Abstract
Novel drug development is a time-consuming process with relatively high debilitating costs. To overcome this problem, computational drug repositioning approaches are being used to predict the possible therapeutic scaffolds against different diseases. In the current study, computational drug repositioning approaches were employed to fetch the promising drugs from the pool of FDA-approved drugs against Ewing sarcoma. The binding interaction patterns and conformational behaviors of screened drugs within the active region of Ewing sarcoma protein (EWS) were confirmed through molecular docking profiles. Furthermore, pharmacogenomics analysis was employed to check the possible associations of selected drugs with Ewing sarcoma genes. Moreover, the stability behavior of selected docked complexes (drugs-EWS) was checked by molecular dynamics simulations. Taken together, astemizole, sulfinpyrazone, and pranlukast exhibited a result comparable to pazopanib and can be used as a possible therapeutic agent in the treatment of Ewing sarcoma.
1. Introduction
Drug development is a time-consuming and overpriced process with particularly low success and relatively high failure rates. To overcome such problems, there are multiple computational drug-designing approaches, including drug repositioning that is being used nowadays.1 Drug repositioning approaches assist in minimizing the cost and time of the drug development process due to their known efficacy and therapeutic potential against other diseases.2 There are various computational methods such as feature-based methods, matrix decomposition-based methods, network-based methods, and reverse transcriptome-based methods for drug repositioning.3,4 However, it has been observed that drug development efforts for the treatment of Ewing sarcoma (ES) have been largely unsuccessful in the last decade.5
ES is a cancerous tumor usually observed in bones and other soft tissues like cartilages and nerve tissues, respectively.6 There are different types of ES based on the position of the tumor within the body, such as extraosseous and bone sarcomas, skin tumor, and peripheral primitive neuroectodermal tumor (pPNET). ES usually occurs in the pelvic region, shoulder blades, ribs, and femur bones.7,8 The major symptoms of ES are long-lasting fever, pain in bones, swelling of muscular and nerve tissues, and stiffness of long bones.9 It has been observed that Ewing tumors account for 10% of malignancies in humans and metastasize to the other parts of the body more frequently like bone marrow and lungs.10 A prior research report showed that the Ewing sarcoma protein (EWS) is the basic target of ES and is directly involved in the formation of ES bone carcinogenesis.11 EWS is an RNA binding protein that binds to Friend leukemia integration 1 transcription factor FLI1 forming EWS/FLI1 fusion protein. The N-terminus of the EWS/FLI1 domain allows EWS/FLI1 to bind with RNA polymerase II and recruit the barrier-to-autointegration factor complex. However, the C-terminus of EWS/FLI1 retains the DNA-binding domain of FLI1 and particularly binds with the ACCGGAAG core sequence. The EWS/FLI1 is preferentially bound to GGAA-repetitive regions, and there is a positive correlation between the GGAA microsatellites, EWS/FLI1 binding, and target gene expression.12
In the current study, a drug repositioning approach is employed to screen the Food and Drug Administration (FDA)-approved drugs against ES. The human Ewing protein is used as a receptor molecule to screen FDA-approved drugs through a shape-based screening approach. Pazopanib was used as a standard template to access similar ligand structures from FDA-approved compounds through the SwissSimilarity approach. The screened hits having similar chemical structures were accessed from the FDA-approved list and underwent molecular docking analysis using PyRx. Moreover, pharmacogenomics analysis (drug–genes interactions) was carried out by checking all possible genes against all selected drugs. The best-selected drugs were again examined by a docking procedure with AutoDock to check their binding affinities against the Ewing sarcoma protein. Finally, the best-generated docked complexes were further analyzed through molecular dynamics simulations to observe the structural stability through RMSD, RMSF, Rg, and SASA graphs.
2. Computational Methodology
2.1. Retrieval of Protein Structure
The three-dimensional (3D) solution structure of the RNA recognition motif of the Ewing Sarcoma (EWS) protein having PDBID 2CPE (https://www.rcsb.org/structure/2CPE) was retrieved from the Protein Data Bank and its energy was minimized with UCSF Chimera 1.10.1 using conjugate gradient algorithm and Amber force field.13 The structural assessment of the Ewing sarcoma protein such as α-helices, β-sheets, coils, and turns was confirmed through the VADAR 1.8 (http://vadar.wishartlab.com/) server. The Discovery Studio 2.1.0 Client was used to view the 3D structure of the target protein and for the generation of Ramachandran graphs.14
2.2. Shape-Based Screening of FDA-Approved Drugs Using SwissSimilarity
The SwissSimilarity15 is an online platform that allows one to identify similar chemical hits from FDA and other libraries with respect to the reference template structure. Pazopanib (Votrient) is an anticancer FDA-approved drug that was used as the reference template structure against ES.16,17 The chemical structure of pazopanib was retrieved from the Drug Bank (DB06589) and used as a template molecule to screen FDA-approved drugs. All of the screened drugs were ranked according to their predicted similarity score values (Table S2). The best-screened drugs were sketched in ACD/ChemSketch and further utilized for docking experiments.
2.3. Prediction of Active Binding Sites of the Ewing Sarcoma Protein
The Prankweb (http://prankweb.cz/) is an online source that explores the probability of amino acids involved in the formation of active binding sites. The binding pocket information was not available in PDB; therefore, active binding site residues of the Ewing sarcoma protein were predicted using Prankweb.
2.4. Molecular Docking Using PyRx and AutoDock
Before conducting our docking experiments, all of the screened drugs were sketched in the ACD/ChemSketch tool and accessed in the mol format. Furthermore, the UCSF Chimera 1.10.1 tool was employed for energy minimization of each ligand having default parameters such as steepest descent and conjugate gradient with 100 steps with a step size of 0.02 (Å), and the update interval was fixed at 10. In the PyRx docking experiment, all screened drugs were docked with the Ewing sarcoma protein using the default procedure.18 Before docking, the binding pocket of the target protein was confirmed from Prankweb and literature data. In docking experiments, the grid box dimension values were adjusted as center – X = −0.8961, Y = −1.6716, and Z = 0.3732, whereas size – X = 37.8273, Y = 36.5416, and Z = 36.5756, respectively, with the default exhaustiveness value = 8. The grid box size was adjusted on binding pocket residues to allow the ligand to move freely in the search space. Furthermore, the generated docked complexes were keenly analyzed to view their binding conformational poses at the active binding site of the Ewing sarcoma protein. Moreover, these docked complexes were evaluated based on the lowest binding energy (kcal/mol) values and binding interaction patterns between ligands and target proteins. The graphical depictions of all of the docked complexes were accomplished with UCSF Chimera 1.10.1 and Discovery Studio (2.1.0).
Furthermore, another docking experiment was employed on best-screened drugs against the Ewing sarcoma protein using the AutoDock 4.2 tool.19 In brief, for the receptor protein, the polar hydrogen atoms and Kollman charges were assigned. For the ligand, the Gasteiger partial charges were designated, and nonpolar hydrogen atoms were merged. All of the torsion angles for screened drugs were set free to rotate through the docking experiment. A grid map of 80 × 80 × 80 Å3 was adjusted on the binding pocket of the Ewing sarcoma protein to generate the grid map and to obtain the best conformational state of docking. A total of 100 runs were adjusted using docking experiments. The Lamarckian genetic algorithm (LGA) and empirical free energy function were applied by taking docking parameters default. All of the docked complexes were further evaluated on the lowest binding energy (kcal/mol) values, and hydrogen and hydrophobic interaction analysis using Discovery Studio (2.1.0) and UCSF Chimera 1.10.1 was performed.
2.5. Designing of Pharmacogenomics Networks
To design the pharmacogenomics network model for best-selected drugs, Drug Gene Interaction Databases (DGIdb) (https://www.dgidb.org/) and Drug Signatures Database (DSigDB) (http://dsigdb.tanlab.org/DSigDBv1.0/) were employed to obtain the possible list of different disease-associated genes. Furthermore, a detailed literature survey was performed against all predicted genes to identify their involvement in ES. Moreover, clumps of different disease-associated genes were sorted based on Ewing sarcoma, and the remaining disease-associated genes were eliminated from the data set.
2.6. Molecular Dynamics (MD) Simulations
The best-screened drug-EWS complexes having good energy values were selected to understand the residual backbone flexibility of protein structure; MD simulations were carried out using the Groningen Machine for Chemicals Simulations (GROMACS) 4.5.4 package20 with GROMOS 96 force field.21 The protein topology was designed by pdb2gmx command by employing GROMOS 96 force field. For ligand topology, all three drugs were separated from docking complexes and retrieved in the mol format using UCSF Chimera. Furthermore, SwissParam (https://www.swissparam.ch/), an online server, was used to generate ligands topologies files. Finally, both generated topologies (protein and ligand coordinates) were merged to run the simulation. Additionally, a simulation box with a minimum distance to any wall of 10 Å (1.0 nm) was generated on the complex by the editconf command. Moreover, the box was filled with solvent molecules using the gmx solvate command by employing the spc216.gro water model. The overall system charge was neutralized by adding ions. The steepest descent approach (1000 ps) for protein structure was applied for energy minimization. For energy minimization, the nsteps = 50 000 were adjusted with an energy step size (emstep) value of 0.01. The Particle Mesh Ewald (PME) method was employed for energy calculation and for electrostatic and van der Waals interactions; cutoff distance for the short-range VdW (rvdw) was set at 14 Å, whereas neighbor list (rlist) and nstlist values were adjusted as 1.0 and 10, respectively, in the em.mdp file.22 This method permits the use of the Ewald summation at a computational cost comparable to that of a simple truncation method of 10 Å or less, and the linear constraint solver (LINCS)23 algorithm was used for covalent bond constraints and the time step was set to 0.002 ps. Finally, the molecular dynamics simulation was carried out at 100 ns with nsteps 50 000 000 in the md.mdp file. Different structural evaluations such as root mean square deviations and fluctuations (RMSD/RMSF), solvent-accessible surface areas (SASA), and radii of gyration (Rg) of backbone residues were analyzed through Xmgrace software (http://plasma-gate.weizmann.ac.il/Grace/) and UCSF Chimera 1.10.1 software.
3. Results and Discussion
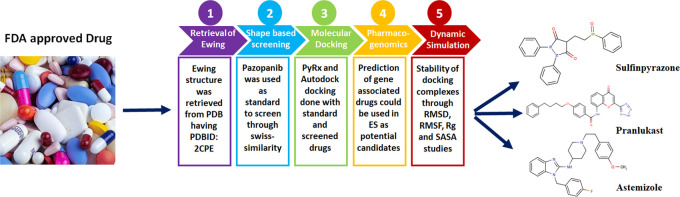
The overall design of the research is depicted in Figure 1, showing the flow starting from screening the FDA database into the best-screened drug having good therapeutic potential against ES. Figure 1 shows the different computational evaluation steps such as protein retrieval, drug screening, docking, pharmacogenomic, and MD simulation studies at both protein and drugs level to find out the keen and best possible chemical hits against ES.
Figure 1.
Drug repositioning of ES.
3.1. Structural Assessment of the Ewing Sarcoma Protein
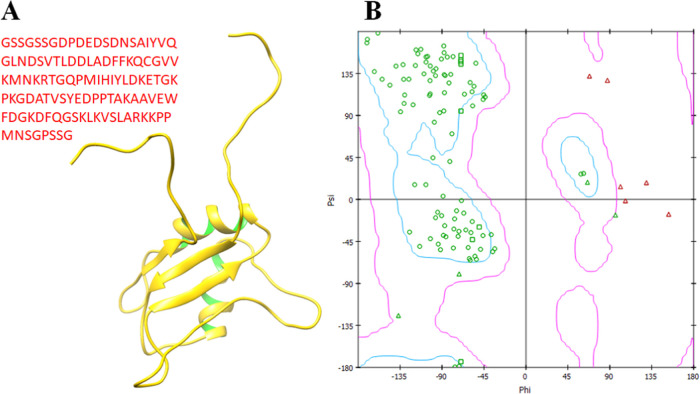
The Ewing sarcoma protein belongs to a class of hydrolases and consists of a single chain and comprises 113 amino acids (346–458 AA). The overall protein structure shows loops, α-helices, and β-sheets. It has been observed that two twisted loop structures were present at the terminal regions of the EWS protein and the central binding cavity of helices. Moreover, VADAR 1.8 structure analysis depicted that EWS consists of 25% α-helices, 30% β-sheets, 43% coils, and 20% turns. The Ramachandran plots and values indicated that 93.5% of amino acids exist in the favored region with good accuracy of phi (φ) and psi (ψ) angles. Moreover, the coordinates of EWS residues were also plunged into the acceptable region. The overall protein structure and Ramachandran graphs are shown in Figure 2A,B.
Figure 2.
(A, B) 3D structure of the Ewing sarcoma protein with Ramachandran graph.
3.2. Shape-Based Screening and Retrieval of Similar Drugs
In the drug repositioning approach, the shape-based screening, pharmacogenomics and molecular docking simulation are considered significant parameters to predict the possible therapeutic effects of known drugs against different targets.1,24 Pazopanib was used as a standard drug against ES25−27 and used as a template to screen FDA-approved drugs having a similar skeleton. In our computational results, SwissSimilarity results showed 100 FDA-approved drugs that were selected from the pool of 220 FDA-approved drugs based on similarity scoring values ranging from 0.005 to 0.998 (Table S1). Of the 100 FDA-approved drugs, 24 were selected based on best scoring values and have been reported in Table 1. Droperidol (0.015), delavirdine (0.010), irbesartan (0.014), tasosartan (0.013), and apixaban (0.010) showed the highest-scoring values as compared to the rest of all drugs. The screened drugs were ranked based on similarity scoring values, ranging from 0 to 1. The 0 value represents dissimilarity between compounds, whereas 1 is used for highly identical compounds in the screening approach.1 The chlorthalidone, mazindol, and progabide showed a unique value of similarity score of 0.005 as compared to the standard value. Therefore, 24 drugs were categorized based on the highest, lowest, and medium scoring values and further employed for the docking procedure to check which drug has good binding potential inside the binding pocket of the target protein. Therefore, the selection of drugs was made based on both similarity and docking energy values (Table 1).
