Abstract
Aims
Desmoplastic infantile astrocytomas and gangliogliomas (DIA/DIGs) are rare brain tumours of infancy. A distinctive feature of their histopathology is a combination of low-grade and high-grade features. Most DIA/DIGs can be surgically resected and have a good prognosis. However, high-grade features often dominate recurrent tumours, some of which have a poor outcome. In this study, we test the hypothesis that low-grade and high-grade areas in DIA/DIGs have distinct molecular characteristics.
Methods
Tissue samples from microdissected low-grade and high-grade areas in 12 DIA/DIGs were analysed by DNA methylation profiling, whole exome sequencing, RNA sequencing and immunohistochemistry to search for potential differences at multiple molecular levels.
Results
Copy number variants among tumours and between the two morphologically distinct areas were infrequent. No recurrent genetic alterations were identified across the tumour series, and high-grade areas did not have additional genetic alterations to explain their distinct morphology or biological behaviour. However, high-grade areas showed relative hypomethylation in genes downstream of the transcription factors SOX9 and LEF1 and evidence of a core SOX9 transcription network alongside activation of the BMP, WNT, and MAPK signalling pathways.
Conclusions
This study contributes to our knowledge of molecular genetic alterations in DIA/DIGs, uncovers molecular differences between the two distinct cell populations in these tumours, and suggests potential therapeutic targets among the more proliferative cell population in DIA/DIGs.
Keywords: Desmoplastic infantile astrocytoma / ganglioglioma, high-grade, low-grade, SOX9, BMP, WNT, MAPK
Graphical Abstract
Introduction
Desmoplastic infantile astrocytoma (DIA) and desmoplastic infantile ganglioglioma (DIG) are rare clinically and histopathologically related tumours of the cerebral hemispheres, which generally present in infancy [1]. A peripheral location and involvement of the meninges are typical [2]. The histopathological features of DIA and DIG are the same, except that DIG has a neoplastic ganglion cell component in addition to the astrocytic component of a DIA. A characteristic finding in DIA/DIGs is a combination of low-grade and high-grade histopathological features. High-grade areas generally manifest as relatively circumscribed foci within larger low-grade areas and contain poorly differentiated cells, sometimes with an embryonal cytology, and mitotic figures. Microvascular proliferation and necrosis can also be present. Despite the presence of high-grade areas, DIA/DIGs are classified as CNS WHO grade 1, reflecting the good prognosis of most cases [3–6]. However, not all DIA/DIGs have a favourable outcome; relapse with more prominent high-grade histopathological features and metastatic disease across the neuraxis suggest that some are not cured by surgical resection alone, which is facilitated by the circumscribed nature and peripheral location of these tumours [7–12].
The rarity of DIA/DIGs has precluded more than limited studies of their molecular alterations [13–17], and no study has determined whether low-grade and high-grade areas differ with respect to their molecular profiles. In the present study, we have analysed the molecular characteristics of 12 DIA/DIGs, including separate analyses of low-grade and high-grade areas, using genome-wide DNA methylation profiling, analysis of copy number variation (CNV), whole exome sequencing (WES), transcriptome analysis (RNA-seq), and immunohistochemistry. Our data contribute to the knowledge of molecular alterations in these tumours, indicating a range of MAPK pathway alterations, and show that activation of the BMP and WNT signalling pathways distinguish high-grade from low-grade areas.
Materials and Methods
Study cohort, histopathological evaluation, and microdissection
Twelve DIA/DIGs with low-grade and high-grade areas were identified through a review of the clinical database in the Department of Pathology, St. Jude Children’s Research Hospital. All were cerebral hemispheric tumours. The median age of the patients at presentation was 5.5 months (range 1 – 14 months). High-grade areas were defined by a mitotic count of >4/10 high-power fields (1.96 mm2). Such areas also showed an increased cell density (Fig. 1). Microvascular proliferation and necrosis were also frequent findings.
Fig. 1. Study design.
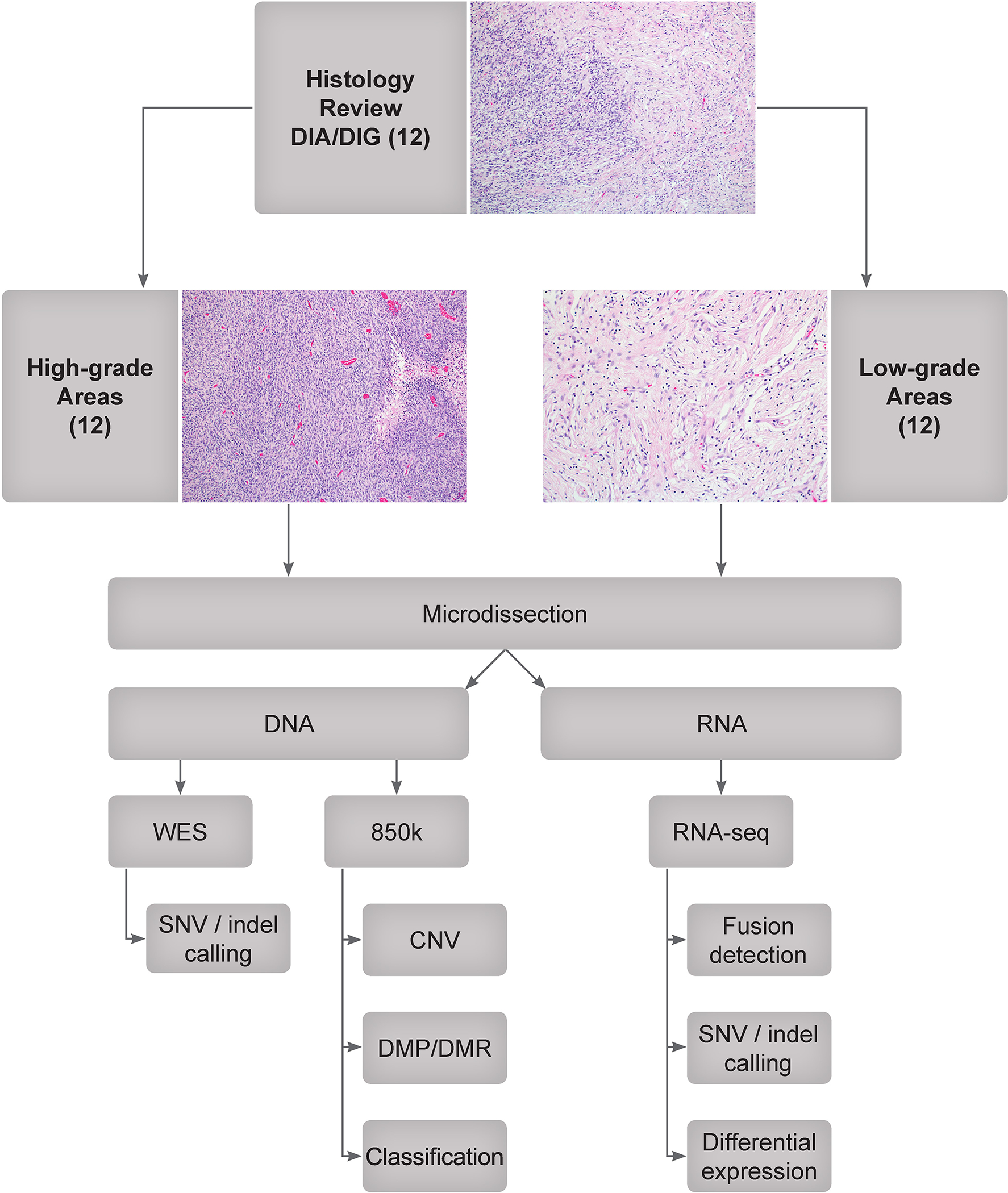
Twelve DIA/DIG tumours were identified by a retrospective data search and pathological review. High-grade and low-grade areas were identified and microdissected. DNA was extracted for whole exome sequencing (WES) and EPIC methylation array (850k) analysis, and RNA was extracted for total RNA sequencing (RNA-seq). CNV: copy number variation; DMP: differentially methylated probe; DMR: differentially methylated region; indel: insertion or deletion; SNV: single-nucleotide variation.
