Abstract
Strictly regulated protein degradation by ubiquitin-proteasome system (UPS) is essential for various cellular processes whose dysregulation is linked to serious diseases including cancer. Skp2, a well characterized component of Skp2-SCF E3 ligase complex, is able to conjugate both K48-linked ubiquitin chains and K63-linked ubiquitin chains on its diverse substrates, inducing proteasome mediated proteolysis or modulating the function of tagged substrates respectively. Overexpression of Skp2 is observed in various human cancers associated with poor survival and adverse therapeutic outcomes, which in turn suggests that Skp2 engages in tumorigenic activity. To that end, the oncogenic properties of Skp2 are demonstrated by various genetic mouse models, highlighting the potential of Skp2 as a target for tackling cancer. In this article, we will describe the downstream substrates of Skp2 as well as upstream regulators for Skp2-SCF complex activity. We will further summarize the comprehensive oncogenic functions of Skp2 while describing diverse strategies and therapeutic platforms currently available for developing Skp2 inhibitors.
Keywords: ubiquitination, Skp2-SCF complex, cancer targeting
1. Introduction
1.1. Ubiquitin-Proteasome System (UPS)
Protein degradation, as a necessary component of protein turnover, is essential for cells to rapidly recycle amino acids in response to various extracellular stimuli. UPS serves as the most critical posttranslational modification machinery that governs protein degradation [1]. Functional UPS mediated proteolysis is responsible for maintaining protein homeostasis by eliminating misfolded protein, which is important for regulating various cellular processes such as apoptosis [2] and cell cycle progression [3]. Moreover, recent studies have suggested that the non-proteasomal ubiquitin signals are also significantly involved in processes such as metabolic regulation [4], DNA repair [5], autophagy [6], signal transduction [7] and immune response [8]. Importantly, dysfunction of UPS may result in serious diseases such as cancer [9], Parkinson disease [10] and Alzheimer's disease [11].
In essence, protein degradation is tightly regulated by two sequential processes: the conjugation of ubiquitin moieties to targeted substrates, followed by proteolysis of ubiquitin tagged protein in the 26S proteasome. The covalent link of ubiquitin moieties to its targets requires several critical components, including ubiquitin, ATP, the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2) and the ubiquitin-protein ligase (E3) [12]. Mechanistically, three reaction steps are involved in the E1-E2-E3 catalyzing cascade. First, the cysteine residue on E1 forms a linkage with the carboxyl group of Glycine residue on ubiquitin through a thioester bond, thus activating ubiquitin in an ATP-consuming manner. Then, activated ubiquitin is transiently transferred to Cysteine residue of E2 through a thioester linkage. Finally, E3 covalently attaches ubiquitin to the amino group of Lysine residue on targeted substrates [13] (Fig. 1). Repetitions of these sequential reactions lead to the consecutive incorporation of ubiquitin moieties onto targeted protein, which is termed polyubiquitination.
Fig. 1.
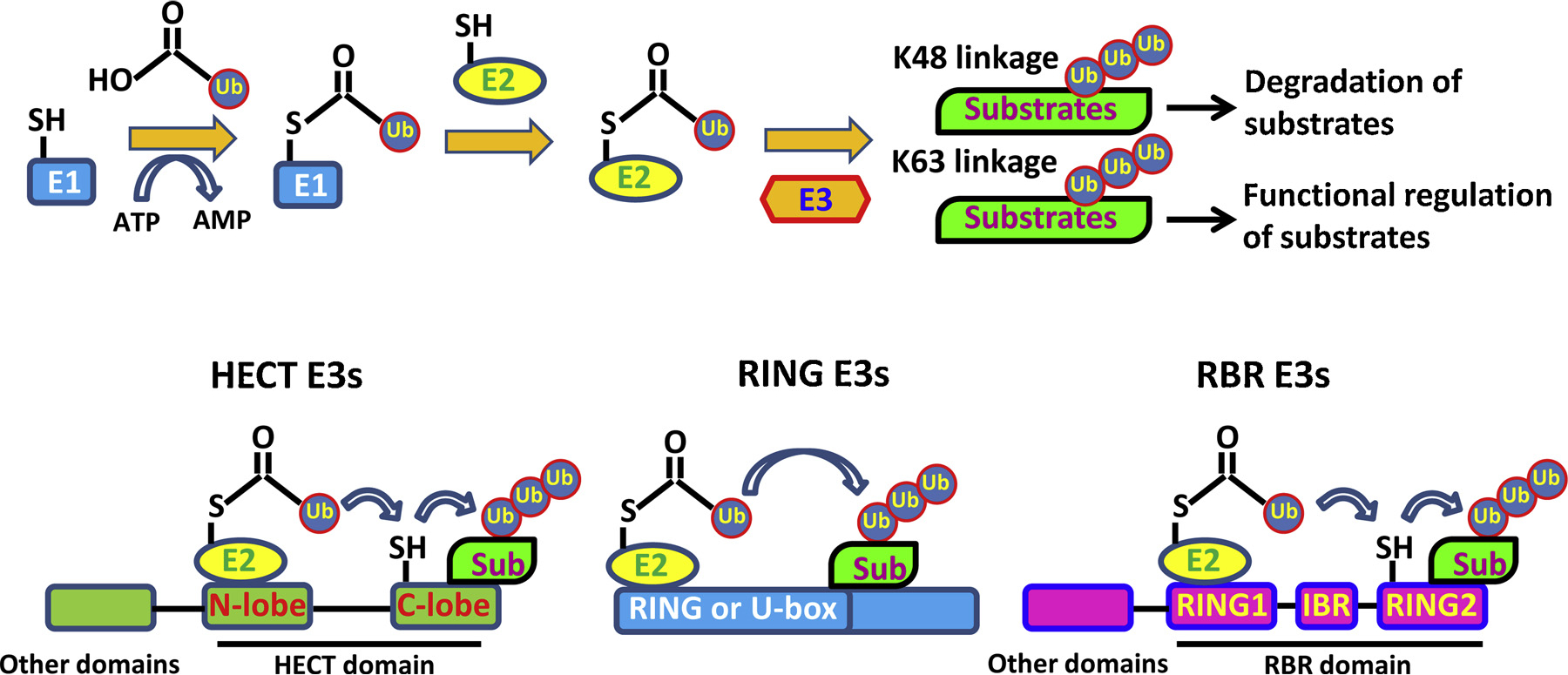
Ubiquitin conjugation reaction and classification of E3 ligases.
1.2. Ubiquitin linkage type
Ubiquitin is a 76 amino-acid protein which is highly conserved among various species and ubiquitously expressed in all types of cells. Notably, seven Lysine residues (K6, K11, K27, K29, K33, K48 and K63) or Methionine residue (M1) on ubiquitin molecules could serve as a receptor to conjugate other ubiquitin resulting in diverse linkages of polyubiquitination to exert different cellular functions [14,15]. Generally, K48-linked polyubiquitin chains often induce 26S proteasome mediated proteolysis of tagged substrates [16]. However, rather than mediating protein degradation, K63-linked polyubiquitin chains regulate protein-protein interaction, protein activity or intracellular protein trafficking involved in numerous biological processes such as immune regulation [17], DNA repair [18], cell proliferation and survival [19,20], stress response [21] and autophagy [22]. In addition to K48- and K63-linked ubiquitin chains, which are referred to as canonical ubiquitin linkage types, other atypical linkage types have begun emerging in recent years [23]. Similar to K48-linked ubiquitination, K11-linked polyubiquitin chains are also recognized by proteasomal receptor for protein degradation, which plays a critical role in cell division [24]. Meanwhile, K27-linked polyubiquitination on histone 2A (H2A), representing the major ubiquitin linkage type on chromatin upon DNA damage, does not trigger protein degradation, but is crucial for the recruitment of DNA repair machinery [25]. Smurf1 mediated K29-linked polyubiquitination on Axin results in its dissociation with Wnt co-receptors LRP5/6, thus leading to the suppression of Wnt/β-Catenin signal [26]. Interestingly, M1-linked linear ubiquitination on NEMO modified by LUBAC is essential for the activation of IKK complex, which serves as a key regulator in NF-κB signal [27]. With the advancement of innovative technologies in proteomics and structural biology, the physiological relevance of these non-canonical ubiquitin linkages will be further delineated.
1.3. Classification of E3 ligases
Among E1-E2-E3 catalyzing cascades, only two E1s (UBA1 and UBA6 [28]) have been well defined so far. However, around 40 E2s and more than 600 E3s encoded by the human genome [29] are identified. Compared with E1 and E2 enzymes, E3 ligases receive greater attention, since the high variability of E3 ligases plays a critical role in determining the specificity of substrates and targeting E3 could enhance drug efficacy by minimizing the off-target effects.
Based on the characteristic domain as well as the mechanism of how ubiquitin is transferred to substrate protein, E3 ligases can be classified into three major families: HECT (homologous to E6-associated protein C terminus) E3s, RBR (RING-in-between-RING) E3s and RING (Really Interesting New Gene) E3s [30]. A summary of the differences among these three families is illustrated in Fig. 1. In mammals, around 30 HECT E3 ligases have been identified. HECT E3 family members share a conserved HECT domain at C terminus, which is featured by a bi-lobar structure: N-terminal lobe, responsible for ubiquitin-charged E2 binding and substrate recognition and C-terminal lobe with the catalytic Cysteine for receiving and transferring ubiquitin. Two lobes are tethered by a flexible hinge, which ensures the free rotation of these two lobes during ubiquitin transfer [31]. Two sequential steps are required for HECT E3s to catalyze substrate ubiquitination: ubiquitin is first transferred from E2s to the catalytic Cysteine of C-lobe and then it is passed from E3 to the Lysine residue of the substrate whose specificity is determined by N-lobe [32]. Intramolecular interactions critically regulate the catalytic activity of HECT E3s by autoinhibition, which is released upon various stimuli. Three subfamilies of HECT E3s have been further categorized based on the characteristic domain on N-terminal: (1) Nedd4 subfamily including 9 members such as Nedd4, Itch and Smurf1, which contain two to four Tryptophan-Tryptophan (WW) domains, (2) HERC (HECT and RCC1-like domain) subfamily including 6 members as HERC1 ~6, which contain one or more chromosome condensation 1 (RCC1)-like domains (RLDs), (3) Other HECT E3s such as E6AP, which contain diverse domains [33].
In recent years, RBR E3s have been gradually identified. This enigmatic subfamily is composed of 14 members, including PARKIN, whose dysregulation is related to the pathogenesis of early-onset Parkinson's disease [10]. As indicated by its name, RBR E3s harbor two RING domains which are separated by an in-between-RING domain (IBR). In general, RING1 is responsible for ubiquitin charged E2 recruitment, while RING2 with a catalytic cysteine residue is involved in receiving ubiquitin from RING1 through a trans-thioesterification reaction [34]. Similar to HECT E3s, RBR E3s catalyze substrate ubiquitination in a two-step manner: ubiquitin is first transferred to the Cysteine residue of E3 from E2 and then passed to a substrate. Other than RBR domains, these E3s also contain additional functional domains, based on which, several subfamilies are further classified: the Ariadne family members harbor featured Ariadne domains including TRIAD1, PARC and others, while the rest contain various other domains including PARKIN, HOIP and others [35].
The remaining large numbers of E3s (more than 600) belong to the RING family. They are characterized by the existence of a zinc-binding RING domain or a U-box domain, which shares a similar RING fold albeit without zinc-binding [36]. Notably, unlike HECT E3s and RBR E3s, RING E3s catalyze a direct transfer of ubiquitin from E2 to targeted substrates [29]. Both RING domain and U-box domain can work as monomers (c-CBL, E4B), homodimers (RNF4, Prp19), or heterodimers (BRCA1-BARD1). Other RING E3s consist of multiple subunits, such as the Cullin-RING ligases (CRLs) and the APC (Anaphase promoting complex), which are well documented and characterized. Importantly, Cullin-RING E3s, representing the largest family of E3 ubiquitin ligases, account for the ubiquitination of approximately 20% of cellular proteins degraded through UPS [37]. These Cullin-RING ligases are featured by the common Cullin scaffold protein and can be further divided into several categories: Skp1/Cullin 1/F-box protein complex (SCF), Cullin 2-Elongin B/C-VHL or SOCS proteins (CRL2), Cullin 3-BTBs (CRL3s), Cullin 4-DDB1-DCAFs (CRL4), Cullin 5-ElonginB/C-SOCS proteins (CRL5) and Cullin 7/FBXW8 (CRL7) [37,38].
The SCF complex consists of Skp1 (S-phase kinase-associated protein 1), Cullin 1, Rbx1 (Ring box protein 1)/Roc1 (Regulator of Cullin 1) and diverse F-box proteins [39]. Two key functional domains are presented in each of the identified F-box proteins: the C-terminal domain responsible for substrate recognition and the F-box motif connecting to other components of SCF complex through Skp1. In the human genome, 69 F-box proteins have been documented. These F-box proteins can be further divided into three categories based on the characterized domain beyond F-box: FBXWs with WD40 repeats, including 10 members such as β-TrCP, FBXLs with Leucine-rich repeats (LRR), including 21 members such as Skp2 and FBXOs with other various domains, including the remaining 38 members such as FBXO4.
Although most F-box proteins have been documented for many years, only a few of them have been well studied, among which Skp2 is the best characterized mammalian F-box protein with various proposed substrates. Because of its pro-oncogenic properties, which are involved in the pathogenesis of multiple cancers, Skp2 has become a promising target for cancer therapy [40]. Thus, in the following discussion, we will focus on the role of Skp2 in the development of cancer as well as the regulation of Skp2 activity during tumorigenesis, with a particular emphasis on its implication in cancer therapy and prevention.
2. Downstream targets of Skp2
As an F-box protein, Skp2 exerts its function mainly through its E3 ligase activity, although the involvement of Skp2 in transcriptional regulation is also defined, which is independent of its E3 ligase activity [41]. Two categories of substrates by Skp2 have been identified based on the ubiquitin linkage types, which are summarized in Fig. 2. Skp2 is initially found capable of inducing K48-linked ubiquitination, leading to proteasome mediated proteolysis of tagged substrates including p27 [42], Foxo [43] and mH2A1 [44]. However, more recent studies also indicate the nonproteolytic function of Skp2 through conjugating K63-linked polyubiquitin chains to targeted proteins, such as Akt [45], LKB1 [46], NBS1 [5], YAP [47], Rag A [48], Bcr-Abl [49], MTH1 [50] and Aurora B [51], opening up a new direction for studying Skp2 in signaling and cancer regulation.
Fig. 2.
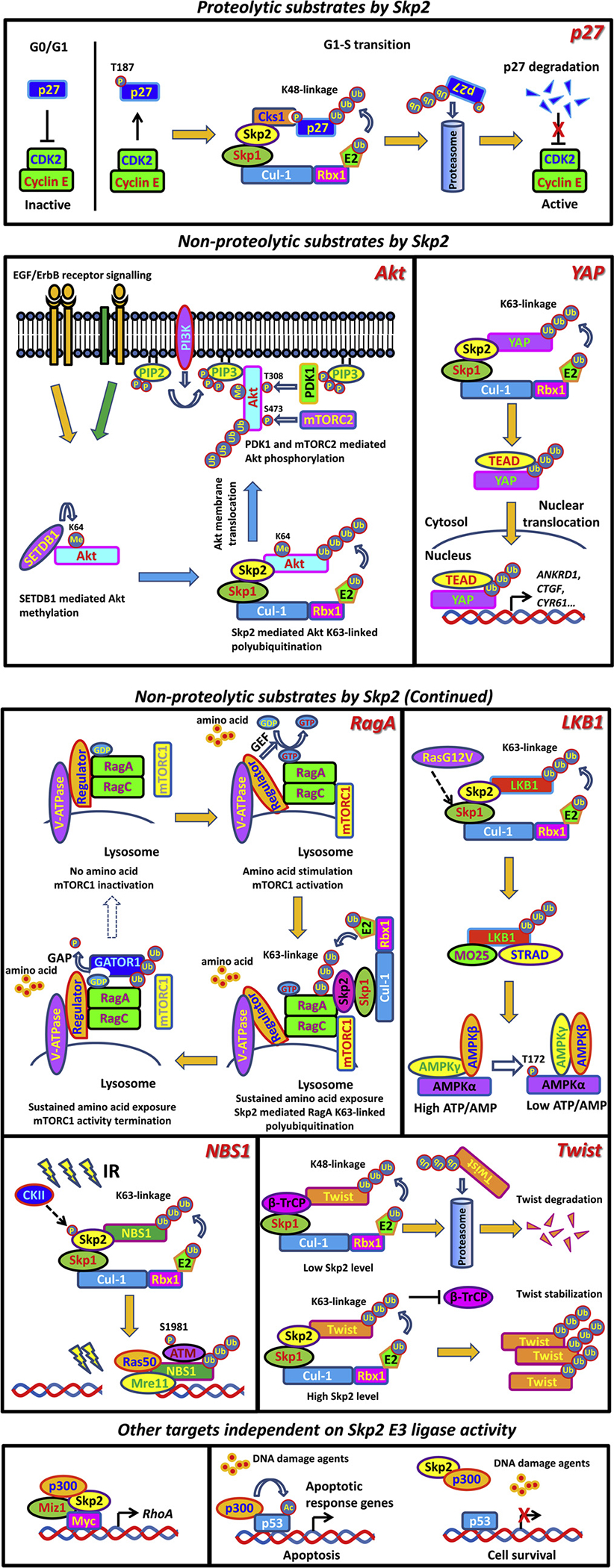
Representative downstream targets of Skp2.
2.1. Proteolytic substrates by Skp2 mediated K48-linked ubiquitination
2.1.1. p27
In the mammalian cell cycle, the transition from G0/G1 phase to S phase is tightly controlled by p27. Generally, p27 is elevated in the G0/G1 phase and undergoes rapid degradation after mitogenic stimulation, thus releasing the inhibitory status of Cyclin E/Cdk2 and allowing for cell cycle progression [52]. Reduction of p27 abolishes the proper control over the transition from G1 to S phase of the cell cycle, resulting in the uncontrolled cellular growth and proliferation, which is linked to the pathogenesis of multiple cancer types including breast, prostate, colon and lung [53–56]. Posttranslational modification, in particular by UPS mediated proteolysis, predominantly regulates the abundance of p27 during normal cell cycles as well as in pathological conditions.
Numerous studies have demonstrated that Skp2 is a bona fide E3 ligase for p27. Overexpression of Skp2 induces K48-linked polyubiquitination of p27, followed by proteasome mediated degradation, while Skp2 deficiency stabilizes p27 [57–59]. It is important to note that Cyclin E/Cdk2 induced p27 phosphorylation at T187 residue is critical for Skp2 mediated degradation of p27. T187A mutant of p27 cannot be targeted by Skp2, leading to enhanced p27 protein level and G1 arrest even upon Cyclin E overexpression [60,61]. Interestingly, the adaptor protein Cks1 is indispensable for the interaction and recognition of p27 by Skp2, as Cks1 depletion prevents Skp2 binding to p27, resulting in p27 accumulation [62–64]. Moreover, mouse embryonic fibroblasts (MEFs) derived from Cks1−/− and Skp2−/− mice consistently display impaired p27 ubiquitination and enhanced p27 stability, accompanied by compromised cell proliferation ability [57,65], further proving the Cks1-dependent recognition of p27 by Skp2. Of note, enhanced Skp2 expression level, correlated with p27 downregulation, is presented in diverse human tumor samples and predicts poor prognosis [66–69]. These studies all indicate that p27 is a major substrate for Skp2 under physiological and pathological conditions.
