Acute myeloid leukemia (AML) is an aggressive blood cancer with poor prognosis.1 AML is thought to be initiated and maintained by a rare population of leukemia stem cells (LSCs) or leukemia-initiating cells (LICs) that have acquired the capacity for self-renewal and is blocked in their ability to differentiate by the accumulation of a series of mutations and/or epigenetic changes.2-3 Clinical studies show that LICs are resistant to conventional chemotherapy and/or targeted therapies.3 Thus, there is an unmet need to elucidate the molecular mechanisms governing LIC self-renewal and develop novel therapeutic approaches that can target LICs and improve leukemia treatment.3
The tumor suppressor p53 is a stress response protein that regulates a large number of genes in response to a variety of cellular insults, including oncogene activation, DNA damage and inflammation.4 These signals activate p53 primarily through post-translational modifications that result in augmented levels of p53 protein and transactivation activity.4 Activated p53 suppresses cellular transformation mainly by inducing growth arrest, apoptosis, DNA repair and differentiation in damaged cells.4 Accordingly, p53 function is always compromised in tumor cells, usually as a result of somatic mutations and deletions, which occur in approximately half of all human cancers.5 The TP53 gene encodes the tumor suppressor p53. The frequency of TP53 mutations in AML is approximately 10%. However, in AML with complex karyotype, the frequency of TP53 mutations and/or deletions is almost 70%.6 While TP53 mutations confer drug resistance and poor prognosis in AML, the role of mutant p53 in the initiation and progression of AML is largely unknown.6-7
We have been investigating the role of tumor suppressor p53 in normal and malignant hematopoiesis. We found that wild type p53 maintains HSC quiescence and inhibits HSC self-renewal.8 Codon 248 of p53 is frequently mutated in AML and p53R248W has been shown to be a gain-of function (GOF) mutant in human cancer cells as well as in animal models.6-7, 9 We recently reported that p53R248W enhances HSC self-renewal in steady state and promotes HSC expansion following genotoxic stresses.10 Of note, homozygous p53−/− and p53R248W/R248W mice develop lymphoid tumors, including lymphoma and thymoma, but not myeloid malignancies,9 suggesting that expression of mutant p53 is not sufficient for inducing myeloid leukemia in mice. This has led to a search for potential second hits that cooperate with mutant p53 in the pathogenesis of myeloid malignancies, primarily focused on using mouse models.
While coexisting mutations with TP53 mutations in AML are limited,6-7 previous studies indicate that TP53 mutations co-occur with AML driver mutations in oncogenic signaling molecules such as FMS-like tyrosine kinase receptor-3 (FLT3).11 Mutations in FLT3 have been identified in myeloid malignancies, including myeloproliferative neoplasms (MPN) and AML.12 Internal tandem duplications in the juxtamembrane domain (FLT3-ITD) and mutations in the activating loop of FLT3 (FLT3-TKD) are seen in 30 to 35% of AML patients.12 Both ITD and TKD mutations of FLT3 lead to constitutive activation of the tyrosine kinase, promoting proliferation and survival of leukemic blasts.12 Given that expression of FLT3-ITD in the hematopoietic compartment results in MPN in mice and that FLT3-ITD impairs HSC self-renewal in vivo,13 we reasoned that mutant p53 might synergize with FLT3-ITD in driving the development of myeloid leukemia through enhancing LIC self-renewal.
