Abstract
Salmonella species are pathogenic bacteria often detected in sewage, freshwater, marine coastal water, and groundwater. Salmonella spp. can survive for long periods in natural waters, and the persistence of specific and epidemic strains is of great concern in public health. However, the diversity of species found in the natural environment remains unknown. The aim of this study was to investigate the diversity of Salmonella strains isolated from different natural aquatic systems within a Mediterranean coastal watershed (river, wastewater, and marine coastal areas). A total of 574 strains isolated from these natural environments were identified by both conventional serotyping and the ribosomal spacer-heteroduplex polymorphism (RS-HP) method (M. A. Jensen and N. Straus, PCR Methods Appl. 3:186–194, 1993). More than 40 different serotypes were found, and some serotypes probably mobilized from widespread animal-rearing activities were detected only during storm events. These serotypes may be good indicators of specific contamination sources. Furthermore, the RS-HP method based on the PCR amplification of the intergenic spacer region between the 16S and 23S rRNA genes can produce amplicon profiles allowing the discrimination of species at both serotype and intraserotype levels. This method represents a powerful tool that could be used for rapid typing of Salmonella isolates.
Salmonella spp. are ubiquitous enteric bacteria. These gram-negative rods are the etiologic agents of food-borne salmonellosis and also the agents that cause typhoid and paratyphoid fevers. Although food products, including shellfish, are the most common sources of salmonellosis, Salmonella is a prime example of a water- and shellfish-transmitted pathogen. Salmonella is a large genus of bacteria including more than 2,300 serotypes, and diagnosis in the majority of laboratories relies on costly and laborious culture screening with both nonselective and selective media (14). Nevertheless, only a limited set of serotypes are prevalent within clinical isolates. For instance, 15 serotypes of clinical origin accounted for 80% of the 22,100 isolates or data collected by the French Agency for Food Safety in 1998. In contrast, environmental isolates represented less than 4.4% of these isolates.
Both human and animal excreta are sources of Salmonella, and many potential routes are used for the transmission of these excreted enteric pathogens. The ability of Salmonella to be transmitted by any of these routes depends largely on its resistance to environmental factors, which control its survival, and its capacity to be carried by water as it moves through the environment. Nevertheless, this survival capacity may depend on species and pollution sources. Although most studies have focused on the determination of Salmonella strain concentrations in some polluted areas, it was recently shown that the annual bacterial loads of this pathogen in rivers and coastal areas can be very important (3). However, information on the diversity and occurrence of Salmonella strains is very scarce (19, 29), and as a consequence, the ecology of these species remains unknown. This is partly due to the laborious methods required for the detection, isolation, and identification of Salmonella strains.
Although serotyping offers a very precise and reliable method for differentiating isolated strains, it remains time-consuming and requires the use of more than 150 specific serum samples (14). Thus, serotyping is not accessible to many laboratories, and this may partly explain why little information is available on the diversity of strains in natural ecosystems. Furthermore, some strains cannot be identified due to the untypeability of the isolate (rough strains). With the development of molecular biology methods, it is now desirable to use alternative methods which provide a higher power of discrimination and allow a more rapid identification attainable by nonspecialized laboratories.
DNA amplication to generate genomically distinct groups of products has begun to play an increasing role in the differentiation of bacterial species, serotypes, and strains. The amplification of the 16S-23S ribosomal spacer regions and the size polymorphism of the resulting products have proven useful in the species-level identification of a broad range of bacteria (12). These sequences are generally found in multiple copies in most bacterial genomes (9, 13). When multiple amplification products, which contain homologous 3′ and 5′ sequences flanking heterologous intervening sequences, are generated, these products can cross-hybridize to form heteroduplex DNA structures (13). Homoduplex double-stranded DNA structures and heteroduplex structures have already been successfully used to identify Salmonella serotypes isolated from clinical samples (13). Thus, the comparison of pattern groups of ribosomal spacer-heteroduplex polymorphism (RS-HP) types may provide a rapid and convenient alternative method for identifying Salmonella serotypes isolated from aquatic environmental samples.
The purpose of the present study was to provide information on the diversity of Salmonella isolates from marine and freshwater sediments, freshwater, and wastewater samples collected within a Mediterranean coastal watershed. Both conventional serotyping and RS-HP typing methods were used and compared to investigate the efficiency of the RS-HP method as an alternative to traditional serotyping and a more convenient method for nonspecialized laboratories.
MATERIALS AND METHODS
Field samples.
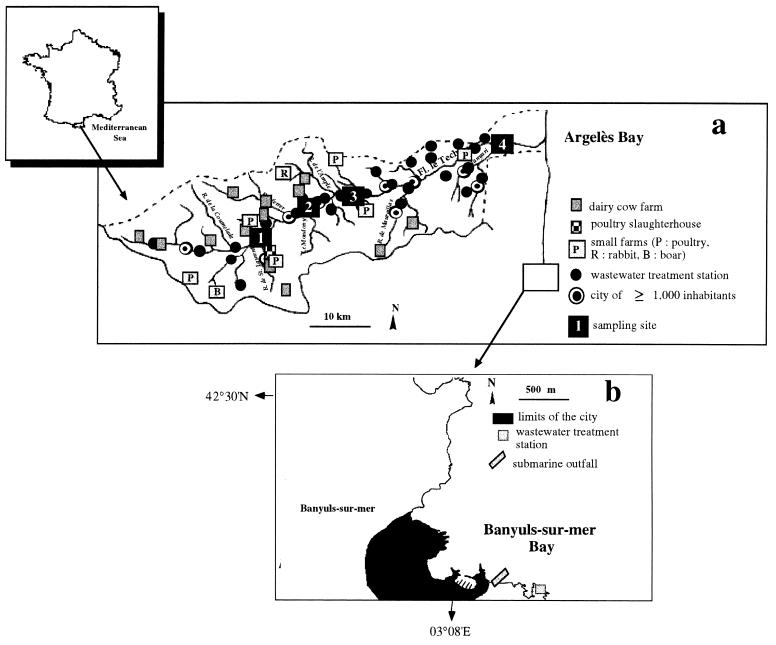
The Tech is a typical coastal river on the western Mediterranean coast with a drainage area of 780 km2. The river's source is located at an altitude of 2,345 m and 78 km from the river's mouth. The upstream area of the watershed is characterized by agriculture and forests with extensive animal-rearing activities (cattle and boar). Some small poultry farms (ducks, chickens, and geese) are also located in this area (Fig. 1a). Numerous wastewater treatment stations are located within the watershed with their outfalls being located in the river and its tributaries.