2.1.2. FOXO1
FOXO1, working as a transcription factor, belongs to the evolutionally conserved FOXO (Forkhead box-containing, O subfamily) family. FOXO proteins are widely believed to possess tumor suppressor properties through their transcriptional regulation of genes that are involved in apoptosis, cell cycle arrest, oxidative stress resistance and DNA repair [70]. The cellular abundance of FOXO1 is largely regulated by UPS. In many cell models, Skp2 is found to specifically bind to FOXO1 and induce polyubiquitination and degradation [43,71]. Importantly, Akt-mediated phosphorylation of FOXO1 at Serine 256 serves as a prerequisite for Skp2 mediated ubiquitination and proteasome mediated degradation [43,72]. Thus, a phosphodegron motif may be needed for Skp2 binding and recognition. This hypothesis is based on the observation that phosphorylation at Serine 256 by Akt and Theronine 187 by Cyclin E/Cdk2 is indispensable for Skp2-induced ubiquitination on FOXO1 and p27 respectively. Interestingly, ectopic expression of Skp2 abolishes apoptotic effects induced by WT FOXO1 but not degradation-resistant mutant of FOXO1, suggesting Skp2 is capable of antagonizing apoptosis partially through promoting degradation of FOXO1. Furthermore, FOXO1 accumulation is observed in malignant T cells upon Skp2 deficiency in a mouse lymphoma model, supporting the inverse correlation between Skp2 and FOXO1 in vivo.
2.1.3. CARM1
Autophagy is critical for cells to maintain homeostasis and survive in response to diverse stimuli such as starvation [73]. CARM1, as a histone arginine methyltransferase, acts as a novel regulator for starvation induced autophagy through elevating histone H3 R17 methylation, followed by enhanced transcription of autophagy and lysosomal-related genes [74,75]. Surprisingly, as a newly identified substrate of Skp2, CARM1 undergoes ubiquitination and proteasomal degradation by Skp2 under nutrition-rich conditions. After glucose deprivation, the expression level of Skp2 is compromised due to its transcriptional repression, in turn resulting in CARM1 stabilization and enhanced autophagy [74,76], suggesting the critical involvement of Skp2 in the regulation of autophagy process through modulating CARM1 ubiquitination and degradation. Downregulation of autophagy is related to many aging-related diseases such as cardiomyocyte dysfunction [77]. Notably, excessive degradation of CARM1 mediated by increased expression of Skp2 largely accounts for the dysfunction of autophagy in aged hearts [78]. Moreover, inverse correlation between Skp2 and CARM1 is also displayed in hepatocellular carcinoma tissues [79]. However, it is still unclear whether downregulation of CARM1 indeed contributes to Skp2 oncogenic function.
Apart from the substrates mentioned above, Skp2 also induces K48-linked ubiquitination and proteasome mediated degradation of many other substrates. Many of these proteins, including p21 [80,81], p57 [82], E2F-1 [83], MEF [84], p130 [85,86], Tob1 [87], Cyclin D [81], Cyclin E [88], Myc [89,90], B-Myb [91], RASSF1A [92], RhoE [93] and Cygb [94] are responsible for cell cycle regulation. In particular, apoptosis regulators such as Myc [89,90] and PDCD4 [95] are also targeted by Skp2. Moreover, other Skp2 substrates are involved in diverse cellular processes including DNA replication factors Orc1p [96] and Cdt1 [97,98], DNA repair factors RAG2 [99] and Brca2 [100], transcriptional elongation factor Cdk9 [101] and viral oncogenesis proteins HPV18 E2 [102] and HPV-E7 [103], as well as multiple signal transducers such as MKP1 involved in ERK signaling [104], UBP43 in Interferon signaling [105] and Smad4 in TGFβ signaling [106]. Additional efforts are needed to determine the physiological significance or clinical relevance of these substrates for Skp2 functions.
2.2. Nonproteolytic substrates by Skp2 mediated K63-linked ubiquitination
2.2.1. Akt
Akt signaling is a central pathway regulating metabolism, proliferation, cell survival and angiogenesis in response to various extracellular cues. The aberrant Akt network accounts for various pathogenesis such as developmental and overgrowth syndromes, cancer, cardiovascular disease, insulin resistance and type-2 diabetes, inflammatory and autoimmune disorders and neurological disorders [107]. Posttranslational modification of Akt is essential for its activation. So far, Akt has been identified as substrate of phosphorylation [108], methylation [109,110], ubiquitination [7,45], hydroxylation [111], SUMOylation [112] and acetylation [113], among which K63-linked nonproteolytic ubiquitination is well characterized for its critical role in Akt activation. TRAF6 is the first E3 ligase identified to induce K63-linked ubiquitination and activation of Akt in response to IGF-1 stimulation [7]. Interestingly, Skp2-SCF complex is another pivotal E3 ligase specific for ErbB-receptor-mediated Akt ubiquitination upon EGF treatment [45]. Of note, Skp2-mediated Akt K63-linked ubiquitination facilitates Akt membrane recruitment, allowing Akt to be phosphorylated at T308 by PDK1 and S473 by mTORC2. Depletion of Skp2 largely abolishes Akt ubiquitination, thus compromising Akt membrane translocation and activation under EGF stimulation. Moreover, in Her2-positive breast cancer patients, enhanced Skp2 expression is positively correlated with Akt activation, which predicts poor prognosis [45]. Of note, SETDB1-mediated methylation on Akt at Lysine 64 is indispensable for the recruitment of Skp2 to elicit K63-linked ubiquitination upon growth factor EGF stimulation [110]. Elimination of Akt methylation by either SETDB1 depletion or introducing methylation dead mutant of Akt (K64R) into the cells largely abrogates the interaction between Akt and Skp2, thus preventing Akt ubiquitination and activation [110]. In addition to regulating membrane recruitment, Skp2-mediated Akt K63-linked ubiquitination also contributes to Akt mitochondria localization [114]. However, it is still unclear how K63-linked ubiquitination of Akt regulates different subcellular localization of Akt.
2.2.2. YAP
The Hippo/YAP pathway is essential for organ size control and tissue homeostasis. Under high cell density conditions, YAP, as the downstream of Hippo signal, is phosphorylated by MST1/2-LATS1/2 cascade, resulting in cytoplasmic retention and β-TRCP-mediated K48-linked ubiquitination, then leading to YAP degradation [115]. Under low cell density conditions, dephosphorylated YAP will translocate to the nucleus and bind to transcription factors such as TEAD to activate proliferation and growth-related gene transcription [115]. Importantly, dysregulation of YAP localization and activity is associated with the pathological conditions such as cancer. Apart from its phosphorylation, YAP is also subjected to K63-linked ubiquitination, which is induced by Skp2. This nonproteolytic ubiquitination does not impact YAP phosphorylation, but instead facilitates the interaction between YAP and its nuclear binding partner TEAD, thus enhancing YAP’s nuclear localization, transcriptional activity and growth-promoting function. Interestingly, elimination of YAP ubiquitination by either Skp2 depletion or introducing ubiquitination dead mutant of YAP into the cells retains YAP in the cytoplasm and impairs its activity, which is independent of Hippo pathway mediated YAP phosphorylation [47]. Although this study sheds light on additional layer of regulation on YAP, whether Skp2 mediated K63-linked ubiquitination of YAP is responsible for YAP-induced organ growth and tumorigenesis in vivo warrants further investigation.
2.2.3. NBS1
The Mre11-Rad50-Nbs1 (MRN) complex is critically involved in the recruitment of ATM to DNA foci to initiate the DNA repair process in response to DNA double strand breaks (DSBs) [116]. However, how the MRN complex regulates ATM recruitment and activation remains elusive until recently. NBS1, as a key component of MRN complex, is found to be a bona fide substrate of Skp2 and undergo K63-linked polyubiquitination by Skp2 upon DSBs [5]. Ubiquitinated NBS1 creates the docking site for ATM with the ubiquitin binding property, thereby facilitating the recruitment of ATM to DNA foci for activation. Importantly, depletion of Ubc13, a major E2 responsible for K63-linked ubiquitination, recapitulates the phenotypes caused by Skp2 deficiency with impaired NBS1 ubiquitination and reduced ATM activation due to the compromised interaction between NBS1 and ATM. Expectedly, both Ubc13 and Skp2 deficiency cells display defects in homologous recombination (HR) repair upon genotoxic stress [5]. These data collectively suggest that Skp2 mediated K63-linked ubiquitination of NBS1 serves as a key event for ATM activation. Thus, Skp2 targeting may be a promising strategy to improve the efficacy of radiotherapy and chemotherapy in cancer treatment.
2.2.4. LKB1
LKB1 exerts its function through directly phosphorylating its substrates such as AMPK and is thereby responsible for various cellular events including energy metabolism, proliferation, apoptosis and cell polarity [117,118]. The maintenance of LKB1 activity relies on the integrity of the heterotrimeric complex composed of LKB1, STRAD and MO25 [119]. Recently, LKB1 is uncovered as a substrate of Skp2. K63-linked LKB1 polyubiquitination by Skp2 is essential for LKB1 activation by maintaining LKB1-STRAD-MO25 complex integrity [46,120]. Loss of Skp2 abrogates LKB1 ubiquitination, resulting in the impairment of LKB1 activity due to compromised interaction between LKB1 and MO25. Notably, Ras hyperactivation facilitates the integrity of Skp2-SCF complex, thereby resulting in elevated K63-linked polyubiquitination of LKB1 and subsequent activation of LKB1/ AMPK signaling. Further, Skp2-mediated ubiquitination of LKB1 is critically involved in the pro-survival function of Ras under metabolic stress, since ablation of Skp2 or LKB1 abolishes Ras-mediated protective effects on cell survival under glucose starvation [46,120]. Thus, targeting Skp2 or LKB1 may serve as a novel therapeutic strategy for tackling Ras-driven cancer.
2.2.5. RagA
mTOR is the key regulator coupling amino acid availability to cell growth and autophagy, whose activation is mediated through an amino acid sensing cascade involving RAGs complex, a heterodimer of GTP binding proteins RagA or RagB with RagC or RagD [121,122]. Importantly, upon amino acid stimulation, GTP charged RagA/RagB recruits mTORC1 to the lysosomal membrane for activation [123]. In contrast, the GATOR1 complex, as a negative regulator, displays GTPase activation protein (GAP) activity to switch RagA/B-bound GTP to GDP, resulting in the inactivation of mTOR signal in response to amino acid deprivation [124]. Interestingly, Skp2 E3 ligase is capable of catalyzing K63-linked ubiquitination of RagA, which enhances the recruitment of GATOR1 to RagA followed by RagA(GTP) hydrolysis, thereby providing a negative feedback loop to compromise mTORC1 lysosomal recruitment and prevent mTORC1 hyperactivation in response to sustained amino acid exposure [48]. Moreover, Skp2-mediated RagA ubiquitination and mTORC1 inhibition are responsible for autophagy activation, but cell size and cilia growth inhibition, suggesting the physiological relevance of Skp2/RagA/mTOR axis.
2.2.6. Twist
Twist transcription factor is a key regulator for epithelial-mesenchymal transition (EMT) process, which is believed to be responsible for tumor metastasis, acquired cancer stemness and drug resistance of advanced cancer [125]. In a recent report, Skp2 is found to interact with Twist and trigger K63-linked polyubiquitin chains on Twist, thus enhancing the stabilization of Twist by antagonizing β-Trcp mediated proteolysis [126]. Notably, Skp2/Twist axis facilitates the EMT process and cancer stem cell acquisition, contributing to the progression of castration-resistant prostate cancer (CRPC) [126]. Chemotherapy resistance is the key challenge for current CRPC treatment. Importantly, inactivation of Skp2 by genetic ablation or pharmacological inhibition confers CRPC cells re-sensitive to chemotherapy treatment, providing the proof of the principle that targeting the Skp2/Twist cascade is a promising strategy to combat CRPC.
As the second most abundant type of ubiquitination in cells, K63-linked polyubiquitination is generally believed to serve as a protein–-protein interaction motif allowing for inducible recruitment of interacting partners with ubiquitin-binding domain (UBD). In addition to the substrates mentioned above, Skp2 also catalyzes K63-linked ubiquitination of Bcr-Abl [49], MTH1 [50], Aurora B [51] and other substrates. However, the detail mechanism for how Skp2-mediated ubiquitinations modulate the function of these substrates and whether they are involved in the oncogenic function of Skp2 remain to be determined.
2.3. Other targets independent on Skp2 E3 ligase activity
Beyond conjugating polyubiquitin chains on its substrates, Skp2 exerts its functions, such as transcription regulation and signal pathway regulation, irrespective of its E3 ligase activity.
2.3.1. RhoA
As a well-known member of the Rho family of GTPases, RhoA is involved in various biological functions and linked to cancer metastasis [127]. However, the transcription machinery responsible for RhoA gene expression remains not well understood. Surprisingly, Skp2 is found to function as a co-transcription factor that binds to Myc and recruits Miz1 and p300 to the RhoA promoter region and this Myc-Skp2-Miz1-p300 transcriptional complex plays an essential role in RhoA transcription. As a result, deficiency of this complex abolishes RhoA expression leading to the impairment of cell migration, invasion and breast cancer metastasis. Of note, Skp2-LRR mutant, which loses the capability of forming Skp2-SCF complex and E3 ligase activity towards promoting ubiquitination of p27 due to lack the N-terminal and F-box domain, also synergizes with Myc to activate RhoA transcription as efficiently as WT Skp2 does, indicative of E3 ligase-independent role of Skp2 in RhoA transcription [41]. Apart from RhoA regulation, Skp2 is also critically involved in the transcription process of other Myc target genes such as AHCY, CAD, CDC2, ODC, Rcl and Cyclin D2. However, in these cases Skp2 E3 ligase activity is indeed required, since Skp2-LRR mutant or Skp2 ΔF mutant (lacking F box domain) fails to cooperate with Myc to induce the transcription of these genes [89,90]. While Skp2 acts as a cofactor for Myc-mediated gene transcription, it also elicits Myc ubiquitination and degradation [89,90,128]. However, the interplay between these two events under diverse cellular contexts remains a mystery. Collectively, these studies underscore the important role of Skp2 in regulating Myc-mediated gene transcription.
2.3.2. p300
p53 is a tumor suppressor that induces cell cycle arrest and apoptosis in response to genotoxic and oncogenic stress. p300 mediated acetylation of p53 is indispensable for the transcriptional activity as well as stability of p53 [129]. Notably, Skp2 is reported to form a complex with p300 through the CH1 and the CH3 domains of p300. These two domains are also critically involved in the interaction between p300 and p53. Ectopic expression of Skp2 sequesters p300 away from its interaction with p53, resulting in impaired acetylation and compromised transcriptional ability of p53 [130]. As a result, Skp2 overexpression renders cancer cells more resistant to DNA damage-induced apoptosis by suppressing the transactivation function of p53, likely accounting for the oncogenic property of Skp2 [130]. Thus, it is worthwhile to determine whether the clinical outcome of radiotherapy will be improved upon simultaneous Skp2 inhibition.
3. Regulation of Skp2 expression and its E3 ligase activity
Aberrant Skp2 expression and dysregulation of its activity are associated with many human cancers [131]. The expression level of Skp2 as well as its E3 ligase activity are tightly regulated under multiple layers including transcription, protein stability and post-translational modifications, as illustrated in Fig. 3.
Fig. 3.
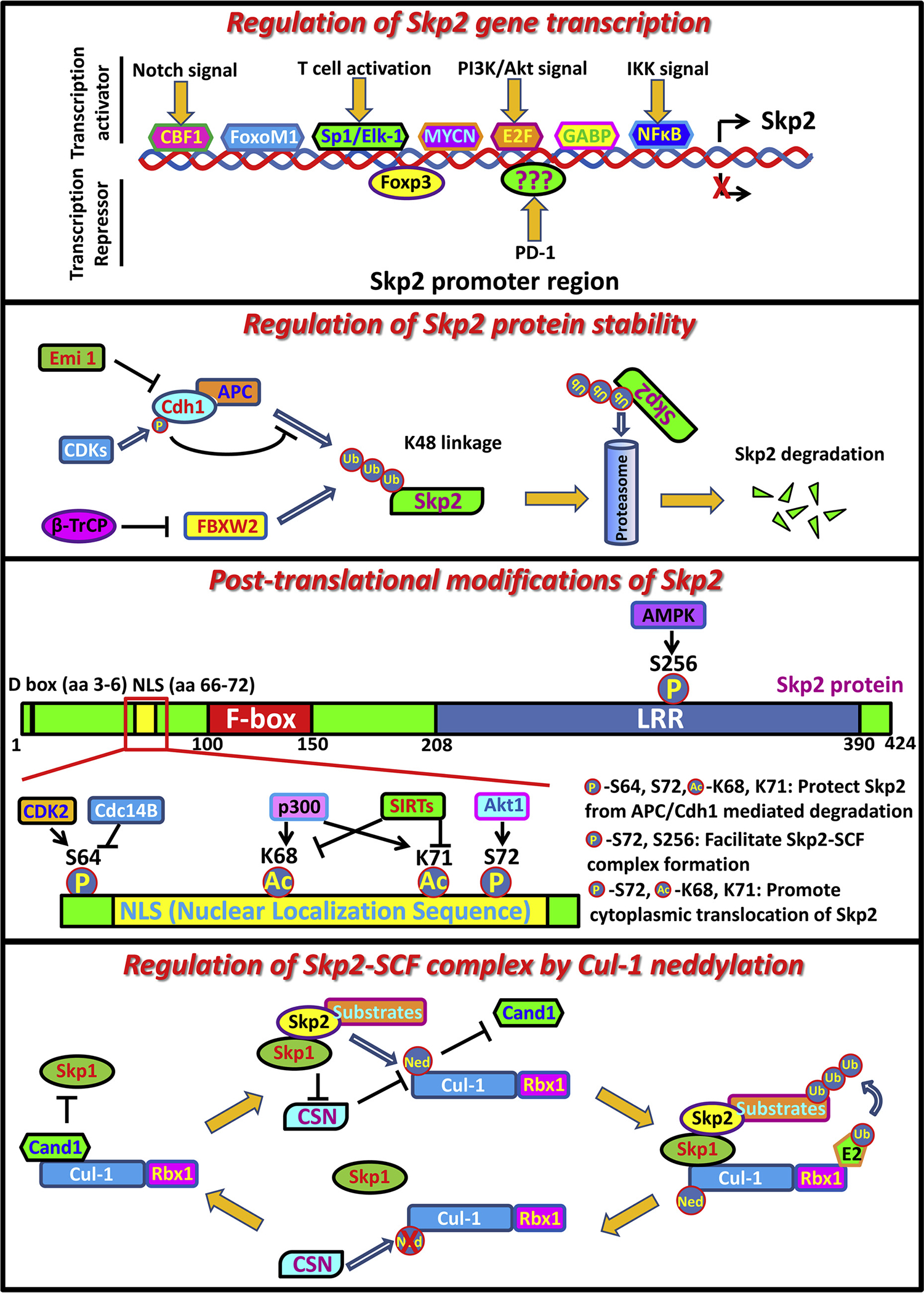
Regulation of Skp2 expression and Skp2-SCF E3 ligase activity.
3.1. Regulation of Skp2 gene transcription
Through analyzing Skp2 promoter region, several transcription factors have been identified and are responsible for Skp2 gene transcription including E2F1 [132], NF-kB [133], CBF1 [134], FoxM1 [135], GABP (GA-binding protein) [136], SP1, Elk1 [137] and MYCN [138]. Activation of the Notch pathway induces Skp2 gene transcription through CBF1, resulting in the degradation of p27 and p21 followed by premature entry into S phase [134]. Notably, similar to Skp2−/− MEFs, FoxM1 deficient cells display cell cycle arrest and polyploidy, indicating FoxM1 may induce the expression of Skp2, thereby promoting cell progression and maintaining the genomic stability [135]. Interestingly, unlike the other components of the Skp2-SCF complex such as Rbx1/Roc1, Cul1 and Skp1 which are constitutively expressed, Skp2 is almost undetectable in resting T cells and is transcriptionally induced upon CD3/CD28 stimulation. Enhancement of Skp2 gene transcription through SP1 and Elk-1 transcription factor in turn induces p27 degradation and allows primary T lymphocytes to enter into S phase for proliferation and activation [137].
