Abstract
Confined by the Boltzmann distribution of the energies of states, motion is inherent to biomolecules. For a detailed understanding of a protein’s function, not only the 3D structure but also the description of its dynamics is thus required. This is particularly important in the elucidation of the nature of protein allostery. Protein allostery is a phenomenon involving the long range coupling between two distal sites in a protein. Here we study the enzyme Pin1, which features two flexibly thethered domains, the binding domain (WW) and the catalytic domain (PPI), that undergo coupled structural rearrangements. We present multi-state structures of the WW domain of the free form and in complex with two antagonizing ligands determined by experimentally-derived exact nuclear Overhauser effect (eNOE) rates. We find that the two ligands respectively strengthen and suppress the inter-domain allostery. In the absence of ligands, the protein undergoes a micro-second exchange between two states, one of which is predisposed to interact with the catalytic domain, while the other one is not. In presence of the positive allosteric ligand, the equilibrium between the two states is shifted towards the mode of ligand action, suggesting conformational selection as proposed by Monod. In contrast, the allostery-suppressing ligand decouples the side-chain arrangement at the interface into anti-correlated orientation and dynamics, thereby reducing the inter-domain interaction. As such, this mechanism is an example of dynamic allostery. The presented distinct modes of action highlight the power of the dynamics-function interplay in the biological activity of proteins.
Keywords: NMR, proteins, protein allostery, correlated dynamics, NOE, nuclear Overhauser effect, structure calculation, structure ensemble, Pin1, WW domain
1. Introduction
Allostery in proteins describes the process by which a signal such as ligand binding on one site of a protein or protein complex is transmitted to another distal functional site thereby regulating biological activities (Motlagh et al., 2014). Several models on the mechanism of allostery have been postulated including the sequential mechanism (Koshland et al., 1966), the conformational selection mechanism (originally termed the symmetric model; Monot et al., 1963), and the dynamic allostery model (Cooper and Dryden, 1984). While the sequential mechanism assumes adaptability of the structure upon ligand binding, the model by Monod is based on the existence of two pre-existing exchanging states whose population equilibrium shifts upon ligand binding since the ligand selects one of the two states. The dynamic allostery model assumes that ligand binding changes the frequency and amplitude of thermal fluctuations within a protein without perturbing the average structure.
Experimental elucidation of allostery as ‘an action at a distance’ phenomenon is challenging (Motlagh et al., 2014). The challenge is due to the availability of mostly low resolution, local data in NMR – including relaxation studies (Kay, 2005; Kay et al., 1989) – or individual determined structures of trapped states (such as free and ligand-bound states, or intermediate states stabilized by for example mutagenesis). Integrating analysis of experimental data with molecular dynamic simulations using for example recently developed statistical methods is however emerging as an interesting and powerful approach (Swope et al., 2004, Noe et al., 2006, Shaw et al., 2013, Olsson & Noe 2017, Olsson et al 2017).
Recent progress in NMR-based methods opened an avenue towards a more holistic description of motion and ensembles of structures. These include residual dipolar coupling (RDC) measurements, relaxation dispersion NMR experiments, cross-correlated relaxation (CCR), paramagnetic relaxation enhancement (PRE), and exact Nuclear Overhauser Enhancement or Effect (eNOE) data in combination with molecular dynamics simulation, structure prediction software, or ensemble-based structure calculations (Tolman et al, 1997; Yao et al., 2008; Vallurupalli et al., 2008; Neudecker et al., 2007; Fuentes et al., 2006; Fuentes et al., 2005; Dhulesia et al., 2008; Clore and Schwieters, 2004a, Clore and Schwieters, 2004b; Lindorff-Larsen, 2005; Bouvignies et al., 2005; Vögeli et. al. 2012; Lange et al., 2008; Chi et al., 2015, Olsson et al, 2016).
Here, we made use of the eNOE approach, which allows for the multi-state structure determination of well behaving proteins because of the high accuracy (i.e. < 0.1 Å) of the ensemble-averaged restraints obtained (Vögeli et. al. 2012; Chi et al., 2015; Nichols et al. 2017; Nichols et al. 2018) and applied it to a variant of the prototypical allostery-comprising WW domain of Pin1 (see Material and Methods section). Pin1 is a peptidyl-prolyl cis-trans isomerase (PPIase). Its biological significance includes amongst others an involvement in the regulation of mitosis (Lu et al., 1996), a protective function against Alzheimer’s disease (Ma et al., 2012), increase of hepatitis C infection (Lim et al., 2011) and it is overexpressed in many human cancer cells (Lu et al., 2003). Pin1 contains an N-terminal WW domain (the name has its origin in the presence of two Trp residues) separated by a flexible linker from the C-terminal catalytic PPIase domain (Ranganathan et al., 1997) (Fig. 1A).
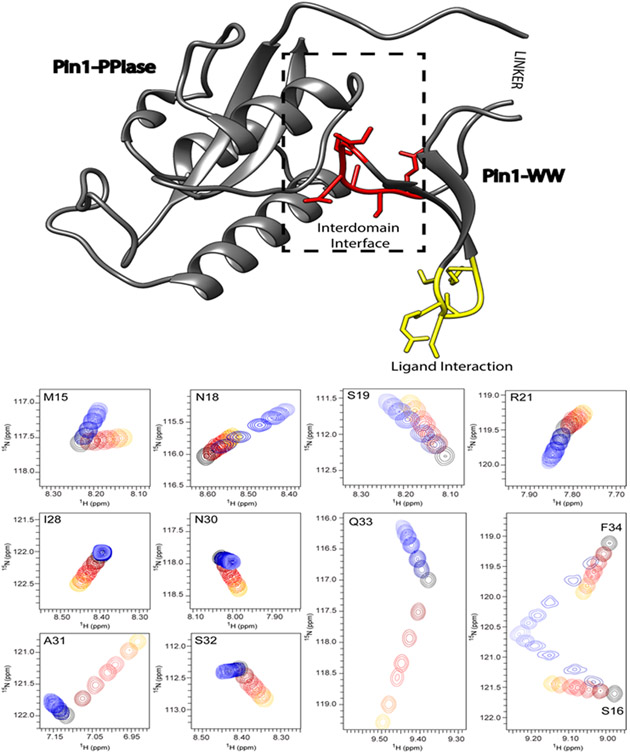
Figure 1: The 3D structure of Pin1 with its postulated allosteric interaction within the variant WW domain.