Table 1. SwissSimilarity Scoring Values of FDA-Screened Drugs.
| drug bank ID | screened drugs | score | drug bank ID | screened drugs | score |
|---|---|---|---|---|---|
| DB00310 | chlorthalidone | 0.005 | DB01029 | irbesartan | 0.014 |
| DB00450 | droperidol | 0.015 | DB01122 | ambenonium | 0.008 |
| DB00496 | darifenacin | 0.006 | DB01138 | sulfinpyrazone | 0.007 |
| DB00546 | adinazolam | 0.007 | DB01342 | forasartan | 0.009 |
| DB00579 | mazindol | 0.005 | DB01349 | tasosartan | 0.013 |
| DB00637 | astemizole | 0.006 | DB01411 | pranlukast | 0.007 |
| DB00643 | mebendazole | 0.006 | DB06589 | pazopanib | 0.998 |
| DB00705 | delavirdine | 0.010 | DB06605 | apixaban | 0.010 |
| DB00808 | indapamide | 0.008 | DB08828 | vismodegib | 0.008 |
| DB00837 | progabide | 0.005 | DB08974 | flubendazole | 0.007 |
| DB00972 | azelastine | 0.006 | DB09003 | clocapramine | 0.006 |
| DB01026 | ketoconazole | 0.008 | DB00280 | disopyramide | 0.007 |
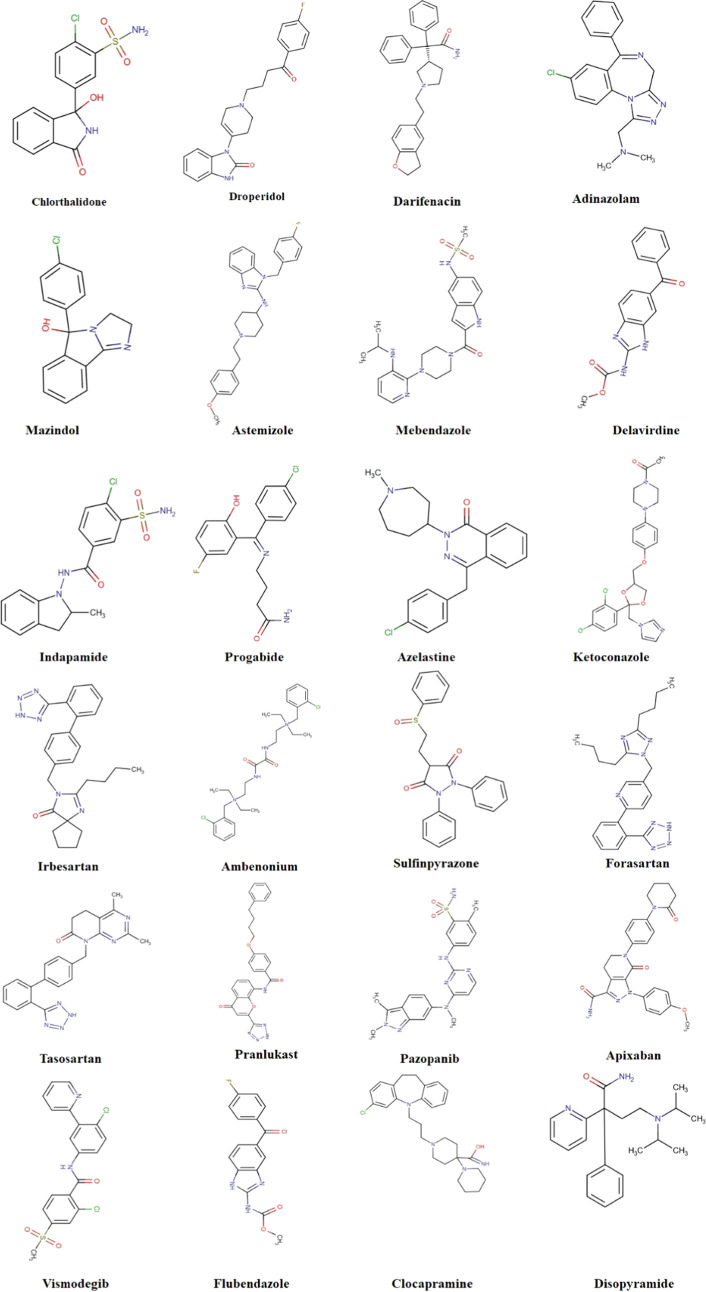
Although SwissSimilarity scoring values were low relative to the reference standard value range, structural moieties were similar at different parts in different screened FDA-approved drugs with respect to the standard drug (pazopanib). Therefore, a detailed docking study was run against all screened 24 drugs to check their binding interactions behavior in comparison with pazopanib. Based on these docking results, drugs were selected for further analysis (Figure 3).
Figure 3.
Screened FDA-approved drugs.
3.3. Binding Pocket Analysis of the EWS Protein
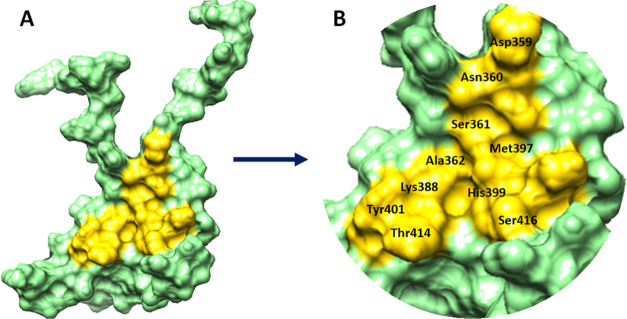
The position of a ligand in the holostructure of a protein most probably determines the binding pocket and channels of the target protein.28 P2Rank is a novel machine learning-based method for the prediction of ligand binding sites inside the protein structure.29 PrankWeb, a web server built upon P2Rank, was used by us to explore the binding pockets of the target protein with different pocket sizes and positions inside the target protein. Four different residue binding pockets were predicted based on scoring values (3.52, 2.80, 1.18, and 0.98). The higher pocket score value is 3.52 and constitutes amino acids (Asp359, Asn360, Ser361, Ala362, Lys388, Met397, His399, Tyr401, Thr414, and Ser416) at the central part of EWS. The Soluble Accessible Surface (SAS) area represents the area having a propensity to interact with neighboring atoms. Pocket 1 showed a good SAS value of 50 as compared to other binding pocket values (37, 25, and 21) with different amino acids of EWS (Figure 4). The graphical representation of the binding pocket of EWS is highlighted and depicted in Figure 5A,B.
Figure 4.
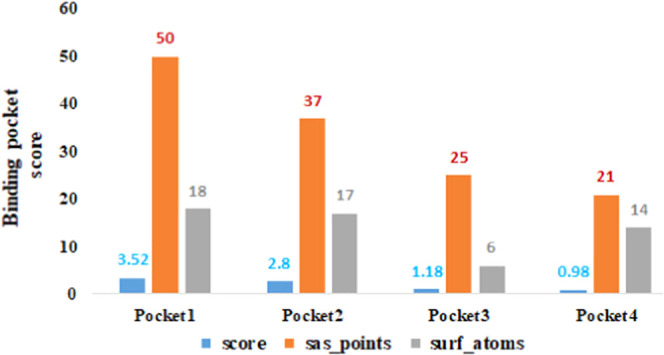
Predicted binding pockets.
Figure 5.
(A, B) Binding pocket of the EWS protein. The EWS protein is represented in cyan color, whereas the binding pocket site is highlighted in yellow color with the labeling of different binding pocket residues.
3.4. Molecular Docking
3.4.1. Binding Affinity Analysis of Screened Drug through PyRx
Molecular docking is a computational approach used to predict the binding conformational behavior of biomolecules, i.e., drugs and proteins.30−34 All of the screened drugs were docked and analyzed based on binding affinity (kcal/mol) (see the Supplementary Data S2). From docking results, it has been observed that from 100 FDA-approved drugs, 24 drugs showed binding affinity values higher than −7 kcal/mol and may have good binding potential inside the binding pocket of the EWS protein. The comparative analysis showed that darifenacin (DB00496) exhibited the highest binding affinity value of −9.2, whereas the rest of the drugs showed values ranging from −7 to −9 kcal/mol (Table 2).
Table 2. Binding Affinities of Screened Docking Complexes.
| accession numbers | drugs complexes | binding affinity (kcal/mol) | accession numbers | drug complexes | binding affinity (kcal/mol) |
|---|---|---|---|---|---|
| DB00310 | chlorthalidone | –7.8 | DB01029 | irbesartan | –7.2 |
| DB00450 | droperidol | –7.2 | DB01122 | ambenonium | –7.2 |
| DB00496 | darifenacin | –9.2 | DB01138 | sulfinpyrazone | –7 |
| DB00546 | adinazolam | –7.7 | DB01342 | forasartan | –7.8 |
| DB00579 | mazindol | –7.4 | DB01349 | tasosartan | –8.9 |
| DB00637 | astemizole | –8.3 | DB01411 | pranlukast | –8 |
| DB00643 | mebendazole | –7.4 | DB06589 | pazopanib | –7.6 |
| DB00705 | delavirdine | –7.5 | DB06605 | apixaban | –7.6 |
| DB00808 | indapamide | –7 | DB08828 | vismodegib | –7.7 |
| DB00837 | progabide | –7.2 | DB08974 | flubendazole | –7.1 |
| DB00972 | azelastine | –8.7 | DB09003 | clocapramine | –8.1 |
| DB01026 | ketoconazole | –7.7 | DB00280 | disopyramide | –7.1 |
3.5. Pharmacogenomics Analysis
3.5.1. Chlorthalidone, Droperidol, Darifenacin, Adinozolam, and Mazindol
The best-screened FDA-approved drugs having good binding affinity results were further analyzed through pharmacogenomics analysis. Pharmacogenomics aims to develop rational means to optimize drug therapy with respect to the patients’ genotype to ensure maximum efficacy with minimal adverse effects.35 In our computational analysis, a couple of pharmacogenomics databases were employed to predict the possible links of screened drugs with their respective genes and their association with diseases. The drug’s predicted genes were ranked based on interaction score values. In the chlorthalidone–genes network, 10 genes (NPPA, SLC12A1, AGT, SLC12A3, CA1, CA14, CA7, ACE, CA4, and MMP3) were observed with different interaction values and their association with multiple diseases (Table 3). Most of the genes like SLC12A1, AGT, SLC12A3, ACE, and MMP3 are directly involved in the osteosarcoma (a type of cancer that produces immature bone). Therefore, chlorthalidone could be used as a repositioned drug against bone cancer by targeting such gene-encoded protein and their associated pathways (Table 3). In droperidol–genes interactions, six genes, DRD2, KCNH2, DRD4, ADRA1A, DRD3, and CYP2D6, have been observed to have close interaction and involvement in different diseases. The highest predicted interaction of droperidol was observed with DRD2, which is directly linked with osteosarcoma of bones.36 Moreover, the predicted HTR2A is also involved in osteosarcoma of bone, especially in childhood.37 Darifenacin showed interactions with CHRM1/2, CHRM3, CHRM4, CHRM5, and CYP2D6 genes with different correlation values. Darifenacin–CHRM3 showed the highest interaction value (0.61) and was involved in the osteosarcoma of bone.37
Table 3. Screened Drugs Chlorthalidone, Droperidol, Darifenacin, Adinozolam, and Mazindol Association with Predicted Genes.
| genes | interaction scores | functions/diseases | references | |
|---|---|---|---|---|
| chlorthalidone | NPPA | 5.68 | osteoarthritis, spine | (39) |
| SLC12A1 | 3.03 | osteosarcoma | (40) | |
| AGT | 0.33 | childhood osteosarcoma | (41) | |
| SLC12A3 | 0.32 | sarcoma, neoplasms | (42) | |
| CA1 | 0.44 | neoplasms | (43) | |
| CA14 | 0.41 | malignant neoplasms | (44) | |
| CA7 | 0.36 | colorectal carcinoma | (45) | |
| ACE | 0.16 | synovial sarcoma | (46) | |
| CA4 | 0.25 | retinitis pigmentosa 17 | (47) | |
| MMP3 | 0.21 | osteosarcoma of bone; primary osteosarcoma | (48, 49) | |
| droperidol | DRD2 | 0.35 | malignant bone neoplasm; osteosarcoma of bone | (36) |
| KCNH2 | 0.04 | malignant neoplasm of prostate | (50) | |
| DRD4 | 0.11 | carcinoma of the lung | (51) | |
| ADRA1A | 0.09 | osteoporosis | (52) | |
| DRD3 | 0.08 | neoplasms | (53) | |
| CYP2D6 | 0.01 | bone cysts, aneurysmal | (54) | |
| HTR2A | 0.04 | osteosarcoma of bone; childhood osteosarcoma | (37) | |
| darifenacin | CHRM3 | 0.61 | osteosarcoma of bone | (37) |
| CHRM1/2 | 0.32 | mental depression | (55) | |
| CHRM4 | 0.46 | schizophrenia | (56) | |
| CHRM5 | 0.45 | systemic scleroderma | (57) | |
| CYP2D6 | 0.03 | eosinophilia-myalgia syndrome | (58) | |
| adinozolam | GABRR3/2 | 0.46 | restless legs syndrome | (59) |
| GABRR1 | 0.43 | migraine disorders | (60) | |
| GABRP | 0.22 | tumor progression | (61) | |
| GABRE | 0.22 | malignant neoplasms | (62) | |
| GABRD | 0.21 | Rett syndrome | (63) | |
| GABRG1 | 0.21 | body height | (64) | |
| mazindol | SLC6A3 | 1.27 | scoliosis | (65) |
| SLC6A2 | 0.97 | childhood osteosarcoma; osteosarcoma of bone | (66) | |
| SLC18A2 | 0.56 | childhood osteosarcoma; osteosarcoma of bone | (66) | |
| SLC6A4 | 0.21 | synovial sarcoma | (67) | |
| NAT1 | 0.16 | neoplasms | (68) | |
| HTT | 0.05 | Huntington Disease | (69) |
The Adinozolam–gene network showed that six genes, GABRR1, GABRR3/2, GABRP, GABRE, GABRD, and GABRG1, have a close association with Adinozolam having different interaction values. Literature data showed that these genes are involved in different disorders (Table 3); however, there was no connection between these genes and ES or bone osteosarcoma. Mazindol interacts with SLC6A3, SLC6A2, SLC18A2, SLC6A4, NAT1, and HTT, having good interaction values of 1.27, 0.97, 0.56, 0.21, 0.16, and 0.05, respectively. It was observed that SLC6A2 and SLC18A2 have a close association with childhood osteosarcoma in bone.38 The osteosarcoma of bone in childhood is the basic characteristic of ES; therefore, our proposed computational research favors the chlorthalidone and mazindol could be used for ES after evaluating and passing through clinical trials.
