Abstract
Plant Nucleotide binding-Leucine rich repeat (NLR) proteins play a significant role in pathogen detection and the activation of effector-triggered immunity. NLR regulation has mainly been studied at a protein level, with large knowledge gaps remaining regarding the transcriptional control of NLR genes. The mis-regulation of NLR gene expression may lead to the inability of plants to recognize pathogen infection, lower levels of immune response activation, and ultimately plant susceptibility. This highlights the importance of understanding all aspects of NLR regulation. Three main mechanisms have been shown to control NLR expression: epigenetic modifications, cis elements which bind transcription factors, and post-transcriptional modifications. In this review, we aim to provide an overview of these mechanisms known to control NLR expression, and those which contribute toward successful immune responses. Furthermore, we discuss how pathogens can interfere with NLR expression to increase pathogen virulence. Understanding how these molecular mechanisms control NLR expression would contribute significantly toward building a complete picture of how plant immune responses are activated during pathogen infection—knowledge which can be applied during crop breeding programs aimed to increase resistance toward numerous plant pathogens.
Keywords: pathogen infection, NLR, epigenetics, transcriptional regulatinon, NB-LRR, cis elements, NLR expression
Introduction
Various pathogens, including bacteria, fungi, oomycetes, and viruses, constantly bombard plant species and may cause large crop losses in agricultural settings. Plants have in turn evolved a complex set of defense mechanisms to combat pathogen infection (Jones and Dangl, 2006). Understanding how these plant defense responses are regulated and activated during pathogen attack will accelerate crop breeding programs and may contribute to the development of transgenic crop species with the desired resistance characteristics (Wang et al., 2019). Research focused on unraveling plant immune responses has, unsurprisingly, been of particular interest for the past decade (Bezerra-Neto et al., 2020). All research studies have contributed to reveal an increasingly more complex system, with thousands of signaling molecules, receptors, and hormones, each playing a role in plant immune responses (Wan et al., 2012; Van den Berg et al., 2018; Adachi et al., 2019).
Jones and Dangl (2006) first explained plant immune responses with the well-known Zig-Zag model. This model explains that pathogens are first recognized when pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) are recognized by pattern recognition receptors (PRRs). After PAMP recognition, PRRs activate a low amplitude immune response, known as PAMP-triggered immunity (PTI). This immune response is often able to overcome infection by suppressing pathogen growth. However, some pathogens can overcome PTI responses. Plant Resistance (R) proteins may then recognize Avirulence effector (Avr) proteins, secreted by pathogens, and trigger effector-triggered immunity (ETI; Davis and Hahlbrock, 1987). A successful ETI response leads to the reactive oxygen species (ROS) and activation of the hypersensitive response (HR)—leading to localized plant cell death and the suppression of pathogen growth. When a R protein is either not present, or unable to recognize a corresponding Avr protein, effector-triggered susceptibility (ETS) is triggered, often leading to plant death (Cui et al., 2015).
This model can, however, be deceivingly simple (Naveed et al., 2020). For example, the model explains that a successful ETI response (able to overcome host-adapted pathogen attack) can only be activated when R proteins recognize a respective Avr protein. However, an increasing amount of evidence has suggested that the expression levels of R genes also contribute toward a successful immune response (Bradeen et al., 2009; Andam et al., 2020). When R gene expression is mis-regulated, the amplitude of ETI activation decreases, and an ETI response strong enough to suppress pathogen growth cannot be triggered (Umadevi and Anandaraj, 2017; Xu et al., 2018). Thus, even when a R protein is able to recognize a corresponding Avr protein, insufficient levels of this protein would lead to susceptibility. Understanding how R gene expression is regulated is thus the first step in untangling the mechanisms behind successful immune responses. In this review, we will focus on the regulatory mechanisms controlling R gene expression during pathogen infection, and how pathogens interfere and highjack these mechanisms for their own advantage.
The Main Character: NLR Proteins
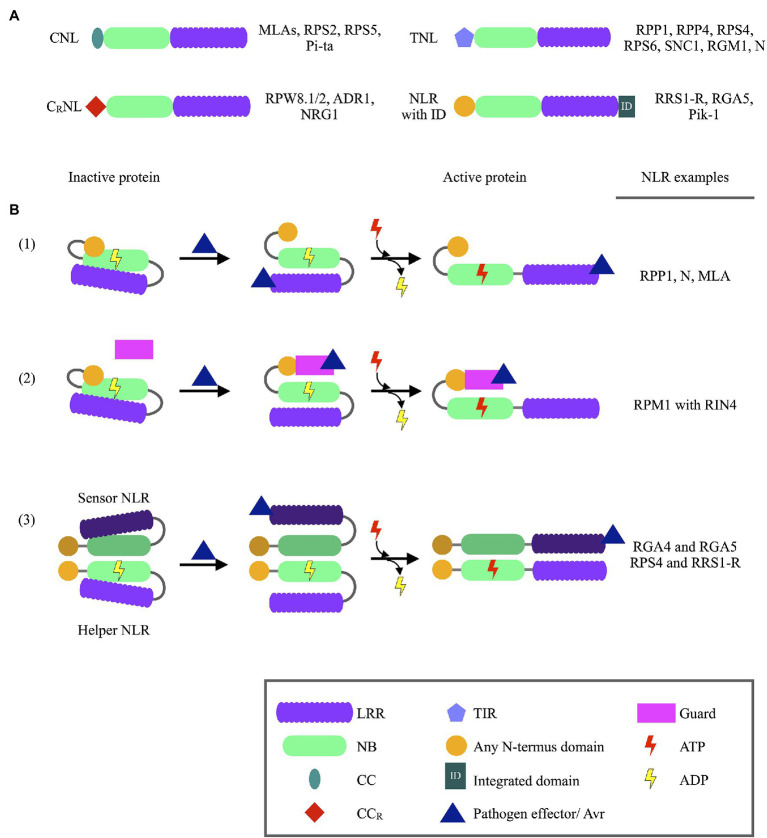
NLR proteins, also known as NB-LRRs, constitute the largest subclass of R proteins and are characterized based on containing a Nucleotide binding (NB/NB-ARC) domain and a Leucine rich repeat (LRR) domain. NLRs can further be classified based on either having a Coiled coil (CC) domain, a CC with an integrated RPW8 domain (CCR), or a Toll/interleukin-1 receptor domain located at the protein’s N-terminus, termed CNLs, CRNLs, and TNLs, respectively (Figure 1; Takken and Goverse, 2012). These N-terminus domains are normally described to control homo- or heterodimerization events between NLRs (Xu et al., 2000; Maekawa et al., 2011). In NLR dimer pairs, one NLR often acts as a “sensor” NLR, able to recognize pathogen Avr proteins (Bonardi et al., 2011). The second NLR acts as a “helper” NLR, triggering the ETI response following activation by the sensor NLR. NB domains remain largely conserved between species and are often used during phylogenetic studies (Maiti et al., 2014). NB domains function as molecular switches for NLR proteins, determining whether the protein is in an active or inactive state. This molecular switch is controlled by ADP and ATP binding to the NB domain P-loop, with ATP binding to activate NLRs following Avr recognition (Steele et al., 2019). The LRR domain, however, is variable in length and shows large sequence variations since this domain is responsible for Avr recognition (Maule et al., 2007). The LRR domain also exerts a negative regulatory effect on the NLR, with loss of this domain leading to increased cell necrosis (Bentham et al., 2017).
Figure 1.
Schematic representation of NLR protein domains and NLR activation, together with NLR protein examples. (A) Different structures of NLR proteins identified in plants, with NLR protein examples listed on the right of each schematic. (B) Models of Nucleotide binding-Leucine rich repeat (NLR) protein recognition of pathogen Avirulence (Avr) proteins. (1) Direct recognition of pathogen Avr protein through binding to LRR domains of NLR proteins. Avr recognition is followed by the exchange of ADP with ATP at the Nucleotide binding (NB) domain, which activates the protein and downstream immune responses. (2) Pathogen Avr proteins may bind to guard proteins which are under the surveillance of NLR proteins. Once Avr binding is recognized the NLR protein is activated through the binding of ATP, ultimately leading to immune response activation. (3) Avr recognition by NLR dimer pairs occurs when an Avr proteins binds to a sensor NLR. Structural changes of the sensor NLR induced by the Avr activates a helper NLR, subsequently leading to immune response activation. The binding of ATP to the sensor NLR is not required for NLR function (ADP—Adenosine diphosphate; ATP—Adenosine triphosphate; CC—Coiled coil; CCR—Coiled coil domain with integrated RPW8 domain; LRR—Leucine rich repeat; NB—Nucleotide binding; TIR—Toll/interleukin-1 receptor).
