We have shown that when a permanent occlusion of left anterior descending coronary artery (LAD) surgery is performed in newborn large mammal (pigs) on postnatal day 1 (P1) 1, the animals can completely recover from a myocardial infarction (MI) that occurs on postnatal day 28 (P28) with no evidence of scarring or decline in contractile performance.2 Our results also suggested that cardiac apical resection (AR) on P1 preserved cardiomyocyte (CM) cell-cycle activity as the animals aged; thus, we analyzed single-nucleus RNA-sequencing (snRNA-seq) datasets to compare the transcriptomes of cardiomyocytes from fetal (embryonic-day 80) pigs and from pigs that underwent AR surgery on P1 (ARP1), AR surgery on P1 followed by LAD occlusion MI induction on P28 (ARP1MIP28), MI on P28 without previous AR (MIP28), or neither surgical procedure (CTL). Hearts from ARP1MIP28 animals were explanted from P30-P56, and hearts from all other groups were explanted on P1, P28, or P56 (Figures A–D). All experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham (UAB) and performed under the National Institutes of Health “Guide for the Care and Use of Laboratory Animals”. UAB is licensed as an animal research facility (64-R-0004) by the USDA and has an Animal Welfare Assurance (A3255-01) on file with the Office of Laboratory Animal Welfare. Data were processed using the Seurat toolkit, latent representations of single nuclei were obtained with scVI-tools, and dimension reduction was performed via Uniform Manifold Approximation and Projection (UMAP).
Figure. snRNA-seq analysis identifies six cardiomyocyte subpopulations in injured and uninjured pig hearts.
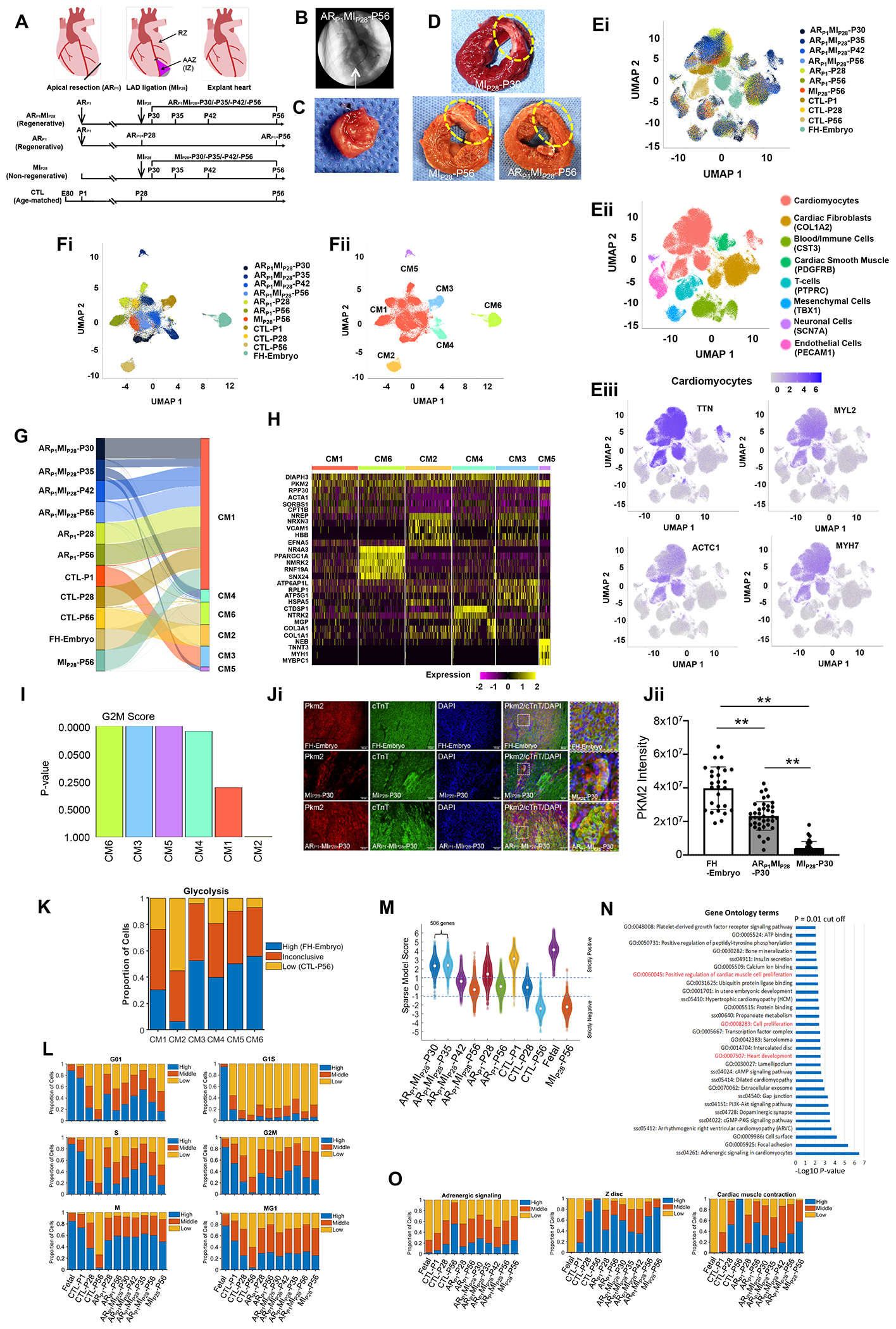
(A) Timeline of experimental procedures. (B) Coronary angiographic image with identification of the ligation site (arrow). (C-D) Images of (C) the resected apex and (D) left-ventricular sections; infarcted region is circled. (E-F) Cluster analysis for snRNA-seq data from (E) all cells and (F) cardiomyocytes; results are presented according to (Ei, Fi) experimental group/time-point, (Eii) cell type (defined by marker expression), (Eiii) abundance of each of four cardiomyocyte-specific genes, and (Fii) cardiomyocyte cluster. (G) Distribution of cardiomyocytes from each experimental group/time-point across CM clusters. (H) Heat map of expression levels for genes that were explicitly associated with each CM cluster. (I) The expression