Abstract
Our ability to prognosticate the clinical course of patients with cancer has historically been limited to clinical, histopathological, and radiographic features. It has long been clear however, that these data alone do not adequately capture the heterogeneity and breadth of disease trajectories experienced by patients. The advent of efficient genomic sequencing has led to a revolution in cancer care as we try to understand and personalize treatment specific to patient clinico-genomic phenotypes. Within prostate cancer, emerging evidence suggests that tumor genomics (e.g. DNA, RNA and epigenetics) can be utilized to inform clinical decision making. In addition to providing discriminatory information about prognosis, it is likely tumor genomics also hold a key in predicting response to oncologic therapies which could be used to further tailor treatment recommendations. Herein we review select literature surrounding the use of tumor genomics within the management of prostate cancer, specifically leaning towards analytically validated and clinically tested genomic biomarkers utilized in radiotherapy and/or adjunctive therapies given with radiotherapy.
Introduction
In the United States, Prostate cancer (PCa) represents the most common solid organ malignancy among men accounting for nearly 250,000 new cases and over 30,000 deaths in 20211. Given the high incidence of disease, it is therefore unsurprising the significant heterogeneity in disease course and outcomes of men diagnosed with PCa. Although many with PCa experience a highly curable and indolent disease course, a subset of patients have more aggressive disease biology with progression to metastatic disease, castration-resistance and ultimately death2. The D’Amico prostate cancer risk stratification for localized disease strove to classify this spectrum by incorporating clinical and pathologic factors, and has subsequently guided the management of localized prostate cancer for decades3. Despite this landmark achievement characterizing the behavior of localized prostate cancer, there is still wide variability not fully explained by clinical factors alone.
Historically, oncologists have been limited to using clinical features such as radiographic and histopathologic findings to infer the disease course despite recognizing the complex and diverse genetic and molecular alterations involved in tumorigenesis. With the identification of driver mutations and subsequent development of targeted therapies, a more personalized approach to treatment became possible4–8. The advent of affordable and efficient next-generation sequencing (NGS) allowed for whole-genome, whole-exome, limited panel, transcriptome, and epigenetic sequencing to become increasingly utilized9. These monumental advancements in sequencing technology over the past two decades allowed for a more nuanced understanding of the tumor biology of an individual and has opened countless doors for greater precision in clinical oncology. NGS has not only allowed for more rapid identification of specific targetable driver mutations, but also for uncovering the milieu of genomic alterations that may be leveraged as prognostic and more importantly predictive biomarkers to help guide treatment decisions10–12.
With an eye towards prostate cancer, tumor genomics hold potential to demystify the continued heterogeneous disease course observed among men and provide greater insight into predicting response to the various available treatments. Herein we review current select literature of genomic markers, somatic and when applicable germline, for radiotherapeutic efficacy across all stages of prostate cancer from localized disease to lethal metastatic castration-resistance. Given the considerable breadth of the topic, we apologize in advance that we cannot include all of the excellent research in this area, but we chose specifically to lean towards analytically validated genomic biomarkers tested for predictive radiotherapy efficacy and/or adjunctive therapies given with radiotherapy
Pan-cancer Genomic Biomarkers of Radiation Sensitivity
Determining the proper radiation dose has commonly been identified through dose escalation/de-escalation trials based on histology. With the adoption of tumor genomic profiling, developing an assay to predict tumor specific radiation sensitivity has garnered greater interest. Initial in vitro work by Torres-Roca et al. developed and validated a radiation sensitivity classifier that predicts the survival fraction of various cancer cell lines to 2 Gy dependent upon gene expression profiles of 3 novel genes (RbAp48, RGS19, and R5PIA)13. Follow-up work validated a clinical radiosensitive index (RSI) modeled as a function of expression of 10 genes extracted from an interaction network across 48 cancer cell lines. This RSI was found to be significantly different in responders versus nonresponders in patients with rectal and esophageal cancer treated with radiation14. This radiation sensitivity index has further demonstrated utility in glioblastoma15, breast16, pancreatic17, and endometrial18 cancer.
Using the gene-expression based RSI and a linear quadratic model, which is used to estimate biologic effective dose of varying radiation fractionation schemes, Scott et al. derived a genome-based model for adjusting radiotherapy dose (GARD) 19 and calculated a GARD score for 8271 tumors across 20 disease sites. The GARD was then validated in a clinical cohort of glioblastoma, breast, pancreatic and lung cancer. GARD values across these samples ranged from 1.66–172.2 with a higher GARD representing a greater biologic effect to radiation. Among the 186 patients with prostate cancer treated with 70 Gy there was significant heterogeneity within GARD values particularly skewed towards higher values. This indicates prostate cancer may be particularly well suited for dose de-escalation dependent upon tumor gene expression and radiation fraction size. Within a pan-cancer analysis, GARD (as a continuous variable) was demonstrated to be associated with overall survival which was dependent upon whether the patient received radiation therapy20. Although RSI/GARD have not yet been utilized in a randomized clinical trial, it has been identified as trial ready by the EORTC21.
Radiation is believed to exert its biologic effect predominately through the generation of free radicals leading to DNA double strand breaks (DSB). Given this mechanism of action, several mutations in DNA damage response (DDR) pathways, specifically those involved in DSB repair, have been implicated in response to radiation. The Ataxia Telangiectasia-Mutated (ATM) protein is linked to DDR as it is recruited to assist in DSB repair by the MRE11-RAD50-NBSS1 (MRN) complex and has been implicated in several malignancies22. Within a pan-cancer assessment of patients with either ATM loss of function or variant of unknown significance (VUS), loss of function was associated with significantly improved 2-year local progression following radiation (13.2% vs 27.5%)23. Despite this preliminary evidence for radiation sensitivity in context of ATM disruption, the Ataxia Telangiectasia and Rad3-related gene (ATR) is able to partially compensate for ATM loss of function due to redundant overlapping function24,25. This has led to interest in utilizing ATR inhibition as a radiosensitizer among patients with DDR mutations26 24,25. Pathogenic mutations within BRCA1/2 (involved in homologous recombination repair) have similarly been implicated in several malignancies and have demonstrated increased dependency upon poly(ADP-ribose) polymerase (PARP) for DNA repair27, including in PCa28, likely due to defects in homologous recombination leading to a synthetic lethal relationship29. This has led to the utilization of PARP inhibition for use as a radiosensitizer in BRCA deficient tumors30,31. Although uncommon in prostate cancer, several mutations have also been identified that appear to confer radioresistance across several malignancies including NRF2, BRAF, and EGFR and may hold promise as targeted radiosensitizers through molecular antagonists32–36.
Molecular Subtypes within Prostate Cancer
Given the parallels of breast and prostate cancer as hormone driven malignancies, the notion of molecular subtyping, pioneered within breast cancer, has recently been applied to prostate cancer37,38. The PAM50 algorithm is a clustering-based genomic classifier based on the expression of a set of 50 genes categorizing patients into “luminal” and “basal” molecular subtypes. Zhao et al. applied this PAM50 classifier to 3782 localized prostate cancer samples which segregated 3 distinct molecular subtypes including luminal A, luminal B, and basal type39. Similar to breast cancer, luminal subtypes have increased hormone receptor expression and downstream signaling. This work further identified Luminal B prostate cancer to demonstrate the worst clinical prognosis and was the only subtype to be significantly associated with postoperative response to androgen deprivation therapy (ADT). The presence of these molecular subtypes has subsequently been identified in patients with metastatic castration-sensitive40 and -resistant prostate cancer41. Within castration-resistant disease, luminal type tumors have demonstrated significantly better survival following treatment with androgen-signaling inhibitors (ARSI) while basal tumors (encompassing 90% of small cell/neuroendocrine PCa) do not benefit in relative comparison41. Given the very recent application of molecular subtyping of prostate cancer, there is a paucity of data on how these subtypes may influence response to radiation therapy. Within breast cancer, Lalani et al. evaluated whether molecular subtypes exhibited differential response to radiation by fractional dose42. This work did not identify any significant interaction between subtype and fractionation regimen. It is not currently known whether molecular subtypes experience differential response to total dose of radiation or to the radiosensitizing effects of ADT.
Localized Prostate Cancer
Genomic Biomarkers within Localized Prostate Cancer at Initial Diagnosis
Within localized prostate cancer, several commercially available genomic biomarkers have been developed aiming to improve risk stratification over clinical factors alone. The Decipher Prostate Biopsy (Veracyte, San Diego, CA, USA) represents the most ubiquitous and well validated genomic classifier (GC). Decipher is a commercially available 22-gene GC that utilizes a whole-transcriptome oligonucleotide analytically validated microarray platform. A random forest algorithm identified the expression of 22 RNA biomarkers related to androgen receptor signaling, cell proliferation, differentiation, motility and immune modulation that comprises the GC43. Although initially intended for use after radical prostatectomy, the Decipher Prostate Biopsy has been approved in the United States for use in the entire spectrum of localized disease. Providing a score ranging from 0–1, it provides an estimated risk of adverse pathologic features at RP (Grade group 3–5, pT3b-T4, lymph node involvement) as well as 5- and 10-year risk of distant metastasis and 15-yr prostate cancer specific mortality. A summary of current data and future trials for the Decipher genomic score within localized prostate cancer can be found in Figure 1. The Oncotype Dx (Exact Sciences, Madison, WI, USA) genomic prostate score test is a 17-gene reverse transcription polymerase chain reaction assay that has been clinically validated to predict the likelihood of adverse pathology (Gleason Grade Group >3 and/or ≥T3a), distant metastases, and prostate-cancer specific mortality. This assay measures 12 cancer related genes across 4 biologic pathways including stromal response, androgen signaling, cellular organization and proliferation along with 5 reference genes44–46 providing likelihood of adverse pathology at radical prostatectomy. The Prolaris test (Myriad Genetics, Salt Lake City, UT, USA) evaluates total RNA expression levels of 31 cell cycle progression genes and reports a CCP score that estimates 10-yr disease specific mortality47,48.
