual review, as clinical judgment outside of the established feature definitions can be used to identify cases not explicitly documented in the EHR. NLP-based extraction, however, will identify features on the basis of how they are described in the established feature definitions and explicitly captured in clinical notes. Given that certain metastatic sites are reported with richer syntactic and semantic variation in clinical notes, these features have slightly lower accuracy by both NLP and manual extraction. Specifically, NLP extracted lymph node metastases less accurately than manual review owing to analogous terms used in radiology reports not included in the feature definitions. Similarly, it is often difficult to determine whether a lung mass is metastatic, resulting in unclear documentation within imaging reports. Here, clinical judgment allowed the manual reviewer to identify lymph node or lung metastases that were not explicitly documented as metastases. Our iterative process used to define features attempts to account for this complexity found across clinical documents, but clinical documentation is often not explicit and varies considerably in content and quality.
Beyond linguistic complexities and unclear documentation, some clinical characteristics rely on knowledge-based inference more than others. For example, dexamethasone was extracted more accurately by manual review than NLP owing to clinical knowledge that many chemotherapy regimens include dexamethasone without explicit mention of this in the EHR. This unique characteristic of dexamethasone administration as part of chemotherapy was not incorporated into the feature definitions for either manual or NLP review. However, manual reviewers with clinical knowledge naturally deviated from the definition to identify cases in which dexamethasone was administered on the basis of concomitant therapies. Another feature requiring clinical interpretation was PD-L1 immunohistochemistry results. During the study time frame, PD-L1 testing was a relatively newer addition to routine biomarker testing in advanced lung cancer patients, with rapidly evolving guidelines defining criteria for positive or negative PD-L1 status. In 2015, at the beginning of this study period, optimal immunohistochemistry cutoffs were uncertain, and it was unclear which patients would benefit from anti–PD-L1 agents.14 Subsequent studies introduced various cutoffs for PD-L1 expression that would determine whether a patient was labeled as positive for PD-L1, ranging from a TPS of greater than 1% to greater than or equal to 50%.15,16 More recently it has been suggested that both PD-L1 positive and negative patients may benefit from therapies targeting PD-L1.17 Given the evolution of PD-L1 threshold requirements, the way these results have been reported in the EHR has shifted over time. To reflect this, two features were developed for PD-L1 in this study: explicit mention of “positive” or “negative” for PD-L1, and TPS (<1%, 1%–49%, and ≥50%) of PD-L1. These two features were not always simultaneously recorded, and when PD-L1 status was not explicitly documented, DARWEN did not infer positivity or negativity on the basis of TPS alone. This resulted in slightly lower accuracy and specificity of NLP-extracted PD-L1 results when compared with manual extraction, which was supplemented by clinical interpretation.
Dynamic variables are also a challenge to capture accurately over time. For example, accurate capture of smoking status goes beyond identifying the terms “smoker” or “nonsmoker” in a patient’s record. The specific definition of smoker status used in this study requires that a “former smoker” has quit for at least 1 year before their date of diagnosis. This, in turn, requires not only identifying the smoking status as above but also determining whether the patient stopped smoking and when. These ideas are often fragmented across multiple notes throughout the patient record, and may be repeated inconsistently or stated imprecisely, with only approximate relative time (e.g., “patient is a smoker who quit about 3 to 4 y ago”). Compound error is the consequence of this fragmentation, imprecision, and inconsistency; error accumulates at multiple levels, creating a messy “picture” of the patient’s true smoking status.
This study has several limitations, including some inherent to the structure of EHRs and the content captured in these documents. EHRs as a source of real-world clinical data include both structured and unstructured fields. Unstructured notes provide clinicians with the opportunity to record clinical information using their preferred language, which can vary widely over time. These unstructured notes can contain semistructured fields, which are formatted to capture clinical data with relatively low syntactic variability. In contrast, unstructured fields that are unformatted can result in linguistic variability, presenting a challenge to manual and NLP-based extraction alike. As this study only includes EHRs of patients with advanced lung cancer from a single cancer center in Canada, it may not be representative of national or global EHR documentation, necessitating varying degrees of tuning for different cohorts of patients. The algorithms evaluated in this study were applied to another hospital site in Alberta, Canada, and achieved comparable results after fine-tuning.18 Sensitivity and specificity were variable across rare biomarkers in this cohort, emphasizing the value of larger sample sizes for training and implementing NLP and the potential benefit of purposefully selected validation cohorts. Finally, despite our iterative process of developing, testing, and modifying feature definitions with input from clinical experts at each stage, unanticipated language was encountered in some patient records. In rare circumstances (e.g., lung and lymph node metastases), this led to relatively lower accuracy and sensitivity for extraction by NLP compared with manual extractors, who could exercise clinical judgment to interpret as they reviewed patient records. When possible, subsequent work should translate this clinical judgment into additional feature definition requirements to improve NLP accuracy. However, clinical judgment can be subjective, and clinicians may disagree. Regardless, NLP-extracted data in a consistent, objective, and accurate manner, and at a much faster and larger scale than can be achieved manually.
In conclusion, NLP-based data extraction from structured and unstructured fields of EHRs is highly accurate and produces results faster than manual methods. Key challenges remain, including inconsistently structured EHRs, and the use of complex, variable, and vague terms to describe clinical information. Despite these challenges, the use of NLP offers a practical alternative to traditional manual extraction, enabling real-world evidence studies at a larger scale than ever before.
CRediT Authorship Contribution Statement
Marie-Pier Gauthier: Data curation, Investigation, Validation, Writing - original draft.
Jennifer H. Law: Conceptualization, Methodology, Project administration, Writing - original draft.
Lisa W. Le: Conceptualization, Methodology, Formal analysis, Visualization, Writing - original draft.
