ABSTRACT
Many successful pathogens cause latent infections, remaining dormant within the host for years but retaining the ability to reactivate to cause symptomatic disease. The human opportunistic fungal pathogen Cryptococcus neoformans establishes latent pulmonary infections in immunocompetent individuals upon inhalation from the environment. These latent infections are frequently characterized by granulomas, or foci of chronic inflammation, that contain dormant and persistent cryptococcal cells. Immunosuppression can cause these granulomas to break down and release fungal cells that proliferate, disseminate, and eventually cause lethal cryptococcosis. This course of fungal latency and reactivation is understudied due to limited models, as chronic pulmonary granulomas do not typically form in mouse cryptococcal infections. A loss-of-function mutation in the Cryptococcus-specific MAR1 gene was previously described to alter cell surface remodeling in response to host signals. Here, we demonstrate that the mar1Δ mutant strain persists long term in a murine inhalation model of cryptococcosis, inducing a chronic pulmonary granulomatous response. We find that murine infections with the mar1Δ mutant strain are characterized by reduced fungal burden, likely due to the low growth rate of the mar1Δ mutant strain at physiological temperature, and an altered host immune response, likely due to inability of the mar1Δ mutant strain to properly employ virulence factors. We propose that this combination of features in the mar1Δ mutant strain collectively promotes the induction of a more chronic inflammatory response and enables long-term fungal persistence within these granulomatous regions.
KEYWORDS: Cryptococcus neoformans, granuloma, GM-CSF, cell wall, Titan cell, cell cycle defects, hypoxia
INTRODUCTION
Granulomas are complex foci of chronic inflammation that form in response to many stimuli, including microbial infections. A hallmark of indolent infections such as tuberculosis disease, granulomas are often characterized by multinucleated giant cells, epithelioid macrophages, and dormant and/or slowly proliferating microorganisms (1–4). The traditional understanding of the granuloma considered it to be a host-directed defense response that restricts microbial access to nutrients and oxygen, resulting in an immune microenvironment that limits microbial proliferation and prevents dissemination (3, 4). However, more recent work has demonstrated that granulomas are a dynamic component of the complex host-microbial “arms race.” In addition to serving as a host-directed protection mechanism, microorganisms can exploit the granuloma as a microniche for long-term survival in the host, where they remain shielded from immune detection until microbial reactivation (3–5). Although most work on granulomas has been conducted in the context of mycobacterial infections, many other infectious microorganisms induce granuloma formation in the human lung (6).
The fungal pathogen Cryptococcus neoformans is a significant cause of pneumonia and fatal meningoencephalitis in immunocompromised populations, resulting in more than 180,000 deaths annually (7). Primary infection occurs upon inhalation of environmental C. neoformans cells and/or spores, often early in life (4, 8). Immunocompetent hosts typically control the primary infection, with fungi remaining dormant but viable within lung-associated granulomas (9). As a result, immunocompetent hosts rarely manifest infection-related symptoms during this stage of latency (10). However, this chronic, latent infection can reactivate when a previously exposed individual becomes immunocompromised, especially in the setting of CD4+ T cell functional deficiency due to HIV infection, organ transplantation, or immunosenescence (2, 4, 7, 11). Breakdown of the cryptococcal granuloma structure results in fungal proliferation and systemic dissemination, including to the central nervous system.
The reactivation of fungal cells from granulomas is an understudied facet of cryptococcal disease, largely due to limited experimental models. Although the mouse is the most well-characterized and commonly used animal model to study Cryptococcus-host interactions, most murine models do not form chronic granulomas in response to clinically relevant isolates of C. neoformans (12). As a result, most murine experiments focus on primary cryptococcal infection and subsequent systemic dissemination. To explore cryptococcal latency and reactivation, investigators have adopted models of cryptococcosis in rabbits (13) and rats (14, 15) or employed less virulent C. neoformans strains in mice (12, 16, 17). Recently, a novel latent model was reported in which pulmonary granulomas form in mice in response to infection with the gcs1Δ mutant strain lacking the glucosylceramide synthase (18–21). Infection with gcs1Δ mutant cells results in chronic granulomas in the lungs containing persistent gcs1Δ mutant cells that can reactivate and disseminate upon immunosuppression (22).
We recently reported the identification and characterization of the C. neoformans MAR1 gene, which is required for cell surface remodeling in response to the host environment (23). The mar1Δ loss-of-function mutant strain displays altered cell surface features when exposed to host physiological conditions, including decreased cell wall glucans and mannans, increased exposure of cell wall chitin, and impaired polysaccharide capsule attachment (23). These cell surface alterations make the mar1Δ mutant strain more immunogenic than the wild-type (WT) strain, resulting in enhanced macrophage activation in vitro and hypovirulence in a murine inhalation model of cryptococcosis (23). We report here that inoculation with the mar1Δ mutant strain induces a pulmonary granulomatous response in mice, resulting in a chronic and persistent infection. Furthermore, we describe both host and fungal factors that contribute to this granulomatous response. From the host perspective, granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling, a known contributor to granuloma formation (17, 24–26), is similarly required for the formation of granulomatous regions of infection in this model. From the fungal perspective, murine infections with the mar1Δ mutant strain are characterized by reduced fungal burden and an altered host immune response. Specifically, during early stages of infection, the mar1Δ mutant strain induces WT strain-like pulmonary cytokine and leukocyte responses, despite its reduction in fungal burden. As infection proceeds, murine lungs infected with the WT strain exhibit progressive increases in pulmonary cytokines and leukocytes, while murine lungs infected with the mar1Δ mutant strain display more stable levels of pulmonary cytokines and leukocytes. In vitro phenotypic studies demonstrate that the mar1Δ mutant strain has slow growth at mammalian body temperature and is impaired in virulence factor employment, specifically Titan cell formation and polysaccharide capsule extension. We propose that the low growth rate of the mar1Δ mutant strain at mammalian body temperature drives its reduced fungal burden throughout the course of infection. Furthermore, we propose that the inability of the mar1Δ mutant strain to properly employ virulence factors drives the altered pulmonary immune response observed throughout the course of infection. These observations suggest that this combination of mar1Δ mutant strain phenotypes is responsible for the observed granulomatous response and the ability of the mar1Δ mutant strain to survive and persist within these granulomatous regions long term. Because MAR1 is a Cryptococcus-specific gene, this model represents a unique addition to the tools available to study chronic, and potentially latent, cryptococcal disease.
RESULTS
Inoculation with the mar1Δ mutant strain induces a granulomatous response in murine lungs.
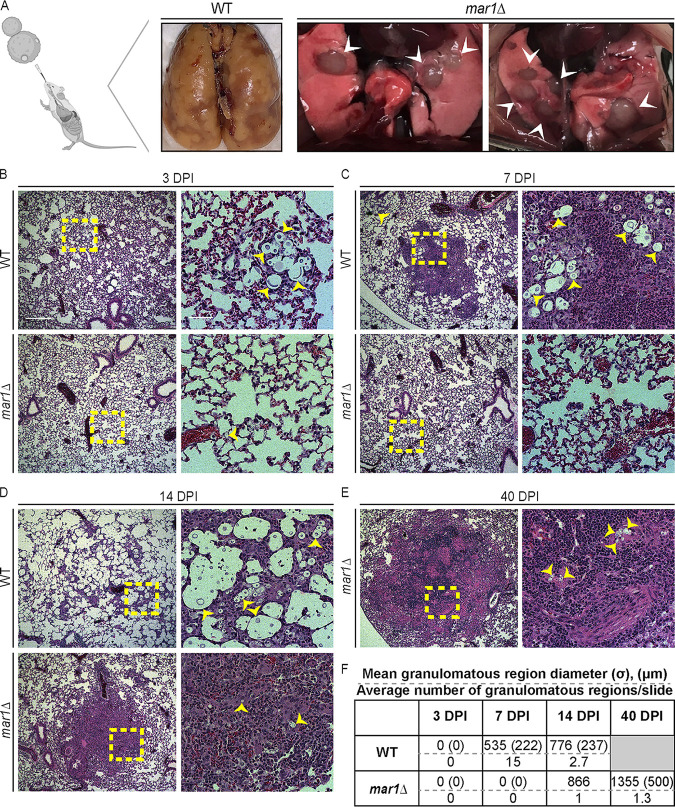
We previously observed that the mar1Δ mutant strain is hypovirulent compared to the wild-type (WT) strain in a murine inhalation model of cryptococcosis, despite having a more immunogenic cell surface than the WT strain (23). Highly immunogenic fungal strains often induce a hyperinflammatory response that is detrimental to the host, resulting in hypervirulence (27–29). We therefore explored in greater detail the mechanisms by which the highly immunogenic mar1Δ mutant strain simultaneously activates and is controlled by the host immune response. As an initial investigation into the interactions between the mar1Δ mutant strain and the host, we assessed the gross appearance of infected lungs from C57BL/6 mice from our previously reported murine inhalation infection experiment (23). In this experiment, C57BL/6 and BALB/c mice inoculated with the WT strain succumbed to cryptococcal infection between 10 and 30 days postinoculation (dpi) (23). In contrast, mice inoculated with the mar1Δ mutant strain displayed prolonged survival, with nearly 40% of the C57BL/6 mice and all the BALB/c mice surviving beyond 40 dpi (23). Gross histological analyses of C57BL/6 mice at the time of sacrifice revealed diffuse injury patterns throughout the entirety of WT strain-inoculated lungs (Fig. 1A). In contrast, mar1Δ mutant strain-inoculated lungs displayed large, well-circumscribed inflammatory foci surrounded by healthy-appearing lung tissue (Fig. 1A). These differences in the gross pathology suggested unique interactions between the mar1Δ mutant strain and the murine lung.
FIG 1.
Pulmonary granulomatous region formation in murine cryptococcal infections. (A) Dissected lungs of female C57BL/6 mice infected with 1 × 105 cells of the WT strain or mar1Δ mutant strain were harvested at clinical endpoints corresponding to imminent mortality. Gross organ examination revealed large, well-circumscribed inflammatory regions (white arrowheads) in mar1Δ mutant strain-inoculated lungs compared to diffuse lung injury in WT strain-inoculated lungs. (Created with BioRender.com). (B to E) Lungs of female C57BL/6 mice inoculated with 1 × 105 cells of the WT strain or the mar1Δ mutant strain sacrificed at the predetermined endpoints of 3 (B), 7 (C), 14 (D), and 40 (E) dpi were harvested for histopathological analyses. Hematoxylin and eosin staining was utilized to visualize microscopic lung pathology (fungal cells [yellow arrowheads], inset [yellow boxes]). The 5× scale bar (left) is 250 μm, and the 10× scale bar (right) is 50 μm. (F) Granulomatous region diameter (in micrometers) was measured using Fiji. Data are summarized from one slide from three mice per strain per time point. σ, standard deviation (micrometers). The gray box indicates no experimental subjects can be assessed at this time point.