Figure 1.
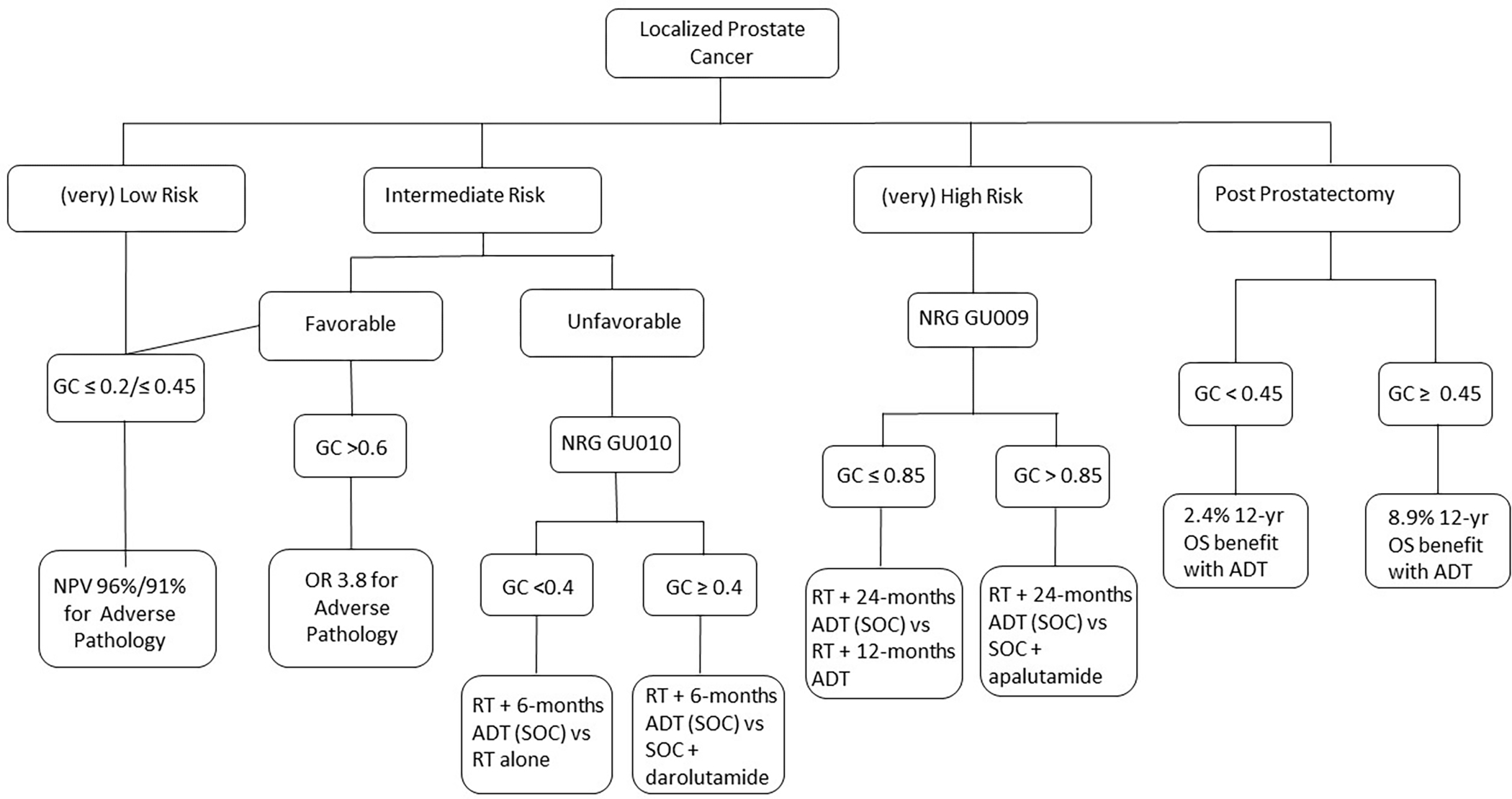
Decipher genomic classifier within localized prostate cancer
GC: Genomic Classifier; OR: Odds Ratio; NPV: Negative Predictive Value; SOC: standard of care; OS: Overall Survival; RT: Radiation Therapy; ADT: Androgen deprivation therapy
Currently, for patients with very low- or low-risk prostate cancer, active surveillance is the favored option given the side effects from radical treatment and without affecting prostate-cancer specific survival49. Although most men undergoing active surveillance can be effectively treated with either radical prostatectomy or RT if they experience disease progression, there is potentially an increased risk of development of distant metastasis compared to those undergoing upfront treatment which cannot be identified with clinical features alone49. Hyung et al. demonstrated that among men with very low-, low-, or favorable intermediate risk prostate cancer, the Decipher biopsy GC can predict for adverse pathologic features (Gleason primary pattern 4 or 5, pTb or greater, or LN involvement) with an odds ratio of 1.29 (95%CI 1.03–1.61) per 10% increase in score and demonstrated a negative predictive value of 96% when Decipher score was ≤ 0.250. This can therefore aid in selecting the appropriate population who can be safely be monitored on active surveillance and conversely which low-risk patients should be recommended for upfront treatment. Herlemann et al. further demonstrated that among men with favorable intermediate-risk disease, in which active surveillance is controversial, only those with Decipher high-risk tumors (score >0.6) had increased risk of adverse pathology upon radical prostatectomy51. This suggests GCs may be integrated into identifying patients with favorable intermediate-risk disease in whom active surveillance is potentially appropriate. Taken together, these studies demonstrate the utility of GC to appropriately select patients for active surveillance, however there remains no consensus on what the threshold score should be and thus mutual decision making between the patient and physician is prudent.
In addition to augmenting our identification of low-risk patients, the GC has identified patients at higher risk of developing metastatic disease following radical treatment. Jairath et al. reported a systematic review of studies evaluating the Decipher GC on biopsy tissue including 18 studies with 19,223 patients ranging from single and multicenter retrospective studies, as well as analyses of prospective clinical trials and prospective registry studies.52 These studies consistently demonstrated increased risk of development of metastatic disease with a multivariate hazard ratio ranging from 1.33–1.72 (per 10% increase in Decipher score) across low-, intermediate- and high-risk disease treated with either radical prostatectomy or radiation therapy (RT) +/− androgen deprivation therapy (ADT) while clinical and pathologic features were inferior 53–58. Additionally, the incorporation of the GC to the regression models based on either NCCN risk grouping, CAPRA59, or Stephenson60 models demonstrated significantly improved AUC and C-index metrics53–57.
Currently, several ongoing prospective clinical trials are aiming to incorporate the GC for greater precision in the management of localized prostate cancer (Table 1). NRG GU009 and GU010 are two parallel phase III randomized clinical trials aiming to de-intensify or intensify high- and unfavorable intermediate-risk prostate cancer, respectively, based on GC risk. GU009 will randomize 2,478 patients with high-risk prostate cancer and those with: (i) low/intermediate GC risk (score ≤ 0.85) to either standard of care with RT + 24 months ADT versus RT + 12 months ADT (Deintensification arm); or, (ii) high GC risk (score >0.85) to either same standard of care versus the addition of apalutamide (Intensification arm). GU010 will randomize 2,050 patients with unfavorable intermediate-risk prostate cancer and those with: (i) low GC risk (score <0.4) to either standard of care with RT + 6 months ADT versus RT alone (Deintensification arm); or, (ii) higher GC risk (score ≥ 0.4) to same standard of care versus the addition of darolutamide. The genomics in Michigan to Adjust Outcomes in Prostate cancer (G-Major) trial is a randomized trial enrolling 900 patients with newly diagnosed favorable intermediate-risk prostate cancer to either standard of care versus integration of GC to further guide management. The investigators hypothesize a greater proportion of patients will be managed with active surveillance within the GC arm.
Table 1.
Select ongoing trials evaluating genomic biomarkers to guide management within prostate cancer.
| Trial Name | Full/formal name of trial | Common name | Setting | Phase | Number of participants | Status (March 2022) |
|---|---|---|---|---|---|---|
| NRG GU009 NCT04513717 |
Two Studies for Patients With High Risk Prostate Cancer Testing Less Intense Treatment for Patients With a Low Gene Risk Score and Testing a More Intense Treatment for Patients With a High Gene Risk Score | PREDICT-RT | Upfront localized | III | 2478 | Recruiting |
| NRG GU010 NCT05050084 |
Two Studies for Patients With Unfavorable Intermediate Risk Prostate Cancer Testing Less Intense Treatment for Patients With a Low Gene Risk Score and Testing a More Intense Treatment for Patients With a Higher Gene Risk Score | GUIDANCE | Upfront localized | III | 2050 | Recruiting |
| NCT04396808 | Genomics in Michigan to AdJust Outcomes in Prostate CanceR or Men With Newly Diagnosed Favorable Risk Prostate Cancer | G-MAJOR | Upfront localized | III | 350 | Recruiting |
| NRG GU006 NCT03371719 |
Biomarker Trial of Apalutamide and Radiation for Recurrent Prostate cancer | BALANCE | Recurrent | II | 311 | Active, not recruiting |
| EA8183 NCT04484818 |
Testing the Addition of Darolutamide to Hormonal Therapy (Androgen Deprivation Therapy [ADT]) After Surgery for Men With High-Risk Prostate Cancer | ERADICATE | Post-prostatectomy | III | 810 | Recruiting |
| NCT02783950 | Genomics in Michigan Impacting Observation or Radiation | G-MINOR | Post-prostatectomy | N/A | 356 | Active, not recruiting |
Outside of the commercially available GC discussed above, several individual and multigene signatures have additionally been implicated in more aggressive localized prostate cancer and radiation resistance though most lack the robust analytical and clinical validation of the work discussed above. Mutations within tumor suppressor genes have been shown to promote metastatic progression61–63. Chipidza et al identified and validated a TP53 mutational signature that was associated with an approximate 20–30% absolute decrease in 5-year metastasis-free survival64. Somatic HOXB13 mutations have also been evaluated within localized disease. HOXB13 protein interacts with the androgen receptor and contributes to the regulation of AR-transcriptomes important for prostate cancer growth65. Weiner et al. demonstrated that patients with localized prostate cancer with the highest quartile of HOXB13 expression demonstrated significantly worse metastasis-free survival compared to the lowest quartile with adjusted hazard ratio ranging 1.46–1.866. BRCA germline mutations have also been implicated in more aggressive localized prostate cancer, demonstrating more advanced disease at diagnosis and worse metastasis free- and cause-specific survival67,68. More generally, Fraser et al. demonstrated that within localized non-indolent prostate cancer, although there appears to be lack of clinically actionable single nucleotide variants, the presence of numerous genomic alterations portends worse clinical outcomes69.