Several oncogenic pathways, such as PI3K/Akt signaling, are known to regulate Skp2 gene expression, although the detailed mechanisms have not been fully characterized. Abrogation of PI3K/Akt signaling by either LY294002 treatment or Akt depletion reduces Skp2 mRNA levels [139,140], suggesting the critical involvement of PI3K/Akt signaling in the regulation of Skp2 gene transcription. E2F is suggested to be responsible for Akt mediated Skp2 expression as evidenced by the fact that inhibition of PI3K/Akt signaling impairs the capability of E2F to bind to the promoter of Skp2 [141]. Additionally, other oncogenic signals deregulated in human cancers, such as BCR-ABL and Her2/Neu, are also found to promote transcriptional activation of Skp2 through the PI3K/Akt pathway [45,142].
Although several transcription factors have been identified to facilitate Skp2 gene transcription, the transcription repressors are less understood thus far. Foxp3 (Forkhead box p3), exhibiting tumor suppressor property in breast and prostate cancer [143], is considered as the transcription repressor of Skp2. The supporting evidence from the earlier study reveals that ectopic expression of Foxp3 in mouse breast cancer cells abolishes Skp2 expression, resulting in the accumulation of p27 and polyploidy [144]. Of note, enhanced expression of Skp2 in breast cancer tissues is correlated with downregulation of Foxp3. Intriguingly, PD-1—a potent T cell inhibitor—also serves as a negative regulator for Skp2 transcription, which in turn leads to p27 stability, thus blocking cell cycle progression in the G1 phase in T lymphocytes [145]. It will be interesting to explore the role of Skp2/p27 cascades in PD-1-mediated immune checkpoints and tumor immune escape.
3.2. Regulation of Skp2 protein stability
Skp2 is originally defined as S-phase kinase-associated protein 2 whose expression level alters periodically with cell cycle progression. Surprisingly, in all phases of the cell cycle, Skp2 mRNA is stably expressed with only minor fluctuations [146]. Skp2 protein is usually undetectable in G0 and G1 phases, but gradually accumulates during G1-S transition and lasts through S phase to mitosis [147]. It is generally believed that different protein stability of Skp2 in G0/G1 phase and S phase accounts for the discrepancy of its expression. To fulfill the rapid turnover of Skp2 protein during cell cycle transition, UPS-mediated proteolysis is critically involved in governing Skp2 protein stability and degradation. APC (anaphase promoting complex)/Cdh1 complex serves as a key E3 ligase responsible for Skp2 ubiquitination and degradation in early G1 phase [148,149], thus preventing unscheduled DNA replication. Cdh1 depletion induces accumulation of Skp2 proteins and unscheduled degradation of p21 and p27, in turn promoting premature S-phase transition. Cdh1 targets Skp2 through recognizing the N-terminal D-box motif of Skp2, while Skp2 with deletion of this motif displays resistance to Cdh1-mediated ubiquitination and degradation [148,149]. In line with the periodical expression of Skp2, the activation of APC/Cdh1 is restricted to the late M and G0/G1 phases [150]. During G1-S transition, the activity of APC/Cdh1 is compromised due to the interaction between Cdh1 and its inhibitory factor Emi1 (Early mitotic inhibitor 1) as well as CDKs mediated phosphorylation on Cdh1 [150–152], leading to the stabilization of Skp2 and cell cycle progression. It is thus conceivable that enhanced expression of Skp2 in aggressive tumors may result from dysregulation of APC/Cdh1 complex. Alternatively, mutations on the N-terminal D-box motif of Skp2 may also prevent its interaction with Cdh1, rendering Skp2 refractory to ubiquitination and degradation by the APC/Cdh1 complex.
It is interesting to note that another F box protein, FBXW2, has also been reported to directly induce ubiquitination and degradation of Skp2. The expression level of FBXW2 is regulated by β-Trcp [153], as a result, β-Trcp could act through FBXW2-Skp2 axis to govern cell cycle progression, proliferation and survival of lung cancer cells [153].
3.3. Regulation of Skp2 function by post-translational modifications
A variety of post-translational modifications on Skp2 have been identified, including phosphorylation and acetylation. These modifications play important roles in regulating protein turnover, cellular localization of Skp2 and the integrity and E3 ligase activity of the Skp2-SCF complex.
3.3.1. Phossphorylation
Phosphorylation is the most abundant modification on Skp2, which occurs on multiple sites catalyzed by different kinases. CDK2 is reported to induce phosphorylation of Skp2 on Ser 64 in the G1 phase [154]. This modification does not impact on the integrity of the Skp2-SCF complex and its E3 ligase activity, but largely protects Skp2 from APC/Cdh1 mediated degradation, resulting in Skp2 stabilization and S phase entry. Since Ser 64 is located close to D-Box motif within Skp2, which is recognized by Cdh1 for its interaction, Ser 64 phosphorylation on Skp2 indeed counteracts the association of Cdh1 due to the space steric effect. Consistently, Skp2 S64A mutant displays enhanced interaction with Cdh1 and compromised protein stability [154]. On the other hand, as a phosphatase, Cdc14B specifically dephosphorylates Skp2 on Ser64 during M-G1 transition and renders it more susceptible to APC/Cdh1 mediated degradation. Of note, Cdc14B depletion significantly accelerates S phase entry, which is completely reversed by further knockdown Skp2 in Cdc14B deficient cells, suggesting Skp2 is a bona fide substrate of Cdc14B to exert its regulation on the cell cycle [154]. Collectively, CDK2 (Kinase) and Cdc14B (Phosphatase) are responsible for the periodic expression of Skp2 during the cell cycle through orchestrating Ser 64 phosphorylation of Skp2, which negatively governs Skp2 degradation by APC/Cdh1.
Akt1, but not Akt2, is another kinase reported to induce Skp2 phosphorylation on Ser 72 by two independent studies [155,156]. Mechanistically, Akt mediated Ser 72 phosphorylation of Skp2 not only stabilizes Skp2 through disrupting the association between APC/Cdh1 and Skp2 [155], but also maintains functional E3 ligase activity through facilitating SCF complex formation [156]. Interestingly, in normal cells Skp2 is mainly localized in the nucleus, while during cancer progression, Skp2 is found to translocate to the cytoplasm [67,68], although its implication in cancer development still remains elusive. Remarkably, both studies suggest that Ser 72 phosphorylation by Akt disrupts the putative NLS (Nuclear Localization Sequence) of Skp2 and promotes its cytoplasmic translocation [155,156]. In contrast, inhibition of Akt activation by LY294002 or Wortmannin restrains Skp2 in the nucleus and phosphorylation dead mutant of Skp2 (S72A) is restricted to the nuclear compartment even upon Akt activation.
Interestingly, two different mechanisms are proposed respectively by these two studies: First, Ser 72 phosphorylation creates a binding site that can be recognized by the 14-3-3 proteins. Thus Akt activation profoundly induces the recruitment of 14-3-3β to Skp2, which in turn masks the NLS of Skp2 and facilitates its relocalization from the nucleus to the cytoplasm [156]. Second, importin proteins are responsible for importing proteins containing the NLS into the nucleus. Phosphorylation of Serine or Threonine residues within the NLS could abrogate the association between the importin complex and the NLS [157]. Importin α5 and importin α7 are found to interact with Skp2, in charge of Skp2 nuclear import. Akt mediated Ser 72 phosphorylation on Skp2 impairs the interaction of Skp2 with importin α5 and importin α7, thus resulting in Skp2 cytosolic retention by preventing its nuclear import [155]. It is possible that both of these mechanisms account for Skp2 cytosolic translocation driven by Akt mediated Ser 72 phosphorylation on Skp2.
Although Skp2 cytosolic localization, correlated with Pten loss and Akt hyperactivation, is well documented in prostate and colon cancer [156], little is known about the potential function of Skp2 in the cytoplasm. Interestingly, restoration of Skp2-NES construct, which enforces cytosolic translocation of Skp2 by fusing NES (nucleus export signal) to the C-terminal of Skp2, fully rescues the impaired cell migration ability upon Skp2 knockout [156], implying the potential function of cytosolic Skp2 in promoting cell migration and cancer metastasis. In agreement with this, cytosolic Skp2 is indeed linked to high lymph node metastasis rate in colon cancers [156]. Surprisingly, the cell migration promoting effect trigged by Skp2 cytoplasm relocalization is independent of its canonical ability to promote p27 degradation, since Skp2-NES fails to form SCF complex with Cul1 or Skp1 and induce p27 degradation [156]. As a result, identification of novel substrates as well as physiological function of cytosolic Skp2 warrants further investigation.
Interestingly, a mutual regulation mode has been revealed when investigating the current knowledge of Skp2 and Akt. On one hand, Akt regulates Skp2 expression and function in many aspects including inducing Skp2 mRNA transcription, protecting Skp2 from APC/Cdh1 mediated proteolysis, facilitating Skp2-SCF complex formation and promoting Skp2 cytoplasm translocation. On the other hand, Skp2 in turn regulates Akt activation through conjugating K63-linked polyubiquitination. Taken together, the interplay between growth factor pathway (PI3K/Akt signaling) and Skp2 mediated ubiquitination pathway likely provides a novel paradigm for cancer therapy.
Recently, the stress sensor AMPK has been demonstrated to induce Skp2 phosphorylation on Ser 256 under various stimuli such as EGF, hypoxia, glucose deprivation and H2O2 treatment [158]. This phosphorylation of Skp2 by AMPK is crucial for maintaining the integrity of Skp2-SCF complex and promoting its E3 ligase activity, leading to the K63-linked polyubiquitination and activation of Akt. Moreover, AMPK/Skp2/Akt cascade is responsible for various oncogenic events including protecting cancer cells from metabolic stress or hypoxia-induced cell death, facilitating vascularization through promoting VEGF expression and enhancing EGF-stimulated glucose metabolism and cancer cell migration. These pro-tumor properties are impaired in AMPK deficient cells due to compromised Skp2 S256 phosphorylation and subsequent attenuated Akt activity, which are rescued by re-introduction of either phospho-mimic Skp2 mutant (Skp2 S256D) or constitutively active form of Akt (Myr-Akt) [158]. The expression levels of pAMPK, pSkp2 (S256) and pAkt are highly upregulated and correlated in late-stage breast cancer, predicting worse breast cancer disease-free survival and metastasis-free survival. Because Akt phosphorylation and activation is linked to acquired drug resistance in many cancer types [159], depletion of AMPK or Skp2 impairs Akt activation and renders Gefitinib-resistant H1975 cells sensitive to Gefitinib treatment [158]. Thus, AMPK/Skp2 axis serves as a novel therapeutic target for tackling acquired resistance to EGFR targeting therapy.
3.3.2. Acetylation
Acetylation of Skp2 is mediated by p300 at Lys 68 and 71 and is neutralized by the SIRT3 deacetylase [160]. These modifications stabilize Skp2 through compromising its interaction with Cdh1, thus rendering Skp2 more resistance to APC/Cdh1-mediated degradation. The removal of Skp2 acetylation, either by p300 deficiency or SIRT3 coexpression, restores the association between Cdh1 and Skp2, leading to pronounced reduction in Skp2 abundance [160]. Interestingly, acetylation of Skp2 on Lys 68 and 71 within NLS also promotes Skp2 cytosolic relocalization through disrupting the interaction of importin proteins with Skp2 [161]. However, this phenomenon is independent of previously reported Akt-mediated Skp2 phosphorylation on Ser 72, since Skp2 S72A mutant also displays cytosolic translocation upon p300 ectopic expression but not response to Akt activation [160]. Importantly, overexpression of the acetylation-mimic mutant of Skp2 significantly promotes in vitro cell migration as well as in vivo tumorigenesis [161], although more efforts are needed to investigate whether acetylation of Skp2 indeed affects in vivo tumor metastasis.
Additionally, it is reported by a recent study that p300 mediated Skp2 acetylation followed by cytosolic retention is induced upon YAP activation, resulting in polyploid cell growth as well as earlier onset of liver tumors [162]. This is mainly achieved by hyper-accumulation of p27 in nucleus, leading to mitotic arrest and subsequently cell polyploidy, as well as excessive degradation of pro-apoptotic factors FoxO1/3, resulting in polyploid cell division, genomic instability and oncogenesis, all of which rely on the sequestration of acetylated Skp2 in cytoplasm [162]. Thus targeting cytosolic Skp2 may be a promising therapeutic strategy to treat liver tumorigenesis resulting from Hippo signaling defects. Although both Akt and p300 govern Skp2 cytosolic translocation through inducing phosphorylation and acetylation of Skp2 respectively, how these two events orchestrate the oncogenic function of Skp2 still needs to be addressed.
3.3.3. Modifications on other components of Skp2-SCF
The Skp2-SCF complex is composed of multiple components other than Skp2, including Skp1, Cullin 1 (Cul-1) and Roc1/Rbx1. Cul-1 acts as a scaffold protein with the N-terminal domain for its interaction with the Skp1 adaptor protein and the C-terminal domain for the binding with Roc1/Rbx1. Nedd8 mediated neddylation of Cul-1, which is promoted by the Skp2/Skp1 complex, is proposed to stabilize Skp2-SCF complex for its functional E3 ligase activity by preventing the interaction of Cul-1 with Cand1, a negative regulator for the Skp2 SCF complex [163]. In contrast, deneddylation of Cul-1 is catalyzed by COP9/signalosome (CSN) complex through cleaving Cul-1-Nedd8 conjugates, which is inhibited by Skp1/Skp2/substrates complex [164]. Cand1, as termed from cullin-associated nedd8-dissociated protein 1, preferentially associates with unneddylated Cul-1 and forms a ternary complex with Cul-1 and Roc1 irrespective of Skp1 and Skp2, thus sequestrating the scaffold protein Cul-1 from Skp2-SCF complex and leading to the deconstruction of Skp2-SCF complex as well as impaired E3 ligase activity [165]. Surprisingly, although Cand1 competes the interaction of Cul-1 with Skp1 and Skp2, the disruption of Cand1 and Cul-1 interaction paradoxically impairs Skp2-SCF E3 ligase activity as revealed by genetic studies in Arabidopsis [166], suggesting that Cand1 is required for optimal SCF E3 ligase activation. It is likely that Cand1 steadily binds to Cul-1/Rbx1 when SCF complex dissembles. As Skp2 and its substrates gradually accumulate during G1-S transition or in response to other stimuli, enhanced Skp1/Skp2/substrate complex induces neddylation of Cul-1 through facilitating Nedd8 conjugation as well as inhibiting CSN complex, resulting in the dissociation of Cand1 from Cul-1 and functional SCF complex assembly. Once the substrates are consumed or Skp2 level decreases, deneddylated Cul-1 by the CSN complex is bound by Cand1, waiting for the next run of the SCF complex assembly. Thus Cand1 is essential for the recycling of Cul-1, whose mechanism is likely to allow various Skp1/F-box protein (Skp2)/substrate subcomplexes to be utilized by the same Cul-1/Rbx1 core in rapid response to diverse cellular events, although in vivo evidence is still needed to fully prove this notion.
4. Evidence of tumor promoting function of Skp2
In 1995, Skp2, together with Skp1, was initially defined as a critical component of cyclin A-CDK2 kinase complex and deemed to be required for the S-phase entry in many transformed cells [167]. This was the first piece of evidence of the oncogenic property of Skp2. In the last 20 years, studies from diverse genetic mouse models, as well as human clinical tumor samples, have further confirmed the pro-tumor activity of Skp2.
4.1. Oncogenic function of Skp2 evidenced by genetic mouse models
Skp2−/− mice are viable but have smaller body size. Other defects of Skp2−/− mice include reduced quiescence of hematopoietic stem cells (HSCs) [168], impaired fertility of female mice due to severely compromised ovary development [169] as well as decreased fat pad mass and adipocyte number [170]. Of note, most abnormalities of Skp2−/− mice can be rescued by the simultaneous depletion of p27 [169,170], suggesting that p27 is a major physiological target of Skp2-SCF complex and accumulation of p27 accounts for these physiological defects upon Skp2 loss. At the cellular level, cells derived from multiple organs of Skp2−/− mice, including liver, lung, kidney and testis, as well as Skp2−/− MEFs, display enlarged nuclei with polyploidy and centrosome overduplication [57]. Moreover, Skp2−/− MEFs show growth retardation and enhanced apoptosis rate accompanied with p27 and Cyclin E accumulation, which can similarly be rescued by simultaneous deletion of p27 [59,171].
To further evaluate the role of Skp2 during tumor development, Skp2−/− mice are crossed with multiple mouse tumor models. Intriguingly, genetic ablation of Skp2 impairs the incidence of sarcoma/lymphoma formation and extends the lifespan of Arf−/− mice. Identically, Skp2 depletion also renders prostate-specific Pten deletion mice more resistant to prostate tumorigenesis [172]. In both models, Skp2 deficiency leads to robust cellular senescence in response to inactivation of tumor suppressors (Arf or Pten), which depends on Atf4, p21 and p27 induction rather than the canonical p19(Arf)-p53 pathway [172], thus preventing malignant transformation. Similarly, in TRAMP (transgenic adenocarcinoma mouse prostate) mice, which develop spontaneous prostate cancer, largely mimicking the disease progression in humans, from prostatic intraepithelial neoplasia (PIN) to metastatic castration-resistant prostate cancer (CRPC), Skp2 deletion restricts prostate cancer formation as well as prevents distal metastasis, resulting in prolonged overall survival of mice [126]. Additionally, genetic inactivation of Skp2 also abrogates MMTV-Neu-induced breast cancer progression and metastasis [45], BCR-ABL-induced leukemogenesis [173] as well as Rb (retinoblastoma protein) heterozygosity elicited spontaneous tumorigenesis [174]. However, Skp2 depletion in mice has no profound effects on Myc-driven proliferation and lymphomagenesis [175], suggesting the different response of Skp2 under various oncogenic contexts. On the other hand, prostate-specific Skp2 transgenic mice display hyperplasia, dysplasia and low-grade carcinoma in the prostate gland accompanied by excessive degradation of p27 [176]. Furthermore, T-lymphoid lineage-specific Skp2 knock-in cooperates with activated Nras transgene to elicit T cell lymphomas with shorter latency and higher penetrance, leading to significant impairment of mice survival [177]. All these results from both Skp2 knockout mice and transgenic mice extensively reveal the causal linkage between Skp2 and tumorigenesis.
4.2. Prevalence of Skp2 in different types of cancer
4.2.1. Breast cancer
High expression level of Skp2 and low expression level of p27 are displayed in estrogen receptor (ER) and Her2/neu negative tumors, which predicts high tumor grade, poor survival and resistance to antiestrogen treatment [178]. In another study including 212 breast cancer tissues, Skp2 overexpression correlated with Akt activation serves as a marker for poor prognosis, short metastasis-free survival and resistance to Herceptin treatment in Her2-positive patients [45]. Of note, cytoplasmic expression of Skp2 is highly detected in patients with invasive ductal carcinomas of the breast, associated with larger tumor size, more advanced histological grade and poor disease-free and overall survival [179].
4.2.2. Prostate cancer
From a tissue microarray study including 622 radical prostatectomy specimens, elevated expressions of Skp2 are only presented in prostate cancer tissues but not in normal prostate tissues, which are positively correlated with preoperative serum prostate-specific antigen (PSA) level, tumor Gleason score as well as shorter biochemical recurrence-free survival time after radical prostatectomy [180]. Remarkably, the reverse correlation of Skp2 with its physiological target p27 and with its putative negative regulator PTEN is also noted in tumor samples [180]. High levels of Skp2 also predict prostate cancer metastasis reported in another study [66].
