Abstract
There are several DNA helicases involved in seemingly overlapping aspects of homologous and homoeologous recombination. Mutations of many of these helicases are directly implicated in genetic diseases including cancer, rapid aging, and infertility. MCM8/9 are recent additions to the catalog of helicases involved in recombination, and so far, the evidence is sparse, making assignment of function difficult. Mutations in MCM8/9 correlate principally with primary ovarian failure/insufficiency (POF/POI) and infertility indicating a meiotic defect. However, they also act when replication forks collapse/break shuttling products into mitotic recombination and several mutations are found in various somatic cancers. This review puts MCM8/9 in context with other replication and recombination helicases to narrow down its genomic maintenance role. We discuss the known structure/function relationship, the mutational spectrum, and dissect the available cellular and organismal data to better define its role in recombination.
Keywords: MCM8, MCM9, DNA replication, DNA repair, homologous recombination, infertility, cancer, helicase
1. Introduction
Cellular survival and propagation of an uncorrupted genome by the replisome is dependent on the ability of the cell to efficiently recognize and remove genetic insults which impede routine nuclear processes and threaten genomic integrity. Further, the origins, whether endogenous or exogenous, and biochemical outcomes of DNA damage are fraught with chemical and physical diversity. These genomic insults can result in replisome stalling and fork collapse giving rise to DNA double-strand breaks (DSBs). The total number of DSBs suggested to form is staggering. Up to ten DSBs are projected to form in any given cell per day, and based on current estimates of cellular abundance (3.72 × 1013) in the human body, 372 trillion DSBs are predicted to form every day [1]. Fortunately, there are a multitude of pathways that the cell has to prevent DSB formation or repair a DSB once formed. A comprehensive assessment of these pathways and mechanisms are beyond the scope of this review, but should the reader seek more information, multiple recent reviews can be easily found [2–5].
Prolonged replisome stalling represents a serious threat to cellular survival due to replication fork collapse and potential DSB formation. In response, the cell has evolved several mechanisms to mediate replisome progression, prevent replication fork collapse, and initiate fork restart to restore replication progression. Should DSBs happen, the global pathways of non-homologous end joining (NHEJ) and homologous recombination (HR), each with its own menagerie of sub-pathways are the most general mechanisms of repair. Choice between NHEJ and HR is mainly influenced by the cell cycle phase. NHEJ serves as the default pathway and predominates during G1. As cells progress into S and then G2, NHEJ is inhibited and the balance shifts toward HR repair [6]. Mitotic HR requires that a homologous sister chromatid be present to serve as a template from which to restore genetic information. The processed 3’ end of the DSB coated with the Rad51 recombinase invades the sister chromatid and searches for homology for downstream synthesis. A single Holliday junction (HJ) intermediate is formed and dissolved after sufficient synthesis to produce primarily non-crossovers (NCO) products.
Meiotic recombination shares many similarities and components to that discussed for mitotic HR with some specific differences [7, 8]. First, meiotic DSB formation is a more highly controlled process catalyzed by Spo11 and other accessory proteins acting primarily during the leptotene stage of meiosis. Instead of Rad51, the meiosis-specific recombinase, Dmc1, catalyzes strand invasion, synapsis, and facilitates more extensive synthesis to promote capture of the free DSB end to create a joint double Holliday junction (dHJ) intermediate. Structure and meiosis-specific nucleases then cleave (or resolve) dHJs to facilitate crossover formation (CO) to promote genetic recombination.
During all these processes, there are a multitude of DNA helicases that act in specific ways and on specific intermediates to prevent replication fork collapse, restart stalled forks, facilitate strand resection, mediate homology search, and promote dissolution of HJs. A newly identified helicase complex that is emerging as an important mediator of DNA repair and guardian of genomic integrity is formed between the minichromosome maintenance (MCM) 8 and 9 proteins [9–12].
2. Roles of MCM8/9 in meiosis, infertility, and cancer
Initial experiments by Rhoda Grell in Drosophila melanogaster in 1984 identified a temperature sensitive recombination gene termed recombination defective (REC) [13]. REC null organisms displayed higher levels of nondisjunction and infertility despite undisrupted homologous chromosome pairing, and so, it was proposed that REC functions to create CO during meiosis. Subsequent deletion, restriction mapping, and sequencing work localized the REC gene to chromosome 3 and was identified as a homolog of MCM8 [14, 15]. Further, MCM8 deficiency reduced CO events by ~95%, were abnormally distributed, and gene conversion tract lengths were ~2-fold shorter. Based on these data, it was proposed that dmMCM8 functions after SPN-A (Rad51 homolog) but before the MEI-9 endonuclease to aid in strand invasion and DNA synthesis to give rise to successful CO events.
Links connecting MCM8 and 9 to infertility and cancer did not come until several years after their initial discovery when Hartford and colleagues, prompted by a report that MCM9 interacts with Cdt1 to assist MCM2–7 loading prior to replication firing, created MCM9 knockout mice using gene trap insertions. MCM9−/− mice were viable and grossly normal with no discernible MCM2–7 loading defects. However, MCM9-mutated mouse embryonic fibroblasts (MEFs) exposed to aphidicolin (a B-family DNA polymerase inhibitor) were delayed in their progression from G0/G1 to S-phase [16]. Male and female MCM9-mutated mice also exhibited reproductive system abnormalities and germ cell depletion. Removal of p53 and p21 through cross-breeding further depleted germ cell abundance and indicated that loss likely occurs through an apoptotic-independent pathway. Female mice also displayed fewer primordial follicles, were infertile, and presented hyperplasias and tubulostromal adenomas consistent with pre-mature ovarian failure (POF). Further, an increase, though minimal, in genome instability was observed along with an increased incidence of sex-specific tumor formation with a higher incidence of hepatocellular carcinoma in males, while females were prone to ovarian tumor formation. Based on these data, it was proposed that MCM8/9 function in genome maintenance during meiotic replication-induced stress.
Currently, multiple reports have corroborated mutations in MCM8 or 9 with POF leading to infertility in humans [17–22]. The genetic basis of POF is complex with ~1% of women under 40 years-of-age affected. Many of the molecular mechanisms which cause POF are unknown, however the disruption of a multitude of genes involved in all stages of oogenesis, folliculogenesis, and DNA repair in establishing and maintaining the ovarian reserve have been implicated [23].
The first report outlining MCM8/9 contributions to POF in humans came in 2014/15 when our colleagues examined daughters from three separate consanguineous families that exhibited primary amenorrhea, short stature, low weight, and genomic instability [17, 18]. A combination of single nucleotide polymorphism (SNP) arrays, comparative genomic hybridization, and whole-exome sequencing identified a C>T nonsense mutation that likely results in an early truncated form of MCM9 and a T>C mutation that disrupted normal splicing between exons 9 and 10 of MCM9. Within the affected MCM8 family, a missense mutation resulted in the amino acid change P149R within the canonical DNA binding domain. For all these mutants, the ability to form damage foci upon treatment with the DNA cross-linker mitomycin C (MMC) was severely compromised and affected patient lymphocytes showed a significant increase in chromosomal breaks and rearrangements.
Since these initial discoveries, multiple mutations in MCM8 and MCM9 have been associated with POF (Figure 1A–C) and direct links to cancer are less clear. Only a single report links mutations in MCM9 with mixed polyposis and early-onset colorectal cancer [22], although another study found it was more rare in hereditary Lynch syndrome patients in a broader Australian population [24]. Even so, there are more than 460 mutations in MCM8 or MCM9 cataloged in The Cancer Genome Atlas and cBioPortal for Cancer Genomics implicating MCM8/9 in cancer transformation or progression that are awaiting characterization. The MCM8/9 mutations identified in POF patients (Figure 1A–C) and the ever expanding database of cancer genomes with MCM8 or 9 mutations all lack sufficient biochemical characterization to detail what molecular and cellular effects these mutations have on contributing to POF or cancer progression.