A multigene DNA-based 100-locus copy number alteration (CNA) genomic signature stratified patients with localized prostate cancer into low- and high-risk of recurrence70. It is comprised of 276 genes and was developed using a ∼27,000 probe array comparative genomic hybridization platform. To better translate to the clinic, the signature has been refined to a 31-locus biomarker that can be assessed on the analytically validated and FDA approved NanoString CNV platform71. Both 100- and 31-locus biomarkers reflect genomic instability and were shown to have high prognostic value in >500 prostate cancer patients. A more recent signature comprising expression of 28-hypoxia based genes has shown to be independently prognostic for relapse and metastasis in eleven cohorts of low- to high-risk prostate cancer patients with localized disease treated with surgery or radiotherapy (definitive and post-operative cohorts)72. Improved prognostication may be achievable by combining the 28- hypoxia gene expression signature and indicators of genome instability such as with the 31-locus biomarker72, but these specific strategies need prospective validation. Several other biologic pathways have also been generally implicated with increased radiation resistance including altered DNA damage repair and increased activation of PI3K-Akt-mTor pathway73–76. Although the genomic biomarkers described previously have been shown to be highly prognostic, unfortunately, there are currently no clinically validated predictive genomic biomarkers to predict response to definitive RT within localized prostate cancer
Genomic Classifiers in the Post-Prostatectomy Setting
Following radical prostatectomy, three randomized clinical trials demonstrated immediate post-prostatectomy RT improved biochemical progression-free survival in patients with adverse pathologic features including positive margins, extracapsular extension, or seminal vesicle invasion77–79. Given the added morbidity associated with post-prostatectomy RT, several trials were performed evaluating adjuvant versus early salvage RT (esRT; defined as PSA 0.1–0.2)80–82. The ARTISTIC meta-analysis of these trials demonstrated no improvement in event free survival with adjuvant RT compared with early salvage83. Given these findings, NCCN guidelines currently allow for consideration of adjuvant RT +/− ADT for patients with adverse pathologic features versus esRT. The Decipher GC has the potential for identifying patients who are at highest risk for disease progression and therefore may have the greatest benefit to aggressive management.
Among the commercially available genomic biomarker test, the Decipher GC has been validated in the post-prostatectomy setting and provides prognostic information regarding 5- and 10-year risk of clinical metastases and 15-year prostate cancer specific mortality. Feng et al. demonstrated the Decipher GC was independently associated with distant metastases (HR 1.17, 95%CI 1.05–1.32), prostate cancer specific mortality (HR 1.39, 95%CI 1.20–1.63), and OS (HR 1.17, 95%CI 1.06–1.29) in patients treated on RTOG 9601 (salvage RT +/− antiandrogen therapy in recurrent prostate cancer)84. Interestingly, this work also identified the benefit of ADT on 12-year OS was nearly 3-fold greater in patients with intermediate/high- (8.9% 12-OS benefit) versus low-risk (2.4% 12-OS benefit) GC. This is particularly noteworthy as there continues to be controversy over which patients should be offered ADT along with salvage RT in the post-prostatectomy setting, especially those eligible for low pre-salvage RT PSA or esRT85–87.
Given the current uncertainty in the management of post-prostatectomy patients with adverse pathologic features, prospective studies have aimed to evaluate the integration of genomic biomarkers, such as the Decipher GC, into management decisions. Marascio et al. evaluated two prospective registries (clinical utility cohort and clinical benefit cohort) of prostate cancer patients treated between 2014–2019 with adverse features following prostatectomy. Within the clinical utility cohort, GC testing altered treatment recommendations in 39% of patients. Within the clinical benefit cohort, patients with high GC risk experienced significantly improved 2-year PSA recurrence with adjuvant RT (3% vs 25%). Additionally, patients with low or intermediate risk score demonstrated similar 2-year PSA recurrence (0% vs 2.8%)88. Further prospective studies similarly demonstrated the utility of the GC in guiding treatment decisions with a number needed to treat ranging from 1.5–4 patients to change management, most commonly in patients with high GC risk89,90. Given the complex interaction between clinical, radiographic, and genomic data there is further interest in applying machine learning methods to integrate these features for improved clinical predictions91,92.
The Post-operative Radiation Therapy Outcomes Score (PORTOS) is a predictive signature of distant metastases risk after RT developed incorporating expression of 24 genes implicated in DNA damage repair and response to radiation. In a validation cohort of 330 patients, those treated with radiotherapy had a decreased incidence of distant metastases within the high PORTOS group (4% vs 35%: HR 0.15, p=0.002) but not the low PORTOS group (32% vs 32%: HR 0.92, p=0.76) with a significant interaction93. PORTOS represents the only clinically validated biomarker predictive for response to radiation therapy in the post-prostatectomy setting.
Similar to the upfront setting, several randomized clinical trials are currently enrolling to further appreciate precisely how to apply these genomic data (Table 1). NRG Oncology GU006 (BALANCE) is a 324-patient phase II randomized trial biomarker stratified by the PAM50 classifier of salvage RT +/− the next-generation anti-androgen apalutamide. . The ERADICATE trial is currently recruiting and intends to randomize 810 patients treated with radical prostatectomy with high GC score (≥0.6) to either 12 months of ADT +/− darolutamide. The genomics in Michigan impacting observation or Radiation (G-Minor) trial will randomize 356 patients treated with radical prostatectomy with adverse pathologic features and undetectable post-op PSA to either receive Decipher GC versus standard of care and evaluate the proportion of patients that receive adjuvant therapy within each group.
Although not ready for clinical use, several emerging biomarkers are being evaluated outside of the commercially available genomic tests. Brady et al. demonstrated decrease/loss of ERG expression levels are associated with immediate biochemical progression following radical prostatectomy with decrease/loss of PTEN expression demonstrating a trend towards immediate biochemical progression94. Circulating tumor DNA (ctDNA) is also being evaluated in predicting rapid progression after prostatectomy though data are conflicting. Lau et al. demonstrated patients with tumor variants in ctDNA after prostatectomy had rapid disease recurrence and progression compared to those where variants were not detected. Detection of TP53 mutations in ctDNA also demonstrated significantly shorter metastasis-free survival95. To the contrary, Hennigan et al. demonstrated ultra-low-pass whole-genome sequencing was unable to detect ctDNA in plasma of patients after RP prior to PSA biochemical recurrence96.
Metastatic Castration-Sensitive Prostate Cancer
Genomic Biomarkers within Oligometastatic Castration-Sensitive Prostate Cancer
Management of metastatic castration sensitive prostate cancer (mCSPC) is largely focused on hormonal and systemic therapies with radiation therapy historically playing a palliative role as needed97–100. This paradigm was challenged by the results of the STAMPEDE Arm H trial which evaluated radiotherapy to the primary tumor within de novo metastatic prostate cancer101. This phase III trial of 2,061 patients randomized to either standard of care systemic therapy or the addition of primary radiotherapy demonstrated an improvement in failure-free survival and overall survival in patients with “low-burden” but not “high-burden” disease. Radiation as metastasis directed therapy (MDT) has also garnered further interest among patients with oligometastasis, typically defined as three to five (or fewer) metastases on conventional imaging102–108.
Several prospective randomized trials within varying histologies have demonstrated that MDT improves progression-free survival (PFS) and overall survival (OS) in oligometastatic disease102,104,105,109. Within metachronous oligometastatic castration-sensitive prostate cancer (omCSPC) specifically, MDT has demonstrated prolonged time to initiation of androgen deprivation therapy (ADT) and PFS compared to observation, with no decrement to quality of life102,103. Our group previously reported the ORIOLE trial102, a randomized phase II trial of 54 patients with omCSPC (≤ 3 lesions) not on ADT, randomized to either stereotactic ablative body radiotherapy (SABR) versus observation (2:1 randomization). At six months, the proportion of patients experiencing progression was lower in the SABR arm compared to the observation arm (19% vs 61%, p = 0.005). This finding translated into median PFS not yet reached for the SABR arm and 5.8 months in the observation arm (HR 0.30, p = 0.002). On the ORIOLE trial, saliva, plasma and matched leukocyte DNA samples were collected in all patients at baseline and mutations in specific genes where analyzed from circulating tumor DNA (ctDNA) using the CAPP-Seq (cancer personalized profiling by deep sequencing) method110. Using this method, we developed a high-risk mutational signature composed of truncating/pathogenic mutations encompassing ATM, BRCA1/2, RB1, or TP53. Among patients with detectable ctDNA or germline mutations, those without a high-risk mutation, PFS was significantly longer among participants receiving SABR than among those in the observation arm. However, in those with a high-risk mutation, no difference in PFS was observed between the SABR and observation group, suggesting genomic profiles can provide predictive information regarding response to SABR MDT. Work is currently ongoing to validate this potentially predictive genomic signature using the STOMP trial103 (Deek et al. unpublished communication).
