Abstract
Background
Hepatitis B virus (HBV) can encode microRNA-HBV-miR-3, which can be detected in both HBV-infected cell lines and peripheral blood exosomes of chronic hepatitis B (CHB) patients. This study was conducted to further evaluate its relationship with the current viral markers and their dynamics during antiviral therapy.
Methods
We used Stem-loop real time quantitative PCR (RT-qPCR) to quantify HBV-miR-3 in the serum exosomes of CHB patients by extracting exosomes using the Supbio exosome separation kit and designing primers and Taqman probes specific for HBV-miR-3. We conducted a cross-sectional study and two cohort studies. In the cross-sectional study, 48 treatment-naive (TN) CHB patients were enrolled. In the nucleoside analogues (NAs) cohort study, 20 hepatitis B e antigen (HBeAg) negative CHB patients with negative HBV DNA on NA therapy were followed up for 96 weeks. In the NAs + pegylated interferon (Peg-IFN) cohort study, 40 patients with hepatitis B surface antigen (HBsAg) <1,500 IU/mL, negative HBV DNA, and HBeAg after NAs treatment were enrolled and were switched to Peg-IFN therapy for 48 weeks. HBV-miR-3 titers and other viral markers were detected at different time points.
Results
HBV-miR-3 only existed in CHB patients with a concentration of 6.41±3.55 log10 copies/mL. HBV-miR-3 was positively correlated with HBV DNA, pregenomic RNA (pgRNA), and HBsAg. In the NAs cohort, HBsAg, pgRNA, and HBV-miR-3 levels showed little fluctuation during the 96 weeks of NA treatment (P>0.05). In the NAs + PEG-IFN cohort, HBsAg, pgRNA, and HBV-miR-3 levels declined significantly during the 48 weeks of sequential therapy (P<0.05).
Conclusions
HBV-miR-3 was positively correlated with HBV DNA, pgRNA, and particularly HBsAg in TN CHB patients. Peg-IFN following NA therapy had a positive impact on HBsAg, pgRNA, and HBV-miR-3 decrease.
Keywords: Chronic hepatitis B (CHB), viral markers, exosome, microRNA, hepatitis B surface antigen (HBsAg)
Introduction
The hepatitis B virus (HBV) engenders an increased risk developing lethal complications for many people globally. As of 2019, 296 million people were living with HBV infection worldwide, with 820,000 deaths caused by HBV-related diseases every year (1,2). There are numerous serum markers of HBV infection, including surface antigen (HBsAg), e antigen (HBeAg), core protein-related antigen (HBcrAg), HBV DNA, HBV pregenomic (pgRNA), and so on. With a more comprehensive understanding of HBV and advances in its treatment, HBsAg has become increasingly important for monitoring the effect of treatment, especially concerning HBV cure. HBsAg has been shown to be closely correlated to covalently closed circular DNA (cccDNA) in liver tissues. Also, HBsAg clearance is an indicator to withdraw nucleoside analogues (NAs) in the current major guidelines [American Association for the Study of Liver Disease (AASLD); European Association for the Study of the Liver (EASL); Asian-Pacific Association for the Study of the Liver (APASL)] for chronic hepatitis B (CHB) (3-5). The 2018 AASLD Hepatitis B Guidance proposed a definition of hepatitis cure; undetectable HBsAg was regarded as the main index of functional cure, while complete cure was defined as negative serum HBsAg and intrahepatic cccDNA simultaneously.
HBcrAg contains three proteins, namely HBeAg, core antigen (HBcAg), and pre-core protein, and has a relative molecular weight of 22 kDa, sharing the same 149 amino acid sequences (6). A previous study showed that HBcrAg was associated with serum HBV DNA and intrahepatic cccDNA other than HBsAg (7). The 3.5 kb HBV pgRNA is a transcript of cccDNA, probably encapsulated in nucleocapsid without retroviral transcription. In naive CHB patients, it was positively correlated with the levels of alanine aminotransferase (ALT), HBsAg, and HBV DNA (8). Experiments have shown that it is also correlated with the intrahepatic cccDNA level. One cross-sectional study showed that the HBV pgRNA positive rate was 74.4% among 47 CHB patients receiving entecavir (ETV) treatment after the clearance of HBV DNA, with a concentration of 2.3–4.8 log10 copies/mL (9). Although the current serum virological markers have advantages for the diagnosis and monitoring of CHB, they cannot replace the detection of cccDNA in liver tissues. Therefore, further research is needed to explore better serological biomarkers.
Exosomes were first found in sheep reticulocytes in 1983 and were previously considered cell waste (10). Later studies have highlighted their pathophysiological importance in different pathways (11-13). In different viral infections, exosomes were considered to be carriers to deliver signal substances to innocent cells (14-19). Moreover, a growing body of research has demonstrated the secretion of exosomes by malignant cells to promote tumor progression or inhibit the immune response against tumors (20-23). Exosomal microRNAs are diagnostic biomarkers in various tumors. Exosomal secretion of hepatocellular carcinoma cells is as high as 4E10/million cells. Exosomes in HBV infection have many clinical significances. Firstly, Specific changes in protein contents were detected in exosomes in HBV infected individuals other than controls (24). Secondly, exosomes containing characteristic molecules could become potential indicators for HBV related advanced liver diseases such as liver cirrhosis and hepatocyte carcinoma (HCC). Thirdly, antiviral agents wrapped in exosomes may enhance the efficacy of anti-HBV treatment (25).
Yang et al. found that HBV can encode HBV-related microRNA (26), and named it HBV micro RNA 3 (HBV-miR-3), which is positioned at 373–393 sites in the HBV genome and can be encoded by three of four message RNAs (mRNAs) [except for hepatitis B X (HBx) mRNA]. HBV-miR-3 was shown to be highly expressed in hepatocellular carcinoma cell lines and was positively correlated with HBV DNA. HBV-miR-3 was reported to have an inhibitory effect on HBV-induced liver injury through activating the innate immune response to suppress HBV replication, thus prevent HBV infection from becoming persistent (27). In this study, we detected various virological markers in CHB patients to better understand their correlation with the current HBV serum markers and to observe their dynamics in CHB patients treated with NAs and pegylated interferon (Peg-IFN). Different from previous studies, we followed up different cohorts of CHB patients which allowed us to observe the dynamics of HBV-miR-3 during antiviral therapy. This would provide us new perspectives on the significance of the new biomarker in the management of CHB. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2119/rc).