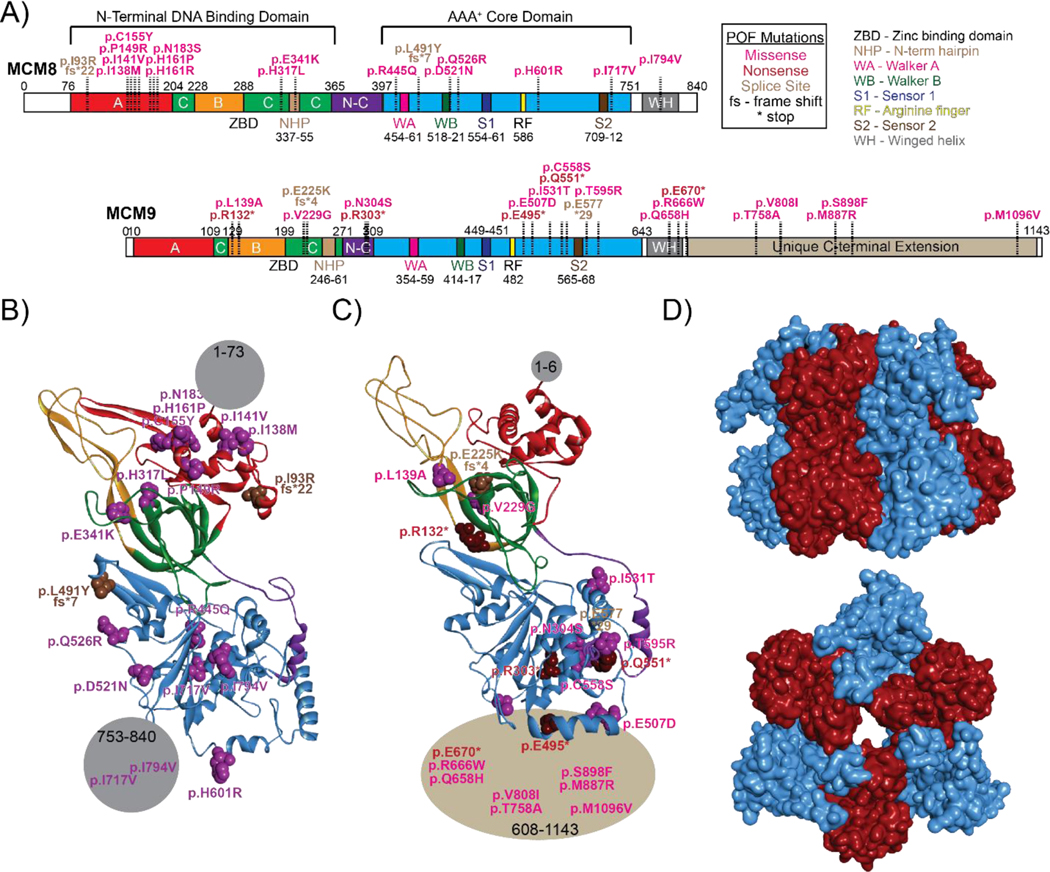
Figure 1:
POF implicated mutations mapped onto the A) primary sequences of MCM8 or MCM9 with indicated domains and motifs for each. Structural homology models shown in ribbon of B) MCM8 or C) MCM9 modelled from the most closely related yeast MCM2 subunit (PDBID:3JA8) with POF mutations mapped as indicated. D) Predicted quaternary homology model of the heterohexameric structure of MCM8/9 built upon the yeast MCM2–7 complex.
3. Identification, conservation, and structural characterization of MCM8/9
Though MCM8 and 9 form a functional complex in vivo, the two enzymes and alternatively spliced isoforms were discovered separately through bioinformatics analyses by multiple groups [25–29]. MCM8 and 9 arose early during eukaryotic evolution and are conserved among many eukaryotic organisms, with yeast and C. elegans as exceptions, which is suggestive of a co-evolutionary function [30]. However, D. melanogaster and related species possess only the MCM8 protein (i.e. REC) in the absence of an MCM9 partner, suggestive of a divergence point during evolution in which MCM8 assumed the role of MCM9 or MCM9 presence was redundant and selected against.
MCM8 and 9 are homologs of the MCM2–7 hetero-hexameric helicase complex, which together with Cdt1 and GINS make up the CMG complex becoming the primary DNA helicase involved in eukaryotic replication. MCM8/9 was first thought to aid in replication initiation [31–33], however subsequent studies have argued instead for a role during replication elongation or recombination [34, 35], although direct interactions with the other MCM2–7 subunits has not been found. At the time of writing this review, there has been no structural information (X-ray or EM) for MCM8 or 9. However, MCM structures from several species (including eukaryotic) have been solved [36–39]. It is likely that the highly conserved helicase domains of MCM8 (Figure 1B) and MCM9 (Figure 1C) adopt similar heterohexameric structures to that of the other MCM complexes (Figure 1D). The precise stoichiometry of the MCM8/9 complex also remains to be definitively addressed. Initial differential centrifugation and gel filtration experiments suggest that MCM8/9 can exist within a range of dimeric to hexameric species [11, 34]. With this data, in combination with structural and stoichiometric information from the primary replicative MCM complexes and other AAA+ proteins from yeast and Archaea, it is attractive to propose a structural model whereby MCM8 and 9 subunits alternate to form the hetero-hexameric ring (Figure 1D) with a central channel that can accommodate DNA. If this is the case, it will be interesting to learn how the ringed hexamer is loaded onto DNA and whether its loading is as tightly controlled as it is for MCM2–7.
There are over 100 mutations in both MCM8 and MCM9 which are implicated in POF and cancer. Mapping all reported mutations to their respective domains has revealed that there is a clear bias in mutational abundance within the ATPase domain (61%) of MCM8 with the remaining 39% localized to the DNA binding domain. The opposite trend was observed for MCM9 with 43% of total reported mutations localized to the DNA binding domain with a nearly even distribution of the remaining mutations at 30% and 27% between the ATPase and C-terminal domains (CTD), respectively. It is expected that most point mutations within these conserved DNA binding and ATPase domains will negatively affect their respective activities (DNA binding and ATPase/helicase) concomitant with replication or repair activity. Additionally, point mutations within the long CTD of MCM9, which likely functions as a regulatory domain for protein-protein interactions (discussed in more detail below), would severely perturb MCM8/9 activity. It is possible that mutations within these domains might also affect the formation of the MCM8/9 complex and subsequent localization to sites of DNA damage.
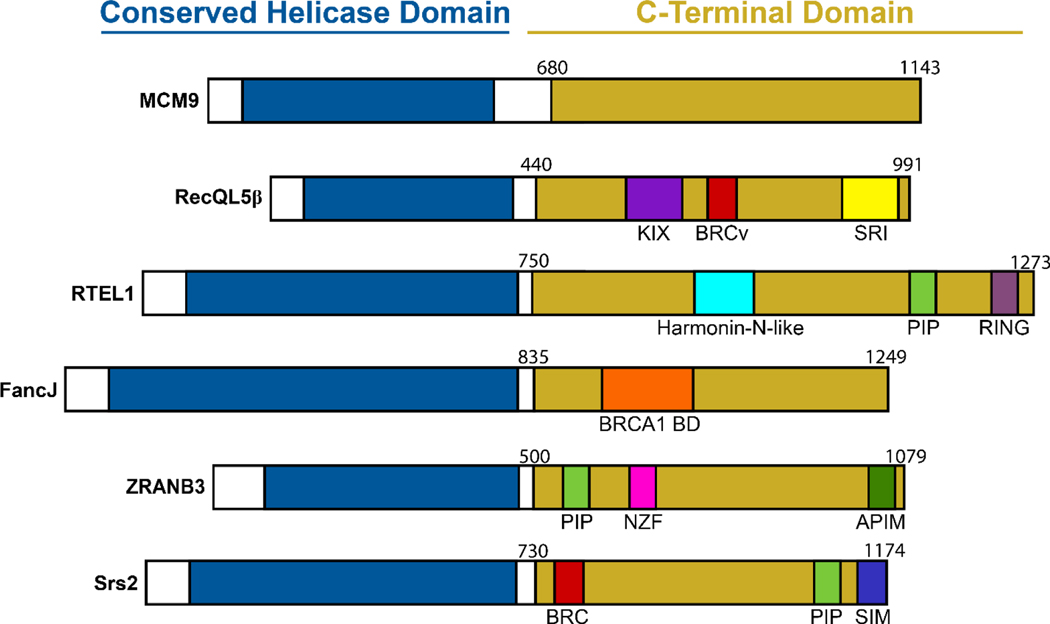
The defining feature separating MCM9 from MCM8 and other MCM proteins is the presence of a unique and long CTD (~530 amino acid residues) (Figure 1A). At the time of writing this review, no structural information for MCM9 CTD has been reported. It is hypothesized that the majority of this CTD is unstructured and disordered. However, structure prediction with the RaptorX-Property software has suggested and number of α-helical and β-sheet regions present within amino acids 643–862 [40]. This CTD is important for MCM9 function as nonsense mutations that result in MCM9 truncation or complete deletion are linked primarily to infertility but also cancer. Strikingly, the layout of MCM9 bears resemblance to that of the yeast Srs2 helicase (despite having low sequence homology) which also contains a long C-terminal extension with multiple motifs that mediate the interaction and regulation of other proteins including Rad51, PCNA, and SUMO [41] (Figure 2). Interestingly, it has been previously demonstrated through immunofluorescence confocal microscopy that MCM9 possesses Rad51 recruitment activity in the presence of DNA damage [12]. These overlapping domain architectures and activities of Srs2 and MCM9 likely point to a function whereby the MCM9 CTD serves as an interaction “hub” to mediate protein-protein interactions.
Figure 2:
Linear representations shown with conserved DNA helicase domains (blue) and extended and uncharacterized C-terminal domains (yellow) from a variety of related helicases with indicated motifs.
