Abstract
The effect of X-ray irradiation on cell survival, induction, and repair of DNA damage was studied by using 10 Chroococcidiopsis strains isolated from desert and hypersaline environments. After exposure to 2.5 kGy, the percentages of survival for the strains ranged from 80 to 35%. In the four most resistant strains, the levels of survival were reduced by 1 or 2 orders of magnitude after irradiation with 5 kGy; viable cells were recovered after exposure to 15 kGy but not after exposure to 20 kGy. The severe DNA damage evident after exposure to 2.5 kGy was repaired within 3 h, and the severe DNA damage evident after exposure to 5 kGy was repaired within 24 h. The increase in trichloroacetic acid-precipitable radioactivity in the culture supernatant after irradiation with 2.5 kGy might have been due to cell lysis and/or an excision process involved in DNA repair. The radiation resistance of Chroococcidiopsis strains may reflect the ability of these cyanobacteria to survive prolonged desiccation through efficient repair of the DNA damage that accumulates during dehydration.
Members of the genus Chroococcidiopsis are characterized by a pronounced ability to withstand the lethal effects of desiccation (7). In nature these cyanobacteria dominate the most extreme arid habitats in hot and cold deserts, where they survive in a desiccated (or frozen) state most of the time (15).
The cytology and ultrastructure of field- and laboratory-desiccated Chroococcidiopsis cells were investigated, and the development of thick multilayered envelopes, rich in polysaccharides, was found to be correlated with desiccation tolerance (8, 9). The molecular mechanisms that contribute to the dehydration resistance of Chroococcidiopsis cells are poorly understood, as are similar mechanisms in most prokaryotes (3, 16, 17), but it is widely accepted that the ability to survive desiccation is correlated with the ability to develop spores and/or the ability to produce extracellular polysaccharides (3, 17).
Because dehydration affects the membranous and proteinaceous components of a cell, as well as its nucleic acids, the ability to survive prolonged desiccation involves a complex array of factors at every level of cell structure and function (3, 16, 17). For example, it appears that at the onset of rehydration, the capacity to repair DNA damage that accumulates during desiccation is critical for desiccation tolerance (5). Desiccation tolerance and radiation resistance in Deinococcus radiodurans have a common basis, as DNA repair-deficient mutants lack both properties (11). On the other hand, in Escherichia coli, dehydration-induced mortality is not correlated with induction of DNA breaks, suggesting that differences between radiation-resistant and radiation-susceptible microorganisms extend beyond the ability of the organisms to repair DNA damage (11).
In order to elucidate the mechanisms of desiccation tolerance in Chroococcidiopsis cells, nine strains isolated from different extreme arid environments and one strain isolated from a hypersaline environment were studied to determine their colony-forming abilities and their capacities to repair DNA damage after exposure to high doses of X rays ranging from 2.5 to 20 kGy (1 kGy = 0.1 megarad).
MATERIALS AND METHODS
Organisms and growth conditions.
Of the 10 Chroococcidiopsis sp. strains used in this study, 8 were isolated from different hot desert areas of the world, 1 was isolated from the Antarctic desert, and 1 was isolated from a hypersaline evaporation pond (Table 1). All of these strains are maintained in the Culture Collection of Microorganisms from Extreme Environments at Florida State University (now located at the University of Oregon, Eugene). The unicellular aquatic cyanobacterium Synechococcus sp. strain UTEX 625 and a non-desiccation-resistant bacterium, E. coli B, were used for comparison. Cyanobacterial strains were grown at 25°C in BG-11 medium (18) and were illuminated with a flux density of 90 μmol m−2 s−1 provided by fluorescent cool white bulbs; a cycle consisting of 16 h of light and 8 h of darkness was used. E. coli was grown in Luria-Bertani medium (19) at 37°C. The cultures used for radiation experiments were 3-month-old cultures of each Chroococcidiopsis strain and 1-month-old cultures of Synechococcus sp. strain UTEX 625 (all of these cultures were in the stationary phase and had a cell density of about 108 cells ml−1), as well as E. coli cultures that had been grown overnight and washed in SM medium (19).
TABLE 1.