4.2.3. Lung cancer
Using a comparative genomic hybridization method, amplification of DNA at 5p13, the region where Skp2, CDH6 and PC4 are located, is frequently observed in both small cell lung cancers (SCLCs) and non-small cell lung cancers (NSCLCs). The DNA copy number aberration accounts for the elevated expression of Skp2 in 83% SCLC samples and 44% NSCLC samples [181,182]. For NSCLC patients with KRas mutation, enhanced Skp2 expression serves as an independent diagnosis marker for poor survival [183]. Notably, a six-gene signature based genomic copy number alteration including Skp2, Eno1, Fhit, Hyal2, p16 and 14-3-3zeta is identified for sputum-based early detection of NSCLCs with 86.7% sensitivity and 93.9% specificity in distinguishing stage I NSCLC patients from the noncancer individuals [184].
Additionally, high expressions of Skp2 are widely observed in various solid tumors or hematological malignancies including head and neck [185], liver [46,71,186], pancreas [187], esophageal [188], gastric [189], colorectal [190], thyroid [191], brain [192], ovary cancer [193], sarcoma [194,195], leukemia [196], lymphoma [197] and melanoma [198], correlated with disease progression and poor survival.
5. Oncogenic property of Skp2
As a putative oncoprotein, Skp2 governs tumor formation and progression in many aspects such as promoting cell cycle, preventing cellular senescence and apoptosis, orchestrating cancer metabolism, maintaining cancer cell stemness, facilitating tumor metastasis, inducing drug resistance and other effects, as summarized in Fig. 4.
Fig. 4.
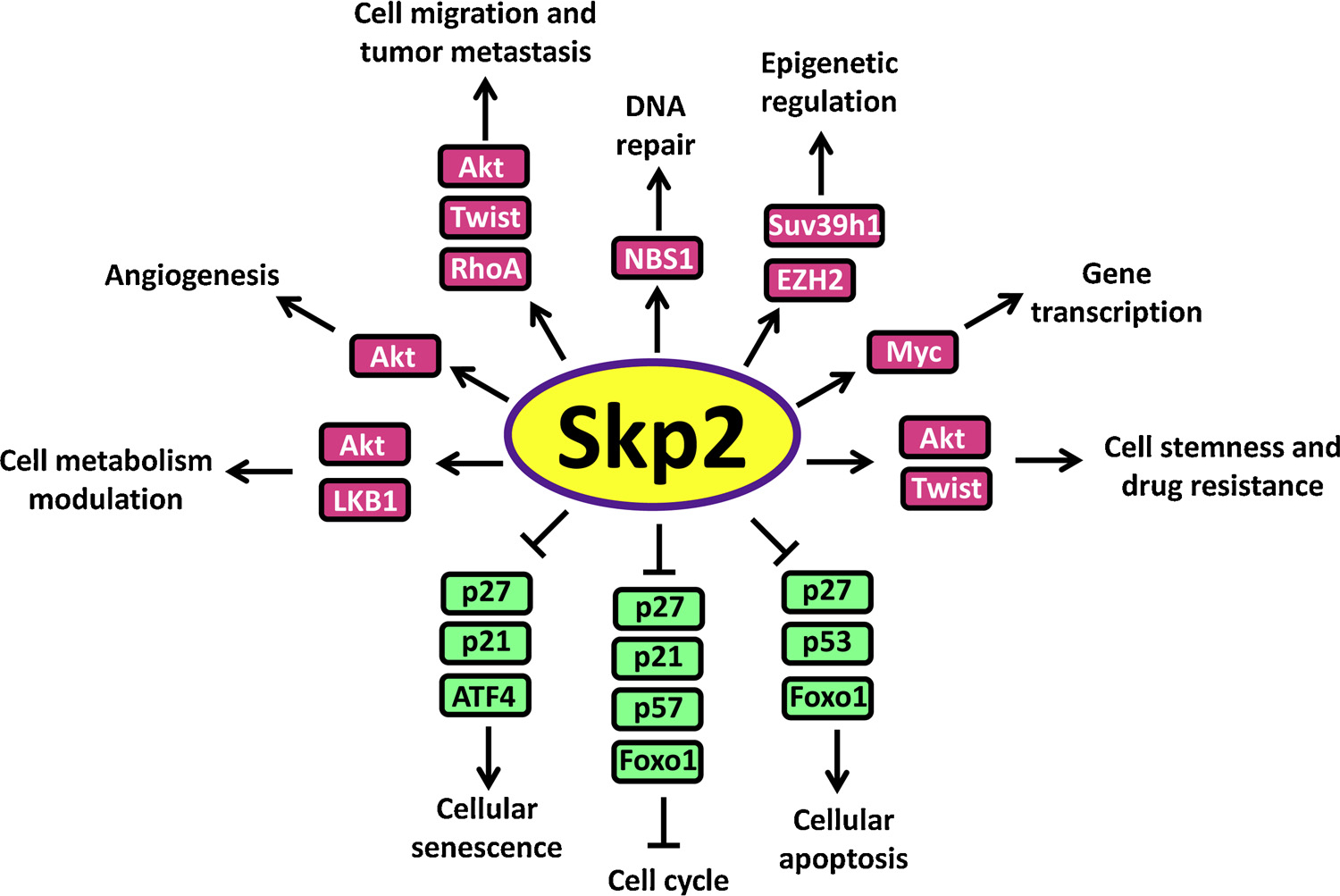
Oncogenic property of Skp2.
5.1. Cell cycle enhancement
Tight regulation of the cell cycle is essential for the prevention of cancer, which is achieved by orchestrating activation or inactivation of CDKs (Cyclin-dependent Kinases) through different phases of the cell cycle. Aberrant cell cycle progression is linked to malignant transformation. p27 and p21, as critical CKIs (CDK inhibitors), are capable of neutralizing the activation of CDKs and inducing cell cycle arrest, thus serving as critical brakes for improper cell cycle progression. In normal cells, Skp2-mediated periodical proteolysis of p27 is indispensable for releasing the inhibitory status of Cyclin A or Cyclin E/Cdk2 complex by p27, allowing for the proper G1-S transition. Genetic ablation of p27 leads to the development of cancer evidenced by multiple p27−/− mouse studies [199]. Notably, ectopic expression of Skp2 in breast epithelial cells (MCF10A) or quiescent fibroblasts (Rat-1) promotes p27 degradation with subsequent CDK activation and profoundly enhances S-phase entry of these cells which would otherwise growth-arrest, suggesting that aberrant activation of Skp2 could result in a deregulated cell cycle [58,200]. Coexpression of a degradation-resistant p27 mutant (p27 T178A) abrogates Skp2-induced CDK activation and S-phase entry [58], suggesting excessive degradation of p27 followed by hyperactivation of CDKs largely accounts for the premature S-phase entry upon Skp2 overexpression. In contrast, Skp2 deficient cells display proliferation inhibition and cell cycle arrest, which could be rescued through co-suppression of p27 [59,171,200]. Interestingly, tumor suppressor Rb protein has been shown to interact with the N-terminal of Skp2 to dissociate Skp2-p27 interaction, thereby inhibiting p27 ubiquitination and resulting in G1 arrest [201]. In a recent study, Skp2 is also reported to facilitate G2/M transition through regulating mH2A1-CDK8 cascade, thus preventing polyploidy and promoting breast cancer progression [44]. Furthermore, other substrates degraded by Skp2 such as p57 [82], p21 [80,81], p130 [85,86] and FOXO1 [43,71] also serve as negative regulators for cell cycle, suggesting Skp2 governs cell division process likely through comprehensive mechanisms.
5.2. Cellular senescence evasion
Cellular senescence serves as a critical barrier to prevent malignant transformation upon oncogenic induction or loss of tumor suppressor [202,203]. Senescence evasion is indispensable for the early-onset of carcinogenesis. Skp2 orchestrates with H-Ras(G12 V) to induce malignant transformation of primary rodent fibroblasts [204], while oncogenic Ras alone mainly provokes cell senescence rather than malignant transformation in primary cells, suggesting the critical role of Skp2 in preventing cellular senescence [205]. Consistently, although genetic ablation of Skp2 alone does not impact on cellular senescence, Skp2 inactivation combined with depletion of tumor suppressor Arf or Pten dramatically triggers senescence, leading to tumor regression [172]. These observations indicate enhanced expression of Skp2 largely protects cells from senescence, thus facilitating malignant transformation upon oncogenic activation or tumor suppressor defects. Although Arf/p53 pathway is critically involved in cellular senescence, emerging evidence suggests that cellular senescence could also be induced in an Arf/p53 independent manner. Surprisingly, the senescence response induced by combined deficiency of Skp2 and Arf or Pten does not rely on p53 activation but instead depends on the accumulation of cell cycle inhibitors p21 and p27 as well as endoplasmic reticulum stress protein ATF4 [172].
5.3. Cellular apoptosis prevention
Evasion from apoptosis is one of the hallmarks of cancer. Skp2 also regulates cell apoptosis process emphasized by the facts that cells upon Skp2 deficiency including Skp2 −/− MEFs display enhanced apoptosis [57,174,206]. p53 is a key nodule mediating cellular apoptotic response. Skp2 associates with p53 to compromise its acetylation and subsequent transcriptional activity through sequestering p300 from p53, thus preventing cellular apoptosis [130]. Of note, Skp2 deficiency potentiates DNA damage induced apoptosis only in p53 +/+ HCT116 cells, but not in p53 −/− HCT116 cells [130], indicating that Skp2 governs cellular apoptosis through its modulation on p53. p27, the notable substrate of Skp2, also functions in regulation of apoptosis in addition to cell cycle [207,208]. Indeed, activation of Skp2 mediated p27 degradation is indispensable for not only protecting aberrantly proliferating Rb1-deficient cells from apoptosis but also spontaneous tumorigenesis in Rb1+/− mice [174]. Remarkably, either depletion of Skp2 or ectopic expression of p27 T187A mutant (resistance to Skp2 mediated degradation) in both Rb1+/− mice and RB1-deficient human retinoblastoma cells induces apoptotic cell death and tumor regression [174], suggesting Rb loss renders cells dependent on Skp2 and sensitive to aberrantly expressed p27. As a result, p27 T187 phosphorylation-dependent function of Skp2 represents a promising target for the prevention and treatment of Rb-deficient tumors.
Furthermore, Skp2 is also found to promote the ubiquitination and degradation of pro-apoptotic protein Foxo1, thus abolishing its transactivation and subsequent apoptotic response in prostate cancer cell model and mouse lymphoma model [43]. Collectively, multiple mechanisms have been unraveled to account for the anti-apoptosis function of Skp2 under different cellular contexts, however, the crosstalk among these events to orchestrate Skp2 oncogenic function remains a mystery.
5.4. Cell migration and tumor metastasis promotion
Tumor metastasis accounts for the major cause of cancer related deaths. Significant correlation between Skp2 overexpression and cancer metastasis has been noted in multiple types of cancers [190,209,210], raising the possibility that Skp2 may also play an essential role in cell migration and tumor metastasis. In line with this, Skp2 deficiency dramatically compromises cell migration and in vivo metastasis, while ectopic expression of Skp2 displays reverse phenotype [126,156,211,212]. Mechanistically, Skp2 orchestrates with Myc, Miz1 and p300 to form a transcriptional complex, which is responsible for RhoA transcription [41]. Depletion of RhoA or deconstruction of the transcriptional complex restrains cell migration, invasion and breast cancer metastasis, while restoration of RhoA expression rescues cell invasion defects upon Skp2 knockdown [41], delineating the Skp2/RhoA cascade in governing cell migration and tumor metastasis. Recently, Skp2 is reported to facilitate EMT (epithelial mesenchymal transition) process through stabilizing Twist [126], which also raises the possibility that Skp2/Twist/EMT axis acts as another arm to drive cell migration and cancer metastasis, although in vivo evidence is still required to address this possibility in the future.
5.5. Cancer metabolism modulation
Tumor progression and metastasis are usually accompanied with metabolic reprogramming to confer cancer cells adaptation to various hostile environments [213]. Cancer cells prefer the usage of glycolysis for energy production and biosynthesis even under sufficient oxygen condition, which is referred as Warburg effect [214]. Akt signaling is critically involved in the regulation of glycolysis through facilitating glucose uptake and glycolytic flux [215]. Importantly, both genetic depletion and pharmacological inactivation of Skp2 restrains Akt activation and subsequent Glut1 transcription, leading to impaired glucose uptake and glycolysis, some of which can be restored upon re-introduction of active form Akt in Skp2 deficient cells [45,216]. Furthermore, Skp2 also protects cancer cells from metabolic stress and hypoxia-induced cell death in both hepatocellular carcinoma and breast cancer, which relies on Skp2 mediated ubiquitination and activation of LKB1 and Akt [46,158].
5.6. Drug resistance
Drug resistance represents the major obstacle for curing cancer [217]. The involvement of Skp2 overexpression in acquired drug resistance has been observed in multiple studies. It is noted that enhanced preoperative expression of Skp2 predicts resistance to cyclophosphamide/doxorubicin/5-fluorouracil therapy in 94% of breast cancer patients from a clinical retrospective study [218]. Similarly, in rectal cancer patients with neoadjuvant chemoradiotherapy followed by surgery, Skp2 overexpression serves as a molecular marker for poor therapeutic response and worse outcomes [219]. In line with these observations, Skp2 depletion renders Her2 positive breast cancer cells more vulnerable to Herceptin treatment, highlighting that Skp2 is an appealing therapeutic target to combine with Herceptin for tackling cancer [45]. Of note, Skp2 silencing or inactivation in the Gefitinib-resistant NSCLC cells restores their sensitivity to Gefitinib treatment, raising the potential of Skp2 targeting to overcome Gefitinib resistance in NSCLS patients [158]. CRPC remains a major clinical challenge due to its resistance toward both hormone depletion and other forms of chemotherapy. Recent studies indicate that genetic ablation or pharmacological inactivation of Skp2 re-sensitizes CRPC cells toward chemotherapies such as paclitaxel or doxorubicin [126]. Although emerging evidence has demonstrated the involvement of Skp2 in drug resistance, the underlying mechanism still remains elusive. The existence of cancer stem cells is regarded as one of the key reasons accounting for acquired drug resistance during cancer treatment [220,221]. Indeed, inactivation of Skp2-SCF complex is found to restrict cancer stem cell traits by multiple independent studies [126,216,222]. However, whether Skp2 mediated drug resistance relies on its regulation on cancer stem cell population warrants further investigation.
Additionally, Skp2 also serves to govern angiogenesis [158], DNA damage response and DNA repair [5], gene transcription [89,90] and epigenetic regulation [223,224]—all contributing to its oncogenic activity. Thus, Skp2-SCF complex acts as an attractive target for combating cancer.
6. Targeting Skp2 for cancer therapy
Giving the critical involvement in governing cancer progression, Skp2 has emerged as an appealing pharmacological target for both cancer prevention and cancer treatment. In the last several decades, multiple strategies have been proposed to limit either Skp2 expression or Skp2-SCF complex activity for cancer therapy, as summarized in Fig. 5.
Fig. 5.
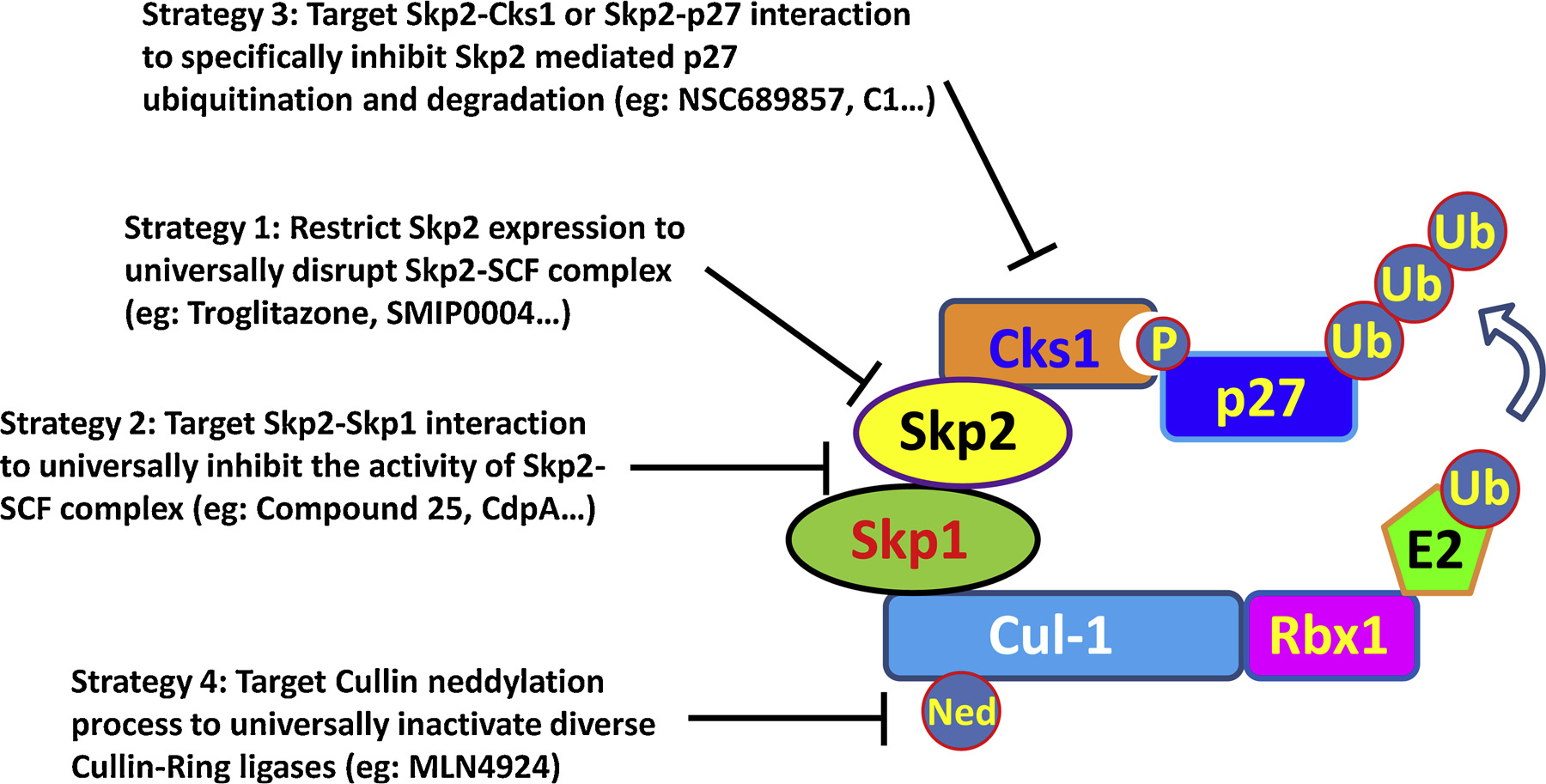
Strategies for developing Skp2 inhibitors.
6.1. Targeting Skp2 expression
PPARγ is a critical negative regulator for Skp2 expression, evidenced by the inverse correlation of PPARγ and Skp2 in human breast cancer specimen [225]. Impaired Skp2 expression level is displayed upon PPARγ ectopic expression, resulting in compromised cell proliferation as well as spontaneous apoptosis [225]. Troglitazone, as a selective PPARγ agonist, is widely used for diabetes and inflammation treatment. Interestingly, Skp2 expression restriction accompanied with p27 induction is observed in cells with Troglitazone treatment, leading to cell cycle arrest in both hepatocellular carcinoma and cervical cancer cell models [226–228]. These observations highlight the unexpected anti-neoplasm function of Troglitazone through targeting Skp2. Of note, using a high throughput platform, a total of 7368 chemical compounds are screened to identify novel Skp2 inhibitors by monitoring p27 level. Compound SMIP0004 has been unraveled to restrain Skp2 expression, thus protecting p27 from degradation followed by G1 arrest and apoptosis in prostate cancer model [229]. Additionally, other chemical compounds such as simvastatin [230], compound-7 g [231], anti-malarial compound quinacrine [232] and caffeic acid phenethyl ester [233] are also found to compromise Skp2 expression and display anti-tumor function, although the detail mechanism involved remains elusive.