The recognition of Avr proteins can either occur through direct binding to LRR domains, with the use of a guard protein, or through sensor NLR proteins (Chiang and Coaker, 2015). The use of a guard, or sensor NLRs, to recognize Avr proteins have been shown to significantly increase the variety of Avr proteins recognized by a particular NLR protein. Furthermore, many guard proteins including RIN4, are under the surveillance of multiple NLR proteins (Su et al., 2018). This increases the chances of Avr recognition and successful ETI activation. Arabidopsis RPP1 proteins serve as an example for NLR proteins which directly recognize Avr proteins (Botella et al., 1998). Some NLR proteins also contain integrated domains (ID), which may resemble Avr targets. For example, a WRKY domain was identified in the Arabidopsis RRS1-R protein which recognizes Ralstonia solanacearum effectors (Deslandes et al., 2002; Grund et al., 2019). Ralstonia solanocearum PopP2 and AvrRps4 effectors normally target WRKY transcription factors (TFs), which abolishes transcriptional activation of defense-related genes. However, when these effectors bind to RRS1-R proteins, the RRS1-R/RPP4 complex is activated and triggers defense responses (Ma et al., 2018). Thus, the WRKY ID acts as a decoy for R. solanocearum effectors.
More Is Better—Sometimes: NLR Expression
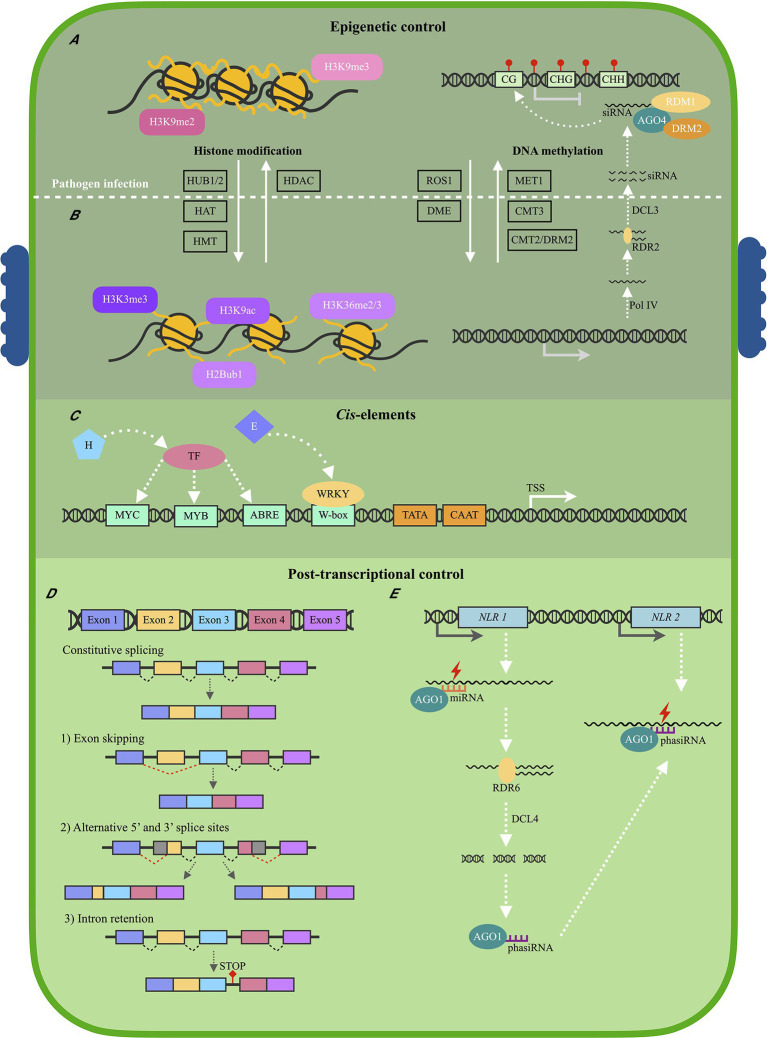
NLR regulation has mainly been studied on protein level, and very little is known regarding NLR transcriptional regulation (Yu et al., 2022). Studies focusing on the expression of NLR genes have highlighted the importance of proper timing and level of NLR expression to activate successful immune responses during pathogen attack (Liu et al., 2020a). The overexpression of NLR genes leads to stunted growth and Avr-independent cell death (Palma et al., 2010; Li et al., 2015). However, rice mutants in which the APIP4 NLR was knocked down showed increased susceptibility when infected with Magnaporthe oryzae (Zhang et al., 2020). Thus, enough NLR proteins need to be activated to trigger successful immune responses during pathogen infection. This indicates that NLR expression needs to be above a certain threshold (Bieri et al., 2004). These factors make it unsurprising to observe differences in NLR expression levels between resistant and susceptible plant genotypes when infected with a pathogen. For example, 22 NLRs were upregulated in Raphanus sativus resistant to Plasmodiophora brassicae, but not in a susceptible genotype during P. brassicae infection (Wang et al., 2022a). Furthermore, significant differences in NLR expression were observed between a partially resistant and susceptible Persea americana rootstocks infected with Phytophthora cinnamomi, especially after 6 h post-inoculation (Fick et al., 2022). The expression of NLR genes is regulated by three main mechanisms: (1) epigenetic mechanisms, (2) cis elements and TFs, and (3) post-transcriptional modifications (Figure 2; Bezerra-Neto et al., 2020). Epigenetic mechanisms include histone modifications and DNA methylation, which influence chromatin density, and subsequently the ability of TFs and transcription machinery to bind to gene promoter sequences. TFs, which bind to cis elements in gene promoter sequences either exert a positive or negative regulatory effect on gene transcription. Furthermore, post-transcriptional modifications include alternative splicing patterns and small RNA, which can introduce stop codons, change protein structure or cause NLR mRNA degradation.
Figure 2.
Schematic illustration of the main regulatory mechanisms of plant NLR expression. (A) Before pathogen infection, a heterochromatin structure is maintained by histone methylation marks H3K9me2 and H3K9me3, which suppresses Nucleotide binding-Leucine rich repeat (NLR) expression. Histone deacetylases (HDAC) also contribute to a heterochromatin structure. The H3K9me3 mark is also associated with DNA methylation of CG, CHG, and CHH sites, established by the RNA-directed DNA methylation pathway. De novo DNA methylation is guided by small interfering RNA (siRNA) in association with Argonaute 4 (AGO4), Domains rearranged methyltransferase 2 (DRM2), and RNA-dependent DNA methylation 1 (RDM1) proteins. DNA methylation is further maintained by DNA methyltransferase 1 (MET1), CMT3 (Chromomethylase 3), and CMT2/DRM2. (B) Following pathogen infection, a euchromatin structure is adopted which allows for the activation of NLR expression. Histone marks H3K3me3 and H3K36me2/3 are established by Histone methyl transferases (HMT), H3K9ac by Histone acetyltransferases (HAT), and H2Bub1 by Histone monoubiquitination 1 and 2 (HUB1/2). Repressor of silencing 1 (ROS1) and DEMETER enzymes (DME) antagonize the RNA-directed DNA methylation pathway, and lower levels of DNA methylation is observed. (C) A euchromatin structure allows for Transcription factors (TFs) to bind to cis elements within NLR promoter sequences located upstream from the Transcription start site (TSS). Most TFs are activated following stress hormone (H) detection, but may also be activated by pathogen effectors (E). The four most common cis elements identified within NLR promoter sequences include W-boxes, ABRE, MYB, and MYC elements. (D) Following NLR expression, alternative splicing (AS) patterns may contribute to different NLR mRNA isoforms, and thus, different levels of NLR proteins. AS may produce mRNAs containing (1) different exons, (2) different Untranslated regions (UTRs), (3) or a retained intron which may code for a stop codon, producing a truncated NLR protein following translation. (E) MicroRNA (miRNA) molecules in association with AGO1 can downregulate NLR expression by binding to NLR mRNAs to either block mRNA translation or cause mRNA degradation. Phased secondary RNA (phasiRNA) molecules can also be produced when diced NLR mRNAs are reverse transcribed by RNA-directed RNA polymerase 6 (RDR6) and diced by Dicer-like 4 (DCL4). These phasiRNA molecules may then target more NLR mRNA molecules to further contribute to NLR suppression (DCL3—Dicer-like 3; Pol IV—RNA polymerase IV).