Chroococcidiopsis sp. strains from the Culture Collection of Microorganisms from Extreme Environments used in this study
| CCMEE designationa |
History |
|---|---|
| (015) N90 | Chroococcidiopsis sp.; endolithic in conglomerate rock, Negev Desert, Israel |
| (028) N690 4 J | Chroococcidiopsis sp.; cryptoendolithic in Nubian sandstone, Negev Desert, Makhtesh Ramon |
| (029) N6904 | Chroococcidiopsis sp.; cryptoendolithic in Nubian sandstone, Negev Desert, Makhtesh Ramon |
| (057) S 6e | Chroococcidiopsis sp.; chasmoendolithic in granite, Sinai Desert, Egypt |
| (084) 70-33 | Chroococcidiopsis sp.; chasmoendolithic in granite rock, Sonoran Desert, Mexico, near Mexicali |
| (101) 71-18 | Chroococcidiopsis sp.; chasmoendolithic in granite rock, Sonoran Desert, near La Joyita |
| (171) A789-2 | Chroococcidiopsis sp.; cryptoendolithic in Beacon sandstone, Ross Desert (McMurdo Dry Valleys), University Valley, Antarctica |
| (313) Au 81-01 | Chroococcidiopsis sp.; hypolithic under stones of desert pavement, Broken Hill, New South Wales, Australia |
| (584) Gobi 91-19 | Chroococcidiopsis sp.; hypolithic under stones of desert pavement, Gobi Desert, Mongolia, between ranges of Gobi Tienshan |
| (662) Solar Pond B | Chroococcidiopsis versatilis Dor; solar evaporation pond near Dead Sea, Israel |
CCMEE, Culture Collection of Microorganisms from Extreme Environments.
Irradiation.
Samples (4 ml) were irradiated at room temperature with photons generated by 3 MeV of electrons striking a water-cooled tungsten disk. Aerobic conditions were maintained by bubbling air through culture samples. Dosimetry was performed with type FTW 60 radiochromic film used as recommended by the manufacturer (Far West Technology, Inc., Goleta, Calif.).
Estimation of survival.
To determine levels of survival, 500-μl aliquots were removed from irradiated samples at various times, and 100-μl aliquots were plated directly or after suitable dilution. Unirradiated samples were diluted and used for plate counting. Each Chroococcidiopsis single cell or cellular aggregate was considered 1 CFU (see below). Colonies derived from surviving CFU were counted 2 to 4 months later, depending on differences in the growth rates of the Chroococcidiopsis strains. The plating efficiencies of 3-month-old Chroococcidiopsis cultures, as deduced by comparing direct cell counts of unirradiated samples with numbers of colonies found, were 70 to 80%. Surviving cells of Synechococcus sp. strain UTEX 625 and E. coli were counted after 3 weeks and 2 days, respectively.
DNA isolation.
To evaluate radiation-induced damage, DNA was extracted from 25-ml samples of unirradiated cultures and from irradiated cultures immediately and 3, 12, and 24 h after irradiation, as previously described (4). Briefly, after lysozyme treatment and osmotic shock, cells were ground with glass beads in the presence of hot phenol. The quality of the chromosomal DNA was assessed by agarose gel electrophoresis.
Excision repair.
The release of DNA from X-ray-irradiated Chroococcidiopsis cells was evaluated by using the method of Lett et al. (10). Chroococcidiopsis cells in the exponential phase of growth were labeled with [3H]thymidine (10 μCi ml−1) for 2 weeks, washed four times with BG-11 medium, and then grown for 1 week in the presence of thymidine (5 μM). Following irradiation with 2.5 kGy of X rays, samples were allowed to grow, and 1-ml aliquots were removed at intervals. After centrifugation at 12,000 × g for 20 min, supernatant fractions were incubated with 5 ml of 10% (wt/vol) trichloroacetic acid (TCA) on ice for 1 h, filtered onto membrane filters, washed once with ice-cold 1% (wt/vol) TCA and three times with 95% (vol/vol) ethanol, and analyzed by scintillation counting. The controls consisted of unirradiated aliquots of Chroococcidiopsis cultures labeled as described above, and the value for the radioactivity detected in the supernatant fraction was considered 100%.
RESULTS AND DISCUSSION
Radiation survival.
