Abstract
The recent FDA approval of several antisense and siRNA drugs illustrates the utility of nucleic acid chemical modifications, but numerous challenges remain for generalized nucleic acid therapeutics, urging the exploration of new modification strategies. Replacing backbone phosphates with amides has shown promise for enhancing siRNA activity, specificity, and nuclease resistance; however, amide-linked RNA has not been fully explored due to lengthy and low yielding manual amide coupling procedures. We have addressed this by automating the assembly of amide-linked RNA using an Expedite 8909 nucleic acid synthesizer and optimizing solid-phase synthesis conditions to achieve 91–95% yields in just 5 min of coupling time. The optimized protocol allowed synthesis of a 21-nucleotide-long siRNA guide strand having six consecutive amide linkages at the 3′-end with an overall yield of ∼1%. Our results show that the steric hindrance caused by bulky 2′-O protecting groups and steric hindrance of the solid support are the key optimization variables for improving the amide couplings.
Introduction
Altering phosphodiester backbone linkages has led to some of the most successful examples of nucleic acid modifications, including phosphorothioate (PS) linkages and phosphorodiamidate morpholino oligomers (PMOs). PS linkages replace one of the phosphate’s non-bridging oxygens with sulfur, retaining a negative charge. Fomivirsen (vitravene), Kynamro (mipomersen), and Tegsedi (inotersen) are examples of FDA-approved antisense drugs using PS modifications. Unlike PS linkages, phosphorodiamidate linkages of PMOs have no backbone charge. The recent FDA approval of the PMO antisense drugs Viltepso (viltolarsen) and Vyondys 53 (golodirsen) to correct exon 53 skipping and the earlier approval of Exondys 51 (eteplirsen) for Duchenne muscular dystrophy patients illustrate the utility of non-ionic backbone modifications.
Our laboratory has explored amide linkages (AM1, Figure 1) as non-ionic backbone modifications for improving the properties of short interfering RNAs (siRNAs).1 AM1 internucleoside linkages were originally developed as isolated modifications in DNA in 19932 and 19943 among many other backbone modifications for antisense oligonucleotides.4 Of the several possible constitutional isomers, the AM1 linkage had been studied the most and had only minor effects on the melting temperature of DNA duplexes, even when long stretches were incorporated with no intervening phosphate linkages, such as in a fully amide-linked 15-mer.4,5 Our laboratory reported that in RNA, AM1 linkages were excellent phosphate mimics that had little impact on the structure and thermal stability of RNA duplexes when incorporated as isolated6,7 or several consecutive8 modifications. Isolated AM1 modifications were well-tolerated in siRNAs9,10 and CRISPR RNAs,11 and, when placed at certain positions, increased siRNA activity and specificity by directing guide-strand loading in the RNAi machinery12 and improved the nuclease resistance of siRNAs.13 Placing several consecutive AM1 linkages in siRNAs decreased the RNAi activity; however, detailed studies on such siRNAs were hampered by lengthy manual procedures and poor yields of solid-phase amide couplings.8,13
Figure 1.
Structures of RNA, amide-linked RNA (AM1), and monomers for the preparation of AM1.
Using 2′-OTBS-protected 5′-azido-monomers 1 (Figure 1), Robins and co-workers reported the solution-phase assembly of an amide-linked pentamer using either N,N′-dicyclohexylcarbodiimide activation or 4-nitrophenyl esters as activated intermediates, but coupling yields did not exceed 77% (including the necessary reduction of 5′-azide to amine) with coupling times as long as 32 h at 65 °C.14 Using solid-phase synthesis, Iwase and co-workers synthesized siRNAs having two consecutive amide linkages using PyAOP activation of uridine monomer 2b with average coupling yields of 82% (the coupling time was not reported).15,16 Using a 2′-OAc-protected monomer 2a, our group reported 8 h HATU-mediated solid-phase couplings with approximately 90% yield; however, monomer 2a was later abandoned in favor of 2′-OTBS-protected 2b due to excessive amide-product hydrolysis during deprotection of the 2′-OAc group.8 Using HATU or PyAOP activation, coupling yields of 2b did not exceed 80–85% using double couplings (a 4 h coupling followed by a 12 h coupling), which allowed us to synthesize an amide-linked RNA having seven consecutive amide linkages, albeit with poor overall yield.13 Collectively, these studies revealed that the yield of amide coupling was inferior to that of >95% expected in solid-phase RNA phosphoramidite synthesis, and taken together with long coupling times, it was the limiting factor for studies on amide-linked RNA.
In the present study, we explored various activating agents, coupling times, and the impacts of steric factors on automated solid-phase synthesis of amide-linked RNA. We optimized rapid and efficient automated assembly of AM1-linked RNA that proceeds in 91–95% stepwise yields for 5 min amide couplings. Using the optimized automated solid-phase protocol, we synthesized a 21-nucleotide-long siRNA guide strand having six consecutive amide linkages at the 3′-end with an overall yield of ∼1%, which was a significant improvement compared to the 0.14% yield achieved using our previously published manual procedures.13 Our results suggested that the main limitation of coupling efficiency was steric hindrance, originating from both bulky 2′-O-TBS and steric crowding on the solid support. These findings will facilitate future studies on AM1 modifications in functional RNAs, such as siRNAs and CRISPR RNAs.
Results and Discussion
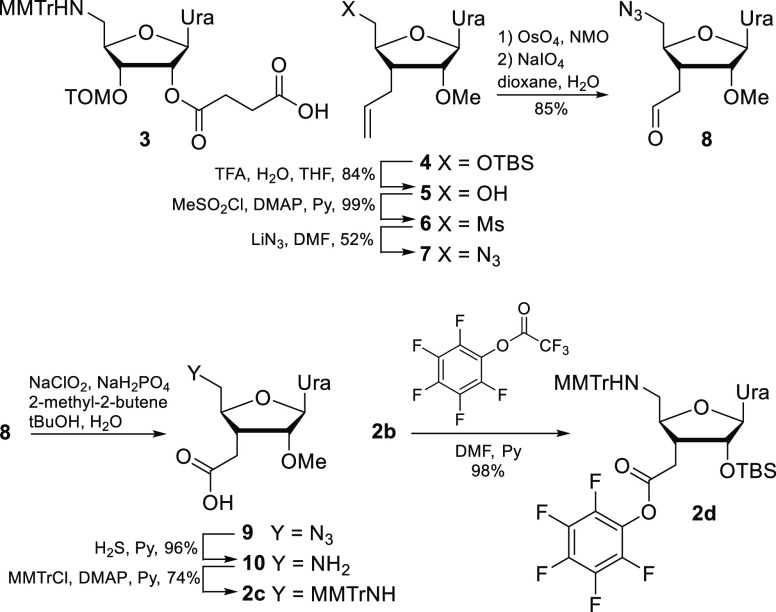
Succinate ester 3(13) and monomer 2b(17) were synthesized as previously reported. Monomer 2c was synthesized from the known intermediate 4(6) following our previously developed procedures (Scheme 1).7 Monomer 2d was prepared by converting monomer 2b to a pentafluorophenyl ester in a single quantitative step.5,18 Succinate 3 was coupled to the alkylamine functionality of CPG resin (1000 Å, Glen Research) using HATU as the activator. Functionalization conditions were varied to produce three solid supports with loading values of 49 (CPG-1), 41 (CPG-2), and 23 (CPG-3) μmol/g, respectively.