3.5.2. Astemizole, Indapamide, Delavirdine, Progabid, Azelastine, and Ketoconazole Pharmacogenomic Analysis
The astemizole pharmacogenomic analysis showed 10 genes, EED, KCNH1, CYP2J2, HPSE, HRH1, KCNH2, PPARD, ABCB1, CYP3A4, and CYP2D6, having different interaction scoring values (Table 4). Our computational results showed that astemizole has the potential to interact with multiple genes that are directly linked with ES through different ways like mutational or crosslinked signaling pathways. astemizole showed the highest interaction value (7.57) with EED as compared to other genes that are directly linked to ES.70 Moreover, astemizole has another interaction with the ABCB1 gene, which possesses a direct role in the etiology of ES. Literature reports also showed that ABCB1 has a good correlation with some diseases such as osteosarcoma of bone, childhood osteosarcoma, sarcoma of soft tissues, fibrosarcoma, adult fibrosarcoma, and peripheral primitive neuroectodermal tumor.71−75 Another report showed that the KCNH1 and HPSE genes are linked with multiple bone-associated diseases such as childhood osteosarcoma, fibrosarcoma, and synovial sarcoma.76−80 Indapamide interacts with SLC12A3, KCNE1, KCNQ1, and APEX1 with different scoring values. Prior data showed that SLC12A3 is involved in different diseases such as sarcoma, neoplasms, chondrosarcoma, and adult synovial sarcoma, respectively.42 Similarly, KCNE1, KCNQ1, and APEX1 are linked with atrial fibrillation,81 adenocarcinomas,82 and adenocarcinoma of the lung.83
Table 4. Astemizole, Indapamide, Delavirdine, Progabid, Azelastine, and Ketoconazole Gene Interactions.
| genes | interaction scores | functions/diseases | references | |
|---|---|---|---|---|
| astemizole | EED | 7.57 | Ewing sarcoma | (70) |
| KCNH1 | 0.95 | osteosarcoma of bone; childhood osteosarcoma | (76, 77) | |
| CYP2J2 | 0.76 | carcinogenesis | (93) | |
| HPSE | 0.58 | childhood osteosarcoma; fibrosarcoma; synovial sarcoma | (78−80) | |
| HRH1 | 0.27 | atherogenesis | (94) | |
| KCNH2 | 0.12 | malignant neoplasm of the prostate | (50) | |
| PPARD | 0.03 | obesity | (95) | |
| ABCB1 | 0.01 | Ewing’s sarcoma; osteosarcoma of bone; childhood osteosarcoma; sarcoma of soft tissue; fibrosarcoma; adult fibrosarcoma; peripheral primitive neuroectodermal tumor | (71−75) | |
| CYP3A4 | 0.01 | osteosarcoma | (96) | |
| CYP2D6 | 0.01 | bone cysts, aneurysmal | (54) | |
| indapamide | SLC12A3 | 3.94 | sarcoma, neoplasms, chondrosarcoma, adult synovial sarcoma | (42) |
| KCNE1 | 7.1 | atrial fibrillation | (81) | |
| KCNQ1 | 5.01 | adenocarcinoma | (82) | |
| APEX1 | 0.08 | adenocarcinoma of the lung | (83) | |
| delavirdine | ABCG2 | 0.36 | childhood osteosarcoma; osteosarcoma of bone | (84) |
| ABCC3 | 0.91 | carcinoma | (85) | |
| ABCC2 | 0.42 | sarcoma, fibrosarcoma | (86) | |
| ABCC1 | 0.42 | Ewing’s sarcoma of bone; osteosarcoma of bone; childhood osteosarcoma | (87, 88) | |
| ABCB1 | 0.09 | osteosarcoma of bone; childhood osteosarcoma | (75, 97, 98) | |
| progabid | GABBR1 | 15.49 | nasopharyngeal carcinoma | (89) |
| GABRA1 | 1.76 | osteochondrosis | (90) | |
| azelastine | LTC4S | 7.1 | myeloid leukemia, chronic | (99) |
| HRH1 | 1.08 | atherogenesis | (94) | |
| HRH2 | 1.58 | skin carcinoma | (100) | |
| PLA2G1B | 0.92 | degenerative polyarthritis | (101) | |
| ketoconazole | CYP21A2 | 4.37 | congenital adrenal hyperplasia | (102) |
| CYP3A43 | 0.62 | carcinogenesis | (103) | |
| CYP4F2 | 0.55 | hypertensive disease | (104) | |
| KCNA10 | 0.31 | brain neoplasms | (105) | |
| CYP17A1 | 0.31 | osteoporosis; carcinoma; spondylarthritis; spondylarthropathies | (106−108) | |
| ABCG2 | 0.07 | childhood osteosarcoma; osteosarcoma of bone | (84) | |
| NR1I3 | 0.16 | childhood osteosarcoma; osteosarcoma of bone; synovial sarcoma; exostoses | (91, 92) | |
| CYP3A4 | 0.02 | childhood osteosarcoma; osteoporosis; osteosarcoma of bone | (96, 109) | |
| NR1I2 | 0.04 | osteosarcoma of bone; adolescent idiopathic scoliosis | (110, 111) | |
| SNCA | 0.07 | Parkinson disease 1 | (112) |
Another screened drug, delavirdine, showed possible interactions with different genes such as ABCG2, ABCC3, ABCC2, ABCC1, and ABCB1 with good interaction scoring values. The literature data reports that all of the genes are involved in the osteosarcoma, osteosarcoma of bone, childhood osteosarcoma, and fibrosarcoma.84−86 However, among all five genes, ABCC1 is directly involved in ES through different pathways.87,88 Therefore, our computational results showed that delavirdine could also be used as a good chemical scaffold for the treatment of ES after in vitro, in vivo, and clinical trials.
Progabid showed interactions with GABBR1 and GABBA1, respectively, which are causative partners of nasopharyngeal carcinoma and osteochondrosis, respectively.89,90 The drug network showed that azelastine showed interactions with LTC4S, HRH1, HRH2, and PLA2G1B with different interaction values. It has been observed that these genes are myeloid leukemia, chronic atherogenesis, skin carcinoma, and degenerative polyarthritis, respectively. Ketoconazole showed interactions with CYP21A2, CYP3A43, CYP4F2, KCNA10, CYP17A1, ABCG2, NR1I3, CYP3A4, NR1I2, and SNCA. Literature data reported that among all 10 genes, four (ABCG2, NR1I3, CYP3A4, and NR1I2) were involved in childhood osteosarcoma84,91,92 (Table 4).
3.5.3. Irbesartan, Ambenonium, Sulfinpyrazone, Forasartan, Tasosartan, Pranlukast, and Gene Interactions
In Irbesartan pharmacogenomic analysis, 10 genes (AGTR1, SLC10A1, AGT, EDN1, APOB, APOE, ACE, JUN, SLC2A4, and CYP2C9) were involved in interactions with different scoring values. Among them, AGTR1, AGT, and SLC2A4 are involved in osteosarcoma of bone, childhood osteosarcoma, and osteoarthritis, respectively. However, the rest of the genes were associated with different diseases (Table 5). Ambenonium is associated with the acetylcholinesterase (AChE) gene, which is directly involved in Alzheimer’s disease.113
Table 5. Screened Drugs Irbesartan, Ambenonium, Sulfinpyrazone, Forasartan, Tasosartan, Pranlukast and Gene Interactions.
| genes | interaction scores | functions/diseases | references | |
|---|---|---|---|---|
| irbesartan | AGTR1 | 1.76 | osteosarcoma of bone; childhood osteosarcoma; osteoarthritis | (114, 115) |
| SLC10A1 | 4.73 | hepatitis B | (119) | |
| AGT | 1.11 | childhood osteosarcoma | (41) | |
| EDN1 | 0.79 | carcinogenesis | (120) | |
| APOB | 0.5 | carcinogenesis | (121) | |
| APOE | 0.47 | Alzheimer’s Disease | (122) | |
| ACE | 0.2 | osteoporosis | (123) | |
| JUN | 0.3 | osteosarcoma | (124) | |
| SLC2A4 | 0.22 | osteosarcoma of bone; childhood osteosarcoma | (125) | |
| CYP2C9 | 0.04 | ankylosing spondylitis | (126) | |
| ambenonium | ACHE | 9.19 | Alzheimer’s disease | (113) |
| sulfinpyrazone | SLC22A12 | 2.37 | renal hypouricemia | (127) |
| ABCC1 | 0.88 | Ewing’s sarcoma of bone osteosarcoma of bone; childhood osteosarcoma | (87, 88) | |
| ABCC2 | 0.79 | sarcoma, fibrosarcoma | (86) | |
| FPR1 | 0.59 | carcinogenesis | (128) | |
| UGT1A9 | 0.5 | carcinoma | (129) | |
| NR1I2 | 0.1 | osteosarcoma of bone; adolescent idiopathic scoliosis | (110, 111) | |
| CYP3A4 | 0.02 | osteosarcoma | (96) | |
| VDR | 0.01 | osteoporosis | (130) | |
| HPGD | 0.02 | carcinogenesis | (131) | |
| forasartan | AGTR1 | 8.91 | osteosarcoma of bone; childhood osteosarcoma; osteoarthritis, knee | (114, 115) |
| tasosartan | AGTR1 | 5.01 | osteosarcoma of bone; childhood osteosarcoma; osteoarthritis, knee | (114, 115) |
| AGTR2 | 7.1 | hypertensive disease | (132) | |
| pranlukast | RNASE3 | 20.29 | Ewing’s sarcoma of bone; osteosarcoma of bone; deformity of bone | (116−118) |
| CYSLTR1 | 6.09 | carcinogenesis; adenocarcinoma | (133) | |
| IL5 | 6.09 | asthma | (134) | |
| MUC2 | 6.09 | carcinoma, signet ring cell | (135) | |
| CYSLTR2 | 1.01 | carcinogenesis | (136) | |
| TNF | 0.37 | rheumatoid arthritis | (137) | |
| NFKB1 | 0.25 | osteosarcoma | (138) |
Sulfinpyrazone showed interactions with SLC22A12, ABCC1 and ABCC2, FPR1, UGT1A9, NR1I2, CYP3A4, VDR, and HPGD, which are involved in renal hypouricemia, fibrosarcoma, carcinogenesis, and different osteosarcomas. Most importantly, the ABCC1 gene is also directly involved in ES in different mechanistic pathways.87,88 Therefore, computational prediction of the literature data justify that sulfinpyrazone could be used as a screened drug against ES by targeting ABCC1-encoded protein and their associated downstream signaling pathways. Furthermore, a couple of other drugs, forasartan and tasosartan, showed interactions with AGTR1 and AGTR2, which are also connected with osteosarcoma of bones.114,115
Pranlukast formed a complex with seven different genes such as RNASE3, CYSLTR1, IL5, MUC2, CYSLTR2, TNF, and NFKB1, which are involved in different diseases. Pranlukast–RNASE3 showed the highest interaction value (20.29) as compared to other pharmacogenomics complexes. Moreover, the literature data showed that RNASE3 is involved in ES through different mechanistic pathways. Therefore, computational prediction and literature mining suggest that pranlukast could also be a good therapeutic agent against ES by targeting RNASE3.116−118
3.5.4. Pazopanib, Apixaban, Vismodegib, Clocapramine, and Disopyramide Pharmacogenomic Analysis
Pazopanib showed interactions with SH2B3, FGF1, ABCG2, and HLA-B, which play a key role in different diseases including leukemia, osteosarcoma of bone, and ankylosing spondylitis, respectively. Apixaban formed a pharmacogenomic complex with three genes such as ABCG2, F10, and CYP3A5, respectively. Literature data showed that these genes are mainly involved in the osteosarcoma of bone in childhood and osteoporosis.84,139 Vismodegib showed interactions with PTCH1, SMO, and SHH with different interaction scoring values. Prior data reported that these genes are involved in rhabdomyosarcoma and osteoarthritis.140,141 Moreover, a couple of other screened drugs, clocapramine and disopyramide, also formed pharmacogenomic complexes with different genes, which are involved in different diseases (Table 6).
Table 6. Screened Drugs Apixaban, Vismodegib, Clocapramine, Disopyramide, and Genomic Interactions.
| genes | interaction scores | functions/diseases | references | |
|---|---|---|---|---|
| pazopanib | SH2B3 | 0.84 | precursor cell lymphoblastic leukemia lymphoma | (142) |
| FGF1 | 0.36 | osteosarcoma of bone; bone neoplasms | (143, 144) | |
| ABCG2 | 0.04 | childhood osteosarcoma of bone | (84) | |
| HLA-B | 0.15 | ankylosing spondylitis | (145) | |
| apixaban | ABCG2 | 0.89 | childhood osteosarcoma of bone | (84) |
| F10 | 2.41 | osteoporosis | (139) | |
| CYP3A5 | 0.54 | neoplasms | (146) | |
| vismodegib | PTCH1 | 31.56 | rhabdomyosarcoma | (140) |
| SMO | 25.25 | osteoarthritis of the hip | (141) | |
| SHH | 12.62 | polydactyly | (147) | |
| clocapramine | PPARD | 0.39 | obesity | (95) |
| disopyramide | KCNH2 | 0.1 | malignant neoplasm of the prostate | (50) |
| CHRM2 | 0.16 | mental depression | (55) | |
| CHRM1 | 0.12 | mental depression | (55) | |
| CHRM3 | 0.09 | osteosarcoma of bone | (37) | |
| CYP2D6 | 0.01 | bone cysts, aneurysmal | (54) |
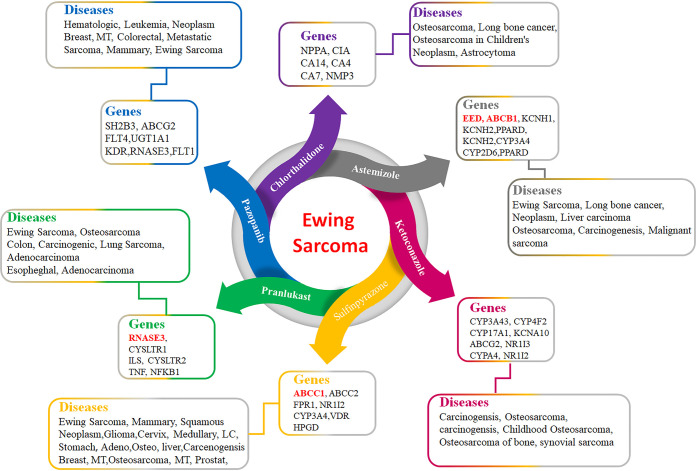
Based on pharmacogenomics analysis and extensive data mining of five screened FDA-approved drugs, chlorthalidone, astemizole, ketoconazole, sulfinpyrazone, and pranlukast were selected for further molecular docking and MD simulation analysis. Figure 6 shows that these five drugs have direct involvement in ES and related bone cancers. Chlorthalidone has genomic interactions with different genes, which are associated with different diseases. Similarly, astemizole has pharmacogenomic interactions with different genes and is associated with long bone cancer (ES), lung cancer, liver, breast, and stomach sarcoma. It has been observed that astemizole may be used as a good therapeutic potential against Ewing sarcoma. The predicted pharmacogenomic results of ketoconazole also showed its therapeutic potential against osteoporosis (bone cancer) and different sarcomas. Sulfinpyrazone also has an association with osteoporosis and lung and stomach cancer. The comparative result showed that a couple of screened FDA-approved drugs have a direct association with bone cancer such as ES, which could be further used to check their efficacy through cellular and clinical evaluations.
Figure 6.
FDA-approved drugs and associations with ES.
3.6. Binding Affinity Evaluations Using AutoDock
Based on PyRx docking energy (kcal/mol) and pharmacogenomics analysis, the best five drugs chlorthalidone (DB00310), astemizole (DB00637), ketoconazole (DB01026), sulfinpyrazone (DB01138), and pranlukast (DB01411) have been selected for further binding conformational analysis. The generated AutoDock results showed the binding energy, drug efficiency, and internal, electrostatic, and torsional energy values (kcal/mol) of best-selected screened drugs (Table 7).
Table 7. Binding Affinities of Screened Docking Complexes.
| drugs | binding energy (kcal/mol) | drug efficacy | internal energy (kcal/mol) | electrostatic (kcal/mol) | torsional energy (kcal/mol) |
|---|---|---|---|---|---|
| chlorthalidone | –4.88 | 0.22 | 5.18 | 0.01 | 0.3 |
| astemizole | –1.20 | 0.04 | 3.59 | 0.01 | 2.39 |
| ketoconazole | –0.66 | 0.02 | 2.15 | 0.15 | 1.49 |
| sulfinpyrazone | –5.18 | 0.18 | 3.39 | 0.13 | 1.79 |
| pranlukast | –11.84 | 0.33 | 9.75 | 0.01 | 2.09 |
3.6.1. Superimposition of Screened Drugs within the Active Region of the EWS Protein
All of the docked structures were superimposed to check the binding configurations of all five screened drugs within the active region of the EWS protein. The binding pocket analysis showed that all of the screened FDA-approved drugs were narrowed in the binding pocket and bound with similar residues with little different conformational poses within the binding pocket of the EWS protein. The binding of all FDA-approved drugs at the same position justified the docking reliability and the accuracy of predicted interactive results (Figure 7).