Epigenetic Control of NLR Genes
Epigenetic modifications regulate whether the chromatin is in a euchromatin (open) or heterochromatin (condensed) structure, thus controlling whether NLR transcription can be activated (Figures 2A,B). Histone modifications and DNA methylation patterns are dynamic molecular mechanisms able to change chromatin structure following pathogen infection (Zhang et al., 2018). Histone modifications have mostly been studied for Arabidopsis NLR genes. One histone modification often associated with transcriptional activation is the trimethylation of lysine 4 of histone H3 (H3K4me3) and is observed to regulate the expression of the Arabidopsis RPP4 and SNC1 NLR genes (Kouzarides, 2007; Xia et al., 2013). This histone mark is established by the histone methyltransferase ATXR7, with the expression of both NLRs being reduced in atxr7 mutants (Xia et al., 2013). Expression of LAZ5, another Arabidopsis NLR gene, is also controlled by histone methylation. Histone methyltransferase SDG8 is responsible for di- or trimethylating H3K36, activating LAZ5 transcription (Palma et al., 2010). The di- and trimethylated H3K36 mark, is interestingly associated with alternative splicing patterns (discussed below) of NLR genes. H3K36me2/me3 levels were significantly higher at the 5’ UTR (untranslated region) of the ARG1 NLR gene in resistant Sorghum bicolor genotypes when infected with Colletotrichum sublineola (Lee et al., 2022). H3K36me2/me3 was shown to increase the expression of ARG1 and regulate alternative splicing patterns to produce a full-length ARG1 mRNA transcript. In the susceptible S. bicolor genotype, lower H3K36me2/me3 marks and expression of ARG1 was observed, together with truncated ARG1 mRNA. This indicates that histone modifications also control NLR expression in an indirect manor at post-transcriptional levels.
Histone acetylation is also associated with active transcription of NLR genes (Luna et al., 2012). Histone acetylation is established by histone acetyltransferases (HATs) and removed by histone deacetylases (HDACs; Bannister and Kouzarides, 2011). Acetyl groups are negatively charged, and histone acetylation would thus result in the chromatin adopting a euchromatin structure (Luna et al., 2012). One HDAC, HDA9, in association with HOS15 was shown to regulate the expression of 62 NLR genes in Arabidopsis, with hda9 and hos15 mutants showing increased NLR expression levels and fewer H3K9ac marks (Yang et al., 2019). Overexpression of another HDAC protein, HDA19, was also shown to enhance Arabidopsis resistance toward the necrotrophic fungus Alternaria brassicicola (Zhou et al., 2005). Since NLR proteins activate the HR, decreased NLR expression could be hypothesized to lead to increased resistance toward necrotrophic pathogens. Histone deacetylation and thus, transcriptional repression might be favorable toward certain types of pathogens. HDA19 expression was also induced by jasmonic acid (JA) and ethylene—signaling hormones produced in response to necrotrophic pathogens (Li et al., 2019). This shows that some histone modifying proteins are activated by either biotrophic- or necrotrophic pathogens, resulting in a different immune response which would prove more suitable toward a specific pathogen. Lastly, histone ubiquitylation is also associated with NLR transcription. HUB1 and HUB2, both E3 ubiquitin ligases, mono-ubiquitylates H2B to H2Bub1 during Pseudomonas syringae pv. tomato DC3000 infection of Arabidopsis. H2Bub1 levels increase, leading to the subsequent increase in expression of RPP4 and SNC1 (Zou et al., 2014).
The H3K9me2 histone mark is associated with transcriptional repression and is seen at the first intron region of the Arabidopsis RPP7 NLR gene (Tsuchiya and Eulgem, 2013). This mark influences alternative polyadenylation patterns of this NLR mRNA, influencing RPP7 protein structure and ultimately governs resistance levels toward Hyaloperonospora arabidopsidis (Eulgem et al., 2007; Tsuchiya and Eulgem, 2013). The H3K9me3 mark is also functionally associated with DNA methylation. DNA methylation of cytosine (position 5; 5mC) occurs at GC, CHG, or CHH (where H is A, C, or T) sites within plants, often aimed at silencing transposable elements (TEs) frequently found within NLR sequences (Miyao et al., 2003; Cuerda-Gil and Slotkin, 2016). De novo DNA methylation is mainly controlled by the RNA-directed DNA methylation pathway (RdDM) in plants (Law and Jacobsen, 2010). Small interfering RNAs (siRNAs) are produced during the canonical RdDM pathway when double-stranded RNA is synthesized by RNA-dependent RNA polymerase 2 (RDR2) and diced by Dicer-like 3 (DCL3). This double-stranded siRNA molecule is then incorporated into Argonaute 4 (AGO4) as single-stranded siRNA. In association with the AGO4-siRNA complex, Domains rearranged methyltransferase 2 (DRM2) and RNA-dependent DNA methylation 1 (RDM1) establishes de novo DNA methylation (Wendte and Pikaard, 2017). Thereafter, DNA methylation is maintained by MET 1 (DNA methyltransferase 1) at CG sites, CMT3 (Chromomethylase 3) at CHG sites, and CMT2/DRM2 at CHH sites. DNA methylation defective Arabidopsis plants showed increased resistance levels toward P. syringae pv. tomato DC3000 and H. arabidopsidis, indicating that decreased levels of DNA methylation may lead to increased NLR expression and immune activation (Dowen Robert et al., 2012; López Sánchez et al., 2016).
The widespread loss of DNA methylation (hypomethylation) at TEs has been observed to occur during the activation of immune responses following pathogen infection (Annacondia et al., 2021). Demethylation of promoters leads to cis elements being more accessible to TFs, ultimately leading to increased NLR gene expression and disease resistance. In poplar trees infected with Lonsdalea populi, hypomethylation occurred at CH sites within promoter regions of defense-related genes (Xiao et al., 2021). Higher levels of hypomethylation were particularly noted in poplar trees with increased resistance toward L. populi when compared to susceptible trees. This observation suggests that hypomethylation results in increased defense-related gene expression which may ultimately lead to increased resistance levels. This could be explained by the observation that AGO4 is repressed after Arabidopsis treatment with PAMP flagellin-22 (flg22), and Aegilops tauschii infection with Blumeria graminis f. sp. tritici (Yu et al., 2013; Geng et al., 2019). The repression of AGO4 leads to lower levels of DNA methylation, which decreases transcriptional repression. Furthermore, ROS1 (Repressor of silencing 1) antagonizes RdDM mediated DNA methylation and promotes resistance toward P. syringae pv. tomato DC3000 (Halter et al., 2021). ROS1 has specifically been implicated in the regulation of some Arabidopsis NLR genes. In ros1 mutants, four NLR genes showed decreased expression levels, due to active demethylation being repressed (Kong et al., 2020). ROS1 was also shown to demethylate promoter regions in which WRKY TFs bind (Halter et al., 2021). In particular, ROS1 demethylated promoter regions of RGM1 TNL after PTI activation by P. syringae pv. tomato DC3000 flagellin proteins. Lastly, DNA demethylation by DEMETER (DME) enzymes also contributes to enhanced resistance toward Verticillium dahlia and P. syringae pv. tomato infection in Arabidopsis (Zeng et al., 2021). In dme mutants, a hypermethylated region was associated with the AtPRX34 TNL gene. This gene showed lower expression levels following P. syringae pv. tomato infection in dme mutants when compared to wild-type plants. These results indicate that DME demethylates NLR sequences in response to bacterial and fungal infection.
Cis Elements of NLR Genes
Cis elements of NLR genes remain largely unknown due to these genes having unusually large promoter sequences (Yu et al., 2022). An NLR promoter sequence has often been defined as the 2 kb region upstream from the NLR transcription start site (Figure 2C). Multiple cis elements are frequently identified within NLR promoter sequences, many being pathogen-inducible cis elements (Table 1). These cis elements may be found in different arrangements within promoter sequences, resulting in complex gene regulatory mechanisms (Wang et al., 2021). Regulatory mechanisms are further complicated by the fact that the TFs which bind to these cis elements either exert a positive or negative regulatory effect on gene expression. Both a positive and negative cis-acting element were identified within the SNC1 NLR gene promoter using CRISPR/Cas9 directed mutations in Arabidopsis (Yu et al., 2022). This study further identified that two other NLR genes, RPP4 and SIKIC2, are also affected by these mutations. This may indicate that these genes share the same cis elements. This hypothesis is supported by the fact that plant NLRs are often found within gene clusters and arranged in a head-to-head configuration (Narusaka et al., 2009; Van Wersch and Li, 2019). Many of these NLRs are often co-expressed following infection, further suggesting that these genes might be under the control of the same promoter, or promoters with the same cis elements (Liang et al., 2019; Yang et al., 2021).
Table 1.