The presence of this C-terminal domain is not confined to MCM9 or Srs2. In fact, multiple human helicases involved in various DNA repair pathways possess CTDs which contain protein-protein interaction motifs or domains (Figure 2). For example, the nuclear isoform of RecQL5 (RecQL5β) also contains a C-terminal extension of ~500 amino acids that houses a triple helix bundle termed a KIX domain and a Set2 Rpb 1-interacting (SRI) domain which interact with the large subunit of RNA pol II [42]. The KIX domain along with the helicase function of RecQL5 is suggested to play an important role in regulation of HR and sister chromatid exchange by disrupting Rad51 nucleoprotein filaments [43]. The FancJ helicase, which is involved in interstrand crosslink repair possesses a BRCA1 binding domain and associates with BRCA1 during replication to protect the replication fork [44]. ZRANB3, like Srs2, also has a PCNA-interacting peptide (PIP) motif along with a secondary PCNA interacting motif termed an AlkB homolog 2 PCNA interacting motif (APIM) and an Npl4 Zinc-finger (NZF) which recognizes polyubiquitinated (pUB) PCNA [45]. Translocase activity and interaction with PCNA by ZRANB3 are important for fork protection and reversal to effect error-free DNA repair [46]. Lastly, the CTD of RTEL1 contains a PIP motif, Harmonin-N-like, and Really interesting new gene (RING) finger domains which are suggested to facilitate replication and stress responses through protein-protein interactions [47–50]. Any sequence motifs or specific protein partners which interact with the CTD of MCM9 remain to be identified, but it would not be surprising to find them.
4. Helicases bridging DNA replication and recombination
MCM8/9 have been linked with DNA elongation processes, upregulated with specific DNA damaging agents, localized with several HR proteins, and mutations are implicated in both infertility and cancer. However, a specific enzymatic role or substrate specificity has not been defined, making assignment of precise function difficult. In fact, there are several analogous DNA helicases which appear to share similar or overlapping roles in various aspects of DNA processing linking replication with interstrand crosslink repair or directly with recombination (Table 1). Specific activities for helicases in these arenas include aiding in fork protection as accessory helicases to the CMG complex, being directly involved in fork reversal for replication restart, promoting resection of reversed forks or double-strand breaks, acting as antirecombinases to remove homeologous synapses formed from microhomologies, or branch migrating and dissolving specific HR intermediates. The RecQ family of helicases (RecQL1, WRN, BLM, RecQL4, and RecQL5) are conserved from bacteria to humans and are critical for many of these aspects of genomic maintenance [51], however they are not alone. Many of these DNA helicases have gene defects linked directly to cancer and other aging related diseases, including Werner syndrome (WRN), Rothmond-Thomson (RecQL4), Bloom syndrome (BLM), and Fanconi anemia (FancJ) [52, 53]. Loss of function in any DNA repair helicase has the potential to lead to disease [54].
Table 1:
Replication/Recombination Helicases
| E. coli | S. cerevisiae | X. laevis / H. sapiens | |
|---|---|---|---|
|
| |||
| Fork Progression | DnaB | MCM2–7 | MCM2–7 |
| Rep | Pif1, Rrm3 | Pif1 | |
| RecQ | RecQL4, RecQL5 | ||
| PriA | FancJ, RTEL | ||
| Mph1 | FancM | ||
| MCM8, MCM9 | |||
|
| |||
| Fork Reversal | SMARCAL1*, ZRANB3* | ||
| HTLF* | |||
| RecG | Sgs1 | BLM, WRN | |
| UvrD | Srs2 | FBH1 | |
| Mph1 | FancM | ||
| MCM8, MCM9 | |||
|
| |||
| Fork Protection | FancJ, FancM | ||
|
| |||
| Fork Restart | RecQ | Sgs1 | RecQL1 |
| FancJ | |||
|
| |||
| Resection | RecQ | Sgs1 | BLM, WRN |
| UvrD | Srs2 | FBH1 | |
| DNA2 | DNA2 | ||
| RecBCD | |||
| FancJ | |||
| MCM8, MCM9 | |||
|
| |||
| Synapsis | Sgs1 | RecQL1, BLM, RecQL5 | |
| FancJ, RTEL1 | |||
| UvrD | Mph1 | FBH1 | |
| MCM8, MCM9 | |||
| Mer3 | HFM1, HelQ | ||
|
| |||
| Branch Migration | RecG, RecQ | Sgs1 | RecQL1, BLM, WRN |
| RuvAB | |||
|
| |||
| Dissolution | RecQ | Sgs1 | BLM |
translocase
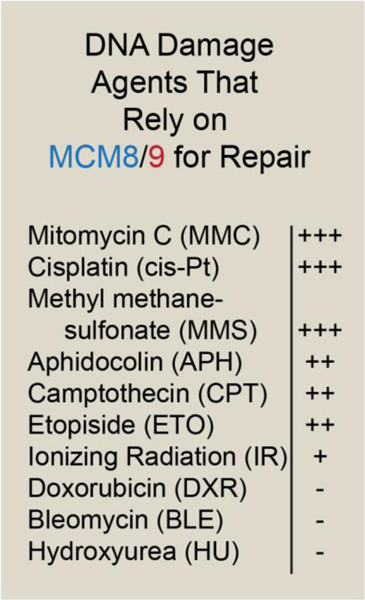
A variety of factors including genotoxic agents that cause DNA damage, DNA-protein crosslinks, protein complex collisions on chromosomes, and difficult to replicate sequences will all stall replication forks [55–58]. Analysis of cellular viability, protein localization/colocalization fluorescence microscopy, and coimmunoprecipitations using a variety of genotoxic agents in both animal and cell models indicate that MCM8/9 are likely involved in only a sub-set of DNA repair pathways dependent on the type of damage (Figure 3). For example, cells deficient for MCM8/9 show increased sensitivity to DNA cross-linking agents MMC and cisplatin (cis-Pt) in addition to the DNA alkylating agent methyl methanesulfonate (MMS) [10–12, 34]. These data indicate that MCM8/9 are important for crosslink repair preferentially encountered during replication and likely interact with members of the Fanconi anemia pathway. A less dramatic effect was observed for the DNA polymerase inhibitor aphidicolin (APH) and the topoisomerase poisons: camptothecin (CPT) and etoposide (ETO). Lastly, little to no effect was observed for ionizing radiation (IR), doxorubicin (DXR), bleomycin (BLE), and hydroxyurea (HU) arguing against a role in direct DSB repair or fork stalling. Instead, MCM8/9 seem to be present after severe replication induced stalling induced primarily by crosslinks or other large lesions that shuttles repair towards HR.
Figure 3:
Cumulative impact of various DNA damage agents on perceived activity, regulation, localization, interactions, and viability of MCM8/9 from organismal and cellular studies.
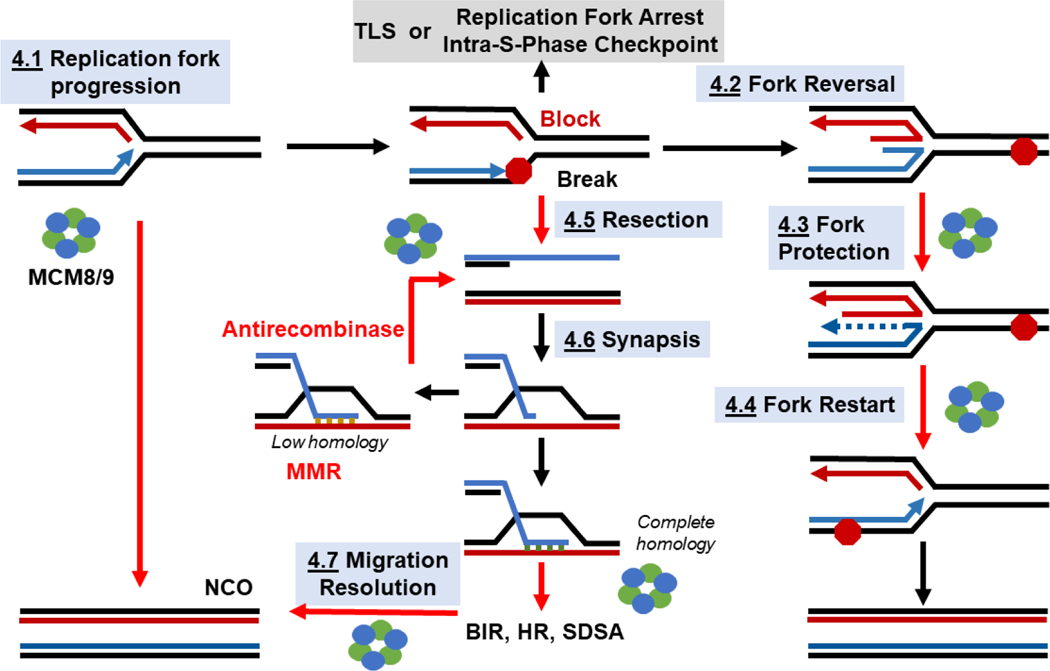
4.1. Fork Progression.
Besides the CMG helicase, other accessory helicases are associated with the replisome and thought to aid in fork progression (Figure 4) over difficult to replicate regions. Pif1 is known to bind to and unwind structures relating to stalled replication forks and is active during replication stress where replication fork slowing is observed [59]. Besides, CMG, RecQL5 and MCM8/9 are the only other DNA helicases enriched at active replication forks over those on general chromatin in the absence of stress [43, 60]. Cells deficient in RecQL5 (either knockout or knockdown) have an increase in spontaneous γ-H2AX foci positive for PCNA and are strongly sensitive to topoisomerase inhibition, but are proficient in HR [61, 62]. This result suggests that RecQL5 is more important in maintaining active DNA replication and preventing fork collapse [42]. FancM, and recently RTEL, have been implicated in aiding MCM2–7 progression past ICLs and unwinding difficult hairpin structures or fragile sites [63, 64], although neither are known to be normally associated with the replisome. Interestingly, a genetic truncating mutation of FancM has also found to be associated with POI and secondary amenorrhea with slightly milder developmental phenotypes than for MCM8 or MCM9 affected patients but with sensitivities to MMC and decreased monoubiquitination (mUb) of FancD2 [65].