To further expand upon these findings, our group investigated high-risk mutations within the spectrum of mCSPC by grouping patients into four categories: biochemical recurrence (micrometastases), metachronous oligometastatic disease, metachronous polymetastatic disease, and de novo metastatic disease111. Driver mutations in high-risk genes were significantly different across metastatic categories and increased in frequency across the spectrum from biochemical recurrence, to polymetastatic and de novo metastatic disease for several genes and pathways. Mutations in DDR genes (IRR 1.61; p<0.001), and TP53 (IRR 1.45; p=0.004) were associated with increasing number of metastatic lesions. Along with the specific high-risk mutations detailed above, additional work in this space may hold potential in further classifying omCSPC patients111–124.
Metastatic Castration-Resistant Prostate Cancer
Within metastatic castration-resistant prostate cancer (mCRPC) much more is understood about the genomic landscape115,125,126 however the role of radiation therapy has thus far been limited as improvements in outcomes have been driven by advancements in systemic therapy and increasingly immunotherapy127–129. Outside of palliation, the integration of systemic radioisotope therapies such as Radium-223 and PSMA targeted radioligand therapy have expanded the utilization of radiation in mCRPC.
Genomic Predictors of Synthetic Lethal Response to Radium-223
Radium-223 is an alpha-particle-emitting bone-targeted therapy that demonstrated consistent improvement in pain and OS in patients with mCRPC harboring bone disease130,131. Alpha particles emitted at the site of disease have high linear energy transfer, resulting in the deposition of energy in the immediate vicinity of the radionuclide decay. This highly localized radiotherapy selectively targets the bone microenvironment and metastatic tumor cells, causing what is suspected to be irreparable DNA DSB and very locally restricted cytotoxic effects132.
Germline mutations in DDR genes are present in 8–12% of mCRPC, whereas the previously estimated prevalence was 4–5% in localized disease133,134. In addition, somatic aberrations in genes linked to DNA repair are seen in 20–25% of mCRPC patients135. Corroborating studies have shown that both germline and somatic homologous recombination repair (HRR) gene mutations are seen in up to one-third of patients with mCRPC136. Synthetic lethality described previously, are observed in tumors with defects in mechanisms of DNA repair that are theoretically more susceptible to therapies that cause DNA damage, such as DSBs. The high prevalence of DDR mutations in mCRPC has led to their validation as prognostic67,68,137,138 and predictive biomarkers135,139.
Velho et al. hypothesized that mCRPC patients who harbored either germline and/or somatic HRR mutations may have a greater clinical benefit from radium-223, due to DSBs going unrepaired because of an underlying HRR in the tumor cells140. Medical records of 190 mCRPC patients for whom germline and/or somatic DNA sequencing data were available recovered 28 men who had also received standard-of-care radium-223. Of these 28 patients, 10 men (35.7%) had a germline/somatic HRR mutation (three in BRCA2, and one each in ATM, ATR, CHEK2, FANCG, FANCI, FANCL, and PALB2) and 18 (64.3%) did not have HRR associated mutations. In this exploratory study, bone-metastatic mCRPC patients with inactivating HRR mutations demonstrated significantly improved alkaline phosphatase responses (80% versus 38%, p = 0.04), time to ALP progression (median 10.4 versus 5.8 mo, hazard ratio [HR] 6.4, p = 0.005), and a trend toward longer OS (median 36.9 versus 19.0 mo, HR 3.3, p = 0.11). Follow-up work by van der Doelen et al. confirmed these findings demonstrating patients with HRR mutations experienced significantly improved OS (36.3 vs 17.0 mo; HR 2.29, p=0.01)141. These provocative results and “synthetic lethality” hypothesis between HRR mutations and radium-223 activity is being prospectively tested in a phase II study (NCT04489719).
Genomic Predictors of Response to PSMA-Targeted Radioligand Therapy
Prostate-specific membrane antigen (PSMA) is a transmembrane glycoprotein expressed on the surface of prostate cancer that has demonstrated overexpression within both local and metastatic prostate cancer lesions with high expression independently associated with poor survival142–144. (177Lu)-PSMA-617 is a PSMA targeted radioligand that delivers beta-particle radiation. The VISION trial was a phase III trial which randomized 831 patients with mCRPC to either (177Lu)-PSMA-617 or standard of care and demonstrated an improvement in median PFS (8.7 versus 3.4 months: HR 0.4, p<0.001) and OS (15.3 versus 11.3 months: HR 0.62, p<0.001)145. Response to (177Lu)-PSMA-617 has previously been associated with detection of androgen receptor (AR) gene amplification in plasma cell-free DNA. De Giorgi et al. evaluated AR copy number in pretreatment plasma samples and correlated detection of AR gene amplification with early progressive disease on (177Lu)-PSMA-617146. Patients who experienced early progressive disease were significantly more likely to have AR gain identified in plasma (OR 16.00, p=0.0007) and patients with plasma AR gain were found to have significantly shorter OS compared to AR-normal patients (7.4 versus 19.1 months, p=0.02). However, the levels of ctDNA in patient plasma are strongly prognostic147,148, so it is difficult to know whether the observed relationship with AR gain detection is independent of the prognostic effect of detecting any ctDNA signal. Mutations in DDR also appear to be implicated in response to PSMA targeted radioligand therapy however preliminary data among small cohorts and case reports149–151 are conflicting. Conteduca et al. analyzed 25 patients with PSMA targeted radioligand therapy who underwent whole exome sequencing152. Patients with BRCA1/2 mutations were found to have significantly improved PFS whereas those with a TP53 mutation demonstrated worse PFS. Conversely, Kratochwil et al. identified 7 patients with a poor response to PSMA-targeting α-radiation therapy who underwent NGS153. Six of the 7 patients were found to have at least 1 genetic alteration negatively affecting the DDR pathway leading the authors to hypothesize DDR mutations may confer resistance to PSMA targeted radioligand therapy. Privé et al. demonstrated no difference in PFS between patients with or without DDR mutations in a cohort of 40 patients with mCRPC treated with PSMA targeted radioligand therapy154. Future work with a more comprehensive approach to detection of genomic alterations applied to larger and ideally prospectively collected set of cohorts will likely be needed to fully understand these relationships.
Conclusions
Growing evidence continues to demonstrate the prognostic implications of genomic biomarkers across the spectrum of prostate cancer. Several commercially available genomic biomarkers are currently in use which have consistently demonstrated improved stratification of patients at higher risk of experiencing a more aggressive clinical course. Though the evidence and clinical utility of prognostic genomic biomarkers is mounting, there unfortunately is a scarcity of biomarkers predicting therapeutic response to radiation and/or adjunctive treatments in prostate cancer patients treated with radiotherapy. Consequently, even less is understood about how genomic markers may be predictive of various radiation techniques and dosing however remains an active area of interest. Prospective randomized integral-biomarker radiotherapy trial data utilizing these genomic tests are currently lacking, however, such trials are active and their results should be available in the coming years in the localized space. Integration of tumor genomics into these current and future clinical trials will allow for a more robust identification and utilization of predictive biomarkers and ultimately allow for precision radiation therapy in the management of patients across the entire spectrum of prostate cancer.