The allostery within the WW domain occurs between the ligand-binding site (indicated in yellow) and the interaction site with its catalytic domain highlighted in red and labeled as the inter-domain interface. This interaction is shown on top of the 3D crystal structure (pdb code 1PIN) represented by a ribbon with the residues of interest also highlighted by side chains. The NMR chemical shift titrations of the 15N-labeled variant WW with the positive allosteric peptide FFpSPR and the negative allosteric peptide pCdc25C are shown for the relevant residues (i.e. M15-R21 form the ligand binding site and I28-Q33 are residues in the inter-domain interface). The black cross peak corresponds to the 15N-1H moiety of the apo form. Upon titration with FFpSPR highlighted by the color code ranging from black over red to yellow, the cross peaks move with increasing concentration away from the apo form. Similarly, the color changes from dark to light blue indicate the chemical shift changes due to the interaction with the ligand pCdc25C. The shift changes indicate a fast exchange regime (i.e. μs time regime). In the binding site the cross peaks move in the same direction for both ligands, while at the interface they shift in opposite directions. While the interface peaks undergo smaller shifts for the negative-allosteric ligand pCdc25C in comparison with FFpSPR, the binding-site peaks show similar shift magnitudes.
The 34-residues-long N-terminal WW domain is thought to be responsible for ligand recognition and binding as evidenced by NMR titration experiments (Fig. 1), the C-terminal domain contains the catalytically active site. The two domains interact loosely via Loop 2 of the WW domain (i.e. residues 27-33) and the extent of interaction depends on the ligand that binds at a distal Loop 1 (Bayer et al., 2003; Wilson et al., 2013; Guo et al., 2015). One family of substrate (such as the peptide pCdc25C of interest here) reduces the inter-domain contact, while other peptide families (such as the peptide FFpSPR of interest here) enhance the inter-domain contact (Jacobs et al. 2003; Wilson et al., 2013; Peng, 2015). These properties require a substantial allosteric cross-talk between Loop 2 and the ligand binding site Loop 1 of the WW domain (Peng, 2015; Peng et al., 2007; Peng et al., 2009; Morcos et al., 2010; Fenwick et al., 2014; Crane et al., 2000; Socolich et al., 2005; Russ et al., 2005) as evidenced here by ligand titration NMR experiments with the isolated WW domain, showing chemical shift changes at the distal site Loop 2 upon ligand binding at Loop 1 (Fig. 1). Furthermore, the two ligands induce distinct chemical shift changes in direction and magnitude on Loop 2 in line with their opposing property in the inter-domain interaction (Peng, 2015). In order to explore the nature of this allosteric coupling at atomic resolution eNOE ensemble structures were determined of the apo-state of the WW domain as well as the WW domain in presence of either of the two peptide ligands FFpSPR and pCdc25C.
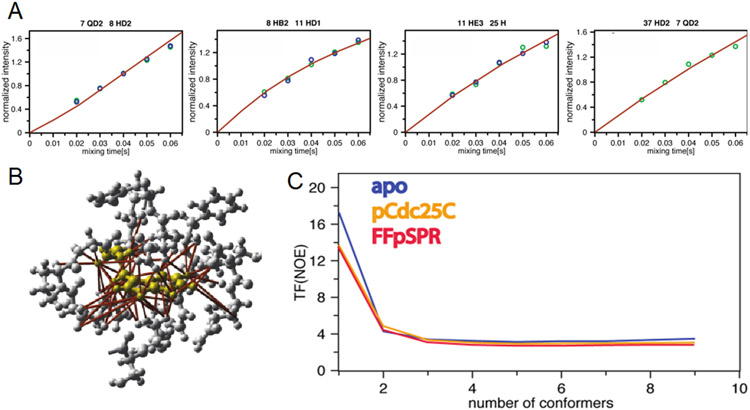
Following an established protocol (Vögeli et. al. 2012; Chi et al., 2015) with the eNORA2 program (Orts et al., 2012, Strotz et al., 2017, CYANA version), ensemble structure calculations were performed for all three systems with eNOE-based distance restraints (Fig. 2A) and scalar couplings (Supplementary Tables S1-S3 and Material for more details). In Figure 2B, the large number of restraints is demonstrated for Trp11 in the apo form of the WW domain, for which ca. 60 distance restraints have been collected, while on average there are roughly 20 eNOE-derived distance restraints per residue. As a measure of the quality of the calculated structures, the CYANA target function (TF), which is a weighted sum of all squared violations of the experimental restraints, is used. It drops significantly from one state to two states and levels off after three states (Fig. 2C). In a ten-state structure calculation, the two states are still observed (as exemplified in the Ramachandran plot for Thr29 in Fig. S1) further supporting the two-state nature of the system. While the TF is an insensitive measure for determining the populations of the two states (Fig. S2A), the details of the two states are preserved (including the correlated/non-correlated configurations of Thr29 and Ala31 discussed below) for a population range 1:1 – ~1:3 for apo WW and WW domain in complex with pCdc25C, while for WW - FFpSPR it is conserved in the range of 1:9 – 4:6 (Fig. S2A).
Figure 2: Distance restraint collection and structure calculation.