Microdissection of high-grade areas was accomplished by laying unstained sections upon a printout of a marked H&E-section and scraping the area of interest with sterile razor blades or 30-gauge needles under a stereomicroscope with 40× magnification.
Whole exome sequencing (WES)
Genomic DNA was extracted from microdissected formalin-fixed paraffin-embedded (FFPE) tissue using a QIAamp DNA FFPE Tissue Kit (Qiagen). At least 250 ng of genomic DNA was used for each sample. DNA quality was assessed on a 4200 TapeStation (Agilent). Genomic DNA libraries were generated using the SureSelectXT Kit (Agilent Technologies), followed by exon enrichment using the SureSelectXT Human All Exon V7 bait set (Agilent Technologies). The resulting exon-enriched libraries were subjected to paired-end, 100-cycle sequencing performed on a HiSeq 4000 (Illumina). Sequencing results were analysed using an institutionally established pipeline for alignment and calling of single nucleotide variants (SNVs) and insertions or deletions (indels), which were annotated and ranked by putative pathogenicity using a workflow named “medal ceremony” and subsequently manually reviewed [18–20].
Transcriptome sequencing (RNA-seq) and differential expression analysis
A PureLink FFPE Total RNA Isolation Kit (Thermo Fisher Scientific) was used for total RNA extraction from microdissected FFPE tissue. Purified RNA was quantified on a Qubit 1.27 fluorometer (Thermo Fisher Scientific) using Qubit™ RNA BR Assay Kit (Thermo Fisher Scientific). Total RNA sequencing was performed using the Illumina Total Stranded RNA protocol with at least 400 ng of total RNA. The quality of the starting materials was checked with the RNA 6000 Nano Assay on a 2100 Bioanalyzer (Agilent) or the RNA Pico Sensitivity Assay on a LabChip GX Touch (PerkinElmer). Libraries were prepared using the TruSeq Stranded Total RNA Sample Prep Kit (Illumina), followed by library quantification through qPCR using Quant-iT™ PicoGreen dsDNA Assay Kits (Thermo Fisher Scientific) or KAPA Library Quantification Kits for Illumina platforms (KAPA Biosystems), and through low pass sequencing on a MiSeq Nano v2 (Illumina). All sequencing data were generated after 100 cycles of paired end runs on an Illumina HiSeq 2500 or HiSeq 4000. The RNA-seq data were aligned to the human reference genome (build hg19), as previously described [19, 21]. Sequencing results were analysed in the same manner as WES [18–20].
Differentially expressed genes between low-grade and high-grade areas in DIA/DIGs were analysed using edgeR [22, 23] in Bioconductor 3.13 (http://bioconductor.org/), NetBID 2.0.3 [24], and SJARACNe 0.2.0 [25] with the default parameters and then queried against MSigDB [26–29]. Briefly, the read counts of gene expression data were preprocessed, normalized, and filtered to remove genes with very low expression values (bottom 5%) in more than 90% samples using edgeR 3.34.0 [22, 23] in R 4.1.0. Bayesian inference approach [24] and SJARACNe-based network reconstruction [25] were then used to infer drivers (transcription factors or signalling factors) from the transcriptomics data by calculating the activity of drivers and gene sets in Ubuntu 18.04 with Python 3.6.1. Differentially expressed genes and differentially activated drivers were then queried against MSigDB v7.4 [26–29], with the aim of finding functional or pathway enrichment.
Genome-wide DNA methylation profiling and copy number variation analysis
Genomic DNA was used for genome-wide methylation profiling and CNV analysis by the Illumina Infinium MethylationEPIC platform [21]. For comparison, publicly available well-characterized reference methylation profiles of brain tumours were downloaded from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). Analysis of methylation profiles, including normalization, filtering, t-distributed stochastic neighbor embedding, identification of differentially methylated probes and regions, and CNV analysis was performed in R 4.1.0 using ChAMP 2.22.0 [30], bumphunter [31], minfi 1.38.0 [32], limma 3.48.0 [33], conumee 1.26.0, and Rtsne 0.15 packages in Bioconductor 3.13 (http://bioconductor.org/) as previously described [21, 34–38]. Genes within differentially methylated regions were queried against MSigDB v7.4 [26–29] to identify functional or pathway enrichment.
Immunohistochemistry
The following antibodies were used on 4 μm FFPE tissue sections: SOX9 (Abcam, ab185966, 1:2000), MEIS1 (Abcam, ab19867, 1:2000), PBX1 (Novus Biologicals, NBP1–85803, 1:500), LEF1 (Abcam, ab137872, 1:100), phospho-ERK1/2 (Cell Signaling, 4370, 1:200), and phospho-Smad1/5/8 (Sigma-Aldrich, AB3848, 1:50).
Results
Clinical features and histopathology review
Twelve tumours containing both low-grade and high-grade areas were identified for this study (Fig. 1). All tumours were large cerebral masses with solid and cystic areas. Patient clinical parameters are listed in Supplementary Table 1. As expected for tumours with a tendency to occur in infancy, the median age of the cohort is 5.5 months (range 1 – 14 months). There is no gender predilection (male-to-female ratio 1:1). Upon histopathological review, all tumours showed the typical morphological features of DIA/DIGs. All contained low-grade areas, which were characterized by elongated tumour cells at relatively low density and pericellular collagen deposition, and high-grade areas, with compact poorly differentiated tumour cells. Endothelial proliferation and areas of necrosis were frequently present.
Matched methylation and CNV profiles and differentially methylated regions across low-grade and high-grade areas in DIA/DIGs
DNA methylation profiling was used to demonstrate that low-grade and high-grade areas from our series of DIA/DIGs formed a single cluster alongside reference cases of DIA/DIG in a t-SNE plot (Fig. 2a), supporting the histopathological classification of the tumour series. Using DNA methylation profiling, we demonstrated that, although high-grade areas appear to have an increased frequency of focal chromosomal losses across the genome, specific alterations only occur in isolated tumours (Fig. 2b). There are no large-scale chromosomal gains or losses, and no single CNV occurs in more than 50% of tumours. Analysis of differentially methylated regions by Bump Hunter [31] and gene set enrichment through MSigDB v7.4 [26–29] identified enrichment of relatively hypomethylated genes downstream of transcription factors SOX9, LEF1, NFAT, NF1, CHX10, ZNF92, ZNF563, and SMN1/SMN2 in high-grade areas (Fig. 2c; Suppl. Fig. 1).
Fig. 2. Methylation profiling, copy number variations, and differentially methylated genes of low-grade and high-grade areas in DIA/DIGs.