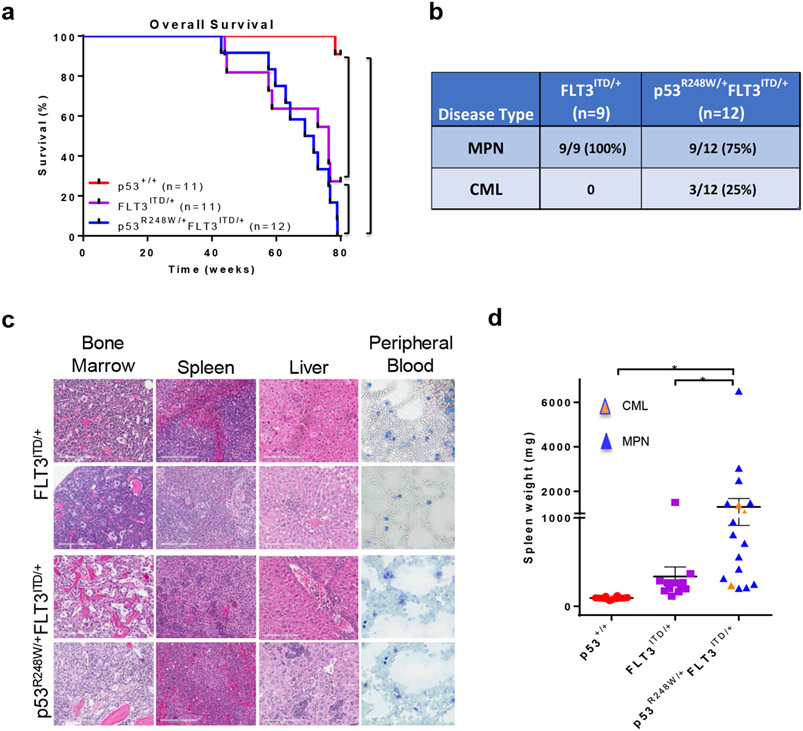
To test this hypothesis, we generated p53R248W/+FLT3ITD/+ mice and monitored overall survival and tumor development of these mice. We observed that both FLT3ITD/+ and p53R248W/+FLT3ITD/+ mice have decreased life span compared to p53+/+ mice (Figure 1a). Some p53R248W/+ mice develop myelodysplastic syndromes (MDS) with age and other p53R248W/+ mice developed lymphoma and sarcoma based upon pathological analysis of bone marrow (BM), spleen, liver, and peripheral blood (PB) (S.C. and Y.L., unpublished data). However, the majority of p53R248W/+FLT3ITD/+ mice developed MPN as seen in FLT3ITD/+ mice (Figures 1b, 1c and data not shown), suggesting that FLT3-ITD-induced MPN development does not depend on mutant p53. Histological observation of spleen sections from MPN mice showed disarray of normal splenic architecture with a reduction and almost total absence of the white pulp in some cases and increased red pulp area with increased extramedullary hematopoiesis (Figure 1c). These features appeared in conjunction with hepatosplenomegaly, variable leukocytosis and overproduction of myeloid cells in bone marrow, spleen and peripheral blood (Figure 1c). We noted that bone marrow cellularity decreased as splenomegaly increased, consistent with increased extramedullary hematopoiesis. Notably, approximately 25% of p53R248W/+FLT3ITD/+ mice developed chronic myeloid leukemia (CML).14 Upon necropsy, mice with CML displayed severe splenomegaly, and some also displayed hepatomegaly. Morphological analysis of peripheral blood smears revealed increased myeloid cells with dysplastic features (Figure 1c). Bone marrow cellularity varied from hypocellular to hypercellular among animals. Increased number of myeloid cells (blast to immature myeloid cells) was observed in bone marrow with extensive spread of myeloid elements in spleen and in a few livers (Figure 1c). While p53R248W/+FLT3ITD/+ mice showed marked splenomegaly compared to p53+/+ and FLT3ITD/+ mice (Figure 1d), this is not likely due to CML development as majority of double-mutant mice developed MPN (Figures 1b and 1c).
Figure 1.
Mutant p53 cooperates with FLT3-ITD in the pathogenesis of myeloid leukemia. (a) FLT3ITD/+ and p53R248W/+FLT3ITD/+ mice show decreased survival compared to p53+/+ mice (n=11, p53+/+; n=11, FLT3ITD/+; n=12, p53R248W/+FLT3ITD/+, **p<0.01, ****p<0.0001). (b) Disease spectrums in FLT3ITD/+ and p53R248W/+FLT3ITD/+ mice were determined by pathological analysis of bone marrow, spleen, liver, and peripheral blood (n=9, FLT3ITD/+; n=12, p53R248W/+FLT3ITD/+). (c) Representative H&E (20X) images of bone marrow, spleen, liver and peripheral blood smears from FLT3ITD/+ mice with MPN and p53R248W/+FLT3ITD/+ mice with CML. (d) Spleen weight of p53+/+, FLT3ITD/+, and p53R248W/+FLT3ITD/+ mice. Mean values (±SEM) are shown (n=12, p53+/+; n=12, FLT3ITD/+; n=17, p53R248W/+FLT3ITD/+, *p<0.05).
Given that patients with homozygous FLT3-ITD mutations have a more severe disease compared to those with heterozygous FLT3-ITD mutations,11-12 we examined whether mutant p53 cooperates with homozygous FLT3-ITD mutant in leukemia development. We transplanted 3 x 106 whole bone marrow cells from p53+/+, FLT3ITD/ITD, or p53R248W/+FLT3ITD/ITD mice into lethally irradiated recipient mice and measured their overall survival. Both FLT3ITD/ITD and p53R248W/+FLT3ITD/ITD recipient mice had decreased life spans compared to p53+/+ recipient mice (Figure S1a). Interestingly, approximately 30% of the p53R248W/+FLT3ITD/ITD transplanted animals developed CML (Figure S1b), similar to that seen in p53R248W/+FLT3ITD/+ animals (Figure 1b). Rest of the p53R248W/+FLT3ITD/ITD mice developed MPN (Figure S1b).