FIG. 1.
Study areas. (a) The Tech River watershed and location of sampling sites. (b) Location of the wastewater treatment station of Banyuls-sur-Mer.
The wastewater treatment station of Banyuls-sur-Mer (Fig. 1b) is a monitoring station performing physical and chemical treatment. The population varies from 5,000 residents out of season to 15,000 inhabitants during the summertime. The residence time of wastewater in the sewage station is 1.5 h. The sewage system combines rainfall waters and wastewaters, and effluents are discharged via a submarine outfall located 500 m offshore and 20 m in depth below the surface.
Sampling strategy.
Four sampling stations (1 to 4) were chosen along the stream (Fig. 1a). Station 1 was located 20 km downstream from the source and was considered a reference station for the upstream watershed. Station 2 was located 27 km downstream from the source and city of Arles-sur-Tech. Stations 2 and 3 were located upstream and downstream, respectively, from the wastewater effluents released by the city of Amélie-les-Bains. Station 4 was located 5 km upstream from the river's mouth. Water samples were taken monthly at each station from June 1996 to October 1997 and twice monthly in September of each year. During flood events, samples were collected downstream at station 4.
For wastewaters, a total of 11 sampling days were selected from May 1996 to November 1997 including 4 summer and 7 nonsummer sampling days.
Sediments were collected at station 4 in the Tech River; in the Bay of Argelès, corresponding to the river discharge area; and in the Bay of Banyuls-sur-Mer near the submarine outfall.
Isolation of Salmonella.
Samples were collected in sterile flasks and analyzed within 4 h after sampling. Salmonella spp. are present in natural waters at low concentrations within a more or less important background of other bacterial genera depending on the type of water (river or wastewater). The enumeration of Salmonella spp. was performed by using the most probable number (MPN) procedure, but two different culturing procedures were used (see below) to optimize the isolation of Salmonella strains (2, 4, 27). Three triplicate samples of three different volumes (1 liter, 100 ml, and 10 ml) were used to determine the MPN. Each sample was filtered through one or more 47-mm-diameter membrane filters (Millipore Corp., Bedford, Mass.), since a set of different filters was sometimes needed to filter one sample. Then, each set of filters was incubated in one tube and all the tubes (three per volume) were used to determine the MPN.
For river samples, Salmonella spp. were isolated after preenrichment in peptone-water broth without indole (Sanofi Diagnostic Pasteur, Marnes-La-Coquette, France) at 37°C for 24 h. Then, selective enrichment in Rappaport-Vassiliadis broth (RV10; Difco Laboratories, Detroit, Mich.) was used with an incubation at 43°C for 24 h. For wastewater samples, Salmonella spp. were isolated using the method proposed by Baleux et al. (2). Briefly, filters were incubated in selective Selenite F broth (BioMérieux, Marcy-l'Etoile, France) modified by the addition of novobiocin (45 μg/ml; Sigma Chemical, St. Louis, Mo.) and pril (200 μg/ml; Henkel France SA, Boulogne, France). After the selective enrichment step, colonies were isolated by spread plating aliquots onto Salmonella-Shigella agar (Sanofi Diagnostic Pasteur) and incubating them at 37°C for 24 to 48 h. Typical colonies of Salmonella spp. were confirmed by biochemical tests. Isolates were tested for their inability to produce oxidase (oxidase test; Difco), their ability to produce H2S and degrade 4-methylumbelliferyl caprilate (Research Organics, Inc., Cleveland, Ohio), and their inability to oxidize and ferment lactose (1, 6). All Salmonella spp. isolates were identified by both serotyping and analysis of RS-HP patterns.
Serotyping.
Serotyping was performed by seroagglutination using both commercial antisera (Sanofi Diagnostic Pasteur) and antisera produced by the Institut Pasteur (Paris, France) to determine the antigenic formula. Polyvalent Salmonella O and H antisera were used to obtain a presumptive diagnosis, and then the definitive antigenic formula was determined using monovalent antisera. Because antigen O or H is not always expressed, sometimes no agglutination was observed for some isolates (approximately 1% of isolates from the environment).
DNA extraction.
DNA was extracted by the method described by Garcia-Pichel et al. (7). Briefly, an aliquot of a colony was picked and suspended in 49 μl of lysis solution. The final DNA extraction was obtained by five successive freeze-heat cycles, in a liquid nitrogen bath and in a water bath at 55°C, respectively. One microliter of proteinase K (10 μg/ml; Sigma) was added to the lysis solution, and the whole was incubated at 55°C for 1 h. The extraction products were stored at −20°C before amplification.
DNA amplification.
Oligonucleotide primers were designed to amplify the 16S-23S ribosomal DNA intergenic spacers. The primer sequences used in the present study, L17 (5′-CAA-GGC-ATC-CAC-CGT-GT-3′) and G17 (5′-GTG-AAG-TCG-TAA-CAA-GG-3′), were previously described by Jensen and Straus (11). Amplification was performed in a final volume of 72 μl with a reaction mixture containing 3 μl of a deoxynucleoside triphosphate mixture (each at a 5 mM concentration; Eurogentec, Herstal, Belgium), 3 μl of primers (each at a 12.5 μM concentration; Eurogentec), 7.5 μl of 10× PCR buffer (500 mM KCl, 100 mM Tris-HCl [pH 8.3 at 25°C], 15 mM MgCl2, 0.003% gelatin, and 55 μl of purified water [Sigma]), and 2 μl of lysis solution. A Hybaid Touchdown thermocycler (Hybaid Ltd., Teddington, United Kingdom) used for thermal amplification was programmed for the following: (i) an initial extensive denaturation step, consisting of treatment at 94°C for 2 min; (ii) an intermediate step to add 2.4 μl of Taq polymerase (SuperTaq; HT Biotechnology, Cambridge, United Kingdom) at a concentration of 1 U/μl, consisting of treatment at 80°C for 1 min; (iii) 25 reaction cycles, with each cycle consisting of treatment at 94°C for 15 s, 55°C for 4 min, and 72°C for 1 min; and (iv) a final extension step, consisting of treatment at 72°C for 30 min.