Figure 7.
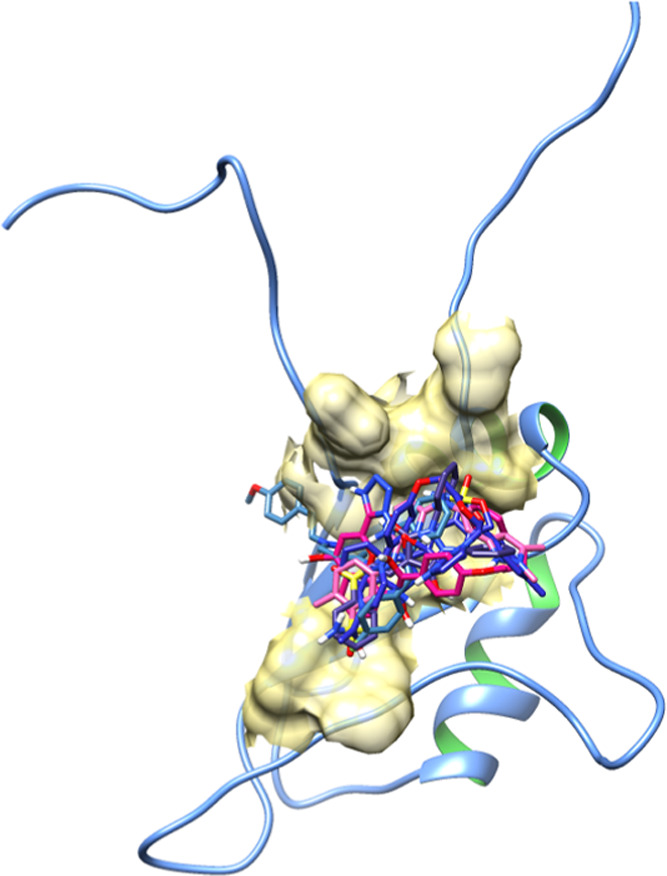
Superimposition of five docking complexes.
3.6.2. Chlorthalidone Hydrogen Binding Analysis
The chlorthalidone–EWS docked complex is analyzed based on the interaction pattern of binding pocket residues of EWS. The chlorthalidone binds with EWS having a good conformational position inside the active region encompassed by His399, Ile398, Leu374, Thr373, Val372, Pro409, and Leu402 residues, respectively. The oxygen atom of the thiol group in chlorthalidone formed hydrophobic interactions with His399 with a bond distance of 4.10 Å. Moreover, another oxygen atom is present in the five-member ring of the drug forming another hydrogen bond with Thr373 with a bond distance of 2.66 Å. In both chlorthalidone docking interactions both bonds provided good stable behavior to the docking complex, and bond distances were comparable to standard values (<5 hydrophobic and <3 Å hydrogen bonds), respectively (Figure 8).
Figure 8.
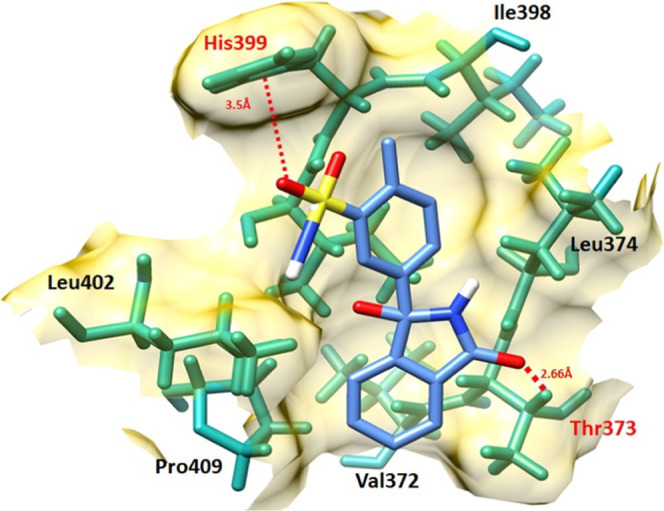
Chlorthalidone–EWS docking complex. The protein structure is represented in gray and purple color, whereas the binding pocket of the EWS protein is highlighted in yellow color in the surface format. The residues are highlighted in dark green color, whereas chlorthalidone is highlighted in blue color and embedded moieties such as oxygen, sulfur, and hydrogen are represented by red, yellow, and light gray colors, respectively.
3.6.3. Astemizole Hydrogen Binding Analysis
In astemizole–EWS docking, astemizole binds within the target site of EWS with an appropriate conformational position through interaction with different residues, His399, Ser416, Met397, Thr393, Gln395, Leu374, Ile400, and Ile398. The nitrogen atom of the amino group attached to a heterocyclic group formed a couple of hydrogen bonds with Ile398 and His399, with bond distances of 2.14 and 3.06 Å, respectively. Both astemizole interactions have good comparable values with standard values (<5 hydrophobic and <3 Å hydrogen bonds), respectively (Figure 9).
Figure 9.
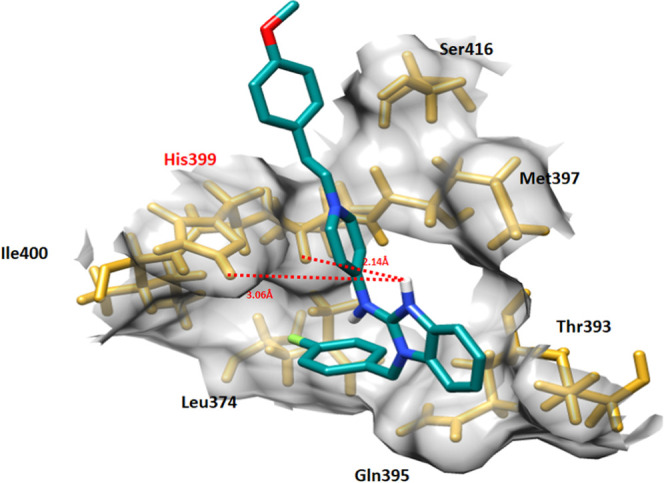
Astemizole–EWS docking complex. The EWS structure is represented in light pink color, whereas the binding pocket of the EWS protein is highlighted in gray color in the surface format. The residues are highlighted in golden color, whereas astemizole is highlighted in blue color and embedded moieties different colors, respectively.
3.6.4. Ketoconazole Hydrogen Binding Analysis
In ketoconazole–EWS docking, the drug binds with site-specific residues with appropriate conformational behavior. Ketoconazole encompassed different Arg392, Asn390, Gln395, Leu374, Thr373, Ile402, Ile400, His399, Ile398, and Met397 residues. Four hydrogen bonds were observed between the ketoconazole–EWS docking complex. The oxygen atom of the benzene ring formed tetrahydrogen bonds at Arg392 and Asn390 with bond distances of 2.04, 2.81, 2.66, and 2.72 Å, respectively. Ketoconazole interactions with EWS exhibited stable behavior in the docking complex (Figure 10).
Figure 10.
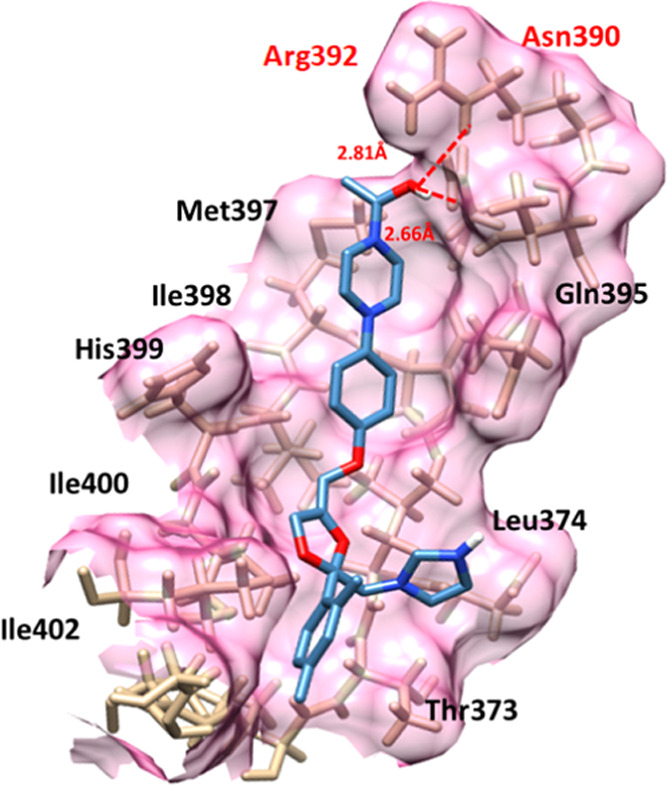
Ketoconazole–EWS docking complex. The interaction residues are highlighted in light pink color, whereas red dotted lines represent the hydrogen bonds in angstrom (Å).
3.6.5. Sulfinpyrazone and Pranlukast Hydrogen Binding Analysis
In sulfinpyrazone–EWS and pranlukast–EWS docking complexes, drugs bind with the binding pocket of EWS at slightly deviant conformational positions. The oxygen atom of sulfinpyrazone forms a single hydrogen bond with Met397 with a bonding distance of 1.86 Å. However, pranlukast forms two hydrogen bonds with EWS at Met397 and Tyr401 with bonding distances of 2.49 and 1.97 Å, respectively (Figures 11 and 12).
Figure 11.
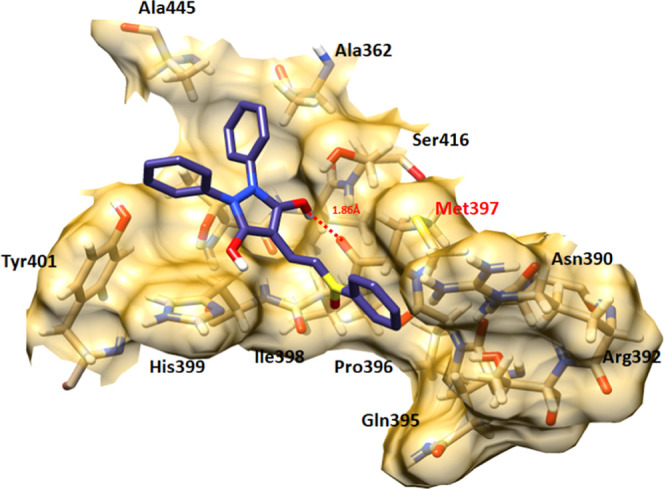
Sulfinpyrazone–EWS docking complex. The interaction residues are highlighted in light pink color, whereas red dotted lines represent the hydrogen bonds in angstrom (Å).
Figure 12.
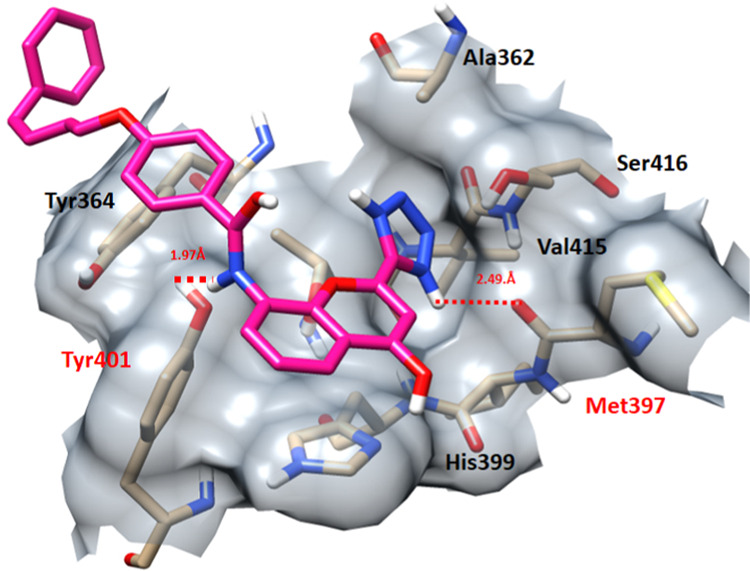
Pranlukast–EWS docking complex. The interaction residues are highlighted in light pink color, whereas red dotted lines represent the hydrogen bonds in angstrom (Å).
3.6.6. Pazopanib Docking Analysis
To check the accuracy of our docking results of screened FDA-approved drugs, the pazopanib–EWS docking complex was analyzed and checked for the interactive behavior against the target protein. The pazopanib-docking results showed similar amino acids such as Leu444, Ala445, Ala362, Ser416, Thr414, Tyr401, Tyr364, Gln366, and Ser443. Two hydrogen bonds were observed between the nitrogen of amino (NH2) and hydrogen atom of the methyl group (CH3) with Tyr401 and Ala445 with bond distances of 3.13 and 2.16 Å, respectively (Figure 13). The comparative analysis showed that the best five screened drugs bind with the EWS protein in a conformational pattern similar to pazopanib–EWS interactions. Therefore, the screened drugs may be used as a therapeutic template for the designing of novel drugs for the treatment of ES.
Figure 13.
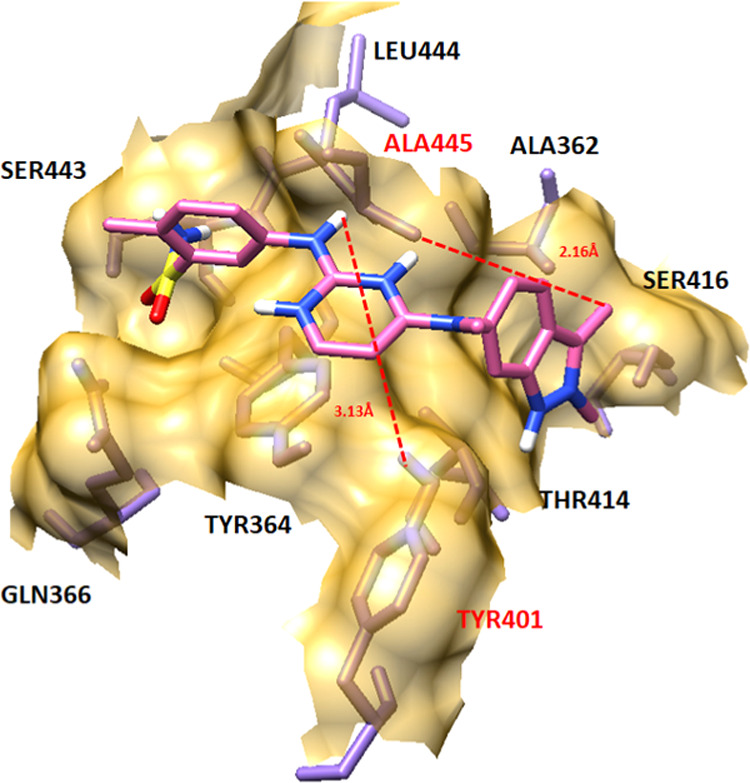
Pazopanib–EWS docking complex.