Other than these chemical compounds, multiple nature compounds have also been identified to restrain Skp2 expression with the decades` efforts. Several phytochemicals including Sulforaphane, Pentagalloylglucose, Curcumin, Lycopene and Quercetin are reported to downregulate Skp2 expression in cancer cells, leading to p27 and Foxo1 accumulation and subsequent cell cycle arrest [234–236]. Interestingly, both Gallic acid and EGCG (Epigallocatechin-3-gallate), two essential extracts from tea leaf, inhibit Skp2 expression and display anti-tumor effects [237,238]. Similarly, in prostate cancer model, Silibinin and Vitamin D3 treatments induce significant reduction of Skp2 expression accompanied with p27 stabilization and cell proliferation inhibition [239,240]. All-trans retinoic acid (ATRA), an active metabolite of Vitamin A, is the first choice drug in the treatment of acute promyelocytic leukemia [241]. Surprisingly, ATRA promotes ubiquitination and degradation of Skp2, resulting in G1 arrest and growth inhibition. Forced expression of Skp2 confers cells resistance to ATRA by preventing p27 accumulation [242]. Given the relatively low toxicity of these natural compounds, it is appealing to combine these agents with conventional chemotherapy to achieve safe treatment outcome.
6.2. Targeting Skp2-SCF complex
Although multiple chemical and natural compounds negatively regulate Skp2 function, the low specificity and unknown side effects prevent these compounds to be further investigated in clinical practice. In recent years, more specific inhibitors for Skp2 have been developed based the structure of Skp2-SCF complex. One strategy is to deconstruct Skp2-SCF complex by abrogating Skp2-Skp1 interaction. Through high-throughput in silico screening of chemical libraries containing 120,000 compounds, Compound 25 (also known as SZL-P1–41) is identified to physically interact with F-box domain of Skp2, thus preventing Skp1 association and Skp2 SCF complex formation [216,243]. Importantly, this inhibitor specifically compromises the E3 ligase activity of Skp2-SCF complex, but not other F-box SCF complexes such as Fbw7 and β-Trcp. Strikingly, compound 25 phenocopies the effects upon Skp2 depletion and displays potent anti-tumor activities through Skp2 targeting by abolishing Akt-mediated glycolysis and cell survival as well as enhancing cellular senescence [216]. Compound 25 thereby appears to be an appealing drug for cancer therapy, although more humanized cancer models such as PDX (Patient derived xenograft) or organoid are needed to further justify the efficacy of compound 25 for human cancer.
In another high-throughput screening using an in vitro reconstituted ubiquitination system, compound A (CdpA) is found to neutralize Skp2-mediated p27 ubiquitination [244], highlighting the potential inhibitory effect of CdpA on Skp2 E3 ligase activity. Interestingly, Skp2 is not presented in the Skp1 immunoprecipitation portion when treated with CdpA, suggesting this inhibitor is likely to diminish Skp2-Skp1 association to exclude Skp2 from SCF complex, although more rigorous evidence is needed to further support this notion. Treatment of CdpA induces G1 arrest and cell death. Moreover, CdpA is able to restore the chemotherapy sensitivity of myeloma cells and active against both the myeloma and leukemia cells [244].
6.3. Targeting Skp2-Cks1 interaction
p27 degradation is the most critical event linking to Skp2 oncogenic activity. Cks1 is indispensable for Skp2 to recognize T187 phosphorylated p27 and subsequent ubiquitination. Cks1 −/− cells display proliferation inhibition due to elevated p27 [65]. Targeting the interaction between Skp2 and Cks1 thereby represents an appealing strategy to restore p27 level by abolishing Skp2-mediated p27 ubiquitination and degradation in cancer cells. A high-throughput screening (HTS) assay based on PerkinElmer AlphaScreen technology yields two compounds named NSC689857 and NSC681152 possessing inhibitory effects on p27 ubiquitination through interrupting Skp2 and Cks1 interaction [245]. Similarly, from HTS conducted by different groups, several other compounds such as 22d [246], Skp2E3LIs (C2, C5, C20) [247], linichlorin A and gentian violet [248] are also identified as potential inhibitors for preventing Skp2 from binding with Cks1, leading to p27 accumulation and cell growth retardation.
Another strategy used for developing Skp2 inhibitors is to disrupt the interaction between Skp2 and its substrate, such as p27. Based on the published Skp2-Cks1-p27 crystal structure, Skp2 and Cks1 jointly form a pocket flanked by residues Skp2-R294, Skp2-Y346, Cks1-R44 and Cks1-Q52, responsible for p27 binding and/or ubiquitination [63]. Inhibitors that fit into the molecular surface pocket at the Skp2-Cks1 interface are screened by an in silico Virtual Ligand Screening from 315,000 diverse compounds [249]. Four compounds (C1, C2, C20, or C16) have been identified to specifically disrupt Skp2-p27 interaction, resulting in p27 induction and cell cycle arrest in multiple cancer cell models [249].
6.4. Targeting other components of SCF complex
Cul-1 functions as a scaffold protein responsible for the integrity of Skp2-SCF complex. Similar as Skp2, Cul-1 overexpression enhances cell proliferation through p27 degradation and enhanced Cul-1 expression is correlated with poor survival of cancer patients [250,251], highlighting the potential of targeting Cul-1 to inhibit Skp2-SCF activity. Previous studies indicate neddylation of Cul-1 is essential for the functional activity of Skp2-SCF complex [163]. MLN4924 specifically inhibits E1 enzyme (Nedd8-activating enzyme, NAE) required for Nedd8 conjugation and compromises Cul-1 neddylation, resulting in deconstruction and deactivation of Skp2-SCF complex as well as p27 accumulation associated cell death [252]. MLN4924 has been advanced into phase I clinical trials as a promising anticancer agent due to its profound anti-neoplasm activity and low toxicity. Since neddylation modification is universally required for the function of all Cullins (cullin 1, 2/5, 3, 4A/4B and 7), MLN4924 is able to inactivate diverse Cullin-Ring ligases (CRLs) and induce the accumulation of various CRL substrates. Thus, whether Skp2-SCF complex is the main target of MLN4924 responsible for its anti-tumor efficacy remains unclear.
7. Conclusion and perspective
Skp2, as an E3 ligase coupling with Skp1, Cul1 and Rbx1/Roc1 to form Skp2-SCF complex, conjugates both K48-linked and K63-linked ubiquitin chains on its substrates, leading to proteasome mediated degradation as well as nonproteolytic functional regulation of the substrates respectively. The expression of Skp2 is under tight regulation during cell cycle progression in normal cells, while Skp2 overexpression is observed in multiple types of cancer such as breast, prostate, lung and others, correlated with poor overall survival, tumor metastasis and drug resistance. Using genetic mouse models and in vitro cell models, the pro-tumor property of Skp2 has been comprehensively demonstrated including cell cycle promotion, cellular senescence and apoptosis prevention, cell migration enhancement, stemness maintenance and many others. So, Skp2appears to be an attractive target for the prevention and/or the treatment of cancer.
Multiple efforts have been made to develop novel inhibitors targeting the Skp2-SCF complex. Disruption of Skp1-Skp2 interaction is the most appealing strategy to restrain the Skp2-SCF complex E3 ligase activity, resulting in the abrogation of ubiquitination on multiple substrates of Skp2 such as p27 and Akt [216]. Moreover, other studies focus on the specific interruption of Skp2 mediated p27 ubiquitination either by blocking Skp2-Cks1 interaction or by directly abrogating Skp2-p27 association [245,249]. In recent years, the development of PROTAC (proteolysis targeting chimera) technology has opened a new area for inducing selective intracellular proteolysis of targeted protein, which serves as a novel anti-cancer strategy through specifically eliminating oncogenic proteins [253,254]. Thus PROTAC based technology could represent another strategy for developing the next generation of Skp2 inhibitors with more potent efficacy in the future.
The potent anti-tumor activities of these Skp2 inhibitors have been documented either in in vitro cell models or pre-clinical mouse models, highlighting the potential therapeutic application of Skp2 inhibitors towards human malignancy. Nevertheless, the potential linkage between Skp2 depletion and genomic instability raises some concerns for the long term benefits of exclusive Skp2 inhibition. First, Skp2 is also found functional in V(D)J (Variable [V], Diversity [D] and Joining [J] gene segments) recombination required for DNA rearrangement, which is indispensable for the maturation of T cell and B cell [255]. During G1/S transition, RAG1 and RAG2 activate V(D)J recombination through recognizing the recombination sequence signal and making double strand break, followed by non-homologous end joining machinery mediated DNA repair. Cell cycle coupled periodic destruction of RAG2 by Skp2 is essential for preventing the potential genomic instability that could otherwise result from excessive or unscheduled RAG-mediated DNA cleavage [99]. Mice with knock-in RAG2 T490A mutant, which is resistant to Skp2 mediated ubiquitination and degradation due to the defects on the prerequisite phosphorylation of T490 by cyclin A-Cdk2, show enhanced frequency of aberrant recombination and cellular apoptosis owing to uncoupled DNA cleavage from cell cycle, which is a similar phenotype observed upon Skp2 depletion [256]. Importantly, mice with combined RAG2 T490A mutation and p53-deficiency display induced lymphoid malignancies characterized by clonal chromosomal translocations [256]. Thus Skp2 mediated periodic RAG2 degradation is fundamental for genomic stability by preventing unscheduled DNA cleavage, representing the unexpected tumor suppressor arm of Skp2. Although RAG1 and RAG2 are exclusively expressed in lymphoid tissue for antigen receptor genes reassembly under physiological condition, aberrant expression of RAG1 and RAG2 is also revealed in cancer cells [257]. Therefore, long term use of Skp2 inhibitors may lead to abnormal accumulation of RAG2 and subsequent genomic instability in both tumor tissues and lymphoid tissues. It is worthwhile to investigate the cellular genotoxicity upon chronic Skp2 inactivation in the future.
Additionally, although accumulation of both p27 and Cyclin E is observed in Skp2 deficient cells [57], Cyclin E/CDK2-associated kinase activity remains unchanged in these cells probably because enhanced abundance of p27, a potent inhibitor of CDK2 activity [258]. However, inhibition of Skp2 in cells with p27 deletion or mutation may induce unrestricted Cyclin E expression and subsequent Cyclin E/CDK2 activation [259]. Accumulated evidences prove the pro-tumor property of Cyclin E overexpression [260,261]. Thus, side effects resulted from enhanced Cyclin E expression upon Skp2 inhibition warrant further evaluation in the future, especially for those cancers with p27 deletion or mutation [262].
Immunotherapy has emerged as a promising strategy for combating cancer [263]. Although Skp2 targeting by Skp2 inhibitors has been proved to restrict cancer development mainly using immune-deficient tumor models, the role of Skp2 targeting in immune regulation remains largely unexplored. Further investigation of the functions of Skp2 in T cell and NK cell activation, macrophage differentiation, as well as antigen recognition and presentation will not only delineate the underlying mechanisms of immune escape during tumorigenesis but also yield more comprehensive understanding of how Skp2 inhibitors work to suppress cancer development.
Conclusively, due to the comprehensive oncogenic function of Skp2, the development of Skp2 specific inhibitor is likely to benefit for cancer therapy. With the further investigation of Skp2, more and more substrates will be unraveled. Exploitation of substrate specific inhibitors for Skp2 would largely limit the potential side effects resulted from deubiquitination of multiple substrates by general Skp2 inhibitors. Moreover, more clinical relevant human tumor models, such as PDX and organoids and genetic mouse models with intact immunity should be included to evaluate the efficacy and potential side effects of Skp2 inhibitors. As multiple molecular signals for diverse biological processes regulated by Skp2 are also essential for the pathogenesis of diseases other than cancer, such as abnormal PI3K/Akt signaling in diabetes [107] and deregulated autophagy during viral infection [264], it will also be interesting to evaluate the efficacy of Skp2 inhibitors in the treatment of diabetes or viral infection. We believe that our review article would draw remarkable attention and ignite further studies to yield new strategies for Skp2 targeting in the treatment of cancer and other diseases.
Acknowledgements
We would like to thank the members in Lin’s laboratory for the inputs and suggestions throughout the course of this study. This work was supported by start-up funds from Wake Forest School of Medicine and NIH grants (R01CA182424 and 1R01CA193813).
Footnotes
Declaration of Competing Interest
There are no conflicts of interest declared by the authors.
References
- [1].Wilkinson KD, Protein ubiquitination: a regulatory post-translational modification, Anti-cancer drug design 2 (2) (1987) 211–229. [PubMed] [Google Scholar]
- [2].Vucic D, Dixit VM, Wertz IE, Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death, Nature reviews, Molecular cell biology 12 (7) (2011) 439–452. [DOI] [PubMed] [Google Scholar]
- [3].Nakayama KI, Nakayama K, Ubiquitin ligases: cell-cycle control and cancer, Nature reviews, Cancer 6 (5) (2006) 369–381. [DOI] [PubMed] [Google Scholar]
- [4].Lee HJ, Li CF, Ruan D, He J, Non-proteolytic ubiquitination of Hexokinase 2 by HectH9 controls tumor metabolism and cancer stem cell expansion, Nature Communications 10 (1) (2019) 2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu J, Zhang X, Zhang L, Wu CY, Rezaeian AH, Chan CH, Li JM, Wang J, Gao Y, Han F, Jeong YS, Yuan X, Khanna KK, Jin J, Zeng YX, Lin HK, Skp2 E3 ligase integrates ATM activation and homologous recombination repair by ubiquitinating NBS1, Molecular cell 46 (3) (2012) 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shi CS, Kehrl JH, TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy, Science signaling 3 (123) (2010) ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, Lin HK, The E3 ligase TRAF6 regulates Akt ubiquitination and activation, Science (New York, N.Y.) 325 (5944) (2009) 1134–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dybas JM, O’Leary CE, Ding H, Spruce LA, Integrative proteomics reveals an increase in non-degradative ubiquitylation in activated CD4(+) T cells, Nature Immunology 20 (6) (2019) 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Senft D, Qi J, Ronai ZA, Ubiquitin ligases in oncogenic transformation and cancer therapy, Nature reviews, Cancer 18 (2) (2018) 69–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Harper JW, Ordureau A, Heo JM, Building and decoding ubiquitin chains for mitophagy, Nature reviews, Molecular cell biology 19 (2) (2018) 93–108. [DOI] [PubMed] [Google Scholar]
- [11].Zhang Y, Chen X, Zhao Y, Ponnusamy M, Liu Y, The role of ubiquitin proteasomal system and autophagy-lysosome pathway in Alzheimer’s disease, Reviews in the neurosciences 28 (8) (2017) 861–868. [DOI] [PubMed] [Google Scholar]
- [12].Ravid T, Hochstrasser M, Diversity of degradation signals in the ubiquitin-proteasome system, Nature reviews, Molecular cell biology 9 (9) (2008) 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ciechanover A, Schwartz AL, The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death, Proceedings of the National Academy of Sciences of the United States of America 95 (6) (1998) 2727–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li W, Ye Y, Polyubiquitin chains: functions, structures and mechanisms, Cellular and Molecular Life Sciences: CMLS 65 (15) (2008) 2397–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Akutsu M, Dikic I, Bremm A, Ubiquitin chain diversity at a glance, Journal of cell science 129 (5) (2016) 875–880. [DOI] [PubMed] [Google Scholar]
- [16].Mallette FA, Richard S, K48-linked ubiquitination and protein degradation regulate 53BP1 recruitment at DNA damage sites, Cell research 22 (8) (2012) 1221–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Okamoto M, Kouwaki T, Fukushima Y, Oshiumi H, Regulation of RIG-I Activation by K63-Linked Polyubiquitination, Frontiers in immunology 8 (2017) 1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lee BL, Singh A, Mark Glover JN, Hendzel MJ, Spyracopoulos L, Molecular Basis for K63-Linked Ubiquitination Processes in Double-Strand DNA Break Repair: A Focus on Kinetics and Dynamics, Journal of molecular biology 429 (22) (2017) 3409–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bellail AC, Olson JJ, Yang X, Chen ZJ, Hao C, A20 ubiquitin ligase-mediated polyubiquitination of RIP1 inhibits caspase-8 cleavage and TRAIL-induced apoptosis in glioblastoma, Cancer discovery 2 (2) (2012) 140–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, Kogel U, Scheffner M, Helin K, Eilers M, The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation, Cell 123 (3) (2005) 409–421. [DOI] [PubMed] [Google Scholar]
- [21].Silva GM, Finley D, Vogel C, K63 polyubiquitination is a new modulator of the oxidative stress response, Nat Struct Mol Biol. 22 (2) (2015) 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kuang E, Qi J, Ronai Z, Emerging roles of E3 ubiquitin ligases in autophagy, Trends in biochemical sciences 38 (9) (2013) 453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rajalingam K, Dikic I, SnapShot: Expanding the Ubiquitin Code, Cell 164 (5) (2016) 1074–1074.e1. [DOI] [PubMed] [Google Scholar]
- [24].Wickliffe KE, Williamson A, Meyer HJ, Kelly A, Rape M, K11-linked ubiquitin chains as novel regulators of cell division, Trends in cell biology 21 (11) (2011) 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gatti M, Pinato S, Maiolica A, Rocchio F, Prato MG, Aebersold R, Penengo L, RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage, Cell reports 10 (2) (2015) 226–238. [DOI] [PubMed] [Google Scholar]
- [26].Fei C, Li Z, Li C, Chen Y, Chen Z, He X, Mao L, Wang X, Zeng R, Li L, Smurf1-mediated Lys29-linked nonproteolytic polyubiquitination of axin negatively regulates Wnt/beta-catenin signaling, Molecular and cellular biology 33 (20) (2013) 4095–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D, Cohen P, Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains, Proceedings of the National Academy of Sciences of the United States of America 110 (38) (2013) 15247–15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Eldridge AG, O’Brien T, Therapeutic strategies within the ubiquitin proteasome system, Cell death and differentiation 17 (1) (2010) 4–13. [DOI] [PubMed] [Google Scholar]