Cis elements identified in promoter sequences of plant NLR genes.
| Cis element | Species | Putative function | Reference |
|---|---|---|---|
| Common | |||
| CAAT | Pinus monticola | Common element |
Liu and Xiang, 2019
Wang et al., 2022b Rampino et al., 2017 |
| Lagenaria siceraria | |||
| Triticum durum | |||
| TATA-box | P. monticola | Core element |
Liu and Xiang, 2019
Wang et al., 2022b Qi et al., 2022 Rampino et al., 2017 |
| L. siceraria | |||
| Tomato | |||
| T. durum | |||
| Pathogen-inducible/stress | |||
| ABRE | Rice | Abscisic acid responsive element |
Ding et al., 2020
Wang et al., 2022b Rampino et al., 2017 |
| L.a siceraria | |||
| T. durum | |||
| AS-1 (TGACG) | Rice | Salicylic acid responsive element |
Kong et al., 2018
Goyal et al., 2021 Diao et al., 2021 |
| Vitis vinifera | |||
| Glycine max | |||
| BIHD-binding site (TGTCA) | P. monticola | Regulation of defense-related genes | Liu and Xiang, 2019 |
| CGCG-box (ACGCGT) | P. monticola | Stress tolerance genes | Liu and Xiang, 2019 |
| CGTCA-motif and TGACG-motif | L. siceraria | Methyl jasmonate responsive element |
Wang et al., 2022b
Rampino et al., 2017 Cui et al., 2017 |
| T. durum | |||
| G. max | |||
| E-box (CANNTG) | Rice | Jasmonic acid responsive element | Miyamoto et al., 2012 |
| ERE-box (ATTTCAAA) | P. monticola | Ethylene responsive element |
Liu and Xiang, 2019
Diao et al., 2021 Wang et al., 2020 Rampino et al., 2017 |
| G. max | |||
| Actinidia chinensis | |||
| T. durum | |||
| G-box | Rice | Regulation of defense-related genes |
Kong et al., 2018
Rampino et al., 2017 |
| T. durum | |||
| GARE-motif, P-box, and TATC-box | L. siceraria | Gibberellin responsive element |
Wang et al., 2022b
Rampino et al., 2017 Cui et al., 2017 |
| T. durum | |||
| G. max | |||
| GCC-box (AGCCGCC) | Rice | Ethylene and pathogen responsive gene |
Kong et al., 2018
Wang et al., 2021 |
| Saccharum | |||
| spontaneum | |||
| GT1-box (GAAAAA) | P. monticola | Pathogen and salt-induced gene expression | Liu and Xiang, 2019 |
| GTTGA | Zea mays | Rhizoctonia solani inducible | Li et al., 2017 |
| H-box (CCTACCN7CT) | Rice | Regulation of defense-related genes | Kong et al., 2018 |
| MYB recognition elements | Rice | Stress responsive elements |
Kong et al., 2018
Ding et al., 2020 Wang et al., 2020 |
| S. spontaneum | |||
| A. chinensis | |||
| Myb1-box (GTTAGTT) | P. monticola | Regulation of defense and drought-related genes |
Liu and Xiang, 2019
Wang et al., 2022b |
| L. siceraria | |||
| MYC elements | Rice | Stress responsive elements | Ding et al., 2020 |
| STRE | Rice | Stress responsive elements | Ding et al., 2020 |
| TATTT | Z. mays | Rhizoctonia solani inducible | Li et al., 2017 |
| TC-rich repeats | V. vinifera | Stress responsive element |
Goyal et al., 2021
Wang et al., 2022b Cui et al., 2017 |
| L. siceraria | |||
| G. max | |||
| TCA element | V. vinifera | Salicylic acid responsive element |
Goyal et al., 2021
Cui et al., 2017 |
| G. max | |||
| TGA element | V. vinifera | Auxin responsive element |
Goyal et al., 2021
Wang et al., 2022b |
| L. siceraria | |||
| W-box (TTTGACY) | P. monticola | Regulation of defense-related genes |
Liu and Xiang, 2019
Wang et al., 2021 Wang et al., 2020 |
| V. vinifera | |||
| Arabidopsis | |||
| S. spontaneum | |||
| A. chinensis | |||
| Other (Growth/development) | |||
| ACACNNG | P. monticola | Abscisic acid induced gene expression | Liu and Xiang, 2019 |
| ARR1-binding site (AGATT) | P. monticola | Cytokinin responsive gene | Liu and Xiang, 2019 |
| MADS-box/ CArG-motif (CCW6GG) | P. monticola | Regulation of plant flowering time and vernalization genes | Liu and Xiang, 2019 |
| Circadian motif (CAAN4ATC) | P. monticola | Circadian gene expression | Liu and Xiang, 2019 |
| NtBBF1 binding site (ACTTTA) | P. monticola | Tissue-specific expression and auxin induction | Liu and Xiang, 2019 |
| SRE (TTATCC) | P.s monticola | Activation of axillary bud outgrowth | Liu and Xiang, 2019 |
| T-box (ACTTTG) | P. monticola | Light activated element | Liu and Xiang, 2019 |
| WUS-binding site (TTAATGG) | P. monticola | Establishment and maintenance of stem cells in shoot and floral meristems | Liu and Xiang, 2019 |
| W-box | Malus domestica | Sorbitol inducible element | Meng et al., 2018 |
It is important to remember that the abundance of TFs and certain arrangements of cis elements also influence gene expression levels, and the simple binding of a specific TF does not necessarily activate gene expression (Hoang et al., 2017). Thus, the identification of NLR cis elements alone cannot be used to predict the level of NLR expression, or when transcription will be activated. In tomato plants, a single nucleotide difference was identified in the promoter region of the Sl5R-1 NLR gene when compared between a Tomato spotted wilt virus (TSWV) resistant and susceptible plant (Qi et al., 2022). This single nucleotide deletion in resistant tomato plants resulted in a new TF binding site to be formed, which increases Sl5R-1 expression and subsequent resistance. Importantly, cis elements are not the only regulatory sequences to control NLR expression—the tobacco N TNL contains two introns which contribute to increased expression levels of this gene during Tobacco mosaic virus (TMV) infection (Ikeda et al., 2021). Transient expression of the N gene without these introns showed lower levels of expression.
Cis elements identified most often in NLR promoter sequences include W-boxes, ABRE, MYB, and MYC elements (Mohr et al., 2010; Ding et al., 2020). W-boxes bind WRKY TFs, which is a large, diverse group of zinc finger TFs (Babu et al., 2006). These TFs are mostly activated by pathogen infection, effectors, and stress hormones, such as salicylic acid (SA) and JA. Following activation, a subset of WRKYs trigger the expression of PTI and ETI-related proteins, and the synthesis of stress hormones (Chen et al., 2019). Interestingly, an apple (Malus domestica) NLR gene, MdNLR16, is under the control of the MdWRKY79 TF which is responsive to sorbitol levels (Meng et al., 2018). Higher sorbitol levels lead to increased MdNLR16 expression and subsequently enhanced resistance levels toward Alternaria alternata. ABRE elements are abscisic acid responsive elements which are recognized by bZIP proteins (Hobo et al., 1999). A single ABRE element, however, is not able to activate transcription, instead multiple elements are needed for transcriptional activation (Shen et al., 1996). Lastly, MYB and MYC elements have been shown to activate gene transcription in response to both abiotic and biotic stressors (Feng et al., 2013; Fang et al., 2018; Wu et al., 2019). NLR cis element identification studies have thus shown that the TFs controlling NLR expression is mostly activated by abiotic and biotic stress. Cis element studies may then also be used to identify putative NLR function. For example, in Pinus monticola Douglas ex D. Don (Western white pine trees), cis element identification of the PmTNL2 gene suggested that this NLR might be important for both plant immune responses, as well as growth and development (Liu and Xiang, 2019).
Post-transcriptional Modifications of NLRs
Alternative splicing (AS) contributes significantly toward the diversity of the NLR transcriptome and NLR proteome—altering levels of different mRNA isoforms in response to developmental and environmental conditions (Kelemen et al., 2013). AS can result in NLR mRNAs to contain different exons, 5′- and 3′ untranslated regions, and introns which may introduce stop codons resulting in truncated NLR proteins (Figure 2D). The example used most often for AS of NLR genes is the N TNL protein associated with resistance toward TMV. The N gene produces either a short N mRNA (NS) or a long N mRNA (NL; Erickson et al., 1999). NL contains an exon which encodes a stop codon, resulting in a truncated protein. NS however is translated into a complete protein. Both these proteins are expressed during TMV infection and needed for full TMV resistance. One rice CNL, Pi-ta, produces up to 11 different protein isoforms as a result of AS (Costanzo and Jia, 2009). In response to M. oryzae infection, a resistant rice genotype showed increased expression levels of a Pi-ta protein with a C-terminus thioredoxin domain, when compared to a susceptible genotype.
In both barley and wheat, AS was seen to regulate whether and which IDs were present in NLR proteins in response to different experimental conditions (Halterman et al., 2003; Andersen et al., 2020). Different IDs may influence where NLR proteins localize to in the plant cell or may even cause the NLR protein to act as a decoy target for Avr proteins (Yang et al., 2014). It is worth noting that AS may also have an impact on proteins guarded by NLRs, suggesting that AS may regulate NLR activity in an indirect manner. For example, a NLR Rpi-vnt1.1 guards the GLYK (Glycerate 3-kinase) protein in potato (Gao et al., 2020). A truncated isoform of GLYK, which does not contain a chloroplast transit peptide-encoding sequence, is expressed in dark conditions, and cannot be recognized by a Phytophthora infestans Avr protein AVRvnt1. Thus, Rpi-vnt1.1 cannot activate immune responses. In light conditions, the full-length GLYK mRNA is expressed, and this protein isoform binds to the Avr during infection, leading to Rpi-vnt1.1 being activated to trigger ETI.