of genes associated with each cell cycle phase was analyzed and used to calculate a G2M-phase probability score for each CM cluster. (Ji) Representative images from each section FH-Embryo, ARP1MIP28-P30, and MIP28-P30 were stained with PKM2 antibody, cardiac Troponin T, and DAPI. (Jii) Quantification of PKM2 intensity by Image J. n=3 each. Square dots are for high magnification. The scale bar is 50 um. **P<0.01. (K) The expression of glycolysis genes in each CM cluster was compared to their expression in high (FH-embryo) and low (CTL-P56) cardiomyocytes. (L) The proportion of cardiomyocytes with high, medium, and low levels of expression for genes associated with each phase of the cell cycle. (M) Violin plot of the sparse model score. (N) Pathways that were significantly upregulated in ARP1MIP28-P30 and -P35 cardiomyocytes were identified via Gene Ontology Analysis. (O) The proportion of cardiomyocytes with high, medium, and low levels of expression for genes associated with adrenergic signaling, Z-disc development, and contractile activity.
All single-cell transcriptomic data, including the raw and processed files, are publicly available at Gene Expression Omnibus, Accession number GSE185289 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185289). A total of 218,945 high-quality nuclei were captured, and Louvain clustering analysis for known lineage markers identified all eight major cardiac cell-types (Figure E). Figure Ei shows that most of the non-cardiomyocytes clusters contain nuclei from all heart samples regardless of the treatment or age, indicating limited variation between different batches of sample collections. Complete gene-nucleus data were available for 94,844 cardiomyocytes, and variations between the anterior apical zone (AAZ) and the remote zone (RZ) within each group were negligible when compared to variations between groups. Cardiomyocytes were distributed among six clusters: CM1-CM6 (Figure F). Three of the clusters were almost entirely composed of cardiomyocytes from a single experimental group: CM2 (96.2% CTL-P56), CM3 (95.7% CTL-P1), and CM6 (92.4% FH-embryo); while clusters CM4 and CM5 contained primarily ARP1MIP28-P35 cardiomyocytes (68.6% and 97.8%, respectively). The CM1 cluster included cardiomyocytes from all other animal groups and time points, as well as a small number of ARP1MIP28-P35 cardiomyocytes, but very few (<1%) fetal or CTL-P56 cardiomyocytes (Figure G).