Electrophoresis and imaging.
A 5-μl aliquot of the PCR mixture was combined with 2.5 μl of loading buffer (15% Ficoll, 5 mM EDTA, 0.1% sodium dodecyl sulfate, 0.1% xylene cyanol). PCR products were separated on a 4% acrylamide-bis (29:1) gel with 1× Tris-borate-EDTA running buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA [pH 8.3]) for 150 min at a field strength of 12.5 V/cm to optimize the separation of DNA fragments. A wide-range molecular-weight DNA marker (100-bp ladder; Life Technologies GibcoBRL, Paisley, United Kingdom) was used on each gel as standard. Electrophoresis was performed with a D-Code system (Bio-Rad, Hercules, Calif.). Following electrophoresis, gels were stained for 30 min in a solution of ethidium bromide at 0.5 μl/ml. PCR products were visualized with UV light and photographed. Polaroid pictures were scanned on a G3 Macintosh computer using Adobe Photo Deluxe software.
Statistical data analysis.
The size of each DNA-amplified fragment was determined using the molecular-weight DNA marker. A total of 44 fragments corresponding to different sizes were recorded and numbered from 1 to 44. Then, each profile was transformed into binary code depending on the presence or absence of each band. A similarity matrix for all pairwise combinations of RS-HP profiles was constructed using the simple matching coefficient of Sokal and Sneath as a measure of proximity (23). Binary code profiles were computed using the R package for numerical taxonomic analysis distributed by Pierre Legendre (University of Montréal, Montréal, Québec, Canada). Then, a dendrogram was constructed from the similarity matrix by using the UPGMA algorithm (unweighted pair group method with arithmetic mean) (23).
RESULTS
Number of isolates.
A total of 574 Salmonella spp. isolates were obtained from the different samples. Table 1 shows the distribution of isolates within the different types of samples. The highest numbers of isolates were obtained from samples collected in the river and in wastewaters. A high proportion of strains isolated at station 4 were found during flood events. In contrast, isolates found in sediments represented only 6.3% of total isolates.
TABLE 1.
Distribution of 574 Salmonella isolates among different serotypes and environmental samples
| Serotype | O group | No. of isolates
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Water samples
|
Sediment samplesb
|
|||||||||
| Wastewater | Rivera
|
RS | AS | BS | |||||||
| St 1 | St 2 | St 3 | St 4 | Flood | |||||||
| Agona | B | 7 | 3 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 |
| Anatum | E1 | 3 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Bardo | C3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Bareilly | C1 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| Bovis morbificans | C2 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Braenderup | G1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Brandenburg | B | 36 | 17 | 0 | 9 | 5 | 4 | 0 | 1 | 0 | 0 |
| Bredeney | B | 4 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 0 | 0 |
| Cerro | K | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Coeln | B | 5 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 |
| Derby | B | 4 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 |
| Enteritidis | D1 | 7 | 0 | 0 | 0 | 1 | 4 | 1 | 1 | 0 | 0 |
| Give | E1 | 4 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | 0 |
| Gold Coast | C2 | 3 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Grumpensis | G2 | 21 | 0 | 0 | 0 | 0 | 2 | 19 | 0 | 0 | 0 |
| Hadar | C2 | 20 | 1 | 4 | 2 | 1 | 1 | 10 | 1 | 0 | 0 |
| Haifa | B | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Indiana | B | 43 | 7 | 2 | 26 | 3 | 2 | 3 | 0 | 0 | 0 |
| Infantis | C1 | 23 | 7 | 1 | 2 | 4 | 1 | 8 | 0 | 0 | 0 |
| Kapemba | D1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Kedougou | G2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Kottbus | C2 | 11 | 0 | 4 | 0 | 2 | 0 | 5 | 0 | 0 | 0 |
| London | E1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Manhattan | C2 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Mbandaka | C1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Mikawasima | C1 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| Montevideo | C1 | 5 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 0 |
| Muenster | E1 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| Newport | C2 | 84 | 55 | 1 | 0 | 0 | 2 | 18 | 4 | 0 | 4 |
| Oranienburg | C1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Panama | D1 | 26 | 0 | 0 | 0 | 0 | 1 | 24 | 1 | 0 | 0 |
| Paratyphi B | B | 10 | 0 | 8 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Richmond | C1 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| Rissen | C1 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| Saint-Paul | B | 20 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Senftenberg | E4 | 12 | 1 | 0 | 5 | 5 | 1 | 0 | 0 | 0 | 0 |
| Typhimurium | B | 156 | 7 | 15 | 18 | 30 | 25 | 49 | 8 | 4 | 0 |
| Veneziana | F | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Virchow | C1 | 18 | 1 | 1 | 0 | 2 | 2 | 11 | 0 | 0 | 1 |
| Wien | B | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| III b.38: z10: z53 (“Salmonella enterica subsp. diarizonae”) | P | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| No. of strains with incomplete serotype | 13 | 0 | 1 | 1 | 0 | 2 | 3 | 2 | 0 | 4 | |
| No. of rough strains | 10 | 0 | 0 | 5 | 4 | 0 | 1 | 0 | 0 | 0 | |
| Total no. of isolates | 574 | 126 | 40 | 73 | 64 | 53 | 183 | 22 | 4 | 10 | |
Sampling stations (St) located on the river (St 1, St 2, St 3, and St 4) and flood event samples from station 4 (Flood).
