Abstract
The Penicillium roqueforti group has recently been split into three species, P. roqueforti, Penicillium carneum, and Penicillium paneum, on the basis of differences in ribosomal DNA sequences and secondary metabolite profiles. We reevaluated the taxonomic identity of 52 livestock feed isolates from Sweden, previously identified by morphology as P. roqueforti, by comparing the sequences of the ribosomal internal transcribed spacer region. Identities were confirmed with random amplified polymorphic DNA analysis and secondary metabolite profiles. Of these isolates, 48 were P. roqueforti, 2 were P. paneum, and 2 were Penicillium expansum. No P. carneum isolates were found. The three species produce different mycotoxins, but no obvious relationship between mold and animal disease was detected, based on medical records. P. roqueforti appears to dominate in silage, but the ecological and toxicological importance of P. carneum and P. paneum as feed spoilage fungi is not clear. This is the first report of P. expansum in silage.
A central issue in the field of feed quality and storage is the problem of mold spoilage. Fungal growth reduces nutritional value and may result in the production of mycotoxins and allergenic spores. One way of preserving grass forage is ensiling, in which organic acids produced by lactic acid bacteria and low oxygen pressure prevent growth of spoilage molds and bacteria. However, nonuniform distribution of acids or failure to maintain a low oxygen pressure, especially when breaking silos and big bales for feedout, often induces the growth of microaerophilic acid-tolerant molds. Members of the genus Penicillium are commonly found in feedstuffs in temperate climates. Penicillium roqueforti, which can grow on organic acids (7), is the dominant fungus in most silage samples (1, 16, 19, 24). Animal health disorders are correlated with the production of toxic metabolites in vitro (6, 14, 26). Recently, Auerbach et al. (1) reported that 21 of 24 visibly moldy silage samples contained roquefortine C. This neurotoxic (29) mycotoxin as well as the mutagenic (23, 28) PR toxin, both produced by P. roqueforti, are believed to be involved in disease symptoms observed in farm animals, i.e., extensive paralysis of cows (12) and bovine abortion and placental retention (26), respectively.
P. roqueforti was recently split into the three species, P. roqueforti, Penicillium carneum, and Penicillium paneum, (collectively referred to as the P. roqueforti group) based on ribosomal DNA sequence comparison, random amplified polymorphic DNA (RAPD) profiles, and secondary metabolite profiles (2). These three species synthesize different mycotoxins. All three produce roquefortine C, only P. roqueforti produces PR toxin, and both P. carneum and P. paneum produce patulin, which is mutagenic, immunotoxic, and neurotoxic (5, 8). PR toxin is the most acutely toxic metabolite produced, with 50% lethal dose values in mice ranging from 1 to 5.8 mg kg of body weight−1 (intraperitoneally [IP]) (5). The equivalent 50% lethal dose values for roquefortine C and patulin are 20 and 5 mg kg of body weight−1 (IP), respectively (5, 25). All three Penicillium species can grow on 0.5% acetic acid (2) and have similar microaerophilic capacity with regard to their ability to grow at low oxygen and high carbon dioxide pressures (20).
Our objectives in this study were: (i) to determine the natural occurrence and/or distribution of P. roqueforti, P. carneum, and P. paneum in animal feed and (ii) to relate, when possible, mold identity to animal disease. Our results are the first indication of the relative importance, as feed spoilage organisms within the P. roqueforti group, of the newly defined species P. carneum and P. paneum.
MATERIALS AND METHODS
Fungal isolates and isolation method.
The National Veterinary Institute (SVA), Uppsala, Sweden, supplied strains isolated between 1988 and 1998 from feed suspected to be the cause of various diseases in livestock. Isolates were picked from either dichloran-glycerol (DG18) (Oxoid, Basingstoke, Hampshire, England) or dichloran rose bengal chloramphenicol (DRBC) (Oxoid) agar plates after incubation at 25°C for 5 to 6 days. Isolates identified as P. roqueforti by microscopy were stored on Czapek yeast (autolysate) extract (CYA) agar (Oxoid) slants at 4°C at the SVA. Type strains of P. roqueforti (IBT 6754) (Institut for Bioteknologi Culture Collection, Technical University of Denmark [DTU], Lyngby, Denmark), P. carneum (IBT 6884), and P. paneum (IBT 12407) were supplied by Jens C. Frisvad, Department of Biotechnology, DTU.