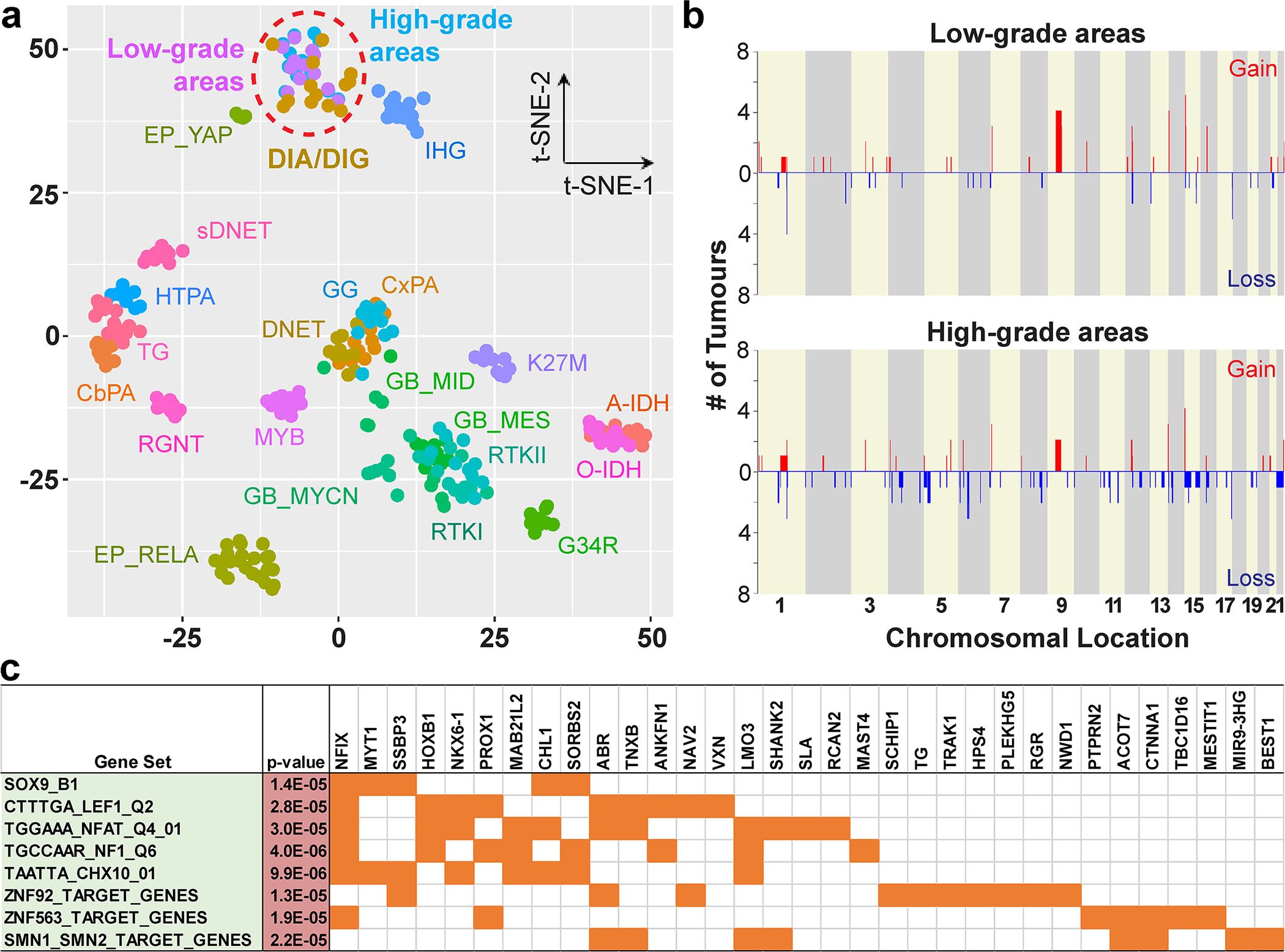
(a) A t-SNE plot shows the methylation profiles of low-grade and high-grade areas in our series of DIA/DIGs and those of reference tumours. A-IDH: IDH1-mutant diffuse astrocytoma; CbPA: cerebellar pilocytic astrocytoma; CxPA: cerebral hemispheric pilocytic astrocytoma; DNET: dysembryoplastic neuroepithelial tumour; EP: ependymoma; GB: glioblastoma; G34R: H3F3A G34R-mutant diffuse glioma; GG: ganglioglioma; HTPA: hypothalamic pilocytic astrocytoma; K27M: H3 K27M-mutant diffuse midline glioma; MYB: MYB-altered diffuse glioma; O-IDH: IDH1-mutant and 1p/19q-codeleted oligodendroglioma; RGNT: rosette-forming glioneuronal tumour; RTK I, RTK II, MES, MID, and MYCN: specific molecular groups of glioblastoma; sDNET: septal dysembryoplastic neuroepithelial tumour / myxoid glioneuronal tumour; TG: tectal glioma. (b) CNV plots of low-grade and high-grade areas in DIA/DIGs. X-axis, chromosomal location; Y-axis, number of tumours containing the gain or loss. (c) Differentially hypomethylated regions in the high-grade areas are enriched for genes downstream of specific transcription factors, including SOX9 and LEF1.
No recurrent genetic alterations in DIA/DIG
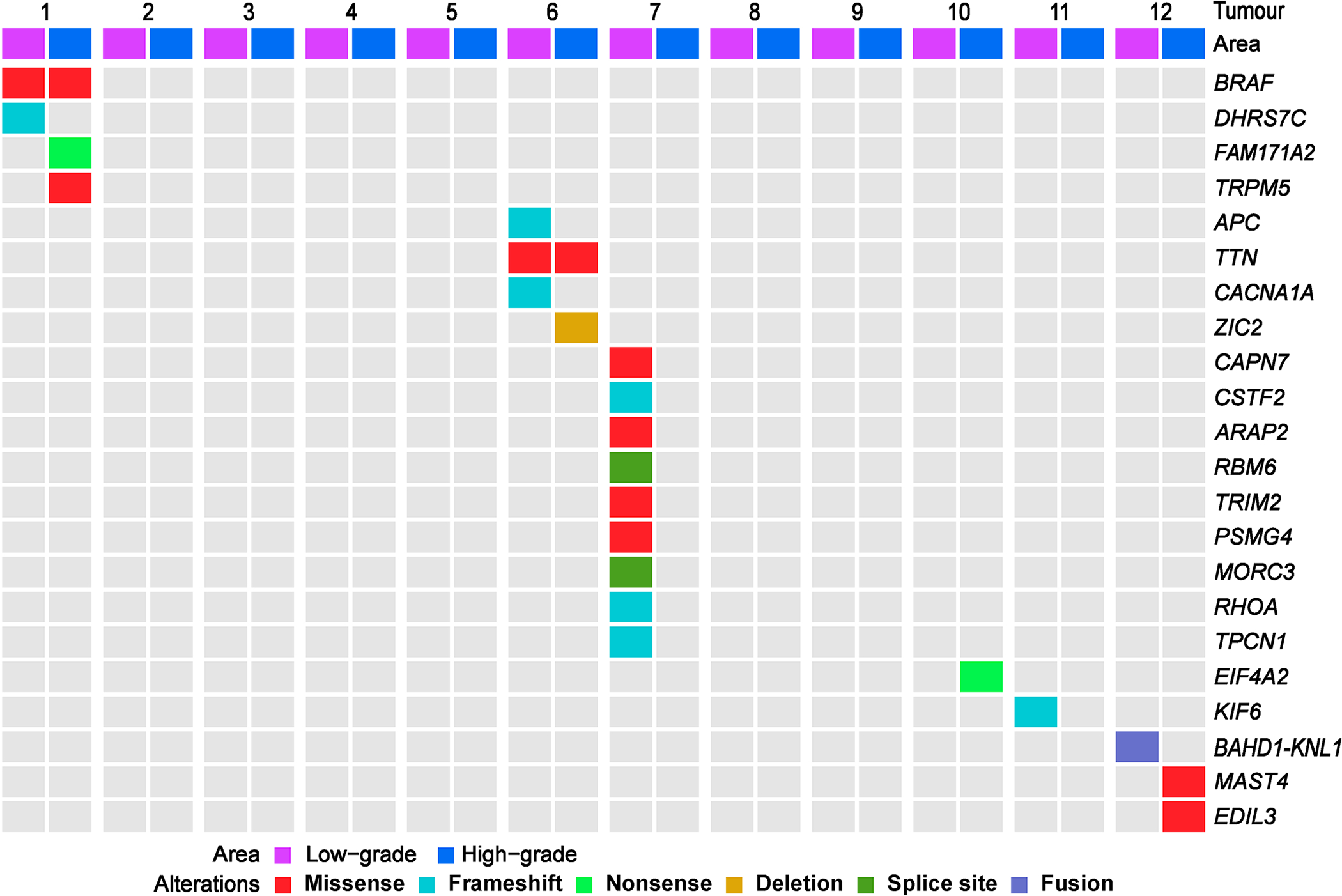
Using WES and RNA-seq, we found no recurrent genetic alterations in our series of DIA/DIGs (Fig. 3). Identified alterations are all subclonal; variant allele frequencies are 3–30% (median 10.5%). All alterations, except two, are not shared between low-grade and high-grade areas. From these data, we suspect that multiple subclones co-exist in DIA/DIGs, especially in low-grade areas from tumour 7. No genetic alterations were identified in six (50%) tumours. Among identified alterations, the BRAF G469A, ARAP2 R1387C, and EDIL3 T343M variants are predicted to be pathogenic or likely pathogenic [39]. The DHRS7C Y23fs, FAM171A2 S476*, APC C1578fs, CACNA1A F453_A454fs, CSTF2 L172fs, RHOA A132fs, EIF4A2 exon 1–8 truncation, KIF6 H545fs, and RBM6 exon 7 and MORC3 exon 4 splice region variants are predicted to generate non-functional proteins. The BAHD1-KNL1 (exon 1 - exon 2) fusion is a novel alteration. The TRPM5 A658T, TTN G6433W, ZIC2 H239del, CAPN7 V739I, TRIM2 D714N, PSMG4 A144T, TPCN1 L842_K843fs, and MAST4 S59L variants are of uncertain significance in brain tumours.