NLR repression can be regulated at a post-transcriptional level using siRNAs and micro RNAs (miRNAs; Figure 2E). miRNAs are non-coding RNAs between 20 and 24 nucleotides in length. They are encoded by miRNA genes, which are transcribed by RNA polymerase II to produce a long, primary miRNA (Xie et al., 2005). After processing, a precursor miRNA (pre-miRNA) is produced which forms a hairpin structure with a self-complementary stem loop. This pre-miRNA molecule is diced by DCL1 or DCL4, and produces a 22 nucleotide double-stranded miRNA, which is exported to the cytoplasm (Sun et al., 2019). An RNA-induced silencing complex (RISC) is then formed when the mature miRNA binds to an AGO1 protein. miRNAs guide AGO1 proteins to target mRNAs, either resulting in endonucleolytic cleavage and degradation or inhibition of translation. The P-loop domain, important for ATP binding and NLR protein activation, is a common target for miRNAs (Zhai et al., 2011; Fei et al., 2015).
NLR mRNA cleavage by miRNAs may also produce phasiRNAs (phased secondary small interfering RNAs), which then target and degrade other mRNAs with the same sequence (Liu et al., 2020b). Three Medicago truncatula miRNA families target mRNA transcripts of 74 NLRs, leading to the production of phasiRNAs which suppress the expression of 324 NLR genes (Zhai et al., 2011). Liu et al. (2014) showed that the barley miR9863 family targets MLA1 CNL transcripts, with the resulting phasiRNAs also leading to MLA1 mRNAs being degraded. The authors suspect that this pathway prevents immune responses from being overloaded, and thus, NLR downregulation may have a positive effect on plant resistance levels. In Arabidopsis miR472 knock-down mutants, increased resistance levels were observed toward P. syringae, and reduced resistance levels were observed when this miRNA was overexpressed (Boccara et al., 2014). This presents an interesting method of pathogen control—transient expression of miRNA targets in host plants may increase resistance levels toward various pathogens. In tomato, transient expression of short tandem target mimic RNAs increased resistance levels toward P. infestans and P. syringae (Canto-Pastor et al., 2019). These mimic RNAs acted as targets for miR482/211b, which resulted in increased NLR expression and enhanced disease resistance.
Oh No You Do Not: How Pathogens Interfere With NLR Expression
With multiple proteins contributing to the regulation of NLR expression, comes multiple opportunities for pathogen interference. Despite this, very few cases are documented in which pathogen Avr proteins influence NLR expression. However, Avr targets remain largely unknown, and it is yet to be discovered how NLR regulation is hijacked by pathogens (Wu et al., 2022). An average of 32% of Avr proteins from bacteria, fungi, and oomycetes localize in the plant cell nucleus, indicating that these Avr proteins may interfere with NLR transcription (Khan et al., 2018). Two cytoplasmic effectors from M. oryzae, MoHTR1, and MoHTR2, bind to effector binding elements (EBE) in rice gene promoters and function as transcription repressors (Kim et al., 2020). These EBEs were present in many defense-related gene promoters, and the binding of these effectors led to significant transcription reprogramming. Transient expression of MoHTR1 and MoHTR2 in rice not only led to increased susceptibility toward M. oryzae, but also to Xanthomonas oryzae pv. oryzae and Cochliobolus miyabeanus. It remains unclear whether these effectors bind host repressor proteins, or whether they interfere with the binding of transcription activators. A Melampsora larici-populina effector, Mlp124478, also interferes with the transcription of WRKY TFs which indirectly inhibits the activation of defense-related gene expression, including the RPP8 NLR (Ahmed et al., 2018). Some pathogen Avr proteins also interfere with the synthesis of stress hormones. SA metabolism is inhibited by Phytophthora sojae PsIsc1 and Verticillium dahliae VdIsc1 enzymatic effectors, which redirects the precursor molecule of SA from the plastid into the cytosol (Liu et al., 2014). The metabolism of SA decreases and thus SA-mediated immune responses cannot be activated. Since some TFs which bind to NLR cis elements are activated by SA, lower SA levels may disrupt the activation of NLR expression (Goyal et al., 2021).
From another perspective, the suppression of NLR expression may not be the ultimate goal of the pathogen. Phytophthora species are hemibiotrophic oomycetes, which switch from a biotrophic to necrotrophic phase during infection (Zuluaga et al., 2016). During the necrotrophic phase, increased NLR expression may be beneficial to the pathogen since NLRs activate the HR and thus, plant cell death. P. sojae RxLR effectors PSR1 and PSR2 suppress plant RNA silencing by interfering with the miRNA synthesis pathway, which increased susceptibility in Nicotiana benthamiana (Qiao et al., 2013). This may lead to higher NLR levels and activation of HR, resulting in a more favorable environment for necrotrophic pathogens. This hypothesis is further supported by the fact that PSR2 is only expressed during the later stages of infection, when P. sojae switches to a necrotrophic stage (Qiao et al., 2013). Furthermore, the V. dahliae VdSSR1 protein was shown to interfere with the nuclear exportation of AGO1-miRNA complexes in N. benthamiana (Zhu et al., 2022). Decreased AGO1-miRNA exportation would subsequently lead to decreased suppression of NLR expression, which may contribute to the observed increased susceptibility in transgenic plants expressing VdSSR1 at higher levels. However, VdSSR1 expression data is needed to indicate whether this protein is only expressed during the necrotrophic stage of this hemibiotrophic fungus. Lastly, a necrotrophic fungus, Botrytis cinerea, is able to translocate siRNAs into plant cells and may redirect host siRNA machinery (Weiberg et al., 2013). B. cinerea siRNAs were associated with AGO1 proteins during infection of Arabidopsis, indicating that B. cinerea may hijack RISC to increase virulence (Ellendorff et al., 2009). It would be interesting to investigate whether these siRNA molecules cause siRNA-directed cleavage and degradation of NLR mRNA transcripts (Qiao et al., 2021). It may also be of interest to investigate whether pathogen-derived siRNAs influence DNA methylation patterns during infection.
Conclusion
NLR proteins play a significant role in activating plant immune responses during pathogen attack. The mis-regulation of NLR-encoding genes considerably impairs the plant’s ability to detect pathogen Avr proteins, which ultimately leads to susceptibility. Thus, a comprehensive understanding of NLR gene regulation is of particular interest. Unfortunately, NLR protein regulation has mainly been studied on a post-translational level, with a large knowledge gap remaining regarding the transcriptional- and post-transcriptional regulation of these proteins. Identifying epigenetic marks, and cis elements which control NLR expression in response to pathogen attack provides the first step in unraveling these complex regulatory mechanisms. These mechanisms can further be compared between susceptible and resistant plant genotypes to understand the factors which contribute to a successful immune response. Furthermore, investigating how pathogens interfere with these mechanisms would provide much needed insight into plant–pathogen interactions. Ultimately, knowledge in these areas may be used during plant breeding programs which aim to produce genotypes with increased resistance toward a variety of pathogens. These mechanisms can also be used to drive the expression of trans-NLR genes in genetically modified crops, with the goal of increasing resistance toward both biotic and abiotic stresses.