Methods
Patients
We conducted a cross-sectional study and two cohort studies. In the cross-sectional study, we enrolled 48 treatment-naïve CHB patients [who were designated as the treatment-naive (TN) group] from the Third Affiliated Hospital of Sun Yat-sen University between September 2016 and March 2017. All CHB patients were diagnosed according to the “China Guidelines for the Prevention and Treatment of Chronic Hepatitis B in 2015”. Twelve healthy individuals, 12 hepatitis C patients, and 12 non-alcoholic fatty liver disease (NAFLD) patients were also enrolled as control subjects, respectively.
In the NAs cohort study, we enrolled 20 CHB patients with negative HBeAg and HBV DNA who received NA therapy for more than 1 year between June 2009 and June 2012. The patients were followed up for 2 years during which there were no relapses. Clinical data and serum samples were collected at baseline and 24, 48, 72, and 96 weeks. In the NAs + Peg-IFN cohort study, 40 CHB patients meeting the same inclusion criteria as the NAs cohort discontinued NA therapy and received Peg-IFN monotherapy for 48 weeks. The clinical data and serum samples of these patients were analyzed at different time points, including at baseline and 12, 24, and 48 weeks.
This study was approved by the Institutional Review Board (IRB) of the Third Affiliated Hospital of Sun Yat-sen University (No. [2016]2-129), and all included participants signed an informed consent form. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was registered at U.S. National Library of Medicine ClinicalTrials (No. NCT02745704).
Blood specimens and clinical tests
Five milliliters of blood were drawn from each patient in a fasting state at baseline and every visit. The prepared serum samples were maintained at −80 ℃ until further utilization. Biochemical tests such as aspartate aminotransferase (AST), ALT, albumin (ALB), total bilirubin (TBIL), direct bilirubin (DBIL), and indirect bilirubin (IBIL) were collected and tested by the clinical laboratory of the hospital (Hitachi 7600 automatic analyzer, Tokyo, Japan). HBsAg was quantitatively detected by the Elecsys HBsAg II (Roche Diagnostics, USA, the minimum detection limit is 0.05 IU/mL). HBsAb, HBeAg, HBeAb, and HBcAb were detected using the Abbott Chemiluminescence method. HBV DNA was quantitatively detected by the Roche Internal Standard method (Cobas Taqman, Roche Molecular Diagnostics, USA, the minimum detection limit is 20 IU/mL).
Serum exosome preparation and total miRNA extraction
The exosome separation kit (SUPI-1406, SUPBIO, China) was used to extract 200 µL of exosome suspension. Electron microscopy (JEM-1200EX, Japan Electronics Corporation, Japan) was used to identify exosomes. The NanoFCM Flow NanoAnalyzer (H-Wayen, China) was used to determine the exosomal diameter. Total mircro RNA (miRNA) was extracted using a commercial kit (DP503 miRcute, TIANGEN, China). HBV pgRNA was quantified using a Hepatitis B Virus pgRNA Detection Kit (SUPBIO, China).
HBV-miR-3 quantification—Stem-loop real time quantitative PCR (RT-qPCR)
For quantification of HBV-miR-3, we used stem-loop RT-qPCR as demonstrated previously (28), using miR-21 as a positive control. The specific primers and Taqman minor groove binder (MGB) probes were as follows: Specific reverse transcription primer for HBV-miR-3 5'-CGCGAGCAC-AGAATTAATACGACTCACTATACGCGAAACGCCGCA-3', forward primer for HBV-miR-3 5'-TGTGCCCGCTGGATGTGTC-3', reverse primer for HBV-miR-3 5'-CGCGAGCACAGAA-TTAATACG-3', Taqman MGB probe for HBV-miR-3 5'-FAM-CTCACTATACGCGAAAC-G-MGB-3', reverse transcription primer for miR-21 5'-GTCGTATCCAGTGCAGGGTCCGAG-GTATTCGCACTGGATACGACTCAACA-3', forward primer for miR-21 5'-GCCCGCTAGCT-TATCAGACTGATG-3', reverse primer for miR-21 5'-GTGCAGGGTCCGAGGT-3', and Taqman MGB probe for miR-21 5'-FAM-CTGGATACGACTCAACA-MGB-3'.
We synthesized the HBV-miR-3 standard (TaKaRa Co., Ltd., China) to construct a standard curve in the range of 6.02E8–6.02E2 copies/µL (10-fold gradient). Reverse transcription was performed in a reaction system of 15 µL (TaqManTM microRNA Reverse Transcription Kit, Applied Biosystems Co., Ltd., USA), which comprised 5 µL of sample and 800 nM of reverse transcription primer. Other reagents and their concentrations, as well as the reverse transcription procedure, were described in the instructions.
Following reverse transcription, 2 µL of product was used to perform qPCR. The reaction system comprised 20 µL of 2× Taqman Universal Master Mix II, no uracil-N-glycosylase (UNG) (Applied Biosystems Co., Ltd., USA), 1.5 µM of forward primer, 0.7 µM of reverse primer, and 0.2 µM of Taqman MGB probe. The reactions were performed using the instrument according to the procedure described in the instructions (95 ℃ for 10 min, 40 cycles of 15 s at 95 ℃, and 1 min at 60 ℃). The quantity of HBV-miR-3 in the samples could then be calculated from the standard curve.
Statistical analysis
We used SPSS 20.0 software (IBM, USA) to establish the database and carry out statistical processing. Normally distributed variables were described by the mean ± standard deviation (SD). The t-test or one-way analysis of variance (ANOVA) was used to compare independent samples. Non-normally distributed measurement data were described by the median (M), and the rank-sum test was used for comparison. P<0.05 was considered statistically significant.