Given that some p53R248W/+FLT3ITD/+ mice develop CML, we next examined the impact of mutant p53 on FLT3-ITD+ hematopoietic stem and progenitor cells (HSPCs) in order to understand the underlying mechanisms. We first analyzed peripheral blood (PB), bone marrow (BM), and spleen of p53+/+, p53R248W/+, FLT3ITD/+ and p53R248W/+FLT3ITD/+ mice (8 to 12 week-old). PB white blood cell (WBC) counts, BM cellularity, and spleen weight were comparable among the four groups of mice (Figures S1c, S1d, and S1e). We then examined the frequency of hematopoietic stem and progenitor cells in the BM of p53R248W/+FLT3ITD/+ mice. While the number of LT-HSCs and ST-HSCs was comparable among these mice, LSKs and MPPs were expanded in the p53R248W/+FLT3ITD/+ mice compared with that of the p53+/+, p53R248W/+ and FLT3ITD/+mice (Figure S2a). We also observed increased frequency of myeloid progenitors (Lin−Kit+ cells) in the bone marrow of p53R248W/+FLT3ITD/+ mice (Figure S2b). These findings suggest that the effects of mutant p53 and FLT3-ITD on myeloid progenitor cell expansion appears additive. However, the number of common lymphoid progenitors (CLPs) was comparable among four group of mice (Figure S2c).
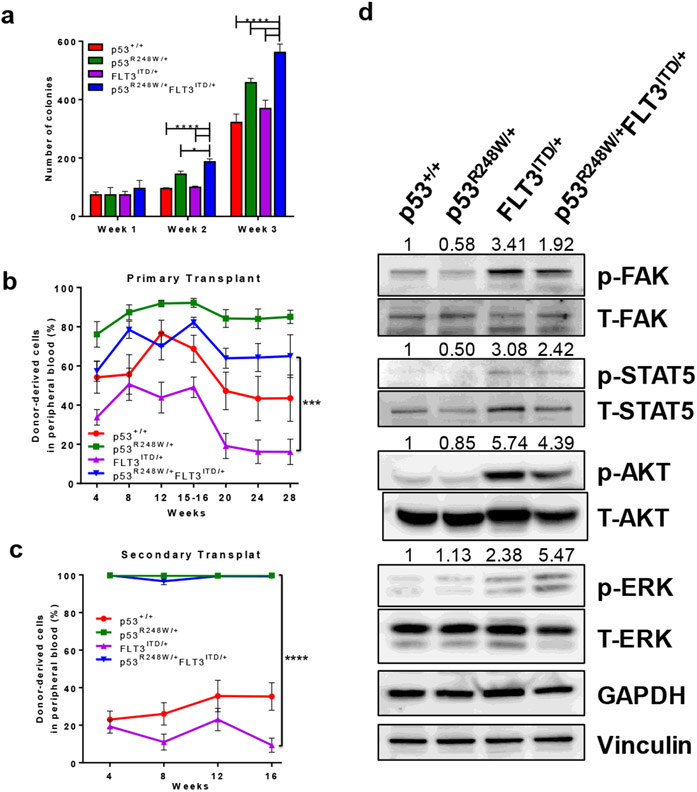
FLT3 mutations have been show to enhance the proliferation of hematopoietic stem and progenitor cells.13 We then examined the cell cycle status of LSKs isolated from p53+/+, p53R248W/+, FLT3ITD/+ and p53R248W/+FLT3ITD/+ mice. We confirmed that FLT3ITD/+ LSKs shown enhanced proliferation compared with p53+/+ LSKs (Figure S2d). However, mutant p53 did not alter the proliferation of FLT3ITD/+ LSKs (Figure S2d). To determine the impact of mutant p53 on myeloid progenitors, we performed serial replating assays using BM cells from p53+/+, p53R248W/+, FLT3ITD/+ and p53R248W/+FLT3ITD/+ mice. While the colony formation potential of p53+/+ and FLT3ITD/+ BM cells was comparable in serial replating assays, p53R248W/+FLT3ITD/+ BM cells show enhanced replating potential compared to p53R248W/+ and FLT3ITD/+ BM cells (Figure 2a), suggesting that expanded myeloid progenitors in p53R248W/+FLT3ITD/+ mice are functional in vitro.
Figure 2.