Acknowledgements
PTT was funded by an anonymous donor, the Movember Foundation-Distinguished Gentlemen’s Ride-Prostate Cancer Foundation, Barbara’s Fund, National Capitol Cancer Research Fund and the NIH/NCI (U01CA212007 and U01CA231776) and DoD (W81XWH-21–1-0296)
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. Jan 2021;71(1):7–33. doi: 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2.Chang AJ, Autio KA, Roach M 3rd, Scher HI. High-risk prostate cancer-classification and therapy. Nat Rev Clin Oncol. Jun 2014;11(6):308–23. doi: 10.1038/nrclinonc.2014.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Amico AV, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. Jama. Sep 16 1998;280(11):969–74. doi: 10.1001/jama.280.11.969 [DOI] [PubMed] [Google Scholar]
- 4.Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. Dec 4 2014;371(23):2167–77. doi: 10.1056/NEJMoa1408440 [DOI] [PubMed] [Google Scholar]
- 5.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. Apr 5 2001;344(14):1031–7. doi: 10.1056/nejm200104053441401 [DOI] [PubMed] [Google Scholar]
- 6.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. Jun 24 2010;362(25):2380–8. doi: 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 7.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. Oct 20 2005;353(16):1659–72. doi: 10.1056/NEJMoa052306 [DOI] [PubMed] [Google Scholar]
- 8.Dotan E, Aggarwal C, Smith MR. Impact of Rituximab (Rituxan) on the Treatment of B-Cell Non-Hodgkin’s Lymphoma. P t. Mar 2010;35(3):148–57. [PMC free article] [PubMed] [Google Scholar]
- 9.Guan YF, Li GR, Wang RJ, et al. Application of next-generation sequencing in clinical oncology to advance personalized treatment of cancer. Chin J Cancer. Oct 2012;31(10):463–70. doi: 10.5732/cjc.012.10216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. Jul 2009;9(7):489–99. doi: 10.1038/nrc2645 [DOI] [PubMed] [Google Scholar]
- 11.Bassullu N, Turkmen I, Dayangac M, et al. The Predictive and Prognostic Significance of c-erb-B2, EGFR, PTEN, mTOR, PI3K, p27, and ERCC1 Expression in Hepatocellular Carcinoma. Hepat Mon. Oct 2012;12(10 hcc):e7492. doi: 10.5812/hepatmon.7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Sun Y, Yao L, et al. Association between CYP2D6 *10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. Aug 2008;19(8):1423–1429. doi: 10.1093/annonc/mdn155 [DOI] [PubMed] [Google Scholar]
- 13.Torres-Roca JF, Eschrich S, Zhao H, et al. Prediction of radiation sensitivity using a gene expression classifier. Cancer Res. Aug 15 2005;65(16):7169–76. doi: 10.1158/0008-5472.Can-05-0656 [DOI] [PubMed] [Google Scholar]
- 14.Eschrich SA, Pramana J, Zhang H, et al. A gene expression model of intrinsic tumor radiosensitivity: prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys. Oct 1 2009;75(2):489–96. doi: 10.1016/j.ijrobp.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed KA, Chinnaiyan P, Fulp WJ, Eschrich S, Torres-Roca JF, Caudell JJ. The radiosensitivity index predicts for overall survival in glioblastoma. Oncotarget. Oct 27 2015;6(33):34414–22. doi: 10.18632/oncotarget.5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres-Roca JF, Fulp WJ, Caudell JJ, et al. Integration of a Radiosensitivity Molecular Signature Into the Assessment of Local Recurrence Risk in Breast Cancer. Int J Radiat Oncol Biol Phys. Nov 1 2015;93(3):631–8. doi: 10.1016/j.ijrobp.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strom T, Hoffe SE, Fulp W, et al. Radiosensitivity index predicts for survival with adjuvant radiation in resectable pancreatic cancer. Radiother Oncol. Oct 2015;117(1):159–64. doi: 10.1016/j.radonc.2015.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohammadi H, Prince A, Figura NB, et al. Using the Radiosensitivity Index (RSI) to Predict Pelvic Failure in Endometrial Cancer Treated With Adjuvant Radiation Therapy. Int J Radiat Oncol Biol Phys. Mar 1 2020;106(3):496–502. doi: 10.1016/j.ijrobp.2019.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott JG, Berglund A, Schell MJ, et al. A genome-based model for adjusting radiotherapy dose (GARD): a retrospective, cohort-based study. Lancet Oncol. Feb 2017;18(2):202–211. doi: 10.1016/s1470-2045(16)30648-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott JG, Sedor G, Ellsworth P, et al. Pan-cancer prediction of radiotherapy benefit using genomic-adjusted radiation dose (GARD): a cohort-based pooled analysis. Lancet Oncol. Sep 2021;22(9):1221–1229. doi: 10.1016/s1470-2045(21)00347-8 [DOI] [PubMed] [Google Scholar]
- 21.Thomas G, Eisenhauer E, Bristow RG, et al. The European Organisation for Research and Treatment of Cancer, State of Science in radiation oncology and priorities for clinical trials meeting report. European Journal of Cancer. 2020;131:76–88. [DOI] [PubMed] [Google Scholar]
- 22.Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacology & therapeutics. 2015;149:124–138. [DOI] [PubMed] [Google Scholar]
- 23.Pitter KL, Casey DL, Lu YC, et al. Pathogenic ATM mutations in cancer and a genetic basis for radiotherapeutic efficacy. JNCI: Journal of the National Cancer Institute. 2021;113(3):266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rafiei S, Fitzpatrick K, Liu D, et al. ATM Loss Confers Greater Sensitivity to ATR Inhibition Than PARP Inhibition in Prostate Cancer. Cancer Res. Jun 1 2020;80(11):2094–2100. doi: 10.1158/0008-5472.can-19-3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neeb A, Herranz N, Arce-Gallego S, et al. Advanced Prostate Cancer with ATM Loss: PARP and ATR Inhibitors. Eur Urol. Feb 2021;79(2):200–211. doi: 10.1016/j.eururo.2020.10.029 [DOI] [PubMed] [Google Scholar]
- 26.Dillon MT, Boylan Z, Smith D, et al. PATRIOT: A phase I study to assess the tolerability, safety and biological effects of a specific ataxia telangiectasia and Rad3-related (ATR) inhibitor (AZD6738) as a single agent and in combination with palliative radiation therapy in patients with solid tumours. Clin Transl Radiat Oncol. Aug 2018;12:16–20. doi: 10.1016/j.ctro.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Ledermann J, Kohn E. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Annals of oncology. 2014;25(1):32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carreira S, Porta N, Arce-Gallego S, et al. Biomarkers Associating with PARP Inhibitor Benefit in Prostate Cancer in the TOPARP-B Trial. Cancer Discov. Nov 2021;11(11):2812–2827. doi: 10.1158/2159-8290.Cd-21-0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandrasekaran A, Elias KM. Synthetic Lethality in Ovarian Cancer. Mol Cancer Ther. Nov 2021;20(11):2117–2128. doi: 10.1158/1535-7163.Mct-21-0500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bi Y, Verginadis II, Dey S, et al. Radiosensitization by the PARP inhibitor olaparib in BRCA1-proficient and deficient high-grade serous ovarian carcinomas. Gynecol Oncol. Sep 2018;150(3):534–544. doi: 10.1016/j.ygyno.2018.07.002 [DOI] [PubMed] [Google Scholar]
- 31.Verhagen CV, de Haan R, Hageman F, et al. Extent of radiosensitization by the PARP inhibitor olaparib depends on its dose, the radiation dose and the integrity of the homologous recombination pathway of tumor cells. Radiother Oncol. Sep 2015;116(3):358–65. doi: 10.1016/j.radonc.2015.03.028 [DOI] [PubMed] [Google Scholar]
- 32.Scarborough JA, Scott JG. Translation of Precision Medicine Research Into Biomarker-Informed Care in Radiation Oncology. Semin Radiat Oncol. Jan 2022;32(1):42–53. doi: 10.1016/j.semradonc.2021.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gopal P, Abazeed M. High-Throughput Phenotyping of BRAF Mutations Reveals Categories of Mutations That Confer Resistance to Radiation. International Journal of Radiation Oncology, Biology, Physics. 2017;99(2):E591. [Google Scholar]
- 34.Chowdhary M, Patel KR, Danish HH, Lawson DH, Khan MK. BRAF inhibitors and radiotherapy for melanoma brain metastases: potential advantages and disadvantages of combination therapy. OncoTargets and therapy. 2016;9:7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deville SS, Luft S, Kaufmann M, Cordes N. Keap1 inhibition sensitizes head and neck squamous cell carcinoma cells to ionizing radiation via impaired non-homologous end joining and induced autophagy. Cell death & disease. 2020;11(10):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krause M, Ostermann G, Petersen C, et al. Decreased repopulation as well as increased reoxygenation contribute to the improvement in local control after targeting of the EGFR by C225 during fractionated irradiation. Radiotherapy and oncology. 2005;76(2):162–167. [DOI] [PubMed] [Google Scholar]
- 37.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. Mar 10 2009;27(8):1160–7. doi: 10.1200/jco.2008.18.1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallden B, Storhoff J, Nielsen T, et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics. Aug 22 2015;8:54. doi: 10.1186/s12920-015-0129-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao SG, Chang SL, Erho N, et al. Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA oncology. 2017;3(12):1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamid AA, Huang HC, Wang V, et al. Transcriptional profiling of primary prostate tumor in metastatic hormone-sensitive prostate cancer and association with clinical outcomes: correlative analysis of the E3805 CHAARTED trial. Ann Oncol. Sep 2021;32(9):1157–1166. doi: 10.1016/j.annonc.2021.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aggarwal R, Rydzewski NR, Zhang L, et al. Prognosis Associated With Luminal and Basal Subtypes of Metastatic Prostate Cancer. JAMA Oncol. Nov 1 2021;7(11):1644–1652. doi: 10.1001/jamaoncol.2021.3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lalani N, Voduc KD, Jimenez RB, et al. Breast Cancer Molecular Subtype as a Predictor of Radiation Therapy Fractionation Sensitivity. Int J Radiat Oncol Biol Phys. Jan 1 2021;109(1):281–287. doi: 10.1016/j.ijrobp.2020.08.038 [DOI] [PubMed] [Google Scholar]