To examine histopathological features of infected murine lungs at specific time points throughout the course of infection, we replicated the experimental approach used in Fig. 1A, inoculating C57BL/6 mice by inhalation with the WT strain or the mar1Δ mutant strain and harvesting infected lungs at predetermined time points. At 3 dpi, an early time point in infection during which all inoculated mice still appeared healthy, both WT strain-inoculated and mar1Δ mutant strain-inoculated lungs displayed a diffuse but minimal pattern of acute inflammation (Fig. 1B). The only notable difference observed at this time point was that fungal cells were much more readily observed in WT strain-inoculated lungs than in mar1Δ mutant strain-inoculated lungs (Fig. 1B). This observation was supported by our previously published experiments in which mar1Δ mutant strain-inoculated lungs had a significant reduction in fungal burden compared to WT strain-inoculated lungs as early as 1 and 4 dpi (23). Additionally, unlike mar1Δ mutant cells, many WT cells had undergone Titan cell transition with cell body enlargement and highly expanded capsules, as previously described (30).
By 7 dpi, WT strain-inoculated mice began to show signs of fungal disease, but mar1Δ mutant strain-inoculated mice still appeared clinically healthy. At this time point, WT strain-inoculated lungs displayed numerous small foci of inflammation (mean diameter, 535 μm) that failed to contain all fungal cells (Fig. 1C and F; see Fig. S1 in the supplemental material). These foci of inflammation also occasionally displayed histologic hallmarks of early granuloma formation, including the presence of epithelioid macrophages (1, 2, 4) (Fig. 1C; Fig. S1). This nascent granulomatous inflammatory response has been reported previously in the C57BL/6 mouse background infected with the C. neoformans serotype D strain 52D (17). In contrast, mar1Δ mutant strain-inoculated lungs had few visible fungal cells and displayed a more uniform pattern of inflammation throughout the lungs at 7 dpi (Fig. 1C). These observations demonstrate that distinct characteristics of the mar1Δ mutant strain pathology emerge early in infection.
At 14 dpi, a time point in infection in which WT strain-inoculated mice began to succumb to fungal infection and mar1Δ mutant strain-inoculated mice still appeared healthy, WT cells, many of which were Titan cells, proliferated throughout the lungs, with a scattered, unorganized inflammatory response with mixed cell infiltrates (Fig. 1D; Fig. S1). Additionally, in these WT strain-inoculated mice, the number of observed granulomatous regions was reduced from 7 dpi because most nascent granulomatous regions had broken down, concurrent with accelerating clinical symptoms and imminent mortality (Fig. 1D and F). In contrast, mar1Δ mutant strain-inoculated lungs began to form granulomatous regions of inflammation by 14 dpi. Specifically, foci of inflammation (mean diameter, 866 μm) contained rare, small (non-Titan) fungal cells. The regions of the lung immediately surrounding these foci of inflammation were mostly normal appearing, without fungal or inflammatory cells (Fig. 1D and F). Additionally, these inflammatory foci contained hallmarks of granulomas, such as epithelioid macrophages surrounded by lymphocytes (1, 2, 4) (Fig. 1D). In contrast to WT strain-inoculated mice, mar1Δ mutant strain-inoculated mice displayed few infection-related symptoms at this time point.
For mar1Δ mutant strain-inoculated mice that survived to 40 dpi, we observed well-circumscribed foci of more mature granulomatous inflammation (mean diameter, 1,355 μm) containing fungal cells, multinucleated giant cells, and palisading epithelioid macrophages (Fig. 1E and F; Fig. S1). Additionally, fungal cells were not observed in lung tissue outside these granulomatous regions. Collectively, these observations suggest differences in the immune response in the context of WT and mar1Δ mutant strain infections. WT strain-inoculated mice showed a consistently robust mixed inflammatory response and Titan cell response with nascent granuloma formation during early stages of infection (7 dpi) (Fig. 1B and D to F; Fig. S1). As described previously (12), this response was ineffective and was quickly overcome by fungal growth, resulting in fungal proliferation throughout the lungs (14 dpi) (Fig. 1B to D). In contrast, mar1Δ mutant strain-inoculated mice showed a minimal inflammatory response, absent Titan cell formation, and minimal granulomatous inflammatory response during early stages of infection (7 dpi), with a more well-circumscribed granulomatous response in mice that survived to later time points in infection (40 dpi) (Fig. 1B to F; Fig. S1). These mar1Δ mutant strain-induced granulomatous regions appeared to be sufficient to contain most fungal proliferation, which was visualized using Movat staining to identify fungal cells with alcian blue (Fig. S2).
We found that mar1Δ mutant strain-inoculated mice that survived to 40 dpi remained healthy appearing up to 100 dpi. When these mice were sacrificed and cultured for viable C. neoformans cells, all isolated fungal colonies displayed phenotypic similarity to the initially inoculated mar1Δ mutant strain, including dry colony morphology on alkaline pH and nourseothricin (NAT) resistance (23) (Fig. S3). These observations indicate that the mar1Δ mutant strain can persist within murine lung granulomatous regions long term without causing any symptoms or signs of disease, creating a chronic state of fungal infection.
Host GM-CSF signaling is required for the pulmonary granulomatous response.
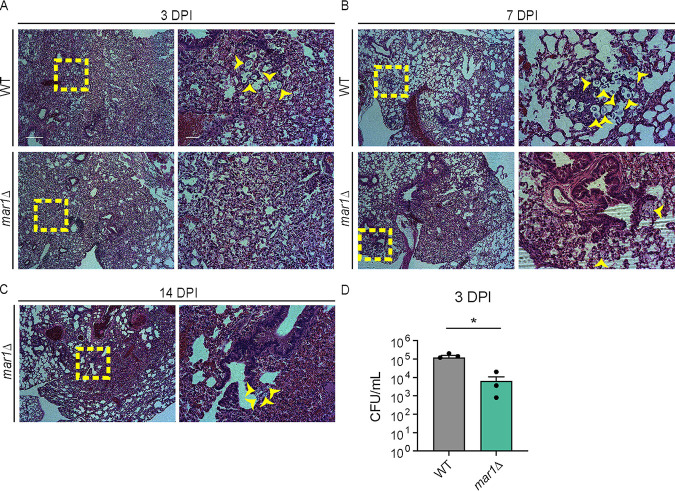
We first explored host features that contribute to the granulomatous response in this model. Granulocyte-macrophage colony-stimulating factor (GM-CSF), a cytokine required for the maturation of myeloid cells, is required for granuloma formation in both mycobacterial (24–26) and cryptococcal (17) infections. We therefore hypothesized that GM-CSF signaling would also be necessary for the formation of pulmonary granulomatous regions in our model. To test this hypothesis, we assessed the progression of infections with the WT strain and the mar1Δ mutant strain in the Csf2rb−/− mouse background, which is defective in GM-CSF signaling due to loss of the functional GM-CSF receptor. We inoculated Csf2rb−/− mice using the inhalation route and harvested lungs for analysis throughout infection. Overall, a similar pattern of inflammation was observed between mice inoculated with the WT strain and mice inoculated with the mar1Δ mutant strain. We observed that well-defined pulmonary granulomatous regions were absent in Csf2rb−/− mice infected with either strain at every tested time point (3, 7, and 14 dpi) (Fig. 2A to C; Fig. S4). Instead, the pattern of inflammation appeared poorly organized and diffuse throughout the entirety of the lungs infected with either fungal strain. Like the C57BL/6 infections, WT fungal cells were abundant throughout the lung, many of which appeared to be Titan cells, while mar1Δ mutant fungal cells were infrequently observed and did not appear to form Titan cells (Fig. 2A to C; Fig. S4). Pulmonary fungal burden assessed at 3 dpi confirmed that mar1Δ mutant strain-inoculated lungs had a significantly lower fungal burden, with a 10-fold reduction compared to WT strain-inoculated lungs, similar to what was observed previously in C57BL/6 infections (23) (Fig. 2D). These data demonstrate that GM-CSF signaling is required for the granulomatous response in both WT and mar1Δ mutant strain infections. However, because loss of GM-CSF signaling did not rescue the reduction of mar1Δ mutant strain fungal burden during early stages of infection, these data also suggest that GM-CSF signaling does not exclusively drive the impaired fitness of mar1Δ mutant cells in the murine lung.
FIG 2.
Contributions of GM-CSF signaling to the pulmonary granulomatous response. (A to C) The lungs of female (n = 2) (shown) and male (n = 2) (not shown) Csf2rb−/− mice inoculated with 1 × 105 cells of the WT strain or the mar1Δ mutant strain sacrificed at the predetermined time points of 3 (A), 7 (B), and 14 (C) dpi were harvested for histopathological analyses. Hematoxylin and eosin staining was utilized to visualize microscopic lung pathology (fungal cells [yellow arrowheads], inset [yellow boxes]). The 5× scale bar (left) is 250 μm, and the 10× scale bar (right) is 50 μm. (D) Pulmonary fungal burden of female (n = 2) and male (n = 2) Csf2rb−/− mice inoculated with 1 × 105 cells of the WT strain or the mar1Δ mutant strain sacrificed at 3 dpi was measured by quantitative cultures. Error bars represent the standard error of the mean (SEM). Statistical significance was determined using Student's t test (*, P < 0.05).
The mar1Δ mutant strain has a reduced fungal burden and induces an altered immune response in vivo.
We next explored fungal features that contribute to the granulomatous response in this model. We previously reported that in C57BL/6 mice, mar1Δ mutant strain-inoculated lungs displayed a significant decrease in fungal burden compared to WT strain-inoculated lungs, with a >10-fold decrease as early as 1 dpi (23). To determine whether chronic granulomatous inflammation was simply a product of a lower persisting fungal burden in mar1Δ mutant strain-inoculated lungs, we inoculated mice with 10-fold fewer cells (1 × 104 CFU) than were used in previous experiments (1 × 105 CFU). We observed similar histological patterns of lung inflammation with this lower inoculum as we did with the higher inoculum. Like our observations with the higher inoculum (Fig. 1; Fig. S1), murine lungs infected with the lower inoculum of the WT strain displayed robust fungal proliferation and a poorly organized inflammatory response by 14 dpi, demonstrating that a 10-fold reduction in the infecting dose was not sufficient to induce chronic granulomatous inflammation (Fig. S5A to D). Also similar to our prior experiments, by 21 dpi murine lungs infected with the lower inoculum of the mar1Δ mutant strain exhibited well-circumscribed regions of granulomatous inflammation with multinucleated giant cells and epithelioid macrophages, as well as rare fungal cells present only in these focal regions of inflammation (Fig. S5E to H). All WT strain-inoculated mice succumbed to fungal infection within 30 dpi with this lower inoculum, similar to the survival kinetics previously observed with the higher inoculum (23) (Fig. S5I). In contrast, mar1Δ mutant strain-inoculated mice displayed 90% survival at 60 dpi with this lower inoculum (Fig. S5I). Because this lower infecting dose resulted in similar histological features of granulomatous inflammation to the higher infecting dose, while maintaining a clearer distinction between uniformly lethal infections due to the WT strain and more indolent infections due to the mar1Δ mutant strain, we utilized this lower inoculum to examine the immune environment of the infected murine lung.