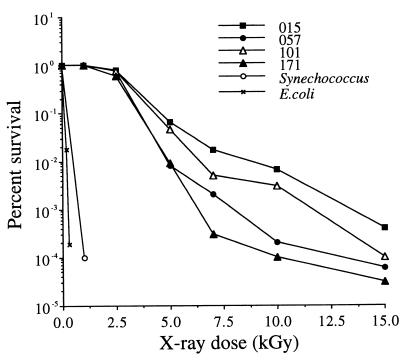
After exposure to 2.5 kGy (0.25 megarad) of X rays, the rates of survival for 10 Chroococcidiopsis strains (Table 1) ranged from about 35% (strain 584) to about 80% (strain 015) (Fig. 1). Three of the most resistant strains (strains 015, 057, and 101, which were chosen to represent different hot and cold deserts) were investigated further. After exposure to 5 kGy of irradiation, the rates of survival of strains 015 and 101 were reduced by 1 order of magnitude, and the rates of survival of 057 and 171 were reduced by 2 orders of magnitude (Fig. 2). When cells were exposed to more than 5 kGy, the rates of survival declined rapidly, and the rates of survival decreased to almost zero after exposure to 20 kGy; that is, about 1 to 10 viable cells per 100 μl of original culture survived (Fig. 2). For comparison, the rate of survival of Synechoccocus sp. strain UTEX 625 irradiated with 1 kGy was reduced 10,000-fold (Fig. 2). After exposure to 2.5 kGy, no viable Synechoccocus sp. strain UTEX 625 cells were detected in 100 μl of culture. The rate of survival of E. coli B was reduced by 2 orders of magnitude after irradiation with 0.2 kGy and by 4 orders of magnitude after irradiation with 0.3 kGy (Fig. 2). No colonies were detected in a 100-μl culture of E. coli B that had been irradiated with 1 kGy.
FIG. 1.
Survival of 10 Chroococcidiopsis strains after irradiation with 2.5 kGy. The values are mean survival rates based on one trial with four replicates.
FIG. 2.
Representative survival curves for four Chroococcidiopsis strains and controls. The values are means based on two independent trials with three replicates per trial.
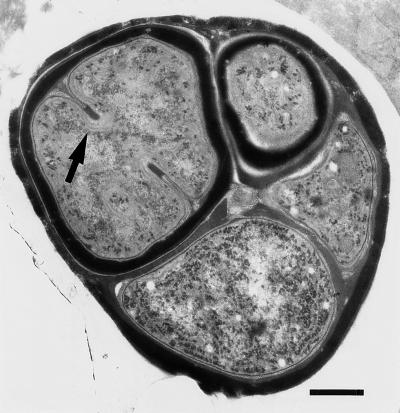
In this study, any Chroococcidiopsis single cell or cellular aggregate was considered 1 CFU. In members of the genus Chroococcidiopsis, single cells form, by division, multicellular aggregates (containing up to about 32 cells) that eventually disperse, and cell size is not a good indicator of the number of cells in an aggregate. In any given population, the ratio of single-celled units to cellular aggregates with different cell numbers varies according to the physiological characteristics of the strain (there are differences in mean generation times). Furthermore, different cells within the same aggregate may be in different physiological states (Fig. 3). It could be argued that survival data are therefore biased, as any CFU may originate from one cell or more than one cell within an aggregate and be dependent on the physiological status of the cell(s). The fact that the cell populations are indeed heterogeneous can be inferred from the shape of the dose-response curves, which initially decrease exponentially and then recurve at higher radiation doses. Such inflection points on survival curves commonly occur in experiments performed with asynchronous cells or in cases in which the irradiated cell population consists of a mixture of single cells and cell aggregates (6). This pattern suggests that the initial part of each survival curve (Fig. 2) is not influenced by the presence of cell aggregates and that the radiation responses of single cells probably follow the pattern observed in the initial low-dose portions (up to 7.5 kGy) of the survival curves. However, not only is this method unavoidable, but it also reflects the life history and adaptive strategy of Chroococcidiopsis cells for survival; in desiccated cultures, survivors are either single-cell units or individual cells that live among many dead cells in multicellular aggregates (8, 9).
FIG. 3.
Electron micrograph of ultrathin section of Chroococcidiopsis sp. strain 029, showing single cells in different developmental and physiological states within the same aggregate. The cells differ in size, in the thickness of the cell envelope, in the presence of a division septum (arrow), and in the abundance of thylakoid membranes in the cytoplasm. Bar = 0.5 μm.
Radiation-induced DNA damage.