Scheme 1. Structures and Synthesis of RNA Monomers for Functionalization of Solid Support (3) and Optimization of Amide Couplings (2b and 2d).
We started with coupling time optimization using an Expedite 8909 nucleic acid synthesizer by creating custom coupling protocols based on a pre-programmed peptide nucleic acid (PNA) coupling protocol (see the Supporting Information). In each trial, we performed two couplings on uridine-functionalized support and estimated coupling yields by using trityl absorbance measurements as well as by determining product distribution by LCMS (see the Experimental Section). For direct comparison with automated couplings, manual double-couplings were performed on the same solid support using monomer 2b as previously described (Table 1, entry 1).13 Interestingly, we obtained similar yields when we repeated this trial using single 30 min manual couplings (Table 1, entry 2), prompting us to explore even shorter coupling times for automated synthesis. Remarkably, 5 min automated couplings similar to the default PNA couplings on the Expedite synthesizer gave yields slightly higher than those obtained using the longer-duration manual couplings (Table 1, entry 3). Increasing the coupling time to 23 or 47 min did not give significant improvement of yield. However, we observed a notable efficiency gain when we switched from monomer 2b to monomer 2c, indicating that the sterically bulky 2′-OTBS might be hampering the coupling efficiency, as has been suggested.19
Table 1. Optimization of Amide Coupling Time on CPG-1, 49 μmol/ga.
| coupling
yield % |
||||||||
|---|---|---|---|---|---|---|---|---|
| trityl |
LCMS |
|||||||
| entry | activator | time | monomer | 1st | 2nd | 1st | 2nd | average |
| 1b | PyAOP | 25 h | 2b | 76 | 75 | 83 | 73 | 77 |
| 2c | PyAOP | 30 min | 2b | 82 | 70 | 85 | 70 | 77 |
| 3 | HATU | 5 min | 2b | 73 | 85 | 77 | 89 | 81 |
| 4 | HATU | 23 min | 2b | 77 | 85 | 83 | 90 | 84 |
| 5 | HATU | 47 min | 2b | 79 | 85 | 84 | 90 | 85 |
| 6 | HATU | 5 min | 2c | 82 | 91 | 88 | 91 | 88 |
| 7 | HATU | 23 min | 2c | 83 | 91 | 92 | 91 | 89 |
Automated couplings (entries 3–7): pre-activation time 150 s (200 s for entries 4, 5, and 7); activator solutions: 0.12 M in DMF; base solutions: 0.2 M DIPEA with 0.3 M 2,6-lutidine in DMF; monomer solutions: 0.2 M in NMP.
Manual double coupling (3 + 22 h).
Manual single coupling.
Based on this observation, we explored if lowering the loading of solid support could improve coupling yields by minimizing steric crowding. Decreasing the loading to 41 μmol/g slightly increased the yield for the 2′-OTBS monomer 2b (Table 2 entries 1 and 2), and lowering of the support loading to 23 μmol/g gave a further notable improvement (c.f., Table 2 entries 1 and 2 with entries 3 and 4). Using either HATU or PyAOP as the activator gave similar results. Using the sterically less hindered 2′-OMe monomer 2c and the 23 μmol/g support gave the best coupling yields of ∼95% (Table 2, entries 7 and 8). Performing a double coupling (2 × 5 min) did not improve this result (Table 2, entry 9).
Table 2. Optimization of Support Loading Using CPG-2 (41 μmol/g) and CPG-3 (23 μmol/g)a.
| coupling yield % |
||||||||
|---|---|---|---|---|---|---|---|---|
| trityl |
LCMS |
|||||||
| entry | activator | support loading μmol/g | monomer | 1st | 2nd | 1st | 2nd | average |
| 1 | HATU | 41 | 2b | 84 | 84 | 89 | 90 | 87 |
| 2b | HATU | 41 | 2b | 81 | 87 | 85 | 91 | 86 |
| 3 | HATU | 23 | 2b | 90 | 90 | 92 | 93 | 91 |
| 4c | HATU | 23 | 2b | 89 | 92 | 92 | 93 | 92 |
| 5 | PyAOP | 23 | 2b | 90 | 91 | 92 | 93 | 92 |
| 6d | PyAOP | 23 | 2b | 89 | 90 | 91 | 92 | 91 |
| 7 | HATU | 23 | 2c | 95 | 94 | 96 | 93 | 95 |
| 8e | HATU | 23 | 2c | 95 | 95 | 96 | 93 | 95 |
| 9e,f | HATU | 23 | 2c | 94 | 95 | 95 | 93 | 94 |
| 10 | PyAOP | 23 | 2c | 94 | 95 | 95 | 93 | 94 |
| 11 | BTFFH | 23 | 2c | 90 | 91 | 94 | 91 | 92 |
| 12 | TCFH | 23 | 2c | 70 | 76 | 83 | 67 | 74 |
Automated 5 min couplings: pre-activation time 150 s; activator solutions: 0.18 M in DMF; base solutions: 0.2 M DIPEA with 0.3 M 2,6-lutidine in DMF; monomer solutions: 0.2 M in NMP.
Activator Solutions: 0.12 M in DMF.
Pre-activation time 300 s.
Monomer solutions: 0.4 M in NMP (double monomer concentration).
Monomer solutions: 0.2 M in DMF.
Double coupling of 2 × 5 min.
A brief screening of other coupling agents reported to enhance sterically inhibited couplings did not lead to further improvement of yields for monomer 2c. PyAOP gave results similar to those of HATU (Table 2, entry 10). Formation of acid fluoride or chloride using bis(tetramethylene)fluoroformamidinium hexafluorophosphate20 (BTFFH) or N,N,N′,N′-tetramethylchloroformamidinium hexafluorophosphate21 (TCFH), respectively, a strategy that has been reported to enhance sterically inhibited couplings due to the small size of the active species, gave less favorable yields than HATU and PyAOP (Table 2, entries 11 and 12). von Matt and co-workers reported 98–99% coupling yields using pre-formed pentafluorophenyl active esters to synthesize amide-linked DNA.5 In our hands, monomer 2d, prepared in one step from 2b, gave moderate yields that progressively increased with longer coupling times, reaching ∼80% in 47 min (Table S1), the longest coupling time we tried. We did not explore longer coupling times because monomer 2d was unstable in solution, and the initial results (Table S1) were clearly less favorable than those for HATU.
Since amide-linking RNA monomers do not have stereocenters at their alpha-position and are therefore not susceptible to epimerization, we also explored the in situ generation of highly active species, such as anhydrides and acid chlorides, using diisopropylcarbodiimide and triphosgene.22 However, this strategy gave considerably lower coupling yields (<40%) than other trials discussed above (Table S2). Thus, the 5 min couplings using HATU, presented in Table 2, gave the best yields for monomers 2b and 2c under the conditions we tested.