Sediment samples from the river (RS), Argelès Bay (AS), and Banyuls Bay (BS).
Distribution of serotypes in the different sample types.
A total of 41 serotypes were found among the 551 isolates for which serotype identification was possible (Table 1). Thirteen isolates were characterized by an incomplete serotype, whereas 10 other isolates formed rough colonies and were not identified by serotyping (Table 1).
In river water samples, 35 different serotypes were found. Nevertheless, the highest diversity of isolates was reported during flood events, during which 26 different serotypes were found. Among these, 10 serotypes were specific to flood events and were never found in other samples (Salmonella enterica serotypes Coeln, Kedougou, Mbandaka, Mikawasima, Muenster, Oranienburg, Richmond, Rissen, Veneziana, and III b.38: z10: z53). Serotype Typhimurium represents 28.4% of total isolates and 33.1% of river isolates. The dominance of this serotype was found at all sampling stations from upstream to downstream and was maintained with time during the sampling period (data not shown). In contrast, only 14 different serotypes were identified in wastewater samples, and serotype Newport represented 43% of total wastewater isolates. The diversity of serotypes found in sediment samples was comparatively low due to the low number of isolated strains.
Banding pattern of Salmonella spp.
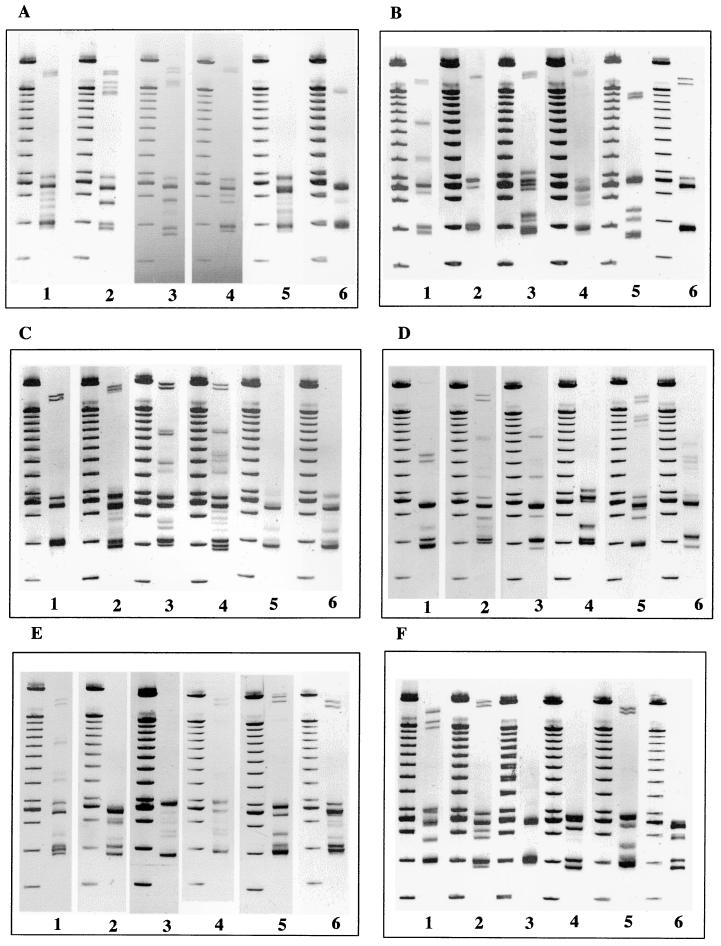
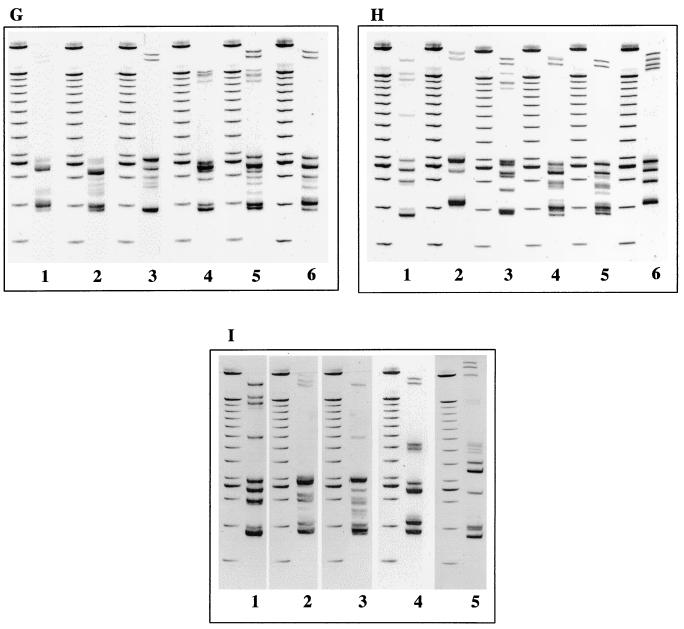
Gel electrophoresis profiles representing the RS-HP types observed for each of the Salmonella serotypes are shown in Fig. 2. These profiles resulted from the amplification of the 16S-23S intergenic spacer regions. Profiles could be discriminated from the number and the position of bands. The number of bands per profile varied from 2 to 16 bands ranging between 370 and 2,000 bp including the primers. The weak intensity of some bands may be attributed to secondary products. This was not considered a problem because the electrophoretic patterns were very constant and reproducible.
FIG. 2.