Figure 4:
Pathways bridging DNA replication, restart, and recombination pathways where helicase and translocases will be required. Numbers and titles correspond to the text, and red arrows indicate possible steps for MCM8/9 contribution.
MCM8/9 do not interact directly with MCM2–7 nor are necessary for pre-replication complex (pre-RC) initiation [34], but rather associate with replisomes later during S-phase. In fact, targeted inactivation of MCM2-delpeted cells maintains DNA synthesis activity primarily through the presumed accessory helicase ability of MCM8/9 [66].
4.2. Fork Reversal.
Replication forks that are stalled or have built up significant ssDNA from unregulated DNA unwinding can be reversed to form ‘chicken foot’-like structures that anneals the nascent DNA strands and resembles a Holliday junction with a one-sided DSB (Figure 4). Fork reversal is fueled primarily by three DNA translocases with certain specificities, HLTF, ZRANB3, and SMARCAL1 [67]. SMARCAL1 seems to target stalled forks with stable RPA- bound ssDNA gaps at the fork junction specifically on the leading strand for direct reversal and annealing. This specificity prevents fork reversal of normal RPA coated lagging-strand gaps that occur normally during replication. ZRANB3 interacts specifically with and stimulates pUB-PCNA found at severely stalled forks [46]. Another translocase, HLTF, also promotes pUB-PCNA and can directly reverse forks via its HIRAN domain [68]. Finally, the major recombinase, Rad51, can participate in the initial phases of fork reversal, particularly when there is significant ssDNA in the reversed end [69, 70]. ssDNA at stalled forks can be engaged by Rad51 to promote pairing with the complementary strand to essentially strand exchange, forming a reversed fork. Other helicases including WRN, BLM, FancM, and FBH1 have also been linked with fork reversal processes [71–73].
Although no direct role for MCM8/9 has been implicated in fork reversal, both are present at the replication fork, known to interact with a subset of Rad51 (that is not bound to BRCA2), and are important for recruitment of Rad51 to DSBs [12]. Therefore, it is plausible that MCM8/9 may have a role in preparing the fork for reversal or recruiting Rad51 to a subset of reversed forks in concert with other translocases. In fact, patient cells deficient in MCM8 or MCM9 show large radial chromosome aberrations [17, 18] indicative of either stalled or reversed forks or other downstream HR intermediates that resulted in translocated and NHEJ processed chromosomes.
4.3. Reversed Fork Protection.
Excessive fork reversal can lead to uncontrolled resection by the various nucleases creating large genomic deletions. When replication forks are remodeled into alternative structures (i.e. reversed forks), they become excellent substrates for a variety of nucleases that can shuttle them into the DSBR pathways. Knockdown of FancJ causes an increase in DNA damage associated with induction of γ-H2AX, slower growth, and microsatellite instability [74, 75]. More recent evidence points to a fork protection role (Figure 4) for FancJ that suppresses HLTF and limits fork degradation [76]. BRCA1 and BRCA2 are now included as fork protectors that stabilize Rad51 filaments on the regressed arms, and when they are absent, forks are robustly degraded by Mre11 [77]. Rad51 can also stabilize the reversed fork by preventing branch migration or preventing access from nucleases [78]. Under situations where G-quadruplex structures are present at reversed/broken forks, the Pif1 helicase can stimulate Exo1, DNA2/BLM, or Mre11 degradation [79].
No role for MCM8/9 in fork protection has been described, but like Pif1, MCM8/9 can stimulate Mre11 resection (discussed below).
4.4. Fork Restart.
The mechanism of restart will likely require a DNA translocase/helicase to convert the four-way one-ended HJ to a three-way replication fork (Figure 4). In the presence of Toposiomerase I (TOP1) inhibited replication forks, RecQ1 can restart forks once PARP1 is degraded [80]. DNA2/WRN degrades reversed replication forks directly to restart unresolved forks in an aberrant manner [81]. ATR phosphorylates BLM to aid in restart of forks after stalling with hydroxyurea (HU) or aphidicolin (APH) [82]. Recently, FancJ was shown to counteract fork reversal by HLTF to reestablish replication forks [76]. Irreversibly damaged forks can only be rescued by endonuclease cleavage to provide substrates for HR. The helicase/nuclease activity of WRN [83] and the BLM dissolvasome complex (BLM/TopIIIα/Rmi1/Rmi2 - BTRR) are probable actors in this pathway. If all else fails, the endonucleases Mus81 or SLX4 can recognize extremely stalled forks and cause DSBs that feed into HR [84, 85].
The evidence for MCM8/9 in fork restart is not direct. Targeted degradation of the MCM2 does not fully halt DNA unwinding. Instead, MCM8/9 is thought to resume some DNA unwinding to process downstream HR intermediates [66]. Whether this unwinding by MCM8/9 occurs from a reversed or regular fork is unknown.
4.5. Resection.
Resection of DSBs or reversed forks that resemble DSBs are substrates for a variety of exonucleases (Figure 4). BLM can stimulate resection of DNA ends by either the helicase/nuclease, DNA2, or the MRN complex in the presence of RPA [86]. In another report, DNA2 was found to cooperate with either BLM or WRN to promote long range end resection [87]. In this role, DNA2 helps to recruit BLM and stimulates unwinding processivity, favoring 5’ strand cleavage by DNA2 resulting in the formation of 3’ ssDNA tails [88]. BLM/DNA2 also interacts with CtIP and/or the Topo IIIα–RMI1-RMI2 (TRR) complex enhancing DNA unwinding processivity for long range end resection [89]. Finally, FancJ (5’−3’) is thought to work together with BLM (3’−5’), each translocating on opposite strands to processively unwind DNA in the presence of RPA for resection by structure specific nucleases DNA2 or Exo1 providing a 3’-end of HR [90–92].
Interestingly, MCM8/9 are also found to be required for efficient end resection of DSBs in concert with the MRN complex [10]. Immunoprecipitation of MCM9 detected the MRN components (and vice versa) showing a direct interaction of these proteins. It may be that the exonuclease activity is stimulated when DNA helicases can provide ssDNA substrates or by enhancing cleaved DNA product release during resection or that the presence of MCM8/9 at a reversed fork can counteract fork protection by recruiting MRN. These possibilities remain to be distinguished.
4.6. Control of Synapsis.
Invasion of duplex DNA by a 3’ ssDNA overhang created after resection is facilitated primarily by Rad51 in combination with BRCA2. The formation of a stable Rad51 (or Dmc1 in meiosis) invaded D-loop structure is a trigger for HR and strand extension by various DNA polymerases (Figure 4). Antagonists to this process will include DNA helicases that can act as antirecombinases or checkpoints for maintaining homology over homeology in mitotic repair. Homeologous synapses are beneficial in creating genetic diversity required in meiosis, however they contribute to genomic instability in mitotic cells. Strand invasion intermediates can either dissociate after extension by a synthesis dependent strand annealing (SDSA) mechanism to create NCO or can be stabilized by more downstream DNA unwinding and subsequent dHJ formation promoting CO. Therefore, there will be a delicate balance between D-loop formation/dissolution, homeology/homology checkpoints, meiotic/mitotic recombination, and SDSA/dHJ. All of these processes will be regulated by various recombinogenic/antirecombinogenic DNA helicases with overlapping and unknown specificities to various intermediates and situations.
Therefore, and not surprisingly, many DNA helicases are involved in regulating synapsis. RecQL5 interacts with Rad51 to displace it from filaments acting as a antirecombinase to suppress D-loops and promote SDSA [61, 93]. Overlapping activities from FancJ, RTEL1, and FBH1 also all strip Rad51 from DNA to disrupt D-loops and suppress HR as an antirecombinase to favor SDSA [44, 94, 95].