3.7. Screened Drugs and Their Possible Repositioned Functions
Based on pharmacogenomics, molecular docking, and detailed literature mining, the selected five drugs, chlorthalidone, astemizole, ketoconazole, sulfinpyrazone, and pranlukast, were keenly observed, and their repositioned functions were proposed by targeting different genes. It has been observed that chlorthalidone is usually used in hypertension; however, their proposed repositioned function is to be used against different osteosarcomas of bones and tissues. Moreover, astemizole is frequently used as an antihistamine and prescribed by medical staff against allergies. However, based on pharmacogenomic study and detailed literature mining, it could be used as a new therapeutic agent against ES by targeting the EED gene. Similarly, ketoconazole is used against Seborrheic dermatitis; however, its proposed repositioned function is osteosarcoma of bone in children. Sulfinpyrazone and pranlukast usually are used against Gouty arthritis and allergic rhinitis and asthma, respectively. However, our computational results showed their significance against ES by targeting ABCC1 and RNASE3 genes, respectively. The comparative results showed that astemizole, sulfinpyrazone, and pranlukast could be used as new drugs against ES by encompassing animal and clinical approaches (Table 8).
Table 8. Selected Drugs and Their Involvement in Diseases.
| no. | drugs | functions | repositioned functions |
|---|---|---|---|
| 1 | chlorthalidone | hypertension | osteosarcoma of bone in childhood |
| 2 | astemizole | allergy | Ewing sarcoma |
| 3 | ketoconazole | seborrheic dermatitis | osteosarcoma of bone in children |
| 4 | sulfinpyrazone | gouty arthritis | Ewing sarcoma |
| 5 | pranlukast | allergic rhinitis and asthma | Ewing sarcoma |
3.8. Molecular Dynamics Simulation
Based on molecular docking, pharmacogenomics, and literature mining results, sulfinpyrazone, chlorthalidone, and astemizole docked structures were selected to evaluate the residual flexibility in the target protein. The MD simulation study was employed at 50 000 ps using Gromacs 4.5.4. tool to generate root mean square deviations (RMSDs), root mean square fluctuations (RMSFs), solvent-accessible surface area (SASA), and radius of gyration (Rg) graphs.
3.8.1. Root Mean Square Deviation and Fluctuations
The RMSD and RMSF graphs were generated to understand the protein backbone behavior in the simulation running time. The RMSD results showed that in docking structures, protein backbone deviation remained steady and stable with the passage of simulation time frame 0–50 000 ps. All of the graph lines exhibited an increasing trend with RMSD values ranging from 0.1 to 0.3 nm from 0 to 30 000 ps.
Initially, all of the graph lines (blue, pink, and red) of wild and both docked complexes showed an increasing trend with RMSD values of 0.1–0.7 nm from 0 to 5000 ps. After 5000–10 000 ps, the graph lines remained stable with a constant RMSD value at 0.6 nm. From 10 000 to 20 000 ps, all three graph lines remained stable; however, little fluctuation was observed in the astemizole structure (red), and the value increased to 0.8 nm. However, with the passage of simulation time, again stable behavior was attained from simulation time of 20 000–30 000 ps. After that, from 30 000 to 50 000 ps, all of the structures exhibited a stable constant RMSD value (0.6 nm). The overall RMSD graphs lines showed that both docked complexes and wild EWS protein remained stable and fluctuated less in the simulation time frame. The generated graph results showed stable behavior in the backbone of all protein complexes. The results showed that the binding of all of these drugs did not affect the structural configurations of EWS and remained stable during the simulation time (Figure 14).
Figure 14.
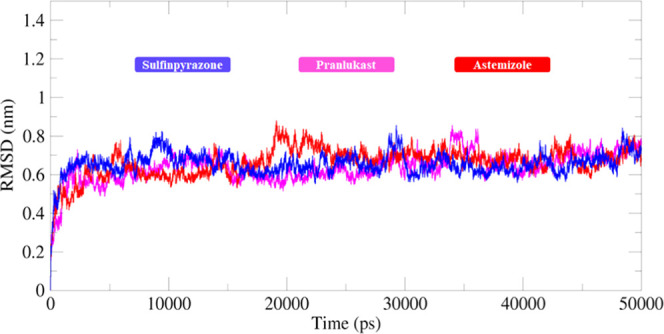
RMSD graph of sulfinpyrazone–EWS (blue), chlorthalidone–EWS (pink), and astemizole–EWS (red) docked structures. In the generated graph, the Y-axis showed RMSD values, whereas the X-axis represents simulation time from 0 to 50 000 ps.
The RMSF results of both docked complexes and standard EWS (blue, pink, and red curves) show that all residues dynamically fluctuated from N to C terminals. The protein structures are composed of 346–458 AA with different structural architectures. A couple of peaks have been observed at both terminal regions. The remaining parts of all protein structures in Figure 14 remained stable throughout the simulation time (0–500 000 ps). Moreover, the central region of the protein, which consists of the binding pocket also showed few variations and fluctuations in the protein molecules. However, these variations do not cause much disturbance in the protein conformations, which ensures that our docking results are much more stable and steady in behavior. Residues from 346 to 401 AA showed fluctuations due to the loop region, whereas from 15 to 30 fewer fluctuations were observed with an RMSF value of 0.25 nm. Moreover, amino acids comprising β-sheets and α-helices (401–445 AA) also remained stable in the simulation graph. After that, a couple of fluctuations peaks were observed from 445 to 460 AA. It has been observed that some interacting residues are also present in this region; after binding with the drug, they may disturb the protein structure, and the RMSF value increased from 0.25 to 0.5 nm. Moreover, from 460 to 490 AA, again smooth and steady peaks were seen, whereas after that again higher fluctuation peaks were observed due to the loop region of the Ewing sarcoma protein (Figure 15).
Figure 15.
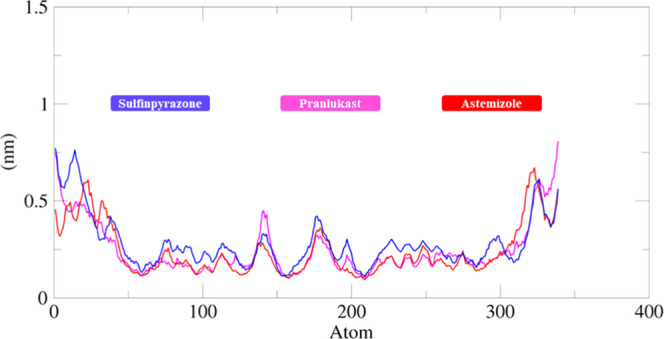
RMSF graph of sulfinpyrazone–EWS, chlorthalidone–EWS, and astemizole–EWS docked structures. The Y-axis shows RMSF (nm) values, whereas X-axis represents residues of EWS.
3.8.2. Solvent-Accessible Surface Area and Radius of Gyration
The structural compactness of protein was calculated by the radius of gyration (Rg). The generated results depicted that Rg values of all of the docked structures showed few variations from 1.5 to 1.7 nm. Initially, the graph lines were not much stable and showed few fluctuations from 0 to 5000 ps, while after that, stable behavior with few fluctuations was observed from 5000 to 10 000 ps time scale. After that, no bigger fluctuations were observed in graph lines and the Rg value also remained stable at 1.6 nm. The solvent-accessible surface areas (SASAs) were also observed and are shown in Figure 16. The results showed that the values of SASA of all five docked complexes were centered on 60 nm2 in the simulation time 0–50 000 ps (Figure 17).
Figure 16.
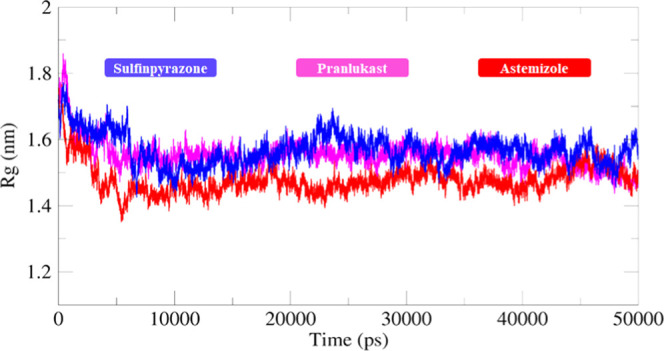
Rg graph of sulfinpyrazone–EWS, chlorthalidone–EWS, and astemizole–EWS docked complexes for a simulation time of 0–50 000 ps.
Figure 17.
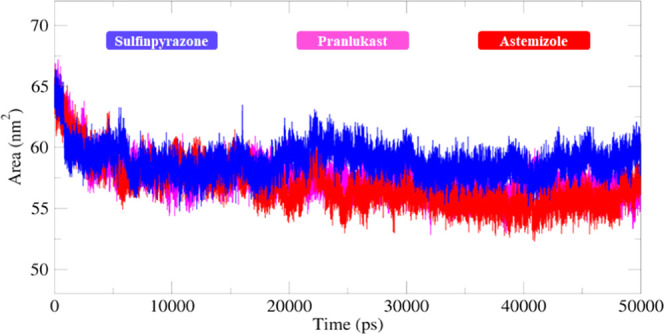
SASA graph of sulfinpyrazone, chlorthalidone, and astemizole–EWS docked structures for a simulation time frame of 0–50 000 ps.
4. Conclusions
Drug repositioning is a computational approach employed for drug discovery. The current study evaluates the repositioning of known drugs for ES using shape-based screening, molecular docking pharmacogenomics, and MD simulation approaches. The computational shaped-based screening results showed that 100 FDA-approved drugs exhibited good structural similarity and scores with standard (pazopanib). Moreover, docking profile and pharmacogenomics evaluations depicted that from the bunch of 24 only five drugs were most active and showed good results compared to other drugs. The detailed pharmacogenomics and extensive data mining showed that three drugs have a direct association with ES by targeting different genes. Moreover, MD simulation results also exposed that these three drugs showed better profiles with respect to their RMSD, RMSF, SASA, and Rg evaluations graphs and steadily stable behavior was observed in all docking complexes. Taken together, it has been concluded that predicted astemizole, sulfinpyrazone, and pranlukast exhibited better repositioning profiles as compared to other screened FDA-approved drugs. Therefore, sulfinpyrazone, pranlukast, and astemizole may be potentially used in the treatment of ES after in vitro and clinical assessment in the future.
Acknowledgments
A.K. acknowledges financial support from NSF Grant DBI 1661391 and NIH Grants R01GM127701 and R01HG012117. M.H. acknowledges the Ohio State University for providing the “President’s Postdoctoral Scholars Program (PPSP)” award and for financial support to complete this computational research.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c00518.
The authors have declared all of the raw data in the supplementary file; furthermore, no paid software and tools were employed and only free available tools were used to evaluate the results (PDF)
Author Contributions
M.H. designed this study and supervised in experiments; M.H. wrote the initial draft of the manuscript; M.Y. and S.S. collected data and performed experiments; and A.K. edited the manuscript and compiled it into the final format.
The authors declare no competing financial interest.
Supplementary Material
References
- Hassan M.; Raza H.; Abbasi M. A.; Moustafa A. A.; Seo S.-Y. The exploration of novel Alzheimer’s therapeutic agents from the pool of FDA approved medicines using drug repositioning, enzyme inhibition and kinetic mechanism approaches. Biomed. Pharmacother. 2019, 109, 2513–2526. 10.1016/j.biopha.2018.11.115. [DOI] [PubMed] [Google Scholar]
- Ghofrani H. A.; Osterloh I. H.; Grimminger F. Sildenafil: from angina to erectile dysfunction to pulmonary hypertension and beyond. Nat. Rev. Drug Discovery 2006, 5, 689–702. 10.1038/nrd2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B.; Hou F.; Ren C.; Bing P.; Xiao X. A Review of Current In Silico Methods for Repositioning Drugs and Chemical Compounds. Front. Oncol. 2021, 11, 711225 10.3389/fonc.2021.711225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarada T. N.; Rokne J. G.; Alhajj R. A review of computational drug repositioning: strategies, approaches, opportunities, challenges, and directions. J. Cheminform. 2020, 12, 46 10.1186/s13321-020-00450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton P. J.; Kurmasheva R. T. Challenges and opportunities for childhood cancer drug development. Pharmacol. Rev. 2019, 71, 671–697. 10.1124/pr.118.016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordóñez J. L.; O D.; Herrero D.; de Álava E.; MadozGúrpide J. Advances in Ewing’s sarcoma research: where are we now and what lies ahead?. Cancer Res. 2009, 69, 7140–7150. 10.1158/0008-5472.CAN-08-4041. [DOI] [PubMed] [Google Scholar]
- Roganović J. Bone tumors in children. Paediatrics Today 2011, 7, 1–9. [Google Scholar]
- Shikhare S. N.; Dubey N.; Peh W. C.. Bone Tumors: Radiographic Pitfalls. In Pitfalls in Musculoskeletal Radiology, Springer, 2017; pp 597–619. [Google Scholar]
- Grünewald T. G. P.; Cidre-Aranaz F.; Surdez D.; Tomazou E. M.; de Álava E.; Kovar H.; Sorensen P. H.; Delattre O.; Dirksen U. Ewing sarcoma. Nat. Rev. Dis. Primers 2018, 4, 5 10.1038/s41572-018-0003-x. [DOI] [PubMed] [Google Scholar]
- Lin P. P.; W Y.; Lozano G. Mesenchymal stem cells and the origin of Ewing’s sarcoma. Sarcoma 2011, 2011, 276463 10.1155/2011/276463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki T. Diagnosis and treatment of Ewing sarcoma of the bone: a review article. J. Orthop. Sci. 2015, 20, 250–263. 10.1007/s00776-014-0687-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cidre-Aranaz F.; Alonso J. EWS/FLI1 target genes and therapeutic opportunities in Ewing sarcoma. Front. Oncol. 2015, 5, 162. 10.3389/fonc.2015.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul Fattah T.; Saeed A.; Channar P. A.; Ashraf Z.; Abbas Q.; Hassan M.; Larik F. A. Synthesis, enzyme inhibitory kinetics, and computational studies of novel 1-(2-(4-isobutylphenyl) propanoyl)-3-arylthioureas as Jack bean urease inhibitors. Chem. Biol. Drug Des. 2018, 91, 434–447. 10.1111/cbdd.13090. [DOI] [PubMed] [Google Scholar]
- Discovery Studio Accelrys [2.1], 2008.