Fig. 3. No recurrent alterations were identified by WES and RNA-seq in low-grade and high-grade areas of DIA/DIGs.
SOX9 core transcription network activation in high-grade areas of DIA/DIG
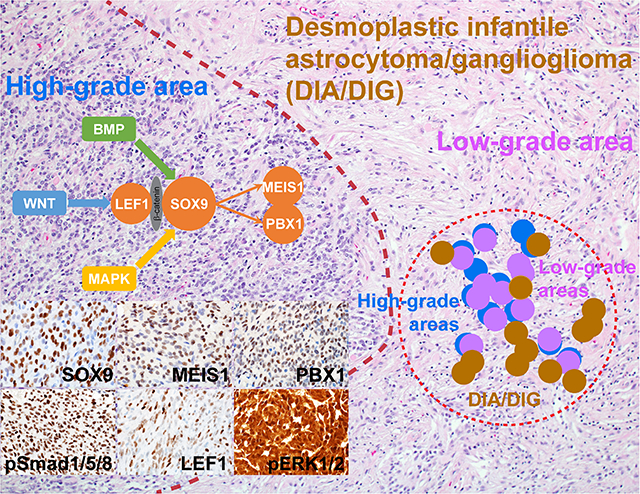
Given that there was enrichment of differentially hypomethylated genes downstream of specific transcription factors in high-grade areas in DIA/DIG, we looked for patterns of differentially expressed genes. Utilizing our transcriptome data and bioinformatics tools, including NetBID [24], SJARACNe [25], and MSigDB [26–29], we identified a core transcription network centred on SOX9 in high-grade areas (Fig. 4). Among eight transcription factors up-regulated in high-grade areas compared to low-grade areas, MEIS1 and PBX1 are co-regulated with and regulated by SOX9 (Fig. 4a). In addition to the SOX9 core transcription network, our differential gene expression analysis identified evidence of BMP and WNT pathway activation in high-grade areas (Fig. 4b), as well as MAPK pathway activation, which has been reported before [14].
Fig. 4. SOX9 core transcription network and BMP, WNT, and MAPK pathway activation in high-grade areas in DIA/DIGs.
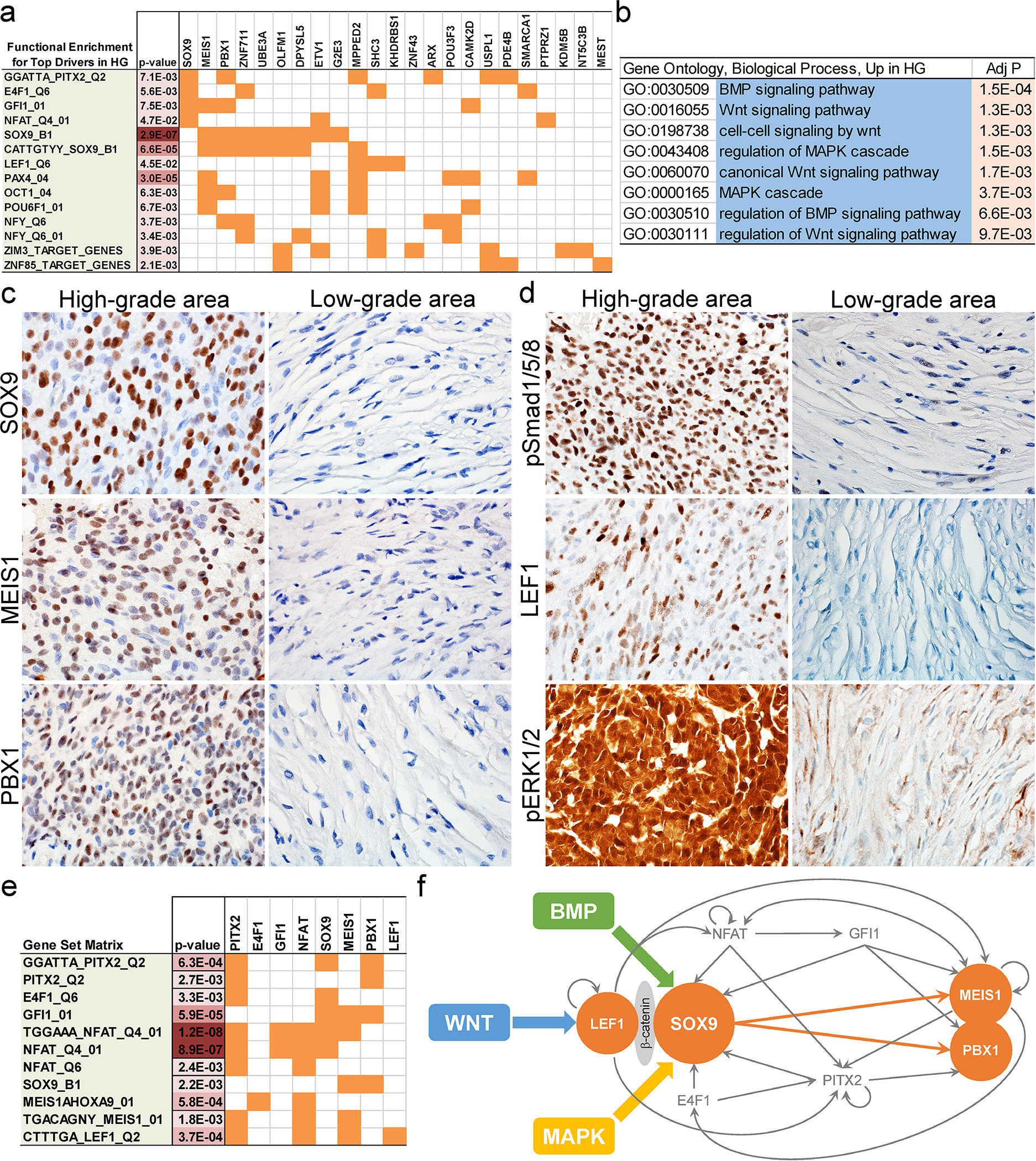
(a) Functional enrichment of differentially expressed genes in high-grade areas in DIA/DIGs. (b) Enrichment of the BMP, WNT, and MAPK pathway genes in high-grade areas in DIA/DIGs. (c) Immunohistochemistry of SOX9, MEIS1, and PBX1 in high-grade and low-grade areas in DIA/DIGs. (d) Enrichment of nuclear phospho-Smad1/5/8, a marker for BMP pathway activation, nuclear LEF1, a central component of the WNT pathway, and nuclear phospho-ERK1/2, a marker for MAPK pathway activation in high-grade areas in DIA/DIGs. (e) Enrichment of reciprocal transcription factor binding sites in genes upstream and downstream of SOX9. (f) Proposed model of an interconnected and self-sustaining SOX9 core transcription network in high-grade areas in DIA/DIGs, in which the BMP, WNT, and MAPK pathways converge on SOX9 through indirect or direct protein-protein interactions.