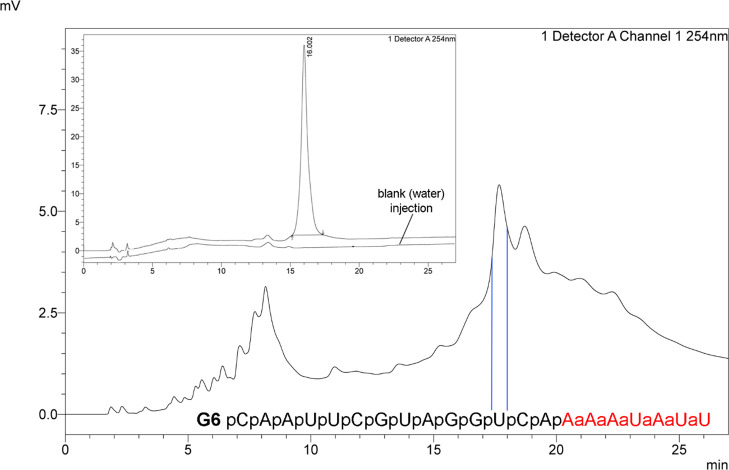
Finally, to demonstrate the utility of our optimized protocol for solid-phase synthesis of longer amide-modified RNAs, we synthesized a 21-nucleotide-long siRNA guide strand (G6, Figure 2) having six consecutive amide linkages at the 3′-end. The overall synthesis, deprotection, and purification followed our previously published procedures,13 except that we used the automated synthesis protocol (Table 2, entry 3, for details, see the Supporting Information) and CPG-3 (23 μmol/g). The major peak, isolated as identified by the blue lines shown in Figure 2, was the target RNA G6 as confirmed by MALDI-TOF, giving the expected mass of 6511 daltons (Figure S5). On a 1 μmol scale, the automated protocol proceeded with an average coupling yield of 87% (89% after the first two lower-yielding couplings, see Table S4) and gave 9.6 nmol of G6 (∼1% overall yield), which was a significant improvement over the previously reported13 80–85% average coupling yields and 1.4 nmol of G6 (0.14% overall yield).
Figure 2.
Sequence (AM1 amide linkages are highlighted in red) and RP-HPLC chromatograms of crude synthesis and purified (inset) siRNA G6. The major peak that contained G6 was isolated as shown by the blue lines. Conditions for all traces: Agilent Bio PLRP-S column (100 Å, 8 μm, 4.6 × 150 mm) at 65 °C, a gradient of acetonitrile in 50 mM triethylammonium acetate buffer, pH 7.2 (buffer A), at 1.0 mL/min. Buffer B was a 40:60 mixture of acetonitrile and buffer A. Gradient method: 0 min–10% B; 5 min–22% B; 25 min–32% B; 30 min–37% B. The slight difference in retention times of crude and purified G6 was likely caused by the presence of residual triethylammonium trihydrofluoride and other salts in the crude synthesis mixture.
Conclusions
Recent studies show that replacement of internucleoside phosphates with amides is well-tolerated in siRNAs.8−10,12,13 Amides have an underexplored potential to improve the properties of siRNAs; however, their study has been limited by low coupling efficiency and tedious and lengthy manual protocols. In the present study, we have developed protocols for rapid (5 min) and efficient (up to 91–95% yields) coupling of amide-linking RNA monomers using an Expedite 8909 nucleic acid synthesizer. The optimized protocol enabled synthesis of an siRNA guide strand having six consecutive amide linkages at the 3′-end in an overall yield of ∼1%. Our results suggest that the main limitations for amide couplings in RNA are steric hindrances caused by bulky 2′-O protecting groups and high loading of first nucleoside on solid support, which may provide avenues for further improvement. Our results will facilitate synthesis of and studies on amide linkages in RNA, which may be a promising chemical modification for the development of nucleic acid therapeutics.
Experimental Section
General Synthetic Procedures
All chemicals were obtained from commercial suppliers and were used without further purification unless stated otherwise. Acetonitrile (MeCN), pyridine, benzene, and N,N-diisopropylethylamine (DIPEA) were dried by refluxing over calcium hydride and then distilling. Dry dichloromethane and toluene were obtained using an MBRAUN solvent purification system. Dry N,N-dimethylformamide (DMF) and N-methyl-2-pyrrolidone (NMP) were purchased from MilliporeSigma. All dry reactions were carried out under an inert atmosphere of nitrogen (N2) gas in oven-dried glassware. Silicycle SiliaPlate 60 Å 10–12 μm silica gel F-254 indicator glass-backed plates were used for TLC analysis. Manual flash chromatography was performed using Silacycle SiliaFlash P60 230–400 mesh silica gels or an Isco CombiFlash Sg 100c chromatography system using disposable columns (details provided where applicable). Synthetic intermediates were characterized using 1H and 13C NMR with a Bruker AVANCE 400 or Bruker 600 MHz spectrometer and electrospray ionization mass spectrometry (MS-ESI) using a Shimadzu LCMS-2020 instrument. HRMS-ESI was performed by the Mass Spectrometry Lab at the University of Illinois Urbana-Champaign School of Chemical Sciences. 1000 Å long chain alkylamine-controlledpore glass (lcaa-CPG) consisting of Fmoc-6-aminohexanoic acid coupled to aminopropyl-controlled pore glass was purchased from Glen Research.
3′-Allyl-2′-O-methyl-3′-deoxyuridine (5)
Compound 4 (2.20 g, 5.55 mmol) was dissolved in 80 mL of tetrahydrofuran (80 mL) and chilled to 0 °C. A chilled (0 °C) mixture of TFA/water (1:1, 40 mL) was slowly added. The reaction mixture was allowed to come to room temperature and stirred at room temperature for 3 h. The mixture was neutralized with saturated aqueous NaHCO3. Water with about 10% brine was then added, bringing the total volume to 800 mL, and the crude product was extracted with 10% MeOH/dichloromethane (8 × 250 mL). The organic phase was dried over Na2SO4 and concentrated under reduced pressure. The residue was dissolved in EtOAc (45 mL). The solution was loaded on a 30 g plug of silica and purified on an Isco CombiFlash Sg 100c chromatography system using a Yamazen Universal Column Premium column (135 g, 25–40 μm particle size, 60 Å pore size); solvent A: hexanes; solvent B: EtOAc; flow rate: 40 mL/min; equilibration: 30% B; elution: 30% B for 10 min, 80% B over 45 min, 80% B for 6 min, 100% B over 14 min, 100% B for 25 min (retention time: 70 min). The fractions containing the target compound were evaporated to afford compound 5 as white crystals (1.32 g, 84% yield, Rf = 0.20 in 70% EtOAc/hexanes). 1H NMR (400 MHz, CDCl3, ppm) δ: 8.54 (1H, s), 8.11 (1H, d, J = 8.3 Hz), 5.88 (1H, s), 5.79 (1H, ddt, J = 17.1, 10.1, 7.0 Hz), 5.69 (1H, dd, J = 8.2, 2.1 Hz), 5.13 (1H, dq, J = 17.0, 1.6 Hz), 5.07 (1H, ddt, J = 10.1, 2.1, 1.1 Hz), 4.14 (1H, dd, J = 12.3, 2.1 Hz), 4.05 (1H, dt, J = 10.5, 2.3 Hz), 3.84–3.74 (2H, m), 3.56 (3H, s), 2.38 (1H, dtd, J = 13.9, 8.0, 6.8, 1.4 Hz), 2.24 (1H, ddt, J = 10.4, 8.8, 5.2 Hz), 2.10 (1H, dt, J = 13.4, 6.5 Hz), 1.90 (1H, s). 13C NMR (101 MHz, CDCl3, ppm) δ: 163.22, 149.97, 140.42, 135.55, 116.99, 101.37, 88.84, 86.05, 85.03, 77.32, 77.00, 76.68, 60.85, 58.19, 39.59, 28.68. MS-ESI (+): Mass calcd for C13H18N2O5 (M + H), 283.1; found, 283.2.