The temporal placement of MCM8/9 in the HR pathway will perhaps provide insight into whether their role is prior to or after D-loop formation with Rad51. Currently, there are conflicting results showing that MCM8/9 acts prior to Rad51 (during resection) [10, 12] as well as downstream of Rad51 in DT40 cells, where knockout of MCM8 or 9 has no effect on DNA damaged induced Rad51 foci formation [11, 66]. A role for MCM8/9 in the regulation of Rad51 activity during HR remains to be definitively addressed. However, we hypothesize that in mammalian cells Rad51 regulation at sites of DNA damage prior to synapsis is dependent on an interaction with the MCM9 CTD. However, Rad51 has multiple functions during DNA replication and repair independent of MCM8/9. This would suggest a context-dependent interaction likely once a stalled replication fork progresses into collapse and subsequent HR repair.
It would be remiss to focus solely on helicases involved in mitotic HR over those that act more specifically in meiosis. For example, HFM1 acts as a meiosis specific DNA helicase (Mer3 homolog in yeast and of the DEAD family) and is a member of the ZMN group of proteins required for formation of the synaptonemal complex and promotion of CO in meiosis. HFM1−/− mice are infertile, stages of meiosis are delayed, and recombination COs are reduced. The initial meiotic recombination steps (including DSB formation) appear unaffected, but synapsis is incomplete, suggesting a role for HFM1 in DNA unwinding downstream of D-loop formation and aiding in dHJ establishment [96]. These defects are highly similar to those observed with MCM8−/− and MCM9−/− mice which are also infertile, meiosis is blocked early in prophase 1, and females having arrested primary follicles and developing ovarian cancers [16, 97]. HELQ-1 is similar (albeit acting just after) in its role for late stage D-loop dissolution in meiosis [98], although it also associates with Rad51 paralogs during ATR-mediated ICL repair [99]. Even though Drosophila lacks an MCM9 homolog, REC is responsible for 95% of meiotic COs and its absence results in incomplete recombination transitioning towards SDSA and NCO products [15]. Further evidence in plants suggests that MCM8 is involved primarily with Rad51 in meiosis promoting NCO when the primarily CO pathway directed by Dmc1 fails [100].
Interestingly, many recombination (RecQ1, WRN, BLM, and FancJ) helicases [101–103] also interact with mismatch repair (MMR) proteins similar to MCM9 [104]. In this situation, DNA helicases will either direct MMR to homeoduplex pairing, remove homeologous duplexes recognized by MMR proteins in mitotic repair for NCO, or promote recombination and CO during meiosis [105, 106]. In mitotic DNA repair, homeologous duplex rejection would be a primary activity for DNA helicases. This is well established in E. coli where the UvrD helicase interacts with MMR proteins and dismantles RecA filaments to regulate homeologous recombination [107]. Both yeast helicases, Sgs1 and Srs2, interact with MMR proteins to suppress CO [108, 109], but the human helicase responsible to homeologous duplex rejection is not currently known. One candidate is BLM which also interacts with MMR proteins [110], but knockout of BLM in mammalian cells has no effect on the degree of homeologous recombination [111]. It is tempting to speculate that MCM8/9 is directed to MMR bound homeologous duplexes to either reject mismatched synapses for mitotic recombination or promote downstream dHJ formation for CO in meiosis.
4.7. HJ Branch Migration and Resolution.
dHJ formation requires the downstream unwinding of the dsDNA target for efficient extension of the D-loop and DNA synthesis (Figure 4). This activity is suppressed in mitotic HR but is promoted in meiotic recombination [112]. Dissolution of dHJ occurs though action of a higher order ‘dissolvasome’ consisting of the BTRR complex to release NCO [113, 114]. Resolution of the dHJ results in both NCO and CO and is processed primarily by two structure specific nuclease complexes: SLX-MUS-EME1 and GEN1 [115, 116]. Both resolvases are constrained by multiple cellular mechanisms until mitosis giving the BTRR complex priority in S-phase for NCO. However, in meiosis, there is sequential activation of SLX-MUS-EME1 followed by GEN1 to enhance CO products [84].
Prior to dissolution or resolution, DNA helicases can migrate synapses and HJs to further DNA synthesis or generate specific structures. RecQL1 can form a tetramer that recognizes and migrates HJs in a coordinated unwinding and annealing mechanism [117]. RecQL5 can also branch migrate HJs in a similar mechanism, although its main cellular role may be elsewhere aiding fork progression [118, 119]. Both BLM and WRN are important in later stages of HR by mediating the topological disentanglement of D-loops or HJ intermediates and suppressing CO products [113]. WRN can also migrate HJ on its own [120], however there is more support for WRN in regulating pathway choice towards alt-NHEJ earlier in this pathway [121]. BLM has enhanced unwinding processivity for branch migration and HJ dissolution within the BTRR complex to yield NCO [122]. Finally, ultra-fine bridges visualized during late stage mitosis are likely unresolved fork stalling or HR products (likely catenanes or hemi-catenanes) that are resolved by BLM/TopIIIα or GEN1 [123].
5. Conclusions and unanswered questions
With all of these DNA helicases (Table 1) acting similarly and in overlapping pathways, there is a question on whether there is redundancy of function or some yet unknown substrate specificity for each. Currently, there are more open questions than there are answers, which predicts an exciting time of research not just for MCM8/9 but also for distinguishing the specificities of the many replication/recombination helicases. There are clear links between germ line mutations in several DNA helicases that give rise to a variety of diseases including cancer, progeria, anemia, reproductive deficiencies, infertility, and an untold number of somatic mutations. The cellular and mechanistic effects of most of these helicase mutations are unknown, except in a few cases [124]. For MCM8/9, mutations can only be classified into categories such as damaging, variant of unknown significance (VUS), or benign [21] based on homology of the mutation, what is known about the structure/function relationship of the MCM family of proteins, and patient outcomes.
Of the numerous unanswered questions regarding the role of MCM8/9 in HR; the biggest of which are characterization of the enzymatic activity, interactions with other HR members, and specificity for particular HR intermediates. The potential for MCM8/9 to act during telomere maintenance as shown for many other related DNA helicases including BLM, WRN, RecQL4, and RTEL [125, 126], remains to be addressed. We are naive about how MCM8/9 is localized or recruited to replication forks or DSBs. It is unknown whether MCM8/9 aids in fork progression past difficult DNA secondary structures or sequences including G-quadruplexes or common fragile sites. Finally, no posttranslational modifications are known to regulate MCM8/9 function as are common with many other human DNA repair proteins.
There are multiple research priorities to pursue that should provide more insight into these scientific gaps of knowledge for MCM8/9 function compared with related DNA helicases. 1) Research should continue to characterize the mitotic DNA damage response and incorporate patient mutations for better functional characterization. 2) Efforts should be made to obtain purified protein complexes for structure/function studies to define substrate specificities. 3) Experiments should move past noting global meiotic deficiencies and instead should focus more specifically on recombination outcomes. 4) Compensation of helicase function should be examined by comparing patient cell lines to cells that are more abruptly knocked down/out. 5) DNA sequencing of mutations correlating with infertility and cancer should continue to be cataloged and functionally characterized. 6) Links between infertility (meiotic recombination) and cancer (DNA damage response) should be explored. These many open research questions will undoubtedly attract significant research attention in the coming years that will provide a much clearer picture for the role of MCM8/9 within the context of many other analogous DNA helicases.
Acknowledgements:
Financial support in our laboratory is provided by Baylor University, the American Cancer Society (RSG-11–049-01-DMC), and NSF-MCB (NSF1613534 to M.A.T.). W.C.G. is supported by a Baylor VPR Postdoctoral Research Fellowship. We thank David McKinzey and Shiva Gomanthingayagam for helpful discussions and the Rajkovic laboratory for continued collaboration.
Footnotes
Conflicts of interest: The authors declare that there are no conflicts of interest.