Results
Identification of exosomes
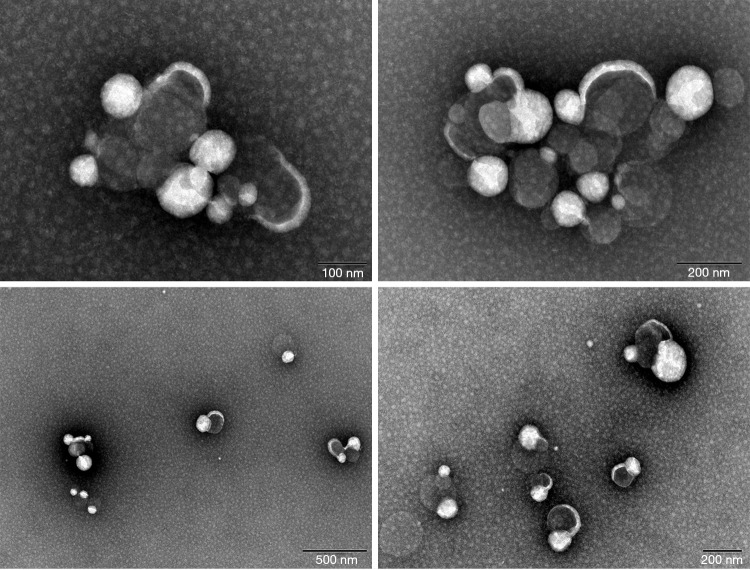
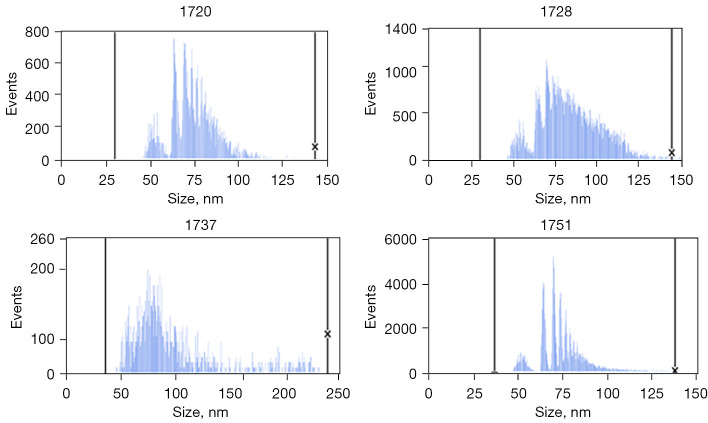
Multiple exosome-like structures could be observed in the samples under electron microscopy (Figure 1). In addition, the particle sizes of the suspensions were determined; the particle sizes of the four tested samples were: sample 1720 (median 73.3 nm, standard deviation 12.1), sample 1728 (median 81.4 nm, standard deviation 17.6), sample 1737 (median 83.8 nm, standard deviation 39.8), and sample 1751 (median 72.4 nm, standard deviation 13) (Figure 2). The concentration of exosomes in these four samples was 3.76E10/mL for sample 1720, 2.74E11/mL for 1728, 4.54E11/mL for sample 1737, and 2.34E11/mL for sample 1751.
Figure 1.
Numerous exosome-like particles were found under electron microscopy.
Figure 2.
Original particle size distribution data.
Expression of HBV-miR-3
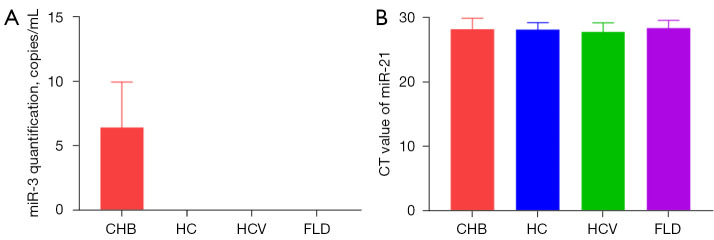
MicroRNAs in the peripheral blood exosomes of the 48 CHB patients, 12 healthy controls (HC), 12 hepatitis C patients (HCV group), and 12 NAFLD patients (FLD group) were detected. HBV-miR-3 was only detected in CHB patients, the concentration of HBV-miR-3 was 6.41±3.55 log10 copies/mL (Figure 3A). Besides, using microRNA21 as an internal reference, no significant difference was found in the cycle threshold (CT) values of the microRNA-21 among the four groups (P>0.05, Figure 3B).
Figure 3.
Detection of HBV-miR-3 in CHB patients. (A) miR-3 quantification, miR-3 was only detected in the CHB group; (B) CT values of miR-21 in the four groups, P>0.05. CHB, chronic hepatitis B; HC, healthy control; HCV, hepatitis C virus; FLD, fatty liver disease; CT, cycle threshold; HBV, hepatitis B virus.
The relationship between HBV-miR-3 and other virological markers
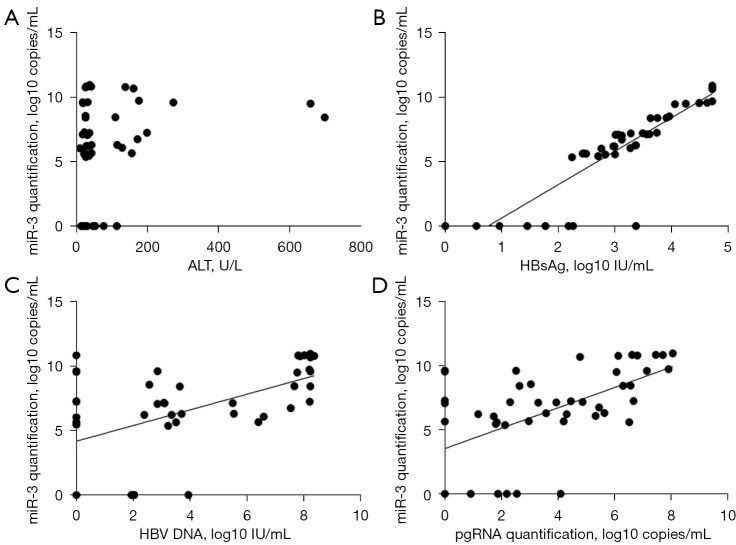
The demographic, serological, and virological data of the 48 CHB patients are displayed in Table 1. The correlation analysis showed that there was no significant correlation between HBV-miR-3 and the ALT level (Figure 4A); however, HBV-miR-3 was positively correlated with HBV DNA, pgRNA, and HBsAg (the highest correlation coefficient R2 was 0.811 for HBsAg, Figure 4B-4D).