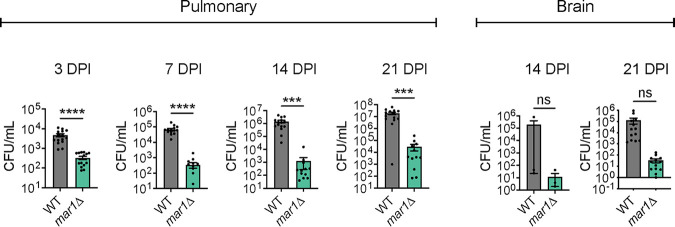
We infected mice with this lower inoculum and assessed fungal burden and pulmonary cytokine and leukocyte responses at time points relevant to the progression of the observed granulomatous regions. At all tested time points (3, 7, 14, and 21 dpi), mar1Δ mutant strain-inoculated lungs had a significantly reduced fungal burden compared to WT strain-inoculated lungs. Specifically, mar1Δ mutant strain-inoculated lungs had a 10-fold reduction in fungal burden at 3 dpi, a 100-fold reduction in fungal burden at 7 dpi, and a >500-fold reduction in fungal burden at 14 and 21 dpi compared to WT strain-inoculated lungs (Fig. 3). These observations support the reduced number of mar1Δ mutant cells observed at these same time points in our histopathology analyses (Fig. 1B to D). As a result of the drastic reduction in pulmonary fungal burden throughout infection, we observed that the mar1Δ mutant strain also had attenuated dissemination to the brain (Fig. 3). Together, these observations indicate that the mar1Δ mutant strain has reduced fungal burden in the murine lung and brain over a range of infecting inocula.
FIG 3.
Fungal burden throughout infection. Pulmonary fungal burden of female C57BL/6 mice (n = 15) inoculated with 1 × 104 cells of the WT strain or the mar1Δ mutant strain was measured by quantitative cultures throughout infection: 3, 7, 14, and 21 dpi. Brain fungal burden of female C57BL/6 mice (n = 15) inoculated with 1 × 104 cells of the WT strain or the mar1Δ mutant strain was measured by quantitative cultures at 14 and 21 dpi. Error bars represent the SEM. Statistical significance was determined using Student's t test (***, P < 0.001; ****, P < 0.0001; ns, not significant).
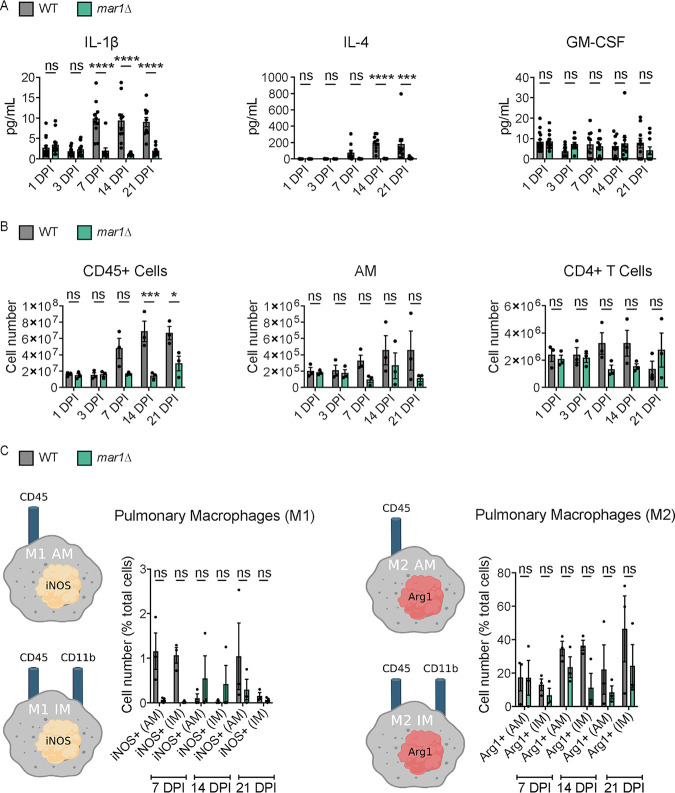
Based on the differences in fungal burden observed between WT strain-inoculated and mar1Δ mutant strain-inoculated lungs, we hypothesized that the immune microenvironments within the lungs would also differ significantly. At early time points in infection, 1 and 3 dpi, we observed similar pulmonary cytokine and leukocyte profiles within WT strain-inoculated lungs and mar1Δ mutant strain-inoculated lungs (Fig. 4A and B; Fig. S6 and S7). Despite the significant reduction in mar1Δ mutant fungal burden at these early time points, the mar1Δ mutant strain induced cytokine and leukocyte responses comparable to those of the WT strain. As infection progressed to 7, 14, and 21 dpi, we observed marked reductions in multiple cytokines (including interleukin-1 β [IL-1β] and IL-4) and leukocytes (including CD45+ cells and alveolar macrophages) in mar1Δ mutant strain-inoculated lungs compared to WT strain-inoculated lungs (Fig. 4A and B; Fig. S6 and S7). While WT strain-inoculated lungs generally displayed increases in most tested cytokines and leukocytes as infection progressed, mar1Δ mutant strain-inoculated lungs generally displayed stable levels of most tested cytokines and leukocytes throughout the course of infection. This observation was further supported by our histopathological observations made at the same time points demonstrating localized regions of inflammation in mar1Δ mutant strain-inoculated lungs (Fig. 1C and D). These observations demonstrate that by mid to late time points in infection, the overall cytokine and leukocyte responses are reduced in mar1Δ mutant strain-inoculated lungs compared to WT strain-inoculated lungs, likely due to the progressive reduction in fungal burden present in mar1Δ mutant strain-inoculated lungs.
FIG 4.
Pulmonary cytokine profile and leukocyte infiltrate associated with the granulomatous response. (A) Pulmonary cytokine responses of female C57BL/6 mice inoculated with 1 × 104 cells of the WT strain or the mar1Δ mutant strain were measured using the Bio-Plex protein array system throughout infection: 1 (n = 15), 3 (n = 15), 7 (n = 10), 14 (n = 10), and 21 (n = 10) dpi. Error bars represent SEM. Statistical significance between strains at each time point was determined using two-way ANOVA (***, P < 0.001; ****, P < 0.0001; ns, not significant). Only a subset of data is shown; refer to Fig. S6 in the supplemental material for full analysis. (B) Pulmonary leukocyte infiltrates of female C57BL/6 mice inoculated with 1 × 104 cells of the WT strain or the mar1Δ mutant strain were measured by flow cytometry throughout infection: 1, 3, 7, and 21 dpi. Data shown are the mean ± SEM of absolute cell numbers from three independent experiments (n = 3) performed using five mice per group per time point per experiment. Error bars represent SEM. Statistical significance between strains at each time point was determined using two-way ANOVA (*, P < 0.05; ***, P < 0.001; ns, not significant). Only a subset of data is shown; refer to Fig. S7 for full analysis. (C) Pulmonary macrophage activation of female C57BL/6 mice (n = 3) inoculated with 1 × 104 cells of the WT strain or the mar1Δ mutant strain was measured by flow cytometry throughout infection: 7, 14, and 21 dpi. Inducible nitrogen oxide synthase (iNOS) was used as a marker for M1 macrophages, and arginase 1 (Arg1) was used as a marker for M2 macrophages. The percentages of total iNOS+ cells and Arg1+ cells are shown. Error bars represent the SEM. Log transformation was used to normally distribute the data for statistical analysis. Statistical significance between strains at each time point was determined using two-way ANOVA. AM, alveolar macrophage (CD45+ CD11b−); IM, interstitial macrophage (CD45+ CD11b+). (Created with BioRender.com).
We further explored macrophage polarization at the same time points to determine whether the reduction in mar1Δ mutant strain fungal burden and the subsequent reduction in the pulmonary immune response were due to differences in macrophage activation (31). At each tested time point (7, 14, and 21 dpi), we observed that the mar1Δ mutant strain-inoculated lungs had a comparable number of or fewer classically activated (M1) and alternatively activated (M2) alveolar and interstitial macrophages compared to WT strain-inoculated lungs (Fig. 4C). These observations demonstrate that the mar1Δ mutant strain does not induce differential macrophage polarization causing the observed reductions in fungal burden and a more protective immune response. Collectively, these data suggest that the mar1Δ mutant strain-induced pulmonary granulomatous response appears to be a largely fungus-driven phenomenon. Despite reductions in fungal burden, the mar1Δ mutant strain induces a WT strain-like immune response early in infection. As infection matures and progresses, there is a relative decrease compared to WT strain-inoculated lungs in many cytokines and leukocytes infiltrating the mar1Δ mutant strain-inoculated lungs that corresponds with the progressive reduction in fungal burden and resulting fungal containment within granulomatous regions.
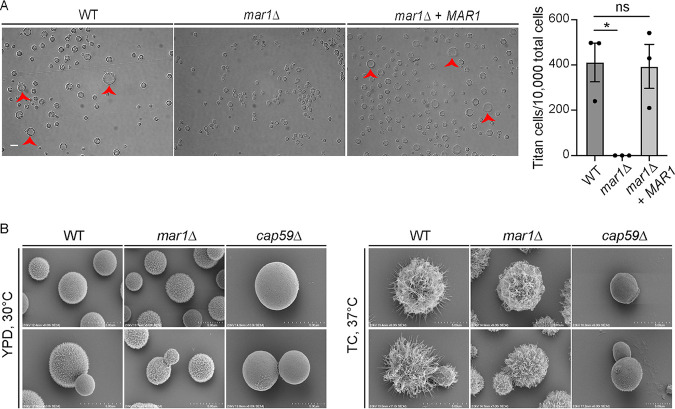
The mar1Δ mutant strain is attenuated in the employment of various virulence factors.
We next explored specific phenotypes of the mar1Δ mutant strain that may drive the observed granulomatous response. In both human and murine infections, a subset of cryptococcal cells form enlarged Titan cells, an important virulence factor that suppresses fungal phagocytosis by host macrophages (30, 32). Using an established in vitro titanization assay (33), we observed that the mar1Δ mutant strain was unable to form Titan cells (Fig. 5A). This observation supported our histopathology experiments, in which Titan cells were absent in mar1Δ mutant strain-inoculated lungs. Additionally, we previously reported that the mar1Δ mutant strain is impaired in the attachment of the polysaccharide capsule, assessed by India ink staining, a defect that was rescued by complementation with the MAR1 gene (23). We utilized high-resolution scanning electron microscopy (SEM) to more rigorously study the mar1Δ mutant strain capsule architecture. Under permissive growth conditions (yeast extract-peptone-dextrose [YPD] medium, 30°C), the capsule of the mar1Δ mutant strain was nearly indistinguishable from that of the WT strain, which contrasted starkly with the acapsular cap59Δ mutant strain (Fig. 5B). However, under capsule-inducing conditions (TC medium, 37°C), the mar1Δ mutant strain lacked the same degree of capsule fiber elongation observed in the WT strain, explaining the reduction in India ink exclusion previously reported for the mar1Δ mutant strain (23) (Fig. 5B). Collectively, these observations demonstrate that the mar1Δ mutant strain is defective in implementing some virulence factors, phenotypes which may contribute to the enhanced immunogenicity of this strain in vivo.