References
- [1].Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, Perez-Amodio S, Strippoli P, Canaider S, An estimation of the number of cells in the human body, Ann. Hum. Biol, 40 (2013) 463–471. [DOI] [PubMed] [Google Scholar]
- [2].Shibata A, Regulation of repair pathway choice at two-ended DNA double-strand breaks, Mutat. Res, 803–805 (2017) 51–55. [DOI] [PubMed] [Google Scholar]
- [3].Chang HHY, Pannunzio NR, Adachi N, Lieber MR, Non-homologous DNA end joining and alternative pathways to double-strand break repair, Nat. Rev. Mol. Cell. Biol, 18 (2017) 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ranjha L, Howard SM, Cejka P, Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes, Chromosoma, (2018). [DOI] [PubMed] [Google Scholar]
- [5].Wright WD, Shah SS, Heyer WD, Homologous recombination and the repair of DNA double-strand breaks, J. Biol. Chem, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Her J, Bunting SF, How cells ensure correct repair of DNA double-strand breaks, J. Biol. Chem, 293 (2018) 10502–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hunter N, Meiotic recombination: The essence of heredity, Cold Spring Harbor perspectives in biology, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lam I, Keeney S, Mechanism and regulation of meiotic recombination initiation, Cold Spring Harbor perspectives in biology, 7 (2014) a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kanemaki MT, The dimeric Mcm8–9 complex of Xenopus laevis likely has a conserved function for resistance to DNA damage, Cell Cycle, 12 (2013) 1338–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lee KY, Im JS, Shibata E, Park J, Handa N, Kowalczykowski SC, Dutta A, MCM8–9 complex promotes resection of double-strand break ends by MRE11-RAD50-NBS1 complex, Nature communications, 6 (2015) 7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nishimura K, Ishiai M, Horikawa K, Fukagawa T, Takata M, Takisawa H, Kanemaki MT, Mcm8 and Mcm9 form a complex that functions in homologous recombination repair induced by DNA interstrand crosslinks, Mol. Cell, 47 (2012) 511–522. [DOI] [PubMed] [Google Scholar]
- [12].Park J, Long DT, Lee KY, Abbas T, Shibata E, Negishi M, Luo Y, Schimenti JC, Gambus A, Walter JC, Dutta A, The MCM8-MCM9 complex promotes RAD51 recruitment at DNA damage sites to facilitate homologous recombination, Mol. Cell Biol, 33 (2013) 1632–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Grell RF, Time of Recombination in the DROSOPHILA MELANOGASTER Oocyte. III. Selection and Characterization of Temperature-Sensitive and -Insensitive, Recombination-Deficient Alleles in Drosophila, Genetics, 108 (1984) 425–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Matsubayashi H, Yamamoto MT, Dissection of chromosome region 89A of Drosophila melanogaster by local transposition of P elements, Genes Genet. Syst, 73 (1998) 95–103. [DOI] [PubMed] [Google Scholar]
- [15].Blanton HL, Radford SJ, McMahan S, Kearney HM, Ibrahim JG, Sekelsky J, REC, Drosophila MCM8, drives formation of meiotic crossovers, PLoS. Genet., 1 (2005) e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hartford SA, Luo Y, Southard TL, Min IM, Lis JT, Schimenti JC, Minichromosome maintenance helicase paralog MCM9 is dispensible for DNA replication but functions in germ-line stem cells and tumor suppression, Proc. Natl. Acad. Sci. U.S.A, 108 (2011) 17702–17707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wood-Trageser MA, Gurbuz F, Yatsenko SA, Jeffries EP, Kotan LD, Surti U, Ketterer DM, Matic J, Chipkin J, Jiang H, Trakselis MA, Topaloglu AK, Rajkovic A, MCM9 mutations are associated with ovarian failure, short stature, and chromosomal instability, Am. J. Hum. Genet, 95 (2014) 754–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].AlAsiri S, Basit S, Wood-Trageser MA, Yatsenko SA, Jeffries EP, Surti U, Ketterer DM, Afzal S, Ramzan K, Faiyaz-Ul Haque M, Jiang H, Trakselis MA, Rajkovic A, Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability, J. Clin. Invest, 125 (2015) 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fauchereau F, Shalev S, Chervinsky E, Beck-Fruchter R, Legois B, Fellous M, Caburet S, Veitia RA, A non-sense MCM9 mutation in a familial case of primary ovarian insufficiency, Clin. Genet, 89 (2016) 603–607. [DOI] [PubMed] [Google Scholar]
- [20].Bouali N, Francou B, Bouligand J, Imanci D, Dimassi S, Tosca L, Zaouali M, Mougou S, Young J, Saad A, Guiochon-Mantel A, New MCM8 mutation associated with premature ovarian insufficiency and chromosomal instability in a highly consanguineous Tunisian family, Fertil. Steril, 108 (2017) 694–702. [DOI] [PubMed] [Google Scholar]
- [21].Desai S, Wood-Trageser M, Matic J, Chipkin J, Jiang H, Bachelot A, Dulon J, Sala C, Barbieri C, Cocca M, Toniolo D, Touraine P, Witchel S, Rajkovic A, MCM8 and MCM9 nucleotide variants in women with primary ovarian insufficiency, J. Clin. Endocrinol. Metab, 102 (2017) 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Goldberg Y, Halpern N, Hubert A, Adler SN, Cohen S, Plesser-Duvdevani M, Pappo O, Shaag A, Meiner V, Mutated MCM9 is associated with predisposition to hereditary mixed polyposis and colorectal cancer in addition to primary ovarian failure, Cancer Genet., 208 (2015) 621–624. [DOI] [PubMed] [Google Scholar]
- [23].Huhtaniemi I, Hovatta O, La Marca A, Livera G, Monniaux D, Persani L, Heddar A, Jarzabek K, Laisk-Podar T, Salumets A, Tapanainen JS, Veitia RA, Visser JA, Wieacker P, Wolczynski S, Misrahi M, Advances in the molecular pathophysiology, genetics, and treatment of primary ovarian insufficiency, Trends Endocrinol. Metab, 29 (2018) 400–419. [DOI] [PubMed] [Google Scholar]
- [24].Liu Q, Hesson LB, Nunez AC, Packham D, Hawkins NJ, Ward RL, Sloane MA, Pathogenic germline MCM9 variants are rare in Australian Lynch-like syndrome patients, Cancer Genet., 209 (2016) 497–500. [DOI] [PubMed] [Google Scholar]
- [25].Gozuacik D, Chami M, Lagorce D, Faivre J, Murakami Y, Poch O, Biermann E, Knippers R, Brechot C, Paterlini-Brechot P, Identification and functional characterization of a new member of the human Mcm protein family: hMcm8, Nucleic Acids Res., 31 (2003) 570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Johnson EM, Kinoshita Y, Daniel DC, A new member of the MCM protein family encoded by the human MCM8 gene, located contrapodal to GCD10 at chromosome band 20p12.3–13, Nucleic Acids Res., 31 (2003) 2915–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lutzmann M, Maiorano D, Mechali M, Identification of full genes and proteins of MCM9, a novel, vertebrate-specific member of the MCM2–8 protein family, Gene, 362 (2005) 51–56. [DOI] [PubMed] [Google Scholar]
- [28].Yoshida K, Identification of a novel cell-cycle-induced MCM family protein MCM9, Biochem. Biophys. Res. Commun., 331 (2005) 669–674. [DOI] [PubMed] [Google Scholar]
- [29].Jeffries EP, Denq WH, Bartko JC, Trakselis MA, Identification, quantification, and evolutionary analysis of a novel isoform of MCM9, Gene, 519 (2013) 41–49. [DOI] [PubMed] [Google Scholar]
- [30].Liu Y, Richards TA, Aves SJ, Ancient diversification of eukaryotic MCM DNA replication proteins, BMC. Evol. Biol., 9 (2009) 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Volkening M, Hoffmann I, Involvement of human MCM8 in prereplication complex assembly by recruiting hcdc6 to chromatin, Mol. Cell. Biol., 25 (2005) 1560–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kinoshita Y, Johnson EM, Gordon RE, Negri-Bell H, Evans MT, Coolbaugh J, Rosario-Peralta Y, Samet J, Slusser E, Birkenbach MP, Daniel DC, Colocalization of MCM8 and MCM7 with proteins involved in distinct aspects of DNA replication, Microsc. Res.Tech., 71 (2008) 288–297. [DOI] [PubMed] [Google Scholar]
- [33].Lutzmann M, Mechali M, MCM9 binds Cdt1 and is required for the assembly of prereplication complexes, Mol. Cell, 31 (2008) 190–200. [DOI] [PubMed] [Google Scholar]
- [34].Gambus A, Blow JJ, Mcm8 and Mcm9 form a dimeric complex in Xenopus laevis egg extract that is not essential for DNA replication initiation, Cell Cycle, 12 (2013) 1225–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Maiorano D, Cuvier O, Danis E, Mechali M, MCM8 is an MCM2–7-related protein that functions as a DNA helicase during replication elongation and not initiation, Cell, 120 (2005) 315–328. [DOI] [PubMed] [Google Scholar]
- [36].Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadi M, Klein MG, Chen XS, Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase, Proc. Natl. Acad. Sci. U. S. A., 105 (2008) 20191–20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Miller JM, Arachea BT, Epling LB, Enemark EJ, Analysis of the crystal structure of an active MCM hexamer, Elife, 3 (2014) e03433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li N, Zhai Y, Zhang Y, Li W, Yang M, Lei J, Tye BK, Gao N, Structure of the eukaryotic MCM complex at 3.8 A, Nature, 524 (2015) 186–191. [DOI] [PubMed] [Google Scholar]
- [39].Zhai Y, Tye BK, Structure of the MCM2–7 double hexamer and its implications for the mechanistic functions of the Mcm2–7 complex, Adv. Exp. Med. Biol., 1042 (2017) 189–205. [DOI] [PubMed] [Google Scholar]