Representative electrophoretic patterns of PCR-amplified bacterial genomic DNA isolated from the indicated Salmonella spp. serotypes, with RS-HP types indicated in brackets. (A) Lane 1, serotype Newport (New4); lane 2, serotype Newport (New3); lane 3, serotype Newport (New2); lane 4, serotype Newport (New1); lane 5, serotype Kottbus (Kott2); lane 6, serotype Kottbus (Kott1). (B) Lane 1, serotype Typhimurium (Typ1); lane 2, serotype Typhimurium (Typ2); lane 3, serotype Oranienburg; lane 4, serotype Bardo; lane 5, serotype Indiana; lane 6, serotype Muenster. (C) Lane 1, serotype Paratyphi B; lane 2, serotype Hadar; lane 3, serotype Senftenberg (Senf2); lane 4, serotype Senftenberg (Senf1); lane 5, serotype Panama (Pana2); lane 6, serotype Panama (Pana1). (D) Lane 1, serotype Derby; lane 2, serotype Gold Coast; lane 3, serotype Wien; lane 4, serotype Veneziana; lane 5, serotype Coeln; lane 6, serotype Mbandaka. (E) Lane 1, serotype Grumpensis; lane 2, serotype Virchow (Vir3); lane 3, serotype Virchow (Vir2); lane 4, serotype Virchow (Vir1); lane 5, serotype Bareilly; lane 6, serotype Braenderup. (F) Lane 1, serotype Saint-Paul; lane 2, serotype Brandenburg (Bran1); lane 3, serotype Brandenburg (Bran2); lane 4, serotype Enteritidis; lane 5, serotype Infantis; lane 6, serotype Richmond. (G) Lane 1, serotype Bredeney (Bre1); lane 2, serotype Bredeney (Bre2); lane 3, serotype Bredeney (Bre3); lane 4, serotype Anatum; lane 5, serotype Agona; lane 6, serotype Montevideo. (H) Lane 1, serotype Give; lane 2, serotype Bovis morbificans; lane 3, serotype Haifa; lane 4, serotype Mikawasima; lane 5, serotype Rissen; lane 6, serotype Kedougou. (I) Lane 1, serotype London; lane 2, serotype Manhattan; lane 3, serotype Cerro; lane 4, serotype Kapemba; lane 5, serotype III b.38: z10: z53 (“Salmonella enterica subsp. diarizonae”). PCR products were run on a 4% acrylamide-bisacrylamide gel (29:1) with a D-code electrophoresis system. Lanes without numbers, molecular-weight DNA markers (100-bp ladder).
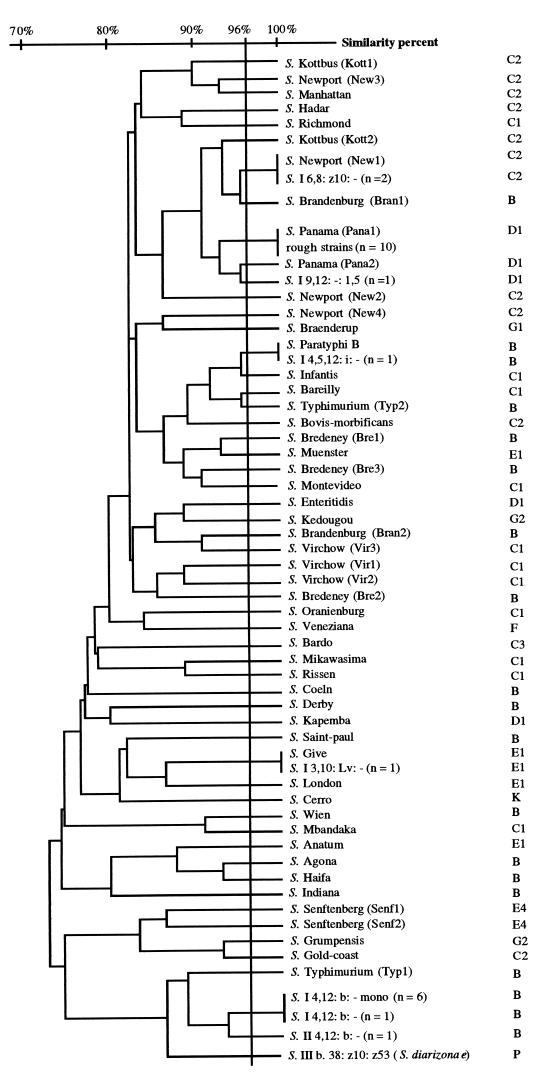
Clustering analysis based on the similarities recorded between the different RS-HP profiles shows that all RS-HP types were discriminated by at least two different bands corresponding to a similarity level of 96% (Fig. 3). Although different RS-HP profiles were found for all the serotypes identified in this study, some serotypes showed different profiles which were clustered at similarities sometimes below 80%. For instance, the two different profiles recorded for serotype Typhimurium isolates clustered at 72.4% similarity. Of the 41 Salmonella serotypes that were studied, 10 serotypes for which at least five isolates were analyzed (serotypes Agona, Coeln, Enteritidis, Grumpensis, Hadar, Indiana, Infantis, Montevideo, Paratyphi B, and Saint-Paul) showed single characteristic RS-HP type patterns. The different isolates which were analyzed for each serotype were found at different sampling times or stations. A single profile was also found for 23 serotypes, but the low number of strains representing each serotype did not allow us to decide if the profile resulted from isolated mutations in the ribosomal DNA spacer or if it corresponded to a single representative of an RS-HP type. The remaining eight serotypes each contained multiple RS-HP types that were unique to that serotype. For instance, of the 36 strains of serotype Brandenburg, 58.4% produced RS-HP type Bran1 and 41.6% produced the type Bran2.
FIG. 3.
UPGMA dendrogram generated from the similarity matrix determined from the different RS-HP banding patterns corresponding to 574 Salmonella spp. strains.
All the 10 isolates forming rough colonies and isolated from different samples showed the same RS-HP type and 100% homology with type Panal (Fig. 3). Four strains which were characterized by an incomplete serotype could be identified by the RS-HP profiles. Two strains clustered at 100% similarity with RS-HP-type New1, one with serotype Paratyphi B and the other with serotype Give. One isolate clustered at 95.5% similarity with Pana2. Eight isolates with an incomplete serological formula were not identified because the profiles clustered at low similarity values. Nevertheless, seven strains among the eight isolates showed a similar RS-HP type (Fig. 3).