The effects of exposure to 2.5 and 5 kGy of X rays on genomic DNA were monitored by using Chroococcidiopsis sp. strains 015, 057, and 171, and comparable results were obtained. After irradiation with 5 kGy, genomic DNA of Chroococcidiopsis sp. strain 171 (Fig. 4A, lane 2) was replaced by a broad smear at a lower molecular weight (Fig. 4A, lane 3), which was still present 12 h after irradiation (Fig. 4A, lane 4) but was not present 24 h after irradiation (Fig. 4A, lane 5). Chroococcidiopsis cells irradiated with 2.5 kGy were able to repair DNA damage within 3 h (data not shown). Part of the DNA degradation evident in Fig. 4A occurred because DNA samples were frozen and thawed once after the extraction, but shearing affected mainly the upper band of the two bands present in the genomic DNA extracts, as shown by a comparison of Fig. 4A, lane 2, and Fig. 4B, lanes 2 and 3. This upper band may represent a megaplasmid, but purification of this band has never been attempted; at the moment, the presence of a plasmid can be discounted only for Chroococcidiopsis sp. strains 029 and 171 (D. Billi, unpublished data). A cycle of freezing and thawing resulted in extensive degradation that produced rRNA and precursors (Fig. 4A), as suggested by the disappearance of the prominent bands between 2 and 0.5 kb, the pattern of which is characteristic (1). This degradation was not observed with samples that were loaded onto the gel immediately after extraction (Fig. 4B). DNA degradation due to irradiation with 5 kGy is shown in Fig. 1C, lane 3; the effect of freezing and thawing on the same sample is shown in lane 2.
FIG. 4.
Ability of Chroococcidiopsis sp. strain 171 to recover from DNA damage following exposure to 5 kGy of irradiation. (A) Lane 1, 1-kb DNA ladder; lane 2, genomic DNA extracted from unirradiated cells; lane 3, genomic DNA extracted immediately after irradiation; lanes 4 and 5, DNA extracted 12 h (lane 4) and 24 h (lane 5) after exposure. (B) Lane 1 contained a 1-kb DNA ladder. When lanes were loaded immediately after extraction, nucleic acids from Chroococcidiopsis sp. strain 015 (lane 2) and strain 171 (lane 3) lacked the shearing in the rRNA and in the upper band (asterisk) of the total DNA. (C) Lane 1, 1-kb DNA ladder; lanes 2 and 3, genomic DNA from irradiated strain 171 loaded immediately after extraction (lane 3) and after a cycle of freezing and thawing (lane 2).
Excision repair.
When release of DNA after irradiation with 2.5 kGy of X rays was studied with Chroococcidiopsis sp. strains 015, 057, and 171 labeled with [3H]thymidine, the amounts of TCA-precipitable radioactivity in supernatants from irradiated cultures were greater than the amounts of TCA-precipitable radioactivity in supernatants from unirradiated cultures (Fig. 5). Thirty minutes after irradiation, the amounts of radioactivity in the supernatants of strains 015 and 057 increased to about 254 and 213% of the amount present in the control and then sharply decreased. The radioactivity in the supernatant of strain 171 increased to about 188% of the control value 2 h after irradiation and then decreased.
FIG. 5.
Release of [3H]thymidine-labeled DNA from three Chroococcidiopsis strains after 2.5 kGy of irradiation.
The radioactivity in the supernatants from irradiated Chroococcidiopsis cultures might have been due to cell lysis and to labeled nucleotides in complexes with proteins of the cell envelope. However, the increases in radioactivity in the supernatants of irradiated cells might have been due to cell lysis and/or to an excision process involved in DNA repair. Export of damaged DNA has been reported in D. radiodurans (10), but its relationship to radiation resistance has never been explored (2). The progressive decrease in the radioactivity detectable in the growth medium of Chroococcidiopsis cells suggests that released nucleotides are reincorporated after DNA synthesis resumes.
Conclusions.
Chroococcidiopsis cells are able to survive doses of X rays as high as 15 kGy, and the rate of survival of these cells is second only to the rate of survival reported for D. radiodurans, the most radiation-resistant organism known (2, 14). Although the resistance of Chroococcidiopsis cells, which decreases sharply after irradiation with 5 kGy or more, is not quite comparable to the resistance of Deinococcus cells, it is still exceptional. When comparable data for survival of other photoprophics after exposure to X rays are available, the radioresistance of Chroococcidiopsis cells will be estimated further. After high doses (up to 15 kGy) of X rays, survivors were still present, but the lack of suitable selectable phenotypes hampered identification of cells with induced mutations.
As our data are based on a study of strains obtained from desert and hypersaline environments, whether Chroococcidiopsis strains in freshwater environments exhibit high levels of radiation resistance is an open question. An evaluation of the 16S rRNA of different Chroococcidiopsis strains is in progress in order to establish the phylogenetic affinities of these organisms. However, the ability to repair DNA damage demonstrated in this study may well be one of the mechanisms that Chroococcidiopsis cells use to withstand prolonged desiccation. We hypothesize that the capacity of these cells to repair radiation-induced DNA damage and probably also desiccation-induced DNA damage is related to their ability to use redundant genetic information, as D. radiodurans does (2, 12, 13).