- 43.Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8(6):e66855. doi: 10.1371/journal.pone.0066855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cullen J, Lynch JA, Klein EA, et al. Multicenter Comparison of 17-Gene Genomic Prostate Score as a Predictor of Outcomes in African American and Caucasian American Men with Clinically Localized Prostate Cancer. J Urol. Apr 2021;205(4):1047–1054. doi: 10.1097/ju.0000000000001484 [DOI] [PubMed] [Google Scholar]
- 45.Van Den Eeden SK, Lu R, Zhang N, et al. A biopsy-based 17-gene genomic prostate score as a predictor of metastases and prostate cancer death in surgically treated men with clinically localized disease. European urology. 2018;73(1):129–138. [DOI] [PubMed] [Google Scholar]
- 46.Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. European urology. 2014;66(3):550–560. [DOI] [PubMed] [Google Scholar]
- 47.Cuzick J, Berney DM, Fisher G, et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer. Mar 13 2012;106(6):1095–9. doi: 10.1038/bjc.2012.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. Mar 2011;12(3):245–55. doi: 10.1016/s1470-2045(10)70295-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamdy FC, Donovan JL, Lane JA, et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N Engl J Med. Oct 13 2016;375(15):1415–1424. doi: 10.1056/NEJMoa1606220 [DOI] [PubMed] [Google Scholar]
- 50.Kim HL, Li P, Huang HC, et al. Validation of the Decipher Test for predicting adverse pathology in candidates for prostate cancer active surveillance. Prostate Cancer Prostatic Dis. Sep 2019;22(3):399–405. doi: 10.1038/s41391-018-0101-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herlemann A, Huang HC, Alam R, et al. Decipher identifies men with otherwise clinically favorable-intermediate risk disease who may not be good candidates for active surveillance. Prostate Cancer Prostatic Dis. Mar 2020;23(1):136–143. doi: 10.1038/s41391-019-0167-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jairath NK, Dal Pra A, Vince R Jr., et al. A Systematic Review of the Evidence for the Decipher Genomic Classifier in Prostate Cancer. Eur Urol. Mar 2021;79(3):374–383. doi: 10.1016/j.eururo.2020.11.021 [DOI] [PubMed] [Google Scholar]
- 53.Klein EA, Haddad Z, Yousefi K, et al. Decipher Genomic Classifier Measured on Prostate Biopsy Predicts Metastasis Risk. Urology. Apr 2016;90:148–52. doi: 10.1016/j.urology.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 54.Klein EA, Yousefi K, Haddad Z, et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol. Apr 2015;67(4):778–86. doi: 10.1016/j.eururo.2014.10.036 [DOI] [PubMed] [Google Scholar]
- 55.Nguyen PL, Haddad Z, Ross AE, et al. Ability of a Genomic Classifier to Predict Metastasis and Prostate Cancer-specific Mortality after Radiation or Surgery based on Needle Biopsy Specimens. Eur Urol. Nov 2017;72(5):845–852. doi: 10.1016/j.eururo.2017.05.009 [DOI] [PubMed] [Google Scholar]
- 56.Tosoian JJ, Birer SR, Jeffrey Karnes R, et al. Performance of clinicopathologic models in men with high risk localized prostate cancer: impact of a 22-gene genomic classifier. Prostate Cancer Prostatic Dis. Dec 2020;23(4):646–653. doi: 10.1038/s41391-020-0226-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berlin A, Murgic J, Hosni A, et al. Genomic Classifier for Guiding Treatment of Intermediate-Risk Prostate Cancers to Dose-Escalated Image Guided Radiation Therapy Without Hormone Therapy. Int J Radiat Oncol Biol Phys. Jan 1 2019;103(1):84–91. doi: 10.1016/j.ijrobp.2018.08.030 [DOI] [PubMed] [Google Scholar]
- 58.Nguyen PL, Martin NE, Choeurng V, et al. Utilization of biopsy-based genomic classifier to predict distant metastasis after definitive radiation and short-course ADT for intermediate and high-risk prostate cancer. Prostate Cancer Prostatic Dis. Jun 2017;20(2):186–192. doi: 10.1038/pcan.2016.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cooperberg MR, Pasta DJ, Elkin EP, et al. The University of California, San Francisco Cancer of the Prostate Risk Assessment score: a straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol. Jun 2005;173(6):1938–42. doi: 10.1097/01.ju.0000158155.33890.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stephenson AJ, Scardino PT, Kattan MW, et al. Predicting the outcome of salvage radiation therapy for recurrent prostate cancer after radical prostatectomy. J Clin Oncol. May 20 2007;25(15):2035–41. doi: 10.1200/jco.2006.08.9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thangavel C, Boopathi E, Liu Y, et al. RB Loss Promotes Prostate Cancer Metastasis. Cancer Res. Feb 15 2017;77(4):982–995. doi: 10.1158/0008-5472.Can-16-1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ecke TH, Schlechte HH, Schiemenz K, et al. TP53 gene mutations in prostate cancer progression. Anticancer Res. May 2010;30(5):1579–86. [PubMed] [Google Scholar]
- 63.Torres-Roca JF, DeSilvio M, Mora LB, et al. Activated STAT3 as a correlate of distant metastasis in prostate cancer: a secondary analysis of Radiation Therapy Oncology Group 86–10. Urology. Mar 2007;69(3):505–9. doi: 10.1016/j.urology.2006.11.006 [DOI] [PubMed] [Google Scholar]
- 64.Chipidza FE, Alshalalfa M, Mahal BA, et al. Development and Validation of a Novel TP53 Mutation Signature That Predicts Risk of Metastasis in Primary Prostate Cancer. Clin Genitourin Cancer. Jun 2021;19(3):246–254.e5. doi: 10.1016/j.clgc.2020.08.004 [DOI] [PubMed] [Google Scholar]
- 65.Norris JD, Chang C-Y, Wittmann BM, et al. The homeodomain protein HOXB13 regulates the cellular response to androgens. Molecular cell. 2009;36(3):405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiner AB, Faisal FA, Davicioni E, et al. Somatic HOXB13 Expression Correlates with Metastatic Progression in Men with Localized Prostate Cancer Following Radical Prostatectomy. Eur Urol Oncol. Dec 2021;4(6):955–962. doi: 10.1016/j.euo.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castro E, Goh C, Leongamornlert D, et al. Effect of BRCA Mutations on Metastatic Relapse and Cause-specific Survival After Radical Treatment for Localised Prostate Cancer. Eur Urol. Aug 2015;68(2):186–93. doi: 10.1016/j.eururo.2014.10.022 [DOI] [PubMed] [Google Scholar]
- 68.Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. May 10 2013;31(14):1748–57. doi: 10.1200/jco.2012.43.1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fraser M, Sabelnykova VY, Yamaguchi TN, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature. Jan 19 2017;541(7637):359–364. doi: 10.1038/nature20788 [DOI] [PubMed] [Google Scholar]
- 70.Lalonde E, Ishkanian AS, Sykes J, et al. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol. Dec 2014;15(13):1521–1532. doi: 10.1016/s1470-2045(14)71021-6 [DOI] [PubMed] [Google Scholar]
- 71.Lalonde E, Alkallas R, Chua MLK, et al. Translating a Prognostic DNA Genomic Classifier into the Clinic: Retrospective Validation in 563 Localized Prostate Tumors. Eur Urol. Jul 2017;72(1):22–31. doi: 10.1016/j.eururo.2016.10.013 [DOI] [PubMed] [Google Scholar]
- 72.Yang L, Roberts D, Takhar M, et al. Development and Validation of a 28-gene Hypoxia-related Prognostic Signature for Localized Prostate Cancer. EBioMedicine. May 2018;31:182–189. doi: 10.1016/j.ebiom.2018.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chaiswing L, Weiss HL, Jayswal RD, Clair DKS, Kyprianou N. Profiles of Radioresistance Mechanisms in Prostate Cancer. Crit Rev Oncog. 2018;23(1–2):39–67. doi: 10.1615/CritRevOncog.2018025946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pungsrinont T, Kallenbach J, Baniahmad A. Role of PI3K-AKT-mTOR Pathway as a Pro-Survival Signaling and Resistance-Mediating Mechanism to Therapy of Prostate Cancer. Int J Mol Sci. Oct 14 2021;22(20)doi: 10.3390/ijms222011088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alberti C Prostate cancer: radioresistance molecular target-related markers and foreseeable modalities of radiosensitization. Eur Rev Med Pharmacol Sci. Aug 2014;18(16):2275–82. [PubMed] [Google Scholar]
- 76.Chang L, Graham PH, Ni J, et al. Targeting PI3K/Akt/mTOR signaling pathway in the treatment of prostate cancer radioresistance. Crit Rev Oncol Hematol. Dec 2015;96(3):507–17. doi: 10.1016/j.critrevonc.2015.07.005 [DOI] [PubMed] [Google Scholar]
- 77.Thompson IM Jr., Tangen CM, Paradelo J, et al. Adjuvant radiotherapy for pathologically advanced prostate cancer: a randomized clinical trial. Jama. Nov 15 2006;296(19):2329–35. doi: 10.1001/jama.296.19.2329 [DOI] [PubMed] [Google Scholar]
- 78.Bolla M, van Poppel H, Tombal B, et al. Postoperative radiotherapy after radical prostatectomy for high-risk prostate cancer: long-term results of a randomised controlled trial (EORTC trial 22911). Lancet. Dec 8 2012;380(9858):2018–27. doi: 10.1016/s0140-6736(12)61253-7 [DOI] [PubMed] [Google Scholar]
- 79.Wiegel T, Bottke D, Steiner U, et al. Phase III postoperative adjuvant radiotherapy after radical prostatectomy compared with radical prostatectomy alone in pT3 prostate cancer with postoperative undetectable prostate-specific antigen: ARO 96–02/AUO AP 09/95. J Clin Oncol. Jun 20 2009;27(18):2924–30. doi: 10.1200/jco.2008.18.9563 [DOI] [PubMed] [Google Scholar]