FIG 5.
Pathogenesis-relevant virulence factor phenotypes of the mar1Δ mutant strain. (A) Titan cell formation was induced in the WT strain, the mar1Δ mutant strain, and the mar1Δ + MAR1 complemented strain. Cells were pregrown in YNB medium at 30°C, and an OD600 of 0.001 was transferred to 10% HI-FBS in PBS incubated at 5% CO2 at 37°C for 96 h. Cells were imaged by DIC microscopy (Zeiss Axio Imager A1). Cell diameter was measured using Fiji, and cells with a diameter of >10 μm were considered Titan cells (red arrowheads). The number of Titan cells per 10,000 cells was calculated for each strain. A minimum of 400 cells were analyzed across three biological replicates (n = 3). Error bars represent the SEM. Statistical significance was determined using a one-way ANOVA (*, P < 0.05; ns, not significant). The 63× scale bar is 10 μm. (B) The WT strain, the mar1Δ mutant strain, and the cap59Δ mutant strain were incubated in YPD medium at 30°C and CO2-independent medium (TC) at 37°C until saturation. Samples were subsequently fixed, mounted, dehydrated, and sputter coated. Samples were imaged with a Hitachi S-4700 scanning electron microscope to visualize capsule organization and elaboration.
The mar1Δ mutant strain displays cell cycle defects that result in a slow-growth phenotype and hypoxia resistance.
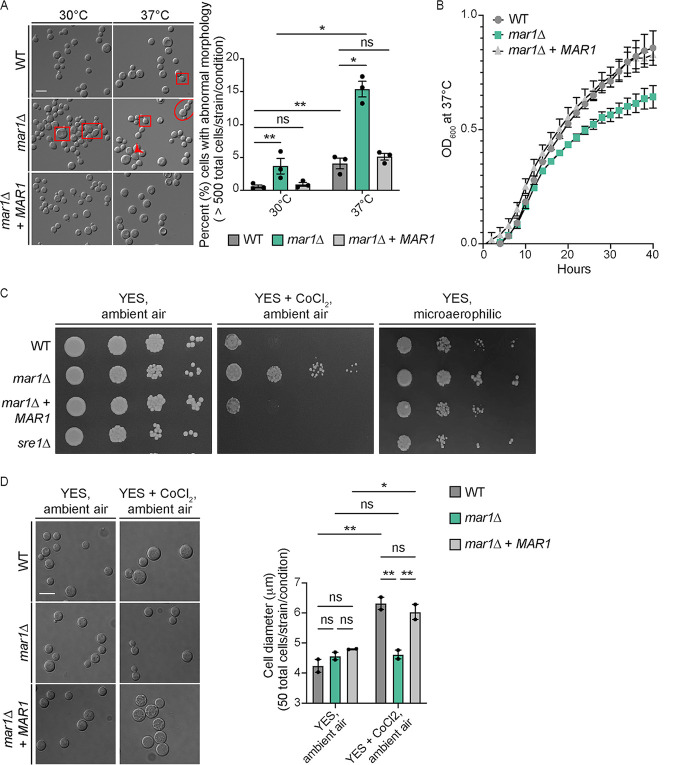
Both Titan cell formation (33, 34) and polysaccharide capsule elaboration (35–37) are mediated by the cell cycle. To explore cell cycle progression in the mar1Δ mutant strain background, we observed mar1Δ mutant cell morphology during the logarithmic growth phase. When incubated at the permissive temperature of 30°C, the mar1Δ mutant strain displayed an increased incidence of cytokinesis defects (such as elongated cells, cells with wide bud necks, and cells that fail to complete cytokinesis), compared to both the WT strain and the mar1Δ + MAR1 complemented strain (Fig. 6A). The frequency of these cytokinesis defects was significantly enhanced at the host physiological temperature of 37°C (Fig. 6A). We next determined the impact of these defects on the growth kinetics of the mar1Δ mutant strain. We observed that the mar1Δ mutant strain displayed a reduction in growth during logarithmic phase at 37°C, compared to both the WT strain and the mar1Δ + MAR1 complemented strain (Fig. 6B). These data demonstrate that the mar1Δ mutant strain has a slow-growth phenotype at mammalian body temperature, a phenotype that is likely driven in part by impaired cell cycle progression resulting in cytokinesis defects.
FIG 6.
Slow-growth phenotypes of the mar1Δ mutant strain. (A) Morphological defects were analyzed in the WT strain, the mar1Δ mutant strain, and the mar1Δ + MAR1 complemented strain through incubation in YPD medium at either 30°C or 37°C. Cells were imaged by DIC microscopy (Zeiss Axio Imager A1) and were subsequently visually inspected for morphological defects, such as elongated cells (red squares), wide bud necks (red arrowhead), and cytokinesis failure (red circle). The percentage of total cells displaying morphological defects was quantified for each strain at each temperature. A minimum of 500 cells were analyzed across three biological replicates (n = 3). Error bars represent the SEM. Log transformation was used to normally distribute the data for statistical analysis. Statistical significance between strains at each time point was determined using two-way ANOVA (*, P < 0.05; **, P < 0.01; ns, not significant). The 63× scale bar is 10 μm. (B) Growth of the WT strain, the mar1Δ mutant strain, and the mar1Δ + MAR1 complemented strain was assessed in YPD medium at 37°C. Growth was tracked for 40 h and was measured by absorbance at OD600. The figure summarizes data across three biological replicates (n = 3). Error bars represent the SEM. (C) Hypoxia resistance was assessed by growth on YES medium supplemented with CoCl2 (0.7 mM) and in a microaerophilic chamber. Serial dilutions of the WT strain, mar1Δ mutant strain, mar1Δ + MAR1 complemented strain, and sre1Δ mutant strain were spotted onto agar plates and incubated at 30°C. Results were compared to the same strains grown on YES medium under ambient air conditions. (D) The WT strain, mar1Δ mutant strain, and mar1Δ + MAR1 complemented strain were incubated on YES medium with and without CoCl2 (0.7 mM) at 30°C under ambient air conditions. After 72 h of growth, cells were isolated and imaged by DIC microscopy (Zeiss Axio Imager A1). Cell diameter (in micrometers) was measured using Fiji. The average cell diameter was quantified for each strain under each condition. A minimum of 50 cells were analyzed across two biological replicates (n = 2). Error bars represent the SEM. Statistical significance was determined using a two-way ANOVA (*, P < 0.05; **, P < 0.01; ns, not significant). The 63× scale bar is 10 μm.
Cell cycle regulation is also known to be related to fungal adaptation to hypoxia (38–40). Because C. neoformans is an obligate aerobe, WT fungal cells undergo G2 arrest in response to hypoxia (41, 42). We assessed the ability of the mar1Δ mutant strain to grow in an environment with reduced oxygen availability by observing growth in the presence of CoCl2 and in a microaerophilic chamber. In both cases, we observed that the mar1Δ mutant strain displayed enhanced growth compared to the WT strain and the mar1Δ + MAR1 complemented strain (Fig. 6C). In these assays, the CoCl2- and hypoxia-sensitive sre1Δ mutant strain was used as a control (43) (Fig. 6C). We then isolated the cells grown in the presence of CoCl2 to examine cell morphology. Consistent with previous literature that has reported that C. neoformans arrests in an unbudded G2 state in response to hypoxia (41, 42), we observed that the WT strain and the mar1Δ + MAR1 complemented strain displayed a significant increase in cell size when incubated in the presence of CoCl2 (Fig. 6D). However, the mar1Δ mutant strain did not display this increase in cell size in response to CoCl2 (Fig. 6D). Collectively, these observations demonstrate that the mar1Δ mutant strain has altered cell cycle dynamics and relative resistance to hypoxia, features that potentially contribute to changes in microbial persistence during infection.
DISCUSSION
Here, we report and characterize the host response to a chronic C. neoformans lung infection, one distinguished by sustained regions of granulomatous inflammation. Using the inhalation route of infection in C57BL/6 mice, we observed a granulomatous response in infections due to both the WT and mar1Δ mutant strains. However, the appearance, development, and maintenance of these granulomatous regions differed significantly. In WT infections, small, immature granulomatous regions formed early in infection. As infection progressed, these nascent granulomatous regions began to degenerate, leading to fungal proliferation throughout the lungs, fungal dissemination to the brain, and eventually murine death. This type of early, immature granulomatous response has been observed previously in murine infections with other C. neoformans WT strains (16, 17). In contrast, in mar1Δ mutant strain infections we observed chronic pulmonary granulomatous regions that developed over several weeks in the absence of overt clinical symptoms. These granulomatous regions differed from those induced by the WT strain because they appeared later in infection, were typically larger, and were more circumscribed. Furthermore, we observed that mar1Δ mutant strain-induced granulomatous regions were maintained throughout infection, from 14 dpi to as late as 100 dpi. Collectively, the mar1Δ mutant strain displays a unique interaction with the host characterized by granulomatous inflammation and chronic infection.
We confirmed the role of GM-CSF as a host driver of cryptococcal granuloma formation. In our model, GM-CSF was modestly elevated in mar1Δ mutant strain-inoculated lungs at 1, 3, and 14 dpi. This increased GM-CSF production may be a result of increased Dectin-1 activation by the mar1Δ mutant strain. We previously reported that the mar1Δ mutant strain is partially recognized by the pathogen recognition receptor Dectin-1, likely due its increased exposed surface β-glucan and chitin (23). Dectin-1 has been shown to be required for normal GM-CSF production in murine macrophages (44). Additionally, GM-CSF production is known to result in an increase in Dectin-1 expression by murine macrophages (44, 45). Here, we found that the granulomatous response in this model is dependent on GM-CSF signaling, as granulomatous regions were absent in Csf2rb−/− mouse background infections with either the WT or mar1Δ mutant strains. These results were expected because GM-CSF plays a significant role in controlling infections due to both Cryptococcus gattii and C. neoformans, as individuals with GM-CSF autoantibodies are unusually susceptible to cryptococcal infection (46–48). Furthermore, previous work in both mycobacterial (24–26) and cryptococcal (17) infections has demonstrated that GM-CSF signaling is required for granuloma formation, likely due to its requirement for macrophage recruitment to the lung during early stages of infection. Our model enables further exploration of the requirement of GM-CSF for granuloma maintenance. For example, future experiments could introduce GM-CSF antibodies into mar1Δ mutant strain-inoculated mice to determine whether GM-CSF is required for the maintenance of mar1Δ mutant strain-induced granulomatous regions. Furthermore, WT strain infections could be supplemented with exogenous GM-CSF to determine whether increased GM-CSF can help maintain WT strain-induced granulomatous regions.