- [40].Wang S, Li W, Liu S, Xu J, RaptorX-Property: a web server for protein structure property prediction, Nucleic Acids Res, 44 (2016) W430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].De Tullio L, Kaniecki K, Kwon Y, Crickard JB, Sung P, Greene EC, Yeast Srs2 helicase promotes redistribution of single-stranded DNA-bound RPA and Rad52 in homologous recombination regulation, Cell reports, 21 (2017) 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Popuri V, Tadokoro T, Croteau DL, Bohr VA, Human RECQL5: Guarding the crossroads of DNA replication and transcription and providing backup capability, Crit. Rev. Biochem. Mol. Biol., 48 (2013) 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P, Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork, Nucleic Acids Res., 34 (2006) 5217–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cantor SB, Guillemette S, Hereditary breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1, Future Oncol., 7 (2011) 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sebesta M, Cooper CDO, Ariza A, Carnie CJ, Ahel D, Structural insights into the function of ZRANB3 in replication stress response, Nature communications, 8 (2017) 15847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Vujanovic M, Krietsch J, Raso MC, Terraneo N, Zellweger R, Schmid JA, Taglialatela A, Huang JW, Holland CL, Zwicky K, Herrador R, Jacobs H, Cortez D, Ciccia A, Penengo L, Lopes M, Replication fork slowing and reversal upon DNA damage require PCNA polyubiquitination and ZRANB3 DNA translocase activity, Mol. Cell, 67 (2017) 882–890 e885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Uringa EJ, Lisaingo K, Pickett HA, Brind’Amour J, Rohde JH, Zelensky A, Essers J, Lansdorp PM, RTEL1 contributes to DNA replication and repair and telomere maintenance, Molecular biology of the cell, 23 (2012) 2782–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Vannier JB, Sandhu S, Petalcorin MI, Wu X, Nabi Z, Ding H, Boulton SJ, RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication, Science, 342 (2013) 239–242. [DOI] [PubMed] [Google Scholar]
- [49].Faure G, Revy P, Schertzer M, Londono-Vallejo A, Callebaut I, The C-terminal extension of human RTEL1, mutated in Hoyeraal-Hreidarsson syndrome, contains harmonin-N-like domains, Proteins, 82 (2014) 897–903. [DOI] [PubMed] [Google Scholar]
- [50].Vannier JB, Sarek G, Boulton SJ, RTEL1: Functions of a disease-associated helicase, Trends Cell. Biol., 24 (2014) 416–425. [DOI] [PubMed] [Google Scholar]
- [51].Bernstein KA, Gangloff S, Rothstein R, The RecQ DNA helicases in DNA repair, Annu. Rev. Genetics, 44 (2010) 393–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Brosh RM Jr., DNA helicases involved in DNA repair and their roles in cancer, Nat. Rev. Cancer, 13 (2013) 542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].LaRocque JR, Stark JM, Oh J, Bojilova E, Yusa K, Horie K, Takeda J, Jasin M, Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells, Proc. Natl. Acad. Sci. U. S. A., 108 (2011) 11971–11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sidorova JM, Monnat RJ, Human RECQ helicases: Roles in cancer, aging and inherited disease, Adv. Genomics Genet., 5 (2015) 19–33. [Google Scholar]
- [55].Yeeles JT, Poli J, Marians KJ, Pasero P, Rescuing stalled or damaged replication forks, Cold Spring Harbor perspectives in biology, 5 (2013) a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cortez D, Preventing replication fork collapse to maintain genome integrity, DNA Repair (Amst), 32 (2015) 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Polleys EJ, House NCM, Freudenreich CH, Role of recombination and replication fork restart in repeat instability, DNA Repair (Amst), 56 (2017) 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Alexander JL, Orr-Weaver TL, Replication fork instability and the consequences of fork collisions from rereplication, Genes Dev., 30 (2016) 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].George T, Wen Q, Griffiths R, Ganesh A, Meuth M, Sanders CM, Human Pif1 helicase unwinds synthetic DNA structures resembling stalled DNA replication forks, Nucleic Acids Res., 37 (2009) 6491–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, Cortez D, The replication checkpoint prevents two types of fork collapse without regulating replisome stability, Mol. Cell, 59 (2015) 998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G, RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments, Genes Dev., 21 (2007) 3073–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hu Y, Lu X, Zhou G, Barnes EL, Luo G, Recql5 plays an important role in DNA replication and cell survival after camptothecin treatment, Mol. Biol. Cell, 20 (2009) 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ, RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity, Cell, 149 (2012) 795–806. [DOI] [PubMed] [Google Scholar]
- [64].Huang M, Kim JM, Shiotani B, Yang KL, Zou L, D’Andrea AD, The FANCM/FAAP24 complex Is required for the DNA interstrand crosslink-induced checkpoint response, Mol. Cell, 39 (2010) 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fouquet B, Pawlikowska P, Caburet S, Guigon C, Makinen M, Tanner L, Hietala M, Urbanska K, Bellutti L, Legois B, Bessieres B, Gougeon A, Benachi A, Livera G, Rosselli F, Veitia RA, Misrahi M, A homozygous FANCM mutation underlies a familial case of non-syndromic primary ovarian insufficiency, Elife, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Natsume T, Nishimura K, Minocherhomji S, Bhowmick R, Hickson ID, Kanemaki MT, Acute inactivation of the replicative helicase in human cells triggers MCM8–9-dependent DNA synthesis, Genes Dev., 31 (2017) 816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Poole LA, Cortez D, Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability, Crit. Rev. Biochem. Mol. Biol., 52 (2017) 696–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kile AC, Chavez DA, Bacal J, Eldirany S, Korzhnev DM, Bezsonova I, Eichman BF, Cimprich KA, HLTF’s ancient HIRAN domain binds 3’ DNA ends to drive replication fork reversal, Mol. Cell, 58 (2015) 1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M, Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells, J. Cell. Biol., 208 (2015) 563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bhat KP, Cortez D, RPA and RAD51: Fork reversal, fork protection, and genome stability, Nat. Struct. Mol. Biol., 25 (2018) 446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Fugger K, Mistrik M, Neelsen KJ, Yao Q, Zellweger R, Kousholt AN, Haahr P, Chu WK, Bartek J, Lopes M, Hickson ID, Sorensen CS, FBH1 catalyzes regression of stalled replication forks, Cell reports, (2015). [DOI] [PubMed] [Google Scholar]
- [72].Machwe A, Xiao L, Groden J, Orren DK, The Werner and Bloom syndrome proteins catalyze regression of a model replication fork, Biochemistry, 45 (2006) 13939–13946. [DOI] [PubMed] [Google Scholar]
- [73].Blackford AN, Schwab RA, Nieminuszczy J, Deans AJ, West SC, Niedzwiedz W, The DNA translocase activity of FANCM protects stalled replication forks, Hum. Mol. Genet., 21 (2012) 2005–2016. [DOI] [PubMed] [Google Scholar]
- [74].Kumaraswamy E, Shiekhattar R, Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase, Mol. Cell. Biol., 27 (2007) 6733–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Matsuzaki K, Borel V, Adelman CA, Schindler D, Boulton SJ, FANCJ suppresses microsatellite instability and lymphomagenesis independent of the Fanconi anemia pathway, Genes Dev., 29 (2015) 2532–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Peng M, Cong K, Panzarino NJ, Nayak S, Calvo J, Deng B, Zhu LJ, Morocz M, Hegedus L, Haracska L, Cantor SB, Opposing roles of FANCJ and HLTF protect forks and restrain replication during stress, Cell reports, 24 (2018) 3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M, Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11, Cell, 145 (2011) 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V, Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis, Nat. Struct. Mol. Biol., 17 (2010) 1305–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Jimeno S, Camarillo R, Mejías-Navarro F, Fernández-Ávila MJ, Soria-Bretones I, Prados-Carvajal R, Huertas P, The helicase PIF1 facilitates resection over sequences prone to forming G4 structures, Cell reports, 24 (2018) 3262–3273.e3264. [DOI] [PubMed] [Google Scholar]
- [80].Berti M, Chaudhuri AR, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, Marino F, Lucic B, Biasin V, Gstaiger M, Aebersold R, Sidorova JM, Monnat RJ, Lopes M, Vindigni A, Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition, Nat. Struct. Mol. Biol., 20 (2013) 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, Zellweger R, Moore H, Lee EH, Hendrickson EA, Cejka P, Stewart S, Lopes M, Vindigni A, DNA2 drives processing and restart of reversed replication forks in human cells, J. Cell. Biol., 208 (2015) 545–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Davies SL, North PS, Hickson ID, Role for BLM in replication-fork restart and suppression of origin firing after replicative stress, Nat. Struct. Mol. Biol., 14 (2007) 677–679. [DOI] [PubMed] [Google Scholar]
- [83].Murfuni I, De Santis A, Federico M, Bignami M, Pichierri P, Franchitto A, Perturbed replication induced genome wide or at common fragile sites is differently managed in the absence of WRN, Carcinogenesis, 33 (2012) 1655–1663. [DOI] [PubMed] [Google Scholar]