3′-Allyl-5′-O-methanesulfonyl-2′-O-methyl-3′-deoxyuridine (6)
Compound 5 (1.30 g, 4.61 mmol) was dried by co-evaporation with dry pyridine (40 mL) and dissolved in dry pyridine (30 mL). To this solution was added DMAP (28 mg, 0.23 mmol, 0.050 equiv), the solution was chilled to 0 °C, and methanesulfonyl chloride (0.90 mL, 12 mmol, 2.5 equiv) was added dropwise. The reaction mixture was allowed to come to room temperature, stirred at room temperature for 15 h, and quenched with saturated aqueous NaHCO3 (50 mL). The mixture was diluted with EtOAc (100 mL), washed with brine (3 × 50 mL), dried over Na2SO4, and concentrated under reduced pressure. The crude product (orange oil) was dissolved in 50% EtOAc/hexanes (20 mL), loaded on a 15 g plug of Celite, and purified on an Isco CombiFlash Sg 100c chromatography system using an Agela Flash Column Silica-CM column (40 g, 40–63 μm particle size, 60 Å pore size); solvent A: hexanes; solvent B: EtOAc; flow rate: 40 mL/min; equilibration: 50% B; elution: 50% B for 14 min, 100% B over 17 min (retention time: 19 min) to afford compound 6 as a white foam (1.65 g, 99% yield, Rf = 0.49 in 100% EtOAc). 1H NMR (400 MHz, CDCl3, ppm) δ: 9.68 (1H, s), 7.70 (1H, d, J = 8.2 Hz), 5.85 (1H, s), 5.83–5.68 (2H, m), 5.18–5.10 (1H, m), 5.08 (1H, dd, J = 10.1, 1.7 Hz), 4.61 (1H, dd, J = 11.9, 2.1 Hz), 4.40 (1H, dd, J = 11.8, 3.4 Hz), 4.20 (1H, ddd, J = 10.5, 3.5, 2.1 Hz), 3.81 (1H, d, J = 4.6 Hz), 3.56 (3H, s), 3.08 (3H, s), 2.45–2.32 (1H, m), 2.11 (2H, dddd, J = 21.1, 10.6, 8.0, 5.6 Hz). 13C NMR (101 MHz, CDCl3, ppm) δ: 163.55, 150.17, 139.21, 134.87, 117.52, 101.84, 89.12, 85.30, 82.09, 77.32, 77.00, 76.68, 67.67, 58.25, 40.87, 37.75, 28.42. MS-ESI (+): Mass calcd for C14H20N2O7S (M + H), 361.1; found, 361.1.
3′-Allyl-5′-azido-2′-O-methyl-3′,5′-dideoxyuridine (7)
Compound 6 (1.07 g, 2.97 mmol) was dissolved in dry DMF (10 mL) which had been purged overnight with N2 gas to remove any dimethyl amine. To this solution was added LiN3 (728 mg, 14.9 mmol, 5 equiv), resulting in a solution with a faint yellow tint. The solution was stirred for 5 h at room temperature and then for 19 h at 40 °C and was slowly cooled to room temperature. The pale yellow solution was concentrated under reduced pressure, redissolved in MeOH, adsorbed onto 30 g of Celite (traces of DMF still present), and purified using an Isco CombiFlash Sg 100c chromatography system using a Yamazen Universal Column Premium (55 g, 25–40 μm particle size, 60 Å pore size); solvent A: hexanes; solvent B: EtOAc; flow rate: 30 mL/min; equilibration: 0% B; elution: 0% B for 15 min, 100% B over 10 min, 100% B for 45 min (retention time: 45 min) to afford compound 7 as a pale yellow oil (476 mg, 52% yield, Rf = 0.48 in 70% EtOAc/hexanes). 1H NMR (400 MHz, CDCl3, ppm) δ: 9.43 (1H, s), 7.84 (1H, d, J = 8.2 Hz), 5.86 (1H, s), 5.83–5.69 (2H, m), 5.18–5.04 (2H, m), 4.09 (1H, dt, J = 10.0, 3.0 Hz), 3.90 (1H, dd, J = 13.7, 2.6 Hz), 3.75 (1H, d, J = 4.6 Hz), 3.59 (1H, dd, J = 13.7, 3.3 Hz), 3.55 (3H, s), 2.38 (1H, dddd, J = 13.1, 10.3, 6.7, 1.5 Hz), 2.13–2.00 (2H, m). 13C NMR (101 MHz, CDCl3, ppm) δ: 163.46, 150.12, 139.47, 135.15, 117.32, 101.83, 88.81, 85.84, 82.57, 77.32, 77.00, 76.68, 58.26, 51.49, 41.59, 28.54. MS-ESI (+): Mass calcd for C13H17N5O4 (M + H), 308.1; found, 308.1.
5′-Azido-3′-CH2COH-2′-O-methyl-3′,5′-dideoxyuridine (8)
Compound 7 (420 mg, 1.37 mmol) was dissolved in dioxane (17 mL). To this solution was added 4-methylmorpholine N-oxide (312 μL, 50 wt % solution in water, 1.50 mmol, 1.1 equiv) followed by OsO4 (174 μL, 4 wt % solution in water, 0.027 mmol 0.020 equiv). The reaction mixture was protected from light and stirred for 23 h at room temperature to affect conversion to a diol intermediate (not isolated, Rf = 0.26 in 10% MeOH/dichloromethane). To the solution was then added 2,6-lutidine (318 μL, 2.73 mmol, 2.0 equiv), yielding a pale-yellow solution, and sodium metaperiodate (1.169 g, 5.47 mmol, 4.0 equiv) in 5.4 mL of water, yielding a white suspension. The reaction mixture was now vigorously stirred at room temperature for 1 h, after which it was diluted with dichloromethane (40 mL). Excess Na2SO4 was added to adsorb the aqueous phase, and the crude product was collected by repeatedly washing the Na2SO4 with dichloromethane (5 × 100 mL). The dichloromethane portions were combined, dried over Na2SO4, filtered through Celite, adsorbed on 10 g of Celite, and purified using an Isco CombiFlash Sg 100c chromatography system using two SiliCycle SiliaSep flash cartridges connected end-to-end (40 g, 40–63 μm particle size, 60 Å pore size); solvent A: hexanes; solvent B: EtOAc; flow rate: 40 mL/min; equilibration: 0% B; elution: 0% B for 10 min, 8% B over 7 min, 8% B for 4 min, 10% B over 2 min, 10% B for 10 min, 22% B over 27 min, 39% B over 13 min, 100% B over 24 min, 100% B for 18 min (retention time: 96 min) to afford compound 8 as white foam (360 mg, 85% yield, Rf = 0.49 in 10% MeOH/dichloromethane). 1H NMR (400 MHz, CDCl3, ppm) δ: 10.29 (1H, s), 9.72 (1H, s), 7.69 (1H, d, J = 8.1 Hz), 5.86 (1H, s), 5.72 (1H, d, J = 8.1 Hz), 4.00 (1H, dt, J = 10.2, 3.4 Hz), 3.89 (1H, d, J = 5.2 Hz), 3.79 (1H, dd, J = 13.7, 3.0 Hz), 3.51 (1H, dd, J = 13.7, 3.6 Hz), 3.41 (3H, s), 2.88–2.75 (1H, m), 2.57–2.41 (2H, m). 13C NMR (101 MHz, CDCl3, ppm) δ: 199.51, 163.75, 150.14, 139.34, 102.02, 88.86, 85.09, 81.86, 77.32, 77.00, 76.68, 58.11, 51.07, 38.62, 36.40. MS-ESI (+): Mass calcd for C12H15N5O5 (M + H), 310.1; found, 310.2.