Of the 48 P. roqueforti isolates, 29 strains were obtained from grass silage and 3 isolates each from ensiled hard-pressed beet fibers, ensiled grain, and whey. The remaining 10 P. roqueforti strains were isolated from various feeds or came from unknown sources. P. paneum was found in grass silage and hard-pressed beet fibers, while Penicillium expansum was only found in grass silage.
Growth media and conditions.
All isolates were grown on DG18 agar (Oxoid) for 7 days at 25°C in the dark before DNA extraction. For determination of tolerance to acetic acid, isolates were inoculated on malt extract agar (Oxoid) with 0.5% (vol/vol) glacial acetic acid added after autoclaving (MEA-HAc) and incubated in the dark for 4 days at 25°C. For secondary metabolite profile analysis, selected isolates were cultured on CYA agar for 7 days at 25°C in the dark (Czapek broth was purchased from Oxoid, and yeast extract was purchased from Difco [Detroit, Mich.]) (22).
Genomic DNA extraction.
DNA was extracted from pure cultures on DG18 by either of two methods. (i) Agar plugs were excised from the plates and DNA was extracted using Fast DNA Prep Kit H (BIO 101, Vista, Calif.). (ii) Spore suspensions were made by adding 500 μl of diluent (0.85 g NaCl, 0.1 g Tween 80, and 0.05 g of agar per 100 ml of H2O) to the plates, followed by slow shaking. Then 50 μl was transferred to an Eppendorf tube, mixed with 50 μl of lysis solution (0.2 M NaOH, 0.2% sodium dodecyl sulfate), heated to 105°C for 30 min, and cooled on ice (18). Samples were neutralized with 1/20 volumes of 5 M potassium acetate (pH 5.5), followed by the addition of 2 volumes of 6 M guanidine thiocyanate (IBI Molecular Biology Certified, Kodak, Rochester, N.Y.), and debris was pelleted for 5 min. The supernatant was transferred to a new tube, 50 μl of silica mixture (2.5 g of silica in 10 ml of 4% Triton X-100 [silica, 0.5 to 10 μm, was purchased from Sigma Chemical Co., St. Louis, Mo.) was added, and the sample was incubated for 5 min at 55°C. After cooling on ice, the silica was pelleted, washed twice in cold 80% ethanol, and dried in a heating block for 1 to 2 min. The silica was resuspended in 50 μl of distilled water, heated for 3 min at 55°C, cooled on ice, and pelleted. The DNA-containing supernatant was transferred to a new tube and stored at −20°C.
PCR conditions.
PCR fragments for sequence analysis were generated using primers ITS4 and ITS5 covering the internal transcribed spacer 1 (ITS1), 5.8S, and ITS2 region of the ribosomal DNA (31). For each 50-μl reaction, a reaction mixture was prepared containing 10 ng of genomic DNA, 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 2.5 mM MgCl2, 0.2 mM of deoxynucleoside triphosphates (Pharmacia Biotech, Uppsala, Sweden), 0.25% Tween 20, 10% dimethyl sulfoxide, 0.3 μM (each) of ITS4 and ITS5, and 2.5 U of Taq DNA polymerase (Amersham Life Science, Buckinghamshire, United Kingdom). Reactions were run on a capillary thermocycler (Rapidcycler; Idaho Technology, Idaho Falls, Idaho) with an initial denaturation of 30 s at 94°C, followed by 30 cycles of 94°C for 15 s, 53°C for 15 s, and 72°C for 30 s, with a final extension at 72°C for 5 min.
Purification of PCR products.
Aliquots of the PCR were electrophoresed on 1.5% (wt/vol) agarose gels in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA), pH 7.7. DNA fragments of 600 bp each were eluted from the gel and then melted at 55°C with 3 volumes of 6 M guanidine thiocyanate. Twenty to thirty microliters of silica mixture (see above) was added, and incubation was continued for 5 min at 55°C. After cooling on ice, the silica was pelleted, washed twice in cold 80% ethanol, and dried in a heating block (55°C) for 1 to 2 min. The PCR fragments were eluted from the silica in 20 to 25 μl of distilled water for 3 min at 55°C. Finally, 5 μl of the eluent was loaded onto a new gel to estimate the DNA concentration.