Janice J. N. Li: Investigation, Visualization, Writing - original draft.
Sajda Zahir, Ryan Chu: Investigation.
Sharon Nirmalakumar, Erin Stewart: Writing - review & editing.
Mike Sung: Data curation.
Christopher Pettengell: Software, Writing - original draft.
Steven Aviv: Software.
Adrian Sacher, Geoffrey Liu, Penelope Bradbury, Frances A. Shepherd: Resources, Writing - review & editing.
Natasha B. Leighl: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing - original draft.
Acknowledgments
Princess Margaret Cancer Foundation (OSI Pharmaceuticals Foundation Chair – Dr. Leighl, Ms. Le, Ms. Zahir, Dr. Chu, Dr. Sung; David and Shapiro Family Fund – Ms. Law). The authors also acknowledge unrestricted support from Roche Canada to the University Health Network for collection of data by natural language processing.
Footnotes
Disclosure: Dr. Pettengell and Mr. Aviv are employees of Pentavere Research Group Inc. The remaining authors declare no conflict of interest.
Cite this article as: Gauthier MP, Law JH, Le LW, et al. Automating access to real-world evidence. JTO Clin Res Rep. 2022;3:100340.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2022.100340.
Supplementary Data
References
- 1.Pons E., Braun L.M., Hunink M.G., Kors J.A. Natural language processing in radiology: a systematic review. Radiology. 2016;279:329–343. doi: 10.1148/radiol.16142770. [DOI] [PubMed] [Google Scholar]
- 2.Petch J., Batt J., Murray J., Mamdani M. Extracting clinical features from dictated ambulatory consult notes using a commercially available natural language processing tool: pilot, retrospective, cross-sectional validation study. JMIR Med Inform. 2019;7 doi: 10.2196/12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somashekhar S., Kumarc R., Rauthan A., et al. Abstract S. 6–07: double blinded validation study to assess performance of IBM artificial intelligence platform, Watson for oncology in comparison with Manipal multidisciplinary tumour board – first study of 638 breast cancer cases. Cancer Res. 2017;77(suppl 4):S6–07. [Google Scholar]
- 4.Wadia R., Akgun K., Brandt C., et al. Comparison of natural language processing and manual coding for the identification of cross-sectional imaging reports suspicious for lung cancer. JCO Clin Cancer Inform. 2018;2:1–7. doi: 10.1200/CCI.17.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y., Weroha S.J., Goode E.L., Liu H., Wang C. Generating real-world evidence from unstructured clinical notes to examine clinical utility of genetic tests: use case in BRCAnes. s. BMC Med Inform Decis Mak. 2021;21:3. doi: 10.1186/s12911-020-01364-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groot O.Q., Bongers M.E.R., Karhade A.V., et al. Natural language processing for automated quantification of bone metastases reported in free-text bone scintigraphy reports. Acta Oncol. 2020;59:1455–1460. doi: 10.1080/0284186X.2020.1819563. [DOI] [PubMed] [Google Scholar]
- 7.Law J., Pettengell C., Chen L., et al. EP1.16-05 real world outcomes of advanced NSCLC patients with liver metastases. J Thorac Oncol. 2019;14:S1066. [Google Scholar]
- 8.Law J, Pettengell C, Le L, et al. Using AI to improve precision medicine: real-world impact of biomarker testing in advanced lung cancer. 2020. Joint Princess Margaret / International Cancer Expert Corps Conference – Cancer AI and Big Data: Success Through Global Collaboration Conference Handbook:20.
- 9.Law J.H., Pettengell C., Le L.W., et al. Generating real-world evidence: using automated data extraction to replace manual chart review. J Clin Oncol. 2019;37(15) [Google Scholar]
- 10.Pettengell C., Law J., Le L., et al. P1.16-07 real world evidence of the impact of immunotherapy in patients with advanced lung cancer. J Thorac Oncol. 2019;14(suppl):S588. [Google Scholar]
- 11.Chilman N., Song X., Roberts A., et al. Text mining occupations from the mental health electronic health record: a natural language processing approach using records from the Clinical Record Interactive Search (CRIS) platform in south London, UK. BMJ Open. 2021;11 doi: 10.1136/bmjopen-2020-042274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajendran S., Topaloglu U. Extracting smoking status from electronic health records using NLP and deep learning. AMIA Jt Summits Transl Sci Proc. 2020;2020:507–516. [PMC free article] [PubMed] [Google Scholar]
- 13.Karhade A.V., Bongers M.E.R., Groot O.Q., et al. Natural language processing for automated detection of incidental durotomy. Spine J. 2020;20:695–700. doi: 10.1016/j.spinee.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Patel S.P., Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 15.Thunnissen E., de Langen A.J., Smit E.F. PD-L1 IHC in NSCLC with a global and methodological perspective. Lung Cancer. 2017;113:102–105. doi: 10.1016/j.lungcan.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Liu X., Guo C.Y., Tou F.F., et al. Association of PD-L1 expression status with the efficacy of PD-1/PD-L1 inhibitors and overall survival in solid tumours: A systematic review and meta-analysis. Int J Cancer. 2020;147:116–127. doi: 10.1002/ijc.32744. [DOI] [PubMed] [Google Scholar]
- 17.Shen X., Zhao B. Efficacy of PD-1 or PD-L1 inhibitors and PD-L1 expression status in cancer: meta-analysis. BMJ. 2018;362:k3529. doi: 10.1136/bmj.k3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung W.Y., Farrer C., Darwish L., Pettengell C., Stewart E.L. 82P Exploring treatment patterns and outcomes of patients with advanced lung cancer (aLC) using artificial intelligence (AI)-extracted data. Ann Oncol. 2021;32(suppl 7):S1407. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.