- 80.Parker CC, Clarke NW, Cook AD, et al. Timing of radiotherapy after radical prostatectomy (RADICALS-RT): a randomised, controlled phase 3 trial. Lancet. Oct 31 2020;396(10260):1413–1421. doi: 10.1016/s0140-6736(20)31553-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kneebone A, Fraser-Browne C, Duchesne GM, et al. Adjuvant radiotherapy versus early salvage radiotherapy following radical prostatectomy (TROG 08.03/ANZUP RAVES): a randomised, controlled, phase 3, non-inferiority trial. Lancet Oncol. Oct 2020;21(10):1331–1340. doi: 10.1016/s1470-2045(20)30456-3 [DOI] [PubMed] [Google Scholar]
- 82.Sargos P, Chabaud S, Latorzeff I, et al. Adjuvant radiotherapy versus early salvage radiotherapy plus short-term androgen deprivation therapy in men with localised prostate cancer after radical prostatectomy (GETUG-AFU 17): a randomised, phase 3 trial. Lancet Oncol. Oct 2020;21(10):1341–1352. doi: 10.1016/s1470-2045(20)30454-x [DOI] [PubMed] [Google Scholar]
- 83.Vale CL, Fisher D, Kneebone A, et al. Adjuvant or early salvage radiotherapy for the treatment of localised and locally advanced prostate cancer: a prospectively planned systematic review and meta-analysis of aggregate data. Lancet. Oct 31 2020;396(10260):1422–1431. doi: 10.1016/s0140-6736(20)31952-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feng FY, Huang HC, Spratt DE, et al. Validation of a 22-Gene Genomic Classifier in Patients With Recurrent Prostate Cancer: An Ancillary Study of the NRG/RTOG 9601 Randomized Clinical Trial. JAMA Oncol. Apr 1 2021;7(4):544–552. doi: 10.1001/jamaoncol.2020.7671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shipley WU, Seiferheld W, Lukka HR, et al. Radiation with or without Antiandrogen Therapy in Recurrent Prostate Cancer. N Engl J Med. Feb 2 2017;376(5):417–428. doi: 10.1056/NEJMoa1607529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dess RT, Sun Y, Jackson WC, et al. Association of Presalvage Radiotherapy PSA Levels After Prostatectomy With Outcomes of Long-term Antiandrogen Therapy in Men With Prostate Cancer. JAMA Oncol. May 1 2020;6(5):735–743. doi: 10.1001/jamaoncol.2020.0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pollack A, Karrison T, Balogh A, et al. Short term androgen deprivation therapy without or with pelvic lymph node treatment added to prostate bed only salvage radiotherapy: the NRG Oncology/RTOG 0534 SPPORT trial. International journal of radiation oncology, biology, physics. 2018;102(5):1605. [Google Scholar]
- 88.Marascio J, Spratt DE, Zhang J, et al. Prospective study to define the clinical utility and benefit of Decipher testing in men following prostatectomy. Prostate cancer and prostatic diseases. 2020;23(2):295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gore JL, du Plessis M, Zhang J, et al. Clinical utility of a genomic classifier in men undergoing radical prostatectomy: The PRO-IMPACT trial. Practical radiation oncology. 2020;10(2):e82–e90. [DOI] [PubMed] [Google Scholar]
- 90.Gore JL, du Plessis M, Santiago-Jiménez M, et al. Decipher test impacts decision making among patients considering adjuvant and salvage treatment after radical prostatectomy: Interim results from the Multicenter Prospective PRO-IMPACT study. Cancer. 2017;123(15):2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goldenberg SL, Nir G, Salcudean SE. A new era: artificial intelligence and machine learning in prostate cancer. Nat Rev Urol. Jul 2019;16(7):391–403. doi: 10.1038/s41585-019-0193-3 [DOI] [PubMed] [Google Scholar]
- 92.Lee G, Singanamalli A, Wang H, et al. Supervised multi-view canonical correlation analysis (sMVCCA): integrating histologic and proteomic features for predicting recurrent prostate cancer. IEEE Trans Med Imaging. Jan 2015;34(1):284–97. doi: 10.1109/tmi.2014.2355175 [DOI] [PubMed] [Google Scholar]
- 93.Zhao SG, Chang SL, Spratt DE, et al. Development and validation of a 24-gene predictor of response to postoperative radiotherapy in prostate cancer: a matched, retrospective analysis. The lancet oncology. 2016;17(11):1612–1620. [DOI] [PubMed] [Google Scholar]
- 94.Brady L, Carlsson J, Baird AM, et al. Correlation of integrated ERG/PTEN assessment with biochemical recurrence in prostate cancer. Cancer Treat Res Commun. 2021;29:100451. doi: 10.1016/j.ctarc.2021.100451 [DOI] [PubMed] [Google Scholar]
- 95.Lau E, McCoy P, Reeves F, et al. Detection of ctDNA in plasma of patients with clinically localised prostate cancer is associated with rapid disease progression. Genome Med. Aug 17 2020;12(1):72. doi: 10.1186/s13073-020-00770-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hennigan ST, Trostel SY, Terrigino NT, et al. Low Abundance of Circulating Tumor DNA in Localized Prostate Cancer. JCO Precis Oncol. 2019;3doi: 10.1200/po.19.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kyriakopoulos CE, Chen YH, Carducci MA, et al. Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer: Long-Term Survival Analysis of the Randomized Phase III E3805 CHAARTED Trial. J Clin Oncol. Apr 10 2018;36(11):1080–1087. doi: 10.1200/jco.2017.75.3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.James ND, de Bono JS, Spears MR, et al. Abiraterone for Prostate Cancer Not Previously Treated with Hormone Therapy. N Engl J Med. Jul 27 2017;377(4):338–351. doi: 10.1056/NEJMoa1702900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chi KN, Agarwal N, Bjartell A, et al. Apalutamide for Metastatic, Castration-Sensitive Prostate Cancer. N Engl J Med. Jul 4 2019;381(1):13–24. doi: 10.1056/NEJMoa1903307 [DOI] [PubMed] [Google Scholar]
- 100.Davis ID, Martin AJ, Stockler MR, et al. Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N Engl J Med. Jul 11 2019;381(2):121–131. doi: 10.1056/NEJMoa1903835 [DOI] [PubMed] [Google Scholar]
- 101.Parker CC, James ND, Brawley CD, et al. Radiotherapy to the primary tumour for newly diagnosed, metastatic prostate cancer (STAMPEDE): a randomised controlled phase 3 trial. Lancet. Dec 1 2018;392(10162):2353–2366. doi: 10.1016/s0140-6736(18)32486-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Phillips R, Shi WY, Deek M, et al. Outcomes of Observation vs Stereotactic Ablative Radiation for Oligometastatic Prostate Cancer: The ORIOLE Phase 2 Randomized Clinical Trial. JAMA Oncol. May 1 2020;6(5):650–659. doi: 10.1001/jamaoncol.2020.0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ost P, Reynders D, Decaestecker K, et al. Surveillance or Metastasis-Directed Therapy for Oligometastatic Prostate Cancer Recurrence: A Prospective, Randomized, Multicenter Phase II Trial. J Clin Oncol. Feb 10 2018;36(5):446–453. doi: 10.1200/jco.2017.75.4853 [DOI] [PubMed] [Google Scholar]
- 104.Iyengar P, Wardak Z, Gerber DE, et al. Consolidative Radiotherapy for Limited Metastatic Non-Small-Cell Lung Cancer: A Phase 2 Randomized Clinical Trial. JAMA Oncol. Jan 11 2018;4(1):e173501. doi: 10.1001/jamaoncol.2017.3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gomez DR, Blumenschein GR Jr., Lee JJ, et al. Local consolidative therapy versus maintenance therapy or observation for patients with oligometastatic non-small-cell lung cancer without progression after first-line systemic therapy: a multicentre, randomised, controlled, phase 2 study. Lancet Oncol. Dec 2016;17(12):1672–1682. doi: 10.1016/S1470-2045(16)30532-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sutera P, Van Der Eecken K, Kishan AU, et al. Definitions of disease burden across the spectrum of metastatic castration-sensitive prostate cancer: comparison by disease outcomes and genomics. Prostate Cancer Prostatic Dis. Jan 11 2022;doi: 10.1038/s41391-021-00484-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Phillips RM, Deek MP, Deweese TL, Tran PT. Metastasis-Directed Therapy in Prostate Cancer. Why, When, and How? Oncology (Williston Park). Oct 28 2019;33(10) [PubMed] [Google Scholar]
- 108.Deek MP, Tran PT. Oligometastatic and Oligoprogression Disease and Local Therapies in Prostate Cancer. Cancer J. Mar/Apr 2020;26(2):137–143. doi: 10.1097/ppo.0000000000000432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Palma DA, Olson R, Harrow S, et al. Stereotactic ablative radiotherapy versus standard of care palliative treatment in patients with oligometastatic cancers (SABR-COMET): a randomised, phase 2, open-label trial. Lancet. May 18 2019;393(10185):2051–2058. doi: 10.1016/s0140-6736(18)32487-5 [DOI] [PubMed] [Google Scholar]
- 110.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. May 2014;20(5):548–54. doi: 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deek MP, Van der Eecken K, Phillips R, et al. The Mutational Landscape of Metastatic Castration-sensitive Prostate Cancer: The Spectrum Theory Revisited. Eur Urol. Jan 5 2021;doi: 10.1016/j.eururo.2020.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ritch E, Fu SYF, Herberts C, et al. Identification of Hypermutation and Defective Mismatch Repair in ctDNA from Metastatic Prostate Cancer. Clin Cancer Res. Mar 1 2020;26(5):1114–1125. doi: 10.1158/1078-0432.Ccr-19-1623 [DOI] [PubMed] [Google Scholar]
- 113.Kumar A, White TA, MacKenzie AP, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A. Oct 11 2011;108(41):17087–92. doi: 10.1073/pnas.1108745108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Van der Eecken K, Vanwelkenhuyzen J, Deek MP, et al. Tissue- and Blood-derived Genomic Biomarkers for Metastatic Hormone-sensitive Prostate Cancer: A Systematic Review. Eur Urol Oncol. Dec 2021;4(6):914–923. doi: 10.1016/j.euo.2021.10.005 [DOI] [PubMed] [Google Scholar]