Author Contributions
AF conceptualized, drafted, and reviewed the manuscript. VS and NB reviewed and assisted in the drafting of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Funding was generously provided by the Hans Merensky Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Adachi H., Derevnina L., Kamoun S. (2019). NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 50, 121–131. doi: 10.1016/j.pbi.2019.04.007, PMID: [DOI] [PubMed] [Google Scholar]
- Ahmed M. B., Santos K. C. G. D., Sanchez I. B., Petre B., Lorrain C., Plourde M. B., et al. (2018). A rust fungal effector binds plant DNA and modulates transcription. Sci. Rep. 8:14718. doi: 10.1038/s41598-018-32825-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andam A., Azizi A., Majdi M., Abdolahzadeh J. (2020). Comparative expression profile of some putative resistance genes of chickpea genotypes in response to ascomycete fungus, Ascochyta rabiei (Pass.) Labr. Rev. Bras. Bot. 43, 123–130. doi: 10.1007/s40415-020-00576-w [DOI] [Google Scholar]
- Andersen E. J., Nepal M. P., Purintun J. M., Nelson D., Mermigka G., Sarris P. F. (2020). Wheat disease resistance genes and their diversification through integrated domain fusions. Front. Genet. 11:898. doi: 10.3389/fgene.2020.00898, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annacondia M. L., Markovic D., Reig Valiente J., Scaltsoyiannes V., Pieterse C., Ninkovic V., et al. (2021). Aphid feeding induces the relaxation of epigenetic control and the associated regulation of the defence response in Arabidopsis. New Phytol. 230, 1185–1200. doi: 10.1111/nph.17226, PMID: [DOI] [PubMed] [Google Scholar]
- Babu M. M., Iyer L. M., Balaji S., Aravind L. (2006). The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 34, 6505–6520. doi: 10.1093/nar/gkl888, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J., Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. doi: 10.1038/cr.2011.22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentham A., Burdett H., Anderson P. A., Williams S. J., Kobe B. (2017). Animal NLRs provide structural insights into plant NLR function. Ann. Bot. 119, mcw171–mcw702. doi: 10.1093/aob/mcw171, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra-Neto J. P., Araújo F. C., Ferreira-Neto J. R. C., Silva R. L. O., Borges A. N. C., Matos M. K. S., et al. (2020). “NBS-LRR genes—plant health sentinels: structure, roles, evolution and biotechnological applications,” in Applied Plant Biotechnology for Improving Resistance to Biotic Stress. eds. Poltronieri P., Hong Y. (Amsterdam, Netherlands: Academic Press; ), 63–120. [Google Scholar]
- Bieri S., Mauch S., Shen Q. H., Peart J., Devoto A., Casais C., et al. (2004). RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16, 3480–3495. doi: 10.1105/tpc.104.026682, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccara M., Sarazin A., Thiébeauld O., Jay F., Voinnet O., Navarro L., et al. (2014). The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 10:e1003883. doi: 10.1371/journal.ppat.1003883, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V., Tang S., Stallmann A., Roberts M., Cherkis K., Dangl Jeffery L. (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. 108, 16463–16468. doi: 10.1073/pnas.1113726108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella M. A., Parker J. E., Frost L. N., Bittner-Eddy P. D., Beynon J. L., Daniels M. J., et al. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10, 1847–1860. doi: 10.1105/tpc.10.11.1847, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradeen J. M., Iorizzo M., Mollov D. S., Raasch J., Kramer L. C., Millett B. P., et al. (2009). Higher copy numbers of the potato RB transgene correspond to enhanced transcript and late blight resistance levels. Mol. Plant-Microbe Interact. 22, 437–446. doi: 10.1094/MPMI-22-4-0437, PMID: [DOI] [PubMed] [Google Scholar]
- Canto-Pastor A., Santos Bruno A. M. C., Valli Adrian A., Summers W., Schornack S., Baulcombe David C. (2019). Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proc. Natl. Acad. Sci. 116, 2755–2760. doi: 10.1073/pnas.1814380116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li C., Wang H., Guo Z. (2019). WRKY transcription factors: evolution, binding, and action. Phytopathol. Res. 1:13. doi: 10.1186/s42483-019-0022-x [DOI] [Google Scholar]
- Chiang Y. H., Coaker G. (2015). Effector triggered immunity: NLR immune perception and downstream defence responses. The Arabidopsis Book 2015:e0183. doi: 10.1199/tab.0183 [DOI] [Google Scholar]
- Costanzo S., Jia Y. (2009). Alternatively spliced transcripts of Pi-ta blast resistance gene in Oryza sativa. Plant Sci. 177, 468–478. doi: 10.1016/j.plantsci.2009.07.012 [DOI] [Google Scholar]
- Cuerda-Gil D., Slotkin R. K. (2016). Non-canonical RNA-directed DNA methylation. Nat. Plants 2:16163. doi: 10.1038/nplants.2016.163 [DOI] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J. E. (2015). Effector triggered immunity: from pathogen perception to robust defence. Ann. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012, PMID: [DOI] [PubMed] [Google Scholar]
- Cui X., Yan Q., Gan S., Xue D., Dou D., Guo N., et al. (2017). Overexpression of gma-miR1510a/b suppresses the expression of a NB-LRR domain gene and reduces resistance to Phytophthora sojae. Gene 621, 32–39. doi: 10.1016/j.gene.2017.04.015, PMID: [DOI] [PubMed] [Google Scholar]
- Davis K. R., Hahlbrock K. (1987). Induction of defence responses in cultured parsley cells by plant cell wall fragments. Plant Physiol. 84, 1286–1290. doi: 10.1104/pp.84.4.1286, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Theulières F., Hirsch J., Feng D. X., Bittner-Eddy P., et al. (2002). Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. 99, 2404–2409. doi: 10.1073/pnas.032485099, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao P., Sun H., Bao Z., Li W., Niu N., Li W., et al. (2021). Expression of an antiviral gene GmRUN1 from soybean is regulated via intron-mediated enhancement (IME). Viruses 13:2032. doi: 10.3390/v13102032, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Xu X., Kong W., Xia X., Zhang S., Liu L. W., et al. (2020). Genome-wide identification and expression analysis of rice NLR genes responsive to the infections of Xanthomonas oryzae pv. oryzae and Magnaporthe oryzae. Physiol. Mol. Plant Pathol. 111:101488. doi: 10.1016/j.pmpp.2020.101488 [DOI] [Google Scholar]
- Dowen Robert H., Pelizzola M., Schmitz Robert J., Lister R., Dowen Jill M., Nery Joseph R., et al. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. 109, E2183–E2191. doi: 10.1073/pnas.1209329109, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellendorff U., Fradin E. F., De Jonge R., Thomma B. P. (2009). RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 60, 591–602. doi: 10.1093/jxb/ern306, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson F. L., Holzberg S., Calderon-Urrea A., Handley V., Axtell M., Corr C., et al. (1999). The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 18, 67–75. doi: 10.1046/j.1365-313x.1999.00426.x, PMID: [DOI] [PubMed] [Google Scholar]
- Eulgem T., Tsuchiya T., Wang X. J., Beasley B., Cuzick A., Tör M., et al. (2007). EDM2 is required for RPP7-dependent disease resistance in Arabidopsis and affects RPP7 transcript levels. Plant J. 49, 829–839. doi: 10.1111/j.1365-313X.2006.02999.x, PMID: [DOI] [PubMed] [Google Scholar]
- Fang Q., Wang Q., Mao H., Xu J., Wang Y., Hu H., et al. (2018). AtDIV2, an R-R-type MYB transcription factor of Arabidopsis, negatively regulates salt stress by modulating ABA signalling. Plant Cell Rep. 37, 1499–1511. doi: 10.1007/s00299-018-2321-6, PMID: [DOI] [PubMed] [Google Scholar]
- Fei Q., Li P., Teng C., Meyers B. C. (2015). Secondary siRNAs from Medicago NB-LRRs modulated via miRNA-target interactions and their abundances. Plant J. 83, 451–465. doi: 10.1111/tpj.12900, PMID: [DOI] [PubMed] [Google Scholar]
- Feng H. L., Ma N. N., Meng X., Zhang S., Wang J. R., Chai S., et al. (2013). A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol. Biochem. 73, 309–320. doi: 10.1016/j.plaphy.2013.09.014, PMID: [DOI] [PubMed] [Google Scholar]
- Fick A., Swart V., Backer R., Bombarely A., Engelbrecht J., Van den Berg N. (2022). Partially resistant avocado rootstock Dusa® shows prolonged upregulation of nucleotide binding-Leucine rich repeat genes in response to Phytophthora cinnamomi infection. Front. Plant Sci. 13:793644. doi: 10.3389/fpls.2022.793644, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Xu H., Huang J., Sun B., Zhang F., Savage Z., et al. (2020). Pathogen manipulation of chloroplast function triggers a light-dependent immune recognition. Proc. Natl. Acad. Sci. 117, 9613–9620. doi: 10.1073/pnas.2002759117, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng S., Kong X., Song G., Jia M., Guan J., Wang F., et al. (2019). DNA methylation dynamics during the interaction of wheat progenitor Aegilops tauschii with the obligate biotrophic fungus Blumeria graminis f. sp. tritici. New Phytol. 221, 1023–1035. doi: 10.1111/nph.15432, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal N., Bhatia G., Garewal N., Upadhyay A., Singh K. (2021). Identification of defence related gene families and their response against powdery and downy mildew infections in Vitis vinifera. BMC Genomics 22:776. doi: 10.1186/s12864-021-08081-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund E., Tremousaygue D., Deslandes L. (2019). Plant NLRs with integrated domains: unity makes strength. Plant Physiol. 179, 1227–1235. doi: 10.1104/pp.18.01134, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T., Wang J., Amesefe D., Lastrucci E., Charvin M., Singla Rastogi M., et al. (2021). The Arabidopsis active demethylase ROS1 cis-regulates defence genes by erasing DNA methylation at promoter-regulatory regions. eLife 10:e62994. doi: 10.7554/eLife.62994, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman D. A., Wei F., Wise R. P. (2003). Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 131, 558–567. doi: 10.1104/pp.014407, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang X. L. T., Nhi D. N. H., Thu N. B. A., Thao N. P., Tran L. S. P. (2017). Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr. Genomics 18, 483–497. doi: 10.2174/1389202918666170227150057, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T., Asada M., Kowyama Y., Hattori T. (1999). ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19, 679–689. doi: 10.1046/j.1365-313x.1999.00565.x, PMID: [DOI] [PubMed] [Google Scholar]