We characterized multiple fungal features associated with chronic inflammation in this model. We observed that throughout the course of infection, mar1Δ mutant strain-inoculated lungs had a significantly lower fungal burden than WT strain-inoculated lungs. Despite this decrease in fungal burden, mar1Δ mutant strain-inoculated lungs induced WT strain-like pulmonary cytokine and leukocyte responses during early stages of infection. As infection progressed, WT strain-inoculated lungs continued to display marked increases in pulmonary cytokines and leukocytes, while mar1Δ mutant strain-inoculated mice displayed more stable levels of pulmonary cytokines and leukocytes. Previous work has implicated classically activated macrophage polarization in enhanced antifungal activity of macrophages (49–51). We found that mar1Δ mutant strain-inoculated lungs had a comparable number of or fewer (depending on the time point) classically activated (M1) and alternatively activated (M2) macrophages compared to WT strain-inoculated lungs, suggesting that differential polarization of macrophages did not contribute to the reduced fungal burden and associated immune response in mar1Δ mutant strain-inoculated lungs.
We also characterized multiple mar1Δ mutant strain phenotypes that may contribute to the granulomatous response and fungal persistence in this model. Titan cell formation is a well-characterized, Cryptococcus-specific persistence mechanism that suppresses fungal phagocytosis by host macrophages (30, 32). Results from an established in vitro titanization assay (33), as well as our histopathological observations, demonstrated that the mar1Δ mutant strain was impaired in the production of Titan cells. As a result, Titan cell transitions do not explain the persistence of the mar1Δ mutant strain in the murine lung. We also observed that the mar1Δ mutant strain was altered in the implementation of another important virulence factor, the polysaccharide capsule. Although the mar1Δ mutant strain had a similar basal level of capsule to the WT strain, it was unable to extend its capsule fibers to the level of the WT strain in response to capsule-inducing signals. In addition to the previously reported defects in the mar1Δ mutant strain cell wall (23), these virulence factor defects likely contribute to the enhanced immunogenicity of the mar1Δ mutant strain.
The expression of many virulence factors is known to be mediated by the cell cycle (37). Furthermore, recent work has proposed that C. neoformans undergoes a unique cell cycle in vivo, the “stress cell cycle,” that regulates the employment of various virulence factors (52). Titan cells are enlarged and polyploid cells that form in both human and mouse lungs during infection (30, 32). This polyploidization and concomitant cell body enlargement is negatively regulated by the transcription factor Usv101, which acts downstream of the cell cycle regulator Swi6 (33, 34). Furthermore, recent work has found that the Cln1 cyclin contributes to Titan cell formation by regulating DNA replication and cell division after G2 arrest in vivo (52). Similarly, capsule elongation is also regulated by the cell cycle, with the majority of capsule elongation occurring in the G1 phase of the cell cycle (36). The dysregulation of these cell-cycle-mediated virulence factors suggested that the mar1Δ mutant strain harbored cell cycle defects.
We indeed observed that the mar1Δ mutant strain displayed a marked increase in cytokinesis defects compared to the WT strain, particularly at 37°C, leading to a decreased growth rate. We furthermore observed that the mar1Δ mutant strain had enhanced growth in hypoxia, potentially because it did not arrest in an unbudded G2 state like the WT strain. In various cell types, including stem cells (53), tumor cells (54), bacteria (55), and fungi (39), a reduction in growth rate is required for survival in the presence of hypoxia. It is possible that its inherent decreased growth rate predisposed the mar1Δ mutant strain to growth in a hypoxic environment. The mammalian environment is known to limit oxygen availability to invading microorganisms, as a stressor used to contain microbial proliferation (38). This important resource is likely even further restricted within the pulmonary granuloma, which is known to have suboptimal oxygen levels in the context of mycobacterial infection (56). Recent work by the Alanio laboratory has demonstrated that cryptococcal dormancy can be induced by a combination of nutrient and oxygen deprivation (40, 57). Furthermore, the Dromer laboratory has found that dormant cryptococcal cells are characterized by reduced metabolic activity and delayed growth (39). With these observations in mind, it is possible that the slow-growth and hypoxia resistance phenotypes of the mar1Δ mutant strain favor its containment and persistence within granulomatous regions in the model described here. Further work will be required to determine whether these phenotypes are necessary and/or sufficient for fungal containment and persistence within granulomatous regions.
Collectively, these observations suggest that the mar1Δ mutant strain-induced granulomatous response is largely a fungus-driven phenomenon. Murine infections with the mar1Δ mutant strain are characterized by reduced fungal burden, likely due to the low growth rate of the mar1Δ mutant strain, and an altered host immune response, likely due to the virulence factor attenuation of the mar1Δ mutant strain. At early time points in infection, the immunogenic mar1Δ mutant strain induces a WT strain-like pulmonary immune response despite a marked reduction in fungal burden. As infection progresses, the WT strain proliferates rapidly throughout the lungs, resulting in a robust pulmonary immune response in WT strain-inoculated lungs that is associated with murine mortality, while the slow-growing mar1Δ mutant strain remains contained within granulomatous regions. Using these approaches, we have defined a detailed timeline of the murine granulomatous response, in both WT strain and mar1Δ mutant strain infections, and characterized multiple fungal factors that contribute to this response and long-term fungal persistence (see Fig. S8 in the supplemental material).
The Del Poeta laboratory has developed a well-characterized murine pulmonary granulomatous response model of cryptococcal disease using the gcs1Δ mutant strain. From the fungal perspective, the gcs1Δ mutant strain lacks the membrane sphingolipid glucosylceramide, making it an obligate intracellular pathogen and, as a result, completely avirulent in a murine inhalation model, the route of infection that most closely replicates the course of human infection (18, 19). It is noteworthy that the gcs1Δ and mar1Δ mutant strains were constructed in the same WT strain background, and as a result, these two mutant strains are comparable and could potentially be used together to explore the complex characteristics of granuloma formation. For example, both strains display cell cycle defects in the presence of physiological stress: the gcs1Δ mutant strain arrests at alkaline pH (18), and the mar1Δ mutant strain displays cytokinesis defects at 37°C. These similarities suggest that a slow-growth phenotype in the host environment may favor fungal containment within regions of granulomatous inflammation. Both strains likely also exist frequently within macrophages in vivo, as the gcs1Δ mutant strain is an obligate intracellular pathogen (18) and the mar1Δ mutant strain has phenotypes, like reduced Titan cell formation and polysaccharide capsule extension, that predispose fungal cells to phagocytosis by macrophages. Virulence potential is a notable difference between the strains. The gcs1Δ mutant strain is unable to initiate infection and disease via the inhalation route of infection (18), categorizing GCS1 as a disease initiation factor (58). In contrast, the mar1Δ mutant strain can establish infection and cause fatal disease in nearly half of the mice infected with 1 × 105 CFU (23), making MAR1 a disease progression factor (58). This may be related to the fact that GCS1 orthologs are found in many pathogenic fungi (18), while MAR1 appears to be a Cryptococcus-specific gene (23). These contrasting features suggest that the granulomatous response is a highly complex process that relies on the interplay between many host and fungal factors.
From the host perspective, gcs1Δ mutant strain-induced granuloma formation requires host sphingosine kinase 1–sphingosine 1-phosphate (SK1-S1P) signaling (20, 21). Most recently, the Del Poeta laboratory has applied this model to explore cryptococcal reactivation. Mimicking human disease, gcs1Δ mutant cells become reactivated from granulomas and disseminate upon immunosuppression with the multiple sclerosis therapeutic FTY720, which suppresses SK1-S1P signaling (22). This model has enabled the first murine reactivation studies of cryptococcal infection. Future work with the mar1Δ mutant strain-induced granulomatous response could similarly explore reactivation in the context of immunosuppression, to better understand the typical course of cryptococcal disease in humans. One of the populations most vulnerable to cryptococcal reactivation includes untreated HIV/AIDS patients (7). In our leukocyte infiltrate analyses, we observed that mar1Δ mutant strain-inoculated lungs had an enhanced CD4+ T cell response compared to WT strain-inoculated lungs at 21 dpi. This observation is particularly striking because mar1Δ mutant strain-inoculated lungs had a decreased or equivalent response compared to WT strain-inoculated lungs for all other leukocytes tested at this time point. CD4+ T cells are present in pulmonary granulomas of immunocompetent humans (59). Furthermore, CD4+ T cells border the periphery of pulmonary granulomas in HIV+ individuals receiving antiretroviral therapy, but they are lost in individuals with advanced HIV/AIDS, suggesting that CD4+ lymphocytes may be involved in granuloma maintenance (2, 59). By inducing CD4+ T cell depletion, and as a result mimicking the HIV/AIDS disease state, we could probe the role of CD4+ T cells in the maintenance of granulomatous regions in this model. Following immunosuppression, we can observe mar1Δ mutant strain-inoculated mice to track the breakdown of granulomatous regions and fungal proliferation with the same approaches used here. Considering both the fungal and host drivers of the granulomatous response outlined here, this model harbors features that make it unique from other existing cryptococcal infection models. Together with the conventional primary murine infection model, chronic murine infection models will advance our understanding of cryptococcal disease progression and define fungal features important for persistence in the human host.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All strains used in this study were generated in the C. neoformans var. grubii H99 (MATα) (13) background and are included in Table 1. Strains were maintained on yeast extract-peptone-dextrose (YPD) medium (1% yeast extract, 2% peptone, 2% dextrose, and 2% agar for solid medium). Unless otherwise indicated, strains were incubated at 30°C.
TABLE 1.
Fungal strains used in this study
Histology analyses.
The murine inhalation model of cryptococcosis was exclusively used in this study (60). For initial histological examination, C57BL/6 female mice were acquired from Charles River Laboratories. Mice were anesthetized with 2% isoflurane utilizing a rodent anesthesia device (Eagle Eye Anesthesia, Jacksonville, FL) and were infected via the intranasal route with 1 × 105 CFU of either the wild-type (WT) (H99) or the mar1Δ mutant (MAK1) strain. Mice were sacrificed at predetermined time points (3, 7, 14, and 40 days postinoculation [dpi]) by CO2 inhalation followed by an approved secondary method of euthanasia. Lungs were perfused with and stored in 10% neutral buffered formalin. Lungs were subsequently paraffin embedded, sectioned, mounted, and stained with hematoxylin and eosin and/or Movat by the Duke University School of Medicine Research Immunohistochemistry Shared Resource. Hematoxylin- and eosin-stained slides were used to identify granulomatous regions and characteristic features of granulomatous inflammation (e.g., multinucleated giant cells and epithelioid macrophages). Granulomatous regions of inflammation were quantified from one slide from three mice per strain per time point. Large cells with multiple nuclei were designated multinucleated giant cells. Mononuclear cells with an ovoid nucleus, dispersed chromatin, and eosinophilic cytoplasm were designated epithelioid macrophages. Movat-stained slides were used to more clearly visualize fungal cells within the lung tissue. All slides were reviewed in a blind manner by a pathologist (J.M.C.) with expertise in pulmonary pathology, including nonneoplastic and inflammatory lung disease.