Focally increased expression of SOX9, MEIS1, and PBX1 was confirmed at the protein level in high-grade areas by immunohistochemistry (Fig. 4c; Suppl. Fig. 2). Nuclear phospho-Smad1/5/8, a marker for BMP pathway activation, nuclear localization of LEF1, a central component of the WNT pathway, and nuclear phospho-ERK1/2, a marker for MAPK pathway activation, are localized to high-grade areas, supporting the findings of our methylation and transcriptome analyses (Fig. 4d; Suppl. Fig. 2). Given the established roles of SOX9 in the MAPK [40–42], WNT [43–47], and BMP [47–51] pathways in tissue development and neoplasia, our findings suggest that activated MAPK, WNT, and BMP pathways in high-grade areas in DIA/DIGs likely converge on SOX9.
Further analysis revealed that transcription factors upstream (PITX2, E4F1, GFI1, and NFAT) and downstream (MEIS1 and PBX1) of SOX9 frequently contain reciprocal transcription factor binding sites 4 kb upstream or downstream of their transcription start sites, supporting the formation of interconnected and self-sustaining positive feedback loops (Fig. 4e). SOX9 has been shown to interact with β-catenin [52], which in turn binds to LEF1 [53]. MEIS1 and PBX1 form a functional heterodimer [54]. Our transcriptome data support the findings of the differential methylation analysis and allow construction of a signalling pathway and transcriptional network model in the cells of high-grade areas, as shown in Fig. 4f.
Discussion
DIA/DIGs are rare cerebral tumours of infancy [1]. Generally, they have a good prognosis, and this is related to their circumscription and peripheral location, which facilitate surgical resection [2–6]. Histopathologically, DIA/DIGs are distinctive biphasic tumours. Their desmoplastic low-grade areas contain bland cells with astrocytic or ganglion cell (DIG) differentiation, and their mitotically active high-grade areas combine cells with astrocytic or embryonal features. When relapse occurs, the histopathology is often dominated by high-grade features [10, 17, 55, 56], suggesting that high-grade areas can influence the biological behaviour and prognosis of these tumours. To understand the molecular pathology of these distinctive biphasic tumours, we performed DNA methylation profiling, including CNV analysis, WES, and RNA-seq on microdissected tissue from low-grade and high-grade areas in a series of 12 DIA/DIGs with typical clinical and histopathological features.
In our study, multiple approaches to the detection of genetic and epigenetic differences between low-grade and high-grade areas in DIA/DIGs initially yielded few meaningful results. Methylation profiling showed that all 12 tumours do form a single cluster with other DIA/DIGs from an established reference series and that profiles from high-grade and low-grade areas also cluster together. We detected no recurrent genetic alteration by WES, RNA-seq, or CNV analysis. This result is consistent with studies that report only rare instances of genetic alterations in DIA/DIGs [13–17] and contrasts with the situation for other paediatric low-grade glioneuronal tumours, which tend to show recurrent genetic alterations [21, 36, 37, 57]. Furthermore, high-grade areas in our series of DIA/DIGs did not contain additional alterations to explain their pathological progression. Homozygous CDKN2A deletion, which is seen in other low-grade neuroepithelial tumours with a tendency for anaplastic progression [58, 59], was not identified in our series of DIA/DIGs. It is possible that methodologies with higher resolution, such as whole genome bisulfite sequencing and whole genome sequencing, which require ample amounts of frozen tissue, might be needed to investigate the molecular pathology of DIA/DIG at a deeper level. Alternatively, the determinants of biological differences between high-grade and low-grade areas in DIA/DIGs might be their distinct intrinsic transcription and protein expression profiles and extrinsic signalling stimuli from the local microenvironment.
Our analysis of differential methylation and gene and protein expression between high-grade and low-grade areas does provide some clues to their distinct histopathological features. We have identified a SOX9 core transcription network in high-grade areas, which is associated with activation of the MAPK, WNT, and BMP pathways. The MAPK, WNT, and BMP pathways have been shown to play roles in gliomagenesis; activation of the MAPK pathway is common in paediatric low-grade gliomas, glioneuronal tumours, and high-grade gliomas [57, 60, 61]. In addition to its role in WNT-activated medulloblastoma, the WNT pathway has been implicated in the progression of glioblastoma and maintenance of glioma stem cells [62–64]. Also, published data support activation of the BMP pathway in diffuse midline glioma [65, 66] and glioma stem cells [67].
Data suggest that activated MAPK, WNT, and BMP pathways likely converge on SOX9 in high-grade areas in DIA/DIGs. The role of SOX9 in the MAPK [40–42], WNT [43–47], and BMP [47–51] pathways has been studied extensively in the context of organ and tissue development and neoplasia. SOX9 is downstream of receptor tyrosine kinase and MAPK signalling [40, 41], and activation of ERK1/2 is required for expression of SOX9 [40, 42, 49]. SOX9 expression requires an active β-catenin–TCF/LEF complex, the transcriptional effector of the WNT pathway [43, 44], and enhances transcriptional activity and nuclear translocation of TCF/LEF [45, 46]. SOX9 physically interacts with β-catenin [52], which in turn binds to LEF1 [53]. Activation of WNT and BMP signalling induces phosphorylation of SOX9, which is essential for its function [47]. The SOX9 promoter contains a BMP-responsive element and one Smad-binding element, and expression of SOX9 is induced by Smad proteins and BMP signalling [48–51]. SOX9 is required for and promotes survival and proliferation of glioma stem cells [68–72], and this could be of functional relevance in the high-grade areas of DIA/DIGs. Thus, up-regulation of SOX9 and its downstream transcription factors MEIS1 and PBX1, producing activation of the MAPK, WNT, and BMP pathways, could provide an explanation for the biology of high-grade areas in DIA/DIGs.
In summary, we have identified a SOX9 core transcription network with activation of the MAPK, WNT, and BMP pathways in the high-grade areas of DIA/DIGs, an entity defined by its unique clinical, histopathologic, and DNA methylome profiles, rather than recurrent genetic alterations.
Supplementary Material
Supplementary Fig. 2 Quantification of nuclear immunoreactivity of SOX9, MEIS1, PBX1, phospho-Smad1/5/8 (pSmad1/5/8), LEF1, and phospho-ERK1/2 (pERK1/2) in high-grade (HG) and low-grade (LG) areas in 12 DIA/DIGs. Five hundred nuclei of each sample were quantified for percentage calculation. Statistical analysis was done by analysis of variance (ANOVA).
Supplementary Fig. 1 Differentially methylated regions in high-grade vs. low-grade areas of DIA/DIGs. Specific probe ID, chromosome location (CHR), corresponding gene, and specific probe location in relation to the corresponding gene are indicated and plotted. Value: beta value.
Key points.
No recurrent genetic alterations were discovered in our series of 12 DIA/DIGs.
Regions of DIA/DIGs with high-grade morphological features do not contain distinctive genetic alterations that explain their progression.
High-grade regions showed relative hypomethylation in genes downstream of the transcription factors SOX9 and LEF1 and evidence of a core SOX9 transcription network alongside activation of the BMP, WNT, and MAPK signalling pathways.
Acknowledgements
This work is supported, in part, by NCI grant P30CA021765 and funding from American Lebanese Syrian Associated Charities (ALSAC).
Abbreviations
- CNV
Copy number variation
- DIA
Desmoplastic infantile astrocytoma
- DIG
Desmoplastic infantile ganglioglioma
- DMP
Differentially methylated probe
- DMR
Differentially methylated region
- FFPE
Formalin-fixed paraffin-embedded
- Indel
Insertion and/or deletion
- RNA-seq
RNA sequencing
- SNV
Single-nucleotide variation
- WES
Whole exome sequencing
Footnotes
Conflicts of Interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee (Institutional Review Board protocol # XPD18–008) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Data Sharing and Data Accessibility
The data that support the findings of this study are available from the Gene Expression Omnibus site – https://www.ncbi.nlm.nih.gov/geo/.