A) Four experimental eNOE buildups of the apo WW domain (green and blue dots) versus time are shown against back-predicted buildups of representative NOEs derived from the calculated two-state (red) ensembles. The NOEs are between residues indicated at the top of each graph. The intensities are normalized to the average value of each buildup. The back-predicted theoretical model fulfills the experimental data very well (see also Fig. S2). The back-predicted buildups were calculated using eNORA2 implemented in CYANA (Orts et al., 2012, Strotz et al., 2017, CYANA version). B) eNOE-derived distance restraints around Trp11 of apo WW are mapped onto the 3D structure indicating the large size of the data set. Residues of and around Trp11 are shown in yellow and ribbon and stick representation in grey, respectively. Over 60 eNOE distance restraints (highlighted in red) were collected contrasting the four degrees of freedom of a Trp. This highlights the high density of information obtained by eNOE-based structure determination. C) CYANA target function (TF) values of various ensemble-based structure calculations demonstrating the importance of the ensemble-based structure. The CYANA TF, which is the (weighted) sum of the squared violations of the conformational restraints versus number of simultaneously calculated states, is shown for all three calculations. The decrease of the TF with an increasing number of states indicates that at least two states are required to describe the experimental data well.
In order to get further insights into the relative population of the two states, we conducted titration experiments with FFpSPR. The broadest signal can be attributed to a 1:1 population allowing a determination of the relative populations of the two states of ~1:3 for apo WW (Fig. S2B). To strengthen this finding a 15N-resolved CEST – [15N,1H]-TROSY experiment was measured for apo WW (Fig. S2C). The CEST data of the allosteric sites Ala31 and Gln33 show the presence of two states, one corresponding to the fully FFpSPR-bound state (Fig. S2C, blue arrow), while the other allosteric state can also be identified (Fig. S2C, cyan arrow) and agrees well with the elucidation of the shifts of the two states by the titration (Figs. S2B and S2C). Together with the knowledge of the chemical shifts of the samples used for the structure determination, it can be estimated that the two states of the apo WW domain are present in a ratio of ~3:1, while in the WW-FFpSPR complex the populations are ~1:4.
Overall, these findings indicate that, in contrast to the single-state structure, multi-state ensembles describe the experimental data well (Tables S1-S3). The agreement of the model with the experimental data is also illustrated by the superposition of experimental NOE data and back-calculated NOE buildups (Fig. 2A). The improvement of the model with respect to the experimental data is shown even more explicitly by comparing back-predictions derived from single-state structures in comparison to the model derived from two-state structures (Fig. S3). Furthermore, a cross-validation test with cross-correlated relaxation data (not used in the structure calculations) fit better with the two-state structural ensembles than the single-state structures (Fig. S4). Finally, a cross-validation test was performed with a jackknife procedure that repeats the structure calculation twenty times with 5% of the experimental input data randomly deleted such that each distance restraint is omitted exactly once. These obtained structures are similar to the original structures including the correlated states between Thr29 and Ala31 of interest below (Fig. S5). As a representative for the following discussion, the two state ensembles described by a structural bundle of 2 x 20 conformers (Fig. 2) are used.
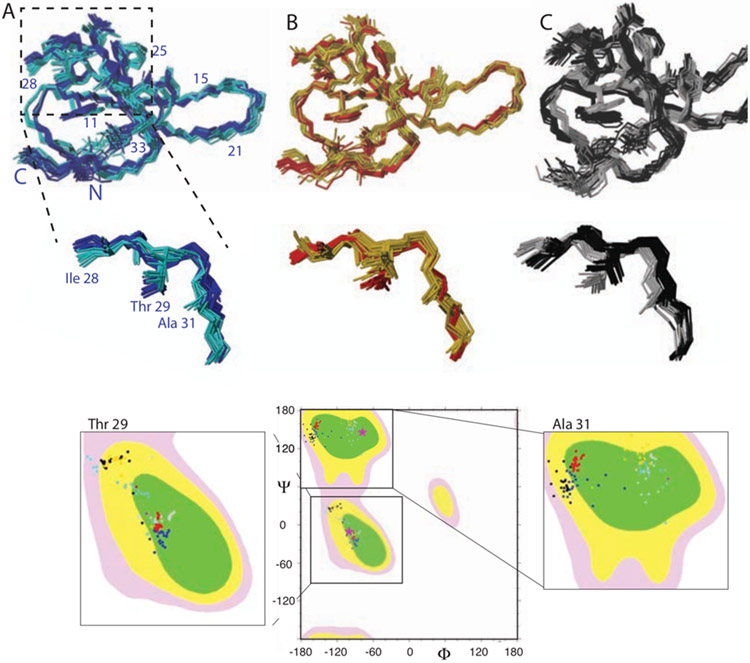
Inspection of the apo WW domain two-states ensemble reveals two spatially well-separated states from the ligand-binding site Loop 1 via the backbone of the β-strand β2 and Asn26 to the inter-domain site Loop 2 (Fig. 3A). The two states are well separated both by the side chains shown (i.e. Phe23, Phe25, Asn26, Ile28, Thr29, Asn30, Ala31) as well as the backbone angles highlighted individually in Ramachandran plots (Fig. 3A). Since only one set of chemical shifts is observed, it is suggested that the two states interchange in the micro-millisecond time range in a concerted fashion.
Figure 3: Two-state structural ensemble of (A) apo WW, WW in complex with the positive (B) and negative (C) allosteric ligand highlighting the presence of two distinct states.
Backbone traces of 20 structural ensembles of the WW domain each representing two different states are shown. In addition, several side chains are shown and labeled. The WW states were color coded with cyan and blue for the apo WW, yellow and red for the positive allosteric ligand FFpSPR complex, and grey and black for the negative allosteric ligand pCdc25C complex, respectively. The two states of the catalytic-domain interacting Loop 2 are enlarged as indicated. In addition, the Ramachandran plots for Ile28, Thr29 and Ala31 are shown with the same color code as in the structures. The Ramachandran angle of the x-ray structure (pdb code 1PIN) is shown with a pink star.