DNA sequencing.
We used an ABI 377 automatic sequencer (Perkin-Elmer Cetus, Branchburg, N.J.) and the Thermo Sequenase dye terminator cycle sequencing premixture kit (Amersham Life Science). For each 10-μl reaction, we used 6 pmol of either ITS4 or ITS5 and 0.5 to 1.0 μg of the purified PCR fragment. Sequences were evaluated with ABI EditView software (Perkin-Elmer Cetus) and compared with previously published sequences of P. roqueforti, P. carneum, and P. paneum (2).
RAPD fingerprinting.
We made RAPD fingerprints of selected isolates as described by Boysen et al. (2) with the universal primers NS2 or NS7 (31). Amplification was performed on a PCT 1196 thermocycler (MJ Research, Watertown, Mass.) with an initial denaturation for 5 min at 94°C and was followed by 40 cycles of 94°C for 5 s, 40°C for 60 s, and 72°C for 60 s, with a final extension at 72°C for 10 min. The type strains of P. roqueforti, P. carneum, and P. paneum were used as controls.
Secondary metabolite profiles.
Secondary metabolite profiles from selected isolates were determined by thin-layer chromatography (TLC) (11). From pure cultures grown on CYA agar (Oxoid) for 7 days at 25°C in the dark, metabolites were transferred to the TLC plate (Merck, Darmstadt, Germany) by consecutively placing three agar plugs per culture, medium side down, for 30 s on each spot. This procedure was repeated on the opposite side of the plate. One side was eluted with chloroform-acetone-isopropanol (85:15:20) and the other side with toluene–ethyl acetate–90% formic acid (5:4:1). Standards of griseofulvin (Merck), patulin, PR toxin, and roquefortine C (Sigma) and extracts from type strains of P. roqueforti, P. carneum, and P. paneum were run in parallel. Results were evaluated under UV light (254 and 365 nm) without spraying. Metabolite band patterns were related to those of the standards.
RESULTS
All isolates were identified as P. roqueforti based on morphological characters at the SVA. Of 34 samples with recorded mold CFU levels, 13 samples had values exceeding 104 CFU g−1, with 5 of these being greater than 107 CFU g−1. In most cases, only one species was found.
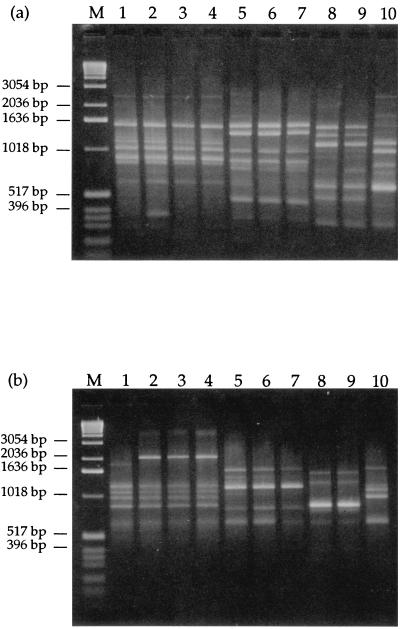
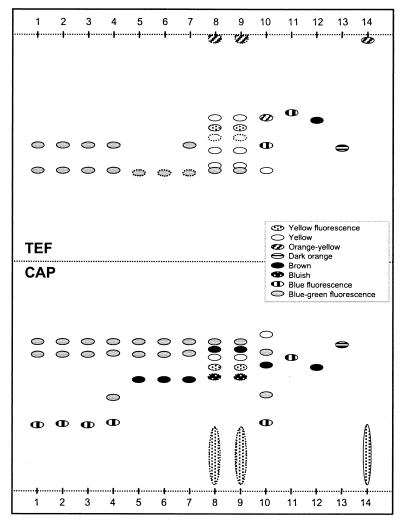
The sequences of the ITS1 and 5.8S regions and parts of the ITS2 region of 52 isolates were compared to previously published sequences of P. roqueforti, P. carneum, and P. paneum (Table 1). Forty-five isolates had sequences identical to P. roqueforti, while three differed at position 180 with either a T or an A replacing the previously reported C (2). The RAPD profiles of these three isolates were similar to those previously published for P. roqueforti (2) (Fig. 1a and b). The secondary metabolite profiles were similar to those of the P. roqueforti type strain (Fig. 2). Two isolates had sequences identical to P. paneum, and two isolates had sequences that differed from those of all members of the P. roqueforti group. No P. carneum isolates were found among the 52 isolates. The identity of the two P. paneum isolates was confirmed by secondary metabolite profiles (Fig. 2) and RAPD analysis (Fig. 1a and b) (2). In a blind test, the SVA included one previously identified isolate of P. roqueforti and two previously identified isolates of P. carneum among the feed isolates. Both DNA sequence analysis and RAPD fingerprinting procedures correctly identified all three isolates (data not shown).
