Abstract
Background/purpose
Several long non-coding RNAs (lncRNAs) harbor miRNA in their genome. MIR31HG harbors miR-31 in its intron and it is speculated that they are co-expressed in tumors. This study addressed whether frequent miR-31 and MIR31HG co-upregulation occurred in oral squamous cell carcinoma (OSCC) and its clinical implications.
Materials and methods
Microarray was performed to retrieve dis-regulated lncRNAs from tissue sample. The ectopic gene expression was carried out to specify the phenotypic influences of selected lncRNA screened from bioinformatic algorithms. The expression of miR-31 and MIR31HG in tissues or scrapped samples was analyzed using qRT-PCR. The implications of gene expression as related to metastasis or survival were further dissected.
Results
Microarray identified disrupted transcripts including MIR31HG and other 152 lncRNAs aberrantly expressed in OSCC tissues. In silico algorithms annotated an eminent involvement of aberrant transcripts in the regulation of cell cycle, extracellular modulation, adhesion, and wound healing. The enhancement of proliferation, wound healing, invasion and anchorage-independent colony formation mediated by MIR31HG was ascertained by ectopic expression in OECM1 cells. Besides, co-upregulation of miR-31 and MIR31HG was conspicuous in OSCC tissues. High expression of miR-31 and MIR31HG designated a trend of worse OSCC prognosis. Interestingly, high MIR31HG expression defined a very poor survival in stage IV diseases. By contrast, high miR-31 expression predicted nodal metastasis in stage I–III diseases.
Conclusion
Assessment of miR-31 and MIR31HG expression in OSCC may enable the prognostic prediction. The candidate lncRNAs isolated from this work can be further validated as crucial factors contributing to OSCC pathogenesis.
Keywords: Carcinoma, miR-31, MIR31HG, Mouth, Oral
Introduction
In human, the coding region comprised only 1.2% of the whole genome.1 The other part of the non-coding regions was previously considered as less important although they make up the majority of genome. The biological function of the non-coding region and their transcriptome draws attention in recent years. As the advancement of whole genome sequencing technology, more and more studies showed that mutations occurred not only in coding region but also in non-coding region, moreover around 80% cancer-related SNPs were found in non-coding region.1 According to size, these non-coding RNA can be divided into two category, long non-coding RNA (lncRNA; >200 bp) and small non-coding RNA(<200 bp). Small non-coding RNAs like microRNAs (miRNA) are thoroughly identified and characterized. And their function in translation repression is also well studied. However, complicated biologic activity of lncRNA is still under investigation. Most lncRNAs are transcribed by RNA polymerase II, hence their structures are similar to mRNA. Studies have shown the comprehensive role of lncRNAs in epigenetic, transcriptional, post-transcriptional, and translation regulation, as well as post-translational modification.2 The interactive functions of lncRNA are more complicated than expected.
In Taiwan, the prevalence of areca chewing accounts for the high incidence of oral squamous cell carcinoma (OSCC) among male adults. The areca quid mainly consists of betel leaf, areca nut and slaked lime. These ingredients, especially areca nut, exert carcinogenesis potential by direct and indirect genotoxicity.3,4 The genomic signature of mismatch repair deficiency has also been identified in areca related OSCC tissues.5 Recent study also showed that areca induced whole transcriptome changes associated with diabetes, obesity and metabolic syndrome in a human monocyte cell line.6 Despites the coding transcriptome change, the importance of non-coding transcriptome aberrance attracts more attention in recent days. miRNAs have high specificity of expression in certain tissues and disease. Studies also demonstrated that miRNA expression profiles have better accuracy in disease classification than mRNA.7,8 Recent microarray profiling identified a set of 105 miRNAs with altered expressed in OSCC and qPCR validation revealed that up-regulation of miR-196a, miR-21, miR-1237 and downregulation of miR-204, miR-144 was associated with poor prognosis of OSCC. The miR-196a/miR-204 ratio served as a good predictor for disease recurrence and survival.9 Our previous studies have demonstrated the important role of miR-21 and miR-31 during oral carcinogenesis.10,11 The upregulation of miR-372/373 