To determine the role of GM-CSF signaling in the granulomatous response in this model, male and female Csf2rb−/− mice (The Jackson Laboratory, no. 005940) were infected via the intranasal route as described above with 1 × 105 CFU of either the WT (H99) or the mar1Δ mutant (MAK1) strain. Lungs were prepared as described above, with a few alterations. Mice were sacrificed at the predetermined time points of 3, 7, and 14 dpi by CO2 inhalation followed by an approved secondary method of euthanasia, and lungs were perfused with phosphate-buffered saline (PBS). The right lung was stored in 10% neutral buffered formalin for future histopathology preparation as described above, while the left lung was used for fungal burden quantification analyses as described below.
Mouse isolate recovery and phenotypic characterization.
C57BL/6 female mice acquired from Charles River Laboratories were infected via the intranasal route as described above with 1 × 105 CFU of either the WT (H99) or the mar1Δ mutant (MAK1) strain. At 61 dpi and 100 dpi, mice were sacrificed by CO2 inhalation followed by an approved secondary method of euthanasia. The lungs were removed and homogenized in 1 mL of sterile PBS as previously described (61). Single fungal colonies were subcultured onto YPD agar medium supplemented with chloramphenicol and subsequently frozen in separate wells of 96-well plates at −80°C. Isolated fungi were stamped onto YPD agar medium incubated at 30°C, YPD agar medium incubated at 37°C, YPD agar medium supplemented with nourseothricin (NAT) (100 μg/mL) incubated at 30°C, and YPD agar medium buffered (150 mM HEPES) to pH 8.15 and incubated at 30°C. All plates were imaged daily. Mouse isolates were determined to be mar1Δ mutant strain isolates based on growth on YPD+NAT medium and dry colony morphology on YPD medium at pH 8.15 (23). The original WT (H99) and mar1Δ mutant (MAK1) strains were included on each plate as controls.
Murine survival studies.
C57BL/6 female mice acquired from Charles River Laboratories were infected via the intranasal route as described above with 1 × 104 CFU of either the WT (H99) or the mar1Δ mutant (MAK1) strain. Mice were monitored over the course of 60 days and sacrificed based on clinical endpoints that predict mortality. Statistically significant differences between survival curves were determined by log rank test with Bonferroni correction (GraphPad Prism).
Fungal burden quantification.
Infected mice were euthanized at predetermined time points by CO2 inhalation followed by an approved secondary method of euthanasia, and lung tissues and/or brain tissues were excised. Tissues were then homogenized in 1 mL of sterile PBS as previously described (61), followed by culture of 10-fold dilutions of each homogenate on YPD agar medium supplemented with chloramphenicol. CFU were enumerated following incubation at 30°C for 48 h. Statistical significance was determined using Student's t test (GraphPad Software, San Diego, CA).
Pulmonary cytokine analyses.
C57BL/6 female mice acquired from Charles River Laboratories were infected via the intranasal route as described above with 1 × 104 CFU of either the WT (H99) or the mar1Δ mutant (MAK1) strain. Mice were euthanized at predetermined time points by CO2 inhalation followed by cervical dislocation. Cytokine levels within the lung homogenates of infected mice were analyzed using the Bio-Plex protein array system (Luminex-based technology; Bio-Rad Laboratories, Hercules, CA). Briefly, lung tissues were excised and homogenized in 1 mL ice-cold sterile PBS. An aliquot (50 μL) was taken to quantify the pulmonary fungal burden, and an antiprotease buffer solution (1 mL) containing PBS, protease inhibitors, and 0.05% Triton X-100 was added to the homogenate. Samples were then clarified by centrifugation (3,500 rpm) for 10 min. Supernatants from pulmonary homogenates were assayed for the presence of IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-17, KC (CXCL1), monocyte chemoattractant protein (MCP-1) (CCL2), macrophage inflammatory protein 1α (MIP-1α) (CCL3), MIP-1β (CCL4), RANTES (CCL5), eotaxin (CCL11), gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), granulocyte-macrophage colony-stimulating factor (GM-CSF), and granulocyte colony-stimulating factor (G-CSF) according to the manufacturer’s instructions. Statistical significance between strains at each time point was determined using two-way analysis of variance (ANOVA) and the Tukey-Kramer test (GraphPad Software, San Diego, CA).
Pulmonary leukocyte isolation.
C57BL/6 female mice acquired from Charles River Laboratories were infected via the intranasal route as described above with 1 × 104 CFU of either the WT (H99) or the mar1Δ mutant (MAK1) strain. Mice were euthanized at predetermined time points by CO2 inhalation followed by cervical dislocation. Lungs of infected mice were excised and processed as previously described (61). Briefly, lungs were digested enzymatically at 37°C for 30 min in 10 mL digestion buffer (RPMI 1640 and 1 mg/mL collagenase type IV [Sigma-Aldrich, St. Louis, MO]) with intermittent (every 10 min) stomacher homogenizations. The digested tissues were then successively filtered through sterile 70- and 40-μm-pore nylon filters (BD Biosciences, San Diego, CA) to enrich for leukocytes, and the cells were then washed three times with sterile Hanks’ balanced salt solution (HBSS). Erythrocytes were lysed by incubation in NH4Cl buffer (0.859% NH4Cl, 0.1% KHCO3, 0.0372% Na2EDTA [pH 7.4]; Sigma-Aldrich) for 3 min on ice followed by a 2-fold excess of sterile PBS.
Flow cytometry analyses.
Pulmonary leukocytes were isolated from mice infected as described above with 1 × 104 CFU of either the WT (H99) or the mar1Δ mutant (MAK1) strain. Standard methodology was employed for the direct immunofluorescence of pulmonary leukocytes (61, 62). Briefly, in 96-well U-bottom plates, 100 μL containing 1 × 106 cells in PBS were incubated with yellow Zombie viability dye (1:1,000 dilution; catalog no. 423104, Biolegend, San Diego, CA) for 15 min at room temperature followed by washing in fluorescence-activated cell sorter (FACS) buffer. Cells were then incubated with Fc block (1:500 dilution [catalog no. 553142, clone 2.4G2; BD Biosciences]) diluted in FACS buffer for 5 min to block nonspecific binding of antibodies to cellular Fc receptors. Cells were then incubated with fluorochrome-conjugated antibodies in various combinations to allow for multistaining for 30 min at 4°C. Cells were washed three times with FACS buffer and fixed in 200 μL of 2% ultrapure formaldehyde (Polysciences, Warrington, PA) diluted in FACS buffer (fixation buffer). Fluorescence minus one (FMO) controls or cells incubated with either FACS buffer alone or single fluorochrome-conjugated antibodies were used to determine positive staining and spillover/compensation calculations, and background fluorescence was determined with FlowJo v.10.8 software (FlowJo, LLC, Ashland, OR). Raw data were collected with a Cell Analyzer LSRII (BD Biosciences) using BD FACSDiva v8.0 software at the University of North Texas Health Sciences Center (UNTHSC) Flow Core, and compensation and data analyses were performed using FlowJo v.10.8 software. Cells were first gated for lymphocytes (SSC-A versus FSC-A) and singlets (FSC-H versus FSC-A). The singlets gate was further analyzed for the uptake of live/dead yellow stain to determine live versus dead cells. From live cells, cells were gated on CD45+ cell expression. For data analyses, 100,000 events (cells) were evaluated from a predominantly leukocyte population identified by back gating from CD45+ stained cells. Statistical significance between strains at each time point was determined using two-way analysis of variance (ANOVA) and the Tukey-Kramer test (GraphPad Software, San Diego, CA).
Macrophage activation analyses.
Intracellular staining of markers of macrophage activation was performed as described previously (62). Leukocytes isolated from mice infected as described above with 1 × 104 CFU of either the WT (H99) or the mar1Δ mutant (MAK1) strain were incubated with cell stimulation cocktail (eBioscience, catalog no. 00-4970-03) according to the manufacturer’s recommendation and incubated at 37°C in 5% CO2 in cRPMI for 2 h in a six-well plate. GolgiPlug (1:100 dilution, brefeldin A; catalog no. 51-2301KZ, BD Biosciences) was added according to the manufacturer’s recommendations, and the mixture was incubated for an additional 4 h (6 h total). Cells were washed with PBS and stained with yellow Zombie viability dye in PBS at room temperature in the dark for 15 min. Cells were then washed with FACS buffer and incubated with Fc block (BD Biosciences) diluted in FACS buffer for 5 min. For nitric oxide (inducible nitric oxide synthase [iNOS]) and arginase 1 (Arg1) production in macrophages, cells were stained for surface markers CD45, CD11b, CD64, F4/80, and CD24, and incubated at 4°C for 30 min. Cells were then washed and fixed with 2% ultrapure formaldehyde (Polysciences, Warrington, PA) for 20 min. Subsequently, cells were washed with 0.1% saponin buffer and stained with antibodies for iNOS and Arg1 for 30 min at 4°C. Finally, cells were washed with saponin buffer and fixed with 2% ultrapure formaldehyde. Samples were processed using a Cell Analyzer LSRII (BD Biosciences) using BD FACSDiva v.8.0 software at the UNTHSC Flow Core, and 100,000 events were collected for analysis using FlowJo v.10.8 software. Statistical significance between strains at each time point was determined using two-way analysis of variance (ANOVA) and the Tukey-Kramer test (GraphPad Software, San Diego, CA).
Titan cell assay and quantification.
A previously described in vitro titanization assay was used here (33). In brief, the WT (H99), mar1Δ mutant (MAK1), and mar1Δ + MAR1 (MAK11) strains were incubated for 18 h at 30°C at 150 rpm in 5 mL yeast nitrogen base (YNB) without amino acids plus 2% glucose prepared according to the manufacturer’s instructions. Cultures were washed six times with PBS. An optical density at 600 nm (OD600) of 0.001 for each strain was transferred to 5 mL 10% heat-inactivated fetal bovine serum (HI-FBS) in PBS and incubated at 37°C with 5% CO2 for 96 h. Cells were imaged by differential interference contrast (DIC) microscopy using a Zeiss Axio Imager A1 microscope equipped with an Axio‐Cam MRm digital camera. Cell diameter was measured using the ImageJ software (Fiji), and cells with a diameter of >10 μm were considered Titan cells. A minimum of 400 cells were analyzed across three biological replicates for each fungal strain. Statistical significance was determined using one-way analysis of variance (ANOVA) and the Tukey-Kramer test (GraphPad Software, San Diego, CA).
SEM polysaccharide capsule visualization.
The WT (H99), the mar1Δ mutant (MAK1), and the cap59Δ mutant (cap59) strains were incubated in YPD medium at 30°C and CO2-independent medium (Gibco) at 37°C until saturation. Samples were fixed with 2.5% glutaraldehyde for 1 h at room temperature and were subsequently washed 3 times with PBS. Each sample was mounted onto 12-mm poly-l-lysine-coated coverslips (Neuvitro Corporation) and subsequently dehydrated by immersing the coverslips in ethanol (30% for 5 min, 50% for 5 min, 70% for 5 min, 95% for 10 min, 100% for 10 min, and 100% for 10 min). Samples were then critical point dried with a Tousimis 931 critical point dryer (Rockville, Maryland) and coated with gold-palladium using a Cressington 108 sputter-coater (Watford, United Kingdom). Coverslips containing the prepared samples were mounted and imaged on a Hitachi S-4700 scanning electron microscope (Tokyo, Japan).