References
- 1.Louis DN OH, Wiestler OD, Cavenee WK, Ellison DW, Figarella-Branger D, Perry A, Reifenberger G, von Deimling A. WHO Classification of Tumours of the Central Nervous System. Revised 4th Edition. International Agency for Research on Cancer, Lyon. 2016: [Google Scholar]
- 2.Trehan G, Bruge H, Vinchon M, Khalil C, Ruchoux MM, Dhellemmes P, Ares GS. MR imaging in the diagnosis of desmoplastic infantile tumor: retrospective study of six cases. AJNR Am J Neuroradiol 2004; 25: 1028–33 [PMC free article] [PubMed] [Google Scholar]
- 3.Taratuto AL, Monges J, Lylyk P, Leiguarda R. Superficial cerebral astrocytoma attached to dura. Report of six cases in infants. Cancer 1984; 54: 2505–12 [DOI] [PubMed] [Google Scholar]
- 4.VandenBerg SR. Desmoplastic infantile ganglioglioma and desmoplastic cerebral astrocytoma of infancy. Brain Pathol 1993; 3: 275–81 [DOI] [PubMed] [Google Scholar]
- 5.Takeshima H, Kawahara Y, Hirano H, Obara S, Niiro M, Kuratsu J. Postoperative regression of desmoplastic infantile gangliogliomas: report of two cases. Neurosurgery 2003; 53: 979–83; discussion 83–4 [DOI] [PubMed] [Google Scholar]
- 6.Sugiyama K, Arita K, Shima T, Nakaoka M, Matsuoka T, Taniguchi E, Okamura T, Yamasaki H, Kajiwara Y, Kurisu K. Good clinical course in infants with desmoplastic cerebral neuroepithelial tumor treated by surgery alone. J Neurooncol 2002; 59: 63–9 [DOI] [PubMed] [Google Scholar]
- 7.Darwish B, Arbuckle S, Kellie S, Besser M, Chaseling R. Desmoplastic infantile ganglioglioma/astrocytoma with cerebrospinal metastasis. J Clin Neurosci 2007; 14: 498–501 [DOI] [PubMed] [Google Scholar]
- 8.Hoving EW, Kros JM, Groninger E, den Dunnen WF. Desmoplastic infantile ganglioglioma with a malignant course. J Neurosurg Pediatr 2008; 1: 95–8 [DOI] [PubMed] [Google Scholar]
- 9.Hummel TR, Miles L, Mangano FT, Jones BV, Geller JI. Clinical heterogeneity of desmoplastic infantile ganglioglioma: a case series and literature review. J Pediatr Hematol Oncol 2012; 34: e232–6 [DOI] [PubMed] [Google Scholar]
- 10.Phi JH, Koh EJ, Kim SK, Park SH, Cho BK, Wang KC. Desmoplastic infantile astrocytoma: recurrence with malignant transformation into glioblastoma: a case report. Childs Nerv Syst 2011; 27: 2177–81 [DOI] [PubMed] [Google Scholar]
- 11.Taranath A, Lam A, Wong CK. Desmoplastic infantile ganglioglioma: a questionably benign tumour. Australas Radiol 2005; 49: 433–7 [DOI] [PubMed] [Google Scholar]
- 12.Uro-Coste E, Ssi-Yan-Kai G, Guilbeau-Frugier C, Boetto S, Bertozzi AI, Sevely A, Lolmede K, Delisle MB. Desmoplastic infantile astrocytoma with benign histological phenotype and multiple intracranial localizations at presentation. J Neurooncol 2010; 98: 143–9 [DOI] [PubMed] [Google Scholar]
- 13.Gessi M, Zur Muhlen A, Hammes J, Waha A, Denkhaus D, Pietsch T. Genome-wide DNA copy number analysis of desmoplastic infantile astrocytomas and desmoplastic infantile gangliogliomas. J Neuropathol Exp Neurol 2013; 72: 807–15 [DOI] [PubMed] [Google Scholar]
- 14.Blessing MM, Blackburn PR, Krishnan C, Harrod VL, Barr Fritcher EG, Zysk CD, Jackson RA, Milosevic D, Nair AA, Davila JI, Balcom JR, Jenkins RB, Halling KC, Kipp BR, Nageswara Rao AA, Laack NN, Daniels DJ, Macon WR, Ida CM. Desmoplastic Infantile Ganglioglioma: A MAPK Pathway-Driven and Microglia/Macrophage-Rich Neuroepithelial Tumor. J Neuropathol Exp Neurol 2019; 78: 1011–21 [DOI] [PubMed] [Google Scholar]
- 15.Clarke M, Mackay A, Ismer B, Pickles JC, Tatevossian RG, Newman S, Bale TA, Stoler I, Izquierdo E, Temelso S, Carvalho DM, Molinari V, Burford A, Howell L, Virasami A, Fairchild AR, Avery A, Chalker J, Kristiansen M, Haupfear K, Dalton JD, Orisme W, Wen J, Hubank M, Kurian KM, Rowe C, Maybury M, Crosier S, Knipstein J, Schuller U, Kordes U, Kram DE, Snuderl M, Bridges L, Martin AJ, Doey LJ, Al-Sarraj S, Chandler C, Zebian B, Cairns C, Natrajan R, Boult JKR, Robinson SP, Sill M, Dunkel IJ, Gilheeney SW, Rosenblum MK, Hughes D, Proszek PZ, Macdonald TJ, Preusser M, Haberler C, Slavc I, Packer R, Ng HK, Caspi S, Popovic M, Faganel Kotnik B, Wood MD, Baird L, Davare MA, Solomon DA, Olsen TK, Brandal P, Farrell M, Cryan JB, Capra M, Karremann M, Schittenhelm J, Schuhmann MU, Ebinger M, Dinjens WNM, Kerl K, Hettmer S, Pietsch T, Andreiuolo F, Driever PH, Korshunov A, Hiddingh L, Worst BC, Sturm D, Zuckermann M, Witt O, Bloom T, Mitchell C, Miele E, Colafati GS, Diomedi-Camassei F, Bailey S, Moore AS, Hassall TEG, Lowis SP, Tsoli M, Cowley MJ, Ziegler DS, Karajannis MA, Aquilina K, Hargrave DR, Carceller F, Marshall LV, von Deimling A, Kramm CM, Pfister SM, Sahm F, Baker SJ, Mastronuzzi A, Carai A, Vinci M, Capper D, Popov S, Ellison DW, Jacques TS, Jones DTW, Jones C. Infant High-Grade Gliomas Comprise Multiple Subgroups Characterized by Novel Targetable Gene Fusions and Favorable Outcomes. Cancer Discov 2020; 10: 942–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greer A, Foreman NK, Donson A, Davies KD, Kleinschmidt-DeMasters BK. Desmoplastic infantile astrocytoma/ganglioglioma with rare BRAF V600D mutation. Pediatr Blood Cancer 2017; 64: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang AC, Jones DTW, Abecassis IJ, Cole BL, Leary SES, Lockwood CM, Chavez L, Capper D, Korshunov A, Fallah A, Wang S, Ene C, Olson JM, Geyer JR, Holland EC, Lee A, Ellenbogen RG, Ojemann JG. Desmoplastic Infantile Ganglioglioma/Astrocytoma (DIG/DIA) Are Distinct Entities with Frequent BRAFV600 Mutations. Mol Cancer Res 2018; 16: 1491–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J, Hedges D, Ma X, Zhou X, Yergeau DA, Wilkinson MR, Vadodaria B, Chen X, McGee RB, Hines-Dowell S, Nuccio R, Quinn E, Shurtleff SA, Rusch M, Patel A, Becksfort JB, Wang S, Weaver MS, Ding L, Mardis ER, Wilson RK, Gajjar A, Ellison DW, Pappo AS, Pui CH, Nichols KE, Downing JR. Germline Mutations in Predisposition Genes in Pediatric Cancer. N Engl J Med 2015; 373: 2336–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qaddoumi I, Orisme W, Wen J, Santiago T, Gupta K, Dalton JD, Tang B, Haupfear K, Punchihewa C, Easton J, Mulder H, Boggs K, Shao Y, Rusch M, Becksfort J, Gupta P, Wang S, Lee RP, Brat D, Peter Collins V, Dahiya S, George D, Konomos W, Kurian KM, McFadden K, Serafini LN, Nickols H, Perry A, Shurtleff S, Gajjar A, Boop FA, Klimo PD Jr., Mardis ER, Wilson RK, Baker SJ, Zhang J, Wu G, Downing JR, Tatevossian RG, Ellison DW. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol 2016; 131: 833–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edmonson MN, Zhang J, Yan C, Finney RP, Meerzaman DM, Buetow KH. Bambino: a variant detector and alignment viewer for next-generation sequencing data in the SAM/BAM format. Bioinformatics 2011; 27: 865–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang JCH, Harreld JH, Tanaka R, Li X, Wen J, Zhang C, Boue DR, Rauch TM, Boyd JT, Chen J, Corbo JC, Bouldin TW, Elton SW, Liu LL, Schofield D, Lee SC, Bouffard JP, Georgescu MM, Dossani RH, Aguiar MA, Sances RA, Saad AG, Boop FA, Qaddoumi I, Ellison DW. Septal dysembryoplastic neuroepithelial tumor: a comprehensive clinical, imaging, histopathologic, and molecular analysis. Neuro Oncol 2019; 21: 800–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012; 40: 4288–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du X, Wen J, Wang Y, Karmaus PWF, Khatamian A, Tan H, Li Y, Guy C, Nguyen TM, Dhungana Y, Neale G, Peng J, Yu J, Chi H. Hippo/Mst signalling couples metabolic state and immune function of CD8alpha(+) dendritic cells. Nature 2018; 558: 141–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khatamian A, Paull EO, Califano A, Yu J. SJARACNe: a scalable software tool for gene network reverse engineering from big data. Bioinformatics 2019; 35: 2165–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015; 1: 417–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberzon A A description of the Molecular Signatures Database (MSigDB) Web site. Methods Mol Biol 2014; 1150: 153–60 [DOI] [PubMed] [Google Scholar]
- 29.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011; 27: 1739–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, Teschendorff AE. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017; 33: 3982–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, Irizarry RA. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol 2012; 41: 200–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fortin JP, Triche TJ Jr., Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 2017; 33: 558–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keenan C, Graham RT, Harreld JH, Lucas JT Jr., Finkelstein D, Wheeler D, Li X, Dalton J, Upadhyaya SA, Raimondi SC, Boop FA, DeCuypere M, Zhang J, Vinitsky A, Wang L, Chiang J. Infratentorial C11orf95-fused gliomas share histologic, immunophenotypic, and molecular characteristics of supratentorial RELA-fused ependymoma. Acta Neuropathol 2020; 140: 963–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang J, Diaz AK, Makepeace L, Li X, Han Y, Li Y, Klimo P Jr., Boop FA, Baker SJ, Gajjar A, Merchant TE, Ellison DW, Broniscer A, Patay Z, Tinkle CL. Clinical, imaging, and molecular analysis of pediatric pontine tumors lacking characteristic imaging features of DIPG. Acta Neuropathol Commun 2020; 8: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang J, Li X, Liu APY, Qaddoumi I, Acharya S, Ellison DW. Tectal glioma harbors high rates of KRAS G12R and concomitant KRAS and BRAF alterations. Acta Neuropathol 2020; 139: 601–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang J, Harreld JH, Tinkle CL, Moreira DC, Li X, Acharya S, Qaddoumi I, Ellison DW. A single-center study of the clinicopathologic correlates of gliomas with a MYB or MYBL1 alteration. Acta Neuropathol 2019; 138: 1091–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu APY, Harreld JH, Jacola LM, Gero M, Acharya S, Ghazwani Y, Wu S, Li X, Klimo P Jr., Gajjar A, Chiang J, Qaddoumi I. Tectal glioma as a distinct diagnostic entity: a comprehensive clinical, imaging, histologic and molecular analysis. Acta Neuropathol Commun 2018; 6: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, Fish P, Harsha B, Hathaway C, Jupe SC, Kok CY, Noble K, Ponting L, Ramshaw CC, Rye CE, Speedy HE, Stefancsik R, Thompson SL, Wang S, Ward S, Campbell PJ, Forbes SA. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019; 47: D941–D7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murakami S, Kan M, McKeehan WL, de Crombrugghe B. Up-regulation of the chondrogenic Sox9 gene by fibroblast growth factors is mediated by the mitogen-activated protein kinase pathway. Proc Natl Acad Sci U S A 2000; 97: 1113–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tew SR, Hardingham TE. Regulation of SOX9 mRNA in human articular chondrocytes involving p38 MAPK activation and mRNA stabilization. J Biol Chem 2006; 281: 39471–9 [DOI] [PubMed] [Google Scholar]
- 42.Ling S, Chang X, Schultz L, Lee TK, Chaux A, Marchionni L, Netto GJ, Sidransky D, Berman DM. An EGFR-ERK-SOX9 signaling cascade links urothelial development and regeneration to cancer. Cancer Res 2011; 71: 3812–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bastide P, Darido C, Pannequin J, Kist R, Robine S, Marty-Double C, Bibeau F, Scherer G, Joubert D, Hollande F, Blache P, Jay P. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol 2007; 178: 635–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blache P, van de Wetering M, Duluc I, Domon C, Berta P, Freund JN, Clevers H, Jay P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol 2004; 166: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma F, Ye H, He HH, Gerrin SJ, Chen S, Tanenbaum BA, Cai C, Sowalsky AG, He L, Wang H, Balk SP, Yuan X. SOX9 drives WNT pathway activation in prostate cancer. J Clin Invest 2016; 126: 1745–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang JQ, Wei FK, Xu XL, Ye SX, Song JW, Ding PK, Zhu J, Li HF, Luo XP, Gong H, Su L, Yang L, Gong LY. SOX9 drives the epithelial-mesenchymal transition in non-small-cell lung cancer through the Wnt/beta-catenin pathway. J Transl Med 2019; 17: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu JA, Wu MH, Yan CH, Chau BK, So H, Ng A, Chan A, Cheah KS, Briscoe J, Cheung M. Phosphorylation of Sox9 is required for neural crest delamination and is regulated downstream of BMP and canonical Wnt signaling. Proc Natl Acad Sci U S A 2013; 110: 2882–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iezaki T, Fukasawa K, Horie T, Park G, Robinson S, Nakaya M, Fujita H, Onishi Y, Ozaki K, Kanayama T, Hiraiwa M, Kitaguchi Y, Kaneda K, Yoneda Y, Takarada T, Guo XE, Kurose H, Hinoi E. The MAPK Erk5 is necessary for proper skeletogenesis involving a Smurf-Smad-Sox9 molecular axis. Development 2018; 145: [DOI] [PubMed] [Google Scholar]
- 49.Pan Q, Yu Y, Chen Q, Li C, Wu H, Wan Y, Ma J, Sun F. Sox9, a key transcription factor of bone morphogenetic protein-2-induced chondrogenesis, is activated through BMP pathway and a CCAAT box in the proximal promoter. J Cell Physiol 2008; 217: 228–41 [DOI] [PubMed] [Google Scholar]
- 50.Raspopovic J, Marcon L, Russo L, Sharpe J. Modeling digits. Digit patterning is controlled by a Bmp-Sox9-Wnt Turing network modulated by morphogen gradients. Science 2014; 345: 566–70 [DOI] [PubMed] [Google Scholar]
- 51.Zehentner BK, Dony C, Burtscher H. The transcription factor Sox9 is involved in BMP-2 signaling. J Bone Miner Res 1999; 14: 1734–41 [DOI] [PubMed] [Google Scholar]