TABLE 1.
Sequence comparison of isolatesa
| Reference species and EMBL accession no. | No. of isolates | Sequence position relative to the 5′ end of the ITS5 primer
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 106 | 128 | 145 | 147 | 150 | 179 | 180 | 181 | 199 | 202 | 211 | 212 | 213 | ||
| P. roquefortiX82358 | A | A | T | T | A | – | C | C | A | C | A | A | C | |
| 48 | • | • | • | • | • | – | Nb | • | • | • | • | • | • | |
| 2 | G | • | C | C | G | C | • | • | T | T | T | G | A | |
| P. paneumX82360 | A | C | C | C | G | – | C | C | A | A | T | G | A | |
| 2 | • | • | • | • | • | – | • | • | • | • | • | • | • | |
| P. carneumX82359 | A | A | T | C | A | – | T | C | A | C | A | A | C | |
| 0 | • | • | • | • | • | – | • | • | • | • | • | • | • | |
Sequence comparison of relevant positions from the ITS1 region of previously published reference isolates (2) and 52 strains isolated from feed and tentatively identified as P. roqueforti by microscopy. Dashes (–) indicate deletions and bullets (•) indicate identical nucleotides.
In two cases, position 180 was T and in one case A, while the remaining 45 positions had a C residue.
FIG. 1.
RAPD fingerprinting using either NS2 (a) or NS7 (b) as a primer. Lanes: 1, P. roqueforti SVA 3631/1996, having a T in position 180 of the ITS1 region; 2, P. roqueforti SVA 8176/1995, isolate 5 (A in position 180); 3, P. roqueforti SVA 2986/1992 (T in position 180); 4, P. roqueforti type strain (IBT 6754); 5, P. paneum SVA 494/1990; 6, P. paneum SVA 7023/1994, isolate 2; 7, P. paneum type strain (IBT 12407); 8, P. expansum SVA 2294/1994, isolate 5; 9, P. expansum SVA 6180/1994 isolate 1; 10, P. carneum type strain (IBT 6884); M, molecular weight marker (1-kb DNA ladder; GIBCO BRL, Gaithersburg, Md.).
FIG. 2.
Schematic illustration of secondary metabolite profiles. TLC plates were eluted in toluene-ethyl acetate-formic acid (TEF) and chloroform-acetone-isopropanol (CAP) and evaluated under UV light (365 nm). Lanes: 1, P. roqueforti SVA 3631/1996, having a T in position 180 of the ITS1 region; 2, P. roqueforti SVA 8176/1995, isolate 5 (A in position 180); 3, P. roqueforti SVA 2986/1992 (T in position 180); 4, P. roqueforti type strain (IBT 6754); 5, P. paneum SVA 494/1990; 6, P. paneum SVA 7023/1994, isolate 2; 7, P. paneum type strain (IBT 12407); 8, P. expansum SVA 2294/1994, isolate 5; 9, P. expansum SVA 6180/1994, isolate 1; 10, P. carneum type strain (IBT 6884). Lanes 11 to 14 are standards: griseofulvin (11), patulin (12), PR toxin (13), and roquefortine C (14). Dotted lines around spots indicate weak spots.