Cellular morphology defect quantification.
The WT (H99), the mar1Δ mutant (MAK1), and the mar1Δ + MAR1 (MAK11) strains were incubated for 18 h in YPD medium at 30°C with shaking at 150 rpm. An OD600 of approximately 0.2 for each strain was transferred to 5 mL of fresh YPD medium and subsequently incubated at either 30°C or 37°C for 18 h with shaking at 150 rpm. Cells were then pelleted, washed with PBS, and imaged by DIC microscopy. DIC images were captured using a Zeiss Axio Imager A1 microscope equipped with an Axio‐Cam MRm digital camera. A minimum of 500 cells were analyzed across three biological replicates for each strain using the ImageJ software (Fiji). Statistical significance was determined using two-way analysis of variance (ANOVA) and the Tukey-Kramer test (GraphPad Software, San Diego, CA).
Growth curve analysis.
The WT (H99), the mar1Δ mutant (MAK1), and the mar1Δ + MAR1 (MAK11) strains were incubated for 18 h in YPD medium at 30°C with 150-rpm shaking. Cultures were normalized to an OD600 of 0.01 in 200 μL of fresh YPD medium and added to wells of a 96-well plate. Growth was then measured at an absorbance of 595 nm every 10 min for 40 h with shaking between readings and incubation at 37°C. Control wells containing YPD medium alone were also included to eliminate any background absorbance.
Hypoxia resistance analyses.
The WT (H99), the mar1Δ mutant (MAK1), mar1Δ + MAR1 (MAK11), and sre1Δ mutant (HEB6) strains were incubated in YPD medium at 30°C until the mid-logarithmic growth phase. Strains were washed once in PBS, normalized to an OD600 of 0.6 in 1 mL PBS, and serially diluted onto YES medium (0.5% [wt/vol] yeast extract, 2% glucose, and 225 μg/mL uracil, adenine, leucine, histidine, and lysine) agar plates with or without cobalt chloride (0.7 mM) (43). Microaerophilic conditions were generated using a sealed chamber (BD GasPak) and two activated GasPak EZ Campy container system sachets (43). Plates were placed in the chamber (microaerophilic) or outside the chamber (ambient air), incubated at 30°C, and imaged daily for 96 h. Cells incubated under ambient air conditions on YES medium and YES medium with cobalt chloride were imaged by DIC microscopy using a Zeiss Axio Imager A1 microscope equipped with an Axio‐Cam MRm digital camera. Cell diameter was measured using the ImageJ software (Fiji). A minimum of 50 cells were analyzed across two biological replicates for each fungal strain. Statistical significance was determined using two-way analysis of variance (ANOVA) and the Tukey-Kramer test (GraphPad Software, San Diego, CA).
Ethical use of animals.
All animal experiments in this article were approved by the University of Texas at San Antonio Institutional Animal Care and Use Committee (IACUC) (protocol no. MU021), the Texas Christian University, the University of North Texas Health Sciences Center (UNTHSC) IACUC (protocol no. 1920-9), and the Duke University IACUC (protocol no. A102-20-05). Mice were handled according to IACUC guidelines.
Data availability.
All fungal strains and reagents are available upon request.
ACKNOWLEDGMENTS
We thank the Duke University School of Medicine for the use of the Research Immunohistochemistry Laboratory Shared Resource, which prepared all histopathology samples. We thank Joseph Heitman and Anna Floyd-Averette for providing the Csf2rb−/− mice. Flow cytometry was performed in the Flow Cytometry and Laser Capture Microdissection Core Facility at The University of North Texas Health Science Center (UNTHSC) (which is supported by National Institutes of Health award ISIORR018999-01A1) and the Cell Analysis Core at The University of Texas at San Antonio. Scanning electron microscopy was performed at the Chapel Hill Analytical and Nanofabrication Laboratory (CHANL), a member of the North Carolina Research Triangle Nanotechnology Network (RTNN), which is supported by National Science Foundation Grant ECCS-1542015, as part of the National Nanotechnology Coordinated Infrastructure (NNCI).
This work was supported by R01 AI074677 from the National Institutes of Health to J.A.A. and F.L.W.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
J. Andrew Alspaugh, Email: andrew.alspaugh@duke.edu.
Mairi C. Noverr, Tulane School of Medicine
REFERENCES
- 1.Adams DO. 1976. The granulomatous inflammatory response. A review. Am J Pathol 84:164–191. [PMC free article] [PubMed] [Google Scholar]
- 2.Shibuya K, Hirata A, Omuta J, Sugamata M, Katori S, Saito N, Murata N, Morita A, Takahashi K, Hasegawa C, Mitsuda A, Hatori T, Nonaka H. 2005. Granuloma and cryptococcosis. J Infect Chemother 11:115–122. 10.1007/s10156-005-0387-x. [DOI] [PubMed] [Google Scholar]
- 3.Ramakrishnan L. 2012. Revisiting the role of the granuloma in tuberculosis. Nat Rev Immunol 12:352–366. 10.1038/nri3211. [DOI] [PubMed] [Google Scholar]
- 4.Ristow LC, Davis JM. 2021. The granuloma in cryptococcal disease. PLoS Pathog 17:e1009342. 10.1371/journal.ppat.1009342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oehlers SH, Cronan MR, Scott NR, Thomas MI, Okuda KS, Walton EM, Beerman RW, Crosier PS, Tobin DM. 2015. Interception of host angiogenic signalling limits mycobacterial growth. Nature 517:612–615. 10.1038/nature13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zumla A, James DG. 1996. Granulomatous infections: etiology and classification. Clin Infect Dis 23:146–158. 10.1093/clinids/23.1.146. [DOI] [PubMed] [Google Scholar]
- 7.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 17:873–881. 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldman DL, Khine H, Abadi J, Lindenberg DJ, Priofski LA, Niang R, Casadevall A. 2001. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics 107:E66. 10.1542/peds.107.5.e66. [DOI] [PubMed] [Google Scholar]
- 9.McDonnell JM, Hutchins GM. 1985. Pulmonary cryptococcosis. Hum Pathol 16:121–128. 10.1016/s0046-8177(85)80060-5. [DOI] [PubMed] [Google Scholar]
- 10.Warr W, Bates JH, Stone A. 1968. The spectrum of pulmonary cryptococcosis. Ann Intern Med 69:1109–1116. 10.7326/0003-4819-69-6-1109. [DOI] [PubMed] [Google Scholar]
- 11.Grebenciucova E, Reder AT, Bernard JT. 2016. Immunologic mechanisms of fingolimod and the role of immunosenescence in the risk of cryptococcal infection: a case report and review of literature. Mult Scler Relat Disord 9:158–162. 10.1016/j.msard.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 12.Normile TG, Bryan AM, Poeta MD. 2020. Animal models of Cryptococcus neoformans in identifying immune parameters associated with primary infection and reactivation of latent infection. Front Immunol 11:581750. 10.3389/fimmu.2020.581750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perfect JR, Lang SD, Durack DT. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol 101:177–194. [PMC free article] [PubMed] [Google Scholar]
- 14.Goldman D, Cho Y, Zhao M, Casadevall A, Lee SC. 1996. Expression of inducible nitric oxide synthase in rat pulmonary Cryptococcus neoformans granulomas. Am J Pathol 148:1275–1282. [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi M, Ito M, Sano K, Koyama M. 2001. Granulomatous and cytokine responses to pulmonary Cryptococcus neoformans in two strains of rats. Mycopathologia 151:121–130. 10.1023/A:1017900604050. [DOI] [PubMed] [Google Scholar]
- 16.Feldmesser M, Casadevall A, Kress Y, Spira G, Orlofsky A. 1997. Eosinophil-Cryptococcus neoformans interactions in vivo and in vitro. Infect Immun 65:1899–1907. 10.1128/iai.65.5.1899-1907.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen GH, Olszewski MA, McDonald RA, Wells JC, Paine R, Huffnagle GB, Toews GB. 2007. Role of granulocyte macrophage colony-stimulating factor in host defense against pulmonary Cryptococcus neoformans infection during murine allergic bronchopulmonary mycosis. Am J Pathol 170:1028–1040. 10.2353/ajpath.2007.060595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rittershaus PC, Kechichian TB, Allegood JC, Merrill AH, Hennig M, Luberto C, Poeta MD. 2006. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Invest 116:1651–1659. 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kechichian TB, Shea J, Poeta MD. 2007. Depletion of alveolar macrophages decreases the dissemination of a glucosylceramide-deficient mutant of Cryptococcus neoformans in immunodeficient mice. Infect Immun 75:4792–4798. 10.1128/IAI.00587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McQuiston T, Luberto C, Poeta MD. 2010. Role of host sphingosine kinase 1 in the lung response against cryptococcosis. Infect Immun 78:2342–2352. 10.1128/IAI.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farnoud AM, Bryan AM, Kechichian T, Luberto C, Poeta MD. 2015. The granuloma response controlling cryptococcosis in mice depends on the sphingosine kinase 1–sphingosine 1-phosphate pathway. Infect Immun 83:2705–2713. 10.1128/IAI.00056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bryan AM, You JK, McQuiston T, Lazzarini C, Qiu Z, Sheridan B, Nuesslein-Hildesheim B, Poeta MD. 2020. FTY720 reactivates cryptococcal granulomas in mice through S1P receptor 3 on macrophages. J Clin Invest 130:4546–4560. 10.1172/JCI136068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esher SK, Ost KS, Kohlbrenner MA, Pianalto KM, Telzrow CL, Campuzano A, Nichols CB, Munro C, Wormley FL, Alspaugh JA. 2018. Defects in intracellular trafficking of fungal cell wall synthases lead to aberrant host immune recognition. PLoS Pathog 14:e1007126. 10.1371/journal.ppat.1007126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Juarrero M, Hattle JM, Izzo A, Junqueira-Kipnis AP, Shim TS, Trapnell BC, Cooper AM, Orme IM. 2005. Disruption of granulocyte macrophage-colony stimulating factor production in the lungs severely affects the ability of mice to control Mycobacterium tuberculosis infection. J Leukoc Biol 77:914–922. 10.1189/jlb.1204723. [DOI] [PubMed] [Google Scholar]