- [84].Matos J, Blanco MG, Maslen S, Skehel JM, West SC, Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis, Cell, 147 (2011) 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Sarbajna S, West SC, Holliday junction processing enzymes as guardians of genome stability, Trends Biochem. Sci., 39 (2014) 409–419. [DOI] [PubMed] [Google Scholar]
- [86].Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC, BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair, Genes Dev., 25 (2011) 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sturzenegger A, Burdova K, Kanagaraj R, Levikova M, Pinto C, Cejka P, Janscak P, DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells, J. Biol. Chem., 289 (2014) 27314–27326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Daley JM, Chiba T, Xue X, Niu H, Sung P, Multifaceted role of the Topo IIIalpha-RMI1-RMI2 complex and DNA2 in the BLM-dependent pathway of DNA break end resection, Nucleic Acids Res., 42 (2014) 11083–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Daley JM, Jimenez-Sainz J, Wang W, Miller AS, Xue X, Nguyen KA, Jensen RB, Sung P, Enhancement of BLM-DNA2-mediated long-range DNA end resection by CtIP, Cell reports, 21 (2017) 324–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Suhasini AN, Brosh RM Jr., Fanconi anemia and Bloom’s syndrome crosstalk through FANCJ-BLM helicase interaction, Trends Genet., 28 (2012) 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Nath S, Somyajit K, Mishra A, Scully R, Nagaraju G, FANCJ helicase controls the balance between short- and long-tract gene conversions between sister chromatids, Nucleic Acids Res., 45 (2017) 8886–8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Brosh RM Jr., Cantor SB, Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi anemia, Front. Genet., 5 (2014) 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Schwendener S, Raynard S, Paliwal S, Cheng A, Kanagaraj R, Shevelev I, Stark JM, Sung P, Janscak P, Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity, J. Biol. Chem., 285 (2010) 15739–15745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, Tissenbaum H, West SC, Rose AM, Boulton SJ, RTEL1 maintains genomic stability by suppressing homologous recombination, Cell, 135 (2008) 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Fugger K, Chu WK, Haahr P, Kousholt AN, Beck H, Payne MJ, Hanada K, Hickson ID, Sorensen CS, FBH1 co-operates with MUS81 in inducing DNA double-strand breaks and cell death following replication stress, Nature communications, 4 (2013) 1423. [DOI] [PubMed] [Google Scholar]
- [96].Guiraldelli MF, Eyster C, Wilkerson JL, Dresser ME, Pezza RJ, Mouse HFM1/Mer3 Is required for crossover formation and complete synapsis of homologous chromosomes during meiosis, PLoS genetics, 9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Lutzmann M, Grey C, Traver S, Ganier O, Maya-Mendoza A, Ranisavljevic N, Bernex F, Nishiyama A, Montel N, Gavois E, Forichon L, de Massy B, Mechali M, MCM8- and MCM9-deficient mice reveal gametogenesis defects and genome instability due to impaired homologous recombination, Mol. Cell, 47 (2012) 523–534. [DOI] [PubMed] [Google Scholar]
- [98].Ward JD, Muzzini DM, Petalcorin MI, Martinez-Perez E, Martin JS, Plevani P, Cassata G, Marini F, Boulton SJ, Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic double-strand break repair, Mol. Cell, 37 (2010) 259–272. [DOI] [PubMed] [Google Scholar]
- [99].Takata K, Reh S, Tomida J, Person MD, Wood RD, Human DNA helicase HELQ participates in DNA interstrand crosslink tolerance with ATR and RAD51 paralogs, Nature communications, 4 (2013) 2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Crismani W, Portemer V, Froger N, Chelysheva L, Horlow C, Vrielynck N, Mercier R, MCM8 is required for a pathway of meiotic double-strand break repair independent of DMC1 in Arabidopsis thaliana, PLoS genetics, 9 (2013) e1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Song L, Yuan F, Zhang Y, Does a helicase activity help mismatch repair in eukaryotes?, IUBMB Life, 62 (2010) 548–553. [DOI] [PubMed] [Google Scholar]
- [102].Doherty KM, Sharma S, Uzdilla LA, Wilson TM, Cui S, Vindigni A, Brosh RM Jr., RECQ1 helicase interacts with human mismatch repair factors that regulate genetic recombination, J. Biol. Chem., 280 (2005) 28085–28094. [DOI] [PubMed] [Google Scholar]
- [103].Saydam N, Kanagaraj R, Dietschy T, Garcia PL, Pena-Diaz J, Shevelev I, Stagljar I, Janscak P, Physical and functional interactions between Werner syndrome helicase and mismatch-repair initiation factors, Nucleic Acids Res., 35 (2007) 5706–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Traver S, Coulombe P, Peiffer I, Hutchins JR, Kitzmann M, Latreille D, Mechali M, MCM9 Is required for mammalian DNA mismatch repair, Mol. Cell, 59 (2015) 831–839. [DOI] [PubMed] [Google Scholar]
- [105].Tham KC, Kanaar R, Lebbink JHG, Mismatch repair and homeologous recombination, DNA Repair, 38 (2016) 75–83. [DOI] [PubMed] [Google Scholar]
- [106].Manhart CM, Alani E, Roles for mismatch repair family proteins in promoting meiotic crossing over, DNA Repair (Amst), 38 (2016) 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA, UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli, EMBO J., 24 (2005) 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE, Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1, Proc. Natl. Acad. Sci. U. S. A., 101 (2004) 9315–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Mitchel K, Lehner K, Jinks-Robertson S, Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes, PLoS genetics, 9 (2013) e1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Pedrazzi G, Bachrati CZ, Selak N, Studer I, Petkovic M, Hickson ID, Jiricny J, Stagljar I, The Bloom’s syndrome helicase interacts directly with the human DNA mismatch repair protein hMSH6, Biol. Chem., 384 (2003) 1155–1164. [DOI] [PubMed] [Google Scholar]
- [111].Larocque JR, Jasin M, Mechanisms of recombination between diverged sequences in wild-type and BLM-deficient mouse and human cells, Mol. Cell Biol., 30 (2010) 1887–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].West SC, Blanco MG, Chan YW, Matos J, Sarbajna S, Wyatt HD, Resolution of recombination intermediates: Mechanisms and regulation, Cold Spring Harb. Symp. Quant. Biol., 80 (2015) 103–109. [DOI] [PubMed] [Google Scholar]
- [113].Wu L, Hickson ID, The Bloom’s syndrome helicase suppresses crossing over during homologous recombination, Nature, 426 (2003) 870–874. [DOI] [PubMed] [Google Scholar]
- [114].Raynard S, Bussen W, Sung P, A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75, J. Biol. Chem., 281 (2006) 13861–13864. [DOI] [PubMed] [Google Scholar]
- [115].Wyatt HD, Sarbajna S, Matos J, West SC, Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells, Mol. Cell, 52 (2013) 234–247. [DOI] [PubMed] [Google Scholar]
- [116].Chan YW, West S, GEN1 promotes Holliday junction resolution by a coordinated nick and counter-nick mechanism, Nucleic Acids Res., 43 (2015) 10882–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Pike AC, Gomathinayagam S, Swuec P, Berti M, Zhang Y, Schnecke C, Marino F, von Delft F, Renault L, Costa A, Gileadi O, Vindigni A, Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: Insights from DNA complex structures, Proc. Natl. Acad. Sci. U. S. A., 112 (2015) 4286–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Garcia PL, Liu YL, Jiricny J, West SC, Janscak P, Human RECQ5 beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide, EMBO J., 23 (2004) 2882–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Islam MN, Fox D 3rd, Guo R, Enomoto T, Wang W, RecQL5 promotes genome stabilization through two parallel mechanisms--interacting with RNA polymerase II and acting as a helicase, Mol. Cell Biol., 30 (2010) 2460–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC, Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest, EMBO Rep., 1 (2000) 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Shamanna RA, Lu HM, de Freitas JK, Tian J, Croteau DL, Bohr VA, WRN regulates pathway choice between classical and alternative non-homologous end joining, Nat. Comm., 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR, BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome, Genes Dev., 22 (2008) 2856–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Chan YW, Fugger K, West SC, Unresolved recombination intermediates lead to ultra-fine anaphase bridges, chromosome breaks and aberrations, Nat. Cell. Biol., 20 (2018) 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Guo M, Vidhyasagar V, Talwar T, Kariem A, Wu Y, Mutational analysis of FANCJ helicase, Methods, 108 (2016) 118–129. [DOI] [PubMed] [Google Scholar]
- [125].Singh DK, Ghosh AK, Croteau DL, Bohr VA, RecQ helicases in DNA double strand break repair and telomere maintenance, Mutat. Res., 736 (2012) 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Uringa EJ, Youds JL, Lisaingo K, Lansdorp PM, Boulton SJ, RTEL1: An essential helicase for telomere maintenance and the regulation of homologous recombination, Nucleic Acids Res., 39 (2011) 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]