5′-Azido-3′-CH2COOH-2′-O-methyl-3′,5′-dideoxyuridine (9)
Compound 8 (800 mg, 2.59 mmol) was dissolved in tBuOH (50 mL) containing a few drops of water using gentle warming (∼40 °C) and stirring. Water (25 mL) was then added dropwise while stirring vigorously, giving a clear solution to which was then added 2-methyl-2-butene (7.76 mL, 2 M solution in THF, 15.5 mmol, 6.0 equiv), resulting in a pale-yellow turbid mixture. NaH2PO4 (776 mg, 6.47 mmol, 2.5 equiv) was then added, followed by the addition of NaClO2 (819 mg, 9.05 mmol, 3.5 equiv), at which point the slightly turbid reaction mixture turned bright yellow and briefly became warm. The reaction mixture was stirred at room temperature for 30 min and then quenched with saturated aqueous sodium thiosulfate (50 mL), causing the mixture to become very turbid and viscous. The crude product mixture was adsorbed on 10 g of Celite under reduced pressure (traces of tBuOH and water still present) and purified using an Isco CombiFlash Sg 100c chromatography system using a Yamazen Universal Column Premium column (55 g, 25–40 μm particle size, 60 Å pore size) solvent A: dichloromethane; solvent B: MeOH; flow rate: 50 mL/min; equilibration: 0% B; elution: 0% B for 15 min, 6% B over 15 min, 6% B for 11 min, 14% B over 11 min, 40% B over 21 min, 40% B for 13 min (retention time: 60 min) to afford a semi-pure product. The initial fractions containing compound 9 were concentrated, yielding crystals containing a non-polar impurity which was removed by washing with dichloromethane, giving 452 mg of pure 9. Later fractions contained a white precipitate and were filtered through Celite, concentrated, re-dissolved in a mixture of dichloromethane/MeOH, adsorbed on 3 g of Celite under reduced pressure, and repurified using an Isco CombiFlash Sg 100c chromatography system using an Agela Flash column Silica-CM (55 g, 40–63 μm particle size, 60 Å pore size) solvent A: dichloromethane; solvent B: MeOH; flow rate: 30 mL/min; equilibration: 2% B; elution: 2% B for 16 min, 4% B over 29 min (retention time: 30 min). The fractions containing compound 9 were concentrated, yielding crystals which after being washed with dichloromethane gave another 110 mg of pure 9. Pure product was combined to afford compound 9 as white crystals (562 mg, 67% yield, Rf = 0.13 in 10% MeOH/dichloromethane). 1H NMR (400 MHz, DMSO-d6, ppm) δ: 12.37 (1H, s), 11.39 (1H, s), 7.72 (1H, d, J = 8.1 Hz), 5.79 (1H, d, J = 1.2 Hz), 5.68 (1H, d, J = 8.1 Hz), 3.95 (2H, dd, J = 8.4, 3.8 Hz), 3.74 (1H, dd, J = 13.7, 2.9 Hz), 3.67 (1H, dd, J = 13.6, 5.7 Hz), 2.50–2.35 (3H, m). 13C NMR (101 MHz, DMSO-d6, ppm) δ: 173.61, 163.60, 150.58, 140.62, 102.28, 89.45, 85.36, 82.45, 58.25, 51.89, 40.63, 40.43, 40.22, 40.01, 39.80, 39.59, 39.46, 39.38, 29.65. HRMS-ESI (+): Mass calcd for C12H15N5O6 (M + H), 326.1; found, 326.2.
5′-Amino-3′-CH2COOH-2′-O-methyl-3′,5′-dideoxyuridine (10)
Compound 9 (530 mg, 1.62 mmol) was dissolved in pyridine (32 mL), and water (8 mL) was added dropwise. Next, H2S gas (generated by adding 25% aq. sulfuric acid dropwise to iron sulfide) was bubbled through the stirred solution for 1 h at room temperature. The reaction mixture, which had turned green and then green-brown, was then sealed with a parafilm, stirred for 15 h at room temperature, and then purged with N2 gas for 3 h, resulting in a color change to orange. The crude product mixture was adsorbed onto 10 g of Celite under reduced pressure (traces of pyridine and water still present) and purified using an Isco CombiFlash Sg 100c chromatography system using a Biotage SNAP Cartridge KP-C18-HS C18 reverse-phase column (60 g, 50 μm particle size, 90 Å pore size) solvent A: water; solvent B: MeOH; flow rate: 40 mL/min; equilibration: 2% B; elution: 2% B for 5 min, 100% B over 60 min; retention time: 6 min to afford a semi-pure product as yellow crystals with a strong H2S odor. The semi-pure product was adsorbed onto 10 g of Celite from a mixture of water and MeOH under reduced pressure (traces of water still present) and re-purified using an Isco CombiFlash Sg 100c chromatography system using a Biotage SNAP Cartridge KP-C18-HS C18 reverse-phase column (60 g, 50 μm particle size, 90 Å pore size) solvent A: water; solvent B: MeOH; flow rate: 40 mL/min; equilibration: 1% B; elution: 1% B for 35 min; retention time: 5 min to afford compound 10 as white crystals (469 mg, 96% yield, Rf = 0.30 in 18:1:1 MeOH/water/acetic acid). 1H NMR (400 MHz, DMSO-d6, ppm) δ: 8.38 (1H, d, J = 8.1 Hz), 5.71 (1H, s), 5.56 (1H, d, J = 8.0 Hz), 3.81 (1H, d, J = 5.2 Hz), 3.75 (1H, dt, J = 10.9, 3.5 Hz), 3.38 (3H, s), 2.92 (1H, dd, J = 14.0, 2.9 Hz), 2.79 (1H, dd, J = 14.0, 4.4 Hz), 2.38 (1H, dq, J = 12.3, 6.5 Hz), 2.23 (1H, dd, J = 16.1, 6.3 Hz), 1.97 (1H, dd, J = 16.0, 7.0 Hz). 13C NMR (101 MHz, DMSO-d6, ppm) δ: 176.43, 163.39, 150.27, 140.98, 100.81, 87.69, 86.83, 85.19, 57.55, 41.70, 40.15, 39.94, 39.73, 39.52, 39.31, 39.10, 38.89, 38.65, 32.51. MS-ESI (+): Mass calcd for C12H17N3O6 (M + H), 300.1; found, 300.2.