Table 1. Demographic and disease characteristics of the CHB patients (n=48).
| Characteristic | N (SD or %) |
|---|---|
| Age, mean (SD), years | 36 (±9) |
| Sex, male | 37 (77%) |
| ALT, mean (SD), U/L | 86 (±138) |
| HBeAg(+) | 17 (35%) |
| HBV DNA, mean (SD), log10 IU/mL | 3.67 (±3.26) |
| HBV DNA >6 log10 IU/mL | 15 (31%) |
| 6 log10 IU/mL > HBV DNA >3 log10 IU/mL | 11 (23%) |
| 3 log10 IU/mL > HBV DNA >20 IU/mL | 6 (13%) |
| HBV DNA, undetected | 16 (33%) |
| HBsAg titre, mean (SD), log10 IU/mL | 3.21 (±1.23) |
| HBsAg ≤100 IU/mL | 6 (13%) |
| 100 IU/mL < HBsAg ≤1,000 IU/mL | 12 (25%) |
| 1,000 IU/mL < HBsAg ≤10,000 IU/mL | 17 (35%) |
| 10,000 IU/mL < HBsAg | 13 (27%) |
| miR-3, mean (SD), log10 copies/mL | 6.41 (±3.55) |
| pgRNA, mean (SD), log10 copies/mL | 3.62 (±2.60) |
CHB, chronic hepatitis B; SD, standard deviation; ALT, alanine aminotransferase; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; HBsAg, hepatitis B surface antigen; pgRNA, pregenomic RNA.
Figure 4.
Correlation analysis. (A) miR-3 quantification was not related to ALT levels; (B) miR-3 quantification was highly related to HBsAg levels, R2=0.811, P<0.001; (C) miR-3 quantification was positively correlated to HBV DNA levels, R2=0.311, P<0.001; (D) miR-3 quantification was positively correlated to pgRNA levels, R2=0.336, P<0.001. ALT, alanine aminotransferase; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; pgRNA, pregenomic RNA.
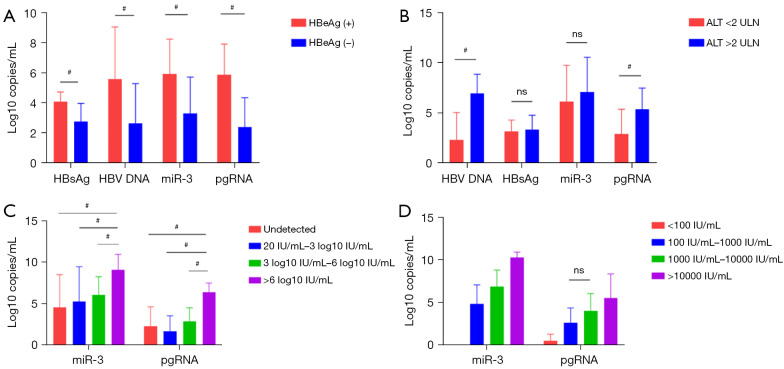
Among the 48 CHB patients, 17 were HBeAg(+) and 31 were HBeAg(−), and there were significant differences between these two groups in ALT (155±210 vs. 49±48 U/L), HBsAg (4.09±0.64 vs. 2.74±1.22 log10 IU/mL), HBV DNA (5.57±3.48 vs. 2.63±2.65 log10 IU/mL), HBV pgRNA quantification (5.93±2.31 vs. 5.88±2.03 log10 copies/mL), and HBV-miR-3 (5.88±2.03 vs. 2.38±1.96 log10 copies/mL) (P<0.05, Figure 5A).
Figure 5.
The relationships between miR-3/pgRNA and other parameters. (A) Concentrations of different viral markers according to HBeAg status; (B) concentrations of different viral markers according to ALT level; (C) concentrations of miR-3 and pgRNA according to HBV DNA level; (D) concentrations of miR-3 and pgRNA according to HBsAg level. ns, not statistically significant P>0.05; #, P<0.05. HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; pgRNA, pregenomic RNA; ALT, alanine aminotransferase; ULN, upper limits of normal.
The 48 patients were divided into two groups according to the level of ALT: (I) ALT <2 upper limits of normal (ULN) group; and (II) ALT >2 ULN group. We observed that HBV DNA and pgRNA were markedly higher in the high ALT group compared to the low ALT group. Meanwhile, there were no differences in HBsAg and HBV-miR-3 between the high and low ALT groups (Figure 5B).
Furthermore, the 48 patients were divided into four groups according to HBV DNA level: (I) high viral load group: HBV DNA >6 log10 IU/mL, 15 cases (31%); (II) medium viral load group: 6 log10 IU/mL > HBV DNA >3 log10 IU/mL, 11 cases (23%); (III) low viral load group: 3 log10 IU/mL > HBV DNA >20 IU/mL, six cases (13%); and (IV) HBV DNA negative group (i.e., HBV DNA was undetectable), 16 cases (33%). The result showed that HBV-miR-3 in high viral load group was significantly higher than those in the other groups (P<0.05), but there was no significant difference between the other three groups (P>0.05). HBV pgRNA also exhibited the same trend (Figure 5C).
Moreover, the 48 patients were also divided into four groups according to HBsAg level: (I) HBsAg >10,000 IU/mL, 13 cases (27%); (II) 1,000 IU/mL < HBsAg ≤10,000 IU/mL, 17 cases (35%); (III) 100 IU/mL < HBsAg ≤1,000 IU/mL, 12 cases (25%); and (IV) HBsAg ≤100 IU/mL, six cases (13%). The results showed a positive correlation between HBsAg and HBV-miR-3, pgRNA (P<0.05, Figure 5D).