For serotypes containing multiple RS-HP types, the number of isolates of each type and their environmental origin are presented in Table 2. For serotypes found in both river and wastewater samples, some RS-HP types (Bran2, New2, New3, New4, Senf1, Typ2, Vir1, and Vir2) were found only in river samples, whereas type Senf2 was found only in wastewater.
TABLE 2.
Distribution of Salmonella strain numbers within the different serotypes and environmental samples
| Serotype | RS-HP type | Rivera | Wastewater |
|---|---|---|---|
| Brandenburg | Bran1 | 4 | 17 |
| Bran2 | 15 | 0 | |
| Bredeney | Bre1 | 1 | 0 |
| Bre2 | 2 | 0 | |
| Bre3 | 1 | 0 | |
| Kottbus | Kott1 | 7 | 0 |
| Kott2 | 4 | 0 | |
| Newport | New1 | 8 | 55 |
| New2 | 3 | 0 | |
| New3 | 1 | 0 | |
| New4 | 13 | 0 | |
| Panama | Pana1 | 17 | 0 |
| Pana2 | 9 | 0 | |
| Senftenberg | Senf1 | 11 | 0 |
| Senf2 | 0 | 1 | |
| Typhimurium | Typ1 | 143 | 7 |
| Typ2 | 2 | 0 | |
| Virchow | Vir1 | 7 | 0 |
| Vir2 | 8 | 0 | |
| Vir3 | 1 | 1 |
Strains isolated from freshwater and sediment samples.
Although the Senf1 profile was found only for 11 isolates from river samples, the Senf2 type was reported for only one isolate in wastewater and serotype Senftenberg did not represent more than 12 isolates. Similarly, New1 was the single and dominant RS-HP type found in wastewater, but it was also found in river samples. Of the 152 strains of serotype Typhimurium, RS-HP type Typ1 was dominant in all samples and the Typ2 pattern was found for only two isolates in freshwater.
DISCUSSION
In a previous study, we quantified Salmonella spp. loads from the Tech River and from the submarine outfall of the wastewater treatment station located in Banyuls-sur-Mer (3). Bacterial loads were estimated during a 16-month period, and a stratified sampling strategy was used to analyze storm events. Salmonella spp. loads from the river were high during storm events, and the annual loads were higher than those estimated from the coastal outfall. Bacterial loads from the river represented 6.9 × 1012 organisms with at least 95% per year occurring during flood events. Those from the submarine outfall represented 4.7 × 1010 organisms per year with a regular temporal discharge. In the present study, we analyzed the diversity of 574 Salmonella strains isolated from these different aquatic biotopes. Identification was performed by traditional serotyping and amplification of the spacer regions between the 16S and 23S rRNA genetic loci.
Pathogens are constantly released at variable concentrations from infected humans, pets, farm animals, and wildlife (8). Municipal sewage and stormwater runoff become the conduits for the passage of pathogens into surface waters (10, 21). In this study, high levels of Salmonella strains were recorded in river and wastewater samples. Nevertheless, the highest diversity of strains was found during flood events, and some serotypes were detected only in these samples. Although wastewater discharge into the river represents a permanent source of pollution, flood events may mobilize a wide variety of Salmonella reservoirs located within the watershed and contribute to the dissemination of a large diversity of species which are transmitted to surface waters (5, 8, 10, 21). Some of the serotypes found only during storm events may correspond to species whose habitat is specific to particular animals and who are not disseminated from the permanent discharge of wastewaters. This is the case for serotypes Mbandaka, Virchow, Hadar, Indiana, Infantis, Saint-Paul, and Senftenberg, which are commonly isolated from poultry farms. Although the origin of these isolates remains unknown, they may originate from poultry breeding sites located within the watershed. In contrast, serotype Brandenburg was found in both wastewater and river samples, whereas this species is commonly isolated from pig breeding sites. Similarly, serotype Grumpensis was found only in river samples whereas this species is commonly found in food. Serotype Typhimurium and serotype Indiana were often isolated at station 2 in the river and outside flood events. Both serotypes are commonly isolated in human pathology and can originate from healthy carriers. Although it was beyond the scope of this work to determine the origin of Salmonella serotypes, it should be useful to identify Salmonella species in the different reservoirs located within the watershed to investigate the potential role of some of these species as indicators of specific pollution sources.
Serotype Typhimurium was dominant in all river samples and in marine and freshwater sediments. A high occurrence of this serotype has already been reported for polluted freshwaters (19, 28). Serotype Typhimurium is the dominant serotype commonly isolated from food and animal reservoirs but preferentially from bovine species. The presence of culturable species in marine sediments collected in the coastal area near the river discharge suggests that this serotype can survive for a long time in the natural environment. However, the absence of other serotypes in marine sediments cannot be interpreted as showing lower survival capacity because of the low number of isolated strains, which favors the selection of dominant species.
In contrast, serotype Newport was the dominant serotype in wastewater, and consequently, it was also found in river samples. Serotype Newport is commonly isolated from poultry breeding sites and food and from both animal and human reservoirs (25). However, this serotype was rarely found at stations 1 to 4 in the river, suggesting that this serotype does not survive well in the natural environment. Similarly, serotype Paratyphi B was found at stations 1 and 2 located upstream in the river and was never found in wastewaters. Although the sampling strategy used here for strain isolation does not allow any quantification of the different serotypes, further investigations should address the survival capacity of these serotypes in freshwaters.
Serological methods play an important role in the identification and characterization of many bacterial species, including those of Salmonella serotypes necessary for epidemiological evaluation. However, alternative techniques have been developed which allow a more rapid and simple identification of isolates. Furthermore, these methods sometimes provide a higher level of differentiation (13). Among these, the PCR ribotyping based on the amplification of the intergenic spacer region sequences between the 16S and 23S rRNA genes has proven very useful (17). Spacer regions within these loci show a significant level of length and sequence polymorphism across both genus and species lines (12, 15, 16, 22, 26, 30). This polymorphism can also result from variable numbers of rrn loci in the Salmonella genome (9, 12).