- Zoete V.; Daina A.; Bovigny C.; Michielin O. SwissSimilarity: a web tool for low to ultra high throughput ligand-based virtual screening. J. Chem. Inf. Model. 2016, 56, 1399–1404. 10.1021/acs.jcim.6b00174. [DOI] [PubMed] [Google Scholar]
- McLaughlin M. M.; Paglione M. G.; Slakter J.; Tolentino M.; Ye L.; Xu C.-F.; Suttle A. B.; Kim R. Y. Initial exploration of oral pazopanib in healthy participants and patients with age-related macular degeneration. JAMA Ophthalmol. 2013, 131, 1595–1601. 10.1001/jamaophthalmol.2013.5002. [DOI] [PubMed] [Google Scholar]
- Van Der Graaf W. T.; Blay J.-Y.; Chawla S. P.; Kim D.-W.; Bui-Nguyen B.; Casali P. G.; Schöffski P.; Aglietta M.; Staddon A. P.; Beppu Y.; et al. Pazopanib for metastatic soft-tissue sarcoma (PALETTE): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet 2012, 379, 1879–1886. 10.1016/S0140-6736(12)60651-5. [DOI] [PubMed] [Google Scholar]
- Dallakyan S.; Olson A. J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- Morris G. M.; Huey R.; Lindstrom W.; Sanner M. F.; Belew R. K.; Goodsell D. S.; Olson A. J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk S.; Páll S.; Schulz R.; Larsson P.; Bjelkmar P.; Apostolov R.; Shirts M. R.; Smith J. C.; Kasson P. M.; Van Der Spoel D.; et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S.-W.; Pandit S. A.; Scott H.; Jakobsson E. An improved united atom force field for simulation of mixed lipid bilayers. J. Phys. Chem. B 2009, 113, 2748–2763. 10.1021/jp807056c. [DOI] [PubMed] [Google Scholar]
- Wang H.; Dommert F.; Holm C. Optimizing working parameters of the smooth particle mesh Ewald algorithm in terms of accuracy and efficiency. J. Chem. Phys. 2010, 133, 034117 10.1063/1.3446812. [DOI] [PubMed] [Google Scholar]
- Amiri S.; Sansom M. S.; Biggin P. C. Molecular dynamics studies of AChBP with nicotine and carbamylcholine: the role of water in the binding pocket. Protein Eng. Des. Sel. 2007, 20, 353–359. 10.1093/protein/gzm029. [DOI] [PubMed] [Google Scholar]
- March-Vila E.; Pinzi L.; Sturm N.; Tinivella A.; Engkvist O.; Chen H.; Rastelli G. On the integration of in silico drug design methods for drug repurposing. Front. Pharmacol 2017, 8, 298. 10.3389/fphar.2017.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggerholm-Pedersen N.; Rossen P.; Rose H.; Safwat A. Pazopanib in the treatment of bone sarcomas: Clinical experience. Transl. Oncol. 2020, 13, 295–299. 10.1016/j.tranon.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi A.; Paioli A.; Palmerini E.; Cesari M.; Abate M. E.; Setola E.; Spinnato P.; Donati D.; Hompland I.; Boye K. Pazopanib in relapsed osteosarcoma patients: report on 15 cases. Acta Oncol. 2019, 58, 124–128. 10.1080/0284186X.2018.1503714. [DOI] [PubMed] [Google Scholar]
- Fox E.; Aplenc R.; Bagatell R.; Chuk M. K.; Dombi E.; Goodspeed W.; Goodwin A.; Kromplewski M.; Jayaprakash N.; Marotti M.; et al. A Phase 1 Trial and Pharmacokinetic Study of Cediranib, an Orally Bioavailable Pan–Vascular Endothelial Growth Factor Receptor Inhibitor, in Children and Adolescents With Refractory Solid Tumors. J. Clin. Oncol. 2010, 28, 5174. 10.1200/JCO.2010.30.9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stank A.; Kokh D. B.; Fuller J. C.; Wade R. C. Protein binding pocket dynamics. Acc. Chem. Res 2016, 49, 809–815. 10.1021/acs.accounts.5b00516. [DOI] [PubMed] [Google Scholar]
- Krivák R.; Hoksza D. In P2RANK: Knowledge-Based Ligand Binding Site Prediction Using Aggregated Local Features, International Conference on Algorithms for Computational Biology, 2015; pp 41–52.
- Hassan M.; Ashraf Z.; Abbas Q.; Raza H.; Seo S.-Y. Exploration of novel human tyrosinase inhibitors by molecular modeling, docking and simulation studies. Interdiscip. Sci.: Comput. Life Sci. 2018, 10, 68–80. 10.1007/s12539-016-0171-x. [DOI] [PubMed] [Google Scholar]
- Hassan M.; Shahzadi S.; Seo S. Y.; Alashwal H.; Zaki N.; Moustafa A. A. Molecular docking and dynamic simulation of AZD3293 and solanezumab effects against BACE1 to treat Alzheimer’s disease. Front. Comput. Neurosci. 2018, 12, 34. 10.3389/fncom.2018.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan M.; Shahzadi S.; Alashwal H.; Zaki N.; Seo S.-Y.; Moustafa A. A. Exploring the mechanistic insights of Cas scaffolding protein family member 4 with protein tyrosine kinase 2 in Alzheimer’s disease by evaluating protein interactions through molecular docking and dynamic simulations. Neurol. Sci. 2018, 39, 1361–1374. 10.1007/s10072-018-3430-2. [DOI] [PubMed] [Google Scholar]
- Hassan M.; Shahzadi S.; Raza H.; Abbasi M. A.; Alashwal H.; Zaki N.; Moustafa A. A.; Seo S.-Y. Computational investigation of mechanistic insights of Aβ42 interactions against extracellular domain of nAChRα7 in Alzheimer’s disease. Int. J. Neurosci. 2019, 129, 666–680. 10.1080/00207454.2018.1543670. [DOI] [PubMed] [Google Scholar]
- Hassan M.; Zahid S.; Alashwal H.; Kloczkowski A.; Moustafa A. A. Mechanistic insights into TNFR1/MADD death domains in Alzheimer’s disease through conformational molecular dynamic analysis. Sci. Rep. 2021, 11, 12256 10.1038/s41598-021-91606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becquemont L. Pharmacogenomics of adverse drug reactions: practical applications and perspectives. Pharmacogenomics 2009, 10, 961–969. 10.2217/pgs.09.37. [DOI] [PubMed] [Google Scholar]
- Dai W.-L.; Liu X.-T.; Bao Y.-N.; Yan B.; Jiang N.; Yu B.-Y.; Liu J.-H. Selective blockade of spinal D2DR by levo-corydalmine attenuates morphine tolerance via suppressing PI3K/Akt-MAPK signaling in a MOR-dependent manner. Exp. Mol. Med. 2018, 50, 1–12. 10.1038/s12276-018-0175-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinman M. A.; Fischer J. A.; Jacobs D. C.; Goodall C. P.; Bracha S.; Chappell P. E. Autocrine production of reproductive axis neuropeptides affects proliferation of canine osteosarcoma in vitro. BMC Cancer 2019, 19, 158 10.1186/s12885-019-5363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Brenner E.; Sifuentes-Rincón A.; Randel R.; Paredes-Sánchez F.; Parra-Bracamonte G.; Arellano Vera W.; Rodríguez Almeida F.; Segura Cabrera A. Association of SNPs in dopamine and serotonin pathway genes and their interacting genes with temperament traits in Charolais cows. J. Appl. Genet. 2017, 58, 363–371. 10.1007/s13353-016-0383-0. [DOI] [PubMed] [Google Scholar]
- Williams F. M. K.; Kato B. S.; Livshits G.; Sambrook P. N.; Spector T. D.; MacGregor A. J. Lumbar disc disease shows linkage to chromosome 19 overlapping with a QTL for hand OA. Ann. Rheum. Dis. 2008, 67, 117–119. 10.1136/ard.2007.073262. [DOI] [PubMed] [Google Scholar]
- Seaayfan E.; Defontaine N.; Demaretz S.; Zaarour N.; Laghmani K. OS9 protein interacts with Na-K-2Cl Co-transporter (NKCC2) and targets its immature form for the endoplasmic reticulum-associated degradation pathway. J. Biol. Chem. 2016, 291, 4487–4502. 10.1074/jbc.M115.702514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L.; Zhang G.; Hou Y.; Chen J.; Yin Y. Elemene inhibits osteosarcoma growth by suppressing the renin-angiotensin system signaling pathway. Mol. Med. Rep. 2018, 17, 1022–1030. 10.3892/mmr.2017.7965. [DOI] [PubMed] [Google Scholar]
- Kito F.; Oyama R.; Takai Y.; Sakumoto M.; Shiozawa K.; Qiao Z.; Uehara T.; Yoshida A.; Kawai A.; Kondo T. Establishment and characterization of the NCC–SS1–C1 synovial sarcoma cell line. Hum. Cell 2018, 31, 167–174. 10.1007/s13577-018-0199-9. [DOI] [PubMed] [Google Scholar]
- Tetteh P. W.; Kretzschmar K.; Begthel H.; Van Den Born M.; Korving J.; Morsink F.; Farin H.; Van Es J. H.; Offerhaus G. J. A.; Clevers H. Generation of an inducible colon-specific Cre enzyme mouse line for colon cancer research. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 11859–11864. 10.1073/pnas.1614057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida S.; Nishimori I.; Tanimura M.; Onishi S.; Shuin T. Effects of von Hippel-Lindau gene mutation and methylation status on expression of transmembrane carbonic anhydrases in renal cell carcinoma. J. Cancer Res. Clin. Oncol. 2002, 128, 561–568. 10.1007/s00432-002-0374-x. [DOI] [PubMed] [Google Scholar]
- Yang G.-Z.; Hu L.; Cai J.; Chen H.-Y.; Zhang Y.; Feng D.; Qi C.-Y.; Zhai Y.-X.; Gong H.; Fu H.; et al. Prognostic value of carbonic anhydrase VII expression in colorectal carcinoma. BMC Cancer 2015, 15, 209 10.1186/s12885-015-1216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou X.-M.; Cheng H.-J.; Meng L.; Zhou L.-L.; Ke Y.-H.; Liu L.-P.; Li Y.-M. Correlations between ACE single nucleotide polymorphisms and prognosis of patients with septic shock. Biosci. Rep. 2017, 37, BSR20170145 10.1042/BSR20170145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandor A.; Ramesar R.; Prince S. Cell-specific differences in the processing of the R14W CAIV mutant associated with retinitis pigmentosa 17. J. Cell. Biochem. 2010, 111, 735–741. 10.1002/jcb.22759. [DOI] [PubMed] [Google Scholar]
- Huang J.-F.; Du W.-X.; Chen J.-J. Elevated expression of matrix metalloproteinase-3 in human osteosarcoma and its association with tumor metastasis. J. BUON 2016, 21, 1279–1286. [PubMed] [Google Scholar]
- Fan H.; Lu S.; Wang S.; Zhang S. Identification of critical genes associated with human osteosarcoma metastasis based on integrated gene expression profiling. Mol. Med. Rep. 2019, 20, 915–930. 10.3892/mmr.2019.10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänze J.; Rexin P.; Jakubowski P.; Schreiber H.; Heers H.; Lingelbach S.; Kinscherf R.; Weihe E.; Hofmann R.; Hegele A. Prostate cancer tissues with positive TMPRSS2-ERG-gene-fusion status may display enhanced nerve density. Urol. Oncol. 2020, 38, 3.e7–3.e15. 10.1016/j.urolonc.2018.07.019. [DOI] [PubMed] [Google Scholar]
- Campa D.; Zienolddiny S.; Lind H.; Ryberg D.; Skaug V.; Canzian F.; Haugen A. Polymorphisms of dopamine receptor/transporter genes and risk of non-small cell lung cancer. Lung Cancer 2007, 56, 17–23. 10.1016/j.lungcan.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Zhao L.-H.; Ma S.; Sutkeviciute I.; Shen D.-D.; Zhou X. E.; de Waal P. W.; Li C.-Y.; Kang Y.; Clark L. J.; Jean-Alphonse F. G.; et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science 2019, 364, 148–153. 10.1126/science.aav7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L.; Shi X.; Chen R.; Wu Z.; Yang Z.; Li Z. Association of Mental Health-Related Proteins DAXX, DRD3, and DISC1 With the Progression and Prognosis of Chondrosarcoma. Front. Mol. Biosci. 2019, 6, 134. 10.3389/fmolb.2019.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karle J.; Bolbrinker J.; Vogl S.; Kreutz R.; Denkert C.; Eucker J.; Wischnewsky M.; Possinger K.; Regierer A. C. Influence of CYP2D6-genotype on tamoxifen efficacy in advanced breast cancer. Breast Cancer Res. Treat. 2013, 139, 553–560. 10.1007/s10549-013-2565-3. [DOI] [PubMed] [Google Scholar]
- Cannon D. M.; Klaver J.; Gandhi S. K.; Solorio G.; Peck S. A.; Erickson K.; N Akula J. S.; Eckelman W.; Furey M.; Sahakian B.; et al. Genetic variation in cholinergic muscarinic-2 receptor gene modulates M2 receptor binding in vivo and accounts for reduced binding in bipolar disorder. Mol. Psychiatry 2011, 16, 407–418. 10.1038/mp.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher C. Minds, Maps, Meanderings. J. Am. Acad. Child Adolesc. Psychiatry 2019, 58, 827–828. 10.1016/j.jaac.2019.05.017. [DOI] [PubMed] [Google Scholar]
- Martin J.-E.; Assassi S.; Diaz-Gallo L.-M.; Broen J. C.; Simeon C. P.; Castellvi I.; Vicente-Rabaneda E.; Fonollosa V.; Ortego-Centeno N.; González-Gay M. A.; et al. A systemic sclerosis and systemic lupus erythematosus pan-meta-GWAS reveals new shared susceptibility loci. Hum. Mol. Genet. 2013, 22, 4021–4029. 10.1093/hmg/ddt248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flockhart D. A.; Clauw D. J.; Sale E. B.; Hewett J.; Woosley R. L. Pharmacogenetic characteristics of the eosinophilia-myalgia syndrome. Clin. Pharmacol. Ther. 1994, 56, 398–405. 10.1038/clpt.1994.154. [DOI] [PubMed] [Google Scholar]
- Chen J.; Luo Q.; Li G.; Huang Y.; Ma J. Genetic Association Study of Restless Legs Syndrome in Chinese Population. Eur. Neurol. 2019, 81, 47–55. 10.1159/000500416. [DOI] [PubMed] [Google Scholar]
- García-Martín E.; Martínez C.; Serrador M.; Alonso-Navarro H.; Navacerrada F.; Esguevillas G.; García-Albea E.; Agúndez J. A.; Jiménez-Jiménez F. J. Gamma-Aminobutyric Acid (Gaba) Receptors Rho (Gabrr) Gene Polymorphisms and Risk for Migraine. Headache 2017, 57, 1118–1135. 10.1111/head.13122. [DOI] [PubMed] [Google Scholar]
- Jiang S.-H.; Zhu L.-L.; Zhang M.; Li R.-K.; Yang Q.; Yan J.-Y.; Zhang C.; Yang J.-Y.; Dong F.-Y.; Dai M.; et al. GABRP regulates chemokine signalling, macrophage recruitment and tumour progression in pancreatic cancer through tuning KCNN4-mediated Ca2+ signalling in a GABA-independent manner. Gut 2019, 68, 1994–2006. 10.1136/gutjnl-2018-317479. [DOI] [PubMed] [Google Scholar]