- [29].Deshaies RJ, Joazeiro CA, RING domain E3 ubiquitin ligases, Annual review of biochemistry 78 (2009) 399–434. [DOI] [PubMed] [Google Scholar]
- [30].Morreale FE, Walden H, Types of Ubiquitin Ligases, Cell 165 (1) (2016) 248–248.e1. [DOI] [PubMed] [Google Scholar]
- [31].Scheffner M, Kumar S, Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects, Biochimica et biophysica acta 1843 (1) (2014) 61–74. [DOI] [PubMed] [Google Scholar]
- [32].Huibregtse JM, Scheffner M, Beaudenon S, Howley PM, A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase, Proceedings of the National Academy of Sciences of the United States of America 92 (7) (1995) 2563–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fajner V, Maspero E, Targeting HECT-type E3 ligases - insights from catalysis, regulation and inhibitors 591 (17) (2017) 2636–2647. [DOI] [PubMed] [Google Scholar]
- [34].Walden H, Rittinger K, RBR ligase-mediated ubiquitin transfer: a tale with many twists and turns, FEBS Lett. 25 (6) (2018) 440–445. [DOI] [PubMed] [Google Scholar]
- [35].Spratt DE, Walden H, Shaw GS, RBR E3 ubiquitin ligases: new structures, new insights, new questions, The Biochemical journal 458 (3) (2014) 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Metzger MB, Pruneda JN, Klevit RE, Weissman AM, RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination, Biochimica et biophysica acta 1843 (1) (2014) 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhao Y, Sun Y, Cullin-RING Ligases as attractive anti-cancer targets, Current pharmaceutical design 19 (18) (2013) 3215–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cardozo T, Pagano M, The SCF ubiquitin ligase: insights into a molecular machine, Nature reviews, Molecular cell biology 5 (9) (2004) 739–751. [DOI] [PubMed] [Google Scholar]
- [39].Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP, Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex, Nature 416 (6882) (2002) 703–709. [DOI] [PubMed] [Google Scholar]
- [40].Wang G, Chan CH, Gao Y, Lin HK, Novel roles of Skp2 E3 ligase in cellular senescence, cancer progression, and metastasis, Chinese journal of cancer 31 (4) (2012) 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chan CH, Lee SW, Li CF, Wang J, Yang WL, Wu CY, Wu J, Nakayama KI, Kang HY, Huang HY, Hung MC, Pandolfi PP, Lin HK, Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis, Nature cell biology 12 (5) (2010) 457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Carrano AC, Eytan E, Hershko A, Pagano M, SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27, Nature cell biology 1 (4) (1999) 193–199. [DOI] [PubMed] [Google Scholar]
- [43].Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, Tindall DJ, Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation, Proceedings of the National Academy of Sciences of the United States of America 102 (5) (2005) 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Xu D, Li CF, Zhang X, Gong Z, Chan CH, Lee SW, Jin G, Rezaeian AH, Han F, Wang J, Yang WL, Feng ZZ, Chen W, Wu CY, Wang J, Chow LP, Zhu XF, Zeng YX, Lin HK, Skp2-macroH2A1-CDK8 axis orchestrates G2/M transition and tumorigenesis, Nature communications 6 (2015) 6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, Hazle JD, Yu D, Wei W, Sarbassov D, Hung MC, Nakayama KI, Lin HK, The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis, Cell 149 (5) (2012) 1098–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lee SW, Li CF, Jin G, Cai Z, Han F, Chan CH, Yang WL, Li BK, Rezaeian AH, Li HY, Huang HY, Lin HK, Skp2-dependent ubiquitination and activation of LKB1 is essential for cancer cell survival under energy stress, Molecular cell 57 (6) (2015) 1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yao F, Zhou Z, Kim J, Hang Q, Xiao Z, Ton BN, Chang L, Liu N, Zeng L, Wang W, SKP2- and OTUD1-regulated non-proteolytic ubiquitination of YAP promotes YAP nuclear localization and activity, Nature Communications 9 (1) (2018) 2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jin G, Lee SW, Zhang X, Cai Z, Gao Y, Chou PC, Rezaeian AH, Han F, Wang CY, Yao JC, Gong Z, Chan CH, Huang CY, Tsai FJ, Tsai CH, Tu SH, Wu CH, Sarbassov dos D, Ho YS, Lin HK, Skp2-Mediated RagA Ubiquitination Elicits a Negative Feedback to Prevent Amino-Acid-Dependent mTORC1 Hyperactivation by Recruiting GATOR1, Molecular cell 58 (6) (2015) 989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Liao Y, Liu N, Xia X, Guo Z, Li Y, Jiang L, Zhou R, Tang D, Huang H, Liu J, USP10 modulates the SKP2/Bcr-Abl axis via stabilizing SKP2 in chronic myeloid leukemia, Cell discovery 5 (2019) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang JY, Liu GZ, Wilmott JS, La T, Feng YC, Yari H, Yan XG, Thorne RF, Scolyer RA, Zhang XD, Jin L, Skp2-Mediated Stabilization of MTH1 Promotes Survival of Melanoma Cells upon Oxidative Stress, Cancer research 77 (22) (2017) 6226–6239. [DOI] [PubMed] [Google Scholar]
- [51].Wu J, Huang YF, Zhou XK, Zhang W, Lian YF, Lv XB, Gao XR, Lin HK, Zeng YX, Huang JQ, Skp2 is required for Aurora B activation in cell mitosis and spindle checkpoint, Cell cycle (Georgetown, Tex.) 14 (24) (2015) 3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bochis OV, Irimie A, Pichler M, Berindan-Neagoe I, The role of Skp2 and its substrate CDKN1B (p27) in colorectal cancer, Journal of gastrointestinal and liver diseases: JGLD 24 (2) (2015) 225–234. [DOI] [PubMed] [Google Scholar]
- [53].He W, Wang X, Chen L, Guan X, A crosstalk imbalance between p27(Kip1) and its interacting molecules enhances breast carcinogenesis, Cancer biotherapy & radiopharmaceuticals 27 (7) (2012) 399–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chu IM, Hengst L, Slingerland JM, The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy, Nature reviews, Cancer 8 (4) (2008) 253–267. [DOI] [PubMed] [Google Scholar]
- [55].Esposito V, Baldi A, De Luca A, Groger AM, Loda M, Giordano GG, Caputi M, Baldi F, Pagano M, Giordano A, Prognostic role of the cyclin-dependent kinase inhibitor p27 in non-small cell lung cancer, Cancer research 57 (16) (1997) 3381–3385. [PubMed] [Google Scholar]
- [56].Loda M, Cukor B, Tam SW, Lavin P, Fiorentino M, Draetta GF, Jessup JM, Pagano M, Increased proteasome-dependent degradation of the cyclin-dependent kinase inhibitor p27 in aggressive colorectal carcinomas, Nature medicine 3 (2) (1997) 231–234. [DOI] [PubMed] [Google Scholar]
- [57].Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, Shirane M, Tsunematsu R, Tsukiyama T, Ishida N, Kitagawa M, Nakayama K, Hatakeyama S, Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication, The EMBO journal 19 (9) (2000) 2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sutterluty H, Chatelain E, Marti A, Wirbelauer C, Senften M, Muller U, Krek W, p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells, Nature cell biology 1 (4) (1999) 207–214. [DOI] [PubMed] [Google Scholar]
- [59].Nakayama K, Nagahama H, Minamishima YA, Miyake S, Ishida N, Hatakeyama S, Kitagawa M, Iemura S, Natsume T, Nakayama KI, Skp2-mediated degradation of p27 regulates progression into mitosis, Developmental cell 6 (5) (2004) 661–672. [DOI] [PubMed] [Google Scholar]
- [60].Tsvetkov LM, Yeh KH, Lee SJ, Sun H, Zhang H, p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27, Current biology: CB 9 (12) (1999) 661–664. [DOI] [PubMed] [Google Scholar]
- [61].Zafonte BT, Hulit J, Amanatullah DF, Albanese C, Wang C, Rosen E, Reutens A, Sparano JA, Lisanti MP, Pestell RG, Cell-cycle dysregulation in breast cancer: breast cancer therapies targeting the cell cycle, Frontiers in bioscience 5 (2000) D938–61. [DOI] [PubMed] [Google Scholar]
- [62].Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A, The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27, Nature cell biology 3 (3) (2001) 321–324. [DOI] [PubMed] [Google Scholar]
- [63].Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP, Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase, Molecular cell 20 (1) (2005) 9–19. [DOI] [PubMed] [Google Scholar]
- [64].Sitry D, Seeliger MA, Ko TK, Ganoth D, Breward SE, Itzhaki LS, Pagano M, Hershko A, Three different binding sites of Cks1 are required for p27-ubiquitin ligation, The Journal of biological chemistry 277 (44) (2002) 42233–42240. [DOI] [PubMed] [Google Scholar]
- [65].Spruck C, Strohmaier H, Watson M, Smith AP, Ryan A, Krek TW, Reed SI, A CDK-independent function of mammalian Cks1: targeting of SCF(Skp2) to the CDK inhibitor p27Kip1, Molecular cell 7 (3) (2001) 639–650. [DOI] [PubMed] [Google Scholar]
- [66].Ben-Izhak O, Lahav-Baratz S, Meretyk S, Ben-Eliezer S, Sabo E, Dirnfeld M, Cohen S, Ciechanover A, Inverse relationship between Skp2 ubiquitin ligase and the cyclin dependent kinase inhibitor p27Kip1 in prostate cancer, The Journal of urology 170 (1) (2003) 241–245. [DOI] [PubMed] [Google Scholar]
- [67].Lim MS, Adamson A, Lin Z, Perez-Ordonez B, Jordan RC, Tripp S, Perkins SL, Elenitoba-Johnson KS, Expression of Skp2, a p27(Kip1) ubiquitin ligase, in malignant lymphoma: correlation with p27(Kip1) and proliferation index, Blood 100 (8) (2002) 2950–2956. [DOI] [PubMed] [Google Scholar]
- [68].Drobnjak M, Melamed J, Taneja S, Melzer K, Wieczorek R, Levinson B, Zeleniuch-Jacquotte A, Polsky D, Ferrara J, Perez-Soler R, Cordon-Cardo C, Pagano M, Osman I, Altered expression of p27 and Skp2 proteins in prostate cancer of African-American patients, Clinical cancer research 9 (7) (2003) 2613–2619. [PubMed] [Google Scholar]
- [69].Fukuchi M, Masuda N, Nakajima M, Fukai Y, Miyazaki T, Kato H, Kuwano H, Inverse correlation between expression levels of p27 and the ubiquitin ligase subunit Skp2 in early esophageal squamous cell carcinoma, Anticancer research 24 (2b) (2004) 777–783. [PubMed] [Google Scholar]
- [70].Huang H, Tindall DJ, Dynamic FoxO transcription factors, Journal of cell science 120 (Pt 15) (2007) 2479–2487. [DOI] [PubMed] [Google Scholar]
- [71].Calvisi DF, Ladu S, Pinna F, Frau M, Tomasi ML, Sini M, Simile MM, Bonelli P, Muroni MR, Seddaiu MA, Lim DS, Feo F, Pascale RM, SKP2 and CKS1 promote degradation of cell cycle regulators and are associated with hepatocellular carcinoma prognosis, Gastroenterology 137 (5) (2009) 1816–26.e1–10. [DOI] [PubMed] [Google Scholar]
- [72].Dehan E, Pagano M, Skp2, the FoxO1 hunter, Cancer cell 7 (3) (2005) 209–210. [DOI] [PubMed] [Google Scholar]
- [73].Levy JMM, Towers CG, Thorburn A, Targeting autophagy in cancer, Nature reviews, Cancer 17 (9) (2017) 528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Shin HJ, Kim H, Oh S, Lee JG, Kee M, Ko HJ, Kweon MN, Won KJ, Baek SH, AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy, Nature 534 (7608) (2016) 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Shin HR, Kim H, Kim KI, Baek SH, Epigenetic and transcriptional regulation of autophagy, Autophagy 12 (11) (2016) 2248–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Xu Z, Klionsky DJ, The AMPK-SKP2-CARM1 axis links nutrient sensing to transcriptional and epigenetic regulation of autophagy, Annals of translational medicine 4 (Suppl 1) (2016) S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Linton PJ, Gurney M, Sengstock D, Mentzer RM Jr, Gottlieb RA, This old heart: Cardiac aging and autophagy, Journal of molecular and cellular cardiology 83 (2015) 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Li C, Yu L, Xue H, Yang Z, Yin Y, Zhang B, Chen M, Ma H, Nuclear AMPK regulated CARM1 stabilization impacts autophagy in aged heart, Biochemical and biophysical research communications 486 (2) (2017) 398–405. [DOI] [PubMed] [Google Scholar]
- [79].Wei X, Li X, Yan W, Zhang X, Sun Y, Zhang F, SKP2 Promotes Hepatocellular Carcinoma Progression Through Nuclear AMPK-SKP2-CARM1 Signaling Transcriptionally Regulating Nutrient-Deprived Autophagy Induction, Cellular physiology and biochemistry 47 (6) (2018) 2484–2497. [DOI] [PubMed] [Google Scholar]
- [80].Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A, Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase, The Journal of biological chemistry 278 (28) (2003) 25752–25757. [DOI] [PubMed] [Google Scholar]
- [81].Yu ZK, Gervais JL, Zhang H, Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins, Proceedings of the National Academy of Sciences of the United States of America 95 (19) (1998) 11324–11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H, Hatakeyama S, Nakayama K, Nakayama KI, Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation, Proceedings of the National Academy of Sciences of the United States of America 100 (18) (2003) 10231–10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Marti A, Wirbelauer C, Scheffner M, Krek W, Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation, Nature cell biology 1 (1) (1999) 14–19. [DOI] [PubMed] [Google Scholar]
- [84].Liu Y, Hedvat CV, Mao S, Zhu XH, Yao J, Nguyen H, Koff A, Nimer SD, The ETS protein MEF is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCFSkp2, Molecular and cellular biology 26 (8) (2006) 3114–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Tedesco D, Lukas J, Reed SI, The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2), Genes & development 16 (22) (2002) 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bhattacharya S, Garriga J, Calbo J, Yong T, Haines DS, Grana X, SKP2 associates with p130 and accelerates p130 ubiquitylation and degradation in human cells, Oncogene 22 (16) (2003) 2443–2451. [DOI] [PubMed] [Google Scholar]
- [87].Hiramatsu Y, Kitagawa K, Suzuki T, Uchida C, Hattori T, Kikuchi H, Oda T, Hatakeyama S, Nakayama KI, Yamamoto T, Konno H, Kitagawa M, Degradation of Tob1 mediated by SCFSkp2-dependent ubiquitination, Cancer research 66 (17) (2006) 8477–8483. [DOI] [PubMed] [Google Scholar]
- [88].Yeh KH, Kondo T, Zheng J, Tsvetkov LM, Blair J, Zhang H, The F-box protein SKP2 binds to the phosphorylated threonine 380 in cyclin E and regulates ubiquitin-dependent degradation of cyclin E, Biochemical and biophysical research communications 281 (4) (2001) 884–890. [DOI] [PubMed] [Google Scholar]
- [89].Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP, Skp2 regulates Myc protein stability and activity, Molecular cell 11 (5) (2003) 1177–1188. [DOI] [PubMed] [Google Scholar]
- [90].von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, Soderberg O, Kerppola TK, Larsson LG, The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription, Molecular cell 11 (5) (2003) 1189–1200. [DOI] [PubMed] [Google Scholar]
- [91].Charrasse S, Carena I, Brondani V, Klempnauer KH, Ferrari S, Degradation of B-Myb by ubiquitin-mediated proteolysis: involvement of the Cdc34-SCF (p45Skp2) pathway, Oncogene 19 (26) (2000) 2986–2995. [DOI] [PubMed] [Google Scholar]
- [92].Song MS, Song SJ, Kim SJ, Nakayama K, Nakayama KI, Lim DS, Skp2 regulates the antiproliferative function of the tumor suppressor RASSF1A via ubiquitin-mediated degradation at the G1-S transition, Oncogene 27 (22) (2008) 3176–3185. [DOI] [PubMed] [Google Scholar]
- [93].Lonjedo M, Poch E, Mocholi E, Hernandez-Sanchez M, Ivorra C, Franke TF, Guasch RM, Perez-Roger I, The Rho family member RhoE interacts with Skp2 and is degraded at the proteasome during cell cycle progression, The Journal of biological chemistry 288 (43) (2013) 30872–30882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].John R, Atri Y, Chand V, Jaiswal N, Raj K, Nag A, Cell cycle-dependent regulation of cytoglobin by Skp2, FEBS letters 591 (21) (2017) 3507–3522. [DOI] [PubMed] [Google Scholar]
- [95].Li C, Du L, Ren Y, Liu X, Jiao Q, Cui D, Wen M, Wang C, Wei G, Wang Y, Ji A, Wang Q, SKP2 promotes breast cancer tumorigenesis and radiation tolerance through PDCD4 ubiquitination, Journal of experimental & clinical cancer research 38 (1) (2019) 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B, Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication, Molecular cell 9 (3) (2002) 481–491. [DOI] [PubMed] [Google Scholar]
- [97].Kondo T, Kobayashi M, Tanaka J, Yokoyama A, Suzuki S, Kato N, Onozawa M, Chiba K, Hashino S, Imamura M, Minami Y, Minamino N, Asaka M, Rapid degradation of Cdt1 upon UV-induced DNA damage is mediated by SCFSkp2 complex, The Journal of biological chemistry 279 (26) (2004) 27315–27319. [DOI] [PubMed] [Google Scholar]
- [98].Li X, Zhao Q, Liao R, Sun P, Wu X, The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation, The Journal of biological chemistry 278 (33) (2003) 30854–30858. [DOI] [PubMed] [Google Scholar]
- [99].Jiang H, Chang FC, Ross AE, Lee J, Nakayama K, Nakayama K, Desiderio S, Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle, Molecular cell 18 (6) (2005) 699–709. [DOI] [PubMed] [Google Scholar]
- [100].Moro L, Arbini AA, Marra E, Greco M, Up-regulation of Skp2 after prostate cancer cell adhesion to basement membranes results in BRCA2 degradation and cell proliferation, The Journal of biological chemistry 281 (31) (2006) 22100–22107. [DOI] [PubMed] [Google Scholar]
- [101].Kiernan RE, Emiliani S, Nakayama K, Castro A, Labbe JC, Lorca T, Nakayama Ki K, Benkirane M, Interaction between cyclin T1 and SCF(SKP2) targets CDK9 for ubiquitination and degradation by the proteasome, Molecular and cellular biology 21 (23) (2001) 7956–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Bellanger S, Tan CL, Nei W, He PP, Thierry F, The human papillomavirus type 18 E2 protein is a cell cycle-dependent target of the SCFSkp2 ubiquitin ligase, Journal of virology 84 (1) (2010) 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Oh KJ, Kalinina A, Wang J, Nakayama K, Nakayama KI, Bagchi S, The papillomavirus E7 oncoprotein is ubiquitinated by UbcH7 and Cullin 1- and Skp2-containing E3 ligase, Journal of virology 78 (10) (2004) 5338–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lin YW, Yang JL, Cooperation of ERK and SCFSkp2 for MKP-1 destruction provides a positive feedback regulation of proliferating signaling, The Journal of biological chemistry 281 (2) (2006) 915–926. [DOI] [PubMed] [Google Scholar]
- [105].Tokarz S, Berset C, La Rue J, Friedman K, Nakayama K, Nakayama K, Zhang DE, Lanker S, The ISG15 isopeptidase UBP43 is regulated by proteolysis via the SCFSkp2 ubiquitin ligase, The Journal of biological chemistry 279 (45) (2004) 46424–46430. [DOI] [PubMed] [Google Scholar]
- [106].Liang M, Liang YY, Wrighton K, Ungermannova D, Wang XP, Brunicardi FC, Liu X, Feng XH, Lin X, Ubiquitination and proteolysis of cancer-derived Smad4 mutants by SCFSkp2, Molecular and cellular biology 24 (17) (2004) 7524–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Manning BD, Toker A, AKT/PKB Signaling: Navigating the Network, Cell 169 (3) (2017) 381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Sarbassov DD, Guertin DA, Ali SM, Sabatini DM, Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex, Science (New York, N.Y.) 307 (5712) (2005) 1098–1101. [DOI] [PubMed] [Google Scholar]