- Ikeda C., Taku K., Miyazaki T., Shirai R., Nelson R. S., Nyunoya H., et al. (2021). Cooperative roles of introns 1 and 2 of tobacco resistance gene N in enhanced N transcript expression and antiviral defence responses. Sci. Rep. 11:15424. doi: 10.1038/s41598-021-94713-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kelemen O., Convertini P., Zhang Z., Wen Y., Shen M., Falaleeva M., et al. (2013). Function of alternative splicing. Gene 514, 1–30. doi: 10.1016/j.gene.2012.07.083, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Seto D., Subramaniam R., Desveaux D. (2018). Oh, the places they’ll go! A survey of phytopathogen effectors and their host targets. Plant J. 93, 651–663. doi: 10.1111/tpj.13780, PMID: [DOI] [PubMed] [Google Scholar]
- Kim S., Kim C. Y., Park S. Y., Kim K. T., Jeon J., Chung H., et al. (2020). Two nuclear effectors of the rice blast fungus modulate host immunity via transcriptional reprogramming. Nat. Commun. 11:5845. doi: 10.1038/s41467-020-19624-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Ding L., Cheng J., Wang B. (2018). Identification and expression analysis of genes with pathogen-inducible cis-regulatory elements in the promoter regions in Oryza sativa. Springer 11:52. doi: 10.1186/s12284-018-0243-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Xia X., Wang Q., Liu L.-W., Zhang S., Ding L., et al. (2020). Impact of DNA demethylases on the DNA methylation and transcription of Arabidopsis NLR genes. Front. Genet. 11:460. doi: 10.3389/fgene.2020.00460, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Law J. A., Jacobsen S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Fu F., Liao C. J., Mewa D. B., Adeyanju A., Ejeta G., et al. (2022). Broad-spectrum fungal resistance in sorghum is conferred through the complex regulation of an immune receptor gene embedded in a natural antisense transcript. Plant Cell 34, 1641–1665. doi: 10.1093/plcell/koab305, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Chen J., Yang F., Wei S., Kong L., Ding X., et al. (2017). Identification of two novel Rhizoctonia solani-inducible cis-acting elements in the promoter of the maize gene, GRMZM2G315431. Sci. Rep. 7:42059. doi: 10.1038/srep42059, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Han X., Feng D., Yuan D., Huang L. J. (2019). Signalling crosstalk between salicylic acid and ethylene/jasmonate in plant defence: do we understand what they are whispering? Int. J. Mol. Sci. 20:671. doi: 10.3390/ijms20030671, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wu J., Yin L., Zhang Y., Qu J., Lu J. (2015). Comparative transcriptome analysis reveals defence-related genes and pathways against downy mildew in Vitis amurensis grapevine. Plant Physiol. Biochem. 95, 1–14. doi: 10.1016/j.plaphy.2015.06.016, PMID: [DOI] [PubMed] [Google Scholar]
- Liang W., Van Wersch S., Tong M., Li X. (2019). TIR-NB-LRR immune receptor SOC3 pairs with truncated TIR-NB protein CHS1 or TN2 to monitor the homeostasis of E3 ligase SAUL1. New Phytol. 221, 2054–2066. doi: 10.1111/nph.15534, PMID: [DOI] [PubMed] [Google Scholar]
- Liu T., Song T., Zhang X., Yuan H., Su L., Li W., et al. (2014). Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5:4686. doi: 10.1038/ncomms5686, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Teng C., Xia R., Meyers B. C. (2020b). PhasiRNAs in plants: their biogenesis, genic sources, and roles in stress responses, development, and reproduction. Plant Cell 32, 3059–3080. doi: 10.1105/tpc.20.00335, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. J., Xiang Y. (2019). Characterization of the western white pine TIR-NBS-LRR (PmTNL2) gene by transcript profiling and promoter analysis. Genome 62, 477–488. doi: 10.1139/gen-2019-0035, PMID: [DOI] [PubMed] [Google Scholar]
- Liu X., Zhao C., Yang L., Zhuang M., Zhang Y., Wang Y., et al. (2020a). A time-resolved dual transcriptome analysis reveals the molecular regulating network underlying the compatible/incompatible interactions between cabbage (Brassica oleracea) and Fusarium oxysporum f. sp. conglutinans. Plant Soil 448, 455–478. doi: 10.1007/s11104-020-04437-z [DOI] [Google Scholar]
- López Sánchez A., Stassen J. H., Furci L., Smith L. M., Ton J. (2016). The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 88, 361–374. doi: 10.1111/tpj.13252, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E., Bruce T. J. A., Roberts M. R., Flors V., Ton J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158, 844–853. doi: 10.1104/pp.111.187468, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Guo H., Hu L., Martinez P. P., Moschou P. N., Cevik V., et al. (2018). Distinct modes of derepression of an Arabidopsis immune receptor complex by two different bacterial effectors. Proc. Natl. Acad. Sci. 115, 10218–10227. doi: 10.1073/pnas.1811858115, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Cheng W., Spiridon L. N., Töller A., Lukasik E., Saijo Y., et al. (2011). Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9, 187–199. doi: 10.1016/j.chom.2011.02.008, PMID: [DOI] [PubMed] [Google Scholar]
- Maiti S., Basak J., Pal A. (2014). “Current understanding on plant R-genes/proteins and mechanisms of defence responses against biotic stresses,” in Review of Plant Pathology. eds. Chakraborty B. N., Chakraborty U. (Jodhpur, India: Scientific Publishers; ), 93–126. [Google Scholar]
- Maule A. J., Caranta C., Boulton M. I. (2007). Sources of natural resistance to plant viruses: status and prospects. Mol. Plant Pathol. 8, 223–231. doi: 10.1111/j.1364-3703.2007.00386.x, PMID: [DOI] [PubMed] [Google Scholar]
- Meng D., Li C., Park H. J., González J., Wang J., Dandekar A. M., et al. (2018). Sorbitol modulates resistance to Alternaria alternata by regulating the expression of an NLR resistance gene in apple. Plant Cell 30, 1562–1581. doi: 10.1105/tpc.18.00231, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K., Shimizu T., Lin F., Sainsbury F., Thuenemann E., Lomonossoff G., et al. (2012). Identification of an E-box motif responsible for the expression of jasmonic acid-induced chitinase gene OsChia4a in rice. J. Plant Physiol. 169, 621–627. doi: 10.1016/j.jplph.2011.12.008, PMID: [DOI] [PubMed] [Google Scholar]
- Miyao A., Tanaka K., Murata K., Sawaki H., Takeda S., Abe K., et al. (2003). Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15, 1771–1780. doi: 10.1105/tpc.012559, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr T. J., Mammarella N. D., Hoff T., Woffenden B. J., Jelesko J. G., McDowell J. M. (2010). The Arabidopsis downy mildew resistance gene RPP8 is induced by pathogens and salicylic acid and is regulated by W-box cis-elements. Mol. Plant-Microbe Interact. 23, 1303–1315. doi: 10.1094/MPMI-01-10-0022, PMID: [DOI] [PubMed] [Google Scholar]
- Narusaka M., Shirasu K., Noutoshi Y., Kubo Y., Shiraishi T., Iwabuchi M., et al. (2009). RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. doi: 10.1111/j.1365-313X.2009.03949.x [DOI] [PubMed] [Google Scholar]
- Naveed Z. A., Wei X., Chen J., Mubeen H., Ali G. S. (2020). The PTI to ETI continuum in Phytophthora-plant interactions. Front. Plant Sci. 11:593905. doi: 10.3389/fpls.2020.593905, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma K., Thorgrimsen S., Malinovsky F. G., Fiil B. K., Nielsen H. B., Brodersen P., et al. (2010). Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6:e1001137. doi: 10.1371/journal.ppat.1001137, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S., Shen Y., Wang X., Zhang S., Li Y., Islam M. M., et al. (2022). A new NLR gene for resistance to tomato spotted wilt virus in tomato (Solanum lycopersicum). Theor. Appl. Genet. 135, 1493–1509. doi: 10.1007/s00122-022-04049-4, PMID: [DOI] [PubMed] [Google Scholar]
- Qiao Y., Liu L., Xiong Q., Flores C., Wong J., Shi J., et al. (2013). Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45, 330–333. doi: 10.1038/ng.2525, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Xia R., Zhai J., Hou Y., Feng L., Zhai Y., et al. (2021). Small RNAs in plant immunity and virulence of filamentous pathogens. Annu. Rev. Phytopathol. 59, 265–288. doi: 10.1146/annurev-phyto-121520-023514, PMID: [DOI] [PubMed] [Google Scholar]