The demonstration of radiation resistance in Chroococcidiopsis strains is another step toward meeting the formidable challenge of elucidating the biology of these desiccation-tolerant cyanobacteria and should be followed by genetic studies of the molecular mechanisms that enable these organisms to live and survive in some of the most extreme environments on Earth.
ACKNOWLEDGMENTS
This work was supported by NASA grant NAGW 4044 to E.I.F.
We thank Inka Dor, Hebrew University, Jerusalem, Israel, for the culture of the hypersaline strain; Powell E. Barber and Gregory A. Brown, Nuclear Service of Florida State University, for help with X rays; Malcolm Potts, Virginia Polytechnic Institute and State University, for critically reading the manuscript; Anne B. Thistle for critical editing; and Ken Womble for help in preparing the graphs.
REFERENCES
- 1.Angeloni S V, Potts M. Polysome turnover in immobilized cells of Nostoc commune (cyanobacteria) exposed to water stress. J Bacteriol. 1986;168:1036–1039. doi: 10.1128/jb.168.2.1036-1039.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battista J R. Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol. 1997;51:203–224. doi: 10.1146/annurev.micro.51.1.203. [DOI] [PubMed] [Google Scholar]
- 3.Billi, D., and M. Potts. Life without water: responses of prokaryotes to desiccation. In K. B. Storey and J. M. Storey (ed.), Cellular and molecular responses to stress, in press. JAI Press, Stamford, Conn.
- 4.Billi D, Grilli Caiola M, Paolozzi L, Ghelardini P. A method for DNA extraction from the desert cyanobacterium Chroococcidiopsis and its application to identification of ftsZ. Appl Environ Microbiol. 1998;64:4053–4056. doi: 10.1128/aem.64.10.4053-4056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dose K, Bieger-Dose A, Labusch M, Gill M. Survival in extreme dryness and DNA-single-strand breaks. Adv Space Res. 1992;12(4):221–229. doi: 10.1016/0273-1177(92)90176-x. [DOI] [PubMed] [Google Scholar]
- 6.Elkind M M, Whitmore G F. The radiobiology of cultured mammalian cells. New York, N.Y: Gordon and Breach, Science Publishers, Inc.; 1967. [Google Scholar]
- 7.Friedmann E I. Extreme environments, limits of adaptation and extinction. In: Guerrero R, Pedrós-Alió C, editors. Trends in microbial ecology. Barcelona, Spain: Spanish Society for Microbiology; 1993. pp. 9–12. [Google Scholar]
- 8.Grilli Caiola M, Billi D, Friedmann E I. Effect of desiccation on envelopes of the cyanobacterium Chroococcidiopsis sp. (Chroococcales) Eur J Phycol. 1996;31:97–105. [Google Scholar]
- 9.Grilli Caiola M, Ocampo-Friedmann R, Friedmann E I. Cytology of long-term desiccation in the desert cyanobacterium Chroococcidiopsis (Chroococcales) Phycologia. 1993;32:315–322. doi: 10.2216/i0031-8884-32-5-315.1. [DOI] [PubMed] [Google Scholar]
- 10.Lett J T, Feldschreiber P, Little J G, Steele K, Dean C J. The repair of X-ray damage to the deoxyribonucleic acid in Micrococcus radiodurans: a study of the excision repair process. Proc R Soc London Ser B. 1967;167:184–201. doi: 10.1098/rspb.1967.0023. [DOI] [PubMed] [Google Scholar]
- 11.Mattimore V, Battista J R. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive desiccation. J Bacteriol. 1996;178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minton K W. DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol Microbiol. 1994;13:9–15. doi: 10.1111/j.1365-2958.1994.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 13.Minton K W, Daly M J. A model for repair of radiation induced DNA double-stand breaks in the extreme radiophile Deinococcus radiodurans. Bioessays. 1995;17:457–464. doi: 10.1002/bies.950170514. [DOI] [PubMed] [Google Scholar]
- 14.Moseley B E B. Photobiology and radiobiology of Micrococcus (Deinococcus) radiodurans. Photochem Photobiol Rev. 1983;7:223–275. [Google Scholar]
- 15.Nienow J A, Friedmann E I. Terrestrial lithophytic (rock) communities. In: Friedmann E I, editor. Antarctic microbiology. New York, N.Y: Wiley-Liss; 1993. pp. 343–412. [Google Scholar]
- 16.Potts M. Desiccation tolerance of prokaryotes. Microbiol Rev. 1994;58:755–805. doi: 10.1128/mr.58.4.755-805.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potts M. The anhydrobiotic cyanobacterial cell. Physiol Plant. 1996;97:788–794. [Google Scholar]
- 18.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]