Mutant p53 enhances the self-renewal potential of FLT3-ITD+ LICs. (a) Serial replating assays of bone marrow cells from young p53+/+, p53R248W/+, FLT3ITD/+ and p53R248W/+FLT3ITD/+ mice. Mean values (±SD) are shown (n=3, *p<0.05, ****p<0.0001). (b) p53R248W enhances the repopulating potential of FLT3ITD/+ hematopoietic cells. Percentage of donor-derived (CD45.2+) cells in the peripheral blood of primary recipient mice post-transplantation, measured at 4-week intervals. Mean values (±SEM) are shown (n=7, ***p<0.001). (c) The percentage of donor-derived cells in the peripheral blood of secondary recipient mice. Mean values (±SEM) shown, (n=7, p53R248W/+ vs FLT3ITD/+ and FLT3ITD/+ vs p53R248W/+FLT3ITD/+, ****p<0.0001). (d) Western blot analysis of activated and total FAK, STAT5, AKT, and ERK protein levels in p53+/+, p53R248W/+, FLT3ITD/+ and p53R248W/+FLT3ITD/+ mononuclear cells differentiated into macrophage progenitors. Loading controls GAPDH and Vinculin are also shown. Quantification of phosphorylated proteins was calculated relative to total protein level and is displayed above each respective phospho-protein.
To examine the impact of mutant p53 on HSCs in vivo, we performed serial competitive bone marrow transplantation assays. We transplanted 5 x 105 donor BM cells (p53+/+, p53R248W/+, FLT3ITD/+ or p53R248W/+FLT3ITD/+, CD45.2+) into lethally irradiated (11 Gy) F1 recipient mice (CD45.1+CD45.2+) along with 5 x 105 competitor BM cells (CD45.1+). Peripheral blood white blood cell counts were comparable among the four groups of mice following transplantation (Figure S2e). While FLT3ITD/+ BM cells showed decreased repopulating ability compared to p53+/+ cells 16 weeks post transplantation, p53R248W/+FLT3ITD/+ BM cells displayed enhanced engraftment compared to FLT3ITD/+ BM cells (Figure 2b). We then sacrificed the recipient mice and examined the frequency of donor-derived hematopoietic stem and progenitor cells in their bone marrow. We found increased number of donor-derived LSKs in the BM of recipient mice repopulated with p53R248W/+ BM cells compared to that of the p53+/+ and FLT3ITD/+ BM cells, whereas the frequency of donor-derived LSKs in the BM of recipient mice repopulated with p53R248W/+ and p53R248W/+FLT3ITD/+ cells was comparable (Figure S2f). We found increased number of donor-derived GMPs in the BM of recipient mice repopulated with p53R248W/+FLT3ITD/+ bone marrow cells compared to that of the FLT3ITD/+ BM cells (Figure S3a). The spleen size was comparable in recipient mice repopulated with four group of BM cells (Figure S3b).
To determine the impact of mutant p53 on the self-renewal potential of FLT3-ITD+ HSCs, we transplanted 3 x 106 BM cells isolated from the primary recipient mice repopulated with p53+/+, p53R248W/+, FLT3ITD/+ or p53R248W/+FLT3ITD/+ cells into lethally irradiated secondary F1 recipients. Sixteen weeks after transplantation, p53R248W/+FLT3ITD/+ cells continued to show increased repopulating ability compared to FLT3ITD/+ BM cells (Figure 2c). These findings suggest that mutant p53 may promote leukemic transformation through enhancing LIC self-renewal.
To examine the impact of mutant p53 on oncogenic signaling pathways, we performed western blot analysis on macrophage progenitor cells derived from p53+/+, p53R248W/+, FLT3ITD/+ or p53R248W/+FLT3ITD/+ bone marrow cells. Consistent with previous studies, cells from FLT3ITD/+ mice had activated FAK, STAT5, and AKT (Figure 2d). Further, expressing FLT3-ITD in a mutant p53 background enhances activated ERK levels but slightly decreases activated FAK and STAT5 levels (Figure 2d). We found increased levels of FLT3 in p53R248W/+FLT3ITD/+ macrophage progenitor cells (Figure S3c). Thus,expressing FLT3-ITD in a mutant p53 background has no effect on FLT3-ITD-induced activation of signaling pathways. However, ERK inhibitor treatment decreased the replating potential of p53R248W/+, FLT3ITD/+ and p53R248W/+FLT3ITD/+ bone marrow cells (Figure S3d). These findings suggest that mutant p53 and FLT3-ITD may function through different signaling pathways in the pathogenesis of hematological malignancies. In the future, we will elucidate the mechanisms by which mutant p53 upregulates FLT3 in HSPCs.