was observed correlated to lymph node metastasis OSCC.12 It is worthy noted that interactions between miRNA and lncRNA are demonstrated and many lncRNAs function as competing endogenous RNAs (ceRNA). LncRNA IUR and MEG3 could sponge miR-21 and neutralize the oncogenic effect of miR-21.13,14 LncRNA FER1L4 is a ceRNA to binding with miR-372, which results in E2F1 up-regulation and proliferative promotion of glioma cells.15 Certain lncRNAs are also identified as host gene for miRNA. The miR-497 and miR-195 were derived from lncRNA MIR497HG. The MIR497HG together with miR-497 and miR-195, were downregulated in bladder cancer, and the expression of MIR497HG could suppress the progression of bladder cancer cells.16 Copy number deletion of the MIR99AHG gene was observed in lung adenocarcinoma, which led to the downregulation of its four transcripts: lncRNA MIR99AHG and the miR-99a/let-7c/miR-125b2 cluster. Further experiments also confirmed the suppressor role of MIR99AHG and miR-99a.17
MIR31HG, a host gene of an important oncomir miR-31, was found in both nucleus and cytoplasm under normal condition, but MIR31HG and miR-31 were exported to cytoplasm following oncogene B-RAF induction. MIR31HG is upregulated in oncogene induced senescence and negatively regulates tumor suppressor p16.18 MIR31HG could also targets HIF1A and p21 and promote cell cycle progression in head and neck cancer cells.19 However, the expression of MIR31HG was negatively correlated to overall survival of lung adenocarcinoma, but it was a favorable prognostic factor in gastrointestinal cancer.20 In this study, we identified the expression profile of lncRNA in areca-related oral carcinogenesis. The expression and clinical implications of MIR31HG in OSCC were also investigated.
Materials and methods
Subjects
The primary OSCC tumors and their paired non-cancerous matched tissue samples were derived from 40 patients (Table 1). Samples were collected after obtaining written informed consent. The study was approved by IRB committee of National Yang-Ming University Hospital and Taipei Mackay Memorial Hospital with approval numbers NYMUH 2014A002 and 17MMHIS053/18MMHIS176, respectively.
Table 1.
Clinical parameters of OSCC samples.
| n = 40 | |
|---|---|
| Age (mean ± SEM) | 57.53 ± 1.58 |
| Gender (male/female) | 36/4 |
| Areca chewing | 30 |
| Tobacco smoking | 30 |
| T1–3 | 15 |
| T4 | 25 |
| N0 | 26 |
| N+ | 14 |
| Stage I–III | 12 |
| Stage IV | 28 |
| Follow-up (mean ± SEM, days) | 2810 ± 227.1 |
| Death | 10 |
| Alive | 30 |
Swabbed samples were collected from lesion and normal looking control mucosa from OSCC patients in National Yang-Ming University Hospital or Taipei MacKay Memorial Hospital with IRB approval numbers of NYMUH2019A013 and 18MMHIS187e, respectively. A Libo specimen collection swab (Cat No. 30221.3, Iron Will, New Taipei City, Taiwan) was used to achieve scrapped cells. The detailed sampling procedures and analysis followed the protocols established in previous study.21
LncRNA microarray
Total RNA was amplified by a Low Input Quick-Amp Labeling kit (Agilent Technologies, Santa Clara, CA, USA) and labeled with Cy3 (CyDye, Agilent Technologies) during the in vitro transcription process. The Cy3-labled cRNA was fragmented to an average size of about 50–100 nucleotides by incubation with fragmentation buffer at 60 °C for 30 min. Corresponding fragmented labeled cRNA was then pooled and hybridized to Agilent SurePrint G3 Human Gene Exp V3 8 × 60K Microarray (Agilent Technologies) at 65 °C for 17 h. After washing and drying by nitrogen gun blowing, microarrays were scanned with an Agilent microarray scanner (Agilent Technologies) at 535 nm for Cy3. Scanned images were analyzed by Feature extraction10.5.1.1 software (Agilent Technologies), an image analysis and normalization software were used to quantify signal and background intensity for each feature.
Bioinformatic analysis
The Venn diagram and gene annotation analysis of microarray data were performed using web-based resources (https://metascape.org/gp/index.html#/main/step1).22 From microarray data, we selected genes with expression change more than and less than two folds for further analysis. Venn diagrams represent the overlapping of genes between samples whereas circoplots demonstrate their relationships. The cluster analysis was also performed using a web-based heatmap visualization and analysis tool clustergrammer.23 The non-supervised cluster analysis was performed on 153 overlapping lncRNAs and also the top 50 dysregulated lncRNAs.