- [109].Guo J, Dai X, Laurent B, Zheng N, Gan W, Zhang J, AKT methylation by SETDB1 promotes AKT kinase activity and oncogenic functions, Nature Cell Biology 21 (2) (2019) 226–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Wang G, Long J, Gao Y, Zhang W, Han F, Xu C, Sun L, Yang SC, Lan J, Hou Z, Cai Z, Jin G, Hsu CC, Wang YH, Hu J, Chen TY, Li H, Lee MG, Lin HK, SETDB1-mediated methylation of Akt promotes its K63-linked ubiquitination and activation leading to tumorigenesis, Nature Cell Biology 21 (2) (2019) 214–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Guo J, Chakraborty AA, Liu P, Gan W, Zheng X, Inuzuka H, Wang B, Zhang J, Zhang L, Yuan M, Novak J, Cheng JQ, Toker A, Signoretti S, Zhang Q, Asara JM, Kaelin WG Jr, Wei W, pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner, Science (New York, N.Y.) 353 (6302) (2016) 929–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Lin CH, Liu SY, Lee EH, SUMO modification of Akt regulates global SUMOylation and substrate SUMOylation specificity through Akt phosphorylation of Ubc9 and SUMO1, Oncogene 35 (5) (2016) 595–607. [DOI] [PubMed] [Google Scholar]
- [113].Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V, Raghuraman H, Cunningham JM, Gupta M, Gupta MP, The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy, Science signaling 4 (182) (2011) ra46. [DOI] [PubMed] [Google Scholar]
- [114].Yu X, Wang R, Zhang Y, Zhou L, Wang W, Liu H, Li W, Skp2-mediated ubiquitination and mitochondrial localization of Akt drive tumor growth and chemoresistance to cisplatin, Oncogene (2019). [DOI] [PubMed] [Google Scholar]
- [115].Yu FX, Guan KL, The Hippo pathway: regulators and regulations, Genes & development 27 (4) (2013) 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Lee JH, Paull TT, ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex, Science (New York, N.Y.) 308 (5721) (2005) 551–554. [DOI] [PubMed] [Google Scholar]
- [117].Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, Alessi DR, LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1, The EMBO journal 23 (4) (2004) 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Alessi DR, Sakamoto K, Bayascas JR, LKB1-dependent signaling pathways, Annual review of biochemistry 75 (2006) 137–163. [DOI] [PubMed] [Google Scholar]
- [119].Zeqiraj E, Filippi BM, Deak M, Alessi DR, van Aalten DM, Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation, Science (New York, N.Y.) 326 (5960) (2009) 1707–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Lee SW, Lin HK, A new mechanism for LKB1 activation, Molecular & cellular oncology 5 (3) (2018) e1035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Jewell JL, Russell RC, Guan KL, Amino acid signalling upstream of mTOR, Nature reviews, Molecular cell biology 14 (3) (2013) 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T, Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B, The Journal of biological chemistry 276 (10) (2001) 7246–7257. [DOI] [PubMed] [Google Scholar]
- [123].Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM, Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1, Cell 150 (6) (2012) 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM, A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1, Science (New York, N.Y.) 340 (6136) (2013) 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zhu QQ, Ma C, Wang Q, Song Y, Lv T, The role of TWIST1 in epithelial-mesenchymal transition and cancers, Tumour biology 37 (1) (2016) 185–197. [DOI] [PubMed] [Google Scholar]
- [126].Ruan D, He J, Li CF, Lee HJ, Liu J, Lin HK, Chan CH, Skp2 deficiency restricts the progression and stem cell features of castration-resistant prostate cancer by destabilizing Twist, Oncogene 36 (30) (2017) 4299–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Jaffe AB, Hall A, Rho GTPases: biochemistry and biology, Annual review of cell and developmental biology 21 (2005) 247–269. [DOI] [PubMed] [Google Scholar]
- [128].von der Lehr N, Johansson S, Larsson LG, Implication of the ubiquitin/proteasome system in Myc-regulated transcription, Cell cycle (Georgetown, Tex.) 2 (5) (2003) 403–407. [PubMed] [Google Scholar]
- [129].Brooks CL, Gu W, The impact of acetylation and deacetylation on the p53 pathway, Protein & cell 2 (6) (2011) 456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Kitagawa M, Lee SH, McCormick F, Skp2 suppresses p53-dependent apoptosis by inhibiting p300, Molecular cell 29 (2) (2008) 217–231. [DOI] [PubMed] [Google Scholar]
- [131].Hao Z, Huang S, E3 ubiquitin ligase Skp2 as an attractive target in cancer therapy, Frontiers in bioscience (Landmark edition) 20 (2015) 474–490. [DOI] [PubMed] [Google Scholar]
- [132].Zhang L, Wang C, F-box protein Skp2: a novel transcriptional target of E2F, Oncogene 25 (18) (2006) 2615–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Schneider G, Saur D, Siveke JT, Fritsch R, Greten FR, Schmid RM, IKKalpha controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression, The EMBO journal 25 (16) (2006) 3801–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Sarmento LM, Huang H, Limon A, Gordon W, Fernandes J, Tavares MJ, Miele L, Cardoso AA, Classon M, Carlesso N, Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation, The Journal of experimental medicine 202 (1) (2005) 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH, Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase, Molecular and cellular biology 25 (24) (2005) 10875–10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Imaki H, Nakayama K, Delehouzee S, Handa H, Kitagawa M, Kamura T, Nakayama KI, Cell cycle-dependent regulation of the Skp2 promoter by GA-binding protein, Cancer research 63 (15) (2003) 4607–4613. [PubMed] [Google Scholar]
- [137].Appleman LJ, Chernova I, Li L, Boussiotis VA, CD28 costimulation mediates transcription of SKP2 and CKS1, the substrate recognition components of SCFSkp2 ubiquitin ligase that leads p27kip1 to degradation, Cell cycle (Georgetown, Tex.) 5 (18) (2006) 2123–2129. [DOI] [PubMed] [Google Scholar]
- [138].Evans L, Chen L, Milazzo G, Gherardi S, Perini G, Willmore E, Newell DR, Tweddle DA, SKP2 is a direct transcriptional target of MYCN and a potential therapeutic target in neuroblastoma, Cancer letters 363 (1) (2015) 37–45. [DOI] [PubMed] [Google Scholar]
- [139].Mamillapalli R, Gavrilova N, Mihaylova VT, Tsvetkov LM, Wu H, Zhang H, Sun H, PTEN regulates the ubiquitin-dependent degradation of the CDK inhibitor p27(KIP1) through the ubiquitin E3 ligase SCF(SKP2), Current biology 11 (4) (2001) 263–267. [DOI] [PubMed] [Google Scholar]
- [140].Auld CA, Caccia CD, Morrison RF, Hormonal induction of adipogenesis induces Skp2 expression through PI3K and MAPK pathways, Journal of cellular biochemistry 100 (1) (2007) 204–216. [DOI] [PubMed] [Google Scholar]
- [141].Reichert M, Saur D, Hamacher R, Schmid RM, Schneider G, Phosphoinositide-3-kinase signaling controls S-phase kinase-associated protein 2 transcription via E2F1 in pancreatic ductal adenocarcinoma cells, Cancer research 67 (9) (2007) 4149–4156. [DOI] [PubMed] [Google Scholar]
- [142].Andreu EJ, Lledo E, Poch E, Ivorra C, Albero MP, Martinez-Climent JA, Montiel-Duarte C, Rifon J, Perez-Calvo J, Arbona C, Prosper F, Perez-Roger I, BCR-ABL induces the expression of Skp2 through the PI3K pathway to promote p27Kip1 degradation and proliferation of chronic myelogenous leukemia cells, Cancer research 65 (8) (2005) 3264–3272. [DOI] [PubMed] [Google Scholar]
- [143].Martin F, Ladoire S, Mignot G, Apetoh L, Ghiringhelli F, Human FOXP3 and cancer, Oncogene 29 (29) (2010) 4121–4129. [DOI] [PubMed] [Google Scholar]
- [144].Zuo T, Liu R, Zhang H, Chang X, Liu Y, Wang L, Zheng P, Liu Y, FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2, The Journal of clinical investigation 117 (12) (2007) 3765–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Patsoukis N, Sari D, Boussiotis VA, PD-1 inhibits T cell proliferation by upregulating p27 and p15 and suppressing Cdc25A, Cell cycle (Georgetown, Tex.) 11 (23) (2012) 4305–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Wirbelauer C, Sutterluty H, Blondel M, Gstaiger M, Peter M, Reymond F, Krek W, The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts, The EMBO journal 19 (20) (2000) 5362–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Bashir T, Pagano M, Don’t skip the G1 phase: how APC/CCdh1 keeps SCFSKP2 in check, Cell cycle (Georgetown, Tex.) 3 (7) (2004) 850–852. [PubMed] [Google Scholar]
- [148].Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M, Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase, Nature 428 (6979) (2004) 190–193. [DOI] [PubMed] [Google Scholar]
- [149].Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG Jr, Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex, Nature 428 (6979) (2004) 194–198. [DOI] [PubMed] [Google Scholar]
- [150].Harper JW, Burton JL, Solomon MJ, The anaphase-promoting complex: it’s not just for mitosis any more, Genes & development 16 (17) (2002) 2179–2206. [DOI] [PubMed] [Google Scholar]
- [151].Hsu JY, Reimann JD, Sorensen CS, Lukas J, Jackson PK, E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1), Nature cell biology 4 (5) (2002) 358–366. [DOI] [PubMed] [Google Scholar]
- [152].Lukas C, Sorensen CS, Kramer E, Santoni-Rugiu E, Lindeneg C, Peters JM, Bartek J, Lukas J, Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex, Nature 401 (6755) (1999) 815–818. [DOI] [PubMed] [Google Scholar]
- [153].Xu J, Zhou W, Yang F, Chen G, Li H, Zhao Y, Liu P, Li H, Tan M, Xiong X, Sun Y, The beta-TrCP-FBXW2-SKP2 axis regulates lung cancer cell growth with FBXW2 acting as a tumour suppressor, Nature communications 8 (2017) 14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Rodier G, Coulombe P, Tanguay PL, Boutonnet C, Meloche S, Phosphorylation of Skp2 regulated by CDK2 and Cdc14B protects it from degradation by APC(Cdh1) in G1 phase, The EMBO journal 27 (4) (2008) 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Gao D, Inuzuka H, Tseng A, Chin RY, Toker A, Wei W, Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction, Nature cell biology 11 (4) (2009) 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, Yang WL, Erdjument-Bromage H, Nakayama KI, Nimer S, Tempst P, Pandolfi PP, Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB, Nature cell biology 11 (4) (2009) 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Zhang F, White RL, Neufeld KL, Cell density and phosphorylation control the subcellular localization of adenomatous polyposis coli protein, Molecular and cellular biology 21 (23) (2001) 8143–8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Han F, Li CF, Cai Z, Zhang X, The critical role of AMPK in driving Akt activation under stress, tumorigenesis and drug resistance, Nat Commun. 9 (1) (2018) 4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Burris HA 3rd, Overcoming acquired resistance to anticancer therapy: focus on the PI3K/AKT/mTOR pathway, Cancer chemotherapy and pharmacology 71 (4) (2013) 829–842. [DOI] [PubMed] [Google Scholar]
- [160].Inuzuka H, Gao D, Finley LW, Yang W, Wan L, Fukushima H, Chin YR, Zhai B, Shaik S, Lau AW, Wang Z, Gygi SP, Nakayama K, Teruya-Feldstein J, Toker A, Haigis MC, Pandolfi PP, Wei W, Acetylation-dependent regulation of Skp2 function, Cell 150 (1) (2012) 179–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Wang Z, Inuzuka H, Zhong J, Liu P, Sarkar FH, Sun Y, Wei W, Identification of acetylation-dependent regulatory mechanisms that govern the oncogenic functions of Skp2, Oncotarget 3 (11) (2012) 1294–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Zhang S, Chen Q, Liu Q, Li Y, Sun X, Hong L, Ji S, Liu C, Geng J, Zhang W, Lu Z, Yin ZY, Zeng Y, Lin KH, Wu Q, Li Q, Nakayama K, Nakayama KI, Deng X, Johnson RL, Zhu L, Gao D, Chen L, Zhou D, Hippo Signaling Suppresses Cell Ploidy and Tumorigenesis through Skp2, Cancer cell 31 (5) (2017) 669–684.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Liu J, Furukawa M, Matsumoto T, Xiong Y, NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases, Molecular cell 10 (6) (2002) 1511–1518. [DOI] [PubMed] [Google Scholar]
- [164].Bornstein G, Ganoth D, Hershko A, Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate, Proceedings of the National Academy of Sciences of the United States of America 103 (31) (2006) 11515–11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H, CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex, Molecular cell 10 (6) (2002) 1519–1526. [DOI] [PubMed] [Google Scholar]
- [166].Zhang W, Ito H, Quint M, Huang H, Noel LD, Gray WM, Genetic analysis of CAND1-CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex, Proceedings of the National Academy of Sciences of the United States of America 105 (24) (2008) 8470–8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Zhang H, Kobayashi R, Galaktionov K, Beach D, p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase, Cell 82 (6) (1995) 915–925. [DOI] [PubMed] [Google Scholar]
- [168].Wang J, Han F, Wu J, Lee SW, Chan CH, Wu CY, Yang WL, Gao Y, Zhang X, Jeong YS, Moten A, Samaniego F, Huang P, Liu Q, Zeng YX, Lin HK, The role of Skp2 in hematopoietic stem cell quiescence, pool size, and self-renewal, Blood 118 (20) (2011) 5429–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Fotovati A, Abu-Ali S, Nakayama K, Nakayama KI, Impaired ovarian development and reduced fertility in female mice deficient in Skp2, Journal of anatomy 218 (6) (2011) 668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Cooke PS, Holsberger DR, Cimafranca MA, Meling DD, Beals CM, Nakayama K, Nakayama KI, Kiyokawa H, The F box protein S phase kinase-associated protein 2 regulates adipose mass and adipocyte number in vivo, Obesity (Silver Spring, Md.) 15 (6) (2007) 1400–1408. [DOI] [PubMed] [Google Scholar]
- [171].Kossatz U, Dietrich N, Zender L, Buer J, Manns MP, Malek NP, Skp2-dependent degradation of p27kip1 is essential for cell cycle progression, Genes & development 18 (21) (2004) 2602–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP, Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence, Nature 464 (7287) (2010) 374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Agarwal A, Bumm TG, Corbin AS, O’Hare T, Loriaux M, VanDyke J, Willis SG, Deininger J, Nakayama KI, Druker BJ, Deininger MW, Absence of SKP2 expression attenuates BCR-ABL-induced myeloproliferative disease, Blood 112 (5) (2008) 1960–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Wang H, Bauzon F, Ji P, Xu X, Sun D, Locker J, Sellers RS, Nakayama K, Nakayama KI, Cobrinik D, Zhu L, Skp2 is required for survival of aberrantly proliferating Rb1-deficient cells and for tumorigenesis in Rb1+/− mice, Nature genetics 42 (1) (2010) 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Old JB, Kratzat S, Hoellein A, Graf S, Nilsson JA, Nilsson L, Nakayama KI, Peschel C, Cleveland JL, Keller UB, Skp2 directs Myc-mediated suppression of p27Kip1 yet has modest effects on Myc-driven lymphomagenesis, Molecular cancer research 8 (3) (2010) 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Shim EH, Johnson L, Noh HL, Kim YJ, Sun H, Zeiss C, Zhang H, Expression of the F-box protein SKP2 induces hyperplasia, dysplasia, and low-grade carcinoma in the mouse prostate, Cancer research 63 (7) (2003) 1583–1588. [PubMed] [Google Scholar]
- [177].Latres E, Chiarle R, Schulman BA, Pavletich NP, Pellicer A, Inghirami G, Pagano M, Role of the F-box protein Skp2 in lymphomagenesis, Proceedings of the National Academy of Sciences of the United States of America 98 (5) (2001) 2515–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Signoretti S, Di Marcotullio L, Richardson A, Ramaswamy S, Isaac B, Rue M, Monti F, Loda M, Pagano M, Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer, The Journal of clinical investigation 110 (5) (2002) 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [179].Liu J, Wei XL, Huang WH, Chen CF, Bai JW, Zhang GJ, Cytoplasmic Skp2 expression is associated with p-Akt1 and predicts poor prognosis in human breast carcinomas, PloS One 7 (12) (2012) e52675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Yang G, Ayala G, De Marzo A, Tian W, Frolov A, Wheeler TM, Thompson TC, Harper JW, Elevated Skp2 protein expression in human prostate cancer: association with loss of the cyclin-dependent kinase inhibitor p27 and PTEN and with reduced recurrence-free survival, Clinical cancer research 8 (11) (2002) 3419–3426. [PubMed] [Google Scholar]
- [181].Yokoi S, Yasui K, Saito-Ohara F, Koshikawa K, Iizasa T, Fujisawa T, Terasaki T, Horii A, Takahashi T, Hirohashi S, Inazawa J, A novel target gene, SKP2, within the 5p13 amplicon that is frequently detected in small cell lung cancers, The American journal of pathology 161 (1) (2002) 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Yokoi S, Yasui K, Mori M, Iizasa T, Fujisawa T, Inazawa J, Amplification and overexpression of SKP2 are associated with metastasis of non-small-cell lung cancers to lymph nodes, The American journal of pathology 165 (1) (2004) 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Zhu CQ, Blackhall FH, Pintilie M, Iyengar P, Liu N, Ho J, Chomiak T, Lau D, Winton T, Shepherd FA, Tsao MS, Skp2 gene copy number aberrations are common in non-small cell lung carcinoma, and its overexpression in tumors with ras mutation is a poor prognostic marker, Clinical cancer research 10 (6) (2004) 1984–1991. [DOI] [PubMed] [Google Scholar]
- [184].Jiang F, Todd NW, Li R, Zhang H, Fang H, Stass SA, A panel of sputum-based genomic marker for early detection of lung cancer, Cancer prevention research (Philadelphia, Pa.) 3 (12) (2010) 1571–1578. [DOI] [PubMed] [Google Scholar]
- [185].Carracedo DG, Astudillo A, Rodrigo JP, Suarez C, Gonzalez MV, Skp2, p27kip1 and EGFR assessment in head and neck squamous cell carcinoma: prognostic implications, Oncology reports 20 (3) (2008) 589–595. [PubMed] [Google Scholar]
- [186].Shin E, Kim SH, Jeong HY, Jang JJ, Lee K, Nuclear expression of S-phase kinase-associated protein 2 predicts poor prognosis of hepatocellular carcinoma, APMIS : acta pathologica, microbiologica, et immunologica Scandinavica 120 (5) (2012) 349–357. [DOI] [PubMed] [Google Scholar]
- [187].Einama T, Kagata Y, Tsuda H, Morita D, Ogata S, Ueda S, Takigawa T, Kawarabayashi N, Fukatsu K, Sugiura Y, Matsubara O, Hatsuse K, High-level Skp2 expression in pancreatic ductal adenocarcinoma: correlation with the extent of lymph node metastasis, higher histological grade, and poorer patient outcome, Pancreas 32 (4) (2006) 376–381. [DOI] [PubMed] [Google Scholar]
- [188].Liang Y, Hou X, Cui Q, Kang TB, Fu JH, Zhang LJ, Luo RZ, He JH, Zeng YX, Yang HX, Skp2 expression unfavorably impacts survival in resectable esophageal squamous cell carcinoma, Journal of translational medicine 10 (2012) 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Masuda TA, Inoue H, Sonoda H, Mine S, Yoshikawa Y, Nakayama K, Nakayama K, Mori M, Clinical and biological significance of S-phase kinase-associated protein 2 (Skp2) gene expression in gastric carcinoma: modulation of malignant phenotype by Skp2 overexpression, possibly via p27 proteolysis, Cancer research 62 (13) (2002) 3819–3825. [PubMed] [Google Scholar]