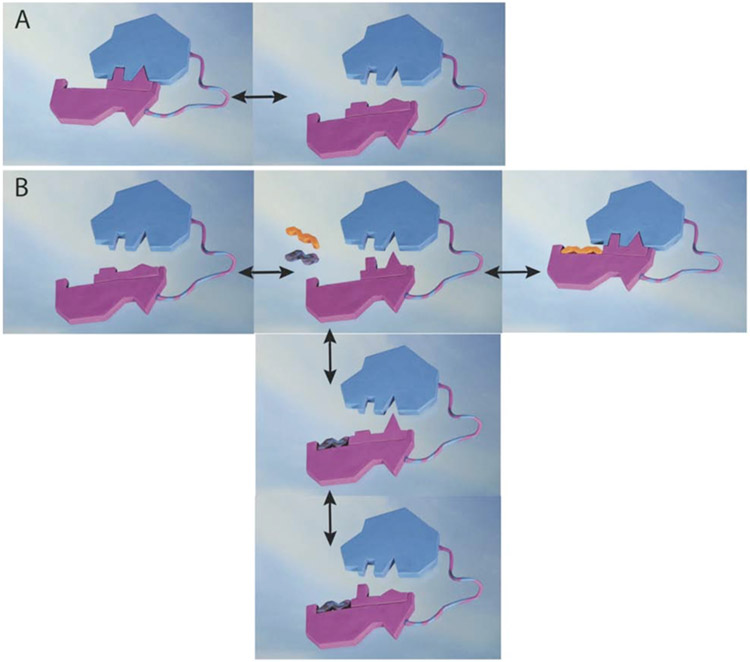
Most interestingly, the two-state ensemble of the WW domain in presence of the positive allosteric ligand FFpSPR just shifts the population of the two states towards the dark blue state. Thus, the FFpSPR peptide appears to select the dark blue state yielding the mode of allosteric action to be conformational selection (Monot et al., 1963). The mechanism of the allosteric coupling between ligand-binding Loop 1 and inter-domain interacting Loop 2 is further illustrated in Figure 4 (clay colored structure). The WW domain possesses two distinct states of similar energy that cover both the ligand binding site (i.e. Loop 1) as well as the inter-domain interacting site (i.e. Loop 2). The interchange between the catalytic domain-binding competent and non-competent states of Loop 2 in the micro-second time range of the apo WW domain thereby perturbs the inter-domain contact. The ligand FFpSPR selects the binding-competent state and enhances the inter-domain interaction.
Figure 4: Allosteric mechanisms of action of the WW domain.
A) The apo form of the WW domain (represented by a pink clay form) is undergoing exchange between two states, one of which (on the left) is able to bind the catalytic domain colored in blue. B) In presence of either the positive allosteric ligand FFpSPR (shown in yellow-red) or the negative allosteric peptide pCdc25C (shown in black) two distinct allosteric mechanisms are active. The positive allosteric ligand FFpSPR selects the catalytic-interacting state enhancing interaction with the catalytic domain (arrow to the right). This mechanism is thus based on the conformational selection as proposed by Monod. The negative allosteric peptide pCdc25C acts via the dynamic allostery model, where the average local structure is not perturbed, but at any given time it is incompatible with interacting with the catalytic side.
In striking contrast to the peptide FFpSPR, peptide pCdc25C influences the inter-domain interaction between the WW domain and its catalytic domain negatively. Based on the finding that the peptide FFpSPR acts by the conformational selection model discussed above, it would seem logical to assume that the pCdc25C peptide selects the binding-incompetent state and thereby interferes with the inter-domain interaction. However, this is not the case as revealed by the two-state structure calculation of the WW domain in presence of the peptide ligand pCdc25C. Still, two states are observed (Fig. 2C). Furthermore, both states are distinct in the backbone (represented by Ramachandran plots in Figure 3C) as well as the side chains of Loop 2 (Fig. 3C). However, the backbone and side-chain states in Loop 2 between residues Ile 28/Thr 29 versus Ala 31 are anti-correlated with each other when compared with the apo structure and the FFpSPR structure (Fig. 3). When Ile 28/Thr 29 are close to the dark blue state of the apo structure, Ala31 is superimposable with the cyan state, while if Ala 31 is close to the cyan apo structure, Ile28/Thr29 align with the dark blue state of the apo structure. While upon local averaging over the two states the apo structure is not distinguishable from the WW domain structure in complex with pCdc25C as supported by minor observed chemical shift changes (Fig. 1), at any given time the side-chain arrangement of Ile28, Thr29 and Ala31 is at odds with an inter-domain fit arrangement and the inter-domain interaction is therefore significantly perturbed as highlighted by the clay figure (Fig. 4B). The pCdc25C peptide – WW domain complex can thus be regarded as an example of a dynamic allostery model, where the local structure is not perturbed on average, but at any given time it is incompatible with interaction with the catalytic domain. This is in contrast to FFpSPR peptide, which performs by the conformational selection model. Thus, depending on the peptide not only the outcome of allostery but also the mechanism of allostery is altered. This is possible since all the structural states involved have similar energies with low activation barriers between them enabling different processes and pathways by small perturbations. The mechanism of action of allostery in the WW domain example shown here highlights the possible multi-dimensional interplay between dynamics and structure that amount to evolutionary selection for fittest performance. It further indicates the astonishing multifaceted possibilities this multi-dimensional dynamic structure landscape possesses.