The dynamics of viral markers during NA therapy
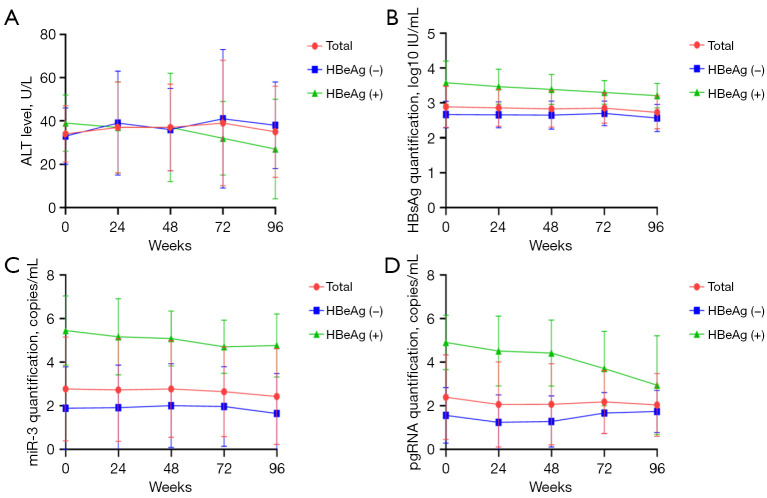
The NAs cohort comprised 20 CHB patients; the baseline demographic, serological, and virological data of these patients are listed in Table 2. Five HBeAg(+) patients and 15 HBeAg(−) patients were enrolled in this study. The HBeAg(+) patients had significantly higher baseline levels of HBsAg (3.58±0.62 vs. 2.67±0.38 log10 IU/mL), HBV-miR-3 (5.46±1.59 vs. 1.89±0.49 log10 copies/mL), and HBV pgRNA (4.91±1.25 vs. 1.56±1.28 log10 copies/mL) than the HBeAg(−) patients (P<0.05) (Figure 6).
Table 2. Baseline characteristics of CHB patients in the NAs cohort (n=20).
| Characteristics | N (SD or %) |
|---|---|
| Age, mean (SD), years | 40 (±9) |
| Sex, male | 19 (95%) |
| NAs types | |
| ETV | 8 (40%) |
| LAM | 7 (35%) |
| ADV | 5 (25%) |
| ALT, mean (SD), U/L | 34 (±13) |
| HBeAg, + | 5 (25%) |
| HBsAg titre, mean (SD), log10 IU/mL | 2.89 (±0.59) |
| miR-3, mean (SD), log10 copies/mL | 4.38 (±3.41) |
| pgRNA, mean (SD), log10 copies/mL | 2.40 (±1.94) |
CHB, chronic hepatitis B; NA, nucleoside analogue; SD, standard deviation; ETV, entecavir; LAM, lamivudine; ADV, adefovir; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; pgRNA, pregenomic RNA.
Figure 6.
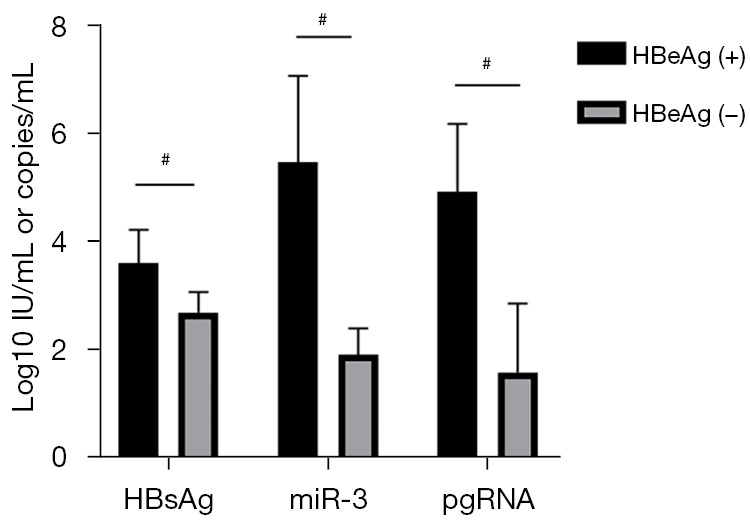
Baseline viral markers according to the HBeAg status. #, P<0.05. HBsAg, hepatitis B surface antigen; pgRNA, pregenomic RNA; HBeAg, hepatitis B e antigen.
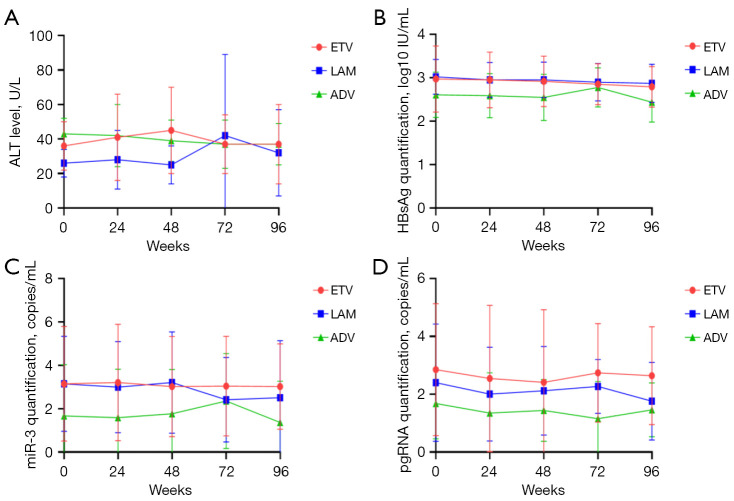
The levels of HBsAg, HBV-miR-3, and pgRNA did not change significantly during the 96 weeks of NA treatment, which exhibited similar dynamics if we divided the patients into subgroups according to HBeAg status (Figure 7). To determine whether NAs had an impact on the dynamics, we divided the patients into different NAs groups: lamivudine (LAM), ETV, and adefovir (ADV). The results showed that there were no significant changes in the above viral marker levels in each group during 2 years of therapy and there were no differences among the three groups at any time point (Figure 8).
Figure 7.
Dynamic change of different serum viral markers according to HBeAg status. ALT, HBsAg, miR-3, and pgRNA were not statistically different during 2 years of NA therapy. (A) Dynamic change of ALT according to HBeAg status; (B) dynamic change of HBsAg according to HBeAg status; (C) dynamic change of miR-3 according to HBeAg status; (D) dynamic change of pgRNA according to HBeAg status. ALT, alanine aminotransferase; HBsAg, hepatitis B surface antigen; pgRNA, pregenomic RNA; HBeAg, hepatitis B e antigen.
Figure 8.
Dynamic change of different serum viral markers according to NAs. ALT, HBsAg, miR-3, and pgRNA showed no statistically significant changes during 2 years of NA therapy. (A) Dynamic change of ALT according to NAs; (B) dynamic change of HBsAg according to NAs; (C) dynamic change of miR-3 according to NAs; (D) dynamic change of pgRNA according to NAs. ALT, alanine aminotransferase; HBsAg, hepatitis B surface antigen; pgRNA, pregenomic RNA; ETV, entecavir; LAM, lamivudine; ADV, adefovir; NA, nucleoside analogue.