3′-CH2COOH-2′-O-methyl-5′-MMTrNH-3′,5′-dideoxyuridine (2c)
Compound 10 (400 mg, 1.34 mmol) was dried azeotropically by co-evaporation with dry pyridine (2 × 20 mL) and suspended in dry pyridine (30 mL). 4-Metoxytritylchloride (MMTrCl, 1.49 g, 3.6 equiv) was then added, causing the reaction mixture to briefly become warm, turn a brown-orange color, and begin decreasing in turbidity. DMAP (50 mg, 0.3 equiv) was then added, and the reaction mixture was stirred at room temperature for 62 h. The brown-red and slightly turbid crude product mixture was then diluted with dichloromethane (50 mL) and filtered through Celite to yield a clear orange solution. The crude product mixture was adsorbed on 10 g of Celite under reduced pressure and purified using an Isco CombiFlash Sg 100c chromatography system using an Agela Flash Silica-CS column (20 g, 40–63 μm particle size, 60 Å pore size) and a SiliCycle SiliaSep Premium Flash Cartridge (40 g, 25 μm spherical particles, 60 Å pore size) connected end-to-end; solvent A: dichloromethane with 0.4% NEt3; solvent B: 10% MeOH/dichloromethane with 0.05% NEt3; flow rate: 35 mL/min; equilibration: 0% B; elution: 0% B for 16 min, 30% B over 10 min, 30% B for 12 min, 30% to 100% B over 20 min, 100% B for 55 min; retention time: 57 min to afford compound 4 containing ∼7 equiv of NEt3. This mixture was partitioned between dichloromethane (100 mL) and water (20 mL), the dichloromethane phase was collected, and the product was further extracted from the aqueous phase with dichloromethane (3 × 20 mL). The dichloromethane portions were combined, dried over Na2SO4, and concentrated, yielding pure compound 4 as an off-white foam of a NEt3 salt (667 mg, 74% yield, Rf = 0.43 in 10% MeOH/dichloromethane). 1H NMR (400 MHz, CDCl3, ppm) δ: 9.28 (1H, s), 7.81 (1H, d, J = 8.1 Hz), 7.45 (4H, dq, J = 6.4, 1.5 Hz), 7.40–7.31 (2H, m), 7.31–7.23 (5H, m), 7.23–7.14 (2H, m), 6.87–6.77 (2H, m), 5.87 (1H, s), 5.49 (1H, d, J = 8.1 Hz), 4.12 (1H, ddd, J = 11.2, 6.2, 2.4 Hz), 3.90 (1H, d, J = 4.9 Hz), 3.78 (3H, s), 3.52 (3H, s), 2.89 (5H, s), 2.79 (1H, dd, J = 13.2, 2.5 Hz), 2.53 (1H, dd, J = 16.7, 8.7 Hz), 2.36–2.24 (1H, m), 2.20 (1H, dd, J = 13.2, 6.3 Hz), 2.10 (1H, dd, J = 16.8, 4.9 Hz). 13C NMR (101 MHz, CDCl3, ppm) δ: 176.97, 163.53, 157.93, 149.98, 146.04, 145.98, 140.10, 137.79, 129.82, 128.57, 128.53, 127.87, 126.34, 113.17, 101.31, 89.21, 86.72, 84.66, 77.32, 77.00, 76.68, 70.27, 58.34, 55.17, 45.44, 44.76, 39.72, 30.91, 8.45. HRMS-ESI (+): Mass calcd for C32H33N3O7 (M + Na), 594.2216; found, 594.2217.
3′-CH2COOPfp-5′-MMTrNH-2′-O-TBS-3′,5′-dideoxyuridine (2d)
Compound 2b(17) (115 mg containing 0.2 equiv of NEt3, 0.166 mmol) was dissolved in dry DMF (1 mL). Pyridine (269 μL, 3.33 mmol, 20 equiv) was then added followed by pentafluorophenyl trifluoroacetate (43 μL, 0.25 mmol, 1.5 equiv), and the reaction mixture was stirred at room temperature for 1 h. The mixture was diluted with EtOAc (40 mL) and washed with saturated aqueous NaHCO3 (3 × 10 mL), water (3 × 10 mL), and brine (10 mL). The organic phase was dried over Na2SO4 and concentrated to afford 2d as off-white foam that was used in coupling trials without further purification (137 mg, 98% yield, Rf = 0.66 in 50% EtOAc/hexanes). 1H NMR (400 MHz, CDCl3, ppm) δ: 7.66 (1H d, J = 8.2 Hz), 7.47 (4H, d, J = 7.5 Hz), 7.43–7.35 (2H, m), 7.30 (4H, dd, J = 8.4, 6.6 Hz), 7.27–7.18 (3H, m), 6.88–6.79 (2H, m), 5.72 (1H, s), 5.59 (1H, d, J = 8.1 Hz), 4.45 (1H, d, J = 4.1 Hz), 4.24 (1H, ddd, J = 10.7, 6.5, 2.6 Hz), 3.80 (3H, s), 3.01 (1H, dd, J = 18.2, 8.8 Hz), 2.86 (1H, dd, J = 13.3, 2.7 Hz), 2.56 (1H, dd, J = 18.1, 4.7 Hz), 2.36–2.19 (2H, m), 0.95 (9H, s), 0.27 (3H, s), 0.11 (3H, s). 13C NMR (101 MHz, CDCl3, ppm) δ: 171.40, 167.97, 164.17, 157.86, 150.22, 145.57, 145.41, 142.19, 142.15, 142.10, 142.06, 142.03, 141.94, 139.51, 139.18, 139.15, 139.10, 139.05, 139.00, 138.98, 138.97, 138.92, 138.86, 138.81, 138.79, 138.77, 137.27, 136.64, 136.61, 136.59, 136.56, 136.51, 136.46, 136.38, 136.33, 136.31, 136.28, 129.57, 128.37, 128.31, 127.94, 126.50, 124.55, 124.52, 124.50, 124.46, 124.41, 124.37, 124.33, 124.26, 124.24, 124.22, 124.20, 113.12, 101.51, 92.07, 83.51, 77.58, 77.32, 77.00, 76.68, 70.26, 60.47, 55.13, 45.24, 39.43, 28.52, 25.74, 21.08, 18.01, 14.12, −4.29, −6.12. 19F NMR (376 MHz, CDCl3) δ −152.08, −152.09, −152.10, −152.15, −152.16, −152.17, −156.85, −156.91, −156.96, −161.46, −161.47, −161.49, −161.54, −161.55, −161.59, −161.61, −161.62. HRMS-ESI (−): Mass calcd for C43H44F5N3O7Si (M – H), 836.2790; found, 836.2795.