Material and Methods
Preparation of samples
The Pin1-WW construct S18N/W34F was used because it behaves experimentally more favorable and it is more resistant to aggregation than the wild-type (Crane et al., 2000, Price et al., 2010). Preparation of the 15N/13C-labeled WW domain variant comprising S18N/W34F was done as follows: The S18N genetic sequence was delivered by GL Biochem Ltd. preassembled in peT32 with a HIS6-tag cleavable by TEV. Transformed E. coli BL21/DE3 cells were grown in pre-cultures started from fresh glycerol stock. In 2L M9 cultures (with either 15N NH4Cl or 15N NH4Cl/ 13C glucose) the cells were grown form OD600 of 0.1 to 0.7 at 37°C and shaken at 120 RPM, then induced with IPTG. Following induction, the temperature was reduced to 25°C and left for expression for 4 hrs before harvesting. Following two-step Ni-column purification the sample was desalted and TEV protease added in 1:50 (m/m) ratio and left over-night at room temperature. A further Ni-column purification step then provided the clean NMR sample. The sample's buffer was exchanged to the NMR buffer (10 mM K2PO4, 100 mM NaCl, 0.02 % NaN3, pH 6.0) using dialysis, then concentrated to 1.2 mM using 2 kDa cutoff concentrator tubes (Sartorius Vivaspin 15R). The gene of full-length Pin1 S18N/W34F variant was bought from genescript, sub-cloned into a pET28a vector containing an N-terminal His-tag with a thrombin cleavage site (MHHHHHHLVPRGS). For expression the cDNA was transformed into E. coli BL21 cells and plated on a kanamycin-containing plate (50 ug/ml). The cells were grown over-night at 37 °C and then used to inoculate a 10 ml pre-culture. The culture was grown at 37 °C for three hours and thereafter used to inoculate a 1-liter culture (kanamycin 50 ug/ml) of M9 medium for 15N/13C- or 15N-labeling. Cells were grown to an OD600 of 0.9. Protein expression was initiated by adding 1 mM IPTG (isopropyl β-D-thiogalactopyranoside). The cells were then allowed to express over night at 18 °C and harvested by spinning at 5,000 g for 15 minutes and re-suspended in purification buffer (10 mM Tris/HCl, 200 mM NaCl). The cells were lyophilized and spun at 40,000 g for 20 minutes. The supernatant was filtered (0.4 μm and 0.2 μm filters) and loaded onto a nickel (II)-charged chelating sepharose FF column (Amersham Biosciences), equilibrated with purification buffer as above and washed with 400 ml of the same buffer. The bound sample was eluted with 250 mM imidazole at pH 7.9, in aliquots of 10 ml. Fractions containing partially pure proteins were pooled, desalted and passed through a DEAE column equilibrated with purification buffer. The sample was collected as flow-through. The purity was checked on SDS PAGE stained with coomassie brilliant blue. The pure protein preparation was concentrated to experimental concentration of 0.4 mM. The concentration was determined by absorption measurements using the molar absorption coefficient calculated.
The phosphorylated ligands pCdc25C (i.e. EQPLpTPVTDL) and FFpSPR were ordered from Bachem AG, Switzerland.
The NMR buffer was 10 mM K2PO4, 100 mM NaCl, 0.02 % NaN3, in 3 % D2O and pH 6.0 with sample concentrations of 1.2 mM WW domain or 0.4 mM full-length Pin1. For the sample in complex with pCdc25C a 4-fold excess of ligand was used, while for the complex with FFpSPR a 10-fold excess of the ligand was used, respectively. Using the isotherms of the chemical shift perturbations for the ligand-binding site residues (excluding allosteric site residues) and two-state exchange models (chemical shift perturbation versus concentration) the pCdc25C affinity (Kd) to the WW domain was determined to be 526 +/− 146 μM in line with isothermal titration calorimetry (ITC) measurements (Supplementary Figure S2) that yielded a similar affinity (Kd) of 158 +/− 70 μM (overall Kd, as opposed to binding site). Correspondingly, the Kd for FFpSPR was determined to be 708 +/− 38 μM. Using the above values the occupancy of the NMR samples were calculated as follows:
Where Lt and Pt are the ligand and protein concentration, respectively, in μM and the Kd was the calculated Kd from NMR titrations (μM).
NMR experiments
All experiments were recorded on a Bruker 700 MHz spectrometer and at 5 °C, except where described otherwise. All spectra were processed and analyzed using the software package NMRPipe (Delaglio et al., 1995), assignment was done in CcpNMR (Vranken et al., 2005). The measurement and analysis of eNOEs using eNORA2 (Orts et al., 2012, Strotz et al., 2017, CYANA version) was described previously in detail (Strotz et al., 2015), in short: series of 3D [15N, 13C]-resolved [1H, 1H]-NOESY-HSQC experiments were recorded to measure NOE buildups (Vögeli et al., 2013). The inter-scan delay was 0.8 s. Simultaneous [15N, 1H]-HSQC and [13C, 1H]-HSQC elements were employed, following indirect proton chemical shift evolution and [1H, 1H]-NOE mixing (τm). Diagonal-peak decays and cross-peak buildups were measured with τm of 20, 30, 40, 50, and 60 ms for all the three samples.
The [15N, 1H]-HSQC for full length Pin1 was recorded with 128(t1) x 1024(t2) real points on a Bruker 700 MHz spectrometer. The maximal evolution times were t1max,1H = 51.5 ms, t2max,15N = 81.1 ms. The time-domain data were multiplied with a squared cosine function in the direct dimension and cosine functions in the indirect dimensions and zero-filled to 1024 x 256 real points. Exponential (EM, with LB 25 Hz) window functions were applied in both dimensions to the free induction decay and zero-filled to 1024 x 256 real points. The final 2D saturation difference spectrum is the difference between the off-resonance and the on-resonance irradiated [15N, 1H]-HSQC spectra collected.
Titration experiments with the 13C,15N-labeled WW domain were done as follows: For a fixed amount of labeled 15N/13C WW domain, increasing amounts of peptides were titrated into the protein solution and the extent of binding was determined by measuring the chemical shift changes in the [15N, 1H]-HSQC spectrum for several resonances and averaged thereafter.
The rotational correlation times τc of the individual samples were determined using 15N-relaxation measurements as described previously in detail (Strotz et al., 2015) yielding a τc for the apo WW domain to be 4.25 ns at 5 °C and 1.2 mM concentration, a τc of 5.67 ns for the WW domain in complex with pCdc25C at 5 °C and 1.2 mM concentration and a τc of 5.13 ns at 5 °C and 1.2 mM concentration for the WW domain in complex with FFpSPR, respectively.