The dynamics of viral markers during sequential therapy (NAs—Peg-IFN)
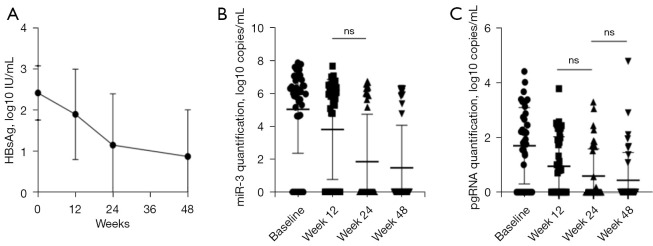
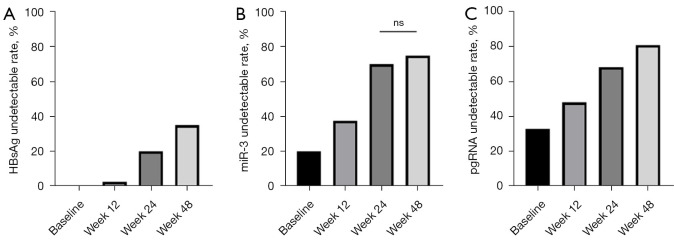
Table 3 shows the baseline demographic, serological, and virological data of 40 CHB patients. We found that the HBsAg level decreased significantly after Peg-IFN treatment for 48 weeks. The quantification of HBsAg at each time point was 2.42±0.66, 1.90±1.10, 1.15±1.25, and 0.87±1.14 log10 IU/mL (P<0.05). The same trend was observed in the changes of HBV-miR-3 and pgRNA, especially from baseline to week 12 (Figure 9). The undetectable rate of three viral parameters increased during the 48-week treatment and reached a maximum for each parameter (35% for HBsAg, 75% for HBV-miR-3, and 80% for pgRNA) (Figure 10).
Table 3. Baseline characteristics of CHB patients in the NAs + Peg-IFN group (n=40).
| Characteristic | N (SD or %) |
|---|---|
| Age, mean (SD), years | 40 (±7) |
| Sex, male | 36 (90%) |
| ALT, mean (SD), U/L | 29 (±17) |
| HBsAg titre, mean (SD), log10 IU/mL | 2.42 (±0.66) |
| 500–1,500 IU/mL | 20 (50%) |
| 200–500 IU/mL | 8 (20%) |
| <200 IU/mL | 12 (30%) |
| miR-3, mean (SD), log10 copies/mL | 5.16 (±3.66) |
| pgRNA, mean (SD), log10 copies/mL | 1.70 (±1.40) |
CHB, chronic hepatitis B; NA, nucleoside analogue; Peg-IFN, pegylated interferon; SD, standard deviation; ALT, alanine aminotransferase; HBsAg, hepatitis B surface antigen; pgRNA, pregenomic RNA.
Figure 9.
Dynamic changes of three different viral markers. (A) HBsAg declined significantly during the 48-week IFN switched therapy; (B) miR-3 declined significantly during 48 weeks of treatment, with little change from week 12 to 24; (C) pgRNA declined significantly through 48 weeks treatment, with little changes from weeks 12 to 24 and weeks 24 to 48. HBsAg, hepatitis B surface antigen; pgRNA, pregenomic RNA; IFN, interferon; ns, not statistically significant, P>0.05.
Figure 10.
Undetectable rate of different viral markers during the 48-week IFN therapy. (A) HBsAg undetectable rate increased over time; (B) miR-3 undetectable rate increased over time except for from week 24 to week 48; (C) pgRNA undetectable rate increased over time. HBsAg, hepatitis B surface antigen; pgRNA, pregenomic RNA; IFN, interferon; ns, not statistically significant, P>0.05.
Discussion
MicroRNA is a non-coding single-stranded RNA (20–24 bp in size) encoded by endogenous genes. Due to their low molecular weights and expression levels, the quantitative study of microRNAs requires extremely sensitive and quantitative analytical tools, including Northern hybridization, Microarray analysis, chip technology, fluorescent quantitative PCR, etc. (29). Microarray is a faster, widely used, and more promising method for studying the expression of microRNAs (30). Liu et al. first published results regarding the high throughput of microlattices (31); their findings revealed the expression markers of tissue-specific microRNAs and were verified by Northern blot and real-time PCR. Although chip technology plays an important role in gene information analysis owing to its high throughput and parallel processing characteristics, a large amount of information does not necessarily equate to higher quality. One of the prominent weaknesses of gene chip technology is its poor stability and repeatability, as well as its inability to achieve quantitative detection. Meanwhile, real-time PCR can accurately quantify the expression of microRNAs (32,33), and is often used to validate predicted microRNAs. In this study, stem-loop RT qPCR was used to quantify the expression of HBV-miR-3. The results showed that HBV-miR-3 only existed in the peripheral blood exosomes of CHB patients, suggesting that HBV-miR-3 was secreted by HBV-infected hepatocytes. HBV-miR-3 is expected to be used in the monitoring of HBV infection.
Regression analysis was used to observe the correlation between HBV-miR-3 and current biochemical and virological indicators. The results showed that HBV-miR-3 was unrelated to ALT level; however, HBV-miR-3 was positively correlated to HBV DNA, HBsAg, and pgRNA. Yang et al. analyzed the relative quantification of HBV-miR-3 levels in CHB patients during the active and convalescent stages. The results showed that the levels of HBV-miR-3 in the active stage were significantly higher than those in the convalescent stage (26). Considering the differences between the active and convalescent stages, the levels of HBsAg and HBV DNA may decrease to some extent with improvement of the patient’s condition at different stages, HBV-miR-3 may also decrease. Our study mainly compared the effects of ALT levels on HBV-miR-3 in different patients horizontally. HBV-miR-3 was positively correlated with HBsAg, HBV DNA, and pgRNA, which was similar to the results of Yang et al. These findings can be further confirmed by enlarging the detection range.