While TP53 and FLT3 mutations are rarely co-occur in MPN and AML,11 the underlying mechanisms are not known. We found that the majority of p53R248W/+FLT3ITD/+ mice developed MPN, as seen in FLT3ITD/+ mice.13 Further, we discovered that mutant p53 and FLT3-ITD cooperate in CML development in mice. Functionally, mutant p53 synergizes with FLT3-ITD to expand the myeloid progenitor cell pool and enhance the self-renewal potential of LICs. TP53 mutations are present in both chronic and blast crisis phase of CML,15 underscoring the importance of mutant p53 in CML pathogenesis. Delineating the role of mutant p53 and FLT3-ITD in LIC self-renewal and pathogenesis of hematological malignancies may facilitate the development of novel therapeutic approaches that can improve leukemia treatment.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs, through the Bone Marrow Failure Research Program - Idea Development Award under Award No. W81XWH-18-1-0265 to YL. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. This work was also supported in part by two NIH R56 Awards (R56DK119524-01 and R56AG05250), a DoD Career Development Award W81XWH-13-1-0187, a Scholar Award from the St. Baldrick’s Foundation, an Elsa Pardee Foundation New Investigator Award, a Leukemia Research Foundation New Investigator Award, a Showalter Trust Fund New Investigator Award, an Alex Lemonade Stand Foundation grant, a Children’s Leukemia Research Association grant, and an American Cancer Society Institutional Research Grant to YL. SCN was supported by a NIH F32 Award 1F32CA203049-01. The authors would like to acknowledge the Flow Cytometry Core and In vivo Therapeutic Core Laboratories, which were sponsored, in part, by the NIDDK Cooperative Center of Excellence in Hematology (CCEH) grant U54 DK106846. This work was supported, in part, by a Project Development Team within the ICTSI NIH/NCRR Grant Number UL1TR001108. We would like to thank Dr. Yang Xu at USCD for providing the p53R248W mice to the study.
Footnotes
CONFLICT OF INTEREST
The authors declared that no conflicts of interest exists.
REFERENCES
- 1.Coombs CC, Tallman MS, Levine RL. Molecular therapy for acute myeloid leukaemia. Nat Rev Clin Oncol 2016;13:305–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat Immunol 2004; 5:738–43. [DOI] [PubMed] [Google Scholar]
- 3.Essers MA, Trumpp A. Targeting leukemic stem cells by breaking their dormancy. Mol Oncol 2010; 4: 443–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov 2014; 13:217–36. [DOI] [PubMed] [Google Scholar]
- 5.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 2009; 9:701–13. [DOI] [PubMed] [Google Scholar]
- 6.Rücker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood 2012; 119: 2114–21. [DOI] [PubMed] [Google Scholar]
- 7.Prokocimer M, Molchadsky A, Rotter V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: projections on diagnostic workup and therapy. Blood 2017;130:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Elf SE, Miyata Y, Sashida G, Liu YH, Huang G et al. p53 Regulates Hematopoietic Stem Cell Quiescence. Cell Stem Cell 2009; 4: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol 2007; 15: 376–88. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Gao R, Yao C, Kobayashi M, Liu SZ et al. Genotoxic stresses promotes the clonal expansion of hematopoietic stem cells expressing mutant p53. Leukemia 2018; 32:850–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garg M, Nagata Y, Kanojia D, Mayakonda A, Yoshida K, Haridas Keloth S et al. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood 2015;126:2491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swords R, Freeman C, Giles F. Targeting the FMS-like tyrosine kinase 3 in acute myeloid leukemia. Leukemia 2012; 26:2176–85. [DOI] [PubMed] [Google Scholar]
- 13.Lee BH, Tothova Z, Levine RL, Anderson K, Buza-Vidas N et al. FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer Cell 2007;12:367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morse HC 3rd, Anver MR, Fredrickson TN, Haines DC, Harris AW, Harris NL, Jaffe ES, Kogan SC, MacLennan IC, Pattengale PK, Ward JM; Hematopathology subcommittee of the Mouse Models of Human Cancers Consortium. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood 2002;100:246–58. [DOI] [PubMed] [Google Scholar]
- 15.Menezes J, Salgado RN, Acquadro F, Gómez-López G, Carralero MC, Barroso A, Mercadillo F, Espinosa-Hevia L, Talavera-Casañas JG, Pisano DG, Alvarez S, Cigudosa JC. ASXL1, TP53 and IKZF3 mutations are present in the chronic phase and blast crisis of chronic myeloid leukemia. Blood Cancer J 2013;3:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.