We also examined the RAPD and secondary metabolite profiles of the two isolates with unknown sequences to determine if they belonged to the P. roqueforti group. Neither RAPD analysis (Fig. 1a and b) nor secondary metabolite profiles (Fig. 2) were consistent with the inclusion of those strains in the P. roqueforti group. Based on morphology (e.g., 3.0- to 3.5-μm conidia and smooth-walled stipes) and physiology (e.g., acid production on creatine sucrose agar), these strains are now classified as P. expansum. Roquefortine C was produced by P. expansum and all isolates produced a number of secondary metabolites (Fig. 2). All 52 isolates were cultured on MEA-HAc, and all but the two P. expansum strains could grow on 0.5% (vol/vol) acetic acid.
Among the 28 P. roqueforti isolates associated with diseased animals, 20 were obtained from feed samples from animals with bovine mastitis, 3 were associated with high or increased rates of mortality, 3 were from animals with fertility problems, and 2 were from animals with general health problems. Both P. paneum isolates came from feed associated with animals with mastitis or general health problems. The remaining isolates came from feed samples without any record of associated animal disease.
DISCUSSION
We reidentified 52 animal feed isolates of P. roqueforti using ITS sequence comparison, RAPD analysis, and secondary metabolite profiles as 48 P. roqueforti, 2 P. paneum, and 2 P. expansum. These results are consistent with previous results (J. C. Frisvad, personal communication), with P. roqueforti dominating and with limited occurrence of both P. carneum (not detected in our study) and P. paneum. Assuming that our sample is unbiased, then the frequency of P. carneum should be less than 6%.
Using a combination of methods for the identification of the isolates makes us confident that we have obtained correct strain identities. The taxonomic information recoverable from highly conserved DNA sequences, such as the ITS regions, can often give sufficient information (3), though it is not advisable to use these sequences as a sole criteria for characterization. However, even conserved regions have small variations, and in those cases we confirmed the identification by analyzing RAPD and secondary metabolite profile patterns. Though RAPD profiles are known to be difficult to reproduce (15), we have obtained consistent results using different DNA extraction methods and different thermocyclers in this and the initial work (2) for the RAPD analysis. The TLC analysis is a simple, fast screening method for analyzing secondary metabolites from mold (10). However, in our hands it was not sufficiently consistent for us to use it as the sole identification tool. Since the method is sensitive, e.g., to the origin of the yeast extract used in the substrates for secondary metabolite production, we could not be certain of an identification based only on individual metabolites. Instead, we used profiles of secondary metabolites, compared to those of reference strains, to complement the morphological and genetic information.
The fresh samples were not analyzed for mycotoxins, so possible relationships between mycotoxin and animal disease could not be evaluated critically. All three species of the P. roqueforti group can produce roquefortine C, the major mycotoxin found in moldy silage (1) or in culture broth of P. roqueforti isolates from silage (17, 30). Thus, mycotoxins could contribute to the animal diseases described in this study.
All three members of the P. roqueforti group have similar morphological and physiological characters, and for practical purposes Pitt and Hocking (21) consider the group a single species, even though some of the species can produce mycotoxins (e.g., PR toxin and patulin) that are more toxic than roquefortine C (2). No PR toxin or patulin production was observed from the tested isolates of the P. roqueforti group (Fig. 2), but as mycotoxins can act synergistically, even low levels of several mycotoxins could be a health hazard (4, 9).
In general, good-quality silage feed contains less than 104 fungal CFU g of silage−1 (13), and acceptable feed should contain less than 105 CFU g−1 (24). To our knowledge, this is the first report of P. expansum being found in high numbers (>107 CFU g−1) in silage. P. expansum is a common cause of apple rot and produces both roquefortine C and patulin (22). P. expansum is common in various fruits but is less common in stored or fresh foods (21). Like P. roqueforti, P. expansum has low oxygen requirements, is psychrophilic, and can grow at low water activity; the minimum water activity required for germination is 0.82 to 0.83 (21). Its ability to grow in acidic environments (e.g., apple, pH ∼3.5) suggests that it may be a potential contaminant of silage. Indeed, preliminary results in our laboratory show that the two P. expansum isolates could grow on at least 0.3% (vol/vol) acetic acid (pH ∼3) or 2% (wt/vol) lactic acid (pH ∼2) in malt extract agar. Growth of P. expansum is inhibited by CO2 levels of >15%, while growth of P. roqueforti is stimulated by CO2 levels up to 15% and can occur even at 80% CO2, provided the O2 level is at least 4.2% (27).