At present, the dynamics of HBsAg during NA treatment are still unclear. Chevaliez et al. of the Paris University of France followed up CHB patients receiving NA therapy for about 102 months, and the dynamic changes of HBV DNA and HBsAg were monitored dynamically for a prolonged period. Their results indicated that there were three situations in the course of anti-HBV treatment. Firstly, the level of HBsAg in serum continuously decreased in both the positive and negative stages of HBV DNA. Secondly, it declined only in the positive stage of HBV DNA, and thirdly, the HBsAg declined only in the HBV DNA negative stage. When the threshold of HBV DNA decline was 3.29±0.49 log10 IU/mL, the serum HBsAg level decreased by 0.007±0.007 log10 IU/mL per month and 0.084 log10 IU/mL per year. It was concluded that the complete elimination of HBsAg in CHB patients would take 52.2 years. Therefore, most CHB patients require lifelong therapy when treated with NAs (34).
In this study, the HBsAg level in CHB patients with negative HBV DNA did not decrease significantly after 96 weeks of NA treatment. The patients were further divided into two groups according to their e-antigen status. Our results suggested that the HBsAg levels of patients with positive e-antigen were significantly higher than those of patients with HBeAg-negative at all follow-up time points. However, there were no significant changes in the HBsAg level after 96 weeks of antiviral treatment regardless of HBeAg status, which was consistent with numerous domestic and foreign studies. We further divided the patients into ETV, LAM, and ADV treatment groups, and the result showed that there were no significant differences among the three groups at all follow-up time points in terms of HBsAg, HBV-miR-3, or pgRNA, suggesting that the types of antiviral drugs had little effect on HBsAg, HBV-miR-3, and pgRNA. It is presumed that NAs mainly act on the reverse transcription process, but exert no inhibitory effect on intrahepatic cccDNA, while HBV-miR-3 and pgRNA are mainly synthesized using cccDNA as a template.
Compared with NAs, interferon can act on cccDNA in hepatocytes, which can reduce the production of its downstream products accordingly. For patients with positive e-antigen, the level of serum HBsAg is closely related to the level of intrahepatic cccDNA, which can reflect the level of hepatic cccDNA. The level of HBsAg can also be monitored during treatment, and its reduction often indicates a better antiviral effect. Janssen et al. reported that a significant reduction of HBsAg is a predictor of sustained virological response (SVR) in HBeAg-positive patients (35). The baseline quantification of HBsAg and its decrease during treatment can predict the serological conversion rate of HBeAg. For patients with negative e-antigen, a lower baseline level of HBsAg or a decrease in HBsAg during treatment often indicates better antiviral efficacy. With the deepening understanding of CHB, many clinical studies have been carried out to improve the HBsAg clearance rate. The new Switch study indicated that 14.4% (22/153) and 20.7% (31/150) of patients sequentially treated with NAs and then Peg-IFNα-2a cleared HBsAg following 48 and 96 weeks of treatment, respectively (36). Moreover, the OSST study showed that the Peg-IFNα-2a combined/sequential treatment group could obtain higher a HBeAg conversion rate and HBsAg clearance rate at week 48 than ETV alone (37).
A total of 40 HBeAg(−) CHB patients with undetectable HBV DNA after NA treatment were enrolled in this study. After sequential Peg-IFN treatment, the levels of HBsAg decreased significantly and the negative rate of HBsAg increased markedly during treatment. Similarly, after 48 weeks of Peg-IFN treatment, the HBV-miR-3 and pgRNA levels decreased significantly. To further explore whether HBV-miR-3 and pgRNA are more representative of cccDNA than HBsAg, this study counted the undetectable rates at different follow-up time points. The results showed that HBsAg could be detected in all patients at baseline, while HBV-miR-3 and pgRNA had higher undetectable rates. During treatment, the undetectable rate of the above three parameters gradually increased, but HBsAg was still the lowest. Compared with HBsAg, the detection sensitivity of HBV-miR-3 and pgRNA was lower, implying their non-superiority to HBsAg in reflecting the level of cccDNA. However, the detection of HBV-miR-3 could still become a supplement of current viral markers on indicating antiviral effect especially for those with Peg-IFN therapy. What’s more, the clearance of HBV-miR-3 may imply a better prognosis. Future studies are still in need to illustrate its mechanism on the progression of HBV infection and its relationship with functional cure.
In conclusion, HBV-miR-3 is a HBV-related miRNA and was positively correlated with HBsAg, HBV DNA, and pgRNA in TN CHB patients. NA monotherapy had little effect on the HBV-miR-3 level when HBV DNA was undetectable. In contrast, Peg-IFN following NA therapy had a positive impact on the decrease of HBsAg, pgRNA, and HBV-miR-3.
Supplementary
The article’s supplementary files as
Acknowledgments
We are grateful to Yuehua Huang and Youming Chen (from Department of Infectious Diseases, the Third Affiliated Hospital of Sun Yat-sen University) for their support and constructive suggestions.