Interestingly, we found that CM3, CM4, CM5, and CM6 clusters have a significantly higher G2M score, representing a more proliferative capability (Figure I). Notably, CM4 had elevated expression of neurotrophic receptor tyrosine kinase 2 (NTRK2), involved in the hippo signaling pathway (Figure H). CM5 was characterized by highly expressed pyruvate kinase M2 (PKM2)(Figure H). Importantly, transient overexpression of Pkm2 has been linked to increases in cardiomyocyte proliferation and improvements in cardiac function after ischemic injury in mouse. Immunofluorescence analyses confirmed that ARP1MIP28-P30 cardiomyocytes expressed PKM2 protein (Figure Ji and Jii) with ample evidence of PKM2 in ARP1MIP28-P30 to P42 cardiomyocytes. Furthermore, glycolytic molecular programs were activated in CM5, almost similar to CM3 and CM6 (Figure K).
To evaluate the proliferative capacity of cardiomyocytes from each group and time-point, we analyzed our snRNA-seq datasets using Support Vector Machine (i.e., an algorithm for dividing a highly dimensional dataset3 into two categories, such as proliferative and nonproliferative) and the linear sparse technique.4, 5 Our results suggested that the proportion of CTL cardiomyocytes in each phase of the CMs cell-cycle declined from P1 to P56, while the proportions for CTL-P1 cardiomyocytes and cardiomyocytes from all other injury groups and at all other time points were broadly similar (Figure L). We also found 506 genes that were positively correlated with highly proliferative (ARP1MIP28-P30 and -P35) cardiomyocytes (Figure M), and Gene Ontology analysis indicated that these genes participated in pathways that regulated heart development, cell proliferation, and cardiomyocyte proliferation (Figure N). Furthermore, assessments of adrenergic signaling, Z-disc development, and contractile activity (Figure O) increased from P1 to P56 in CTL cardiomyocytes, while measurements in cardiomyocytes from all other injury groups and time points were ~40-60% of the measurements in CTL-P56 cardiomyocytes.
In conclusion, the results from our snRNA-seq analyses identified six distinct cardiomyocyte subpopulations, and two of the subpopulations (CM4 and CM5) were cardiomyocytes with increased cell-cycle activity and proliferation were very active by combined ARP1MIP28 injury, which contributed to the remuscularization of the injured LV. However, the interpretation of our results is limited by the multinuclear nature of cardiomyocytes and the lack of data for cytoplasmic RNA transcripts. Furthermore, it will be interesting to examine these two distinct cell populations from ARP1MIP28 injury in future studies. Collectively, these observations provide a foundation for future investigations of the molecular mechanisms and signaling molecules that regulate the injury-induced preservation of cardiomyocyte cell-cycle activity in newborn large mammals.
Acknowledgments:
We thank the UAB Flow Cytometry and Single Cell Core, and the UAB Genomics Core.
Sources of Funding:
This work was supported by the following funding sources: NIH RO1s HL114120, HL 131017, HL149137, and UO1 HL134764.
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability:
No any restrictions on materials and data access. All data is available in the main text or the supplementary materials.
References
- 1.Zhu W, Zhang E, Zhao M, Chong Z, Fan C, Tang Y, Hunter JD, Borovjagin AV, Walcott GP, Chen JY, et al. Regenerative Potential of Neonatal Porcine Hearts. Circulation. 2018;138:2809–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao M, Zhang E, Wei Y, Zhou Y, Walcott GP and Zhang J. Apical Resection Prolongs the Cell Cycle Activity and Promotes Myocardial Regeneration After Left Ventricular Injury in Neonatal Pig. Circulation. 2020;142:913–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haghverdi L, Buettner F and Theis FJ. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 2015;31:2989–98. [DOI] [PubMed] [Google Scholar]
- 4.Bi J, Bennett K, Embrechts M, Breneman C and Song M. Dimensionality reduction via sparse support vector machines. Journal of Machine Learning Research. 2003;3:1229–1243. [Google Scholar]
- 5.Chkifa A, Cohen A and Schwab C. Breaking the curse of dimensionality in sparse polynomial approximation of parametric PDEs. Journal de Mathématiques Pures et Appliquées. 2015;103:400–428. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No any restrictions on materials and data access. All data is available in the main text or the supplementary materials.