Cell culture, plasmid construction and phenotypic assays
The OECM1 OSCC cell line was cultured as previously described.24 The lentiviruses containing MIR31HG transcript cloned in the pLV-EF1a-GFP vector and control virus are gifts from Professor Shu-Chun Lin. OECM1 was infected with virus and selected to achieve stable cell sublines with MIR31HG expression.25 Cell proliferation, wound closure, transwell invasion and anchorage-independent colony formation assays, and were carried out according to previously published protocols.24,25
Quantitative polymerase chain reaction (qPCR) analysis
TaqMan miRNA assay kits and qPCR probes obtained from Apply Biosystems (Waltham, MA, USA) were used to quantify the expression of MIR31HG and miR-31 using GAPDH or RNU6B as internal controls.24,25 The Cat. No. of each probe was Hs01107339_gl, 002279, Hs00266705_gl and 001093, respectively. −ΔCt is the difference in threshold cycle number across the test gene and the internal control. −ΔΔCt indicates the difference in −ΔCt across the test and the control group. 2−ΔΔCt designates the fold change in gene expression relative to control.24
Statistics
Mann–Whitney tests, t-tests, two-way ANOVA test, linear correlation analysis and Kaplan–Meier survival analysis were performed. A receiver operating characteristic (ROC) curve was used to acquire area under curve (AUC), sensitivity and specificity of variables to evaluate separation power.21
Results
Gene expression profile from areca-associated OSCCs
To determine the gene expression profile of areca-related OSCC, normal and cancer tissue pairs from three patients were utilized for microarray analysis. The Agilent SurePrint G3 Human Gene Exp V3 8 × 60K Microarray could detect 30,600 lncRNAs and 26,100 coding genes. Among three cases, two were stage I tumors and one was stage IV disease with lymph node metastasis in neck (Fig. 1A). The gene expression level from normal to tumor were analyzed and genes with more than or less than two-fold changes were counted. The Venn diagram demonstrated the overlap of counted genes between three cases and around 2050 common genes were identified in all three cases (Fig. 1B). Case 1 and case 2 are stage I disease and they shared more common genes than either one comparing to case 3 advanced tumor. The discrepancies were also noted in Circos plot (Fig. 1C). Gene ontology (GO) analysis performed on 2050 common genes annotated the most significant involved pathway was cycle related. It was followed by extracellular matrix related, cell adhesion pathways or responses to wounding, which were important pathways during carcinogenesis (Fig. 1D).
Figure 1.
Microarray analysis of OSCC tumors. (A) The clinical parameters of cases. (B) Venn diagram to show the genes aberrantly expressed for at least two folds. (C). Circos plot to show the higher similarity between case 1 and case 2 in the gene dis-regulation, which separates from those in case 3. (D) GO annotation to show the disrupted pathways.
lncRNA expression profile from areca-associated OSCCs
The microarray comprised 30600 lncRNAs and only lncRNA with more than or less than two-fold expression change from normal to tumor counterpart were selected. There are 478 altered lncRNAs identified in case 1, 677 in case 2, and 481 in case 3. A total of 153 common lncRNAs existing in three cases were showed in Venn diagram (Fig. 2A). The Circo plot analysis revealed the pattern of correlation between 3 cases and the lncRNA expression profile are more similar between case 1 and case 2, which resembles the profile of the whole gene set as mentioned above (Fig. 2B). The heatmap demonstrated the up-regulated (red) and down-regulated (blue) lncRNAs from normal to tumor. Non-supervised cluster analysis confirmed the different expression profile between case 3 and the remains (Fig. 2C). Moreover, if we select the top 50 dys-regulated lncRNAs among three cases, down-regulated lncRNAs outnumber the up-regulated ones (Fig. 3A). Most of dysregulated lncRNAs are RNA genes but some belong to pseudogenes and some are miRNA host genes. Among up-regulated lncRNAs, for example, LOC344887, KIAA0125 were reported correlated with oncogenic activity and stemness.26,27 Interestingly, the non-supervised cluster analysis identified MIR31HG as significant up-regulated lncRNA among top 50 dysregulated lncRNA (Fig. 3B).
Figure 2.
The dis-regulation of lncRNAs in microarray analysis. (A) Venn diagram to show the lncRNAs aberrantly expressed for at least two folds. (B) Circos plot to show the higher similarity and interaction in the lncRNA dis-regulation between case 1 and case 2, comparing to case 3. (C) Heatmap of cluster analysis according to 153 lncRNAs commonly present in three tumors. The similarity between case 1 and case 2 is still higher than case 3. Blue, Down-regulated; red. Up-regulated.
Figure 3.