- [190].Li JQ, Wu F, Masaki T, Kubo A, Fujita J, Dixon DA, Beauchamp RD, Ishida T, Kuriyama S, Imaida K, Correlation of Skp2 with carcinogenesis, invasion, metastasis, and prognosis in colorectal tumors, International journal of oncology 25 (1) (2004) 87–95. [PubMed] [Google Scholar]
- [191].Pratheeshkumar P, Siraj AK, Divya SP, Parvathareddy SK, Begum R, Melosantos R, Al-Sobhi SS, Al-Dawish M, Al-Dayel F, Al-Kuraya KS, Downregulation of SKP2 in Papillary Thyroid Cancer Acts Synergistically With TRAIL on Inducing Apoptosis via ROS, The Journal of clinical endocrinology and metabolism 103 (4) (2018) 1530–1544. [DOI] [PubMed] [Google Scholar]
- [192].Schiffer D, Cavalla P, Fiano V, Ghimenti C, Piva R, Inverse relationship between p27/Kip.1 and the F-box protein Skp2 in human astrocytic gliomas by immunohistochemistry and Western blot, Neuroscience letters 328 (2) (2002) 125–128. [DOI] [PubMed] [Google Scholar]
- [193].Shigemasa K, Gu L, O’Brien TJ, Ohama K, Skp2 overexpression is a prognostic factor in patients with ovarian adenocarcinoma, Clinical cancer research 9 (5) (2003) 1756–1763. [PubMed] [Google Scholar]
- [194].Oliveira AM, Okuno SH, Nascimento AG, Lloyd RV, Skp2 protein expression in soft tissue sarcomas, Journal of clinical oncology 21 (4) (2003) 722–727. [DOI] [PubMed] [Google Scholar]
- [195].Huang HY, Kang HY, Li CF, Eng HL, Chou SC, Lin CN, Hsiung CY, Skp2 overexpression is highly representative of intrinsic biological aggressiveness and independently associated with poor prognosis in primary localized myxofibrosarcomas, Clinical cancer research 12 (2) (2006) 487–498. [DOI] [PubMed] [Google Scholar]
- [196].Min YH, Cheong JW, Lee MH, Kim JY, Lee ST, Hahn JS, Ko YW, Elevated S-phase kinase-associated protein 2 protein expression in acute myelogenous leukemia: its association with constitutive phosphorylation of phosphatase and tensin homologue protein and poor prognosis, Clinical cancer research 10 (15) (2004) 5123–5130. [DOI] [PubMed] [Google Scholar]
- [197].Seki R, Okamura T, Koga H, Yakushiji K, Hashiguchi M, Yoshimoto K, Ogata H, Imamura R, Nakashima Y, Kage M, Ueno T, Sata M, Prognostic significance of the F-box protein Skp2 expression in diffuse large B-cell lymphoma, American journal of hematology 73 (4) (2003) 230–235. [DOI] [PubMed] [Google Scholar]
- [198].Li Q, Murphy M, Ross J, Sheehan C, Carlson JA, Skp2 and p27kip1 expression in melanocytic nevi and melanoma: an inverse relationship, Journal of cutaneous pathology 31 (10) (2004) 633–642. [DOI] [PubMed] [Google Scholar]
- [199].Bloom J, Pagano M, Deregulated degradation of the cdk inhibitor p27 and malignant transformation, Seminars in cancer biology 13 (1) (2003) 41–47. [DOI] [PubMed] [Google Scholar]
- [200].Fujita T, Liu W, Doihara H, Date H, Wan Y, Dissection of the APCCdh1-Skp2 cascade in breast cancer, Clinical cancer research 14 (7) (2008) 1966–1975. [DOI] [PubMed] [Google Scholar]
- [201].Ji P, Jiang H, Rekhtman K, Bloom J, Ichetovkin M, Pagano M, Zhu L, An Rb-Skp2-p27 pathway mediates acute cell cycle inhibition by Rb and is retained in a partial-penetrance Rb mutant, Molecular cell 16 (1) (2004) 47–58. [DOI] [PubMed] [Google Scholar]
- [202].Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, Beach D, Serrano M, Tumour biology: senescence in premalignant tumours, Nature 436 (7051) (2005) 642. [DOI] [PubMed] [Google Scholar]
- [203].Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T, Restoration of p53 function leads to tumour regression in vivo, Nature 445 (7128) (2007) 661–665. [DOI] [PubMed] [Google Scholar]
- [204].Gstaiger M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J, Krek W, Skp2 is oncogenic and overexpressed in human cancers, Proceedings of the National Academy of Sciences of the United States of America 98 (9) (2001) 5043–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW, Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a, Cell 88 (5) (1997) 593–602. [DOI] [PubMed] [Google Scholar]
- [206].Yokoi S, Yasui K, Iizasa T, Takahashi T, Fujisawa T, Inazawa J, Down-regulation of SKP2 induces apoptosis in lung-cancer cells, Cancer science 94 (4) (2003) 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Wang X, Gorospe M, Huang Y, Holbrook NJ, p27Kip1 overexpression causes apoptotic death of mammalian cells, Oncogene 15 (24) (1997) 2991–2997. [DOI] [PubMed] [Google Scholar]
- [208].Kudo Y, Takata T, Ogawa I, Kaneda T, Sato S, Takekoshi T, Zhao M, Miyauchi M, Nikai H, p27Kip1 accumulation by inhibition of proteasome function induces apoptosis in oral squamous cell carcinoma cells, Clinical cancer research 6 (3) (2000) 916–923. [PubMed] [Google Scholar]
- [209].Tosco P, La Terra Maggiore GM, Forni P, Berrone S, Chiusa L, Garzino-Demo P, Correlation between Skp2 expression and nodal metastasis in stage I and II oral squamous cell carcinomas, Oral diseases 17 (1) (2011) 102–108. [DOI] [PubMed] [Google Scholar]
- [210].Sui L, Dong Y, Watanabe Y, Yamaguchi F, Sugimoto K, Tokuda M, Clinical significance of Skp2 expression, alone and combined with Jab1 and p27 in epithelial ovarian tumors, Oncology reports 15 (4) (2006) 765–771. [PubMed] [Google Scholar]
- [211].Zhang Y, Zvi YS, Batko B, Zaphiros N, O’Donnell EF, Wang J, Sato K, Yang R, Geller DS, Koirala P, Zhang W, Du X, Piperdi S, Liu Y, Zheng D, Down-regulation of Skp2 expression inhibits invasion and lung metastasis in osteosarcoma, Scientific Reports 8 (1) (2018) 14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Wang XC, Wu YP, Ye B, Lin DC, Feng YB, Zhang ZQ, Xu X, Han YL, Cai Y, Dong JT, Zhan QM, Wu M, Wang MR, Suppression of anoikis by SKP2 amplification and overexpression promotes metastasis of esophageal squamous cell carcinoma, Molecular cancer research 7 (1) (2009) 12–22. [DOI] [PubMed] [Google Scholar]
- [213].Celia-Terrassa T, Kang Y, Distinctive properties of metastasis-initiating cells, Genes & development 30 (8) (2016) 892–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Koppenol WH, Bounds PL, Dang CV, Otto Warburg’s contributions to current concepts of cancer metabolism, Nature reviews, Cancer 11 (5) (2011) 325–337. [DOI] [PubMed] [Google Scholar]
- [215].Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB, Akt stimulates aerobic glycolysis in cancer cells, Cancer research 64 (11) (2004) 3892–3899. [DOI] [PubMed] [Google Scholar]
- [216].Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, Stagg LJ, Ladbury JE, Cai Z, Xu D, Logothetis CJ, Hung MC, Zhang S, Lin HK, Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression, Cell 154 (3) (2013) 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Kuczynski EA, Sargent DJ, Grothey A, Kerbel RS, Drug rechallenge and treatment beyond progression–implications for drug resistance, Nature reviews, Clinical oncology 10 (10) (2013) 571–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Davidovich S, Ben-Izhak O, Shapira M, Futerman B, Hershko DD, Over-expression of Skp2 is associated with resistance to preoperative doxorubicin-based chemotherapy in primary breast cancer, Breast cancer research 10 (4) (2008) R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Tian YF, Chen TJ, Lin CY, Chen LT, Lin LC, Hsing CH, Lee SW, Sheu MJ, Lee HH, Shiue YL, Huang HY, Pan HY, Li CF, Chen SH, SKP2 overexpression is associated with a poor prognosis of rectal cancer treated with chemoradiotherapy and represents a therapeutic target with high potential, Tumour biology 34 (2) (2013) 1107–1117. [DOI] [PubMed] [Google Scholar]
- [220].Lytle NK, Barber AG, Reya T, Stem cell fate in cancer growth, progression and therapy resistance, Nature reviews, Cancer 18 (11) (2018) 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Dean M, Fojo T, Bates S, Tumour stem cells and drug resistance, Nature reviews, Cancer 5 (4) (2005) 275–284. [DOI] [PubMed] [Google Scholar]
- [222].Wang J, Huang Y, Guan Z, Zhang JL, Su HK, Zhang W, Yue CF, Yan M, Guan S, Liu QQ, E3-ligase Skp2 predicts poor prognosis and maintains cancer stem cell pool in nasopharyngeal carcinoma, Oncotarget 5 (14) (2014) 5591–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Lu W, Liu S, Li B, Xie Y, Izban MG, Ballard BR, Sathyanarayana SA, Adunyah SE, Matusik RJ, Chen Z, SKP2 loss destabilizes EZH2 by promoting TRAF6-mediated ubiquitination to suppress prostate cancer, Oncogene 36 (10) (2017) 1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Santos-Barriopedro I, Bosch-Presegue L, Marazuela-Duque A, de la Torre C, Colomer C, Vazquez BN, Fuhrmann T, Martinez-Pastor B, Lu W, Braun T, Bober E, Jenuwein T, Serrano L, Esteller M, Chen Z, Barcelo-Batllori S, Mostoslavsky R, Espinosa L, Vaquero A, SIRT6-dependent cysteine monoubiquitination in the PRE-SET domain of Suv39h1 regulates the NF-kappaB pathway, Nature Communications 9 (1) (2018) 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Meng J, Ding Y, Shen A, Yan M, He F, Ji H, Zou L, Liu Y, Wang Y, Lu X, Wang H, Overexpression of PPARgamma can down-regulate Skp2 expression in MDA-MB-231 breast tumor cells, Molecular and cellular biochemistry 345 (1–2) (2010) 171–180. [DOI] [PubMed] [Google Scholar]
- [226].Koga H, Harada M, Ohtsubo M, Shishido S, Kumemura H, Hanada S, Taniguchi E, Yamashita K, Kumashiro R, Ueno T, Sata M, Troglitazone induces p27Kip1-associated cell-cycle arrest through down-regulating Skp2 in human hepatoma cells, Hepatology (Baltimore, Md.) 37 (5) (2003) 1086–1096. [DOI] [PubMed] [Google Scholar]
- [227].Motomura W, Takahashi N, Nagamine M, Sawamukai M, Tanno S, Kohgo Y, Okumura T, Growth arrest by troglitazone is mediated by p27Kip1 accumulation, which results from dual inhibition of proteasome activity and Skp2 expression in human hepatocellular carcinoma cells, International journal of cancer 108 (1) (2004) 41–46. [DOI] [PubMed] [Google Scholar]
- [228].Ye J, Yin L, Xie P, Wu J, Huang J, Zhou G, Xu H, Lu E, He X, Antiproliferative effects and molecular mechanisms of troglitazone in human cervical cancer in vitro, OncoTargets and therapy 8 (2015) 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Rico-Bautista E, Yang CC, Lu L, Roth GP, Wolf DA, Chemical genetics approach to restoring p27Kip1 reveals novel compounds with antiproliferative activity in prostate cancer cells, BMC biology 8 (2010) 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Wang ST, Ho HJ, Lin JT, Shieh JJ, Wu CY, Simvastatin-induced cell cycle arrest through inhibition of STAT3/SKP2 axis and activation of AMPK to promote p27 and p21 accumulation in hepatocellular carcinoma cells, Cell death & disease 8 (2) (2017) e2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Chen HY, He LJ, Li SQ, Zhang YJ, Huang JH, Qin HX, Wang JL, Li QY, Yang DL, A Derivate of Benzimidazole-Isoquinolinone Induces SKP2 Transcriptional Inhibition to Exert Anti-Tumor Activity in Glioblastoma Cells, Molecules (Basel, Switzerland) 24 (15) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Jung D, Khurana A, Roy D, Kalogera E, Bakkum-Gamez J, Chien J, Shridhar V, Quinacrine upregulates p21/p27 independent of p53 through autophagy-mediated downregulation of p62-Skp2 axis in ovarian cancer, Scientific reports 8 (1) (2018) 2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Lin HP, Lin CY, Huo C, Hsiao PH, Su LC, Jiang SS, Chan TM, Chang CH, Chen LT, Kung HJ, Wang HD, Chuu CP, Caffeic acid phenethyl ester induced cell cycle arrest and growth inhibition in androgen-independent prostate cancer cells via regulation of Skp2, p53, p21Cip1 and p27Kip1, Oncotarget 6 (9) (2015) 6684–6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Chung YK, Chi-Hung Or R, Lu CH, Ouyang WT, Yang SY, Chang CC, Sulforaphane down-regulates SKP2 to stabilize p27(KIP1) for inducing antiproliferation in human colon adenocarcinoma cells, Journal of bioscience and bioengineering 119 (1) (2015) 35–42. [DOI] [PubMed] [Google Scholar]
- [235].Huang HC, Lin CL, Lin JK, 1,2,3,4,6-penta-O-galloyl-beta-D-glucose, quercetin, curcumin and lycopene induce cell-cycle arrest in MDA-MB-231 and BT474 cells through downregulation of Skp2 protein, Journal of agricultural and food chemistry 59 (12) (2011) 6765–6775. [DOI] [PubMed] [Google Scholar]
- [236].Su J, Zhou X, Wang L, Yin X, Wang Z, Curcumin inhibits cell growth and invasion and induces apoptosis through down-regulation of Skp2 in pancreatic cancer cells, American journal of cancer research 6 (9) (2016) 1949–1962. [PMC free article] [PubMed] [Google Scholar]
- [237].Hsu JD, Kao SH, Ou TT, Chen YJ, Li YJ, Wang CJ, Gallic acid induces G2/M phase arrest of breast cancer cell MCF-7 through stabilization of p27(Kip1) attributed to disruption of p27(Kip1)/Skp2 complex, Journal of agricultural and food chemistry 59 (5) (2011) 1996–2003. [DOI] [PubMed] [Google Scholar]
- [238].Huang HC, Way TD, Lin CL, Lin JK, EGCG stabilizes p27kip1 in E2-stimulated MCF-7 cells through down-regulation of the Skp2 protein, Endocrinology 149 (12) (2008) 5972–5983. [DOI] [PubMed] [Google Scholar]
- [239].Roy S, Kaur M, Agarwal C, Tecklenburg M, Sclafani RA, Agarwal R, p21 and p27 induction by silibinin is essential for its cell cycle arrest effect in prostate carcinoma cells, Molecular cancer therapeutics 6 (10) (2007) 2696–2707. [DOI] [PubMed] [Google Scholar]
- [240].Yang ES, Burnstein KL, Vitamin D inhibits G1 to S progression in LNCaP prostate cancer cells through p27Kip1 stabilization and Cdk2 mislocalization to the cytoplasm, The Journal of biological chemistry 278 (47) (2003) 46862–46868. [DOI] [PubMed] [Google Scholar]
- [241].Guruvayoorappan Siddikuzzaman C, Berlin Grace VM, All trans retinoic acid and cancer, Immunopharmacology and immunotoxicology 33 (2) (2011) 241–249. [DOI] [PubMed] [Google Scholar]
- [242].Dow R, Hendley J, Pirkmaier A, Musgrove EA, Germain D, Retinoic acid-mediated growth arrest requires ubiquitylation and degradation of the F-box protein Skp2, The Journal of biological chemistry 276 (49) (2001) 45945–45951. [DOI] [PubMed] [Google Scholar]
- [243].Chan CH, Morrow JK, Zhang S, Lin HK, Skp2: a dream target in the coming age of cancer therapy, Cell cycle (Georgetown, Tex.) 13 (5) (2014) 679–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, Webb DR, Mercurio F, Nakayama KI, Nakayama K, Orlowski RZ, Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy, Blood 111 (9) (2008) 4690–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Ungermannova D, Lee J, Zhang G, Dallmann HG, McHenry CS, Liu X, High-throughput screening AlphaScreen assay for identification of small-molecule inhibitors of ubiquitin E3 ligase SCFSkp2-Cks1, Journal of biomolecular screening 18 (8) (2013) 910–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Singh R, Sran A, Carroll DC, Huang J, Tsvetkov L, Zhou X, Sheung J, McLaughlin J, Issakani SD, Payan DG, Shaw SJ, Developing structure-activity relationships from an HTS hit for inhibition of the Cks1-Skp2 protein-protein interaction, Bioorganic & medicinal chemistry letters 25 (22) (2015) 5199–5202. [DOI] [PubMed] [Google Scholar]
- [247].Pavlides SC, Huang KT, Reid DA, Wu L, Blank SV, Mittal K, Guo L, Rothenberg E, Rueda B, Cardozo T, Gold LI, Inhibitors of SCF-Skp2/Cks1 E3 ligase block estrogen-induced growth stimulation and degradation of nuclear p27kip1: therapeutic potential for endometrial cancer, Endocrinology 154 (11) (2013) 4030–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Ooi LC, Watanabe N, Futamura Y, Sulaiman SF, Darah I, Osada H, Identification of small molecule inhibitors of p27(Kip1) ubiquitination by high-throughput screening, Cancer science 104 (11) (2013) 1461–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ, Specific small molecule inhibitors of Skp2-mediated p27 degradation, Chemistry & biology 19 (12) (2012) 1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Chen G, Li G, Increased Cul1 expression promotes melanoma cell proliferation through regulating p27 expression, International journal of oncology 37 (5) (2010) 1339–1344. [DOI] [PubMed] [Google Scholar]
- [251].Min KW, Kim DH, Do SI, Sohn JH, Chae SW, Pyo JS, Park CH, Oh YH, Jang KS, Kim HL, Kim M, Diagnostic and prognostic relevance of Cullin1 expression in invasive ductal carcinoma of the breast, Journal of clinical pathology 65 (10) (2012) 896–901. [DOI] [PubMed] [Google Scholar]
- [252].Chen L, Tweddle DA, p53, SKP2 and DKK3 as MYCN Target Genes and Their Potential Therapeutic Significance, Frontiers in Oncology 2 (2012) 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Guo J, Liu J, Wei W, Degrading proteins in animals: "PROTAC"tion goes in vivo, Cell Res. 29 (3) (2019) 179–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].You I, Erickson EC, Donovan KA, Eleuteri NA, Fischer ES, Gray NS, Toker A, Discovery of an AKT Degrader with Prolonged Inhibition of Downstream Signaling, Cell chemical biology 27 (1) (2020) 66–73.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Schatz DG, Ji Y, Recombination centres and the orchestration of V(D)J recombination, Nature reviews, Immunology 11 (4) (2011) 251–263. [DOI] [PubMed] [Google Scholar]
- [256].Zhang L, Reynolds TL, Shan X, Desiderio S, Coupling of V(D)J recombination to the cell cycle suppresses genomic instability and lymphoid tumorigenesis, Immunity 34 (2) (2011) 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Chen Z, Gu J, Immunoglobulin G expression in carcinomas and cancer cell lines, FASEB journal 21 (11) (2007) 2931–2938. [DOI] [PubMed] [Google Scholar]
- [258].Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE, Cyclin E-CDK2 is a regulator of p27Kip1, Genes & development 11 (11) (1997) 1464–1478. [DOI] [PubMed] [Google Scholar]
- [259].Koutsami M, Velimezi G, Kotsinas A, Evangelou K, Papavassiliou AG, Kittas C, Gorgoulis VG, Is exclusive Skp2 targeting always beneficial in cancer therapy? Blood 112 (12) (2008) 4777–4779. [DOI] [PubMed] [Google Scholar]
- [260].Ma Y, Fiering S, Black C, Liu X, Yuan Z, Memoli VA, Robbins DJ, Bentley HA, Tsongalis GJ, Demidenko E, Freemantle SJ, Dmitrovsky E, Transgenic cyclin E triggers dysplasia and multiple pulmonary adenocarcinomas, Proceedings of the National Academy of Sciences of the United States of America 104 (10) (2007) 4089–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Muller-Tidow C, Metzger R, Kugler K, Diederichs S, Idos G, Thomas M, Dockhorn-Dworniczak B, Schneider PM, Koeffler HP, Berdel WE, Serve H, Cyclin E is the only cyclin-dependent kinase 2-associated cyclin that predicts metastasis and survival in early stage non-small cell lung cancer, Cancer research 61 (2) (2001) 647–653. [PubMed] [Google Scholar]
- [262].Dong JT, Chromosomal deletions and tumor suppressor genes in prostate cancer, Cancer metastasis reviews 20 (3–4) (2001) 173–193. [DOI] [PubMed] [Google Scholar]
- [263].Chen DS, Mellman I, Elements of cancer immunity and the cancer-immune set point, Nature 541 (7637) (2017) 321–330. [DOI] [PubMed] [Google Scholar]
- [264].Choi Y, Bowman JW, Jung JU, Autophagy during viral infection - a double-edged sword, Nature reviews, Microbiology 16 (6) (2018) 341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]