- 52.Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, McCrea PD, de Crombrugghe B. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev 2004; 18: 1072–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hurlstone A, Clevers H. T-cell factors: turn-ons and turn-offs. EMBO J 2002; 21: 2303–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bruckmann C, Tamburri S, De Lorenzi V, Doti N, Monti A, Mathiasen L, Cattaneo A, Ruvo M, Bachi A, Blasi F. Mapping the native interaction surfaces of PREP1 with PBX1 by cross-linking mass-spectrometry and mutagenesis. Sci Rep 2020; 10: 16809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prakash V, Batanian JR, Guzman MA, Duncavage EJ, Geller TJ. Malignant transformation of a desmoplastic infantile ganglioglioma in an infant carrier of a nonsynonymous TP53 mutation. Pediatr Neurol 2014; 51: 138–43 [DOI] [PubMed] [Google Scholar]
- 56.Loh JK, Lieu AS, Chai CY, Howng SL. Malignant transformation of a desmoplastic infantile ganglioglioma. Pediatr Neurol 2011; 45: 135–7 [DOI] [PubMed] [Google Scholar]
- 57.Chiang JC, Ellison DW. Molecular pathology of paediatric central nervous system tumours. J Pathol 2017; 241: 159–72 [DOI] [PubMed] [Google Scholar]
- 58.Mistry M, Zhukova N, Merico D, Rakopoulos P, Krishnatry R, Shago M, Stavropoulos J, Alon N, Pole JD, Ray PN, Navickiene V, Mangerel J, Remke M, Buczkowicz P, Ramaswamy V, Guerreiro Stucklin A, Li M, Young EJ, Zhang C, Castelo-Branco P, Bakry D, Laughlin S, Shlien A, Chan J, Ligon KL, Rutka JT, Dirks PB, Taylor MD, Greenberg M, Malkin D, Huang A, Bouffet E, Hawkins CE, Tabori U. BRAF mutation and CDKN2A deletion define a clinically distinct subgroup of childhood secondary high-grade glioma. J Clin Oncol 2015; 33: 1015–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brat DJ, Aldape K, Colman H, Figrarella-Branger D, Fuller GN, Giannini C, Holland EC, Jenkins RB, Kleinschmidt-DeMasters B, Komori T, Kros JM, Louis DN, McLean C, Perry A, Reifenberger G, Sarkar C, Stupp R, van den Bent MJ, von Deimling A, Weller M. cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol 2020; 139: 603–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arabzade A, Zhao Y, Varadharajan S, Chen HC, Jessa S, Rivas B, Stuckert AJ, Solis M, Kardian A, Tlais D, Golbourn BJ, Stanton AJ, Chan YS, Olson C, Karlin KL, Kong K, Kupp R, Hu B, Injac SG, Ngo M, Wang PR, De Leon LA, Sahm F, Kawauchi D, Pfister SM, Lin CY, Hodges HC, Singh I, Westbrook TF, Chintagumpala MM, Blaney SM, Parsons DW, Pajtler KW, Agnihotri S, Gilbertson RJ, Yi J, Jabado N, Kleinman CL, Bertrand KC, Deneen B, Mack SC. ZFTA-RELA Dictates Oncogenic Transcriptional Programs to Drive Aggressive Supratentorial Ependymoma. Cancer Discov 2021: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P, Reimand J, Warnatz HJ, Ryzhova M, Mack S, Ramaswamy V, Capper D, Schweizer L, Sieber L, Wittmann A, Huang Z, van Sluis P, Volckmann R, Koster J, Versteeg R, Fults D, Toledano H, Avigad S, Hoffman LM, Donson AM, Foreman N, Hewer E, Zitterbart K, Gilbert M, Armstrong TS, Gupta N, Allen JC, Karajannis MA, Zagzag D, Hasselblatt M, Kulozik AE, Witt O, Collins VP, von Hoff K, Rutkowski S, Pietsch T, Bader G, Yaspo ML, von Deimling A, Lichter P, Taylor MD, Gilbertson R, Ellison DW, Aldape K, Korshunov A, Kool M, Pfister SM. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 2015; 27: 728–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim KH, Seol HJ, Kim EH, Rheey J, Jin HJ, Lee Y, Joo KM, Lee J, Nam DH. Wnt/beta-catenin signaling is a key downstream mediator of MET signaling in glioblastoma stem cells. Neuro Oncol 2013; 15: 161–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature 2005; 434: 843–50 [DOI] [PubMed] [Google Scholar]
- 64.Denysenko T, Annovazzi L, Cassoni P, Melcarne A, Mellai M, Schiffer D. WNT/beta-catenin Signaling Pathway and Downstream Modulators in Low- and High-grade Glioma. Cancer Genomics Proteomics 2016; 13: 31–45 [PubMed] [Google Scholar]
- 65.Taylor KR, Mackay A, Truffaux N, Butterfield Y, Morozova O, Philippe C, Castel D, Grasso CS, Vinci M, Carvalho D, Carcaboso AM, de Torres C, Cruz O, Mora J, Entz-Werle N, Ingram WJ, Monje M, Hargrave D, Bullock AN, Puget S, Yip S, Jones C, Grill J. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet 2014; 46: 457–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, Bechet D, Faury D, De Jay N, Ramkissoon LA, Corcoran A, Jones DT, Sturm D, Johann P, Tomita T, Goldman S, Nagib M, Bendel A, Goumnerova L, Bowers DC, Leonard JR, Rubin JB, Alden T, Browd S, Geyer JR, Leary S, Jallo G, Cohen K, Gupta N, Prados MD, Carret AS, Ellezam B, Crevier L, Klekner A, Bognar L, Hauser P, Garami M, Myseros J, Dong Z, Siegel PM, Malkin H, Ligon AH, Albrecht S, Pfister SM, Ligon KL, Majewski J, Jabado N, Kieran MW. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet 2014; 46: 462–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gargiulo G, Cesaroni M, Serresi M, de Vries N, Hulsman D, Bruggeman SW, Lancini C, van Lohuizen M. In vivo RNAi screen for BMI1 targets identifies TGF-beta/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell 2013; 23: 660–76 [DOI] [PubMed] [Google Scholar]
- 68.Aldaz P, Otaegi-Ugartemendia M, Saenz-Antonanzas A, Garcia-Puga M, Moreno-Valladares M, Flores JM, Gerovska D, Arauzo-Bravo MJ, Sampron N, Matheu A, Carrasco-Garcia E. SOX9 promotes tumor progression through the axis BMI1-p21(CIP). Sci Rep 2020; 10: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sabelstrom H, Petri R, Shchors K, Jandial R, Schmidt C, Sacheva R, Masic S, Yuan E, Fenster T, Martinez M, Saxena S, Nicolaides TP, Ilkhanizadeh S, Berger MS, Snyder EY, Weiss WA, Jakobsson J, Persson AI. Driving Neuronal Differentiation through Reversal of an ERK1/2-miR-124-SOX9 Axis Abrogates Glioblastoma Aggressiveness. Cell Rep 2019; 28: 2064–79 e11 [DOI] [PubMed] [Google Scholar]
- 70.Wang Z, Xu X, Liu N, Cheng Y, Jin W, Zhang P, Wang X, Yang H, Liu H, Tu Y. SOX9-PDK1 axis is essential for glioma stem cell self-renewal and temozolomide resistance. Oncotarget 2018; 9: 192–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu F, Hon GC, Villa GR, Turner KM, Ikegami S, Yang H, Ye Z, Li B, Kuan S, Lee AY, Zanca C, Wei B, Lucey G, Jenkins D, Zhang W, Barr CL, Furnari FB, Cloughesy TF, Yong WH, Gahman TC, Shiau AK, Cavenee WK, Ren B, Mischel PS. EGFR Mutation Promotes Glioblastoma through Epigenome and Transcription Factor Network Remodeling. Mol Cell 2015; 60: 307–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rani SB, Rathod SS, Karthik S, Kaur N, Muzumdar D, Shiras AS. MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro Oncol 2013; 15: 1302–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 2 Quantification of nuclear immunoreactivity of SOX9, MEIS1, PBX1, phospho-Smad1/5/8 (pSmad1/5/8), LEF1, and phospho-ERK1/2 (pERK1/2) in high-grade (HG) and low-grade (LG) areas in 12 DIA/DIGs. Five hundred nuclei of each sample were quantified for percentage calculation. Statistical analysis was done by analysis of variance (ANOVA).
Supplementary Fig. 1 Differentially methylated regions in high-grade vs. low-grade areas of DIA/DIGs. Specific probe ID, chromosome location (CHR), corresponding gene, and specific probe location in relation to the corresponding gene are indicated and plotted. Value: beta value.