The top 50 dis-regulated lncRNAs. (A) Heatmap of cluster analysis. (B). The illustration of aberrant lncRNAs. Blue, Down-regulated; red. Up-regulated.
MIR31HG influences the oncogenicity of OECM1 cell
To determine the phenotype exerted by MIR31HG, OECM1 OSCC cell line, an areca-chewing related cell line, was selected for MIR31HG overexpression experiments. Lentivirus carrying MIR31HG expression construct was introduced into OECM1 cells through infection. The high infection efficiency was noted in the selected subline (Fig. 4A), and this subline had a stable and robust MIR31HG expression (Fig. 4B). Proliferation assay demonstrated the growth advantage in MIR31HG expression cells (Fig. 4C). Enhanced migration ability was also noted in wound healing assay and invasion assay (Fig. 4D and E). Anchorage-independent colony formation assay revealed the increased in vitro oncogenic potential as related to MIR31HG expression (Fig. 4F). The results suggest an oncogenic role of MIR31HG in OSCC, and the expression profile of MIR31HG and its hosted miR-31 in OSCC tissue was further studied thereafter.
Figure 4.
Exogenous MIR31HG expression in OECM1 cells. (A) Both cell subline with ectopic MIR31HG expression and control cell subline exhibit green fluorescence (×200). VA, vector alone control. OE, MIR31HG expression. (B) MIR31HG expression drastically increases in OE relative to VA. (C–F) OE cells exhibit an increased proliferation (in C), wound closure rate (in D), invasion (in E) and anchorage-independent colony formation ability (in F) relative to VA cells. ns, not significant; ∗, p < 0.05, ∗∗, p < 0.01 and ∗∗∗, p < 0.001. Mann–Whitney test or two-way Anova test.
Validation of MIR31HG expression in OSCCs
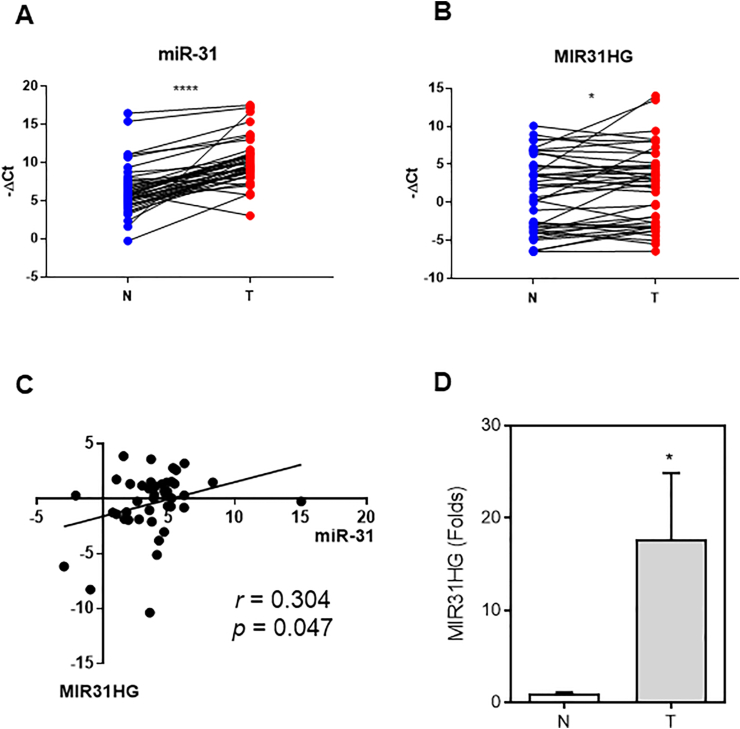
Among 40 OSCC patients enrolled for tissue study 75% (30/40) patients had areca chewing habits (Table 1). The expression of MIR31HG and miR-31 were determined by qPCR. A significantly up-regulated in MIR31HG expression from normal to tumor were found, and the upregulation of miR-31 expression was more conspicuous (Fig. 5A and B). Correlation analysis also showed a significantly positive correlation between MIR31HG and miR-31 expression (Fig. 5C). In a series of brushing samples from 10 OSCC patients, the expression of MIR31HG was significantly up-regulated from normal to tumor counterpart (Fig. 5D). The qPCR results were in line with lncRNA microarray findings, which substantiate the expression of MIR31HG could be a biomarker like miR-31 being previously detected in OSCC.