This is the first report of the recovery of species within the P. roqueforti group from natural samples. We think that the closed and/or special microaerophilic environment and the organic acid substrate of silage favor growth of P. roqueforti and probably that of the entire P. roqueforti group. Further studies in other environments known to favor the P. roqueforti group (e.g., airtight stored grain or rye bread) are needed before concluding that P. roqueforti should be considered the primary or exclusive hazard of the three species.
ACKNOWLEDGMENTS
This study was supported by the Foundation for Strategic Environmental Research (MISTRA).
We thank Per Häggblom, Head of Department of Feed Hygiene, National Veterinary Institute, for constructive discussions initiating this project, Inger Ohlsson for technical assistance, Jens C. Frisvad, Department of Biotechnology, DTU, for assisting with the identification of P. expansum strains, and Gerhart E. Wagner and Jan Stenlid for critically reading the manuscript.
REFERENCES
- 1.Auerbach H, Oldenburg E, Weissbach F. Incidence of Penicillium roqueforti and roquefortine C in silages. J Sci Food Agr. 1998;76:565–572. [Google Scholar]
- 2.Boysen M, Skouboe P, Frisvad J, Rossen L. Reclassification of the Penicillium roqueforti group into three species on the basis of molecular genetic and biochemical profiles. Microbiology. 1996;142:541–549. doi: 10.1099/13500872-142-3-541. [DOI] [PubMed] [Google Scholar]
- 3.Bruns T D, White T J, Taylor J W. Fungal molecular systematics. Annu Rev Ecol Syst. 1991;22:525–564. [Google Scholar]
- 4.Coffey M T, Hagler W M, Jones E E, Cullen J M. Interactive effects of multiple mycotoxin contamination of swine diets. In: Llewellyn G C, O'Rear C E, editors. Biodeterioration research. Vol. 3. New York, N.Y: Plenum Press; 1990. pp. 117–128. [Google Scholar]
- 5.Cole R J, Cox R H. Handbook of toxic fungal metabolites. New York, N.Y: Academic Press; 1981. [Google Scholar]
- 6.Counter D E. An outbreak of mycotic abortion apparently due to mould-infested sugar beet pulp. Vet Rec. 1973;93:425. doi: 10.1136/vr.93.15.425. [DOI] [PubMed] [Google Scholar]
- 7.Engel G, Teuber M. Simple aid for the identification of Penicillium roqueforti Thom. Growth in acetic acid. Eur J Appl Microbiol Biotechnol. 1978;6:107–111. [Google Scholar]
- 8.Engel G, Teuber M. Patulin and other small lactones. In: Betina V, editor. Mycotoxins. Production, isolation, separation, and purification. Vol. 8. Amsterdam, The Netherlands: Elsevier; 1984. pp. 291–314. [Google Scholar]
- 9.Eriksen G S, Alexander J E. Fusarium toxins in cereals—a risk assessment. TemaNord report 1998:502. Copenhagen, Denmark: Nordic Council of Ministers; 1998. [Google Scholar]
- 10.Filtenborg O, Frisvad J C, Svendsen J A. Simple screening method for molds producing intracellular mycotoxins in pure cultures. Appl Environ Microbiol. 1983;45:581–585. doi: 10.1128/aem.45.2.581-585.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filtenborg O, Frisvad J C, Thrane U, Lund F. Screening methods for secondary metabolites produced by fungi in pure culture. In: Samson R A, Hoekstra E S, Frisvad J C, Filtenborg O, editors. Introduction to food-borne fungi. 4th ed. Baarn, The Netherlands: Centraalbureau voor Schimmelcultures; 1995. pp. 270–274. [Google Scholar]
- 12.Häggblom P. Isolation of roquefortine C from feed grain. Appl Environ Microbiol. 1990;56:2924–2926. doi: 10.1128/aem.56.9.2924-2926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensenska Z. Micromycetes in foodstuffs and feedstuffs. Vol. 28. Amsterdam, The Netherlands: Elsevier; 1993. [PubMed] [Google Scholar]