Functionalization of Solid Support CPG-1
Lcaa-CPG resin (1000 Å, Glen Research) with a loading value of 87 μm/g (1.16 g, 101 μmol) was loaded into a sealable fritted column to which was then added a solution of TMS-Cl and dry pyridine (12 mL, 1:2 v/v), a small portion of which was allowed to drain out (until no longer turbid). The column was then sealed for 2 h at room temperature, with occasional gentle agitation. The resin was then washed with pyridine, dichloromethane, MeOH, and DMF (2 × 9 mL each). The resin was then washed with piperidine in DMF (20 mL, 20% v/v), filled with this same solution (12 mL), and sealed for 10 min at room temperature to affect Fmoc-deprotection. The resin was then washed with DMF, dichloromethane, MeCN, and diethyl ether (3 × 9 mL each) and purged with N2 gas for 1 h. A coupling cocktail was then prepared by adding diisopropylethylamine (220 μL, 1.26 mmol, 1.0 equiv) to a solution of HATU (430 mg, 1.13 mmol, 0.9 equiv) in dry DMF (6 mL). The resulting solution (which had turned yellow and then brown-orange) was added to a solution of monomer 3 (1.14 g, 1.26 mmol, 1.0 equiv) in dry DMF (6 mL). The resulting coupling cocktail (12 mL) with HATU, DIPEA, and monomer concentrations of 94, 105, and 105 mM, respectively, was agitated for 1 min and added to the resin, making sure that the dry beads were fully wetted. The column was then sealed and agitated gently for 26 h at room temperature to affect coupling, after which the solid support was washed with DMF, MeCN, and diethyl ether (3 × 9 mL each) and dried by purging with N2 gas for 3 h. Capping of uncoupled amine-sites was then performed by treating the solid support with a mixture of 2,6-lutidine/N-methylimidazole/acetic anhydride/dry MeCN (12 mL, 1:1:1:7 v/v/v/v) for 10 min at room temperature with gentle agitation. The solid support was then washed with MeCN, MeOH, dichloromethane, and diethyl ether (3 × 9 mL each) and dried by purging overnight with N2 gas. The solid support loading values were calculated using Beer–Lambert’s law, and the absorbance of the MMTr-cation (diluted in 3% trichloroacetic acid/dichloromethane w/v) obtained by cleaving the MMTr-protection 3% trichloroacetic acid in dichloromethane (25 mL, w/v) from three samples (3.7, 6.1, and 8.8 mg) of the solid support. The calculated values were averaged to give the final loading value estimate for CGP-1 of 49 μmol/g.
Functionalization of Solid Support CPG-2
The process was conducted in a similar way to that of solid support CPG-1, but with the following changes: (1) loading value of the lcaa-CPG resin used was 84 μm/g; (2) final concentrations of HATU, DIPEA, and 3 in the coupling cocktail were 18, 40, and 20 mM, respectively; (3) coupling cocktail was agitated for 5 min and did not change color beyond turning yellow before being added to the resin; and (4) coupling duration was 90 min with only occasional agitation performed. The loading value was 40 μmol/g.
Functionalization of Solid Support CPG-3
The process was conducted in a similar way to that of solid support CPG-1, but with the following changes: (1) loading value of the lcaa-CPG resin used was 84 μm/g; (2) HATU was added to the monomer solution, followed by DIPEA; (3) final concentrations of HATU, DIPEA, and 3 in the coupling cocktail were 13, 32, and 16 mM, respectively; (4) the coupling cocktail was agitated for 5 min and did not change color beyond turning yellow before being added to the resin; (5) only 2 mL of the coupling cocktail was added to the resin. This did not prove to be enough to fully wet the dry resin beads, so a small amount of additional DMF was added. To ensure homogeneity of the final product, the dry beads were tumbled thoroughly together before estimating their loading value; and (6) coupling duration was 40 min with only occasional agitation performed. The loading value was 23 μmol/g.
Automated amide couplings using an Expedite 8909 synthesizer were performed using 1 μmol of solid supports CPG-1, CPG-2, or CPG-3. The protocols used were based on the Expedite’s standard and extended to 2 μmol PNA coupling cycles, with the position 5 cycle being modified and used. A 30 s wait was introduced in the deblocking step to facilitate manual trityl cation collection. For details, see Supporting Information Tables S3 and S4.
Manual Coupling Procedures
-
1
Deblocking: Over the course of about 1–2 min, a syringe was used to push trichloroacetic acid in dichloromethane (10 mL, 3% w/v) through a synthesis column containing 1 μmol of solid support.
-
2
Coupling: The solid support was washed with dichloromethane and DMF (3 × 3 mL each). A coupling cocktail was then prepared by adding DIPEA in DMF (26 μL, 0.19 M, 5.0 μmol, 1.0 equiv) to HATU in DMF (11 μL, 0.4 M, 4.5 μmol, 0.9 equiv) and adding the now-yellow solution to monomer 2b in DMF (13 μL, 0.4 M, 5.0 μmol, 1.0 equiv). The coupling cocktail (50 μL) with HATU, DIPEA, and monomer concentrations of 90, 100, and 100 mM, respectively, was gently agitated for either 1 min (Table 1, entry 1) or a few seconds (Table 1, entry 2) or then added to the solid support using a 22G-bore needle inserted through a hole that had been pre-made in one of the column frits. The column was then sealed and agitated for either 3 h (Table 1, entry 1) or 30 min (Table 1, entry 2). For the double coupling (Table 1, entry 1), the coupling cocktail was flushed out with N2 gas, fresh coupling cocktail was added (as with the initial coupling), and the column was sealed and agitated for 22 h. The solid support was then washed with dry DMF (3 × 3 mL).
-
3
Capping: Capping of uncoupled amine-sites was performed by treating the solid support with a solution of 2,6-lutidine/N-methyl imidazole/acetic anhydride/dry MeCN (12 mL, 1:1:1:7 v/v/v/v) for 10 min at room temperature with gentle agitation. The solid support was then washed with MeCN, DMF, and dichloromethane (3 × 3 mL each) and immediately subjected to the next round of deblocking.
Coupling Yield Estimation Based on Trityl Cation Absorbency
Deblocking solutions from the starting solid support and from after each coupling were each collected and diluted to 25 mL in a volumetric flask using more 3% (w/v) trichloroacetic acid in dichloromethane. The absorbance of 4-monomethoxytrityl cation was then measured at 478 nm to estimate the coupling efficiency.
Coupling Yield Estimation Based on LCMS Analysis of Products
Synthesis products were cleaved from the solid support and desilylated at their 2′-O- positions following protocols recommended by Glen Research for 2′-O-TOM RNA synthesis. First products were cleaved from solid support by treating with 2 mL of a pre-mixed 1:1 (v/v) solution of 33 wt % ethanolic methylamine and 40 wt % aqueous methylamine for 3 h at room temperature using a syringe connected to each end of the column and pushing fresh solution along after the first hour. The crude product solution was then frozen and lyophilized. To desilylate, the residue was dissolved in DMSO (100 μL) while heating at 65 °C for 5 min to get a clear solution. Then, NEt3·3HF (125 μL) was added, and the reaction mixture was incubated at 65 °C for 3 h. The crude deprotected product mixture was cooled to room temperature, 20 μL of triethylamine was added, and the mixture was frozen and lyophilized. The resulting residue was dissolved in 100 μL of DMSO, filtered through a sub-micron membrane, and analyzed by LCMS. Peak identities were determined by their observed masses. Peak integration values were then adjusted using the relative extinction coefficients of the products at 260 nm, and the resulting values were used to calculate coupling conversions. Representative examples of LC chromatograms are shown in Figures S1 and S2.