Figure 5.
qPCR analysis. (A–B) Analysis of OSCC tissue pairs. (A, B) Before–after plots of miR-31 and MIR31HG, respectively. Y axis, −ΔCt. (C). Linear correlation analysis according to −ΔΔCt. (C) Analysis of MIR31HG expression in swabbed samples. Bar chart, mean ± SE. (A, B, D) Paired-t-test. N, normal control; T, OSCC. ∗, p < 0.05 and ∗∗∗∗, p < 0.0001.
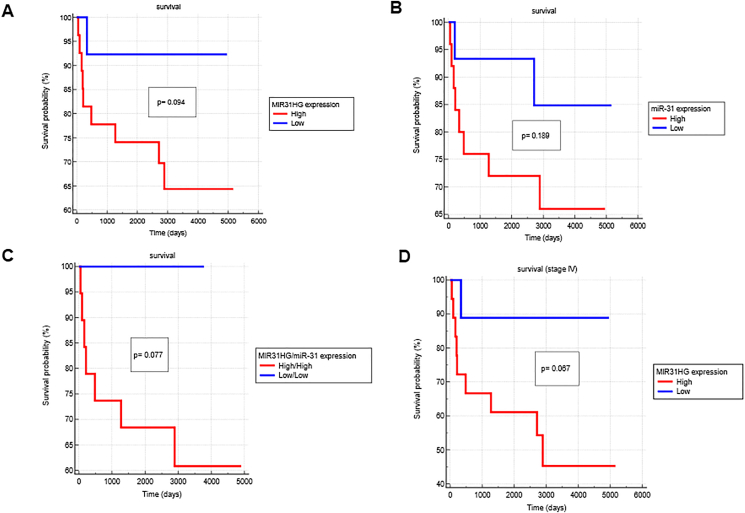
Prognostic value of MIR31HG and miR-31 expression in OSCCs
We assessed the prognosis value of MIR31HG and miR-31 in our patients who had been followed for an average of 2810 days (Table 1). Among them, 10 subjects were dead during follow-up. Initially, the ROC analysis was performed with the classification variable as survival to yield a cut-off value, which could discriminate the expression states of MIR31HG or miR-31 into high or low. The Kaplan–Meier analysis revealed a trend that patients with higher MIR31HG expression had poor survival, however, the differences was not statistically significant (Fig. 6A). Similar trend was found in miR-31 expression (Fig. 6B). Further dissection revealed that patients had both high MIR31HG and high miR-31 expression exhibiting worsened survival (Fig. 6C). Moreover, in stage IV patients, the expression of MIR31HG seemed to be a potent prognosis marker (Fig. 6D).
Figure 6.
Kaplan–Meier survival analysis. (A–C) All tumors. (A) MIR31HG, (B) miR-31, (C) both MIR31HG and miR-31. (D) Stage IV tumors/MIR31HG. Cut off values of −ΔΔCt achieved from preliminary tests is used to divide tumors with high or low expression.
The neck nodal metastasis prediction is a critical issue in OSCC treatment. We performed ROC analysis to determine whether MIR31HG or miR-31 expression could predict nodal metastasis. Interesting, miR-31 has an AUC of 0.65 to diagnose nodal metastasis, which is much better than MIR31HG (Fig. 7A and B). Of note, in stage I–III diseases that neck dissection is sometimes optional, the prediction of nodal metastasis appeared useful as that miR-31 expression could significantly distinguish the nodal metastasis among stage I–III patients (Fig. 7C). The findings warranted a future clinical application in terms of miR-31 expression.
Figure 7.
Receiver Operating Characteristic curves of −ΔΔCt to distinguish nodal metastasis of tumors. (A, B) All tumors. (A) MIR31HG, (B) miR-31. (C) Stage I–III tumors/miR-31. miR-31 expression enables the separation of metastasis vs non-metastasis in non-stage IV tumors.
Discussion
Although the biological functions of lncRNA are not very diverse, their abundance in transcriptome warrants the potential role in carcinogenesis. Our lncRNA microarray results revealed a panel of dysregulated lncRNAs with novelty. Among up-regulated lncRNAs, HAGLROS is a STAT3-induced lncRNA that contributes to the malignant progression of gastric cancer.28 HAGLROS could sponge miR-152 and up-regulate ROCK1 expression in osteosarcoma.29 In lung cancer, LINC00460 promotes epithelial–mesenchymal transition and cell migration.30 LINC00460 also enhances bladder carcinoma cell proliferation and migration by modulating miR-612/FOXK1 Axis.31 KRT42P is a pseudogene with unknown function. LOC100506100, LOC101928710, LOC101559451, LOC100240735 and MGC16025 are all uncharacterized lncRNAs. Intriguingly, LOC100506100 is the lncRNA most profoundly up-regulated in our samples. The down-regulated lncRNAs outnumber the up-regulated lncRNAs in our data, which might imply that more lncRNA may drive suppressive influences. Study has shown that approximately 60% of the protein-coding genes are targeted by miRNA, whereas 70% protein coding genes are targeted by lncRNA.32 Since lncRNA could act as miRNA sponge, the interplay between coding RNAs and non-coding RNAs is far more complicated than expected.