- Rampino P., De Pascali M., De Caroli M., Luvisi A., De Bellis L., Piro G., et al. (2017). Td4IN2: a drought-responsive durum wheat (Triticum durum Desf.) gene coding for a resistance like protein with serine/threonine protein kinase, nucleotide binding site and leucine rich domains. Plant Physiol. Biochem. 120, 223–231. doi: 10.1016/j.plaphy.2017.10.010, PMID: [DOI] [PubMed] [Google Scholar]
- Shen Q., Zhang P., Ho T. H. (1996). Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8, 1107–1119. doi: 10.1105/tpc.8.7.1107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele J. F. C., Hughes R. K., Banfield M. J. (2019). Structural and biochemical studies of an NB-ARC domain from a plant NLR immune receptor. PLoS One 14:e0221226. doi: 10.1371/journal.pone.0221226, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Spears B. J., Kim S. H., Gassmann W. (2018). Constant vigilance: plant functions guarded by resistance proteins. Plant J. 93, 637–650. doi: 10.1111/tpj.13798, PMID: [DOI] [PubMed] [Google Scholar]
- Sun X., Lin L., Sui N. (2019). Regulation mechanism of microRNA in plant response to abiotic stress and breeding. Mol. Biol. Rep. 46, 1447–1457. doi: 10.1007/s11033-018-4511-2 [DOI] [PubMed] [Google Scholar]
- Takken F. L., Goverse A. (2012). How to build a pathogen detector: structural basis of NB-LRR function. Curr. Opin. Plant Biol. 15, 375–384. doi: 10.1016/j.pbi.2012.05.001, PMID: [DOI] [PubMed] [Google Scholar]
- Tsuchiya T., Eulgem T. (2013). An alternative polyadenylation mechanism co-opted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. 110, E3535–E3543. doi: 10.1073/pnas.1312545110, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umadevi P., Anandaraj M. (2017). Genotype specific host resistance for Phytophthora in black pepper (Piper nigrum L.). Physiol. Mol. Plant Pathol. 100, 237–241. doi: 10.1016/j.pmpp.2017.10.011 [DOI] [Google Scholar]
- Van den Berg N., Mahomed W., Olivier N. A., Swart V., Crampton B. G. (2018). Transcriptome analysis of an incompatible Persea americana-Phytophthora cinnamomi interaction reveals the involvement of SA- and JA-pathways in a successful defence response. PLoS One 13:e0205705. doi: 10.1371/journal.pone.0205705, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wersch S., Li X. (2019). Stronger when together: clustering of plant NLR disease resistance genes. Trends Plant Sci. 24, 688–699. doi: 10.1016/j.tplants.2019.05.005, PMID: [DOI] [PubMed] [Google Scholar]
- Wan H., Yuan W., Ye Q., Wang R., Ruan M., Li Z., et al. (2012). Analysis of TIR- and non-TIR-NBS-LRR disease resistance gene analogous in pepper: characterization, genetic variation, functional divergence and expression patterns. BMC Genomics 13:502. doi: 10.1186/1471-2164-13-502, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Hu T., Wang W., Hu H., Wei Q., Yan Y., et al. (2022a). Comparative transcriptome analysis reveals distinct responsive biological processes in radish genotypes contrasting for Plasmodiophora brassicae interaction. Gene 817:146170. doi: 10.1016/j.gene.2021.146170, PMID: [DOI] [PubMed] [Google Scholar]
- Wang T., Jia Z. H., Zhang J. Y., Liu M., Guo Z. R., Wang G. (2020). Identification and analysis of NBS-LRR genes in Actinidia chinensis genome. Plan. Theory 9:1350. doi: 10.3390/plants9101350, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Xu F., Ren H., Lu G., Que Y., Xu L. (2021). Genome-wide characterization of NLRs in Saccharum spontaneum L. and their responses to leaf blight in Saccharum. Agronomy 11:153. doi: 10.3390/agronomy11010153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yang C., Wu X., Wang Y., Wang B., Wu X., et al. (2022b). Genome-wide characterization of NBS-LRR family genes and expression analysis under powdery mildew stress in Lagenaria siceraria. Physiol. Mol. Plant Pathol. 118:101798. doi: 10.1016/j.pmpp.2022.101798 [DOI] [Google Scholar]
- Wang L., Zhao L., Zhang X., Zhang Q., Jia Y., Wang G., et al. (2019). Large-scale identification and functional analysis of NLR genes in blast resistance in the Tetep rice genome sequence. Proc. Natl. Acad. Sci. 116, 18479–18487. doi: 10.1073/pnas.1910229116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F. M., Zhao H., Zhang Z., Kaloshian I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendte J. M., Pikaard C. S. (2017). The RNAs of RNA-directed DNA methylation. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 140–148. doi: 10.1016/j.bbagrm.2016.08.004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Jiang Y., Liang Y., Chen L., Chen W., Cheng B. (2019). Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 137, 179–188. doi: 10.1016/j.plaphy.2019.02.010, PMID: [DOI] [PubMed] [Google Scholar]
- Wu Y., Xie L., Jiang Y., Li T. (2022). Prediction of effector proteins and their implications in pathogenicity of phytopathogenic filamentous fungi: a review. Int. J. Biol. Macromol. 206, 188–202. doi: 10.1016/j.ijbiomac.2022.02.133, PMID: [DOI] [PubMed] [Google Scholar]
- Xia S., Cheng Y. T., Huang S., Win J., Soards A., Jinn T. L., et al. (2013). Regulation of transcription of nucleotide binding-Leucine rich repeat encoding genes SNC1 and RPP4 via H3K4 trimethylation. Plant Physiol. 162, 1694–1705. doi: 10.1104/pp.113.214551, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D., Zhou K., Yang X., Yang Y., Ma Y., Wang Y. (2021). Crosstalk of DNA methylation triggered by pathogen in poplars with different resistances. Front. Microbiol. 12:750089. doi: 10.3389/fmicb.2021.750089, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S. A., Carrington J. C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138, 2145–2154. doi: 10.1104/pp.105.062943, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Liu F., Zhu S., Li X. (2018). The maize NBS-LRR gene ZmNBS25 enhances disease resistance in rice and Arabidopsis. Front. Plant Sci. 9:1033. doi: 10.3389/fpls.2018.01033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Tao X., Shen B., Horng T., Medzhitov R., Manley J. L., et al. (2000). Structural basis for signal transduction by the toll/interleukin-1 receptor domains. Nature 408, 111–115. doi: 10.1038/35040600, PMID: [DOI] [PubMed] [Google Scholar]
- Yang S., Tang F., Zhu H. (2014). Alternative splicing in plant immunity. Int. J. Mol. Sci. 15, 10424–10445. doi: 10.3390/ijms150610424, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wang Z., Hua J. (2021). A meta-analysis reveals opposite effects of biotic and abiotic stresses on transcript levels of Arabidopsis intracellular immune receptor genes. Front. Plant Sci. 12:625729. doi: 10.3389/fpls.2021.625729, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xiangsong C., Wang Z., Sun Q., Hong A., Zhang A., et al. (2019). HOS15 and HDA9 negatively regulate immunity through histone deacetylation of intracellular immune receptor NLR genes in Arabidopsis. New Phytol. 226, 507–522. doi: 10.1111/nph.16380, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Lepère G., Jay F., Wang J., Bapaume L., Wang Y., et al. (2013). Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defence. Proc. Natl. Acad. Sci. U. S. A. 110, 2389–2394. doi: 10.1073/pnas.1211757110, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Yang L., Li Z., Sun F., Li B., Guo S., et al. (2022). In situ deletions reveal regulatory components for expression of an intracellular immune receptor gene and its co-expressed genes in Arabidopsis. Plant Cell Environ. 45, 1862–1875. doi: 10.1111/pce.14293, PMID: [DOI] [PubMed] [Google Scholar]
- Zeng W., Huang H., Lin X., Zhu C., Kosami K. I., Huang C., et al. (2021). Roles of DEMETER in regulating DNA methylation in vegetative tissues and pathogen resistance. J. Integr. Plant Biol. 63, 691–706. doi: 10.1111/jipb.13037, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Jeong D. H., De Paoli E., Park S., Rosen B. D., Li Y., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defence gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553. doi: 10.1101/gad.177527.111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Fang H., Shi X., He F., Wang R., Fan J., et al. (2020). A fungal effector and a rice NLR protein have antagonistic effects on a Bowman-Birk trypsin inhibitor. Plant Biotechnol. J. 18, 2354–2363. doi: 10.1111/pbi.13400, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lang Z., Zhu J. K. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z [DOI] [PubMed] [Google Scholar]
- Zhou C., Zhang L., Duan J., Miki B., Wu K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signalling of pathogen response in Arabidopsis. Plant Cell 17, 1196–1204. doi: 10.1105/tpc.104.028514, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Liu J. H., Zhao J. H., Liu T., Chen Y. Y., Wang C. H., et al. (2022). A fungal effector suppresses the nuclear export of AGO1-miRNA complex to promote infection in plants. Proc. Natl. Acad. Sci. 119:e2114583119. doi: 10.1073/pnas.2114583119, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou B., Yang D. L., Shi Z., Dong H., Hua J. (2014). Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis. Plant Physiol. 165, 309–318. doi: 10.1104/pp.113.227801, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuluaga A. P., Vega-Arreguín J. C., Fei Z., Ponnala L., Lee S. J., Matas A. J., et al. (2016). Transcriptional dynamics of Phytophthora infestans during sequential stages of hemibiotrophic infection of tomato. Mol. Plant Pathol. 17, 29–41. doi: 10.1111/mpp.12263, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]