- 25.Szeliga J, Daniel DS, Yang CH, Sever-Chroneos Z, Jagannath C, Chroneos ZC. 2008. Granulocyte-macrophage colony stimulating factor-mediated innate responses in tuberculosis. Tuberculosis (Edinb) 88:7–20. 10.1016/j.tube.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benmerzoug S, Marinho FV, Rose S, Mackowiak C, Gosset D, Sedda D, Poisson E, Uyttenhove C, Van Snick J, Jacobs M, Garcia I, Ryffel B, Quesniaux VFJ. 2018. GM-CSF targeted immunomodulation affects host response to M. tuberculosis infection. Sci Rep 8:8652. 10.1038/s41598-018-26984-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Bader N, Vanier G, Liu H, Gravelat FN, Urb M, Hoareau CMQ, Campoli P, Chabot J, Filler SG, Sheppard DC. 2010. Role of trehalose biosynthesis in Aspergillus fumigatus development, stress response, and virulence. Infect Immun 78:3007–3018. 10.1128/IAI.00813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ost KS, O'Meara TR, Huda N, Esher SK, Alspaugh JA. 2015. The Cryptococcus neoformans alkaline response pathway: identification of a novel Rim pathway activator. PLoS Genet 11:e1005159. 10.1371/journal.pgen.1005159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zacharias CA, Sheppard DC. 2019. The role of Aspergillus fumigatus polysaccharides in host–pathogen interactions. Curr Opin Microbiol 52:20–26. 10.1016/j.mib.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Zaragoza O, García-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodríguez-Tudela JL, Casadevall A. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog 6:e1000945. 10.1371/journal.ppat.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McQuiston TJ, Williamson PR. 2012. Paradoxical roles of alveolar macrophages in the host response to Cryptococcus neoformans. J Infect Chemother 18:1–9. 10.1007/s10156-011-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cruickshank JG, Cavill R, Jelbert M. 1973. Cryptococcus neoformans of unusual morphology. Appl Microbiol 25:309–312. 10.1128/am.25.2.309-312.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dambuza IM, Drake T, Chapuis A, Zhou X, Correia J, Taylor-Smith L, LeGrave N, Rasmussen T, Fisher MC, Bicanic T, Harrison TS, Jaspars M, May RC, Brown GD, Yuecel R, MacCallum DM, Ballou ER. 2018. The Cryptococcus neoformans Titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Pathog 14:e1006978. 10.1371/journal.ppat.1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hommel B, Mukaremera L, Cordero RJB, Coelho C, Desjardins CA, Sturny-Leclère A, Janbon G, Perfect JR, Fraser JA, Casadevall A, Cuomo CA, Dromer F, Nielsen K, Alanio A. 2018. Titan cells formation in Cryptococcus neoformans is finely tuned by environmental conditions and modulated by positive and negative genetic regulators. PLoS Pathog 14:e1006982. 10.1371/journal.ppat.1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaragoza O, Telzak A, Bryan RA, Dadachova E, Casadevall A. 2006. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol Microbiol 59:67–83. 10.1111/j.1365-2958.2005.04928.x. [DOI] [PubMed] [Google Scholar]
- 36.García-Rodas R, Cordero RJB, Trevijano-Contador N, Janbon G, Moyrand F, Casadevall A, Zaragoza O. 2014. Capsule growth in Cryptococcus neoformans is coordinated with cell cycle progression. mBio 5:e00945-14. 10.1128/mBio.00945-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelliher CM, Leman AR, Sierra CS, Haase SB. 2016. Investigating conservation of the cell-cycle-regulated transcriptional program in the fungal pathogen, Cryptococcus neoformans. PLoS Genet 12:e1006453. 10.1371/journal.pgen.1006453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grahl N, Shepardson KM, Chung D, Cramer RA. 2012. Hypoxia and fungal pathogenesis: to air or not to air? Eukaryot Cell 11:560–570. 10.1128/EC.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alanio A, Vernel-Pauillac F, Sturny-Leclère A, Dromer F. 2015. Cryptococcus neoformans host adaptation: toward biological evidence of dormancy. mBio 6:e02580-14. 10.1128/mBio.02580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hommel B, Sturny-Leclère A, Volant S, Veluppillai N, Duchateau M, Yu CH, Hourdel V, Varet H, Matondo M, Perfect JR, Casadevall A, Dromer F, Alanio A. 2019. Cryptococcus neoformans resists to drastic conditions by switching to viable but non-culturable cell phenotype. PLoS Pathog 15:e1007945. 10.1371/journal.ppat.1007945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohkusu M, Raclavsky V, Takeo K. 2001. Deficit in oxygen causes G2 budding and unbudded G2 arrest in Cryptococcus neoformans. FEMS Microbiol Lett 204:29–32. 10.1111/j.1574-6968.2001.tb10857.x. [DOI] [PubMed] [Google Scholar]
- 42.Ohkusu M, Raclavsky V, Takeo K. 2004. Induced synchrony in Cryptococcus neoformans after release from G2-arrest. Antonie Van Leeuwenhoek 85:37–44. 10.1023/B:ANTO.0000020272.19569.15. [DOI] [PubMed] [Google Scholar]
- 43.Brown HE, Telzrow CL, Saelens JW, Fernandes L, Alspaugh JA. 2020. Sterol-response pathways mediate alkaline survival in diverse fungi. mBio 11:e00719-20. 10.1128/mBio.00719-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walachowski S, Tabouret G, Fabre M, Foucras G. 2017. Molecular analysis of a short-term model of β-glucans-trained immunity highlights the accessory contribution of GM-CSF in priming mouse macrophages response. Front Immunol 8:1089. 10.3389/fimmu.2017.01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willment JA, Lin H-H, Reid DM, Taylor PR, Williams DL, Wong SYC, Gordon S, Brown GD. 2003. Dectin-1 expression and function are enhanced on alternatively activated and GM-CSF-treated macrophages and are negatively regulated by IL-10, dexamethasone, and lipopolysaccharide. J Immunol 171:4569–4573. 10.4049/jimmunol.171.9.4569. [DOI] [PubMed] [Google Scholar]
- 46.Rosen LB, Freeman AF, Yang LM, Jutivorakool K, Olivier KN, Angkasekwinai N, Suputtamongkol Y, Bennett JE, Pyrgos V, Williamson PR, Ding L, Holland SM, Browne SK. 2013. Anti-GM-CSF autoantibodies in patients with cryptococcal meningitis. J Immunol 190:3959–3966. 10.4049/jimmunol.1202526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saijo T, Chen J, Chen SCA, Rosen LB, Yi J, Sorrell TC, Bennett JE, Holland SM, Browne SK, Kwon-Chung KJ. 2014. Anti-granulocyte-macrophage colony-stimulating factor autoantibodies are a risk factor for central nervous system infection by Cryptococcus gattii in otherwise immunocompetent patients. mBio 5:e00912-14. 10.1128/mBio.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viola GM, Malek AE, Rosen LB, DiNardo AR, Nishiguchi T, Okhuysen PC, Holland SM, Kontoyiannis DP. 2021. Disseminated cryptococcosis and anti-granulocyte-macrophage colony-stimulating factor autoantibodies: an underappreciated association. Mycoses 64:576–582. 10.1111/myc.13247. [DOI] [PubMed] [Google Scholar]
- 49.Arora S, Hernandez Y, Erb-Downward JR, McDonald RA, Toews GB, Huffnagle GB. 2005. Role of IFN-gamma in regulating T2 immunity and the development of alternatively activated macrophages during allergic bronchopulmonary mycosis. J Immunol 174:6346–6356. 10.4049/jimmunol.174.10.6346. [DOI] [PubMed] [Google Scholar]
- 50.Hardison SE, Ravi S, Wozniak KL, Young ML, Olszewski MA, Wormley FL. 2010. Pulmonary infection with an interferon-gamma-producing Cryptococcus neoformans strain results in classical macrophage activation and protection. Am J Pathol 176:774–785. 10.2353/ajpath.2010.090634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leopold Wager CM, Wormley FL. 2014. Classical versus alternative macrophage activation: the Ying and the Yang in host defense against pulmonary fungal infections. Mucosal Immunol 7:1023–1035. 10.1038/mi.2014.65. [DOI] [PubMed] [Google Scholar]
- 52.Altamirano S, Li Z, Fu MS, Ding M, Fulton SR, Yoder JM, Tran V, Nielsen K. 2021. The cyclin Cln1 controls polyploid Titan cell formation following a stress-induced G2 arrest in Cryptococcus. mBio 12:e02509-21. 10.1128/mBio.02509-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Latil M, Rocheteau P, Châtre L, Sanulli S, Mémet S, Ricchetti M, Tajbakhsh S, Chrétien F. 2012. Skeletal muscle stem cells adopt a dormant cell state post mortem and retain regenerative capacity. Nat Commun 3:903. 10.1038/ncomms1890. [DOI] [PubMed] [Google Scholar]
- 54.Kinoshita M, Johnson DL, Shatney CH, Lee YL, Mochizuki H. 2001. Cancer cells surviving hypoxia obtain hypoxia resistance and maintain anti-apoptotic potential under reoxygenation. Int J Cancer 91:322–326. . [DOI] [PubMed] [Google Scholar]
- 55.Bagchi G, Das TK, Tyagi JS. 2002. Molecular analysis of the dormancy response in Mycobacterium smegmatis: expression analysis of genes encoding the DevR-DevS two-component system, Rv3134c and chaperone alpha-crystallin homologues. FEMS Microbiol Lett 211:231–237. 10.1111/j.1574-6968.2002.tb11230.x. [DOI] [PubMed] [Google Scholar]
- 56.Qualls JE, Murray PJ. 2016. Immunometabolism within the tuberculosis granuloma: amino acids, hypoxia, and cellular respiration. Semin Immunopathol 38:139–152. 10.1007/s00281-015-0534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alanio A. 2020. Dormancy in Cryptococcus neoformans: 60 years of accumulating evidence. J Clin Invest 130:3353–3360. 10.1172/JCI136223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cramer RA, Kowalski CH. 2021. Is it time to kill the survival curve? A case for disease progression factors in microbial pathogenesis and host defense research. mBio 12:e03483-20. 10.1128/mBio.03483-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shibuya K, Coulson WF, Naoe S. 2002. Histopathology of deep-seated fungal infections and detailed examination of granulomatous response against cryptococci in patients with acquired immunodeficiency syndrome. Nihon Ishinkin Gakkai Zasshi 43:143–151. 10.3314/jjmm.43.143. [DOI] [PubMed] [Google Scholar]
- 60.Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect Immun 68:443–448. 10.1128/IAI.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leopold Wager C, Hole CR, Wozniak KL, Olszewski MA, Mueller M, Wormley FL. 2015. STAT1 signaling within macrophages is required for antifungal activity against Cryptococcus neoformans. Infect Immun 83:4513–4527. 10.1128/IAI.00935-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hole CR, Leopold WC, Castro-Lopez N, Campuzano A, Cai H, Wozniak KL, Wang Y, Wormley FL. 2019. Induction of memory-like dendritic cell responses in vivo. Nat Commun 10:2955. 10.1038/s41467-019-10486-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA. 2013. Cryptococcus neoformans Rim101 Is associated with cell wall remodeling and evasion of the host immune responses. mBio 4:e00522-12. 10.1128/mBio.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplemental material. Download iai.00580-21-s0001.pdf, PDF file, 5.9 MB (5.9MB, pdf)
Data Availability Statement
All fungal strains and reagents are available upon request.