In the perspective of biomarker detection, up-regulation molecule is a better candidate than down-regulation ones. We thus focus on the lncRNAs which were up-regulated in cancer tissues and identify MIR31HG as an important oncogenic lncRNA. MIR31HG genome located on 9p21 and the gene locus is chr9:21,439,475-21,591,766(GRCh38/hg38) consisting of 152,292 nucleotides. The 2166 bp MIR31HG transcript composed of four exons is identified later on a non-coding RNA. In RNAseq analysis, MIR31HG is abundant in gastrointestinal tract compared to other part of human body,33 whereas it seems that the survival prediction of MIR31HG may vary according to cancer type.34 Our data signifies that MIR31HG expression might correlate with overall survival of OSCC patients, especially in late-stage disease. Similar result has been noted in laryngeal SCC that MIR31HG overexpression represents poor prognosis, and MIR31HG could negatively target p21 to facilitate the progression of cell cycle.19 Our in vitro results showed the enrichment in cell proliferation after MIR31HG expression. Furthermore, since our annotations of microarray data revealed that cell cycle related pathway are the most involved ones, the findings on proliferative enhancement together with the promotion on other phenotypes mediated by MIR31HG expression is compatible with in silico prediction. Through the alternative splicing, MIR31HG transcript comprises 4 exons, and miR-31 is localized in intron 1. It is speculated that these two non-coding transcripts could be generated together in cells. In colorectal carcinoma samples, a strong correlation between miR-31 and MIR31HG was shown (Spearman's r > 0.80).35 Both miR-31 and MIR31HG have been known up-regulated in oral precancerous lesions.21,25 In our OSCC patient cohort, the co-upregulation of them was also demonstrated (Spearman's r > 0.3). Since bioinformatics studies predicted that approximately 20% of intronic miRNAs target host mRNA transcripts in a feedback loop,36 miR-31 and MIR31HG might share the coordinative modulation in oncogenesis.
Although the power in survival prediction is not as good as MIR31HG, miR-31 expression seems to correlated more with neck nodal metastasis. Distinct gene expression profile among nodal positive and nodal negative cases was reported.37 Similar result was preliminarily noticed in our microarray data. Only case 3 has nodal metastasis. Although case 2 and case 3 are all tongue cancers, cluster analysis of gene expression profile separated case 3 from case 1 and case 2. Most importantly, our data showed that miR-31 expression could predict the nodal metastasis in stage I–III patients (AUC:0.8, sensitivity 100%, specificity 78%). The findings could assist the therapeutic decision for neck lymph node dissection in early-stage patients. Novel downstream target of MIR31HG such as LBH has been identified recently,25 whereas, the upstream promoter control of MIR31HG could be as complicated as downstream effects. Previous study has shown that EGF up-regulates miR-31 through the C/EBPβ signal cascade.38 MIR31HG could also be negatively regulated by miR-193b.39 However, the regulatory mechanism of and MIR31HG and miR-31co-upregulation in areca-related OSCC needs further elucidation. The interactive roles that MIR31HG and miR-31 co-plays in OSCC pathogenesis require specification. To resolute the functions of uncharacterized lncRNAs may bestow novel mechanistic insights in neoplastic process.
Declaration of competing interest
The authors have no conflicts of interest relevant to this article.
Acknowledgments
We acknowledge the helps from Professors Shu-Chun Lin and Kuo-Wei Chang. This work is supported by grants RD2015003, RD2016003 and RD2018003 from NYMUH and MOST107-2314-B-010-026-MY3 and MOST107-2314-B-010-031 from Ministry of Science and Technology, Taiwan.