In this study, the size of DNA products ranged from 370 to 2,000 bases. This range is consistent with the range reported by Jensen and Hubner (13) using the same set of primers. Furthermore, the RS-HP types corresponding to serotypes Typhimurium (Typ1), Virchow (Vir1), Saint-Paul, Enteritidis, Infantis, and Derby were similar to those reported by these authors. All RS-HP profiles were characterized by (i) a first group of DNA fragments in the size range of 370 to 580 bases corresponding to homoduplex products and (ii) a second group of fragments in the size range of 680 to 2,000 bases corresponding to heteroduplex DNA structure formed by cross-hybridization of products (13).
All the serotypes found in this study were easily discriminated by RS-HP profiles. An examination of the RS-HP type groupings shows that the pattern types clustered into sets that accurately differentiated the Salmonella serotypes. This is congruent with the preliminary results obtained with only 28 different serotypes by Jensen and Hubner (13). Of 41 serotypes tested, 10 serotypes for which at least five isolates were analyzed showed a single characteristic RS-HP pattern and 8 showed multiple pattern types that were also unique to a given serotype characteristic RS-HP pattern.
The occurrence of multiple RS-HP profiles was more important for the strains isolated in the river samples. Inversely, those found in wastewater corresponded to a single profile which was sometimes found in the river, likely due to wastewater discharge in the river. Nevertheless, these results suggest that Salmonella cells in the natural environment are subject to higher genetic diversity and/or variability. This is emphasized by the high occurrence of rough strains and strains with an incomplete serotype which can result from genetic modifications. Although the heterogeneity within some serotypes is not yet explained, it could be due to mutations, any kind of genetic recombination, and/or horizontal transfer of nonhomologous DNA sequences as suggested by Luz et al. (18). Taddei et al. (24) have already demonstrated that environmental stresses increase the mutation rate of Escherichia coli strains and consequently could explain the genetic diversity of isolates.
Of the 156 serotype Typhimurium isolates, 98.7% were clustered with RS-HP type Typ1. This dominance of an RS-HP type within this serotype was not reported by Jensen and Hubner (13). However, this serotype is known to present high genetic diversity of the intergenic spacer region (17, 20). We have analyzed 10 additional strains isolated from viscera and animals and observed that these species were distributed within the same RS-HP types whereas one isolate showed a new profile (Typ3). However, the occurrence of Typ2 isolates was more significant (n = 4) than in aquatic samples (data not shown). It suggests that the diversity of profiles within each serotype may increase when the diversity of biotopes is increased.
We are not aware of any other study providing information on the diversity of Salmonella strains in the natural aquatic environment. Although Salmonella is commonly present in sewage effluents and surface waters, most studies on the distribution of this pathogenic genus have focused on its detection and quantification at the genus level. The identification of isolated strains by RS-HP patterns has proven to be a rapid and relatively low-cost method that allows the screening of a large number of strains. In some situations, this method was more sensitive than traditional serotyping and allowed the identification of strains with an incomplete serological formula. Furthermore, it was possible to discriminate species within a single serotype, suggesting genetic diversity and/or variability within a given serotype. Further investigations will be needed to understand the significance of this genetic heterogeneity.
In this study, it was shown that there is a large diversity of Salmonella serotypes within the natural aquatic environment. Furthermore, some serotypes were found only during storm events, suggesting that they were mobilized from widespread animal-rearing activities within the watershed, and for this reason, they may be good indicators of contamination sources. The RS-HP method may be a powerful tool that can be used for the identification of Salmonella isolates. Further studies should also address the relative survival capacity of these serotypes in the natural environment to better explain which factors control their survival and distribution in aquatic ecosystems.
ACKNOWLEDGMENTS
We thank Nicole Batailler, Joël Grabulos, Céline Fajon, Laetita Bernard, and Philippe Catala for their technical assistance during storm sampling and the Direction Départementale de l'Agriculture et des Forêts and the Direction Départementale de l'Equipement for providing permanent assistance during sampling.
This work was supported by a grant from the Agence de l'Eau Rhône-Méditerranée-Corse and the French Ministry of the Environment.