- 115.Abida W, Armenia J, Gopalan A, et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol. Jul 2017;2017doi: 10.1200/po.17.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hamid AA, Gray KP, Shaw G, et al. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. European urology. 2019;76(1):89–97. [DOI] [PubMed] [Google Scholar]
- 117.Gilson C, Ingleby F, Gilbert DC, et al. Genomic profiles of de novo high-and low-volume metastatic prostate cancer: Results from a 2-stage feasibility and prevalence study in the STAMPEDE trial. JCO Precision Oncology. 2020;4:882–897. [DOI] [PubMed] [Google Scholar]
- 118.Kohli M, Tan W, Zheng T, et al. Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer. EBioMedicine. 2020;54:102728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vandekerkhove G, Struss WJ, Annala M, et al. Circulating tumor DNA abundance and potential utility in de novo metastatic prostate cancer. European urology. 2019;75(4):667–675. [DOI] [PubMed] [Google Scholar]
- 120.Shenderov E, Isaacsson Velho P, Awan AH, et al. Genomic and clinical characterization of pulmonary-only metastatic prostate cancer: A unique molecular subtype. The Prostate. 2019;79(13):1572–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Swami U, Velho PI, Nussenzveig R, et al. Association of SPOP mutations with outcomes in men with de novo metastatic castration-sensitive prostate cancer. European Urology. 2020;78(5):652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stopsack KH, Nandakumar S, Wibmer AG, et al. Oncogenic genomic alterations, clinical phenotypes, and outcomes in metastatic castration-sensitive prostate cancer. Clinical Cancer Research. 2020;26(13):3230–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mateo J, Seed G, Bertan C, et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. The Journal of clinical investigation. 2020;130(4):1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fan L, Fei X, Zhu Y, et al. Comparative Analysis of Genomic Alterations across Castration Sensitive and Castration Resistant Prostate Cancer via Circulating Tumor DNA Sequencing. The Journal of Urology. 2021;205(2):461–469. [DOI] [PubMed] [Google Scholar]
- 125.Hamid AA, Gray KP, Shaw G, et al. Compound Genomic Alterations of TP53, PTEN, and RB1 Tumor Suppressors in Localized and Metastatic Prostate Cancer. Eur Urol. Jul 2019;76(1):89–97. doi: 10.1016/j.eururo.2018.11.045 [DOI] [PubMed] [Google Scholar]
- 126.Abida W, Cyrta J, Heller G, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A. Jun 4 2019;116(23):11428–11436. doi: 10.1073/pnas.1902651116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.de Wit R, de Bono J, Sternberg CN, et al. Cabazitaxel versus Abiraterone or Enzalutamide in Metastatic Prostate Cancer. N Engl J Med. Dec 26 2019;381(26):2506–2518. doi: 10.1056/NEJMoa1911206 [DOI] [PubMed] [Google Scholar]
- 128.de Bono J, Mateo J, Fizazi K, et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med. May 28 2020;382(22):2091–2102. doi: 10.1056/NEJMoa1911440 [DOI] [PubMed] [Google Scholar]
- 129.Antonarakis ES, Piulats JM, Gross-Goupil M, et al. Pembrolizumab for Treatment-Refractory Metastatic Castration-Resistant Prostate Cancer: Multicohort, Open-Label Phase II KEYNOTE-199 Study. J Clin Oncol. Feb 10 2020;38(5):395–405. doi: 10.1200/jco.19.01638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hoskin P, Sartor O, O’Sullivan JM, et al. Efficacy and safety of radium-223 dichloride in patients with castration-resistant prostate cancer and symptomatic bone metastases, with or without previous docetaxel use: a prespecified subgroup analysis from the randomised, double-blind, phase 3 ALSYMPCA trial. Lancet Oncol. Nov 2014;15(12):1397–406. doi: 10.1016/s1470-2045(14)70474-7 [DOI] [PubMed] [Google Scholar]
- 131.Nilsson S, Franzén L, Parker C, et al. Bone-targeted radium-223 in symptomatic, hormone-refractory prostate cancer: a randomised, multicentre, placebo-controlled phase II study. Lancet Oncol. Jul 2007;8(7):587–94. doi: 10.1016/s1470-2045(07)70147-x [DOI] [PubMed] [Google Scholar]
- 132.Ritter MA, Cleaver JE, Tobias CA. High-LET radiations induce a large proportion of non-rejoining DNA breaks. Nature. Apr 14 1977;266(5603):653–5. doi: 10.1038/266653a0 [DOI] [PubMed] [Google Scholar]
- 133.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med. Aug 4 2016;375(5):443–53. doi: 10.1056/NEJMoa1603144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. May 21 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mateo J, Carreira S, Sandhu S, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. Oct 29 2015;373(18):1697–708. doi: 10.1056/NEJMoa1506859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Leith A, Ribbands A, Kim J, et al. Real-world homologous recombination repair mutation testing in metastatic castration-resistant prostate cancer in the USA, Europe and Japan. Future Oncol. Jan 19 2022;doi: 10.2217/fon-2021-1113 [DOI] [PubMed] [Google Scholar]
- 137.Na R, Zheng SL, Han M, et al. Germline Mutations in ATM and BRCA1/2 Distinguish Risk for Lethal and Indolent Prostate Cancer and are Associated with Early Age at Death. Eur Urol. May 2017;71(5):740–747. doi: 10.1016/j.eururo.2016.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Murphy DG, Risbridger GP, Bristow RG, Sandhu S. The Evolving Narrative of DNA Repair Gene Defects: Distinguishing Indolent from Lethal Prostate Cancer. Eur Urol. May 2017;71(5):748–749. doi: 10.1016/j.eururo.2017.01.025 [DOI] [PubMed] [Google Scholar]
- 139.Mateo J, Boysen G, Barbieri CE, et al. DNA Repair in Prostate Cancer: Biology and Clinical Implications. Eur Urol. Mar 2017;71(3):417–425. doi: 10.1016/j.eururo.2016.08.037 [DOI] [PubMed] [Google Scholar]
- 140.Isaacsson Velho P, Qazi F, Hassan S, et al. Efficacy of Radium-223 in Bone-metastatic Castration-resistant Prostate Cancer with and Without Homologous Repair Gene Defects. Eur Urol. Aug 2019;76(2):170–176. doi: 10.1016/j.eururo.2018.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van der Doelen MJ, Isaacsson Velho P, Slootbeek PHJ, et al. Impact of DNA damage repair defects on response to radium-223 and overall survival in metastatic castration-resistant prostate cancer. Eur J Cancer. Sep 2020;136:16–24. doi: 10.1016/j.ejca.2020.05.001 [DOI] [PubMed] [Google Scholar]
- 142.Ross JS, Sheehan CE, Fisher HA, et al. Correlation of primary tumor prostate-specific membrane antigen expression with disease recurrence in prostate cancer. Clin Cancer Res. Dec 15 2003;9(17):6357–62. [PubMed] [Google Scholar]
- 143.Rajasekaran AK, Anilkumar G, Christiansen JJ. Is prostate-specific membrane antigen a multifunctional protein? Am J Physiol Cell Physiol. May 2005;288(5):C975–81. doi: 10.1152/ajpcell.00506.2004 [DOI] [PubMed] [Google Scholar]
- 144.Vlachostergios PJ, Niaz MJ, Sun M, et al. Prostate-Specific Membrane Antigen Uptake and Survival in Metastatic Castration-Resistant Prostate Cancer. Frontiers in Oncology. 2021;11:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sartor O, de Bono J, Chi KN, et al. Lutetium-177-PSMA-617 for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med. Sep 16 2021;385(12):1091–1103. doi: 10.1056/NEJMoa2107322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.De Giorgi U, Sansovini M, Severi S, et al. Circulating androgen receptor gene amplification and resistance to (177)Lu-PSMA-617 in metastatic castration-resistant prostate cancer: results of a Phase 2 trial. Br J Cancer. Oct 2021;125(9):1226–1232. doi: 10.1038/s41416-021-01508-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Annala M, Fu S, Bacon JVW, et al. Cabazitaxel versus abiraterone or enzalutamide in poor prognosis metastatic castration-resistant prostate cancer: a multicentre, randomised, open-label, phase II trial. Ann Oncol. Jul 2021;32(7):896–905. doi: 10.1016/j.annonc.2021.03.205 [DOI] [PubMed] [Google Scholar]
- 148.Annala M, Vandekerkhove G, Khalaf D, et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov. Apr 2018;8(4):444–457. doi: 10.1158/2159-8290.Cd-17-0937 [DOI] [PubMed] [Google Scholar]
- 149.Ahmadzadehfar H, Gaertner F, Lossin PS, Schwarz B, Essler M. BRCA2 Mutation as a Possible Cause of Poor Response to 177Lu-PSMA Therapy. Clin Nucl Med. Aug 2018;43(8):609–610. doi: 10.1097/rlu.0000000000002141 [DOI] [PubMed] [Google Scholar]
- 150.Crumbaker M, Emmett L, Horvath LG, Joshua AM. Exceptional Response to (177)Lutetium Prostate-Specific Membrane Antigen in Prostate Cancer Harboring DNA Repair Defects. JCO Precis Oncol. Dec 2019;3:1–5. doi: 10.1200/po.18.00237 [DOI] [PubMed] [Google Scholar]
- 151.Satapathy S, Das CK, Parihar AS, Sood A, Mittal BR. Response to Concomitant Enzalutamide and 177Lu-PSMA-617 Radioligand Therapy in ATM-Mutated Metastatic Castration Resistant Prostate Cancer. Clin Nucl Med. Jul 1 2021;46(7):582–583. doi: 10.1097/rlu.0000000000003541 [DOI] [PubMed] [Google Scholar]
- 152.Conteduca V, Oromendia C, Vlachostergios PJ, et al. Clinical and molecular analysis of patients treated with prostate-specific membrane antigen (PSMA)-targeted radionuclide therapy. American Society of Clinical Oncology; 2019. [Google Scholar]
- 153.Kratochwil C, Giesel FL, Heussel CP, et al. Patients Resistant Against PSMA-Targeting α-Radiation Therapy Often Harbor Mutations in DNA Damage-Repair-Associated Genes. J Nucl Med. May 2020;61(5):683–688. doi: 10.2967/jnumed.119.234559 [DOI] [PubMed] [Google Scholar]
- 154.Privé BM, Slootbeek PHJ, Laarhuis BI, et al. Impact of DNA damage repair defects on response to PSMA radioligand therapy in metastatic castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. Jul 12 2021;doi: 10.1038/s41391-021-00424-2 [DOI] [PubMed] [Google Scholar]