References
- 1.Dunham I., Kundaje A., Aldred S.F., et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X., Wang W., Zhu W., et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci. 2019;20:5573–5602. doi: 10.3390/ijms20225573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen P.-H., Mahmood Q., Mariottini G.L., Chiang T.-A., Lee K.-W. Adverse health effects of betel quid and the risk of oral and pharyngeal cancers. BioMed Res Int. 2017;2017:3904098. doi: 10.1155/2017/3904098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ko Y.C., Huang Y.L., Lee C.H., Chen M.J., Lin L.M., Tsai C.C. Betel quid chewing, cigarette smoking and alcohol consumption related to oral cancer in taiwan. J Oral Pathol Med. 1995;24:450–453. doi: 10.1111/j.1600-0714.1995.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 5.Yang W.F., Qin N., Song X., et al. Genomic signature of mismatch repair deficiency in areca nut-related oral cancer. J Dent Res. 2020;99:1252–1261. doi: 10.1177/0022034520930641. [DOI] [PubMed] [Google Scholar]
- 6.Cardoso S., Ogunkolade B.W., Lowe R., et al. Areca catchu-(betel-nut)-induced whole transcriptome changes associated with diabetes, obesity and metabolic syndrome in a human monocyte cell line. BMC Endocr Disord. 2021;21:165–176. doi: 10.1186/s12902-021-00827-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J., Getz G., Miska E.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Søkilde R., Persson H., Ehinger A., et al. Refinement of breast cancer molecular classification by miRNA expression profiles. BMC Genom. 2019;20:503–514. doi: 10.1186/s12864-019-5887-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajan C., Roshan V.G.D., Khan I., et al. MiRNA expression profiling and emergence of new prognostic signature for oral squamous cell carcinoma. Sci Rep. 2021;11:7298–7309. doi: 10.1038/s41598-021-86316-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu E.H., Tu H.F., Wu C.H., Yang C.C., Chang K.W. MicroRNA-21 promotes perineural invasion and impacts survival in patients with oral carcinoma. J Chin Med Assoc. 2017;80:383–388. doi: 10.1016/j.jcma.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Hung K.F., Liu C.J., Chiu P.C., et al. MicroRNA-31 upregulation predicts increased risk of progression of oral potentially malignant disorder. Oral Oncol. 2016;53:42–47. doi: 10.1016/j.oraloncology.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 12.Tu H.F., Chang K.W., Cheng H.W., Liu C.J. Upregulation of miR-372 and -373 associates with lymph node metastasis and poor prognosis of oral carcinomas. Laryngoscope. 2015;125:E365–E370. doi: 10.1002/lary.25464. [DOI] [PubMed] [Google Scholar]
- 13.Wang B., Hua P., Zhang L., Li J., Zhang Y. LncRNA-IUR up-regulates PTEN by sponging miR-21 to regulate cancer cell proliferation and apoptosis in esophageal squamous cell carcinoma. Esophagus. 2020;17:298–304. doi: 10.1007/s10388-020-00724-x. [DOI] [PubMed] [Google Scholar]
- 14.Jia H.Y., Zhang K., Lu W.J., Xu G.W., Zhang J.F., Tang Z.L. LncRNA MEG3 influences the proliferation and apoptosis of psoriasis epidermal cells by targeting miR-21/caspase-8. BMC Mol Cell Biol. 2019;20:46–58. doi: 10.1186/s12860-019-0229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia L., Nie D., Wang G., Sun C., Chen G. FER1L4/miR-372/E2F1 works as a cerna system to regulate the proliferation and cell cycle of glioma cells. J Cell Mol Med. 2019;23:3224–3233. doi: 10.1111/jcmm.14198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang C., Liu Y., Fu S., et al. Silencing of lncRNA MIR497HG via CRISPR/CAS13d induces bladder cancer progression through promoting the crosstalk between Hippo/Yap and TGF-β/Smad signaling. Front Mol Biosci. 2020;7:616768. doi: 10.3389/fmolb.2020.616768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han C., Li H., Ma Z., et al. MIR99AHG is a noncoding tumor suppressor gene in lung adenocarcinoma. Cell Death Dis. 2021;12:424–439. doi: 10.1038/s41419-021-03715-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montes M., Nielsen M.M., Maglieri G., et al. The lncRNA MIR31HG regulates p16INK4A expression to modulate senescence. Nat Commun. 2015;6:6967–6981. doi: 10.1038/ncomms7967. [DOI] [PubMed] [Google Scholar]