- 14.Le Bars J, Le Bars P. Especes fongiques des ensilages de mais. Risques mycotoxiques. Rec Med Vet. 1989;165:433–439. [Google Scholar]
- 15.MacPherson J M, Eckstein P E, Scoles G J, Gajadhar A A. Variability of the random amplified polymorphic DNA assay among thermal cyclers, and effects of primer and DNA concentration. Mol Cell Probes. 1993;7:293–299. doi: 10.1006/mcpr.1993.1043. [DOI] [PubMed] [Google Scholar]
- 16.Nout M J R, Bouwmeester H M, Haaksma J, Van Dijk H. Fungal growth in silages of sugarbeet press pulp and maize. J Agr Sci. 1993;121:323–326. [Google Scholar]
- 17.Ohmomo S, Kitamoto H K, Nakajima T. Detection of roquefortines in Penicillium roqueforti isolated from maize silage. J Sci Food Agr. 1994;64:211–215. [Google Scholar]
- 18.Olsson J, Schnürer J, Pedersen L H, Rossen L. A rapid and efficient method for DNA extraction from fungal spores and mycelium for PCR-based detection. J Food Mycol. 1999;2:251–260. [Google Scholar]
- 19.Pelhate J. Mycoflore des mais-fourrages ensilés. Determinisme de son évolution. Rev Mycol. 1975;39:65–95. [Google Scholar]
- 20.Petersson S. Yeast/mold interactions during airtight storage of high-moisture feed grain. Ph.D. thesis. Uppsala, Sweden: Swedish University of Agricultural Sciences; 1998. [Google Scholar]
- 21.Pitt J I, Hocking A D. Fungi and food spoilage. 2nd ed. London, England: Blackie Academic & Professional, Chapman & Hall; 1997. [Google Scholar]
- 22.Samson R A, Hoekstra E S, Frisvad J C, Filtenborg O. Introduction to food-borne fungi. 4th ed. Baarn, The Netherlands: Centraalbureau voor Schimmelcultures; 1995. [Google Scholar]
- 23.Scott P M. PR toxin. In: Betina V, editor. Mycotoxins. Production, isolation, separation, and purification. Vol. 8. Amsterdam, The Netherlands: Elsevier; 1984. pp. 469–474. [Google Scholar]
- 24.Skaar I. Mycological survey and characterisation of the mycobiota of big bale grass silage in Norway. Ph.D. thesis. Oslo, Norway: Norwegian College of Veterinary Medicine; 1996. [Google Scholar]
- 25.Smith J E, Solomons G L. Mycotoxins in human nutrition and health (EUR 16048 EN). Brussels, Belgium: European Commission, Agro-Industrial Research Division; 1994. [Google Scholar]
- 26.Still P E, Wei R, Smalley E B, Strong F M. A mycotoxin from Penicillium roqueforti isolated from toxic cattle feed. Fed Proc. 1972;31:733. [Google Scholar]
- 27.Taniwaki M H. Growth and mycotoxin production by fungi under modified atmosphere. Ph.D. thesis. Sydney, Australia: University of New South Wales; 1995. [Google Scholar]
- 28.Ueno Y. Biochemical mode of action of mycotoxins. In: Smith J E, Henderson R S, editors. Mycotoxins and animal foods. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 437–453. [Google Scholar]
- 29.Wagener R E, Davis N D, Diener U L. Penitrem A and roquefortine production by Penicillium commune. Appl Environ Microbiol. 1980;39:882–887. doi: 10.1128/aem.39.4.882-887.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei R D, Still P E, Smalley E B, Schones H K, Strong F M. Isolation and partial characterization of a mycotoxin from Penicillium roqueforti. Appl Microbiol. 1973;25:111–114. doi: 10.1128/am.25.1.111-114.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White T J, Bruns T D, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 315–322. [Google Scholar]