- Kristensen H.; Haldrup C.; Strand S.; Mundbjerg K.; Mortensen M. M.; Thorsen K.; Ostenfeld M. S.; Wild P. J.; Arsov C.; Goering W.; et al. Hypermethylation of the GABRE∼ miR-452∼ miR-224 promoter in prostate cancer predicts biochemical recurrence after radical prostatectomy. Clin. Cancer Res. 2014, 20, 2169–2181. 10.1158/1078-0432.CCR-13-2642. [DOI] [PubMed] [Google Scholar]
- Okamoto N.; Miya F.; Tsunoda T.; Kato M.; Saitoh S.; Yamasaki M.; Shimizu A.; Torii C.; Kanemura Y.; Kosaki K. Targeted next-generation sequencing in the diagnosis of neurodevelopmental disorders. Clin. Genet. 2015, 88, 288–292. 10.1111/cge.12492. [DOI] [PubMed] [Google Scholar]
- Kichaev G.; Bhatia G.; Loh P.-R.; Gazal S.; Burch K.; Freund M. K.; Schoech A.; Pasaniuc B.; Price A. L. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 2019, 104, 65–75. 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffenberger E. G.; Jinks R. N.; Sougnez C.; Cibulskis K.; Willert R. A.; Achilly N. P.; Cassidy R. P.; Fiorentini C. J.; Heiken K. F.; Lawrence J. J.; et al. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS One 2012, 7, e28936 10.1371/journal.pone.0028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Brenner E.; Sifuentes-Rincón A.; Randel R.; Paredes-Sánchez F.; Parra-Bracamonte G.; Vera W. A.; Almeida F. R.; Cabrera A. S. Association of SNPs in dopamine and serotonin pathway genes and their interacting genes with temperament traits in Charolais cows. J. Appl. Genet. 2017, 58, 363–371. 10.1007/s13353-016-0383-0. [DOI] [PubMed] [Google Scholar]
- Eddahibi S.; Chaouat A.; Morrell N.; Fadel E.; Fuhrman C.; Bugnet A.-S.; Dartevelle P.; Housset B.; Hamon M.; Weitzenblum E.; Adnot S. Polymorphism of the serotonin transporter gene and pulmonary hypertension in chronic obstructive pulmonary disease. Circulation 2003, 108, 1839–1844. 10.1161/01.CIR.0000091409.53101.E8. [DOI] [PubMed] [Google Scholar]
- Savci-Heijink C. D.; Halfwerk H.; Hooijer G.; Koster J.; Horlings H.; Meijer S.; van de Vijver M. Epithelial-to-mesenchymal transition status of primary breast carcinomas and its correlation with metastatic behavior. Breast Cancer Res. Treat. 2019, 174, 649–659. 10.1007/s10549-018-05089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowska M.; Olejniczak M. Gene therapy for Huntington’s disease using targeted endonucleases. Methods Mol. Biol. 2020, 2056, 269–284. 10.1007/978-1-4939-9784-8_17. [DOI] [PubMed] [Google Scholar]
- Burdach S.; Plehm S.; Unland R.; Borkhardt A.; Staege M. S.; Müller-Tidow C.; Richter G. H. Epigenetic maintenance of stemness and malignancy in peripheral neuroectodermal tumors by EZH2. Cell Cycle 2009, 8, 1991–1996. 10.4161/cc.8.13.8929. [DOI] [PubMed] [Google Scholar]
- Zwelling L. A.; Slovak M. L.; Doroshow J. H.; Hinds M.; Chan D.; Parker E.; Mayes J.; Sie K. L.; Meltzer P. S.; Trent J. M. HT1080/DR4: a P-glycoprotein-negative human fibrosarcoma cell line exhibiting resistance to topoisomerase II-reactive drugs despite the presence of a drug-sensitive topoisomerase II. J. Natl. Cancer Inst. 1990, 82, 1553–1561. 10.1093/jnci/82.19.1553. [DOI] [PubMed] [Google Scholar]
- Scotlandi K.; Manara M. C.; Hattinger C. M.; Benini S.; Perdichizzi S.; Pasello M.; Bacci G.; Zanella L.; Bertoni F.; Picci P.; Serra M. Prognostic and therapeutic relevance of HER2 expression in osteosarcoma and Ewing’s sarcoma. Eur. J. Cancer 2005, 41, 1349–1361. 10.1016/j.ejca.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Okada T.; Tanaka K.; Nakatani F.; Sakimura R.; Matsunobu T.; Li X.; Hanada M.; Nakamura T.; Oda Y.; Tsuneyoshi M.; Iwamoto Y. Involvement of P-glycoprotein and MRP1 in resistance to cyclic tetrapeptide subfamily of histone deacetylase inhibitors in the drug-resistant osteosarcoma and Ewing’s sarcoma cells. Int. J. Cancer 2006, 118, 90–97. 10.1002/ijc.21297. [DOI] [PubMed] [Google Scholar]
- Kretschmer N.; Rinner B.; Stuendl N.; Kaltenegger H.; Wolf E.; Kunert O.; Boechzelt H.; Leithner A.; Bauer R.; Lohberger B. Effect of costunolide and dehydrocostus lactone on cell cycle, apoptosis, and ABC transporter expression in human soft tissue sarcoma cells. Planta Med. 2012, 78, 1749–1756. 10.1055/s-0032-1315385. [DOI] [PubMed] [Google Scholar]
- Cheng F.; Zhao Z.; Liu W. Long non-coding RNA ROR regulated ABCB1 to induce cisplatin resistance in osteosarcoma by sponging miR-153-3p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7256–7265. 10.26355/eurrev_201909_18828. [DOI] [PubMed] [Google Scholar]
- Wu J.; Zeng T.; Wu X.; Gao Q.; Zhai W.; Ding Z. Ether a go-go 1 silencing in combination with TRAIL overexpression has synergistic antitumor effects on osteosarcoma. Cancer Biother. Radiopharm. 2013, 28, 65–70. 10.1089/cbr.2012.1283. [DOI] [PubMed] [Google Scholar]
- Wu X.; Chen Z.; Zeng W.; Zhong Y.; Liu Q.; Wu J. Silencing of Eag1 gene inhibits osteosarcoma proliferation and migration by targeting STAT3-VEGF pathway. BioMed Res. Int. 2015, 2015, 617316 10.1155/2015/617316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panny S.; Scott A.; Smith K.; Phillips J. 3rd; Kazazian H. Jr.; Talbot C. Jr.; Boehm C. D. Population heterogeneity of the Hpa I restriction site associated with the beta globin gene: implications for prenatal diagnosis. Am. J. Hum. Genet. 1981, 33, 25–35. [PMC free article] [PubMed] [Google Scholar]
- Simizu S.; Suzuki T.; Muroi M.; Lai N. S.; Takagi S.; Dohmae N.; Osada H. Involvement of disulfide bond formation in the activation of heparanase. Cancer Res. 2007, 67, 7841–7849. 10.1158/0008-5472.CAN-07-1053. [DOI] [PubMed] [Google Scholar]
- Luo C.; Yang Z.; Wang L. Heparanase participates in the growth and invasion of human U-2OS osteosarcoma cells and its close relationship with hypoxia-inducible factor-1α in osteosarcoma. Neoplasma 2010, 57, 562–571. 10.4149/neo_2010_06_562. [DOI] [PubMed] [Google Scholar]
- Weigl I.; Geschwill P.; Reiss M.; Bruehl C.; Draguhn A.; Koenen M.; Sedaghat-Hamedani F.; Meder B.; Thomas D.; Katus H. A.; Schweizer P. A. The C-terminal HCN4 variant P883R alters channel properties and acts as genetic modifier of atrial fibrillation and structural heart disease. Biochem. Biophys. Res. Commun. 2019, 519, 141–147. 10.1016/j.bbrc.2019.08.150. [DOI] [PubMed] [Google Scholar]
- Than B. L. N.; Goos J.; Sarver A. L.; O’Sullivan M. G.; Rod A.; Starr T. K.; Fijneman R. J.; Meijer G. A.; Zhao L.; Zhang Y.; et al. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene 2014, 33, 3861–3868. 10.1038/onc.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G.-S.; Li M.; Xu C.-X.; Wang D. APE1 stimulates EGFR-TKI resistance by activating Akt signaling through a redox-dependent mechanism in lung adenocarcinoma. Cell Death Dis. 2018, 9, 1111 10.1038/s41419-018-1162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundhill E. A.; Jabri S.; Burchill S. A. ABCG1 and Pgp identify drug resistant, self-renewing osteosarcoma cells. Cancer Lett. 2019, 453, 142–157. 10.1016/j.canlet.2019.03.011. [DOI] [PubMed] [Google Scholar]
- Kocher O.; Comella N.; Gilchrist A.; Pal R.; Tognazzi K.; Brown L. F.; Knoll J. PDZK1, a novel PDZ domain-containing protein up-regulated in carcinomas and mapped to chromosome 1q21, interacts with cMOAT (MRP2), the multidrug resistance-associated protein. Lab. Invest. 1999, 79, 1161–1170. [PubMed] [Google Scholar]
- Arlanov R.; Porter A.; Strand D.; Brough R.; Karpova D.; Kerb R.; Wojnowski L.; Schwab M.; Lang T. Functional characterization of protein variants of the human multidrug transporter ABCC2 by a novel targeted expression system in fibrosarcoma cells. Hum. Mutat. 2012, 33, 750–762. 10.1002/humu.22041. [DOI] [PubMed] [Google Scholar]
- Oda Y.; Dockhorn-Dworniczak B.; Jürgens H.; Roessner A. Expression of multidrug resistance-associated protein gene in Ewing’s sarcoma and malignant peripheral neuroectodermal tumor of bone. J. Cancer Res. Clin. Oncol. 1997, 123, 237–239. 10.1007/BF01240323. [DOI] [PubMed] [Google Scholar]
- Kun-Peng Z.; Xiao-Long M.; Chun-Lin Z. LncRNA FENDRR sensitizes doxorubicin-resistance of osteosarcoma cells through down-regulating ABCB1 and ABCC1. Oncotarget 2017, 8, 71881. 10.18632/oncotarget.17985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Fu L.; Wong A. M. G.; Fan Y.-H.; Li M.-X.; Bei J.-X.; Jia W.-H.; Zeng Y.-X.; Chan D.; Cheung K. M.; et al. Identification of genes with allelic imbalance on 6p associated with nasopharyngeal carcinoma in southern Chinese. PLoS One 2011, 6, e14562 10.1371/journal.pone.0014562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks C.; Mömke S.; Drögemüller C.; Leeb T.; Chowdhary B.; Distl O. A high-resolution comparative radiation hybrid map of equine chromosome 4q12–q22. Anim. Genet. 2006, 37, 513–517. 10.1111/j.1365-2052.2006.01510.x. [DOI] [PubMed] [Google Scholar]
- Gu W.; Ogose A.; Kawashima H.; Ito M.; Ito T.; Matsuba A.; Kitahara H.; Hotta T.; Tokunaga K.; Hatano H.; et al. High-level expression of the coxsackievirus and adenovirus receptor messenger RNA in osteosarcoma, Ewing’s sarcoma, and benign neurogenic tumors among musculoskeletal tumors. Clin. Cancer Res. 2004, 10, 3831–3838. 10.1158/1078-0432.CCR-03-0345. [DOI] [PubMed] [Google Scholar]
- Köksal H.; Müller E.; Inderberg E. M.; Bruland Ø.; Wälchli S. Treating osteosarcoma with CAR T cells. Scand. J. Immunol. 2019, 89, e12741 10.1111/sji.12741. [DOI] [PubMed] [Google Scholar]
- Allison S. E.; Chen Y.; Petrovic N.; Zimmermann S.; Moosmann B.; Jansch M.; Cui P. H.; Dunstan C. R.; Mackenzie P. I.; Murray M. Activation of the pro-migratory bone morphogenetic protein receptor 1B gene in human MDA-MB-468 triple-negative breast cancer cells that over-express CYP2J2. Int. J. Biochem. Cell Biol. 2016, 80, 173–178. 10.1016/j.biocel.2016.10.004. [DOI] [PubMed] [Google Scholar]
- Raveendran V. V.; Smith D. D.; Tan X.; Sweeney M. E.; Reed G. A.; Flynn C. A.; Tawfik O. W.; Milne G.; Dileepan K. N. Chronic ingestion of H1-antihistamines increase progression of atherosclerosis in apolipoprotein E-/-mice. PLoS One 2014, 9, e102165 10.1371/journal.pone.0102165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Garf S.; Murdaca J.; Mothe-Satney I.; Sibille B.; Le Menn G.; Chinetti G.; Neels J. G.; Rousseau A.-S. Complementary immunometabolic effects of exercise and PPARβ/δ agonist in the context of diet-induced weight loss in obese female mice. Int. J. Mol. Sci. 2019, 20, 5182. 10.3390/ijms20205182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.; Xia C.; Sun Q.; Sheng F.; Xiong J.; Wang S. Variants of FasL and ABCC5 are predictive of outcome after chemotherapy-based treatment in osteosarcoma. J. Bone Oncol. 2018, 12, 44–48. 10.1016/j.jbo.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T.; Fei Z.; Su H.; Xie R.; Chen L. Polydatin inhibits proliferation and promotes apoptosis of doxorubicin-resistant osteosarcoma through LncRNA TUG1 mediated suppression of Akt signaling. Toxicol. Appl. Pharmacol. 2019, 371, 55–62. 10.1016/j.taap.2019.04.005. [DOI] [PubMed] [Google Scholar]
- Torreggiani E.; Roncuzzi L.; Perut F.; Zini N.; Baldini N. Multimodal transfer of MDR by exosomes in human osteosarcoma. Int. J. Oncol. 2016, 49, 189–196. 10.3892/ijo.2016.3509. [DOI] [PubMed] [Google Scholar]
- Roos C.; Sjölinder M.; Stenke L.; Tornhamre S. Abnormal LTC4 synthase RNA degradation in neutrophils from CML patients. Br. J. Haematol. 2004, 124, 739–745. 10.1111/j.1365-2141.2004.04848.x. [DOI] [PubMed] [Google Scholar]
- Welsh M. M.; Karagas M. R.; Kuriger J. K.; Houseman A.; Spencer S. K.; Perry A. E.; Nelson H. H. Genetic determinants of UV-susceptibility in non-melanoma skin cancer. PLoS One 2011, 6, e20019 10.1371/journal.pone.0020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovetz M.; Baekelandt M.; Berner A.; Trope C. G.; Davidson B.; Reich R. The clinical role of phospholipase A2 isoforms in advanced-stage ovarian carcinoma. Gynecol. Oncol. 2006, 103, 831–840. 10.1016/j.ygyno.2006.06.042. [DOI] [PubMed] [Google Scholar]
- Kamenický P.; Blanchard A.; Lamaziere A.; Piedvache C.; Donadille B.; Duranteau L.; Bry H.; Gautier J.-F.; Salenave S.; Raffin-Sanson M.-L.; et al. Cortisol and aldosterone responses to hypoglycemia and Na depletion in women with non-classic 21-hydroxylase deficiency. J Clin. Endocrinol. Metab. 2020, 105, 55–64. 10.1210/clinem/dgz005. [DOI] [PubMed] [Google Scholar]
- Daly A. K. Significance of the minor cytochrome P450 3A isoforms. Clin. Pharmacokinet. 2006, 45, 13–31. 10.2165/00003088-200645010-00002. [DOI] [PubMed] [Google Scholar]