3JHN,Hα scalar coupling were measured as described previously in detail (Strotz et al., 2015). 3JHα,Hβ scalar couplings were obtained from 3D 13Cα-separated Hα-Hβ in-phase COSY (HACAHB-COSY) experiments (Grzesiek et al., 1995) in D2O. The experiment was recorded with 50(MQ[Cα], t1) × 54(Hβ, t2) × 2048(Hα, t3) complex points, giving t1max, 13C = 22.5 ms, t2max,1H = 10.8 ms, t3max,1H = 204.8 ms. The time domain data were multiplied with a square cosine function in the direct dimension and cosine functions in the indirect dimensions and zero-filled to 256 × 512 × 2048 complex points. The Karplus parameters used in structure calculations were from (Pérez et al., 2001). 3JC’,Cγ and 3JN,Cγ scalar couplings for aromatic side chains were obtained from 13C’-{13Cγ} and 15N-{13Cγ } spin-echo difference [15N,1H]-HSQC experiments (Hu et al., 1997) performed on a Bruker 600 MHz spectrometer. The experiments were recorded with 100(15N, t1) or 200(15N, t1) × 512(1HN, t2) complex points, giving t1max,15N = 50 ms or t1max,15N = 100 ms and t2max,1H = 51.2 ms, respectively. The time domain data were multiplied with a square cosine function in the direct dimension and cosine functions in the indirect dimensions and zero-filled to 512 × 2048 complex points. The Karplus parameters used in structure calculations were from (Pérez et al., 2001).
Cross-correlated relaxation rates ΓHNiNi/HαiCαi + ΓHαiNi/HNiCαi were obtained from two experiments performed on a Bruker 600 MHz spectrometer equipped with a z-axis gradient cryogenic probe. A DIAI (double in-phase/anti-phase inter-conversion) method was realized with a pair of 3D HNCA pulse sequences (“reference” and “trans”) (Pelupessy et al., 1999) for the first experiment. A 3D ct-HNCA MMQ (mixed multi-quantum, with zero- and double-quantum coherence evolution averaged) experiment was used for the second experiment. The ZQ (zero quantum) and DQ (double quantum) coherences were superimposed, resulting in four components to be evaluated (Fenwick et al., 2016; B. Vögeli, 2017). The experiments were recorded with τMQ = 31.0 ms or τMQ = 33.5 ms, 50(MQ[N,Cα], t1) or 55(MQ[N, Cα], t1) × 36(N, t2) × 512(HN, t3) complex points, t1max = 25.0 or 27.5 ms, t2max,15N = 18.0 ms, t3max,1H = 51.2 ms. The time domain data were multiplied with a square cosine function in the direct dimension and cosine functions in the indirect dimensions and zero-filled to 256 × 128 × 2048 complex points. The back-calculation of the cross-correlated relaxation rates followed the procedure described previously in detail (Vögeli, 2010).
The 2D CEST-[15N,1H]-TROSY spectra (Vallurupalli et al., 2012) were recorded on a Bruker 700 MHz spectrometer for both 15N-labeled apo WW. The continuous wave 5% truncated Gaussian pulse used with a length of 75 ms showed an excitation profile of 8.5 Hz. The saturation was obtained by 6 such pulses. The experiments were recorded with 32(15N, t1) × 512(1HN, t2) complex points, giving t1max,15N = 16 ms and t2max,1H = 51.2 ms. Along the 15N-CEST dimension 90 saturations were measured around the Ala31 and Gln33 resonances in steps of 8 Hz.
Structure calculation
The structure calculation ollowed the established ensemble-based protocol (Vögeli et al., 2012, Vögeli, 2014) using the software packages eNORA2 (Orts et al., 2012, Strotz et al., 2017, CYANA version) and CYANA (Guntert et al., 1997, Guntert, 2009). As input for the structure calculation we used upper and lower distance restraints from eNOEs together with backbone, Hβ and aromatic side-chain scalar couplings and conservative Φ and Ψ dihedral angle restraints derived from 13Cα chemical shifts (Tables S1-S3) (Luginbuhl et al., 1995). The weight of the dihedral angle restraints was reduced to zero in the final steps of the structure calculation. Calculations were done with 50’000 torsion angle dynamics steps for 100 conformers with random torsion angles by simulated annealing. The multi-states structural ensembles were each calculated simultaneously and averaged. A weak harmonic well potential with bottom width of 1.2 Å was used to keep identical heavy atoms from the different states together (Vögeli et al., 2012, Vögeli et al., 2013). The 20 conformers with the lowest final target function values were selected and analyzed. The calculated coordinates and the complete data set consisting of the eNOEs together with the upper/lower distance limit tables are deposited in the PDB under 6SVC, 6SVE and 6SVH.
Supplementary Material
References
- Bayer E, Goettsch S, Mueller JW, Griewel B, Guiberman E, Mayr LM, Bayer P (2003) Structural analysis of the mitotic regulator hpin1 in solution: insights into domain architecture and substrate binding. J Biol Chem 278:26183–93 [DOI] [PubMed] [Google Scholar]
- Bouvignies G, Bernadó P, Meier S, Cho K, Grzesiek S, Brüschweiler R, Blackledge M (2005) Identification of slow correlated motions in proteins using residual dipolar and hydrogen-bond scalar couplings. Proc Nat Ac of Sc USA 102(39):13885–13890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüschweiler R, Roux B, Blackledge M, Griesinger, Karplus M, Ernst RR (1992), Influence of Rapid Intramolecular Motion on NMR Cross-Relaxation Rates. A Molecular Dynamics Study of Antamanide in Solution, J Am Chem Soc 114:2289–2302 [Google Scholar]
- Chi CN, Strotz D, Riek R, Vögeli B (2015). Extending the eNOE data set of large proteins by evaluation of NOEs with unresolved diagonals. J Biomol NMR [DOI] [PubMed] [Google Scholar]
- Clore GM, Schwieters CD (2004a) How much backbone motion in ubiquitin is required to acount for dipolar coupling data measured in mutliple alignment media as assessed by independent cross-validation?. J Am Chem Soc 126:2923–2938 [DOI] [PubMed] [Google Scholar]