Acknowledgments
This work was supported by the National Institutes of Health (R35 GM130207 to E.R.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c02742.
Tables of coupling optimization and customized Expedite synthesis protocols; representative LCMS traces; synthesis details for G6; copies of NMR spectra (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Kotikam V.; Rozners E. Amide-Modified RNA: Using Protein Backbone to Modulate Function of Short Interfering RNAs. Acc. Chem. Res. 2020, 53, 1782–1790. 10.1021/acs.accounts.0c00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idziak I.; Just G.; Damha M. J.; Giannaris P. A. Synthesis and Hybridization Properties of Amide-Linked Thymidine Dimers Incorporated into Oligodeoxynucleotides. Tetrahedron Lett. 1993, 34, 5417–5420. 10.1016/s0040-4039(00)73923-2. [DOI] [Google Scholar]
- De Mesmaeker A.; Waldner A.; Lebreton J.; Hoffmann P.; Fritsch V.; Wolf R. M.; Freier S. M. Amides as a new type of backbone modifications in oligonucleotides. Angew. Chem., Int. Ed. Engl. 1994, 33, 226–229. 10.1002/anie.199402261. [DOI] [Google Scholar]
- Freier S.; Altmann K. H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997, 25, 4429–4443. 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Matt P.; De Mesmaeker A.; Pieles U.; Zürcher W.; Altmann K.-H. 2’-deoxyribo-PNAs: a structurally novel class of polyamide nucleic acids with good RNA and DNA binding affinity. Tetrahedron Lett. 1999, 40, 2899–2902. 10.1016/s0040-4039(99)00389-5. [DOI] [Google Scholar]
- Rozners E.; Katkevica D.; Bizdena E.; Strömberg R. Synthesis and Properties of RNA Analogs Having Amides as Interuridyl Linkages at Selected Positions. J. Am. Chem. Soc. 2003, 125, 12125–12136. 10.1021/ja0360900. [DOI] [PubMed] [Google Scholar]
- Selvam C.; Thomas S.; Abbott J.; Kennedy S. D.; Rozners E. Amides as Excellent Mimics of Phosphate Linkages in RNA. Angew. Chem., Int. Ed. 2011, 50, 2068–2070. 10.1002/anie.201007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanui P.; Kennedy S. D.; Lunstad B. D.; Haas A.; Leake D.; Rozners E. Synthesis, biophysical studies and RNA interference activity of RNA having three consecutive amide linkages. Org. Biomol. Chem. 2014, 12, 1207–1210. 10.1039/c3ob42532k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutisya D.; Selvam C.; Lunstad B. D.; Pallan P. S.; Haas A.; Leake D.; Egli M.; Rozners E. Amides are excellent mimics of phosphate internucleoside linkages and are well tolerated in short interfering RNAs. Nucleic Acids Res. 2014, 42, 6542–6551. 10.1093/nar/gku235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutisya D.; Hardcastle T.; Cheruiyot S. K.; Pallan P. S.; Kennedy S. D.; Egli M.; Kelley M. L.; van Brabant Smith A.; Rozners E. Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs. Nucleic Acids Res. 2017, 45, 8142–8155. 10.1093/nar/gkx558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotikam V.; Gajula P. K.; Coyle L.; Rozners E. Amide Internucleoside Linkages Are Well Tolerated in Protospacer Adjacent Motif-Distal Region of CRISPR RNAs. ACS Chem. Biol. 2022, 17, 509–512. 10.1021/acschembio.1c00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle T.; Novosjolova I.; Kotikam V.; Cheruiyot S. K.; Mutisya D.; Kennedy S. D.; Egli M.; Kelley M. L.; van Brabant Smith A.; Rozners E. A Single Amide Linkage in the Passenger Strand Suppresses Its Activity and Enhances Guide Strand Targeting of siRNAs. ACS Chem. Biol. 2018, 13, 533–536. 10.1021/acschembio.7b01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotikam V.; Viel J. A.; Rozners E. Synthesis and biological activity of short interfering RNAs having several consecutive amide internucleoside linkages. Chem.—Eur. J. 2020, 26, 685–690. 10.1002/chem.201903754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins M. J.; Doboszewski B.; Nilsson B. L.; Peterson M. A. Synthesis of Amide-Linked [(3′)CH2CO-NH(5′)] Nucleoside Analogues of Small Oligonucleotides. Nucleosides, Nucleotides Nucleic Acids 2000, 19, 69–86. 10.1080/15257770008032997. [DOI] [PubMed] [Google Scholar]
- Iwase R.; Miyao H.; Toyama T.; Nishimori K. Synthesis and properties of modified siRNA having amide-linked oligoribonucleosides at their 3’ overhang regions. Nucleic Acids Symp. Ser. 2006, 50, 175–176. 10.1093/nass/nrl087. [DOI] [PubMed] [Google Scholar]
- Iwase R.; Toyama T.; Nishimori K. Solid-Phase Synthesis of Modified RNAs Containing Amide-Linked Oligoribonucleosides at Their 3’-End and Their Application to siRNA. Nucleosides, Nucleotides Nucleic Acids 2007, 26, 1451–1454. 10.1080/15257770701542389. [DOI] [PubMed] [Google Scholar]
- Kotikam V.; Rozners E. Concurrent Hydrogenation of Three Functional Groups Enables Synthesis of C3’-Homologated Nucleoside Amino Acids. Org. Lett. 2017, 19, 4122–4125. 10.1021/acs.orglett.7b01934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisfaludy L.; Schön I. Preparation and Applications of Pentafluorophenyl Esters of 9-Fluorenylmethyloxycarbonyl Amino Acids for Peptide Synthesis. Synthesis 1983, 1983, 325–327. 10.1055/s-1983-30327. [DOI] [Google Scholar]
- Somoza Á. Protecting groups for RNA synthesis: an increasing need for selective preparative methods. Chem. Soc. Rev. 2008, 37, 2668–2675. 10.1039/b809851d. [DOI] [PubMed] [Google Scholar]
- Due-Hansen M. E.; Pandey S. K.; Christiansen E.; Andersen R.; Hansen S. V. F.; Ulven T. A protocol for amide bond formation with electron deficient amines and sterically hindered substrates. Org. Biomol. Chem. 2016, 14, 430–433. 10.1039/c5ob02129d. [DOI] [PubMed] [Google Scholar]
- Tulla-Puche J.; Torres Á.; Calvo P.; Royo M.; Albericio F. N,N,N′,N′-Tetramethylchloroformamidinium Hexafluorophosphate (TCFH), a Powerful Coupling Reagent for Bioconjugation. Bioconjugate Chem. 2008, 19, 1968–1971. 10.1021/bc8002327. [DOI] [PubMed] [Google Scholar]
- Fuse S.; Mifune Y.; Takahashi T. Efficient Amide Bond Formation through a Rapid and Strong Activation of Carboxylic Acids in a Microflow Reactor. Angew. Chem., Int. Ed. 2014, 53, 851–855. 10.1002/anie.201307987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.