Funding: This study was funded by the National Science and Technology Major Project of China (No. 2018ZX10302204-002) and the National Natural Science Foundation of China (No. 81672701).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study obtained ethical approval from the Institutional Review Board (IRB) of the Third Affiliated Hospital of Sun Yat-sen University (No. [2016]2-129), and informed consent was signed by all patients and healthy controls. All chronic hepatitis B patients were diagnosed in accordance with the “China Guidelines for the Prevention and Treatment of Chronic Hepatitis B in 2015” and the study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2119/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2119/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2119/coif). The authors have no conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Fact sheets: Hepatitis B: World Health Organization; 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b
- 2.Global hepatitis report: World Health Organization; 2017. Available online: https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/
- 3.Terrault NA, Lok ASF, McMahon BJ, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018;67:1560-99. 10.1002/hep.29800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Association for the Study of the Liver . Electronic address: easloffice@easloffice.eu; European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol 2017;67:370-98. 10.1016/j.jhep.2017.03.021 [DOI] [PubMed] [Google Scholar]
- 5.Sarin SK, Kumar M, Lau GK, et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int 2016;10:1-98. 10.1007/s12072-015-9675-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe T, Inoue T, Tanaka Y., Hepatitis B. Core-Related Antigen and New Therapies for Hepatitis B. Microorganisms 2021;9:2083. 10.3390/microorganisms9102083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuaypen N, Posuwan N, Chittmittraprap S, et al. Predictive role of serum HBsAg and HBcrAg kinetics in patients with HBeAg-negative chronic hepatitis B receiving pegylated interferon-based therapy. Clin Microbiol Infect 2018;24:306.e7-306.e13. 10.1016/j.cmi.2017.07.016 [DOI] [PubMed] [Google Scholar]
- 8.van Campenhout MJH, van Bömmel F, Pfefferkorn M, et al. Host and viral factors associated with serum hepatitis B virus RNA levels among patients in need for treatment. Hepatology 2018;68:839-47. 10.1002/hep.29872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Shen T, Huang X, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol 2016;65:700-10. 10.1016/j.jhep.2016.05.029 [DOI] [PubMed] [Google Scholar]
- 10.Johnstone RM, Adam M, Hammond JR, et al. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 1987;262:9412-20. 10.1016/S0021-9258(18)48095-7 [DOI] [PubMed] [Google Scholar]
- 11.Pirisinu M, Pham TC, Zhang DX, et al. Extracellular vesicles as natural therapeutic agents and innate drug delivery systems for cancer treatment: Recent advances, current obstacles, and challenges for clinical translation. Semin Cancer Biol 2022;80:340-55. 10.1016/j.semcancer.2020.08.007 [DOI] [PubMed] [Google Scholar]
- 12.Kim HJ, Kim G, Lee J, et al. Secretome of Stem Cells: Roles of Extracellular Vesicles in Diseases, Stemness, Differentiation, and Reprogramming. Tissue Eng Regen Med 2022;19:19-33. 10.1007/s13770-021-00406-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Askenase PW. Exosomes provide unappreciated carrier effects that assist transfers of their miRNAs to targeted cells; I. They are 'The Elephant in the Room'. RNA Biol 2021;18:2038-53. 10.1080/15476286.2021.1885189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mack M, Kleinschmidt A, Brühl H, et al. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat Med 2000;6:769-75. 10.1038/77498 [DOI] [PubMed] [Google Scholar]
- 15.Rozmyslowicz T, Majka M, Kijowski J, et al. Platelet- and megakaryocyte-derived microparticles transfer CXCR4 receptor to CXCR4-null cells and make them susceptible to infection by X4-HIV. AIDS 2003;17:33-42. 10.1097/00002030-200301030-00006 [DOI] [PubMed] [Google Scholar]
- 16.Chai N, Chang HE, Nicolas E, et al. Properties of subviral particles of hepatitis B virus. J Virol 2008;82:7812-7. 10.1128/JVI.00561-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spehner D, Drillien R. Extracellular vesicles containing virus-encoded membrane proteins are a byproduct of infection with modified vaccinia virus Ankara. Virus Res 2008;137:129-36. 10.1016/j.virusres.2008.07.003 [DOI] [PubMed] [Google Scholar]
- 18.Masciopinto F, Giovani C, Campagnoli S, et al. Association of hepatitis C virus envelope proteins with exosomes. Eur J Immunol 2004;34:2834-42. 10.1002/eji.200424887 [DOI] [PubMed] [Google Scholar]
- 19.Gastaminza P, Dryden KA, Boyd B, et al. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J Virol 2010;84:10999-1009. 10.1128/JVI.00526-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res 2011;71:3792-801. 10.1158/0008-5472.CAN-10-4455 [DOI] [PubMed] [Google Scholar]
- 21.Zhang HG, Grizzle WE. Exosomes and cancer: a newly described pathway of immune suppression. Clin Cancer Res 2011;17:959-64. 10.1158/1078-0432.CCR-10-1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melo SA, Sugimoto H, O'Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014;26:707-21. 10.1016/j.ccell.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev 2013;32:623-42. 10.1007/s10555-013-9441-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X, Wu Y, Duan J, et al. Quantitative proteomic analysis of exosome protein content changes induced by hepatitis B virus in Huh-7 cells using SILAC labeling and LC-MS/MS. J Proteome Res 2014;13:5391-402. 10.1021/pr5008703 [DOI] [PubMed] [Google Scholar]
- 25.Li J, Liu K, Liu Y, et al. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat Immunol 2013;14:793-803. 10.1038/ni.2647 [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Li H, Sun H, et al. Hepatitis B Virus-Encoded MicroRNA Controls Viral Replication. J Virol 2017;91:e01919-16. 10.1128/JVI.01919-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X, Sun L, Mu T, et al. An HBV-encoded miRNA activates innate immunity to restrict HBV replication. J Mol Cell Biol 2020;12:263-76. 10.1093/jmcb/mjz104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang S, Cao X, Tian X, et al. High-Throughput Sequencing Identifies MicroRNAs from Posterior Intestine of Loach (Misgurnus anguillicaudatus) and Their Response to Intestinal Air-Breathing Inhibition. PLoS One 2016;11:e0149123. 10.1371/journal.pone.0149123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jing H, Song QX, Zhou GH. Advances in approaches for the quantitative detection of microRNAs. Yi Chuan 2010;32:31-40. 10.3724/SP.J.1005.2010.00031 [DOI] [PubMed] [Google Scholar]
- 30.Yan N, Lu Y, Sun H, et al. A microarray for microRNA profiling in mouse testis tissues. Reproduction 2007;134:73-9. 10.1530/REP-07-0056 [DOI] [PubMed] [Google Scholar]
- 31.Liu CG, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A 2004;101:9740-4. 10.1073/pnas.0403293101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res 2007;67:6092-9. 10.1158/0008-5472.CAN-06-4607 [DOI] [PubMed] [Google Scholar]
- 33.Moschos SA, Williams AE, Perry MM, et al. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics 2007;8:240. 10.1186/1471-2164-8-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chevaliez S. Is HBsAg quantification ready, for prime time? Clin Res Hepatol Gastroenterol 2013;37:559-63. 10.1016/j.clinre.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 35.Janssen HL, van Zonneveld M, Senturk H, et al. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet 2005;365:123-9. 10.1016/S0140-6736(05)17701-0 [DOI] [PubMed] [Google Scholar]
- 36.Hu P, Shang J, Zhang W, et al. HBsAg Loss with Peg-interferon Alfa-2a in Hepatitis B Patients with Partial Response to Nucleos(t)ide Analog: New Switch Study. J Clin Transl Hepatol 2018;6:25-34. 10.14218/JCTH.2017.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ning Q, Han M, Sun Y, et al. Switching from entecavir to PegIFN alfa-2a in patients with HBeAg-positive chronic hepatitis B: a randomised open-label trial (OSST trial). J Hepatol 2014;61:777-84. 10.1016/j.jhep.2014.05.044 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as