- Clore GM, Schwieters CD (2004b) Amplitudes of Protein Backbone Dynamics and Correlated Motions in a Small α/β Protein: Correspondence of Dipolar Coupling and Heteronuclear Relaxation Measurements. Biochemistry 43:10678–10691 [DOI] [PubMed] [Google Scholar]
- Crane JC, Koepf EK, Kelly JW, Gruebele M (2000) Mapping the transition state of the WW domain beta-sheet. J Mol Biol 298:283–292 [DOI] [PubMed] [Google Scholar]
- Cooper A, Dryden DTF (1984) Allostery without conformational change. Eur Biophys J 11:103–109 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6(3):277–293. [DOI] [PubMed] [Google Scholar]
- Dhulesia A, Gsponer J, Vendruscolo M (2008) Mapping of Two Networks of Residues That Exhibit Structural and Dynamical Changes upon Binding in a PDZ Domain Protein. J Am Chem Soc 130:8931–8939 [DOI] [PubMed] [Google Scholar]
- Fenwick RB, Orellana L, Esteban-Martín S, Orozco M, Salvatella X (2014) Correlated motions are a fundamental property of β-sheets. Nature communications 5:4070. [DOI] [PubMed] [Google Scholar]
- Fenwick RB, Schwieters CD, Vögeli B (2016) Direct Investigation of Slow Correlated Dynamics in Proteins via Dipolar Interactions. JACS 138(27):8412–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes EJ, Der CJ, Lee AL (2005) Ligand-dependent Dynamics and Intramolecular Signaling in a PDF Domain. J Mol Biol 335:1105–1115 [DOI] [PubMed] [Google Scholar]
- Fuentes EJ, Gilmore SA, Mauldin RV, Lee AL (2006) Evaluation of Energetic and Dynamic Coupling Networks in a PDZ Domain Protein. J Mol Biol 364:337–351 [DOI] [PubMed] [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273(1):283–298 [DOI] [PubMed] [Google Scholar]
- Güntert P (2009) Automated structure determination from NMR spectra. Eur Biophys J 38(2):129–143 [DOI] [PubMed] [Google Scholar]
- Guo J, Pang X, Zhou HX (2015) Two pathways mediate interdomain allosteric regulation in pin1. Structure 23:237–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiek S, Kuboniwa H, Hinck AP, Bax A (1995) Multiple-Quantum Line Narrowing for Measurement of Hα-Hβ J Couplings in Isotopically Enriched Proteins, J Am Chem Soc 117:5312–5315 [Google Scholar]
- Hu JS, Grzesiek SS, Bax A (1997) Two-dimensional NMR methods for determining Xi angles of aromatic residues in proteins from thee-bond JCCy and JNCy couplings. J Am Chem Soc 119:1803–1804 [Google Scholar]
- Jacobs D, Saxena K, Vogtherr M, Bernado P, Pons M, Fiebig KM (2003) Peptide binding induces large scale changes in inter-domain mobility in human Pin1. J Biol Chem 278:26174–26182 [DOI] [PubMed] [Google Scholar]
- Koshland DE, Nemeth G, Filmer D (1966) Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5 365–385 [DOI] [PubMed] [Google Scholar]
- Kay LE, Torchia DA, Bax A (1989) Backbone Dynamics of Proteins as Studied by Nitrogen-15 Inverse Detected Heteronuclear NMR Spectroscopy: Application to Staphylococcal Nuclease. Biochemistry 28:8972–8979 [DOI] [PubMed] [Google Scholar]
- Kay LE (2005), NMR studies of protein structure and dynamics, J Magn Reson 173:193–207 [DOI] [PubMed] [Google Scholar]
- Lange OF, Lakomek NA, Farès C, Schröder GF, Walter KFA, Becker S, Meiler J, Grubmüller H, Griesinger C, de Groot BL (2008) Self-consistent residual dipolar coupling based model-free analysis for the robust determination of nanosecond to microsecond protein dynamics. Science 320:1471–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K, Best RB, Depristo MA, Dobson CM, Vendruscolo M (2005) Nature 433:12832. [DOI] [PubMed] [Google Scholar]
- Lim YS, Tran HTL, Park SJ, Yim SA, Hwang SB (2011) Peptidyl-prolyl isomerase Pin1 is a cellular factor required for Hepatitis C virus propagation. J Virol 85:8777–8788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T, (1996) A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380:544–547 [DOI] [PubMed] [Google Scholar]
- Lu KP (2003) Prolyl isomerase Pin1 as a molecular target for cancer diagnostics and therapeutics. Cancer Cell 4:175–180 [DOI] [PubMed] [Google Scholar]
- Luginbuhl P, Szyperski T, Wüthrich K (1995) Statistical basis for the use of C-13-alpha chemical shifts in protein structure determination. J Magn Reson Ser B 109:229–233 [Google Scholar]
- Ma SL, Pastorino L, Zhou XZ, Lu KP (2012) Prolyl isomerase Pin1 promotes amyloid precursor protein (APP) turnover by inhibiting glycogen synthase kinase-3 (GSK3) activity: Novel mechanism for Pin1 to protect against Alzheimer disease. J Biol Chem 287:6969–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J, Changeux JP, Jacob F (1963) Allosteric proteins and cellular control systems. J. Mol. Biol 6:306–329 [DOI] [PubMed] [Google Scholar]
- Morcos F, Chatterjee S, McClendon CL, Brenner PR, López-Rendón R, Zintsmaster J, Ercsey-Ravasz M, Sweet CR, Jacobson MP, Peng JW (2010) Modeling conformational ensembles of NMR relaxation dispersion: Molecular dynamics study of slow functional motions in Pin1-WW. PLoS Comput Biol 6, e1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motlagh HN, Wrabl JO, Li J, Hilser VJ (2014) The ensemble nature of allostery. Nature doi: 10.1038/nature13001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudecker P, Zarrine-Afsar A, Davidson AR, Kay LE (2007) Phi-value analysis of a three-state protein folding pathway by NMR relaxation dispersion spectroscopy. Proc Natl Acad Sci USA 104:15717–15722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols P, Born A, Henen MA, Strotz D, Orts J, Olsson S, Güntert P, Chi CN, Vögeli B (2017) The exact NOE: Recent advances. Molecule 22:1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols P, Born A, Henen MA, Strotz D, Chi CN, Güntert P, Vögeli B (2018) Extending the applicability of exact nuclear Overhauser enhancements to large proteins and RNA. ChemBioChem, 19:1695–1701 [DOI] [PubMed] [Google Scholar]