- 19.Wang R., Ma Z., Feng L., et al. LncRNA MIR31HG targets HIF1A and p21 to facilitate head and neck cancer cell proliferation and tumorigenesis by promoting cell-cycle progression. Mol Cancer. 2018;17:162–167. doi: 10.1186/s12943-018-0916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu C., Ren X., He J., et al. The predictive value of lncRNA MIR31HG expression on clinical outcomes in patients with solid malignant tumors. Cancer Cell Int. 2020;20:115–127. doi: 10.1186/s12935-020-01194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S.C., Liu C.J., Ji S.H., et al. The upregulation of oncogenic miRNAs in swabbed samples obtained from oral premalignant and malignant lesions. Clin Oral Investig. 2021 doi: 10.1007/s00784-021-04108-y. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523–1532. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez N.F., Gundersen G.W., Rahman A., et al. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci Data. 2017;4:170151–170162. doi: 10.1038/sdata.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C.J., Tsai M.M., Hung P.S., et al. Mir-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010;70:1635–1644. doi: 10.1158/0008-5472.CAN-09-2291. [DOI] [PubMed] [Google Scholar]
- 25.Chang K.W., Hung W.W., Chou C.H., et al. LncRNA MIR31HG drives oncogenicity by inhibiting the limb-bud and heart development gene (LBH) during oral carcinoma. Int J Mol Sci. 2021;22:8383. doi: 10.3390/ijms22168383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu B., Zhang X.J., Li X.G., Jiang L.S., He F. Long non-coding RNA LOC344887 is a potential prognostic biomarker in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017;21:3808–3812. [PubMed] [Google Scholar]
- 27.Hung S.Y., Lin C.C., Hsu C.L., et al. The expression levels of long non-coding RNA KIAA0125 are associated with distinct clinical and biological features in myelodysplastic syndromes. Br J Haematol. 2021;192:589–598. doi: 10.1111/bjh.17231. [DOI] [PubMed] [Google Scholar]
- 28.Chen J.F., Wu P., Xia R., et al. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 2018;17:6–21. doi: 10.1186/s12943-017-0756-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou K., Xu J., Yin X., Xia J. Long noncoding RNA HAGLROS promotes cell invasion and metastasis by sponging miR-152 and upregulating ROCK1 expression in osteosarcoma. Comput Math Methods Med. 2020;2020:7236245. doi: 10.1155/2020/7236245. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Li K., Sun D., Gou Q., et al. Long non-coding RNA LINC00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. doi: 10.1016/j.canlet.2018.01.060. [DOI] [PubMed] [Google Scholar]
- 31.Li J., Huang S., Zhang Y., Zhuo W., Tong B., Cai F. LINC00460 enhances bladder carcinoma cell proliferation and migration by modulating miR-612/FOXK1 axis. Pharmacology. 2021;106:79–90. doi: 10.1159/000509255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Y., Wu H., Pavlosky A., et al. Regulatory non-coding RNA: new instruments in the orchestration of cell death. Cell Death Dis. 2016;7:e2333. doi: 10.1038/cddis.2016.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fagerberg L., Hallström B.M., Oksvold P., et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteom. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y., Fan Y., Zhou X., et al. Significance of lncRNA MIR31HG in predicting the prognosis for Chinese patients with cancer: a meta-analysis. Biomark Med. 2020;14:303–316. doi: 10.2217/bmm-2019-0145. [DOI] [PubMed] [Google Scholar]
- 35.Eide P.W., Eilertsen I.A., Sveen A., Lothe R.A. Long noncoding RNA MIR31HG is a bona fide prognostic marker with colorectal cancer cell-intrinsic properties. Int J Cancer. 2019;144:2843–2853. doi: 10.1002/ijc.31998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinske L.C., Galante P.A., Kuo W.P., Ohno-Machado L. A potential role for intragenic miRNAs on their hosts' interactome. BMC Genom. 2010;11:533–545. doi: 10.1186/1471-2164-11-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roepman P., Wessels L.F., Kettelarij N., et al. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet. 2005;37:182–186. doi: 10.1038/ng1502. [DOI] [PubMed] [Google Scholar]
- 38.Lu W.C., Kao S.Y., Yang C.C., et al. EGF up-regulates miR-31 through the C/EBPβ signal cascade in oral carcinoma. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang H., Liu P., Zhang J., et al. Long noncoding RNA MIR31HG exhibits oncogenic property in pancreatic ductal adenocarcinoma and is negatively regulated by miR-193b. Oncogene. 2016;35:3647–3657. doi: 10.1038/onc.2015.430. [DOI] [PMC free article] [PubMed] [Google Scholar]