REFERENCES
- 1.Aguirre P M, Cacho J B, Folgeira L, Lopez M, Garcia J, Velasco A. Rapid fluorescence method for screening Salmonella spp. from enteric differential agars. J Clin Microbiol. 1990;28:148–149. doi: 10.1128/jcm.28.1.148-149.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baleux B, Alibou J, Trousselier M, Got P. Utilisation du bouillon Sélénite F modifié pour dénombrer Salmonella dans les milieux aquatiques. Rev Sci Eau. 1988;3:401–408. [Google Scholar]
- 3.Baudart J, Grabulos J, Barusseau J-P, Lebaron P. Salmonella spp. and fecal coliform loads in coastal waters from a point vs. nonpoint source of pollution. J Environ Qual. 2000;29:241–250. [Google Scholar]
- 4.Busse M. Media for Salmonella. Int J Food Microbiol. 1995;26:117–131. doi: 10.1016/0168-1605(93)e0030-u. [DOI] [PubMed] [Google Scholar]
- 5.Coyne M S, Howell J M. Agricultural impacts on fecal contamination of shallow groundwaters in the Bluegrass region of Kentucky. Soil Sci News Views. 1994;15:1–3. [Google Scholar]
- 6.Freydiere A M, Gille Y. Detection of salmonellae by using Rambach agar and by a C8 esterase spot test. J Clin Microbiol. 1991;29:2357–2359. doi: 10.1128/jcm.29.10.2357-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Pichel F, Prufert-Bebout L, Muyzer G. Phenotypic and phylogenetic analyses show Microcoleus chthonoplastes to be a cosmopolitan cyanobacterium. Appl Environ Microbiol. 1996;62:3284–3291. doi: 10.1128/aem.62.9.3284-3291.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geldreich E E. Pathogenic agents in freshwater resources. Hydrol Proc. 1996;10:315–333. [Google Scholar]
- 9.Gürtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 10.Irvine K N, Pettibone G W, Droppo I G. Indicator bacteria-sediment relationships: implications for water quality modeling and monitoring. In: James W, editor. Modern methods for modeling the management of stormwater impacts. Guelph, Ontario, Canada: Computational Hydraulics International; 1995. pp. 205–230. [Google Scholar]
- 11.Jensen M A, Straus N. Effect of PCR conditions on the formation of heteroduplex and single-stranded DNA products in the amplification of bacterial ribosomal DNA spacer regions. PCR Methods Appl. 1993;3:186–194. doi: 10.1101/gr.3.3.186. [DOI] [PubMed] [Google Scholar]
- 12.Jensen M A, Webster J A, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen M A, Hubner R J. Use of homoduplex ribosomal DNA spacer amplification products and heteroduplex cross-hybridization products in the identification of Salmonella serovars. Appl Environ Microbiol. 1996;62:2741–2746. doi: 10.1128/aem.62.8.2741-2746.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilger G, Grimont P A D. Differentiation of Salmonella phase 1 flagellar antigen types by restriction of the amplified fliC gene. J Clin Microbiol. 1993;31:1108–1110. doi: 10.1128/jcm.31.5.1108-1110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumari D N, Keer V, Hawkey P M, Parnell P, Joseph N, Richardson J F, Cookson B. Comparison and application of ribosome spacer DNA amplicon polymorphisms and pulsed-field gel electrophoresis for differentiation of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 1997;35:881–885. doi: 10.1128/jcm.35.4.881-885.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kur J, Burkiewicz A, Samet A, Sienkiewicz I. Identification of Serratia marcescens on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Acta Microbiol. 1995;44:219–225. [PubMed] [Google Scholar]
- 17.Lagatolla C, Dolzani L, Tonin E, Lavenia A, Di Michele M, Tommasini T, Monti-Bragadin C. PCR ribotyping for characterizing Salmonella isolates of different serotypes. J Clin Microbiol. 1996;34:2440–2443. doi: 10.1128/jcm.34.10.2440-2443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luz S P, Rodriguez-Valera F, Lan R, Reeves P R. Variations of the ribosomal operon 16S-23S gene spacer region in representatives of Salmonella enterica subspecies. J Bacteriol. 1998;180:2144–2151. doi: 10.1128/jb.180.8.2144-2151.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moriñigo M A, Borrego J J, Romero P. Comparative study of different methods for detection and enumeration of Salmonella spp. in natural waters. J Appl Bacteriol. 1986;61:169–176. doi: 10.1111/j.1365-2672.1986.tb04272.x. [DOI] [PubMed] [Google Scholar]
- 20.Nastasi A, Mammina C. Epidemiological evaluation by PCR ribotyping of sporadic and outbreak-associated strains of Salmonella enterica serotype Typhimurium. Res Microbiol. 1995;146:99–106. doi: 10.1016/0923-2508(96)80274-9. [DOI] [PubMed] [Google Scholar]
- 21.O'Shea M L, Field R. Detection and disinfection of pathogens in storm-generated flows. Can J Microbiol. 1992;38:267–276. doi: 10.1139/m92-045. [DOI] [PubMed] [Google Scholar]
- 22.Saruta K, Matsunaga T, Kono M, Hoshina S, Ikawa S, Sakai O, Machida K. Rapid identification and typing of Staphylococcus aureus by nested PCR amplified ribosomal DNA spacer region. FEMS Microbiol Lett. 1997;146:271–278. doi: 10.1111/j.1574-6968.1997.tb10204.x. [DOI] [PubMed] [Google Scholar]
- 23.Sokal R R, Sneath P H A. Principles of numerical taxonomy. W. H. San Francisco, Calif: Freeman; 1963. [Google Scholar]
- 24.Taddei F, Vulic M, Radman M, Matic I. Genetic variability and adaptation to stress. In: Bijlsma R, Loeschcke V, editors. Environmental stress, adaptation and evolution. Basel, Switzerland: Birkhaüser Press; 1997. pp. 271–290. [DOI] [PubMed] [Google Scholar]
- 25.Todd E D, Guzewich J J, Bryan F L. Surveillance of foodborne disease. IV. Dissemination and uses of surveillance data. J Food Prot. 1997;60:715–723. doi: 10.4315/0362-028X-60.6.715. [DOI] [PubMed] [Google Scholar]
- 26.Tyrrell G J, Bethune R N, Willey B, Low D E. Species identification of enterococci via intergenic ribosomal PCR. J Clin Microbiol. 1997;35:1054–1060. doi: 10.1128/jcm.35.5.1054-1060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vassiliadis P. The Rappaport-Vassiliadis (RV) enrichment medium for the isolation of Salmonellas: an overview. J Appl Bacteriol. 1983;54:69–76. doi: 10.1111/j.1365-2672.1983.tb01302.x. [DOI] [PubMed] [Google Scholar]
- 28.Venkateswaran K, Hashimoto H. Influence of indicator bacteria on the incidence of Salmonella in aquatic environment. Nippon Suisan Gakkaishi. 1988;54:253–258. [Google Scholar]
- 29.Venkateswaran K, Takai T, Navarro I M, Nakano H, Hashimoto H, Siebeling R J. Ecology of Vibrio cholerae non-O1 and Salmonella spp. and role of zooplankton in their seasonal distribution in Fukuyama coastal waters, Japan. Appl Environ Microbiol. 1989;55:1591–1598. doi: 10.1128/aem.55.6.1591-1598.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whiley R A, Duke B, Hardie J M, Hall L M. Heterogeneity among 16S-23S rRNA intergenic spacers of species within the ‘Streptococcus milleri group.’. Microbiology. 1995;141:1461–1467. doi: 10.1099/13500872-141-6-1461. [DOI] [PubMed] [Google Scholar]