- Noe F, Krachtus D, Smith JC, Fischer S (2006) Transition networks for the comprehensive characterization of complex conformational change in proteins. Journal of Chemical Theory and Computation 2:840–857 [DOI] [PubMed] [Google Scholar]
- Olsson S, Wu H, Paul F, Clementi C, Noe F (2017) Combining Experimental and Simulation Data of Molecular Processes via Augmented Markov Models Proc. Natl. Acad. Sci. U.S.A 114(31), pp. 8265–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson S & Noe F (2017) Mechanistic models of chemical exchange induced relaxation in protein NMR J. Am. Chem. Soc, 139 (1), pp 200–210. [DOI] [PubMed] [Google Scholar]
- Olsson S, Strotz D, Vogeli B, Riek R, Cavalli A (2016) The dynamic basis for signal propagation in human Pin1-WW. Structure, 24, pp. 1464–1475 [DOI] [PubMed] [Google Scholar]
- Orts J, Vögeli B, Riek R (2012) Relaxation Matrix Analysis of Spin Diffusion for the NMR Structure Calculation with eNOEs. J Chem Theory Comput 8:3483–3492 [DOI] [PubMed] [Google Scholar]
- Pelupessy P, Chiarparin E, Ghose R, Bodenhausen G (1999) Simultaneous determination of ψ and Φ angles in proteins from measurements of cross-correlated relaxation effects. J Biomol NMR 14:277–280 [Google Scholar]
- Peng T, Zintsmaster JS, Namanja AT, Peng JW (2007) Sequence-specific dynamics modulate recognition specificity in WW domains. Nature structural & molecular biology 14(4):325–331 [DOI] [PubMed] [Google Scholar]
- Peng JW, Wilson BD, Namanja AT (2009) Mapping the dynamics of ligand reorganization via 13CH3 and 13CH2 relaxation dispersion at natural abundance. Journal of Biomolecular NMR 45(1-2):171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J (2015) Investigating Dynamic Interdomain Allostery in Pin1, J Biophys Rev 7:239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez C, Löhr F, Rüterjans H, Schmidt JM (2001) Self-Consistent Karplus Parametrization of 3JCouplings Depending on the Polypeptide Side-Chain Torsion χ 1. J Am Chem Soc 123(29):7081–7093 [DOI] [PubMed] [Google Scholar]
- Price JL, Shental-Bechor D, Dhar A, Turner MJ, Powers ET, Gruebele M, Levy Y, Kelly JW (2010) Context-dependent effects of asparagine glycosylation on Pin WW folding kinetics and thermodynamics. J Am Chem Soc 132:15359–15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan R, Lu K, Hunter T, Noel J (1997) Structural and functional analysis of the mitotic rotamase pin1 suggests substrate recognition is phosphorylation dependent. Cell 89: 875–886 [DOI] [PubMed] [Google Scholar]
- Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R (2005) Natural-like function in artificial WW domains. Nature 437(7058):579–583 [DOI] [PubMed] [Google Scholar]
- Shaw DE, Maragakis Lindorff-Larsen PK, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W (2011) Atomic-level characterization of the structural dynamics of proteins. Science 133:341–346 [DOI] [PubMed] [Google Scholar]
- Strotz D, Orts J, Minges M, Vögeli B (2015) The experimental accuracy of the uni-directional exact NOE. J Magn Reson, doi: 10.1016/j.jmr.2015.07.007 [DOI] [PubMed] [Google Scholar]
- Strotz D, Orts J, Chi CN, Riek R, Vögeli B (2017) eNORA2 Exact NOE Analysis Program. J Chem Theory Comput 13(9):4336–4346 [DOI] [PubMed] [Google Scholar]
- Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R (2005) Evolutionary information for specifying a protein fold. Nature 437(7058):512–518. [DOI] [PubMed] [Google Scholar]
- Swope WC, Pitera JW, Suits F (2004) Describing protein folding kinetics by molecular dynamics simulations. 1. theory. The Journal of Physical Chemistry B 108:6571–6581 [Google Scholar]
- Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH (1997) NMR evidence for slow collective motions in cyanometmyoglobin. Nat Struct Biol 4:292–297 [DOI] [PubMed] [Google Scholar]
- Vallurupalli P, Hansen DF, Kay LE (2008) Structures of invisible, excited protein states by relaxation dispersion NMR spectroscopy. Proc Natl Acad Sci USA. 105:11766–11771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallurupalli P, Bouvignies G, Kay LE (2012) Studying “Invisible” Excited Protein States in Slow Exchange with a Major State Conformation. J Am Chem Soc 134(19):8148–8161 [DOI] [PubMed] [Google Scholar]
- Vögeli B (2010) Comprehensive description of NMR cross-correlated relaxation under anisotropic molecular tumbling and correlated local dynamics on all time scales. J Chem Phys 133(1):014501. [DOI] [PubMed] [Google Scholar]
- Vögeli B, Kazemi S, Güntert P, Riek R (2012) Spatial elucidation of motion in proteins by ensemble-based structure calculation using exact NOEs. Nat Struct Mol Biol 19(10):1053–1057 [DOI] [PubMed] [Google Scholar]
- Vögeli B, Güntert P, Riek R (2013) Multiple-state ensemble structure determination from eNOE spectroscopy. Mol Phys 111(3):437–454 [Google Scholar]
- Vögeli B (2014) The nuclear Overhauser effect from a quantitative perspective. Prog Nucl Magn Reson Spectrosc 78(C):1–46 [DOI] [PubMed] [Google Scholar]
- Vögeli B (2017) Cross-correlated relaxation rates between protein backbone H-X dipolar interactions. J Biomol NMR 67:211–232 [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59(1):687–696 [DOI] [PubMed] [Google Scholar]
- Wilson KA, Bouchard JJ, Peng JW (2013) Interdomain interactions support interdomain communication in human pin1. Biochemistry 52:6968–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Vögeli B, Torchia DA, Bax A (2008) Simultaneous NMR Study of Protein Structure and Dynamics Using Conservative Mutagenesis. J. Phys. Chem. B 112:6045–6056 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.