- Shekhar S.; Varghese K.; Li M.; Fan L.; Booz G. W.; Roman R. J.; Fan F. Conflicting roles of 20-HETE in hypertension and stroke. Int. J. Mol. Sci. 2019, 20, 4500. 10.3390/ijms20184500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S.; Kaluz S.; Devi N. S.; Jabbar A. A.; de Noronha R. G.; Mun J.; Zhang Z.; Boreddy P. R.; Wang W.; Wang Z.; et al. Arylsulfonamide KCN1 inhibits in vivo glioma growth and interferes with HIF signaling by disrupting HIF-1α interaction with cofactors p300/CBP. Clin. Cancer Res 2012, 18, 6623–6633. 10.1158/1078-0432.CCR-12-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somner J.; McLellan S.; Cheung J.; Mak Y.; Frost M. L.; Knapp K. M.; Wierzbicki A. S.; Wheeler M.; Fogelman I.; Ralston S. H.; Hampson G. N. Polymorphisms in the P450 c17 (17-hydroxylase/17, 20-Lyase) and P450 c19 (aromatase) genes: association with serum sex steroid concentrations and bone mineral density in postmenopausal women. J. Clin. Endocrinol. Metab. 2004, 89, 344–351. 10.1210/jc.2003-030164. [DOI] [PubMed] [Google Scholar]
- Hofland J.; Timmerman M. A.; De Herder W. W.; Van Schaik R.; De Krijger R.; De Jong F. H. Expression of activin and inhibin subunits, receptors and binding proteins in human adrenocortical neoplasms. Clin. Endocrinol. 2006, 65, 792–799. 10.1111/j.1365-2265.2006.02668.x. [DOI] [PubMed] [Google Scholar]
- Mori K.; Kizawa H.; Ushiyama T.; Chano T.; Inoue H.; Tsuchiya N.; Okabe H.; Matsusue Y.; Ikegawa S. Association of CYP17 with HLA-B27-negative seronegative spondyloarthropathy in Japanese males. Am. J. Med. Genet. A 2004, 130, 169–171. 10.1002/ajmg.a.30259. [DOI] [PubMed] [Google Scholar]
- Kang Y.; Park S.; Yim C.; Kwak H.; Gajendrarao P.; Krishnamoorthy N.; Yun S. C.; Lee K.; Han K. The CYP3A4* 18 genotype in the cytochrome P450 3A4 gene, a rapid metabolizer of sex steroids, is associated with low bone mineral density. Clin. Pharm. Ther. 2009, 85, 312–318. 10.1038/clpt.2008.215. [DOI] [PubMed] [Google Scholar]
- Koutsounas I.; Patsouris E.; Theocharis S. E. Pregnane X receptor and human malignancy. Histol. Histopathol. 2013, 28, 405–420. 10.14670/HH-28.405. [DOI] [PubMed] [Google Scholar]
- Liu J.; Zhou Y.; Liu S.; Song X.; Yang X.-Z.; Fan Y.; Chen W.; Akdemir Z. C.; Yan Z.; Zuo Y.; et al. The coexistence of copy number variations (CNVs) and single nucleotide polymorphisms (SNPs) at a locus can result in distorted calculations of the significance in associating SNPs to disease. Hum. Genet. 2018, 137, 553–567. 10.1007/s00439-018-1910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsigelny I. F.; Sharikov Y.; Kouznetsova V. L.; Greenberg J. P.; Wrasidlo W.; Overk C.; Gonzalez T.; Trejo M.; Spencer B.; Kosberg K.; Masliah E. Molecular determinants of α-synuclein mutants’ oligomerization and membrane interactions. ACS Chem. Neurosci. 2015, 6, 403–416. 10.1021/cn500332w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Işık M.; Beydemir Ş. AChE mRNA expression as a possible novel biomarker for the diagnosis of coronary artery disease and Alzheimer’s disease, and its association with oxidative stress. Arch. Physiol. Biochem. 2022, 128, 352–359. 10.1080/13813455.2019.1683584. [DOI] [PubMed] [Google Scholar]
- Yamagishi K.; Tsukamoto I.; Nakamura F.; Hashimoto K.; Ohtani K.; Akagi M. Activation of the reninangiotensin system in mice aggravates mechanical loading induced knee osteoarthritis. Eur. J. Histochem. 2018, 62, 2930 10.4081/ejh.2018.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Xu K.; Liu P. Post-transcriptional control of angiotensin II type 1 receptor regulates osteosarcoma cell death. Cell. Physiol. Biochem. 2018, 45, 1581–1589. 10.1159/000487719. [DOI] [PubMed] [Google Scholar]
- McKeon C.; Thiele C. J.; Ross R. A.; Kwan M.; Triche T. J.; Miser J. S.; Israel M. A. Indistinguishable patterns of protooncogene expression in two distinct but closely related tumors: Ewing’s sarcoma and neuroepithelioma. Cancer Res. 1988, 48, 4307–4311. [PubMed] [Google Scholar]
- Chao C.; Li F.; Tan Z.; Zhang W.; Yang Y.; Luo C. miR-195 inhibited abnormal activation of osteoblast differentiation in MC3T3-E1 cells via targeting RAF-1. Exp. Cell Res. 2018, 362, 293–301. 10.1016/j.yexcr.2017.11.030. [DOI] [PubMed] [Google Scholar]
- Lu J.; Chen W.; Liu H.; Yang H.; Liu T. Transcription factor CEBPB inhibits the proliferation of osteosarcoma by regulating downstream target gene CLEC5A. J. Clin. Lab. Anal. 2019, 33, e22985 10.1002/jcla.22985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.-R.; Huang Y.-Y.; Hsiao J.-K. Role of sodium taurocholate cotransporting polypeptide as a new reporter and drug-screening platform: implications for preventing hepatitis B virus infections. Mol. Imaging Biol. 2020, 22, 313–323. 10.1007/s11307-019-01373-y. [DOI] [PubMed] [Google Scholar]
- Yuan W.; Qian M.; Li Z.-X.; Zhao C.-L.; Zhao J.; Xiao J.-R. Endothelin-1 activates the Notch signaling pathway and promotes tumorigenesis in giant cell tumor of the spine. Spine 2019, 44, E1000–E1009. 10.1097/BRS.0000000000003044. [DOI] [PubMed] [Google Scholar]
- Gao J.; Choudhry H.; Cao W. Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like family genes activation and regulation during tumorigenesis. Cancer Sci. 2018, 109, 2375–2382. 10.1111/cas.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman E. M.; Daw E. W.; Yu C.-E.; Payami H.; Steinbart E. J.; Nochlin D.; Conlon E. M.; Bird T. D.; Schellenberg G. D. Evidence for a novel late-onset Alzheimer disease locus on chromosome 19p13. 2. Am. J. Hum. Genet. 2004, 75, 398–409. 10.1086/423393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H.; Nakagami H.; Osako M. K.; Nakagami F.; Kunugiza Y.; Tomita T.; Yoshikawa H.; Rakugi H.; Ogihara T.; Morishita R. Prevention of osteoporosis by angiotensin-converting enzyme inhibitor in spontaneous hypertensive rats. Hypertens. Res. 2009, 32, 786–790. 10.1038/hr.2009.99. [DOI] [PubMed] [Google Scholar]
- Fromigué O.; Hamidouche Z.; Marie P. J. Blockade of the RhoA-JNK-c-Jun-MMP2 cascade by atorvastatin reduces osteosarcoma cell invasion. J. Biol. Chem. 2008, 283, 30549–30556. 10.1074/jbc.M801436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzenberg-Bar-Yoseph F.; Armoni M.; Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004, 64, 2627–2633. 10.1158/0008-5472.CAN-03-0846. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Yi X.-D.; Lu H.-L. Influence of CYP2C9 and COX-2 genetic polymorphisms on clinical efficacy of non-steroidal anti-inflammatory drugs in treatment of ankylosing spondylitis. Med. Sci. Monit. 2017, 23, 1775. 10.12659/MSM.900271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuto Y.; Kawamura M.; Namikawa A.; Takahashi H.; Shibuya Y.; Mori T.; Sohara E. Non-urate transporter 1, non-glucose transporter member 9-related renal hypouricemia and acute renal failure accompanied by hyperbilirubinemia after anaerobic exercise: a case report. BMC Nephrol. 2019, 20, 1–8. 10.1186/s12882-019-1618-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapkov I.; Öqvist C. O.; Figenschau Y.; Kogner P.; Johnsen J. I.; Sveinbjørnsson B. The role of formyl peptide receptor 1 (FPR1) in neuroblastoma tumorigenesis. BMC Cancer 2016, 16, 490 10.1186/s12885-016-2545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaghavan V. L.; Sindhwani P.; Hinds T. D. Jr Glucuronidation and UGT isozymes in bladder: new targets for the treatment of uroepithelial carcinomas?. Oncotarget 2017, 8, 3640. 10.18632/oncotarget.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.; Zhou D.; Shen G.; Cui Y.; Lv Q.; Wei F. Association of VDR and OPG gene polymorphism with osteoporosis risk in Chinese postmenopausal women. Climacteric 2019, 22, 208–212. 10.1080/13697137.2018.1554643. [DOI] [PubMed] [Google Scholar]
- Park Y. S.; Lee J. H.; Jung D.-B.; Kim H.-B.; Jung J.-H.; Pak S.; Ryu Y.-M.; Park H. J.; Park Y.-Y.; Jung H.-Y.; Myung S. J. MicroRNA-21 induces loss of 15-hydroxyprostaglandin dehydrogenase in early gastric tubular adenocarcinoma. Sci. Rep. 2018, 8, 17717 10.1038/s41598-018-36139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W. D.; Steffens D. C.; Ashley-Koch A.; Payne M. E.; MacFall J. R.; Potocky C. F.; Krishnan K. R. R. Angiotensin receptor gene polymorphisms and 2-year change in hyperintense lesion volume in men. Mol. Psychiatry 2010, 15, 816–822. 10.1038/mp.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piromkraipak P.; Sangpairoj K.; Tirakotai W.; Chaithirayanon K.; Unchern S.; Supavilai P.; Power C.; Vivithanaporn P. Cysteinyl leukotriene receptor antagonists inhibit migration, invasion, and expression of MMP-2/9 in human glioblastoma. Cell. Mol. Neurobiol. 2018, 38, 559–573. 10.1007/s10571-017-0507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R. K.; Brickner H.; Panwar B.; Ramírez-Suástegui C.; Herrera-de La Mata S.; Liu N.; Diaz D.; Alexander L. E. C.; Ay F.; Vijayanand P.; et al. Human eosinophils express a distinct gene expression program in response to IL-3 compared with common β-chain cytokines IL-5 and GM-CSF. J. Immunol. 2019, 203, 329–337. 10.4049/jimmunol.1801668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai K.; Satake M.; Tokunaga O. Gastric adenocarcinoma of fundic gland type with signet-ring cell carcinoma component: a case report and review of the literature. World J. Gastroenterol. 2018, 24, 2915. 10.3748/wjg.v24.i26.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venerito M.; Helmke C.; Jechorek D.; Wex T.; Rosania R.; Antweiler K.; Weigt J.; Malfertheiner P. Leukotriene receptor expression in esophageal squamous cell cancer and non-transformed esophageal epithelium: a matched case control study. BMC Gastroenterol. 2016, 16, 85 10.1186/s12876-016-0499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B. S.; Jo S.; Kwon E.; Shin J. H.; Hur J.-W.; Kim T.-H. Effect of biologics in the level of cytokines in the synovial fluid of patients with ankylosing spondylitis. Korean J. Intern. Med. 2020, 35, 465. 10.3904/kjim.2018.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.; Fang W.; Huang Z.; Lu J.; Wang Y.; Tang Q.; Song G.; Kang Y.; Zhu X.; Zou C. Metformin reduces SATB2-mediated osteosarcoma stem cell-like phenotype and tumor growth via inhibition of N-cadherin/NF-κB signaling. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4516–4528. [PubMed] [Google Scholar]
- Somjen D.; Sharfman Z. T.; Katzburg S.; Sharon O.; Maman E.; Salai M.; Stern N.; Dolkart O. Rivaroxaban significantly inhibits the stimulatory effects of bone-modulating hormones: In vitro study of primary female osteoblasts. Connect. Tissue Res. 2017, 58, 215–220. 10.1080/03008207.2016.1220942. [DOI] [PubMed] [Google Scholar]
- Pazzaglia S. Ptc1 heterozygous knockout mice as a model of multi-organ tumorigenesis. Cancer Lett. 2006, 234, 124–134. 10.1016/j.canlet.2005.03.047. [DOI] [PubMed] [Google Scholar]
- Styrkarsdottir U.; Lund S. H.; Thorleifsson G.; Zink F.; Stefansson O. A.; Sigurdsson J. K.; Juliusson K.; Bjarnadottir K.; Sigurbjornsdottir S.; Jonsson S.; et al. Meta-analysis of Icelandic and UK data sets identifies missense variants in SMO, IL11, COL11A1 and 13 more new loci associated with osteoarthritis. Nat. Genet. 2018, 50, 1681–1687. 10.1038/s41588-018-0247-0. [DOI] [PubMed] [Google Scholar]
- Baughn L.; Meredith M.; Oseth L.; Smolarek T.; Hirsch B. SH2B3 aberrations enriched in iAMP21 B lymphoblastic leukemia. Cancer Genet. 2018, 226–227, 30–35. 10.1016/j.cancergen.2018.05.004. [DOI] [PubMed] [Google Scholar]
- Lee C.-H.; Su S.-Y.; Sittampalam K.; Chen P. C.-H.; Petersson F.; Kao Y.-C.; Carpenter T. O.; Hsieh T.-H.; Konishi E.; Tsai J.-W.; et al. Frequent overexpression of klotho in fusion-negative phosphaturic mesenchymal tumors with tumorigenic implications. J. Mod. Hum. Pathol. 2020, 33, 858–870. 10.1038/s41379-019-0416-4. [DOI] [PubMed] [Google Scholar]
- Kostas M.; Haugsten E. M.; Zhen Y.; Sørensen V.; Szybowska P.; Fiorito E.; Lorenz S.; Jones N.; de Souza G. A.; De Souza G. A.; Wiedlocha A. Protein tyrosine phosphatase receptor type G (PTPRG) controls fibroblast growth factor receptor (FGFR) 1 activity and influences sensitivity to FGFR kinase inhibitors. Mol. Cell. Proteomics 2018, 17, 850–870. 10.1074/mcp.RA117.000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels L. E.; Pedersen A. B.; Kristensen N. R.; Vilstrup H.; Stengaard-Pedersen K.; Dahlerup J. F. A positive Helicobacter pylori test is associated with low spondylarthritis incidence in a Danish historical cohort study. Rheumatol. Int. 2020, 40, 359–366. 10.1007/s00296-019-04487-2. [DOI] [PubMed] [Google Scholar]
- Gahmberg C. G.; Sekki A.; Kosunen T. U.; Holsti L. R.; Mäkelä O. Induction of aryl hydrocarbon hydroxylase activity and pulmonary carcinoma. Int. J. Cancer 1979, 23, 302–305. 10.1002/ijc.2910230305. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Yin N.; Gong L.; Tan Z.; Yin B.; Yang Y.; Luo C. Microduplication of 7q36. 3 encompassing the SHH long-range regulator (ZRS) in a patient with triphalangeal thumb-polysyndactyly syndrome and congenital heart disease. Mol. Med. Rep. 2017, 15, 793–797. 10.3892/mmr.2016.6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.